Abstract
Microbiologists have been using agar growth medium for over 120 years. It revolutionized microbiology in the 1890s when microbiologists were seeking effective methods to isolate microorganisms, which led to the successful cultivation of microorganisms as single clones. But there has been a disparity between total cell counts and cultivable cell counts on plates, often referred to as the “great plate count anomaly,” that has long been a phenomenon that still remains unsolved. Here, we report that a common practice microbiologists have employed to prepare agar medium has a hidden pitfall: when phosphate was autoclaved together with agar to prepare solid growth media (PT medium), total colony counts were remarkably lower than those grown on agar plates in which phosphate and agar were separately autoclaved and mixed right before solidification (PS medium). We used a pure culture of Gemmatimonas aurantiaca T-27T and three representative sources of environmental samples, soil, sediment, and water, as inocula and compared colony counts between PT and PS agar plates. There were higher numbers of CFU on PS medium than on PT medium using G. aurantiaca or any of the environmental samples. Chemical analysis of PT agar plates suggested that hydrogen peroxide was contributing to growth inhibition. Comparison of 454 pyrosequences of the environmental samples to the isolates revealed that taxa grown on PS medium were more reflective of the original community structure than those grown on PT medium. Moreover, more hitherto-uncultivated microbes grew on PS than on PT medium.
INTRODUCTION
An enigma termed the “great plate count anomaly” is a central issue in microbiology; only a small proportion of viable cells in microbial communities can be grown using agar (or other gelling agents) growth media (1). Investigators have been attempting to identify factors limiting cultivation of most microbes on agar plates for a long time (2, 3), but more research is necessary since numerous phylogenetic groups of environmental microbes still lack cultivated representatives. With the advent of state-of-the-art sequencing technologies (4, 5), researchers have been trying to circumvent the limitation and laboriousness of isolation work by sequencing genomes of entire communities to reveal potential novel functions and phylogenetic relationships (6–8). Nonetheless, the isolation of microorganisms is still necessary for microbiologists to study organisms genetically and physiologically in depth in the laboratory.
A number of different strategies have been used to successfully increase the number of cells cultivated from environmental samples as well as to isolate novel bacterial taxa. Some examples of strategies that have been successfully used to improve cultivation efficiency include mimicking the natural environment (9, 10), physically separating cells to decrease competition or growth inhibitors (11, 12), and modifying growth media to be more reflective of the natural environment (12, 13). In some cases investigators have subsequently identified specific factors required to cultivate previously uncultivated taxa (14, 15). A number of studies have empirically shown increases in CFU counts when catalase or pyruvate was added to the medium (16–22), indicating that oxidative stress can inhibit colony growth. The variety of factors that may be limiting laboratory cultivation is vast, making it a challenge to identify specific growth medium components required for, or inhibitory to, cell growth.
In addition to chemical components and growth conditions, the method used for medium preparation can also impact cultivation. Detrimental and beneficial chemical reactions between medium components have been found to occur during autoclaving of liquid medium. Early studies demonstrated both growth inhibition (23) and stimulation (24) of some bacterial species when phosphate and glucose were autoclaved together. Maillard reaction products have also been shown to inhibit bacterial growth (25). More recently, furan-2-carboxylic acids, which can inhibit bacterial swarming on agar surfaces, were found in various amounts in commercial agars (26). But studies on specific factors in agar leading to bacterial growth inhibition are limited.
Gemmatimonas aurantiaca T-27T was isolated in our laboratory as the first cultivated representative of the phylum Gemmatimonadetes (27). This organism had eluded cultivation for many years because it grew poorly (very low CFU counts and slow growth) on agar plates, but it was finally found to grow well on medium solidified with gellan gum (28). Since the only difference between the growth media was the gelling agents used, we hypothesized that the agar or products produced from the agar interacting with some other medium components were inhibiting G. aurantiaca growth. The first objective of the study was to identify differences in G. aurantiaca cultivation on agar growth media prepared using two different procedures, i.e., with phosphate and agar autoclaved separately (PS method, where P is phosphate and S is separately) or together (PT method, where T is together). Hydrogen peroxide concentrations were measured in these media to test if a reactive oxygen species (ROS) was potentially contributing to growth inhibition. The second objective of this research was to determine differential colony growth on these agar medium preparations inoculated with samples collected from three different environmental sources. This paper scrutinizes the negative effect of autoclaving phosphate together with agar on colony formation of bacteria to determine if this pitfall of agar medium preparation has been a long-overlooked factor contributing to reduced cultivation efficiency from environmental samples.
MATERIALS AND METHODS
Gemmatimonas aurantiaca cultivation conditions.
G. aurantiaca T-27T (27) was routinely grown in liquid PYG medium (Polypeptone [Nihon Pharmaceutical Co., Ltd., Tokyo, Japan], yeast extract, and glucose; 0.5 g liter−1 of each) at 30°C with shaking. Growth inhibition of G. aurantiaca was tested in PYG medium with the addition of different concentrations of phosphate buffer (ranging from 0.5 to 10 mM Na2HPO4-KH2PO4, pH 7.0) and agar (15 g liter−1). Media were prepared by autoclaving the phosphate buffer and agar either together (T) or separately (S). The PYG medium (20× concentrated) component was always autoclaved separately from the agar and phosphate buffer. Diluted G. aurantiaca cultures (100 μl; optical density at 600 nm [OD600] of ∼10−5) were spread onto the agar media and incubated at 30°C for 11 days.
To test if colony formation could be restored by catalase, 10 μl of filter-sterilized bovine liver catalase solution (10 mg/ml, dissolved in 100 mM phosphate buffer; Wako Pure Chemical Industries, Ltd., Osaka Japan) (T-Cat) was placed in the center of agar plates prepared by autoclaving phosphate buffer (3 mM) and agar together as described above. Controls had 10 μl of 100 mM phosphate buffer (T-PB) added to plates instead of the catalase solution. Plates were incubated at 30°C for 14 days. All media were autoclaved for 15 min at 121°C.
Hydrogen peroxide analyses.
