Abstract
The white rot basidiomycete Ceriporiopsis subvermispora delignifies wood selectively and has potential biotechnological applications. Its ability to remove lignin before the substrate porosity has increased enough to admit enzymes suggests that small diffusible oxidants contribute to delignification. A key question is whether these unidentified oxidants attack lignin via single-electron transfer (SET), in which case they are expected to cleave its propyl side chains between Cα and Cβ and to oxidize the threo-diastereomer of its predominating β-O-4-linked structures more extensively than the corresponding erythro-diastereomer. We used two-dimensional solution-state nuclear magnetic resonance (NMR) techniques to look for changes in partially biodegraded lignin extracted from spruce wood after white rot caused by C. subvermispora. The results showed that (i) benzoic acid residues indicative of Cα—Cβ cleavage were the major identifiable truncated structures in lignin after decay and (ii) depletion of β-O-4-linked units was markedly diastereoselective with a threo preference. The less selective delignifier Phanerochaete chrysosporium also exhibited this diastereoselectivity on spruce, and a P. chrysosporium lignin peroxidase operating in conjunction with the P. chrysosporium metabolite veratryl alcohol did likewise when cleaving synthetic lignin in vitro. However, C. subvermispora was significantly more diastereoselective than P. chrysosporium or lignin peroxidase-veratryl alcohol. Our results show that the ligninolytic oxidants of C. subvermispora are collectively more diastereoselective than currently known fungal ligninolytic oxidants and suggest that SET oxidation is one of the chemical mechanisms involved.
INTRODUCTION
The use of mild biological agents to delignify lignocellulose with minimal loss of its valuable cellulose has been a long-standing technological goal. This process occurs in nature when certain white rot fungi delignify wood selectively, growing on its hemicelluloses before they consume the cellulose that is exposed later via delignification (1–4). In part, timing of gene expression underlies this strategy, with the one-electron oxidative systems that depolymerize lignin via free radical intermediates being expressed before cellulases (5). However, temporal regulation alone cannot explain the observed selectivity, because some fungal ligninolytic oxidants are nonselective reactive oxygen species (e.g., Fenton reagent) that attack cellulose as well as lignin (6); that is, the cellulose will be depolymerized if the ligninolytic oxidants deployed before cellulases are of this nonspecific type, whereas selective delignification will occur instead if the oxidants specifically target the lignin. Thus, it may be inferred simply from the existence of selective white rot that some relatively specific oxidants have a ligninolytic role in this process.
The only fungal ligninolytic oxidants currently known to exhibit the required selectivity are lignin peroxidases (LiPs) and the closely related versatile peroxidases. These enzymes, although absent from some white rot fungi (7), provide a mechanistic paradigm for efficient biological ligninolysis. They oxidize the principal nonphenolic structures of lignin via single-electron transfer (SET), generating aryl cation radical intermediates whose aliphatic side chains then cleave spontaneously between Cα and Cβ (see structure A in Fig. 1a) (8–10). This approach to ligninolysis is exemplified by the well-studied but relatively nonselective delignifier Phanerochaete chrysosporium, which produces LiPs and generates Cα—Cβ cleavage products in lignin during wood decay (11–13). LiPs also oxidize the secreted P. chrysosporium metabolite veratryl alcohol (3,4-dimethoxybenzyl alcohol) to a diffusible aryl cation radical that may then act as a selective SET oxidant of lignin within the intact wood cell wall, where enzymes cannot infiltrate (14–16).
FIG 1.
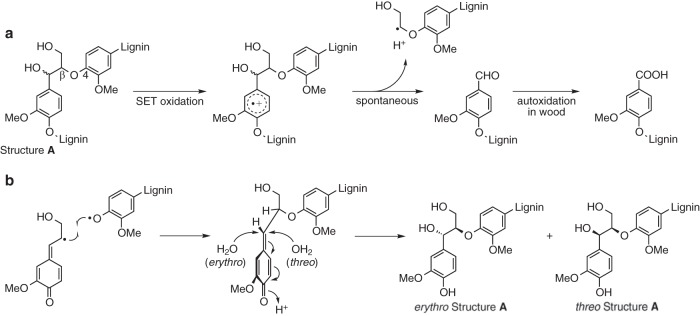
Chemical structures and reactions discussed in the text. (a) Cleavage of lignin structure A via fungal SET oxidation, followed by autooxidation of the resulting lignin benzaldehydes. Stereochemistry is not shown for structure A. (b) Formation of structure A in the lignifying plant cell wall via free radical coupling and nucleophilic addition of water, showing routes for production of the erythro- and threo-diastereomers. OMe and MeO, methoxyl.
Ceriporiopsis subvermispora is a selective delignifier that has been much studied for biotechnological applications (2, 17–19). It removes lignin but not cellulose almost uniformly throughout the wood cell wall during early decay, before the substrate porosity has increased enough to admit enzymes the size of peroxidases (ca. 40 kDa) (3). These results indicate not only that C. subvermispora produces a highly selective ligninolytic agent but also that the proximal oxidant is likely a low-molecular-weight species.
However, C. subvermispora has never been found to produce LiP or versatile peroxidase, the selective ligninolytic oxidants that are currently known from fungi, nor has it been found to secrete small aromatic redox mediators, such as veratryl alcohol, that might work in conjunction with these enzymes to infiltrate and delignify the wood cell wall (5, 20). On the other hand, C. subvermispora carries two genes that were previously shown via heterologous expression to encode enzymes with LiP activity (21, 22). Although the corresponding RNA transcripts were produced at low levels and mass spectrometric analysis failed to detect the encoded proteins during growth of the fungus on ball-milled wood, the situation has not yet been evaluated for biodegradation of solid wood, and therefore, some role for LiPs cannot yet be ruled out.
To complicate the picture further, although previous work with lignin model compounds suggested that C. subvermispora cleaves lignin between Cα and Cβ, consistent with the operation of a SET oxidant (23), a later analysis of wood degraded by the fungus did not reveal Cα—Cβ cleavage products in the residual lignin (24). Thus, not only the nature of the ligninolytic oxidants but also the chemical route for ligninolysis remains in question for this biotechnologically promising fungus.
In principle, chemical changes in the lignin remaining after wood decay could provide clues about the poorly understood ligninolytic system of C. subvermispora. The major structures of lignin are well characterized (25, 26) and might differ from each other in susceptibility to particular oxidants. However, analyses of white-rotted wood have a limitation: the ability of the fungi to mineralize lignin efficiently results in very low steady-state accumulations of partially degraded structures such that almost all of the lignin present throughout decay consists of an intact polymer that has not yet been attacked (27–29). To detect degraded structures, the lignin must first be extracted with polar solvents that dissolve little of the sound polymer, after which oxidative changes can be identified in the small quantity of partially biodegraded lignin that dissolves (28). This fractionation of the lignin could lead to artifacts because some lignin structures are probably not distributed uniformly in sound wood, and it is unlikely that all parts of the wood are attacked to the same extent by the fungus or extracted with equal efficiency by the solvents. To address this issue, analyses of white-rotted lignin should ideally focus on structures that are thought to be uniformly distributed in all wood cell wall fractions.
Two structures that likely meet this criterion are the diastereomers of the predominant arylglycerol-β-aryl ether (β-O-4) unit of lignin (structure A) (Fig. 1b). When wood is formed, structure A is biosynthesized in two steps. First, a phenoxyl radical at the end of the growing lignin polymer couples with a 4-hydroxycinnamyl alcohol phenoxyl radical (shown here in its β-carbon-centered resonance form) to give a new β-O-4-linked unit in which the terminal ring exists as a quinone methide and the β-carbons with their substituents occur in a 1:1 racemic mixture of enantiomers (i.e., the coupling reaction that generates a new chiral center is not stereospecific). The quinone methide then rearomatizes by reacting with water at its α-carbon to give an additional asymmetric center such that the newly formed structure A subunit exists as a racemic mixture of its two diastereomeric pairs of enantiomers, which can be designated RR/SS (also denoted syn or threo) and RS/SR (also denoted anti or erythro) (26).
