Abstract
The phylogenetic diversity of picocyanobacteria in seven alkaline lakes on the Tibetan Plateau was analyzed using the molecular marker 16S-23S rRNA internal transcribed spacer sequence. A total of 1,077 environmental sequences retrieved from the seven lakes were grouped into seven picocyanobacterial clusters, with two clusters newly described here. Each of the lakes was dominated by only one or two clusters, while different lakes could have disparate communities, suggesting low alpha diversity but high beta diversity of picocyanobacteria in these high-altitude freshwater and saline lakes. Several globally distributed clusters were found in these Tibetan lakes, such as subalpine cluster I and the Cyanobium gracile cluster. Although other clusters likely exhibit geographic restriction to the plateau temporally, reflecting endemicity, they can indeed be distributed widely on the plateau. Lakes with similar salinities may have similar genetic populations despite a large geographic distance. Canonical correspondence analysis identified salinity as the only environmental factor that may in part explain the diversity variations among lakes. Mantel tests suggested that the community similarities among lakes are independent of geographic distance. A portion of the picocyanobacterial clusters appear to be restricted to a narrow salinity range, while others are likely adapted to a broad range. A seasonal survey of Lake Namucuo across 3 years did not show season-related variations in diversity, and depth-related population partitioning was observed along a vertical profile of the lake. Our study emphasizes the high dispersive potential of picocyanobacteria and suggests that the regional distribution may result from adaptation to specified environments.
INTRODUCTION
Picocyanobacteria are globally important primary producers in freshwater, brackish, and marine ecosystems (1, 2). They are generally unicellular cyanobacteria smaller than 2 μm. The genera Synechococcus, Prochlorococcus, and Cyanobium are major groups of picocyanobacteria found in nature (3). Synechococcus spp. are polyphyletic and highly genetically diverse and are ubiquitous in inland and marine aquatic environments (4–10). In contrast, Prochlorococcus spp. are obligate marine organisms (11), and Cyanobium spp. are mostly found in freshwater and brackish environments (3).
Synechococcus spp. comprise five major clusters (1 to 5), which were established through phylogenetic analysis based on the 16S rRNA gene sequences (3). Cluster 5 primarily comprises the marine Synechococcus spp., with three known subclusters, 5.1, 5.2 (3), and 5.3 (12). Prochlorococcus, the cluster 5 Synechococcus, and Cyanobium form a tightly clustered phylogenetic group (3, 13), sometimes called the “Syn/Pro clade” (14), the “PS clade” (13), or “group 6b” (5). However, many nonmarine picocyanobacterial lineages (clusters) found in freshwater or brackish environments fell into this group (5, 15–17) (Table 1 shows a summary). A few of these clusters were widely found in both freshwater lakes and the brackish Baltic Sea, such as the C. gracile cluster, subalpine cluster I, subalpine cluster II, group I, and the LBP1 cluster (5, 15, 16, 18, 19), supporting the high dispersive potential of microbes. Some other studies found several novel picocyanobacterial phylogenetic clusters in Tibetan lakes (20), Mazurian lakes (21), and Lake Superior (22), which had not been described elsewhere. Later, Felföldi and colleagues (23) found that some of these picocyanobacterial lineages, such as group M (21) and LS II (22), could be detected in other aquatic environments, emphasizing their ubiquitous dispersal.
TABLE 1.
Picocyanobacterial lineages of Synechococcus and Cyanobium determined through phylogenetic analysis using the 16S-23S rRNA gene ITS sequencea
| Lineage | Distribution area or source reported | Reference(s) |
|---|---|---|
| Cyanobium gracile cluster (group A) | Ponds in United States and Europe, stream in United States, marshland in Europe, Bylot Island tundra pond in Arctic, lakes in Europe and Japan, Baltic Sea, Tibetan lakes | 5, 15, 17, 19, 21, this study |
| Subalpine cluster I (group B), including Tibetan cluster I | Lakes in Japan and Europe, Baltic Sea, Tibetan lakes (only for Tibetan cluster I) | 5, 15–21, this study |
| Subalpine cluster II | Lakes in Europe, Baltic Sea, Long Island Sound, Tibetan lakes | 15, 18, 27, 49, this study |
| Bornholm Sea cluster | Baltic Sea | 15, 18 |
| Lake Biwa cluster | Lake Biwa (Japan) | 15 |
| Group Cz (Group C) | Lakes in Japan, Great Mazurian Lakes (Europe) | 5, 21 |
| LBP1 cluster (Group H) | Lake Biwa, Lake Mondsee (Europe), freshwater lakes in United Kingdom, Baltic Sea | 16, 19, 27 |
| Group I | Lake Mondsee, Lake Sagami (Japan), Bylot Island tundra pond in Arctic, Baltic Sea, Tibetan lakes | 16, 18, 19, this study |
| Group M | Lake Okutama (Japan), Green Lake (United States), Great Mazurian Lakes | 21, 51, 52 |
| Tibetan cluster II | Tibetan lakes | 20 |
| Tibetan cluster III-V | Tibetan lakes | 20, this study |
| Tibetan cluster VI | Tibetan lakes | This study |
| Tibetan cluster VII | Tibetan lakes | This study |
| Marine subcluster 5.1 | Marine environments (estuary, coastal water, and open ocean) | 6–8, 10, 53 |
| Marine subcluster 5.2 | Marine environments (estuary, coastal water) | 9, 53, 54 |
| Marine subcluster 5.3 | Marine environments (coastal water and open ocean) | 9, 54, 55 |
The lineages detected in this study are shown in boldface.
Picocyanobacteria have been serving as an important model system for microbial ecology study (2, 24). Prochlorococcus and the marine Synechococcus have remarkable biogeographies along nutrient, temperature, and light gradients, giving rise to ecotypes confined to specific ecological niches (8, 25, 26). Differing from the broadly connecting marine ecosystem, inland aquatic ecosystems are island-like in nature, which may constrain the global dispersal of these microbial taxa (27). However, some have argued that the geographical isolation in the microbial world could be limited due to the ubiquitous dispersal potential of the small single-cell organisms (28), supporting the “everything is everywhere” hypothesis (29). Indeed, there have been controversial reports that emphasized the role of geographic isolation (30, 31) or, alternatively, the ubiquitous dispersal (32, 33) in shaping microbial community composition and biogeography. Thus, it is interesting to test the distribution boundaries of nonmarine picocyanobacterial lineages within and across inland geographic regions.
The Tibetan Plateau is the world's highest and largest plateau, with an average elevation exceeding 4,000 m, representing a relatively isolated region of the world. Lakes are widespread on the plateau. Two-thirds of them are freshwater lakes, while the remainder have various saline and alkaline conditions (34). The abundances of picocyanobacteria in Tibetan lakes are generally within the range of 104 to 105 cells ml−1 (20), reflecting a significant contribution to the primary production in these typically oligotrophic lakes (chlorophyll a concentration, <0.5 μg liter−1, mostly <0.1 μg liter−1) (35). Several picocyanobacterial clusters were recently found in freshwater and saline lakes on the plateau, most of which were thought to be endemic lineages (20).
In this study, we investigated the genetic diversity of picocyanobacteria in seven Tibetan lakes using the molecular marker of 16S-23S rRNA internal transcribed spacer (ITS) sequences. The picocyanobacterial communities in six of the seven lakes have not been described before. A seasonal time series study across 3 years was conducted for one of the lakes, and vertical profiles were investigated for two lakes. Using previously published environmental picocyanobacterial sequences in the Tibetan lakes involved (20), we compared the Tibetan picocyanobacterial communities among themselves and to those in other regions of the world. By doing so, we attempted to assess the dispersal potential of picocyanobacterial lineages and to test whether and to what extent their distributions are affected by geographic distance or other environmental factors.
MATERIALS AND METHODS
Samples and study area.