The agar-solidified medium was frozen at −80°C and then thawed at room temperature in order to cause syneresis. The remaining gel solids were squeezed, and the liquid fraction was collected for analysis. The H2O2 concentration in the liquid fraction was measured according to the method of Jiang et al. (29), with minor modifications. The sample (0.5 ml) was mixed with 10 μl of 50 mM phosphate buffer (pH 7.0), and then 0.5 ml of freshly prepared 2× assay reagent (200 mM sorbitol, 200 μM xylenol orange, 500 μM ferrous ammonium sulfate, 50 mM H2SO4) was added. A sample blank was prepared by eliminating the H2O2 in the sample prior to the addition of the 2× assay reagent; 50 mM phosphate buffer that contains bovine liver catalase (100 μg/ml) was used instead of the plain buffer. Absorbance was read at 560 nm after a 45-min incubation at room temperature. The concentration of the H2O2 standard solution was determined using the extinction coefficient of 43.6 M−1 · cm−1 at 240 nm.
To determine if H2O2 generation after autoclaving was dependent on phosphate concentration, the effect of increasing phosphate was tested. Solutions of 0.75 g of agar suspended in 50 ml of buffer with phosphate concentrations ranging from 0 (i.e., pure H2O) to 30 mM (Na2HPO4-KH2PO4, pH 7.0) were autoclaved together or separately. For the separate preparation, phosphate buffer (100 mM Na2HPO4-KH2PO4 stock, pH 7.0) was autoclaved and then added to achieve final concentrations ranging from 1 to 30 mM. At least five independent experiments were performed.
Environmental sample sources and collection.
Sources used for cultivation experiments were soil, sediment, and water. Samples were collected in triplicate from three different locations in the city of Sapporo, Hokkaido, Japan (43°7′N, 141°34′E). Soil and sediment samples were collected within Hokkaido University campus. Soil samples from a depth of 5 to 10 cm below the surface were collected from a small forest near the Faculty of Agriculture building. Sediment samples were taken from the bottom of a shallow pond (0 to 10 cm) located in the Hokkaido University campus. Water samples were taken from the surface of the Toyohira River flowing through the center of the city of Sapporo. Reproducibility and variability in results were determined using triplicate samples collected within a square-meter area at each location. Samples were immediately placed on ice after collection and stored at 4°C in the laboratory until used in cultivation experiments within 3 h of collection. Subsamples were stored at −20°C until DNA extractions were performed.
Environmental sample growth medium preparation.
A bacterial growth medium similar to the one used in the G. aurantiaca experiments, but with some additional components, was used to increase the diversity of environmental isolates (30). The medium constituents were grouped into three, and each solution (solutions A, B, and C) was prepared beforehand with final concentrations as follows: for solution A, 2.27 mM (NH4)2SO4, 0.2 mM MgSO4, 45 μM CaCl2, and 15 g liter−1 Bacto agar; for solution B, 10 mM KH2PO4 and 10 mM Na2HPO4·12H2O; for solution C, 0.1 g liter−1 Bacto peptone, 0.1 g liter−1 Bacto yeast extract, and 0.1 g liter−1 glucose. Using these solutions, three medium/preparation methods, designated PT, PS, and PW (where W is without), were tested. The third component was always autoclaved separately to prevent glucose from reacting with phosphate during autoclaving. The PT method allowed phosphate and agar to interact during autoclaving as solutions A and B were mixed together prior to autoclaving, and then the separately autoclaved solution C was added prior to pouring the medium. With the PS method, phosphate and agar were not exposed together during autoclaving; solutions A, B, and C were autoclaved separately and then mixed. For the PW method, solution B was excluded from the medium and solutions A and C were autoclaved separately and mixed to make the medium theoretically phosphate free. But inductively coupled plasma-atomic emission spectrometry (ICP-AES) and Optima 4300DV (31) analysis of acid-hydrolyzed PW medium indicated that a trace amount of phosphate (ca. 6 μM) was still present, likely originating from trace P contamination in the chemicals used to make the medium. All media were autoclaved for 15 min at 121°C.
Cultivation using environmental samples.
Three independent samples for each environmental source were suspended in sterile distilled water and diluted in a 10-fold series from 10−1 to 10−6 using sterile distilled water. Aliquots (100 μl) from each dilution were inoculated onto five replicate plates (90 by 20 mm) of PT, PS, and PW media and incubated at 25°C under dark conditions. This provided both biological and technical replication to determine major sources of variation in the results. The number of CFU on each medium was determined from 1 week of daily colony counts. Only plates with 30 to 300 CFU were included in the cultivation results reported. A total of 6,528 colonies were randomly picked, representing all environmental source-cultivation medium combinations, and identified based on 16S rRNA gene sequences.
DNA extraction.
Total DNA from soil and sediment samples was extracted using an Extrap Soil DNA Plus kit, version 2 (Nippon Steel and Sumikin Eco-Tech Corporation) according to the manufacturer's instructions. Total DNA from river samples was extracted using the protocol of Fuhrman et al. (32) with the exception that a membrane prefilter (10-μm pore size; Advantec) was used instead of a glass filter. The extracted DNA was dissolved in 50 μl of Tris-EDTA (TE) buffer, quality checked by a 1% agarose gel stained with ethidium bromide, quantified by spectrophotometry at an OD260, and stored at −20°C until use.
DNA Sanger sequencing and 454 pyrosequencing.
After a week, colonies from the agar plates were transferred to PCR tubes containing lysis solution, and chromosomal DNA to be used as the template for sequencing was extracted by heating tubes in a microwave (50 s). A partial fragment of the 16S rRNA gene was PCR amplified using a 1:1 mixture of 10 pmol/liter of universal primers 27F (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 1492R (5′-TAC GGY TAC CTT GTT ACG ACT T-3′) (33) and a KOD FX Neo kit (Toyobo). PCR was carried out in 25-μl reaction volumes using Perkin-Elmer GeneAmp System 9700 (Perkin-Elmer) with the following thermal cycling program: initial denaturation at 94°C for 2 min, followed by 30 cycles of 98°C for 10 s, 55°C for 30 s, and 68°C for 90 s, with a final hold at 4°C. The PCR products were purified using a NucleoSpin Gel and PCR Cleanup kit (TaKaRa) and then used as templates for sequencing. Sanger sequencing was performed using primer 357F (5′-CTC CTA CGG GAG GCA GCA G-3′) by the TaKaRa Bio Company.