Since local stereochemistry alone determines which face of the quinone methide reacts faster with water (Fig. 1b), the resulting threo/erythro ratio in structure A units is unlikely to vary depending on where the units are located in the wood cell wall. Rather, the threo/erythro ratio is simply characteristic of the particular 4-hydroxycinnamyl alcohols from which the lignin is biosynthesized. For softwood structure A units formed from coniferyl alcohol, this ratio is close to 1:1 and is generally thought to be invariant throughout the wood (30). Thus, although the fraction of total lignin extracted as a partially degraded polymer from a sample of white-rotted softwood is small, the threo/erythro ratio of the structure A units that this fraction had before biodegradation was likely representative of the entire sound sample. Moreover, as long as the lignin fraction analyzed is polymeric, the two diastereomers cannot be artifactually separated by solvent extraction, because they are randomly distributed and covalently linked to each other.
An analysis of the threo/erythro ratio for structure A units remaining in lignin after white rot is potentially useful because diastereoselectivity can serve as one indicator of an oxidant's tendency to discriminate between chemical targets. Moreover, some relatively selective SET oxidants, including LiPs, have been shown to exhibit diastereoselectivity on structure A. Although the chemical mechanism for this phenomenon is not fully understood, it has been demonstrated repeatedly for dimeric lignin model compounds that represent structure A. When LiPs were used directly on these low-molecular-weight compounds, the threo-diastereomer was oxidized more rapidly (22, 31, 32). When a LiP was used in conjunction with the mediator veratryl alcohol to oxidize a small structure A model, the diastereoselectivity was preserved (31). An inorganic SET oxidant, ceric ammonium nitrate, gave a similar result, whereas oxidation of the structure A model by Fenton reagent, a highly nonselective oxidant, was not diastereoselective (31).
These results suggest that an examination of structure A threo/erythro ratios in residual lignin after attack by C. subvermispora could provide information on the chemical selectivity of the poorly understood ligninolytic oxidants that this fungus employs. However, until now, the issue of diastereoselectivity in delignification has not been addressed for any fungus on a natural lignocellulosic substrate. Here we have used two-dimensional solution-state nuclear magnetic resonance (NMR) analyses of biodegraded lignin to address three pertinent questions: (i) whether C. subvermispora cleaves lignin in wood between Cα and Cβ, consistent with a SET ligninolytic mechanism; (ii) whether the known SET oxidant LiP is diastereoselective in vitro on structure A when it cleaves polymeric lignin, as opposed to the low-molecular-weight models previously employed; and (iii) whether C. subvermispora removes structure A diastereoselectively when it cleaves lignin in wood.
MATERIALS AND METHODS
Fungal cultures.
Ceriporiopsis subvermispora 90031-sp (ATCC 90467) and Phanerochaete chrysosporium ME-446 (ATCC 34541) were obtained from the Center for Forest Mycology, U.S. Forest Products Laboratory, and were maintained on malt agar. To obtain biodegraded lignins from colonized wood, the fungi were grown on wafers of white spruce sapwood (Picea glauca) (12 by 12 by 3 mm, with the large face perpendicular to the grain, ∼150 mg [dry weight]). The wafers were first oven dried at 55°C for 3 days, and the dry weights for 10% of the group were recorded. The wafers were then autoclaved twice at 121°C for 50 min with an interval of 3 days between sterilizations and dried under a vacuum in a sterile desiccator jar. Sterile 20% potato dextrose broth (150 μl; 20%, wt/vol) was added to each wafer, and after the liquid was absorbed, the wafers (100 for each fungus per setup) were transferred onto 150-mm-diameter petri plates. Each plate contained 50 ml of Kirk's B-III agar (1.5%, wt/vol) medium without glucose (33) and was overlaid with sterile nylon mesh. The wood wafers were inoculated with agar plugs containing fungal mycelium and incubated in humidified growth chambers. The cultivation temperature for P. chrysosporium was 38°C, and that for C. subvermispora was 28°C. After 6 weeks, the wafers were harvested, and excess mycelium was gently scraped from their surfaces with a razor blade. The subset of wafers for which dry weights had been recorded was oven dried as described above, and the dry weight losses for these replicates were recorded.
Isolation of lignins.
The biodegraded spruce wood wafers from six separate culture setups, totaling 600 wafers for each fungus, were pooled and ground to a 40-mesh size in a Wiley mill. The milled samples were solvent extracted, using a modification of a procedure reported previously (12), by stirring them successively at room temperature in 1 liter of petroleum ether (1 day), chloroform (1 day), acetone (1 day), methanol (2 days), and dioxane-water (96:4) (2 days). Between extractions, the solvents were recovered by vacuum filtration through glass fiber filters, and the extracted wood was resuspended in the next solvent. Finally the dioxane-water extracts were each vacuum evaporated, dissolved in N,N-dimethylacetamide, and precipitated into 100 ml of water (pH 3, adjusted with dilute formic acid) to remove low-molecular-weight components. The precipitates were stirred for 30 min, collected by centrifugation (27,000 × g for 15 min), and lyophilized. The final extract yields based on the dry weight of degraded wood were 1.20 mg/g and 1.89 mg/g for C. subvermispora and P. chrysosporium, respectively.
To provide a nondegraded control sample, we used macromolecular milled wood lignin from white spruce, prepared by extracting ball-milled spruce wood with dioxane-water (96:4), as described previously (28). This control lignin and the two macromolecular biodegraded lignins were divided into three portions for analysis. One portion of each was assayed for noncondensed structure A units by DFRC (derivatization followed by reductive cleavage) analysis (34). The structure A yields were quantified based on the total lignin content of each sample, which was determined by the acetyl bromide method (35). The second portion was set aside without derivatization for quantitative 13C NMR analysis. The third portion, destined for two-dimensional NMR analyses, was acetylated with acetic anhydride-pyridine (1:1) overnight. The residual acetic anhydride, acetic acid, and pyridine were then removed by rotary evaporation, followed by azeotropic coevaporation with ethanol. In addition to the above-described procedure, three additional acetylated milled spruce wood lignins were added as controls for the two-dimensional NMR analyses.
Preparation of dimeric lignin model I.