In order to cover the Tibetan lakes in broad ranges of both geographic distance and environmental gradient, seven lakes were selected in this study (Fig. 1 and Table 2). To test the spatial and temporal variability of the picocyanobacterial community, vertical samples of water columns for two lakes (Lake Zigetangcuo [ZGTC] and Lake Namucuo [NMC]) and seasonal samples across 3 years for one lake (Lake NMC) were included. Each of the other five lakes (Dongcuo [DC], Dazecuo [DZC], Pengcuo [PE], Anggucuo [AGC], and Pumuyongcuo [PMYC]) was represented by a single surface water sample. For each sample, 0.5 to 1 liter of water was filtered through 0.22-μm-pore-size polycarbonate filters (Millipore). During the late autumn, winter, and early spring seasons (i.e., ice season) (Table 2), water samples were collected beside the ice or beneath the ice cover. The filters were stored at −80°C till DNA isolation. Water temperature, pH, dissolved oxygen (DO), and total dissolved solids (TDS) were measured by using a Hydrolab DS5 Water Quality Multiprobe (Hach, Loveland, CO, USA). Total prokaryotic cell abundance was measured by using flow cytometry (Altra II; Beckman Coulter, Epics, Miami, FL, USA).
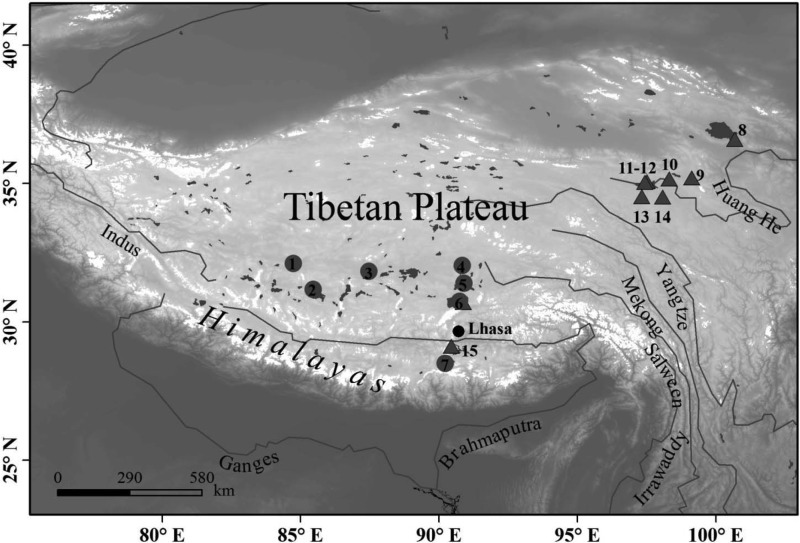
FIG 1.
Locations of the Tibetan lakes. The seven lakes studied here are represented by numbers (1 to 7) in gray circles, and those studied previously (20) are shown by gray triangles with numbers (8 to 15) next to them. The numbering of lakes is as follows: 1, DC; 2, AGC; 3, DZC; 4, ZGTC; 5, PE; 6, MNC; 7, PMYC; 8, Qinghai; 9, Kuhai; 10, Tusuhai; 11, Zhaling; 12, Shuiku; 13, Eling; 14, Xinxinhai; 15, Yanghu. (Base map from the U.S. Geological Survey.)
TABLE 2.
Information on sampling locations
| Lake | Latitude, longitude | Altitude (m) | Area (km2) | Sampling date | Sample identifier (ID) | Sampling depth (m) | pH | TDS (g/liter) | Temp (°C) | DO (mg/liter) | Prokaryotic abundance (106 cells/ml) | No. of sequences |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DC | 32°07′N, 84°44′E | 4,396 | 88 | 25 August 2009 | DC | 0 | 8.8 | 38.8 | 18.7 | 4.83 | 1.62 | 49 |
| DZC | 31°50′N, 87°28′E | 4,379 | 245 | 8 July 2010 | DZC | 0 | 9.8 | 15.8 | 14.1 | 10.2 | 0.06 | 48 |
| ZGTC | 32°03′N, 90°50′E | 4,561 | 191 | 15 August 2009 | ZGTC-0 m | 0 | 10.1 | 14.3 | 14.5 | 4.13 | 1.47 | 45 |
| ZGTC-12 m | 12 | 14.3 | 14.0 | 3.9 | 1.20 | 47 | ||||||
| ZGTC-25 m | 25 | 11.8 | 2.9 | 0.13 | 2.24 | 48 | ||||||
| PE | 31°24′N, 90°54′E | 4,540 | 136 | 8 October 2008 | PE | 0 | 10.3 | 8.7 | 7.8 | 9.62 | 1.25 | 50 |
| AGC | 31°11′N, 85°27′E | 4,658 | 23 | 27 October 2009 | AGC | 0 | 9.1 | 1.7 | 15.1 | 5.87 | 1.00 | 47 |
| NMC | 30°45′N, 90°44′E | 4,728 | 1,961 | 1 October 2009 | NMC-4 m | 4 | 9.2 | 1.2 | 11.7 | 5.61 | 0.58 | 49 |
| NMC-12 m | 12 | 8.8 | 1.2 | 11.6 | 5.62 | 0.75 | 46 | |||||
| NMC-20 m | 20 | 8.0 | 1.4 | 6.9 | 6.51 | 0.51 | 42 | |||||
| NMC-44 m | 44 | 7.2 | 1.5 | 4.5 | 6.87 | 0.42 | 43 | |||||
| NMC-72 m | 72 | 7.5 | 1.5 | 3.9 | 6.54 | 0.34 | 44 | |||||
| NMC-92 m | 92 | 7.9 | 1.5 | 3.8 | 6.31 | 0.20 | 45 | |||||
| 11 November 2006 | NMC-0611 | 0 (ice) | 0.9 | 1.5 | 0.39 | 48 | ||||||
| 18 August 2007 | NMC-0708 | 0 | 1.0 | 12.3 | 1.36 | 50 | ||||||
| 15 December 2007 | NMC-0712 | 0 (ice) | 0.7 | 0.5 | 1.23 | 46 | ||||||
| 16 March 2008 | NMC-0803 | 0 (ice) | 0.7 | 3.0 | 0.26 | 46 | ||||||
| 15 June 2008 | NMC-0806 | 0 | 0.8 | 13.5 | 0.74 | 45 | ||||||
| 15 October 2008 | NMC-0810 | 0 | 0.7 | 10.5 | 1.61 | 46 | ||||||
| 5 January 2009 | NMC-0901 | 0 (ice) | 0.6 | 1.3 | 1.50 | 49 | ||||||
| 15 March 2009 | NMC-0903 | 0 (ice) | 0.5 | 0.7 | 1.13 | 48 | ||||||
| 16 April 2009 | NMC-0904 | 0 (ice) | 0.4 | 5.0 | 1.93 | 47 | ||||||
| PMYC | 28°30′N, 90°13′ E | 5,030 | 290 | 8 October 2008 | PMYC | 0 | 9.2 | 0.2 | 7.6 | 4.64 | 1.69 | 49 |
DNA isolation, PCR amplification, and sequencing.
DNA was isolated from the filter samples by using the phenol-chloroform method. Briefly, a filter was sheared into pieces and transferred into sucrose-Tris-EDTA (STE) buffer and then successively treated with lysozyme (final concentration, 1.0 mg ml−1) and proteinase K (final concentration, 0.5 mg ml−1) for 1 and 2 h, respectively. Then, the solution was successively extracted using phenol-chloroform-isoamyl alcohol (25:24:1) and chloroform-isoamyl alcohol (24:1). Finally, nucleic acid was precipitated using isopropyl alcohol and stored at −80°C.