For 454 pyrosequencing PCR amplification was performed using the 16S rRNA gene universal primers 519F (5′-CAG CMG CCG CGG TAA TWC-3′) and 907R (5′-CCG YCA ATT CMT TTR AGT T-3′) with a Roche sequencing adapter added to the forward primer. Samples were multiplexed using 10-nucleotide-long bar codes. PCRs were carried out in 20-μl volumes using a Phusion High Fidelity DNA polymerase kit (Finnzymes) according to the manufacturer's instructions, with the following thermal cycling program: initial denaturation at 98°C for 30 s, followed by 25 cycles of 98°C for 8 s, 70°C for 20 s, and 72°C for 30 s, with a 5-min final extension step at 72°C. The concentration of template used for PCR was 60 ng · μl−1 for soil and sediment samples and 10 ng · μl−1 for river samples. Amplicons from eight independent PCRs were combined into one tube and then purified using a Wizard SV Gel and PCR Clean-Up System (Promega). Pyrosequencing was performed by Hokkaido System Science Co., Ltd., using a Genome Sequencer FLX and GS FLX Titanium reagents (Roche).
Sequence analyses.
The 16S rRNA gene sequences, determined using the Sanger sequencing method, from all isolated bacteria were checked for chimeras using Decipher (34), and quality was checked using an ABI 3730xl base caller (Applied Biosystems). Possible chimeric sequences and sequences less than 400 bases were deleted. To determine the novelty of taxa isolated, all sequences were taxonomically classified using the Ribosomal Database Project (RDP) Classifier at a confidence threshold of 80%. Isolates were considered novel if they had less than a 97% match to the RDP database using the Classifier program (35). Diversity analysis was performed using the QIIME pipeline (36).
Pyrosequence data were processed using the QIIME pipeline (36) (version 1.7). Pyrosequences were first denoised (37) and then clustered into operational taxonomic units (OTUs) along with the isolate sequences at the 97% similarity level using UCLUST (38). The OTUs were aligned using PyNAST (39) against the Greengenes core reference alignment (40). Chimeric sequences were removed using Chimera Slayer (41). The taxonomic identity of each phylotype was determined using RDP Classifier (35) based on Greengenes taxonomy and a Greengenes reference database (42) (version 13_5). Alpha diversity was analyzed using the following metrics: total observed species, Chao1, Shannon, and phylogenetic distance (PD). Principal coordinate analysis (PCoA; beta-diversity analysis) was conducted using Euclidean distances (Bray-Curtis) and phylogenetic distances (weighted and unweighted UniFrac) (43).
Statistical analysis.
Data were presented as means with standard errors (± SE) or standard deviations (SD). Significant differences among different medium preparations were determined using a Student t test. Statistical analysis (e.g., perANOVA) comparing taxa between communities was performed using software available through the QIIME pipeline. All differences were considered significant at a P value of <0.05.
RESULTS
Gemmatimonas aurantiaca cultivation on agar medium.
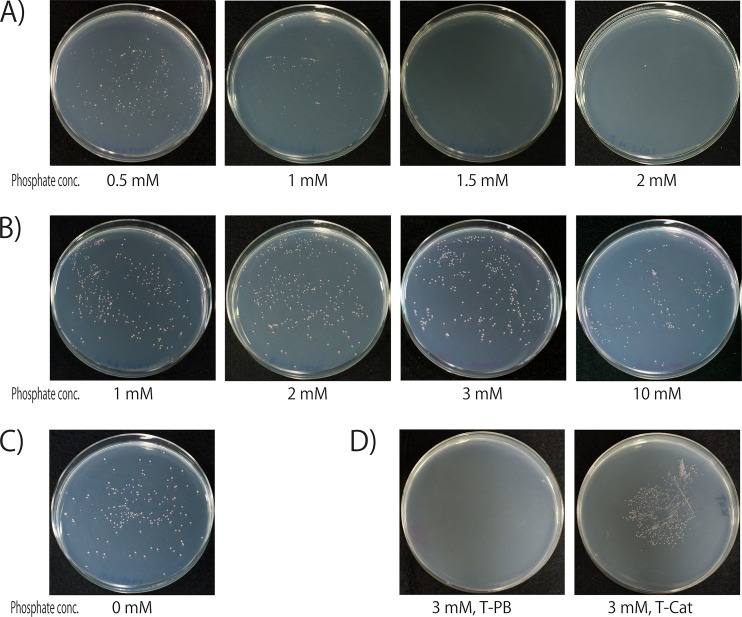
There was a distinct difference in colony numbers on solid medium when agar and phosphate were autoclaved separately versus together (Fig. 1A and B). When agar was autoclaved separately, equivalent colony numbers (∼300 CFU) grew on plates with phosphate concentrations ranging from 0.5 to 3 mM. There was a slight reduction when 10 mM phosphate was used (∼180 CFU). Growth was even seen in control plates with no phosphate added (Fig. 1C), suggesting that there was sufficient phosphate contamination in the chemicals used to make the medium to support growth of G. aurantiaca. In contrast, when phosphate and agar were autoclaved together, differences in colony numbers were seen. Colonies were still present on medium made using 0.5 mM phosphate (∼230 CFU), and a reduced number was present on 1 mM phosphate plates (∼100 CFU). However, colony formation was completely inhibited using phosphate concentrations of 1.5 mM and higher (0 CFU). By adding catalase to the plates made by autoclaving phosphate and agar together, colony growth inhibition could be counteracted (Fig. 1D).
FIG 1.
Inhibitory effect of growth medium preparation method on Gemmatimonas aurantiaca T-27T colony formation. Liquid precultures [OD600 of 0.055, diluted (5.5 × 103)-fold] were spread onto PYG medium with phosphate buffer and agar autoclaved together (A) or separately (B) or with no phosphate addition (C). (D) Restoration of colony formation on PYG plates made by autoclaving phosphate and agar together (3 mM; T-PB) and by adding catalase to the center of plates (3 mM; T-Cat). A culture of G. aurantiaca (OD600 of 0.045, diluted 103-fold) was spread onto agar medium and then either filter-sterilized bovine liver catalase solution (10 mg/ml) (T-Cat) was placed onto the center of the plate or just phosphate buffer was used for controls (T-PB). Plates were incubated at 30°C for 14 days.