A lignin model dimer containing a benzoic acid residue on its β-O-4-linked aromatic ring, 4-{[1,3-bis(acetyloxy)-1-(3,4-dimethoxyphenyl)propan-2-yl]oxy}-3-methoxybenzoic acid (model I), was synthesized in six steps: (i) reaction of 3,4-dimethoxyacetophenone with Br2 in CHCl3 at room temperature to give 2-bromo-1-(3,4-dimethoxyphenyl)ethan-1-one, (ii) coupling with methyl 4-hydroxy-3-methoxybenzoate in refluxing acetone in the presence of K2CO3 to give methyl 4-[2-(3,4-dimethoxyphenyl)-2-oxoethoxy]-3-methoxybenzoate, (iii) aldol coupling with formaldehyde in 1,4-dioxane in the presence of K2CO3 at 38°C to give methyl 4-{[1-(3,4-dimethoxyphenyl)-3-hydroxy-1-oxopropan-2-yl]oxy}-3-methoxybenzoate, (iv) reduction with NaBH4 in methanol at 0°C to give methyl 4-{[1-(3,4-dimethoxyphenyl)-1,3-dihydroxypropan-2-yl]oxy}-3-methoxybenzoate, (v) hydrolysis with LiOH in tetrahydrofuran-H2O (3:1) at room temperature to give 4-{[1-(3,4-dimethoxyphenyl)-1,3-dihydroxypropan-2-yl]oxy}-3-methoxybenzoic acid, and (vi) acetylation with acetic anhydride-pyridine (1:1) at room temperature to give a 76:24 erythro/threo mixture of model I in a final yield of 27%. 1H NMR (700 MHz, dimethyl sulfoxide [DMSO]-d6) δ 7.53 to 7.48 (m, 1H, Ar-H), 7.47 to 7.45 (m, 1H, Ar-H), 7.20 to 7.15 (m, 1H, Ar-H), 7.05 to 7.01 (m, 1H, Ar-H), 6.96 to 6.95 (m, 1H, Ar-H), 6.92 to 6.89 (m, 1H, Ar-H), 5.96 to 5.89 (m, 1H, α-CH-), 5.06 to 5.00 (m, 1H, β-CH-), 4.22 to 3.94 (m, 2H, γ-CH2-), 3.80 to 3.78 (m, 3H, -OCH3), 3.74 to 3.72 (m, 6H, -OCH3), 1.97 to 1.91 [m, 6H, -C(O)CH3]; 13C NMR (176 MHz, DMSO-d6) δ 170.14, 170.07, 169.38, 169.28, 167.00, 166.96, 151.61, 150.78, 149.42, 149.32, 148.95, 148.78, 148.66, 148.47, 128.56, 128.48, 124.41, 124.26, 122.83, 122.80, 119.97, 119.85, 115.44, 115.39, 112.93, 112.82, 111.47, 111.32, 110.99, 110.83, 78.71, 77.74, 74.54, 73.08, 62.58, 62.11, 59.81, 55.72, 55.67, 55.52, 55.47, 55.43, 20.79, 20.75, 20.52, 20.50.
Production and enzymatic cleavage of synthetic lignin.
An α-13C-labeled synthetic lignin (dehydrogenation polymer [DHP]) was prepared from [α-13C]coniferyl alcohol (>99 atom% 13C) by using horseradish peroxidase, and a high-molecular-weight fraction of it was isolated by gel permeation chromatography as described previously (36). A colloidal dispersion of this lignin (1.5 mg) in sodium glycolate (10 mM; pH 4.5)–2-methoxyethanol (7:3) was then treated with P. chrysosporium LiP (isozyme H1, 130 nmol) in the presence of H2O2 (0.15 mmol) and [α-12C]veratryl alcohol (>99.98 atom%; 0.20 mmol), as described previously (36), in a final reaction volume of 500 ml. A control reaction was run under the same conditions without LiP. 13C-labeled lignin was required in this experiment to provide sufficient NMR signal amplitude, because ligninolysis by LiP in vitro can be performed only on dilute dispersions of lignin. Veratryl alcohol was included as a cofactor because P. chrysosporium LiP requires it to depolymerize synthetic lignins in vitro (36). The DHP lignins were recovered from the completed reactions and acetylated as described previously (36).
NMR analysis.
Quantitative, inverse-gated, decoupled, nuclear Overhauser effect-suppressed 13C NMR spectra were acquired on a 500-MHz Bruker Avance instrument equipped with a cryogenically cooled 13C/15N{1H} probe (13C/15N coils closest to the sample) by using Bruker's zgig30 pulse program with a D1 delay of 4 s, a sweep width of 220 ppm, and 65,536 data points. Apodization using a line broadening of 5 Hz was used for processing. Approximately 70 mg of nonderivatized material was dissolved in 500 μl of dimethyl sulfoxide-d6 for all quantitative 13C experiments.
One-bond adiabatic 1H-13C correlation (HSQC) and two- or three-bond 1H-13C correlation (HMBC) spectra were acquired on a 500-MHz Bruker Avance instrument equipped with a cryogenically cooled TCI 1H/{13C/15N} gradient probe with inverse geometry (1H coils closest to the sample) by using Bruker's hsqcetgpsisp2.2 and hmbclplrndqf pulse programs. Approximately 15 mg of acetylated material was dissolved in 500 μl of dimethyl sulfoxide-d6 for two-dimensional analyses of each wood lignin. For 13C-labeled DHP lignins, the entire sample recovered from the reaction performed in vitro (approximately 1 mg) was dissolved and analyzed.
HSQC experiments had the following parameters: a spectral width from 10 to 0 ppm (5,000 Hz) in F2 (1H) using 1,998 data points for an acquisition time (AQ) of 200 ms and an interscan delay (D1) of 1.5 s and a spectral width from 200 to 0 ppm (25,154 Hz) in F1 (13C) using 400 increments (F1 AQ of 8 ms) with 24 scans, giving a total AQ of 4 h 50 min. Data were processed as described previously (37). HMBC experiments were performed by using an 80-ms long-range coupling delay, and data were processed as described previously (38).
Structural assignments were obtained from the NMR database of lignin and cell wall model compounds (version 2004; S. A. Ralph, J. Ralph, and L. L. Landucci [http://ars.usda.gov/Services/docs.htm?docid=10491]) and from assigned spectra in the literature, which were reviewed previously (39).
Measurements of diastereoselectivity.
Quantifications of diastereoselectivity on structure A in lignins were obtained by numerical integration of the well-dispersed threo and erythro contours for Cα of acetylated structure A in the HSQC spectra by using Bruker Biospin TopSpin v. 3.0 software for Macintosh. Diastereoselectivity was expressed as the ratio of the threo-diastereomer integral to the erythro-diastereomer integral. Although HSQC spectra are not strictly quantitative, no systematic error in threo/erythro ratio determination is expected because the chemical environments of the Hα—Cα bonds in threo structure A and erythro structure A are nearly identical. Also, we employed adiabatic pulse sequences, as described previously (40), to improve uniformity over the whole spectrum and to minimize effects due to coupling constant differences.
Measurements of structure A disappearance.
Extents of structure A depletion in white-rotted lignins relative to the structure A content of sound milled wood lignin were determined by DFRC analysis, as described above. The extent of structure A depletion in DHP lignin after treatment with LiP-veratryl alcohol relative to the structure A content in the control reaction mixture without LiP was determined by HSQC analysis. This was done by integrating the HSQC signals for structure A into the two product mixtures and then normalizing them to their total methoxyl contents determined in the same spectra. This normalization was possible because the quantities of methoxyl groups in the samples were effectively constant, since they were almost entirely attributable to the large excess (relative to lignin) of veratryl alcohol added or to its oxidation product veratraldehyde.
Statistical analyses.
For statistical analysis of the threo/erythro ratios and extents of structure A depletion, there were two major sources of variance to consider: biological variance in ligninolytic reactions and measurement variance in quantifying lignin structures.
Given the laborious procedure for sample production and lignin isolation and also the necessity to pool wood samples from many experiments to obtain enough lignin for analysis, it was not feasible to obtain biological replicates for the threo/erythro ratios or for the extents of structure A depletion. Instead, we used the variance in weight loss of the wood as a measurement of biological variance. Since weight loss in biodegrading wood involves processes in addition to ligninolysis (e.g., hemicellulose removal) that contribute additional variance, our use of weight loss variance as a proxy for variance in the threo/erythro ratio change and for variance in structure A removal likely overestimates the biological variance in our experiments. Our analysis indicated that the mean weight losses for the 600-wafer samples that we employed for lignin extraction were, with 95% confidence, within ±2% and ±3% of the mean weight losses that we determined for subsets of the wood degraded by C. subvermispora and P. chrysosporium, respectively.