Picocyanobacterial ITS sequences were amplified using the primer set Picocya16S-F (5′-TGGATCACCTCCTAACAGGG-3′) and Picocya23S-R (5′-CCTTCATCGCCTCTGTGTGCC-3′) and the PCR program described in a previous study (36). The PCR products were excised and purified using the TaKaRa Agarose Gel DNA purification kit (TaKaRa) and cloned using the TaKaRa pMD19-T Vector Cloning Kit (TaKaRa), following the manufacturer's instructions. Fifty clones were sequenced for each sample on the ABI 3730 genetic analyzer (Applied Biosystems) at the Major Biotech Co., Ltd., Shanghai, China.
Phylogenetic and diversity analyses.
The environmental sequences from a clone library were aligned by using Clustal X2 (37) software. The resulting alignments were screened to filter the cloning plasmid and 16S rRNA and 23S rRNA sequences. Then, the pure ITS sequence alignments were scanned to remove chimeric artifacts. After passing this quality control process, the remaining sequences in each clone library were aligned, and the alignment was input to DNADIST (a program in the PHYLIP software package) (38) to generate a distance matrix. The matrix then was used to calculate a batch of diversity indexes (such as Chao and Shannon indexes) and an operational taxonomic unit (OTU) list by using DOTUR (39).
Distance and maximum-likelihood (ML) optimality criterion methods were used to construct and evaluate phylogenetic trees comprising current known clusters based on ITS sequences. Reference sequences of picocyanobacterial lineages (listed in Table 1) were downloaded from GenBank. Representative sequences of our environmental sequences were picked from each OTU, and the selected sequences were aligned with reference sequences. Then, the alignment was input to construct the phylogenies using PAUP* (40) and PHYML (41). Distance tree construction and bootstrap tests (heuristic searching based on the Jukes-Cantor model) were performed using PAUP*. The optimal model for the likelihood method was estimated using PAUP* and ModelTest (42), and the best model for our alignment was GTR + I + Γ (a general time-reversible model with a proportion of invariable sites and a gamma-shaped distribution of rates across sites). ML phylogeny and bootstrap tests were analyzed using PHYML with the model GTR + I + Γ. MEGA 6 (43) was used to visualize, compare, and edit the trees. Cluster-specific trees were built with MEGA 6 using the neighbor-joining method.
Statistical analyses.
Canonical correspondence analysis (CCA) implemented in CANOCO (version 4.5) was performed to estimate the correlation level between picocyanobacterial community compositions and environmental and biological factors (pH, TDS, temperature, DO, and prokaryotic abundance). A Monte Carlo permutation test (499 permutations) was used to determine the significance of these factors for explaining the community similarity pattern. Nonmetric multidimensional scaling (NMDS) and cluster analysis based on Bray-Curtis similarity were performed to analyze the community similarity among the seasonal and vertical samples, respectively, from Lake NMC, using PRIMER 5. Because of the extremely low alpha diversity within a lake and the high beta diversity across lakes, different distance cutoffs were selected to cluster OTUs. The OTU relative abundance matrices were generated by DOTUR at a 0.05 distance cutoff for the CCA that involved seven lakes and at a 0.01 distance cutoff for the NMDS and cluster analyses that involved seasonal and vertical samples from Lake NMC. An analysis of similarity (ANOSIM) test with 999 permutations was used to test the significance of the respective groupings of the ice season samples and non-ice season samples from Lake NMC. Mantel tests were performed to determine the relationship between community similarities and geographic distances or environmental and biological factors. The geographic distance between lakes was calculated based on their coordinates, and the explanatory variables were Z-score transformed and then input to calculate the Euclidean distance matrix.
Nucleotide sequence accession numbers.
The sequences obtained in this study were deposited in GenBank with accession numbers KM025424 to KM026500.
RESULTS
Environmental parameters of the Tibetan lakes.
The seven lakes (Fig. 1, no. 1 to 7) we studied are all located in the Tibetan Plateau at altitudes >4,000 m above sea level (Table 2 and Fig. 1). These lakes have various sizes from 23 to 1,961 km2. All the surface waters are alkaline or extremely alkaline, with pHs from 8.8 to 10.3. According to the measured TDS densities, the lakes could be freshwater (PMYC), oligosaline (NMC and AGC), polysaline (PE, ZGTC, and DZC), or hypersaline (DC), with TDS ranging from nearly zero to 38.8 g liter−1 (Table 2). Except for Lake NMC, the lakes were sampled only once, in summer or autumn. Seasonal samples across 3 years from 2006 to 2009 were collected in Lake NMC, covering all four seasons. It is notable that in winter and spring the lake is frozen on the surface, and water samples were collected from beneath the ice. In addition, vertical samples throughout a whole water column were collected in Lakes ZGTC and NMC. For comparison, the locations of eight other Tibetan lakes where picocyanobacterial communities were investigated previously (20) are also shown (Fig. 1, no. 8 to 15). Lake NMC (no. 6) was also included in the previous study (20).
Phylogenetic lineages of picocyanobacteria in the Tibetan lakes.
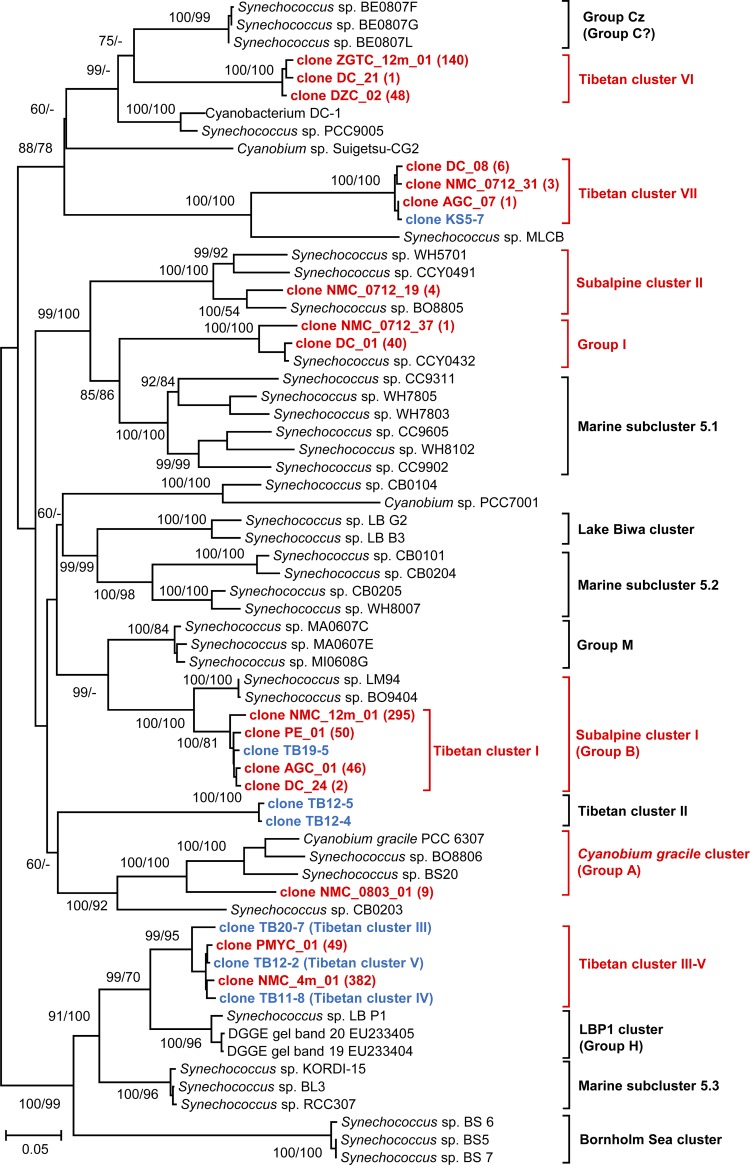
To determine the phylogenetic positions of our picocyanobacterial 16S-23S rRNA gene ITS sequences, the sequences, together with those from known clusters (Table 1), were pooled to construct a phylogenetic tree (Fig. 2). The 1,077 sequences retrieved in this study fell into seven clusters, among which two clusters, Tibetan clusters VI and VII, have not been described before by using ITS sequences. Consistent with a previous description (20), Tibetan cluster I was affiliated with subalpine cluster I but still represented an independent subbranch within the cluster (Fig. 2). Contrary to the polyphyletic relationship among Tibetan clusters III, IV, and V when they were first described (20), our phylogenetic analysis showed that the three clusters formed a monophyletic group (Fig. 2) separate from the LBP1 cluster. Therefore, they are referred to as Tibetan cluster III-V in this study. Four of the seven clusters, Tibetan clusters I, III-V, VI, and VII, comprised only sequences from the Tibetan lakes (the unpublished environmental sequence KS5-7 in Tibetan cluster VII was recovered from the Tibetan Lake Kusai) (Fig. 2). However, differing from its seemingly locally distributed subbranch Tibetan cluster I, subalpine cluster I can be found in various aquatic environments and in very distant regions throughout the world (Table 1). The other three clusters, subalpine cluster II, the C. gracile cluster, and group I, are also distributed in different continents. For instance, the C. gracile cluster was found widely in Europe, Japan, the United States, the Baltic Sea, and the Arctic (Table 1).