Analysis of hydrogen peroxide in agar media.
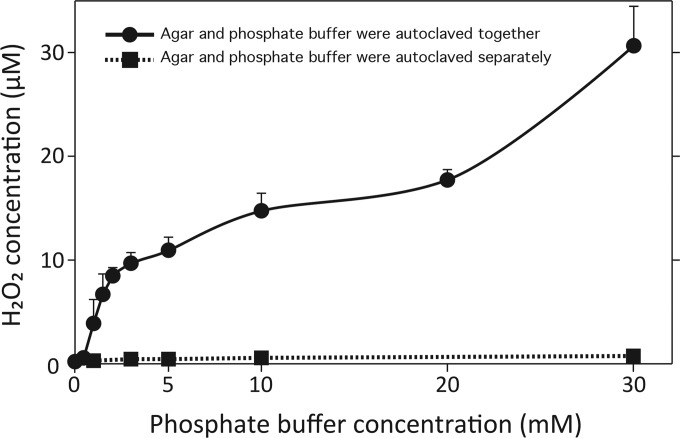
Since the addition of catalase onto agar medium increased the number of CFU, the hydrogen peroxide concentration was measured. Hydrogen peroxide (H2O2) was produced in PT and not PS medium (Fig. 2). Hydrogen peroxide concentration in the medium was dependent upon phosphate concentration, but the trend was not linear. Hydrogen peroxide remained in the agar media even after weeks of storage (data not shown).
FIG 2.
Hydrogen peroxide (H2O2) generation from agar with increasing phosphate concentrations after autoclaving agar and phosphate either together or separately. Two preparation methods are represented: 0.75 g of Bacto agar was suspended in 50 ml of buffer with phosphate concentrations ranging from 0 (i.e., pure H2O) to 30 mM (Na2HPO4-KH2PO4, pH 7.0), and agar and phosphate were autoclaved together; alternatively, 0.75 g of agar was suspended in 50 ml of water and autoclaved, and then autoclaved phosphate buffer (100 mM Na2HPO4-KH2PO4 stock, pH 7.0) was added to final concentrations ranging from 1 to 30 mM. Means of at least five independent experiments with standard deviations are shown.
Comparison of CFU counts on different medium preparations using three sources of environmental inocula.
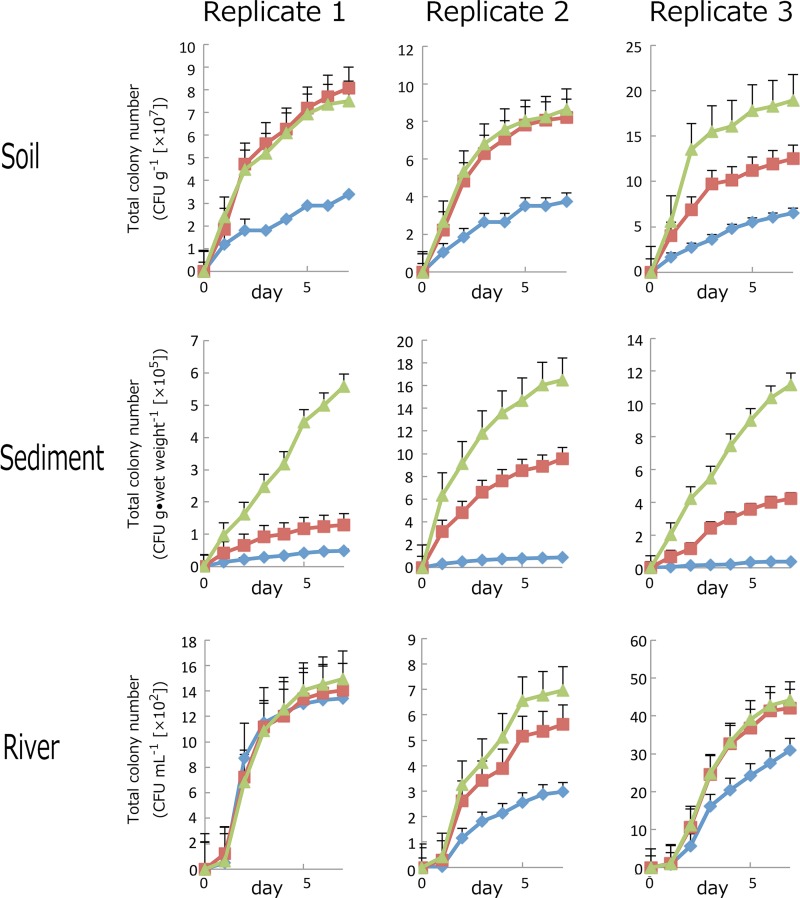
The number of CFU formed among the triplicate samples from each source was highly variable, particularly for the river water samples, due to the heterogeneous nature of these environments. Water samples are known to be highly variable because of the patchiness of bacterial distribution (44). However, for the three media, PT, PS, and PW, the lowest number of CFU formed on PT medium, which was a common trend seen within each sample with the exception of one river sample (Fig. 3). Using sediment and soil samples, the number of CFU on PT was almost one order of magnitude lower than on PS medium. This indicates that high concentrations of phosphate (20 mM phosphate in this experiment) did not inhibit colony formation but likely resulted from a reaction that occurred when phosphate was autoclaved with agar. In most samples, the numbers of CFU on PW medium were similar to or higher than CFU counts on PS medium. This trend of higher CFU counts on PW medium was most consistent among the replicate sediment samples. These results strongly indicate that only a trace amount of phosphate suffices for colony formation on agar medium, and this trace amount does not interact with agar to inhibit colony growth. Subsequent laboratory propagation of organisms frequently isolated from PS and not PT medium confirmed that these isolates formed colonies on PS but not on PT medium (see Fig. S1 in the supplemental material).
FIG 3.