To estimate measurement variance in integrating the HSQC spectra to obtain threo/erythro ratios, we performed each integration five times for each sample to obtain a mean value with an error estimate. The biodegraded samples consisted of the acetylated dioxane-water lignins from 600 pooled spruce wafers for each fungus, whereas the control samples for the biodegradation study consisted of four separately prepared and acetylated milled wood lignins. The DHP lignin samples did not include replicates because the limited amount of isotope-labeled substrate available precluded this approach. However, the DHP experiment involved biochemical oxidation in vitro, not a biotreatment, so the anticipated variance between reactions is low. Our results indicated that the 95% confidence intervals for signal integration were ±2% for sound milled wood lignin, ±3% for nondegraded DHP lignin, ±7% for DHP lignin degraded by LiP, ±17% for wood lignin degraded by P. chrysosporium, and ±28% for wood lignin degraded by C. subvermispora. The relatively high variances for signal integration in biodegraded lignins reflect the marked deletion of structure A in these samples, which resulted in low signal-to-noise ratios.
To estimate measurement variance in the DFRC analyses that determined the extents of structure A depletion in biodegraded wood, we performed each analysis in duplicate. The 95% confidence intervals were found to be ±2% for wood lignin degraded by P. chrysosporium and ±5% for wood lignin degraded by C. subvermispora, in agreement with previous work that demonstrated good reproducibility for this procedure (34). To estimate measurement variance in the HSQC integrations done to determine the extents of structure A depletion in DHP lignin after treatment with LiP, we performed each integration five times. The 95% confidence interval was found to be ±10%.
As the above-described confidence intervals show, the variance in our data was chiefly attributable to errors in HSQC integration, for which replicate analyses were available. The other sources of variance, being much smaller, had a negligible effect on the composite variance estimates. We used the above-described values and Student's t formalism (41) to calculate overall 95% confidence intervals for threo/erythro ratios and extents of structure A depletion.
RESULTS AND DISCUSSION
Lignin isolation.
C. subvermispora and P. chrysosporium degraded wafers of spruce wood to similar extents in our experiments, giving mean dry weight losses of 25% ± 4% and 25% ± 7%, respectively, after 6 weeks in the subset of wafers that we analyzed. To obtain partially biodegraded lignins from the colonized wood wafers, we extracted them with a series of solvents, as described in Materials and Methods. As reported previously for P. chrysosporium (12), we found most of the extractable lignin in the methanol and dioxane-water fractions, as shown by 13C NMR analysis. These analyses also revealed relatively higher levels of low-molecular-weight material in the methanol fractions, and therefore, we restricted further work to the dioxane-water extracts, which we precipitated into water to afford macromolecular degraded lignins. To provide a nondegraded control for comparison, we employed high-molecular-weight milled wood lignin from intact spruce.
Quantitative 13C NMR spectra of the isolated lignins confirmed that all three were macromolecular, as shown by the broad signals obtained (Fig. 2). To assess the extent to which the white-rotted lignins had been degraded, we derivatized them and also the sound milled wood lignin with acetyl bromide, followed by reductive cleavage with zinc and quantification of the resulting 4-acetoxycinnamyl acetates (DFRC analysis). The DFRC method quantifies the noncondensed structure A units (i.e., those present in the unbranched linear polymer) that predominate in lignin, thus capturing a high proportion of all lignin subunits (34). The results showed that the lignins degraded by C. subvermispora and P. chrysosporium were 89% and 90% depleted, respectively, in noncondensed structure A units (Table 1).
FIG 2.
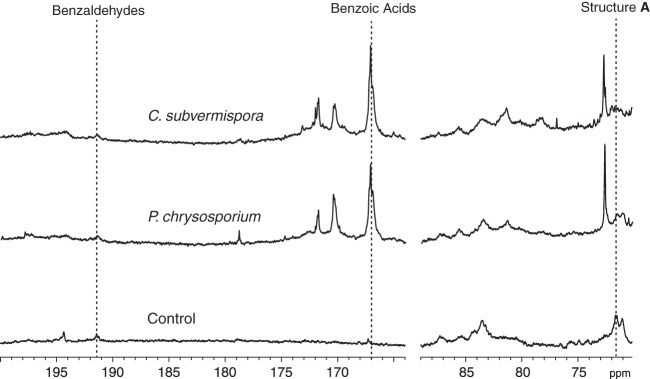
Quantitative 13C NMR spectra of nonacetylated spruce lignins showing signals for the degraded and control samples. The vertical dashed lines indicate chemical shift values for structure A, benzoic acids, and benzaldehydes in the lignins.
TABLE 1.
Yields of product released from structure A as determined by DFRC analysis of sound and decayed spruce wood
| Wood sample and expt | Yield of 4-acetoxy-3-methoxycinnamyl acetatea |
|
|---|---|---|
| g/g lignin | μmol/g lignin | |
| Nondecayed | ||
| 1 | 0.1664 | 630.4 |
| 2 | 0.1662 | 629.5 |
| Decayed by C. subvermispora | ||
| 1 | 0.0190 | 72.0 |
| 2 | 0.0205 | 77.7 |
| Decayed by P. chrysosporium | ||
| 1 | 0.0169 | 64.2 |
| 2 | 0.0172 | 65.3 |
Duplicate determinations for each pooled sample are shown.
Major products of ligninolysis.
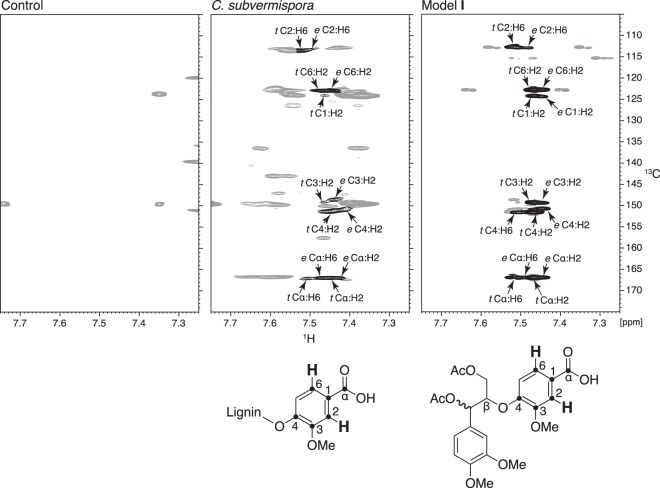
Quantitative 13C NMR spectroscopy showed that benzoic acid residues were the major identifiable degraded structures present after biodegradation by both fungi (Fig. 2). Benzoic acids were found previously in lignin degraded by P. chrysosporium (12, 13) but were not reported in an analysis of lignin degraded by C. subvermispora (24). Therefore, we performed an additional 1H-13C HMBC experiment after acetylation of the C. subvermispora and control samples. This two-dimensional technique associates protons in a molecule with carbons two or three bonds distant and is thus suited for the characterization of nonprotonated functional groups such as benzoic acid carboxyls (42). The results confirmed the 13C NMR data, showing a close correspondence between cross-peaks in the degraded lignin and in a threo/erythro mixture of a model compound that represents a softwood lignin benzoic acid in a β-O-4 linkage with the polymer (model I) (Fig. 3).
FIG 3.