FIG 2.
Phylogenetic analyses of picocyanobacteria. A distance tree is shown. Environmental sequences recovered in this study are shown in red, and other Tibetan sequences from the previous study (20) or from an unpublished data set in GenBank (clone KS5-7; accession no. KC841418) are shown in blue. The bootstrap support numbers indicate distance/maximum likelihood. For each cluster detected in this study, only one sequence from each of the lakes was used to build the phylogenetic trees, and the number in parentheses indicates the number of recovered sequences in each lake.
Low alpha diversity and high beta diversity of the picocyanobacterial communities.
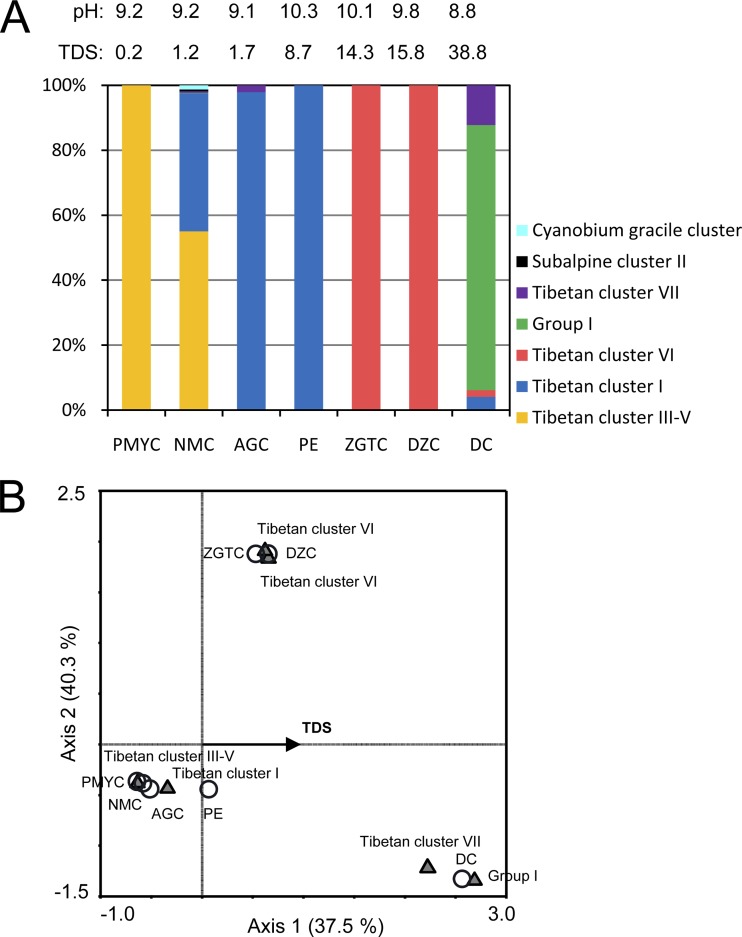
Four of the seven clusters, Tibetan clusters I (393 total sequences), III-V (431 total sequences), and VI (189 total sequences) and group I (41 total sequences), comprised many more sequences than the other three clusters, which contained ≤10 sequences (Fig. 2 and 3A). Remarkably, the picocyanobacterial communities in six of the seven lakes (except Lake NMC) were monotonically dominated by one of the four major clusters (Fig. 2). At the extreme, each of the communities in lakes PMYC, PE, ZGTC, and DZC was comprised of only one picocyanobacterial lineage (Fig. 3A). This result indicates the extremely low alpha diversity of picocyanobacterial communities at the cluster level in each of the Tibetan lakes. The observed OTU abundance, estimated OTU richness (Chao index), and Shannon diversity index of each community also support the low species richness of this prokaryotic photoautotroph group in these high-altitude lakes (Table 3).
FIG 3.
(A) Picocyanobacterial population composition in the Tibetan lakes. The TDS density and pH are shown for the surface water of each lake. (B) CCA diagram showing the correlation between picocyanobacterial community compositions and environmental factors. In order to avoid possible bias, for Lake NMC and Lake ZGTC, only surface water samples that were collected in non-ice seasons were included in this analysis. Therefore, OTUs of two picocyanobacterial lineages, the C. gracile cluster and subalpine cluster II, are not shown. Also, note that Tibetan cluster VI contained two OTUs. Among the environmental factors tested, only TDS significantly explained the species-environment correlation (P < 0.05). Axes 1 and 2 explained 37.5% and 40.3% of the variations in diversity, respectively. On the diagram, lakes are indicated by circles and OTUs are indicated by triangles, and the affiliation of each OTU is shown.
TABLE 3.
Parameters showing alpha diversities at different phylogenetic-distance cutoffsa
| Lake | No. of OTUs observed |
OTU richness estimator (Chao index) |
Diversity estimator (Shannon index) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 0.05 | 0.1 | 0.2 | 0.05 | 0.1 | 0.2 | 0.05 | 0.1 | 0.2 | |
| AGC | 2 | 2 | 2 | 2 | 2 | 2 | 0.10 | 0.10 | 0.10 |
| DC | 4 | 4 | 4 | 4 | 4 | 4 | 0.63 | 0.63 | 0.63 |
| DZC | 2 | 1 | 1 | 2 | 1 | 1 | 0.69 | 0 | 0 |
| PE | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| PMYC | 2 | 1 | 1 | 2 | 1 | 1 | 0.17 | 0 | 0 |
| ZGTCb | 2 | 1 | 1 | 2 | 1 | 1 | 0.69 | 0 | 0 |
| NMCb | 3 | 3 | 3 | 4 | 3 | 3 | 0.63 | 0.62 | 0.62 |
Three phylogenetic-distance cutoffs (0.05, 0.1, and 0.2), representing three different sequence dissimilarity levels, were selected.
The values are averages of multiple samples from Lake ZGTC and Lake NMC.