Total colony counts from three environmental sources (soil, sediment, and river water; three replicates at each site) on three different media (diamond, PT; square, PS; triangle, PW). The numbers of CFU on PT, PS, and PW media were counted daily for 7 days. CFU counts reported were averages from 1 to 5 plates on which 30 to 300 colonies were formed. Plates were excluded from colony counts if overgrown by a few colonies spreading across the entire plate. Error bar shows standard errors of averages calculated from two to five plates.
Composition of a cultivated microbial community using 16S rRNA gene sequences.
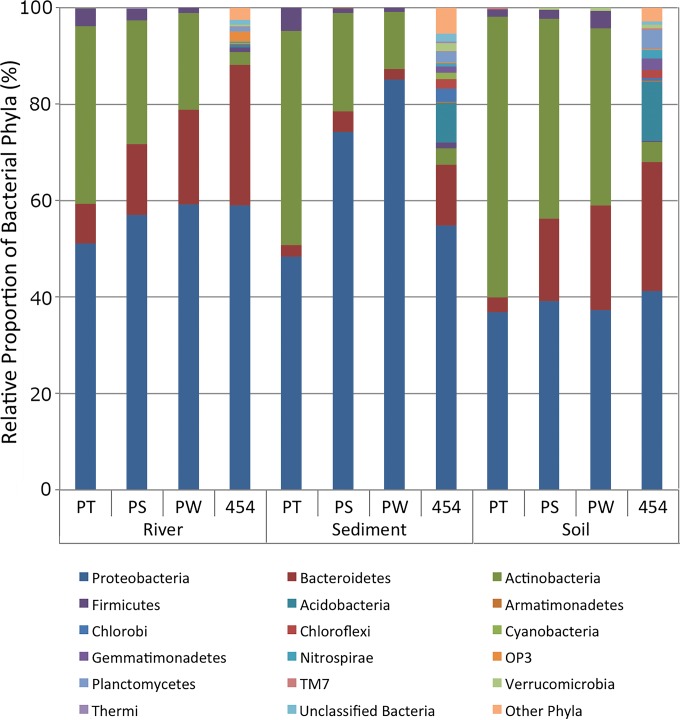
A total of 6,528 colonies were picked from agar plates for Sanger sequencing, but after quality screening 4,763 were kept to determine their phylogenetic positions using partial 16S rRNA gene sequences (1,134 from soil, 1,573 from sediment, and 2,056 from river, consisting of 1,258 from PT, 1,863 from PS, and 1,642 from PW media). Comparisons at the phylum level of the composition of isolates obtained on the three different media showed two consistent trends across the environmental sources (Fig. 4). A greater proportion of isolates belonging to the phylum Actinobacteria were obtained on PT medium than on PS and PW media. The difference in composition was mainly attributed to growth on PS and PW media of a greater proportion of Bacteroidetes from river water and soil and of Proteobacteria from sediment. This was also evident at the genus level using the alpha diversity measures (Chao1, observed species, and PD whole tree); the diversity of isolates from PT medium differed significantly (P < 0.05) from that of PS and PW media for each environmental source (Table 1). Examination of genera contributing to the differences in the phylum Bacteroidetes on the different media was most evident in the soil, where the proportions of Flavobacterium colonies recovered from PS and PW media were large while no colonies were recovered from PT medium (see Table S1 in the supplemental material). Compositional differences were also shown in comparisons of beta diversity measures in which a clear separation of communities in the three environmental sources were seen using Euclidean distances (see Fig. S2 in the supplemental material), a measure of shared taxa, whereas using phylogenetic distances (see Fig. S3), there was an overlap between sediment and soil communities. There was also separation of samples by the growth media used to obtain isolates. Using Euclidean distance, the clearest difference was within the sediment samples, where isolates from PT medium clustered separately from those from PS and PW media. Using unweighted UniFrac, however, there was a general trend for all communities isolated on PT medium to separate from those from PW and PS media along the second PCoA axis.
FIG 4.
Comparison of means of relative proportions of phyla of isolates from river, sediment, and soil samples using PT, PS, and PW media and 454 pyrosequencing of directly extracted DNA. All phyla represented by isolates or comprising >1% of pyrosequences in at least one environmental source are included. An additional 40 phyla from pyrosequencing are included as “other” phyla.
TABLE 1.
Comparison of average alpha diversity values of isolates obtained on PT, PS, and PW media
| Sample source | Diversity metric | Avg value on mediuma |
||
|---|---|---|---|---|
| PT | PS | PW | ||
| River | Chao1 | 87.2 A | 134.9 B | 132.9 B |
| Observed species | 29.7 A | 37.5 B | 37.0 B | |
| PD whole tree | 2.4 A | 2.9 B | 2.9 B | |
| Sediment | Chao1 | 70.9 A | 121.9 B | 103.6 B |
| Observed species | 29.4 A | 35.9 B | 33.7 C | |
| PD whole tree | 2.3 A | 2.9 B | 2.8 B | |
| Soil | Chao1 | 59.6 A | 120.1 B | 113.6 B |
| Observed species | 27.6 A | 35.6 B | 36.1 B | |
| PD whole tree | 2.0 A | 2.8 B | 2.9 B | |
All averages of diversity measures were calculated using 10 iterations of data rarefied to the lowest number of isolates obtained on a single growth medium. Significant differences (P < 0.05) indicated by letters across rows were calculated using Student's t tests of all the iterated data for replicates for each sample source-medium combination.
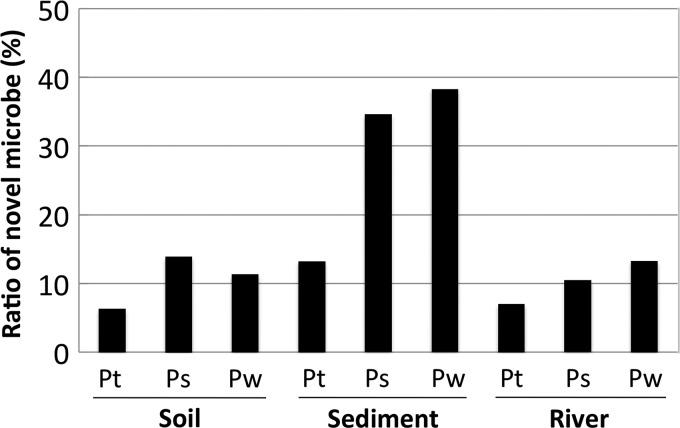
One of the most important findings from this work is the difference in the novelty of microorganisms obtained from each medium. At the genus level, the ratio of novel isolates grown on PS and PW media was higher than on PT medium from all three environmental sources tested (Fig. 5). In particular, isolates obtained from the sediment grown on PS and PW media were found to harbor over 30% hitherto-uncultivated organisms. This is notable compared to the ratio of novel taxa isolated from soil and river samples.