1H-13C HMBC NMR spectra obtained on acetylated lignins from control and degraded spruce wood and from a threo/erythro mixture of lignin model I, showing correlations between aromatic H2/H6 and carbons two to three bonds away. Assigned cross-peaks are shown in black and are labeled to associate them with particular carbons and protons in lignin benzoic acids and in model I, which are depicted below their respective spectra. Each assignment states whether the contour is attributed to the threo (t)- or erythro (e)-diastereomer, followed by the numbered positions of the carbons and protons responsible. Unassigned contours are shown in gray. OMe, methoxyl.
A key question, apparently not yet discussed in the literature, is whether the presence of benzoic acid residues in white-rotted lignin actually signifies cleavage of the polymer. Although benzoic acids have not been found in sound lignin, low levels of benzaldehydes are already present (Fig. 2). In principle, some of these could be oxidized by the fungus to benzoic acids without ligninolysis and then become concentrated in the sample during solvent fractionation. However, the preexisting benzaldehydes in sound lignin are terminal rather than internal units of the lignin polymer (43). It is unlikely that many such terminal units or products derived from them remain in the sample after 90% of the original structure A units have been destroyed, which was the case for our biodegraded lignins. Therefore, the benzoic acids observed after extensive fungal decay likely result from Cα—Cβ cleavage of the lignin.
Previous work has shown that the SET oxidant LiP operating in conjunction with the mediator veratryl alcohol also catalyzes Cα—Cβ cleavage in vitro when DHP lignin is used as the substrate, although in this case, the products are benzaldehydes (36). The presence of the truncated residues as acids rather than aldehydes during white rot of wood (Fig. 1a) presumably reflects the instability of benzaldehydes in oxidizing environments (44), in combination with the fact that wood biodegradation experiments require much longer times than in vitro DHP depolymerizations (weeks versus hours). Our data thus provide evidence that SET oxidation of lignin aromatic rings contributes to ligninolysis by C. subvermispora as well as P. chrysosporium.
Diastereoselectivity of attack on structure A.
To compare the relative extents of fungal attack on threo and erythro structure A in spruce wood, we obtained two-dimensional 1H-13C HSQC spectra of the acetylated samples to associate protons in the lignin with the carbons directly attached to them (42). The results showed that threo structure A had been depleted more than erythro structure A in the white-rotted lignins, with this effect being much more pronounced in the sample degraded by C. subvermispora (Fig. 4). Using the same NMR method, we also found that P. chrysosporium LiP operating in conjunction with veratryl alcohol on α-13C-labeled DHP lignin depleted threo structure A more than it depleted erythro structure A (Fig. 5).
FIG 4.
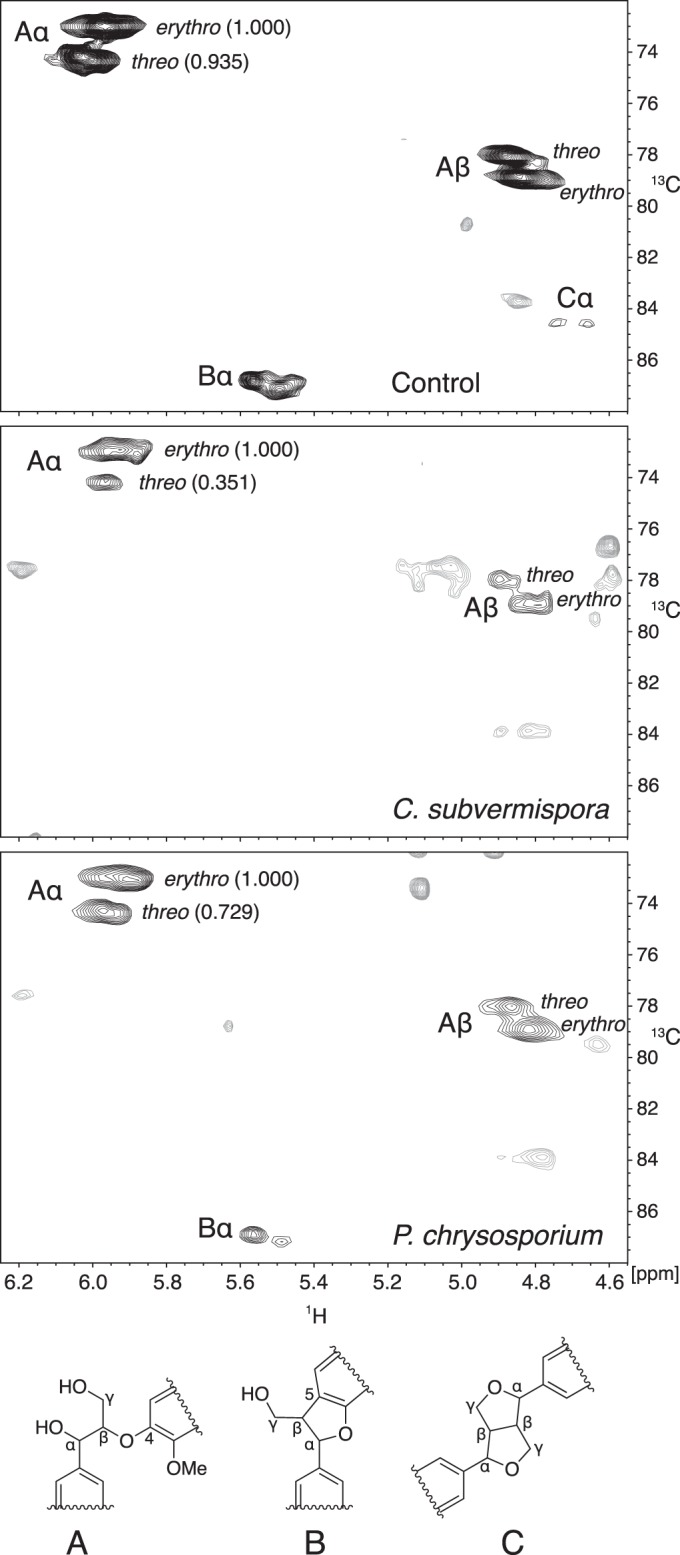
1H-13C HSQC NMR spectra obtained on acetylated lignins from control and degraded spruce wood. Labels next to the black contours associate them with particular carbons in the three lignin structures shown. Numbers in parentheses next to the Aα contours are relative integrals of the contour volumes. Unassigned contours are shown in gray.
FIG 5.
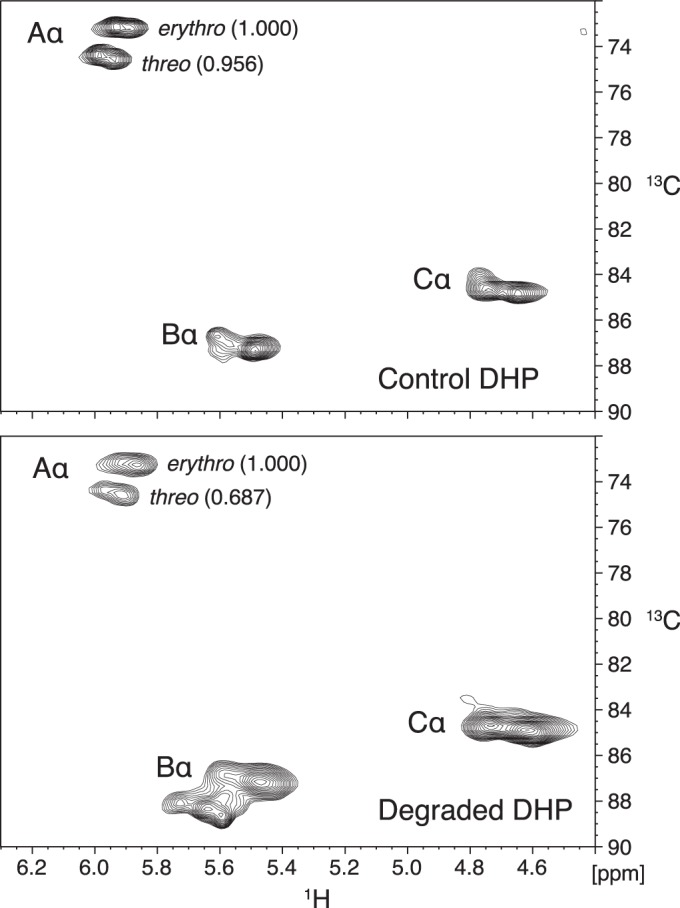
1H-13C HSQC NMR spectra obtained on acetylated α-13C-labeled DHP lignins treated in vitro with and without LiP in the presence of H2O2 and veratryl alcohol. Labels next to the black contours associate them with particular α-carbons in the lignin structures shown at the bottom of Fig. 4. Numbers in parentheses next to the Aα contours are relative integrals of the contour volumes.