On the other hand, as the dominant lineages varied among the lakes, their picocyanobacterial communities appeared as discrete patterns (Fig. 3A), indicating a high beta diversity across the lakes. It is interesting that a transition of the picocyanobacterial populations appears to exist along the salinity gradient from Lakes PMYC, NMC, and AGC to PE (Fig. 3A). In the freshwater Lake PMYC, Tibetan cluster III-V was the sole lineage. In the oligosaline Lake NMC, Tibetan clusters III-V and I cooccurred, but only the latter occurred in Lake AGC and Lake PE, which have higher TDS densities. Moreover, in the two polysaline lakes, ZGTC and DZC, only Tibetan cluster VI was detected. The hypersaline Lake DC was dominated by group I, which can be found in either freshwater lakes or brackish environments, such as the Baltic Sea (Table 1). In order to predict possible factors that may explain the community composition variations across lakes, a CCA was conducted. Among the five environmental and biological factors tested (pH, TDS, temperature, DO, and prokaryotic abundance), only TDS showed a significant correlation with the community variations among the seven lakes (Monte Carlo permutation test, P < 0.05) (Fig. 3B). Nevertheless, axis 1, which represents the TDS gradient here, explained only 37.5% of the variations in diversity, while axis 2 could explain 40.3%. However, it is not clear what environmental gradient(s) the second axis represents. We also assessed the correlation between community compositions and environmental and biological factors using a Mantel test. Although all five factors individually or together were not significantly correlated with the community compositions, TDS still emerged as the most “related” one among them (i.e., it had the highest r value, 0.227, and the lowest P value, 0.125) (Table 4).
TABLE 4.
Mantel test summary statistics
| Parameter | Seven lakes |
Lake NMC seasonal sample |
Lake NMC depth profile |
|||
|---|---|---|---|---|---|---|
| rb | Pc | r | P | r | P | |
| Geographic distance | 0.072 | 0.326 | ||||
| TDS | 0.227 | 0.125 | −0.338 | 0.969 | 0.017 | 0.312 |
| Temp | 0.020 | 0.411 | −0.042 | 0.594 | 0.161 | 0.228 |
| pH | 0.033 | 0.400 | −0.140 | 0.665 | ||
| Sampling depth | 0.674 | 0.035 | ||||
| DO | 0.035 | 0.480 | −0.214 | 0.791 | ||
| Prokaryotic abundance | 0.121 | 0.707 | −0.059 | 0.631 | 0.313 | 0.135 |
| Combinationa | 0.223 | 0.858 | −0.153 | 0.841 | 0.175 | 0.197 |
A combination of environmental parameters, including TDS, temperature, pH, DO, prokaryotic abundance, and sampling depth, if any.
r reflects the correlation level between the two matrices.
A P value of less than 0.05 represents statistical significance of the correlation (shown in boldface).
Microdiversity within dominant picocyanobacterial lineages.
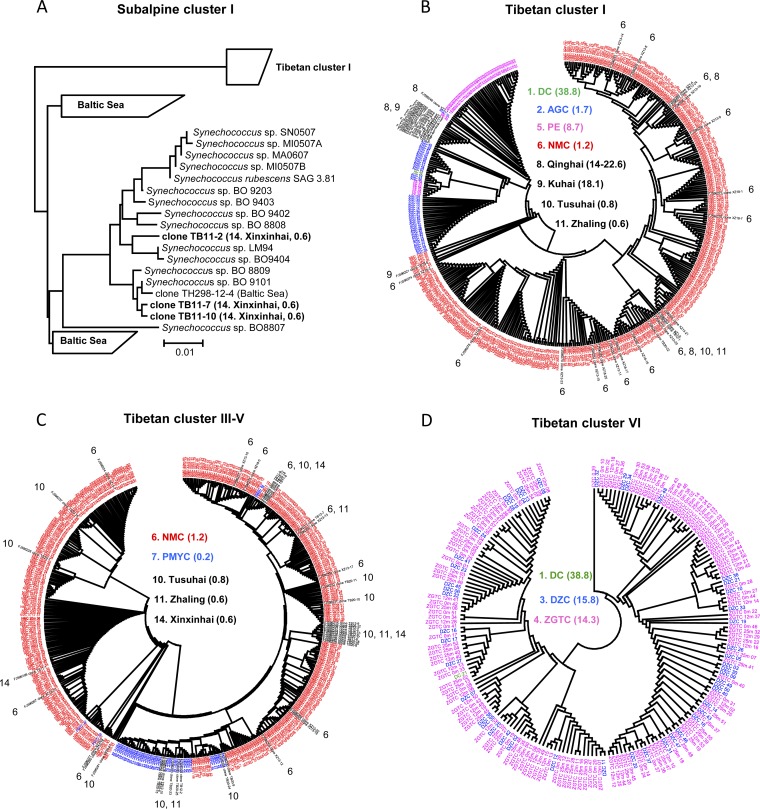
Cluster-specific phylogenies were built in order to address microdiversity for the major lineages, subalpine cluster I (including Tibetan cluster I) and Tibetan clusters III-V and VI (Fig. 4). Subalpine cluster I comprised four major subclusters (Fig. 4A). Similar to the above-mentioned result, within this cluster, Tibetan cluster I was separated from the other three subclusters. Two of these three subclusters consisted solely of Baltic Sea environmental sequences (19), and the remaining one contained Synechococcus strains isolated from lakes (15, 21) and environmental sequences recovered from the Tibetan Lake Xinxinhai (20) and the Baltic Sea (19). Remarkably, Tibetan cluster I comprised environmental sequences derived from lakes with a fairly broad salinity range (TDS densities, 0.6 to 38.8 g/liter). These environmental sequences were clustered mainly corresponding to salinity, that is, sequences from lakes with more similar salinities have closer phylogenetic positions (Fig. 4B). Specifically, sequences from the lower-salinity Lake NMC were tightly grouped together, while those from Lakes AGC, PE, DC, Qinghai, and Kuhai, with higher TDS densities, were more closely related (Fig. 4B). In contrast to Tibetan cluster I, all the sequences within Tibetan cluster III-V were derived from low-salinity lakes (TDS densities, 0.6 to 1.2 g/liter) (Fig. 4C). In this cluster, our sequences from Lakes NMC and PMYC were generally separated. However, sequences retrieved from other Tibetan lakes (Tusuhai, Zhaling, and Xinxinhai) at a considerable geographic distance (Fig. 1) (20) were clustered together with ours (Fig. 4C). Similarly, ZGTC and DZC sequences were deeply mixed within Tibetan cluster VI. These microdiversity patterns indicate little distribution restriction for the two lineages (Tibetan clusters III-V and VI) caused by geographic distance on the plateau (Fig. 4D). No obvious vertical pattern was found along the ZGTC depth profile (Fig. 4D).
FIG 4.
Lineage-specific phylogenetic trees for subalpine cluster I (A), Tibetan cluster I (B), Tibetan cluster III-V (C), and Tibetan cluster VI (D). Sequences recovered in this study are shown in color, and those from the previous study (20) are shown in black. The numbers accompanying the sequences or lake names follow the numbering of lakes in Fig. 1. The TDS density of each lake is shown in parentheses.
Seasonal and vertical variation of picocyanobacterial lineages in Lake NMC.