FIG 5.
The ratio of novel microbes isolated on the three different growth media from each environmental sample. The isolates were defined as novel microbes if their sequences had less than 97% similarity to known species. Taxonomic identity of each phylotype was determined using the RDP Classifier at 80% confidence.
Noncultivation-based determination of microbial community composition using 16S rRNA gene sequences.
Using a cultivation-independent approach, pyrosequencing produced a total of 974,399 high-quality reads of 16S rRNA gene sequences from the three types of environmental samples (number of reads were as follows: soil sample replicate 1, 114,616 reads; replicate 2, 117,502; and replicate 3, 106,076; sediment replicate 1, 119,340; replicate 2, 114,714; and replicate 3, 137,867; and river replicate 1, 128,505; replicate 2, 135,779; and replicate 3, no reads as insufficient DNA was extracted for pyrosequencing). Pyrosequencing revealed the presence of many more phyla (40 in soil, 56 in sediment, and 53 in river water) than was evident by cultivation (6 in soil, 5 in sediment, and 6 in river). Based on the small fraction of isolates obtained compared to number of sequence reads (<0.2%), it is not surprising that all of the phyla were not captured by cultivation. Of the total of 60 different bacterial phyla found by pyrosequencing, only 17 represented more than 1% of the community in at least one environmental source (Fig. 4). Isolates were not obtained from only one phylum, Acidobacteria, which accounted for greater than 5% of the taxa in at least one environmental substrate, suggesting that the media tested do not support growth of this phylum. However, isolates from other rarely cultivated phyla were obtained, for instance, Armatimonadetes and Verrucomicrobia that accounted for less than 2.5% of the taxon proportion of a community (Fig. 4). Unfortunately these rare taxa could not be propagated in the laboratory after sample storage at −80°C in glycerol.
Cultivated and noncultivated microbial community comparisons.
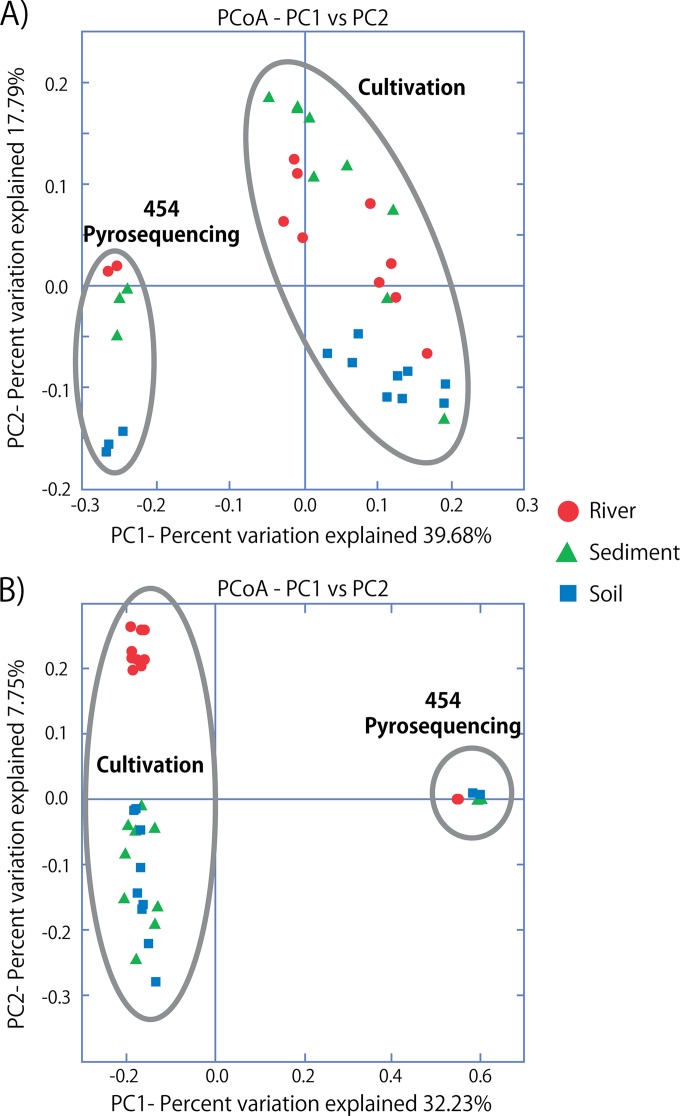
Beta-diversity analysis using principal coordinate analyses of phylogenetic (weighted and unweighted UniFrac) distances revealed that the communities obtained by pyrosequencing separated from the cultivated isolates along the first PCoA axis (Fig. 6). Differences in phylogenetic composition of bacteria at the phylum level were evident between the noncultivation-based pyrosequencing results and culture-dependent analyses (Fig. 4). There were far more Actinobacteria bacteria cultivated from environmental samples than indicated by pyrosequencing. The highest ratio of Actinobacteria was cultivated from PT medium, and the lowest was from PW medium. Pyrosequencing indicated that the phylum Bacteroidetes was the second largest population in the three environments (29, 13, and 27% in the river, sediment, and soil samples, respectively), and proportional trends were similar in the isolates obtained on PS (15, 4, and 17% in the river, sediment, and soil samples, respectively) and PW (20, 2, and 22%) media but quite different from the proportion of isolates cultivated on PT medium (8, 2, and 3% in the river, sediment, and soil samples, respectively). These results indicate that the cultivable microbial communities obtained on PS and PW media were more similar to 454-based community analyses than the bacteria obtained on PT medium regardless of the environmental sample type, suggesting that PS and PW media may tend to be less biased in cultivation than the PT medium (Fig. 4).
FIG 6.