Table 2 shows the observed structure A threo/erythro concentration ratios and associated extents of structure A removal, with 95% confidence intervals included, for the two biodegraded lignins compared with nondegraded wood lignin and for the DHP lignin treated in vitro with LiP-veratryl alcohol compared with DHP lignin from a control reaction without LiP. Although the structure A threo/erythro ratios during these delignification experiments are expected to decrease as a function of structure A removal, the ratios for degraded lignins in Table 2 can be directly compared because the extents of structure A removal were not significantly different among the three oxidative treatments.
TABLE 2.
Diastereoselectivity of structure A removal
| Sample | Structure A depletion (%) ± 95% confidence interval | Structure A threo/erythro ratio ± 95% confidence interval |
|---|---|---|
| Nondecayed spruce lignin | 0 ± 0 | 0.94 ± 0.02 |
| Lignin from spruce decayed by C. subvermispora | 89 ± 5 | 0.35 ± 0.10 |
| Lignin from spruce decayed by P. chrysosporium | 90 ± 3 | 0.73 ± 0.13 |
| Nondegraded DHP lignin | 0 ± 0 | 0.96 ± 0.03 |
| DHP lignin degraded by LiP-veratryl alcohol | 85 ± 9 | 0.69 ± 0.07 |
The results show that all three ligninolytic processes were diastereoselective, giving threo/erythro ratios at approximately 90% structure A depletion that were significantly lower than the initial values of 0.94 to 0.96. The ratios of 0.69 to 0.73 for lignins degraded by P. chrysosporium or by P. chrysosporium LiP in the presence of veratryl alcohol were not significantly different from each other, whereas removal of structure A by C. subvermispora was significantly more diastereoselective than the other two processes, with a ratio of 0.35. The similar diastereoselectivities exhibited by P. chrysosporium on wood and by LiP-veratryl alcohol on DHP lignin appear consistent with a ligninolytic role for this enzyme-mediator system in fungi that produce it. By contrast, the diastereoselectivity exhibited by C. subvermispora on wood is too high to be accounted for by LiP-veratryl alcohol.
Depletion of structures B and C in decayed lignin fractions.
The HSQC analyses also showed that structures B (phenylcoumarans) and C (pinoresinols) were markedly depleted relative to structure A in the white-rotted lignins (Fig. 4). However, this result is of uncertain significance because the relative levels of structures A, B, and C are probably not constant throughout the wood cell wall, in contrast to the uniform relative levels expected for threo and erythro structure A. The current view of lignin biosynthesis is that the secondary wood cell wall lignifies chiefly via endwise polymerization, whereas the middle lamella between wood cells lignifies partly via bulk polymerization (26, 45). Condensed units such as structures B and C are relatively more abundant in lignins formed via bulk polymerization (46) and are consequently likely to be concentrated in the middle lamella. We cannot rule out the possibility that middle lamella lignin was underrepresented in our extracted biodegraded lignins relative to the milled wood lignin control.
Conclusion.
Our results show that the ligninolytic oxidants of C. subvermispora are collectively more diastereoselective on structure A than are currently known fungal agents. This finding is consistent with a role in selective white rot, as it shows that the C. subvermispora oxidants exhibit specificity for their chemical targets, unlike nonselective fungal oxidants such as Fenton reagent. Identification of these selective agents will be a significant challenge. One possibility is that a secreted C. subvermispora enzyme, perhaps a peroxidase or laccase, oxidizes an unknown mediator to a diffusible oxidant that can infiltrate the sound wood cell wall. If so, the responsible ligninolytic radical must be more diastereoselective on structure A than the veratryl alcohol cation radical is, because our results show that the diastereoselectivity of structure A removal by C. subvermispora on wood was significantly higher than the diastereoselectivity on synthetic lignin exhibited in vitro by the LiP-veratryl alcohol system.
Recent work suggests three possible enzyme-mediator systems that could contribute to selective ligninolysis by C. subvermispora. (i) A LiP acts in concert with an oxidizable fungal metabolite. The low transcript levels found for the encoding genes do not favor this hypothesis (5), but it cannot yet be excluded. (ii) A laccase or manganese peroxidase oxidizes wood-derived phenols to produce phenoxyl radical oxidants that attack lignin. However, currently known phenolic mediator systems oxidize lignin model compounds in very low yields (47), so their diastereoselectivity cannot be tested. (iii) A manganese peroxidase generates ligninolytic oxidants from unsaturated lipids (21, 48–51). Although this system cleaves both synthetic lignin and lignin model compounds, the proximal oxidants that are responsible have not yet been identified (52), and whether they show diastereoselectivity on lignin remains unclear.
ACKNOWLEDGMENTS
This work was supported by the U.S. Department of Energy Office of Science, Biological Environmental Research (grants DE-AI02-07ER64480 to K.E.H. and J.R. and DE-SC0006929 to K.E.H., J.R., and C.J.H.). J.R. and F.L. were partially funded by the U.S. Department of Energy Great Lakes Bioenergy Research Center (grant DE-FC02-07ER64494).
Some of the NMR spectra reported here were acquired by using the National Magnetic Resonance Facility at the University of Wisconsin—Madison.