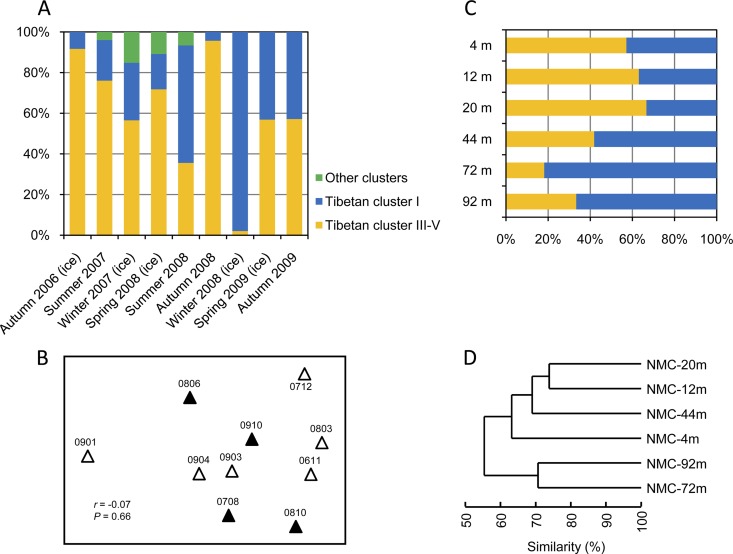
Tibetan cluster III-V and cluster I coexist in Lake NMC, and overall, they are equally abundant in the lake (Fig. 3A). Despite this, the two cooccurring lineages indeed exhibit variations in relative abundance among the seasonal samples (Fig. 5A). However, unexpectedly, no clear seasonal pattern was observed between the ice and non-ice seasons throughout a 3-year time series (Fig. 5A). The NMDS diagram also showed a random pattern of ice season and non-ice season samples, and the ANOSIM analysis did not support respective groupings of ice season and non-ice season samples (r = −0.07; P = 0.66) (Fig. 5C). Moreover, in the Mantel tests, none of the factors (pH, TDS, and prokaryotic abundance) showed significant correlation with the seasonal community variations (Table 4). The vertical pattern of picocyanobacterial population compositions is more apparent, with Tibetan cluster III-V dominating the upper layers and cluster I occupying the deep waters (Fig. 5B). The cluster analysis based on relative abundances of OTUs (determined at a 0.01 sequence dissimilarity cutoff) indicated partitioning of the vertical samples, forming two clusters (4 to 44 m and 72 to 92 m) that correspond to depth (Fig. 5D). Mantel tests also predicted sampling depth to be the only factor that significantly correlated with the vertical community compositions (r = 0.674; P < 0.05) (Table 4).
FIG 5.
(A) Seasonal variation of picocyanobacterial community compositions in Lake NMC across 3 years. (B) NMDS showing the similarities among seasonal picocyanobacterial communities of Lake NMC. Late autumn, winter, and spring samples (ice seasons) are indicated by open triangles and summer and early autumn samples (non-ice seasons) by filled triangles. The numbers accompanying the triangles indicate the sampling years and months, for example, 0806 represents June 2008. The results of the ANOSIM test (r = −0.07; P = 0.66) are shown. (C) Vertical variation of picocyanobacterial community compositions in Lake NMC along a depth profile of six layers. (D) Clustering of the picocyanobacterial communities along a vertical profile of Lake NMC.
DISCUSSION
Our phylogenetic analysis based on ITS sequences (Fig. 2) supports the notion that picocyanobacteria of the Syn/Pro clade over the world's continental and marine aquatic environments are highly diverse (15, 16). It was also shown that nonmarine picocyanobacteria comprise more diverse lineages than their marine counterparts (Fig. 2). However, surprisingly, the picocyanobacterial community in each of the Tibetan lakes was dominated by only one or two phylogenetic clusters (Fig. 3), irrespective of their different sizes, salinities, and alkalinities. More samples from vertical depth profiles and/or from seasonal time series could not increase the alpha diversity in a lake, resulting in a very monotonic cluster level community structure of a lake. Similar low taxa richness of picocyanobacterial (20) and the total bacterial (44) communities in Tibetan lakes was also described. These results contrast with previous observations of diverse picocyanobacterial lineages in other lakes at lower elevations, such as Lake Constance (∼400 m above sea level) (17), Lake Mondsee (∼500 m) (16), Lake Superior (∼180 m) (22), and the Great Mazurian Lakes (∼110 m) (21), or in the brackish Baltic Sea (18, 19). Evidence also showed that individual water bodies can harbor a diverse range of picocyanobacteria (27). However, the environmental stresses in the Tibetan lakes, such as high levels of solar radiation, freezing in winter, oligotrophic nutrient conditions, and high salt concentrations (34), may strongly limit the alpha diversity of picocyanobacteria in each lake. It is likely that, during evolutionary history, only one or a few picocyanobacterial lineages have adapted to a particular lake on the plateau, the highest geographic region in the world.
In contrast to the extremely low alpha diversity, a high level of beta diversity of picocyanobacteria was observed among the Tibetan lakes, which is evidenced by the distinct picocyanobacterial communities when comparing lakes (Fig. 3A). We tried to demonstrate the possible factors that could explain the community composition variations among these lakes. We also attempted to look into the question of whether the picocyanobacteria are randomly dispersed among these high mountain lakes or follow a certain niche-based distribution, testing the well-known hypothesis that “everything is everywhere, but the environment selects” (29).
First, the CCA identified TDS as a factor that could in part explain the variations in picocyanobacterial diversity among these lakes (Fig. 3B). However, it is very possible that some other unknown factor(s) could also contribute to the explanation. The Mantel tests showed little correlation between the picocyanobacterial community composition and the geographic distances of lake pairs (r = 0.072; P = 0.326) (Table 4), suggesting that the picocyanobacterial taxa may not be randomly dispersed on the plateau. However, the Mantel tests failed to identify any environmental or biological factor that may constrain the distribution of picocyanobacteria on the plateau. We consider that this may result from the extremely monotonic community of each lake and the completely distinct communities across lakes, which lack shared OTUs as connections to measure the community composition similarities between lakes. Besides the statistical tests, direct inference could suggest a more obvious relationship among community composition, geographic distance, and some environmental factor. On one hand, lakes within the same range of salinity but in distant locations can have similar genetic populations, such as Lakes DZC and ZGTC (Fig. 1, no. 3 and 4) or Lakes AGC and PE (Fig. 1, no. 2 and 5). On the other hand, lakes with very different salinities may have disparate picocyanobacterial communities despite their small geographic distances, such as Lakes NMC, PE, and ZGTC (Fig. 1, no. 4 to 6). Moreover, the community shift from PMYC and NMC to PE and AGC also reflects a clear salinity gradient (Fig. 3). It appears that some physicochemical characteristics of the environment, such as salinity, rather than geographic distance have a predominant impact on the similarity or dissimilarity between picocyanobacterial communities.
Second, several globally widespread picocyanobacterial clusters can also inhabit the Tibetan lakes, including both rare (C. gracile cluster and subalpine cluster II) and dominant (subalpine cluster I and group I) clusters. These lineages can dwell in both freshwater and brackish water (Table 1), suggestive of a ubiquitous dispersal and a broad adaptive radiation (15, 16). Such cosmopolitan distribution implies that “everything is everywhere.” However, several recently or newly found clusters, such as Tibetan clusters III-V, VI, and VII, have plausible geographic restriction to the plateau. The absence of these lineages in other geographic regions could result from undersampling (23), and we speculate that they may also reside in similar environments outside the plateau. Moreover, many picocyanobacterial genotypes were defined by using other molecular markers, such as the 16S rRNA gene sequences and phycobiliprotein-encoding genes (cpc and cpe) (5, 16). We also cannot exclude the possibility that these Tibetan clusters have been found elsewhere using other gene markers, since direct comparison between phylogenies built using different markers is not possible without enough cultured strains.
Furthermore, the Tibetan picocyanobacterial clusters displayed clear salinity-based partitioning. Tibetan cluster III-V likely can only stand low TDS, while cluster I can exist in low- to high-TDS waters but not in freshwater (Lake PMYC). Group I exhibited a broad salinity adaptation, as picocyanobacteria in the cluster could thrive in the hypersaline Tibetan Lake DC, as well as in freshwater lakes (16) and in the brackish Baltic Sea (18, 19). Tibetan cluster VI may prefer polysaline conditions. Thus, the Tibetan picocyanobacteria likely comprise both stenohaline and euryhaline types, which are adapted to a narrow or a broad range of salt concentrations, respectively (45). At the microdiversity scale, geographic-location-based partitioning could be found within Tibetan cluster I (Fig. 4B). However, such differentiation is more likely a salinity-based partitioning within this broad salinity-adapted lineage. In contrast, differentiation within Tibetan cluster III-V or within cluster VI was not observed; instead, sequences in these clusters from different lakes lacked recognizable phylogenetic distance that can discriminate lakes (Fig. 4C and D). This suggests a lack of geographic coherence for each of the two lineages and further implies their wide distribution on the plateau.