Beta diversity analysis of isolates obtained from river, sediment, and soil samples using PT, PS, and PW media. (A) Jackknife analysis of Euclidean distances using Bray-Curtis with data rarefied to 50 isolates. (B) Jackknife analysis of phylogenetic distances using unweighted UniFrac with data rarefied to 50 isolates. PC1 and PC2, first and second principal coordinate axes, respectively.
DISCUSSION
This study demonstrates that a modification of the approach used for medium preparation can lead to an increase in CFU counts of a species that had been recalcitrant to cultivation, e.g., G. aurantiaca and a diversity of bacteria isolated from various environmental sources. By either removing phosphate from the medium (PW) or autoclaving phosphate and agar separately (PS), we obtained ∼50 times more CFU than on PT medium. The hydrogen peroxide that was produced when agar was autoclaved together with phosphate was likely a factor inhibiting growth of some taxa. Reactive oxygen species are known inhibitors of growth of many bacteria (45). Unlike other method modifications that have been used to increase cultivation efficiency (9–12), this modification, in which medium components are autoclaved separately and then mixed, is relatively simple to implement in any laboratory.
A number of studies have shown that replacing agar with gellan gum can increase the number of CFU and/or lead to the isolation of novel bacteria from environmental samples (13, 28, 46). But the reason for colony growth differences on agar versus gellan gum was not investigated in those studies. In this study, only medium preparation methods differed; the exclusion of phosphate or the permutation of medium components that were autoclaved together indicated that inhibitory growth compounds were produced from autoclaving phosphate with agar. High concentrations of hydrogen peroxide in PT and not PS medium indicated that it was likely a contributing factor to the growth inhibition of G. aurantiaca and some taxa in the environmental samples. Although there are reports of growth-inhibitory compounds produced from autoclaving phosphate with sugars (23) and glucose with proteins (47), we have not found reports about chemical interactions with agar associated with autoclaving. More research is needed to understand the detailed mechanism for generating peroxide or other radicals from agar and phosphate during autoclaving.
Cultivation using different environmental sources demonstrated the bias in results that can occur as a result of the method used for medium preparation. The genus Flavobacterium in the phylum Bacteroidetes appeared to be most sensitive to the inhibitory compounds present in the PT medium. There were higher proportions of Bacteroidetes isolates from PS and PW media than from PT medium for the soil and river water samples. Tamaki et al. (28) also found more Bacteroidetes strains isolated from freshwater sediments using media solidified with gellan gum than with agar. There have been no studies demonstrating specific sensitivity of Flavobacterium to hydrogen peroxide, but growth of a fastidious fish pathogen, Flavobacterium psychrophilum, was improved by the addition of charcoal to absorb inhibitors in agar growth medium (48). Also, the proportions of Betaproteobacteria isolates from sediment and river samples were higher on PS and PW media than on PT medium. These findings indicate that the medium used should be taken into account when the differences in the composition of cultivated bacterial communities from similar environments are interpreted.
These findings demonstrate the importance of considering growth inhibitors in agar medium in microbial ecology and environmental microbiology. To capture a greater diversity, far larger numbers, and more novel microorganisms using traditional agar plate-dependent cultivation techniques, one should take our findings into consideration: (i) phosphate in high concentrations should be autoclaved separately from agar to maximize the number of CFU; (ii) low phosphate concentrations, as in the PW medium (μM that corresponds to 0.01× or even lower concentrations than in commonly used media), may suffice for the growth of many environmental bacteria. These new insights must be considered in medium preparation and can contribute to greater culture-dependent exploitation of microbial resources from natural environments.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Institute of Fermentation, Osaka, Japan.
We thank Y. Sekiguchi for assistance in 454 pyrosequencing analysis, Nippon Steel and Sumikin Eco-Tech Corporation for phosphorus analysis, and K. Asano and T. Sone for their valuable suggestions.
Author contributions are as follows: T.T., K.K., and S.D. performed research. W.K., K.Y., H.T., and C.H.N. analyzed data. C.H.N., M.T., and Y.K. designed experiments. Y.K. supervised and supported the entire study. Y.K. and C.H.N. wrote the paper.
Footnotes
Published ahead of print 3 October 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02741-14.
REFERENCES
- 1.Staley JT, Konopka A. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39:321–346. 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 2.Stewart EJ. 2012. Growing unculturable bacteria. J. Bacteriol. 194:4151–4160. 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puspita ID, Kamagata Y, Tanaka M, Asano K, Nakatsu CH. 2012. Are uncultivated bacteria really uncultivable? Microbes Environ. 27:356–366. 10.1264/jsme2.ME12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen ZT, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer MLI, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu PG, Begley RF, Rothberg JM. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380. 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ronaghi M, Uhlén M, Nyrén P. 1998. A sequencing method based on real-time pyrophosphate. Science 281:363–365. 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 6.Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809. 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 7.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 8.Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AK, Kent AD, Daroub SH, Camargo FA, Farmerie WG, Triplett EW. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283–290. 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaeberlein T, Lewis K, Epstein SS. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127–1129. 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- 10.Ferrari BC, Winsley T, Gillings M, Binnerup S. 2008. Cultivating previously uncultured soil bacteria using a soil substrate membrane system. Nat. Protoc. 3:1261–1269. 10.1038/nprot.2008.102. [DOI] [PubMed] [Google Scholar]