Footnotes
Published ahead of print 26 September 2014
REFERENCES
- 1.Blanchette RA. 1984. Selective delignification of wood by white-rot fungi. Appl. Biochem. Biotechnol. 9:323–324. 10.1007/BF02798957. [DOI] [Google Scholar]
- 2.Blanchette RA, Burnes TA, Eerdmans MM, Akhtar M. 1992. Evaluating isolates of Phanerochaete chrysosporium and Ceriporiopsis subvermispora for use in biological pulping processes. Holzforschung 46:109–116. 10.1515/hfsg.1992.46.2.109. [DOI] [Google Scholar]
- 3.Blanchette RA, Krueger EW, Haight JE, Akhtar M, Akin DE. 1997. Cell wall alterations in loblolly pine wood decayed by the white-rot fungus, Ceriporiopsis subvermispora. J. Biotechnol. 53:203–213. 10.1016/S0168-1656(97)01674-X. [DOI] [Google Scholar]
- 4.Mendonça R, Ferraz A, Kordsachia O, Koch G. 2004. Cellular UV-microspectrophotometric investigations on pine wood [Pinus taeda and Pinus elliottii] delignification during biopulping with Ceriporiopsis subvermispora (Pilát) Gilbn. & Ryv. and alkaline sulfite/anthraquinone treatment. Wood Sci. Technol. 38:567–575. 10.1007/s00226-004-0252-6. [DOI] [Google Scholar]
- 5.Hori C, Gaskell J, Igarashi K, Kersten P, Mozuch M, Samejima M, Cullen D. 2014. Temporal alterations in the secretome of the selective ligninolytic fungus Ceriporiopsis subvermispora during growth on aspen wood reveal this organism's strategy for degrading lignocellulose. Appl. Environ. Microbiol. 80:2062–2070. 10.1128/AEM.03652-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammel KE, Kapich AN, Jensen KA, Jr, Ryan ZC. 2002. Reactive oxygen species as agents of wood decay by fungi. Enzyme Microb. Technol. 30:445–453. 10.1016/S0141-0229(02)00011-X. [DOI] [Google Scholar]
- 7.Riley R, Salamov AA, Brown DW, Nagy LG, Floudas D, Held BW, Levasseur A, Lombard V, Morin E, Otillar R, Lindquist EA, Sun H, LaButti KM, Schmutz J, Jabbour D, Luo H, Baker SE, Pisabarro AG, Walton JD, Blanchette RA, Henrissat B, Martin F, Cullen D, Hibbett DS, Grigoriev IV. 2014. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc. Natl. Acad. Sci. U. S. A. 111:9923–9928. 10.1073/pnas.1400592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammel KE, Cullen D. 2008. Role of fungal peroxidases in biological ligninolysis. Curr. Opin. Plant Biol. 11:349–355. 10.1016/j.pbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Martínez AT. 2002. Molecular biology and structure-function of lignin-degrading heme peroxidases. Enzyme Microb. Technol. 30:425–444. 10.1016/S0141-0229(01)00521-X. [DOI] [Google Scholar]
- 10.Ruiz-Dueñas FJ, Martínez AT. 2009. Microbial degradation of lignin: how a bulky recalcitrant polymer is efficiently recycled in nature and how we can take advantage of this. Microb. Biotechnol. 2:164–177. 10.1111/j.1751-7915.2008.00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alfaro M, Oguiza JA, Ramírez L, Pisabarro AG. 2014. Comparative analysis of secretomes in basidiomycete fungi. J. Proteomics 102:28–43. 10.1016/j.jprot.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Chua MGS, Chen C-L, Chang H-M, Kirk TK. 1982. 13C NMR spectroscopic study of spruce lignin degraded by Phanerochaete chrysosporium. Holzforschung 36:165–172. 10.1515/hfsg.1982.36.4.165. [DOI] [Google Scholar]
- 13.Robert D, Chen C-L. 1989. Biodegradation of lignin in spruce wood by Phanerochaete chrysosporium: quantitative analysis of biodegraded spruce lignins by 13C NMR spectroscopy. Holzforschung 43:323–332. 10.1515/hfsg.1989.43.5.323. [DOI] [Google Scholar]
- 14.Bietti M, Baciocchi E, Steenken S. 1998. Lifetime, reduction potential and base-induced fragmentation of the veratryl alcohol radical cation in aqueous solution. Pulse radiolysis studies on a ligninase “mediator.” J. Phys. Chem. A 102:7337–7342. [Google Scholar]
- 15.Harvey PJ, Schoemaker HE, Palmer JM. 1986. Veratryl alcohol as a mediator and the role of radical cations in lignin biodegradation by Phanerochaete chrysosporium. FEBS Lett. 195:242–246. [Google Scholar]
- 16.Hunt CG, Houtman CJ, Jones DC, Kitin P, Korripally P, Hammel KE. 2013. Spatial mapping of extracellular oxidant production by a white rot basidiomycete on wood reveals details of ligninolytic mechanism. Environ. Microbiol. 15:956–966. 10.1111/1462-2920.12039. [DOI] [PubMed] [Google Scholar]
- 17.Christov LP, Akhtar M, Prior BA. 1998. The potential of biosulfite pulping in dissolving pulp production. Enzyme Microb. Technol. 23:70–74. 10.1016/S0141-0229(98)00017-9. [DOI] [Google Scholar]
- 18.Dorado J, Almendros G, Camarero S, Martínez AT, Vares T, Hatakka A. 1999. Transformation of wheat straw in the course of solid-state fermentation by four ligninolytic basidiomycetes. Enzyme Microb. Technol. 25:605–612. 10.1016/S0141-0229(99)00088-5. [DOI] [Google Scholar]
- 19.Ferraz A, Guerra A, Mendonça R, Masarin F, Vicentim MP, Aguiar A, Pavan PC. 2008. Technological advances and mechanistic basis for fungal biopulping. Enzyme Microb. Technol. 43:178–185. 10.1016/j.enzmictec.2007.10.002. [DOI] [Google Scholar]
- 20.Rüttimann-Johnson C, Salas L, Vicuña R, Kirk TK. 1993. Extracellular enzyme production and synthetic lignin mineralization by Ceriporiopsis subvermispora. Appl. Environ. Microbiol. 59:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernández-Fueyo E, Ruiz-Dueñas FJ, Ferreira P, Floudas D, Hibbett DS, Canessa P, Larrondo LF, James TY, Seelenfreund D, Lobos S, Polanco R, Tello M, Honda Y, Watanabe T, Watanabe T, San RJ, Kubicek CP, Schmoll M, Gaskell J, Hammel KE, St John FJ, Wymelenberg AV, Sabat G, BonDurant SS, Syed K, Yadav JS, Doddapaneni H, Subramanian V, Lavín JL, Oguiza JA, Perez G, Pisabarro AG, Ramirez L, Santoyo F, Master E, Coutinho PM, Henrissat B, Lombard V, Magnuson JK, Kuës U, Hori C, Igarashi K, Samejima M, Held BW, Barry KW, LaButti KM, Lapidus A, Lindquist EA, Lucas SM, Riley R, et al. 2012. Comparative genomics of Ceriporiopsis subvermispora and Phanerochaete chrysosporium provide insight into selective ligninolysis. Proc. Natl. Acad. Sci. U. S. A. 109:5458–5463. 10.1073/pnas.1119912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernández-Fueyo E, Ruiz-Dueñas FJ, Miki Y, Martínez MJ, Hammel KE, Martínez AT. 2012. Lignin-degrading peroxidases from genome of selective ligninolytic fungus Ceriporiopsis subvermispora. J. Biol. Chem. 287:16903–16916. 10.1074/jbc.M112.356378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srebotnik E, Jensen KA, Kawai S, Hammel KE. 1997. Evidence that Ceriporiopsis subvermispora degrades nonphenolic lignin structures by a one-electron-oxidation mechanism. Appl. Environ. Microbiol. 63:4435–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerra A, Mendonça R, Ferraz A, Lu F, Ralph J. 2004. Structural characterization of lignin during Pinus taeda wood treatment with Ceriporiopsis subvermispora. Appl. Environ. Microbiol. 70:4073–4078. 10.1128/AEM.70.7.4073-4078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boerjan W, Ralph J, Baucher M. 2003. Lignin biosynthesis. Annu. Rev. Plant Biol. 54:519–546. 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 26.Ralph J, Brunow G, Harris PJ, Dixon RA, Schatz PF, Boerjan W. 2008. Lignification: are lignins biosynthesized via simple combinatorial chemistry or via proteinaceous control and template replication?, p 36–66 In Daayf F, Lattanzio V. (ed), Recent advances in polyphenol research, vol 1 Wiley-Blackwell, Hoboken, NJ. [Google Scholar]