Similarly, salinity was also inferred to be a strong factor controlling picocyanobacterial (20), bacterial (46, 47), and archaeal (35, 48) communities in the Tibetan lakes. Salt concentration is thought to be an essential factor that separates freshwater, marine, and hypersaline cyanobacteria or that divides stenohaline and euryhaline cyanobacteria, specific strains or genetic lineages of which have different growth requirements for Na+, Mg+, and Ca2+ (3, 45, 49). Thus, salinity likely reflects a fundamental niche for these alkaline Tibetan lakes, and adaptation to different ranges of salinity may be related to the widespread or restricted distribution of picocyanobacterial lineages.
We were surprised that no clear seasonal pattern could be inferred when comparing the relative abundances of Tibetan cluster I and cluster III-V in Lake NMC (Fig. 5A and C). Although Tibetan cluster I and cluster III-V picocyanobacteria likely favor different salinity ranges, comparison of their relative abundances appeared to result in a random temporal pattern in Lake NMC. On the other hand, we indeed observed a depth-related pattern in a vertical profile of the lake (Fig. 5B and D and Table 4). It seems that Tibetan clusters III-V and I took over the communities in upper and deep waters, respectively, in this single case. Nevertheless, it is unclear why this particular lake allows the coexistence of two overall equally abundant picocyanobacterial lineages that have a distant phylogenetic relationship and disparate salinity adaptations, whereas other lakes appear to be dominated by only one lineage. It will be interesting to further explore the variation and succession between these two lineages on fine temporal and spatial scales in Lake NMC.
In summary, our study emphasizes the role of dispersal for picocyanobacteria and supports the notion that niche barriers rather than geographic isolation may limit the dispersive potential of picocyanobacteria among inland aquatic ecosystems. The endemicity of some lineages may result from environment specification and niche adaptation. Our study provides evidence for the idea that both dispersal and adaptation contribute to forming the biogeography of microbes (50) and support the hypothesis that “everything is everywhere, but the environment selects” (29).
ACKNOWLEDGMENTS
Y. Liu was supported by NSFC grant 41171050 and S. Huang by NSFC grant 41206131.
Footnotes
Published ahead of print 3 October 2014
REFERENCES
- 1.Weisse T. 1993. Dynamics of autotrophic picoplankton in marine and freshwater ecosystems, p 327–370 In Jones JG. (ed), Advances in microbial ecology, vol 13 Springer, New York, NY. [Google Scholar]
- 2.Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, Post AF, Hagemann M, Paulsen I, Partensky F. 2009. Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 73:249–299. 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herdman M, Castenholz R, Iteman I, Waterbury J, Rippka R. 2001. Subsection I (formerly Chroococcales Wettstein 1924, emend. Rippka, Deruelles, Waterbury, Herdman and Stanier 1979), p 493-514 In Boone DR, Castenholz RW, Garrity GM. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 1 Springer, New York, NY. [Google Scholar]
- 4.Honda D, Yokota A, Sugiyama J. 1999. Detection of seven major evolutionary lineages in cyanobacteria based on the 16S rRNA gene sequence analysis with new sequences of five marine Synechococcus strains. J. Mol. Evol. 48:723–739. 10.1007/PL00006517. [DOI] [PubMed] [Google Scholar]
- 5.Robertson BR, Tezuka N, Watanabe MM. 2001. Phylogenetic analyses of Synechococcus strains (cyanobacteria) using sequences of 16S rDNA and part of the phycocyanin operon reveal multiple evolutionary lines and reflect phycobilin content. Int. J. Syst. Evol. Microbiol. 51:861–871. 10.1099/00207713-51-3-861. [DOI] [PubMed] [Google Scholar]
- 6.Rocap G, Distel DL, Waterbury JB, Chisholm SW. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 68:1180–1191. 10.1128/AEM.68.3.1180-1191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zwirglmaier K, Heywood JL, Chamberlain K, Woodward EM, Zubkov MV, Scanlan DJ. 2007. Basin-scale distribution patterns of picocyanobacterial lineages in the Atlantic Ocean. Environ. Microbiol. 9:1278–1290. 10.1111/j.1462-2920.2007.01246.x. [DOI] [PubMed] [Google Scholar]
- 8.Zwirglmaier K, Jardillier L, Ostrowski M, Mazard S, Garczarek L, Vaulot D, Not F, Massana R, Ulloa O, Scanlan DJ. 2008. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ. Microbiol. 10:147–161. 10.1111/j.1462-2920.2007.01440.x. [DOI] [PubMed] [Google Scholar]
- 9.Huang S, Wilhelm S, Harvey H, Taylor K, Jiao N, Chen F. 2012. Novel lineages of Prochlorococcus and Synechococcus in the global oceans. ISME J. 6:285–297. 10.1038/ismej.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuller NJ, Marie D, Partensky F, Vaulot D, Post AF, Scanlan DJ. 2003. Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Appl. Environ. Microbiol. 69:2430–2443. 10.1128/AEM.69.5.2430-2443.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chisholm SW, Frankel SL, Goericke R, Olson RJ, Palenik B, Waterbury JB, West-Johnsrud L, Zettler ER. 1992. Prochlorococcus marinus nov. gen. nov. sp.: an oxyphototrophic marine prokaryote containing divinyl chlorophyll a and b. Arch. Microbiol. 157:297–300. 10.1007/BF00245165. [DOI] [Google Scholar]
- 12.Dufresne A, Ostrowski M, Scanlan DJ, Garczarek L, Mazard S, Palenik BP, Paulsen IT, de Marsac NT, Wincker P, Dossat C, Ferriera S, Johnson J, Post AF, Hess WR, Partensky F. 2008. Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 9:R90. 10.1186/gb-2008-9-5-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marin B, Nowack EC, Glockner G, Melkonian M. 2007. The ancestor of the Paulinella chromatophore obtained a carboxysomal operon by horizontal gene transfer from a Nitrococcus-like gamma-proteobacterium. BMC Evol. Biol. 7:85. 10.1186/1471-2148-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez-Baracaldo P, Hayes P, Blank C. 2005. Morphological and habitat evolution in the Cyanobacteria using a compartmentalization approach. Geobiology 3:145–165. 10.1111/j.1472-4669.2005.00050.x. [DOI] [Google Scholar]
- 15.Ernst A, Becker S, Wollenzien UI, Postius C. 2003. Ecosystem-dependent adaptive radiations of picocyanobacteria inferred from 16S rRNA and ITS-1 sequence analysis. Microbiology 149:217–228. 10.1099/mic.0.25475-0. [DOI] [PubMed] [Google Scholar]
- 16.Crosbie ND, Pockl M, Weisse T. 2003. Dispersal and phylogenetic diversity of nonmarine picocyanobacteria, inferred from 16S rRNA gene and cpcBA-intergenic spacer sequence analyses. Appl. Environ. Microbiol. 69:5716–5721. 10.1128/AEM.69.9.5716-5721.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becker S, Singh AK, Postius C, Boger P, Ernst A. 2004. Genetic diversity and distribution of periphytic Synechococcus spp. in biofilms and picoplankton of Lake Constance. FEMS Microbiol. Ecol. 49:181–190. 10.1016/j.femsec.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Haverkamp TH, Schouten D, Doeleman M, Wollenzien U, Huisman J, Stal LJ. 2009. Colorful microdiversity of Synechococcus strains (picocyanobacteria) isolated from the Baltic Sea. ISME J. 3:397–408. 10.1038/ismej.2008.118. [DOI] [PubMed] [Google Scholar]
- 19.Haverkamp T, Acinas SG, Doeleman M, Stomp M, Huisman J, Stal LJ. 2008. Diversity and phylogeny of Baltic Sea picocyanobacteria inferred from their ITS and phycobiliprotein operons. Environ. Microbiol. 10:174–188. 10.1111/j.1462-2920.2007.01442.x. [DOI] [PubMed] [Google Scholar]