- 11.Zengler K, Toledo G, Rappé M, Elkins J, Mathur EJ, Short JM, Keller M. 2002. Cultivating the uncultured. Proc. Natl. Acad. Sci. U. S. A. 99:15681–15686. 10.1073/pnas.252630999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connon SA, Giovannoni SJ. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878–3885. 10.1128/AEM.68.8.3878-3885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391–2396. 10.1128/AEM.68.5.2391-2396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nichols D, Lewis K, Orjala J, Mo S, Ortenberg R, O'Connor P, Zhao C, Vouros P, Kaeberlein T, Epstein SS. 2008. Short peptide induces an “uncultivable” microorganism to grow in vitro. Appl. Environ. Microbiol. 74:4889–4897. 10.1128/AEM.00393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tripp HJ, Kitner JB, Schwalbach MS, Dacey JWH, Wilhelm LJ, Giovannoni SJ. 2008. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452:741–744. 10.1038/nature06776. [DOI] [PubMed] [Google Scholar]
- 16.Andrews GP, Martin SE. 1979. Catalase activity during the recovery of heat-stressed Staphylococcus aureus MF-31. Appl. Environ. Microbiol. 38:390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aruoma OI, Halliwell B. 1987. Action of hypochlorous acid on the antioxidant protective enzymes superoxide dismutase, catalase and glutathione peroxidase. Biochem. J. 248:973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogosian G, Aardema ND, Bourneuf EV, Morris PJL, O'Neil JP. 2000. Recovery of hydrogen peroxide-sensitive culturable cells of Vibrio vulnificus gives the appearance of resuscitation from a viable but nonculturable state. J. Bacteriol. 182:5070–5075. 10.1128/JB.182.18.5070-5075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calabrese JP, Bissonnette GK. 1990. Improved membrane filtration method incorporating catalase and sodium pyruvate for detection of chlorine stressed coliform bacteria Appl. Environ. Microbiol. 56:3558–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marthi B, Shaffer BT, Lighthart B, Ganio L. 1991. Resuscitation effects of catalase on airborne bacteria. Appl. Environ. Microbiol. 57:2775–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin SE, Flowers RS, Ordal ZJ. 1976. Catalase: its effect on microbial enumeration. Appl. Environ. Microbiol. 32:731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imazaki I, Kobori Y. 2010. Improving the culturability of freshwater bacteria using FW70, a low-nutrient solid medium amended with sodium pyruvate. Can. J. Microbiol. 56:333–341. 10.1139/W10-019. [DOI] [PubMed] [Google Scholar]
- 23.Finkelstein RA, Lankford CE. 1957. A bacteriotoxic substance in autoclaved culture media containing glucose and phosphate. Appl. Microbiol. 5:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramsey HH, Lankford CE. 1956. Stimulation of growth initiation by heat degradation products of glucose. J. Bacteriol. 72:511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Einarsson H, Snygg BG, Eriksson C. 1983. Inhibition of bacterial growth by Maillard reaction products. J. Agric. Food Chem. 31:1043–1047. 10.1021/jf00119a031. [DOI] [Google Scholar]
- 26.Hara S, Isoda R, Tahvanainen T, Hashidoko Y. 2012. Trace amounts of furan-2-carboxylic acids determine the quality of solid agar plates for bacterial culture. PLoS One 7:e41142. 10.1371/journal.pone.0041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Sekiguchi Y, Hanada S, Hugenholtz P, Kim H, Kamagata Y, Nakamura K. 2003. Gemmatimonas aurantiaca gen. nov., sp. nov., a Gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int. J. Syst. Evol. Microbiol. 53:1155–1163. 10.1099/ijs.0.02520-0. [DOI] [PubMed] [Google Scholar]
- 28.Tamaki H, Hanada S, Sekiguchi Y, Tanaka Y, Kamagata Y. 2009. Effect of gelling agent on colony formation in solid cultivation of microbial community in lake sediment. Environ. Microbiol. 11:1827–1834. 10.1111/j.1462-2920.2009.01907.x. [DOI] [PubMed] [Google Scholar]
- 29.Jiang ZY, Woollard ACS, Wolff SP. 1990. Hydrogen peroxide production during experimental protein glycation. FEBS Lett. 268:69–71. 10.1016/0014-5793(90)80974-N. [DOI] [PubMed] [Google Scholar]
- 30.Kamagata Y, Fulthorpe RR, Tamura K, Takami H, Forney LJ, Tiedje JM. 1997. Pristine environments harbor a new group of oligotrophic 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl. Environ. Microbiol. 63:2266–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dahlquist RL, Knoll JW. 1978. Inductively coupled plasma-atomic emission spectrometry: analysis of biological materials and soils for major, trace, and ultra-trace elements. Appl. Spectrosc. 32:1–30. 10.1366/000370278774331828. [DOI] [Google Scholar]
- 32.Fuhrman JA, Comeau DE, Hagström Å, Chan AM. 1988. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl. Environ. Microbiol. 54:1426–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heuer H, Hartung K, Wieland G, Kramer I, Smalla K. 1999. Polynucleotide probes that target a hypervariable region of 16S rRNA genes to identify bacterial isolates corresponding to bands of community fingerprints. Appl. Environ. Microbiol. 65:1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright ES, Yilmaz LS, Noguera DR. 2012. DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl. Environ. Microbiol. 78:717–725. 10.1128/AEM.06516-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reeder J, Knight R. 2010. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat. Methods 7:668–669. 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 39.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Human Microbiome Consortium. Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21:494–504. 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6:610–618. 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235. 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmer FE, Methot RD, Staley JT. 1976. Patchiness in the distribution of planktonic heterotrophic bacteria in lakes. Appl. Environ. Microbiol. 31:1003–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vatansever F, de Melo WC, Avci P, Vecchio D, Sadasivam M, Gupta A, Chandran R, Karimi M, Parizotto NA, Yin R, Tegos GP, Hamblin MR. 2013. Antimicrobial strategies centered around reactive oxygen species—bactericidal antibiotics, photodynamic therapy, and beyond. FEMS Microbiol. Rev. 37:955–989. 10.1111/1574-6976.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamaki H, Sekiguchi Y, Hanada S, Nakamura K, Nomura N, Matsumura M, Kamagata Y. 2005. Comparative analysis of bacterial diversity in freshwater sediment of a shallow eutrophic lake by molecular and improved cultivation-based techniques. Appl. Environ. Microbiol. 71:2162–2169. 10.1128/AEM.71.4.2162-2169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakashima T, Seki T, Matsumoto A, Miura H, Sato E, Niwano Y, Kohno M, Omura S, Takahashi Y. 2010. Generation of reactive oxygen species from conventional laboratory media. J. Biosci. Bioeng. 110:304–307. 10.1016/j.jbiosc.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 48.Alvarez B, Guijarro JA. 2007. Recovery of Flavobacterium psychrophilum viable cells using a charcoal-based solid medium. Lett. Appl. Microbiol. 44:569–572. 10.1111/j.1472-765X.2007.02126.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.