- 27.Hata K. 1966. Investigations on lignins and lignification. XXXIII. Studies on lignins isolated from spruce wood decayed by Poria subacida BII. Holzforschung 20:142–147. [Google Scholar]
- 28.Kirk TK, Chang H-M. 1974. Decomposition of lignin by white-rot fungi. I. Isolation of heavily degraded lignins from decayed spruce. Holzforschung 28:217–222. [Google Scholar]
- 29.Kirk TK, Lundquist K. 1970. Comparison of sound and white-rotted sapwood of sweetgum with respect to properties of the lignin and composition of extractives. Sven. Papperstidn. 73:294–306. [Google Scholar]
- 30.Akiyama T, Goto H, Nawawi DS, Syafii W, Matsumoto Y, Meshitsuka G. 2005. Erythro/threo ratio of β-O-4-structures as an important structural characteristic of lignin. Part 4: variation in the erythro/threo ratio in softwood and hardwood lignins and its relation to syringyl/guaiacyl ratio. Holzforschung 59:276–281. 10.1515/HF.2005.045. [DOI] [Google Scholar]
- 31.Bohlin C, Andersson PA, Lundquist K, Jonsson LJ. 2007. Differences in stereo-preference in the oxidative degradation of diastereomers of the lignin model compound 1-(3,4-dimethoxyphenyl)-2-(2-methoxyphenoxy)-1,3-propanediol with enzymic and non-enzymic oxidants. J. Mol. Catal. B 45:21–26. 10.1016/j.molcatb.2006.11.001. [DOI] [Google Scholar]
- 32.Cho DW, Parthasarathi R, Pimentel AS, Maestas GD, Park HJ, Yoon UC, Dunaway-Mariano D, Gnanakaran S, Langan P, Mariano PS. 2010. Nature and kinetic analysis of carbon-carbon bond fragmentation reactions of cation radicals derived from SET-oxidation of lignin model compounds. J. Org. Chem. 75:6549–6562. 10.1021/jo1012509. [DOI] [PubMed] [Google Scholar]
- 33.Kirk TK, Croan S, Tien M, Murtagh KE, Farrell RL. 1986. Production of multiple ligninases by Phanerochaete chrysosporium: effect of selected growth conditions and use of a mutant strain. Enzyme Microb. Technol. 8:27–32. 10.1016/0141-0229(86)90006-2. [DOI] [Google Scholar]
- 34.Lu FC, Ralph J. 1997. Derivatization followed by reductive cleavage (DFRC method), a new method for lignin analysis: protocol for analysis of DFRC monomers. J. Agric. Food Chem. 45:2590–2592. 10.1021/jf970258h. [DOI] [Google Scholar]
- 35.Fukushima RS, Hatfield RD. 2004. Comparison of the acetyl bromide spectrophotometric method with other analytical lignin methods for determining lignin concentration in forage samples. J. Agric. Food Chem. 52:3713–3720. 10.1021/jf035497l. [DOI] [PubMed] [Google Scholar]
- 36.Hammel KE, Jensen KA, Mozuch MD, Landucci LL, Tien M, Pease EA. 1993. Ligninolysis by a purified lignin peroxidase. J. Biol. Chem. 268:12274–12281. [PubMed] [Google Scholar]
- 37.Yelle DJ, Ralph J, Lu FC, Hammel KE. 2008. Evidence for cleavage of lignin by a brown rot basidiomycete. Environ. Microbiol. 10:1844–1849. 10.1111/j.1462-2920.2008.01605.x. [DOI] [PubMed] [Google Scholar]
- 38.Ralph J, Akiyama T, Kim H, Lu FC, Schatz PF, Marita JM, Ralph SA, Reddy MSS, Chen F, Dixon RA. 2006. Effects of coumarate 3-hydroxylase down-regulation on lignin structure. J. Biol. Chem. 281:8843–8853. 10.1074/jbc.M511598200. [DOI] [PubMed] [Google Scholar]
- 39.Ralph J, Landucci LL. 2010. NMR of lignins, p 137–234 In Heitner C, Dimmel DR, Schmidt JA. (ed), Lignin and lignans: advances in chemistry. Taylor and Francis, Boca Raton, FL. [Google Scholar]
- 40.Kupce E, Freeman R. 2007. Compensated adiabatic inversion pulses: broadband INEPT and HSQC. J. Magn. Res. 187:258–265. 10.1016/j.jmr.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Box GEP, Hunter WG, Hunter JS. 1978. Statistics for experimenters. Wiley, New York, NY. [Google Scholar]
- 42.Claridge TDW. 1999. High-resolution NMR techniques in organic chemistry, p 222–257 In Baldwin JE, Williams RM. (ed), Tetrahedron organic chemistry series, vol 19 Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 43.Lapierre C, Pilate G, Pollet B, Mila I, Leple JC, Jouanin L, Kim H, Ralph J. 2004. Signatures of cinnamyl alcohol dehydrogenase deficiency in poplar lignins. Phytochemistry 65:313–321. 10.1016/j.phytochem.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Sankar M, Nowicka E, Carter E, Murphy DM, Knight DW, Bethell D, Hutchings GJ. 2014. The benzaldehyde oxidation paradox explained by the interception of peroxy radical by benzyl alcohol. Nat. Commun. 5:3332. 10.1038/ncomms4332. [DOI] [PubMed] [Google Scholar]
- 45.Syrjänen K, Brunow G. 2000. Regioselectivity in lignin biosynthesis. The influence of dimerization and cross-coupling. J. Chem. Soc. Perkin Trans. 1 2000:183–187. 10.1039/A907919J. [DOI] [Google Scholar]
- 46.Freudenberg K. 1956. Beiträge zur Erforschung des Lignins. Angew. Chem. 68:508–512. 10.1002/ange.19560681603. [DOI] [Google Scholar]
- 47.Nousiainen P, Maijala P, Hatakka A, Martínez AT, Sipilä J. 2009. Syringyl-type simple plant phenolics as mediating oxidants in laccase catalyzed degradation of lignocellulosic materials: model compound studies. Holzforschung 63:699–704. 10.1515/HF.2009.066. [DOI] [Google Scholar]
- 48.Bao W, Fukushima Y, Jensen KA, Moen MA, Hammel KE. 1994. Oxidative degradation of non-phenolic lignin during lipid peroxidation by fungal manganese peroxidase. FEBS Lett. 354:297–300. 10.1016/0014-5793(94)01146-X. [DOI] [PubMed] [Google Scholar]
- 49.Cunha GGS, Masarin F, Norambuena M, Freer J, Ferraz A. 2010. Linoleic acid peroxidation and lignin degradation by enzymes produced by Ceriporiopsis subvermispora grown on wood or in submerged liquid cultures. Enzyme Microb. Technol. 46:262–267. 10.1016/j.enzmictec.2009.11.006. [DOI] [Google Scholar]
- 50.Enoki M, Watanabe T, Nakagame S, Koller K, Messner K, Honda Y, Kuwahara M. 1999. Extracellular lipid peroxidation of selective white-rot fungus, Ceriporiopsis subvermispora. FEMS Microbiol. Lett. 180:205–211. 10.1111/j.1574-6968.1999.tb08797.x. [DOI] [PubMed] [Google Scholar]
- 51.Kapich A, Hofrichter M, Vares T, Hatakka A. 1999. Coupling of manganese peroxidase-mediated lipid peroxidation with destruction of nonphenolic lignin model compounds and 14C-labeled lignins. Biochem. Biophys. Res. Commun. 259:212–219. 10.1006/bbrc.1999.0742. [DOI] [PubMed] [Google Scholar]
- 52.Kapich AN, Korneichik TV, Hatakka A, Hammel KE. 2010. Oxidizability of unsaturated fatty acids and of a non-phenolic lignin structure in the manganese peroxidase-dependent lipid peroxidation system. Enzyme Microb. Technol. 46:136–140. 10.1016/j.enzmictec.2009.09.014. [DOI] [Google Scholar]