- 20.Wu QL, Xing P, Liu WT. 2010. East Tibetan lakes harbour novel clusters of picocyanobacteria as inferred from the 16S-23S rRNA internal transcribed spacer sequences. Microb. Ecol. 59:614–622. 10.1007/s00248-009-9603-z. [DOI] [PubMed] [Google Scholar]
- 21.Jasser I, Krolicka A, Karnkowska-Ishikawa A. 2011. A novel phylogenetic clade of picocyanobacteria from the Mazurian lakes (Poland) reflects the early ontogeny of glacial lakes. FEMS Microbiol. Ecol. 75:89–98. 10.1111/j.1574-6941.2010.00990.x. [DOI] [PubMed] [Google Scholar]
- 22.Ivanikova NV, Popels LC, McKay RM, Bullerjahn GS. 2007. Lake Superior supports novel clusters of cyanobacterial picoplankton. Appl. Environ. Microbiol. 73:4055–4065. 10.1128/AEM.00214-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felföldi T, Somogyi B, Márialigeti K, Vörös L. 2011. Notes on the biogeography of non-marine planktonic picocyanobacteria: re-evaluating novelty. J. Plankton Res. 33:1622–1626. 10.1093/plankt/fbr051. [DOI] [Google Scholar]
- 24.Coleman ML, Chisholm SW. 2007. Code and context: Prochlorococcus as a model for cross-scale biology. Trends Microbiol. 15:398–407. 10.1016/j.tim.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Moore LR, Rocap G, Chisholm SW. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464–467. 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 26.Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EM, Chisholm SW. 2006. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311:1737–1740. 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez-Baracaldo P, Handley BA, Hayes PK. 2008. Picocyanobacterial community structure of freshwater lakes and the Baltic Sea revealed by phylogenetic analyses and clade-specific quantitative PCR. Microbiology 154:3347–3357. 10.1099/mic.0.2008/019836-0. [DOI] [PubMed] [Google Scholar]
- 28.Fenchel T, Finlay BJ. 2004. The ubiquity of small species: patterns of local and global diversity. Bioscience 54:777–784. 10.1641/0006-3568(2004)054[0777:TUOSSP]2.0.CO;2. [DOI] [Google Scholar]
- 29.De Wit R, Bouvier T. 2006. ‘Everything is everywhere, but, the environment selects'; what did Baas Becking and Beijerinck really say? Environ. Microbiol. 8:755–758. 10.1111/j.1462-2920.2006.01017.x. [DOI] [PubMed] [Google Scholar]
- 30.Whitaker RJ, Grogan DW, Taylor JW. 2003. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301:976–978. 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- 31.Papke RT, Ramsing NB, Bateson MM, Ward DM. 2003. Geographical isolation in hot spring cyanobacteria. Environ. Microbiol. 5:650–659. 10.1046/j.1462-2920.2003.00460.x. [DOI] [PubMed] [Google Scholar]
- 32.Finlay BJ. 2002. Global dispersal of free-living microbial eukaryote species. Science 296:1061–1063. 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 33.Finlay BJ, Clarke KJ. 1999. Ubiquitous dispersal of microbial species. Nature 400:828. 10.1038/23616. [DOI] [Google Scholar]
- 34.Zheng M, Tang J, Liu J, Zhang F. 1993. Chinese saline lakes. Hydrobiologia 267:23–26. 10.1007/BF00018789. [DOI] [Google Scholar]
- 35.Hu A, Yao T, Jiao N, Liu Y, Yang Z, Liu X. 2010. Community structures of ammonia-oxidising archaea and bacteria in high-altitude lakes on the Tibetan Plateau. Freshwat. Biol. 55:2375–2390. [Google Scholar]
- 36.Cai H, Wang K, Huang S, Jiao N, Chen F. 2010. Distinct patterns of picocyanobacterial communities in winter and summer in the Chesapeake Bay. Appl. Environ. Microbiol. 76:2955–2960. 10.1128/AEM.02868-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larkin M, Blackshields G, Brown N, Chenna R, McGettigan P, McWilliam H, Valentin F, Wallace I, Wilm A, Lopez R. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 38.Felsenstein J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164–166. [Google Scholar]
- 39.Schloss PD, Handelsman J. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501–1506. 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swofford DL. 2002. PAUP: phylogenetic analysis using parsimony (* and other methods), version 4. Sinauer Associates, Sunderland, MA. [Google Scholar]
- 41.Guindon S, Lethiec F, Duroux P, Gascuel O. 2005. PHYML Online—a Web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557–W559. 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818. 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 43.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30:2725–2729. 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xing P, Hahn MW, Wu QL. 2009. Low taxon richness of bacterioplankton in high-altitude lakes of the eastern Tibetan Plateau, with a predominance of Bacteroidetes and Synechococcus spp. Appl. Environ. Microbiol. 75:7017–7025. 10.1128/AEM.01544-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Golubic S. 1980. Halophily and halotolerance in cyanophytes. Orig. Life 10:169–183. 10.1007/BF00928667. [DOI] [Google Scholar]
- 46.Wu QL, Zwart G, Schauer M, Kamst-van Agterveld MP, Hahn MW. 2006. Bacterioplankton community composition along a salinity gradient of sixteen high-mountain lakes located on the Tibetan Plateau, China. Appl. Environ. Microbiol. 72:5478–5485. 10.1128/AEM.00767-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang H, Dong H, Yu B, Liu X, Li Y, Ji S, Zhang CL. 2007. Microbial response to salinity change in Lake Chaka, a hypersaline lake on Tibetan Plateau. Environ. Microbiol. 9:2603–2621. 10.1111/j.1462-2920.2007.01377.x. [DOI] [PubMed] [Google Scholar]
- 48.Jiang H, Dong H, Deng S, Yu B, Huang Q, Wu Q. 2009. Response of archaeal community structure to environmental changes in lakes on the Tibetan Plateau, northwestern China. Geomicrobiol. J. 26:289–297. 10.1080/01490450902892662. [DOI] [Google Scholar]
- 49.Waterbury JB, Watson SW, Valois FW, Franks DG. 1986. Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus. Can. Bull. Fish. Aquat. Sci. 214:71–120. [Google Scholar]
- 50.Martiny JBH, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102–112. 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 51.Katano T, Fukui M, Watanabe Y. 2001. Identification of cultured and uncultured picocyanobacteria from a mesotrophic freshwater lake based on the partial sequences of 16S rDNA. Limnology 2:213–218. 10.1007/s10201-001-8038-0. [DOI] [Google Scholar]
- 52.Jones SE, Newton RJ, McMahon KD. 2009. Evidence for structuring of bacterial community composition by organic carbon source in temperate lakes. Environ. Microbiol. 11:2463–2472. 10.1111/j.1462-2920.2009.01977.x. [DOI] [PubMed] [Google Scholar]
- 53.Chen F, Wang K, Kan J, Suzuki MT, Wommack KE. 2006. Diverse and unique picocyanobacteria in Chesapeake Bay, revealed by 16S-23S rRNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 72:2239–2243. 10.1128/AEM.72.3.2239-2243.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi DH, Noh JH. 2009. Phylogenetic diversity of Synechococcus strains isolated from the East China Sea and the East Sea. FEMS Microbiol. Ecol. 69:439–448. 10.1111/j.1574-6941.2009.00729.x. [DOI] [PubMed] [Google Scholar]
- 55.Ahlgren NA, Rocap G. 2006. Culture isolation and culture-independent clone libraries reveal new marine Synechococcus ecotypes with distinctive light and N physiologies. Appl. Environ. Microbiol. 72:7193–7204. 10.1128/AEM.00358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]