Abstract
Bacillus thuringiensis is an entomopathogenic bacterium that has been used as an efficient biopesticide worldwide. Despite the fact that this bacterium is usually described as an insect pathogen, its life cycle in the environment is still largely unknown. B. thuringiensis belongs to the Bacillus cereus group of bacteria, which has been associated with many mobile genetic elements, such as species-specific temperate or virulent bacteriophages (phages). Temperate (lysogenic) phages are able to establish a long-term relationship with their host, providing, in some cases, novel ecological traits to the bacterial lysogens. Therefore, this work focuses on evaluating the potential influence of temperate tectiviruses GIL01 and GIL16 on the development of different life traits of B. thuringiensis. For this purpose, a B. thuringiensis serovar israelensis plasmid-cured (nonlysogenic) strain was used to establish bacterial lysogens for phages GIL01 and GIL16, and, subsequently, the following life traits were compared among the strains: kinetics of growth, metabolic profiles, antibiotics susceptibility, biofilm formation, swarming motility, and sporulation. The results revealed that GIL01 and GIL16 lysogeny has a significant influence on the bacterial growth, sporulation rate, biofilm formation, and swarming motility of B. thuringiensis. No changes in metabolic profiles or antibiotic susceptibilities were detected. These findings provide evidence that tectiviruses have a putative role in the B. thuringiensis life cycle as adapters of life traits with ecological advantages.
INTRODUCTION
Bacillus thuringiensis is a Gram-positive, aerobic, endospore-forming bacterium whose distinctive characteristic of forming proteinaceous crystalline inclusions (crystals) during sporulation distinguishes it from other closely related bacteria clustered in the Bacillus cereus group (1, 2). These parasporal crystals are mainly composed of insecticidal toxin proteins, also known as Cry toxins, which are responsible for the entomopathogenic activity that has propelled B. thuringiensis to be the main biological agent used against insects representing agricultural pests (3). Although B. thuringiensis is an insect pathogen, the ecology of this bacterium is still obscure (2, 4). B. thuringiensis is usually referred to as a soil-dwelling microorganism, but it has also been isolated from different environmental habitats, including phylloplane, rhizosphere, endophytic, and aquatic environments (4, 5). Due to this ubiquitous distribution, B. thuringiensis might have a hidden ecological role beyond a bacterium-insect interaction and pathogenicity.
The great diversity of B. thuringiensis strains is often associated with the genetic plasticity bestowed by the presence of many plasmids, which can vary in number (up to 17) and in size (from 2 to >300 kb) (6–8). Specifically, B. thuringiensis serovar israelensis (H14) contains at least eight double-stranded DNA (dsDNA) extrachromosomal molecules, including three small circular plasmids (5.4, 6.7, and 7.6 kb), four large circular plasmids (128, 145, 240, and 350 kb) (8–13), and one linear plasmid of 14,931 bp delimited by imperfect 73-bp terminal repeats (14). Remarkably, the linear molecule is a temperate phage, named GIL01, which is able to reside and replicate independently as a linear plasmid inside the host cell and to produce viable phage particles after induction by DNA-damaging treatments (14). Phage GIL01 and its closest fully sequenced siblings, Bam35, GIL16, AP50, and Wip1, belong to the family Tectiviridae, whose members are icosahedral phages characterized by the presence of an internal lipid membrane/vesicle that, upon adsorption, can act as a tail-like structure for genome delivery (15, 16). Beside these 5 fully sequenced representatives, 52 other novel putative tectiviruses infecting the B. cereus group have recently been discovered, among which are various GIL01- and GIL16-like phages infecting B. thuringiensis strains (17, 18). Also, tectiviruses found in Gram-positive bacteria exhibit a strong similarity to the 15,274-bp cryptic linear plasmid pBClin15 from B. cereus ATCC 14579 (19, 20).
Although dsDNA temperate phages may enter the lytic pathway that leads to host cell death, they normally execute a lysogenic cycle in which there is a tight repression of the lytic behavior and the phage genome replicates with that of the host cell (21). Therefore, lysogeny can be seen as the perpetuation of prophages as part of the bacterial replicating system, where the bacterium (lysogen) becomes immune against further infections with phages of the same type (22). Lysogenic phages establish a stable relationship with their host bacteria and can in some cases confer novel properties, such as virulence traits (e.g., lysogenic conversion) or increased survival/ecological fitness, to the bacterial lysogens (23).
In this work, the question of whether or not temperate tectiviruses influence some life traits in B. thuringiensis serovar israelensis was addressed. For this purpose, the kinetics of growth, metabolic profiles, antibiotic susceptibility, biofilm formation, swarming motility, and sporulation rates were compared between a plasmid-cured B. thuringiensis serovar israelensis strain (referred to here as the nonlysogenic strain) and two isogenic lysogens, derived from the same strain, harboring either the GIL01 or GIL16 tectivirus. It was found that GIL01 and GIL16 lysogeny had a significant influence on the bacterial growth, sporulation rate, biofilm formation, and swarming motility of B. thuringiensis, all of which are traits involved in the survival and colonization of this bacterium in different environmental habitats. These findings suggest that tectiviruses play a role in the lifestyle of B. thuringiensis.
MATERIALS AND METHODS
Bacterial strains, phages, and culture conditions.
B. thuringiensis serovar israelensis plasmid-cured derivative strain GBJ002 was used as a nontectiviral lysogenic bacterial strain (i.e., this strain contains some chromosomal prophages). To simplify, strain GBJ002 is referred to here as the nonlysogenic strain. This strain was used to propagate phages GIL01 and GIL16 and establish lysogens GBJ002/GIL01 and GBJ002/GIL16 (see below). Strain GBJ002 has chromosomal resistance to nalidixic acid (Nal) (13). The natural host of phage GIL01 is B. thuringiensis serovar israelensis strain NB31 (14), while phage GIL16 was isolated from B. thuringiensis strain B16 (19). Bacteria were routinely cultured in lysogeny broth (LB) medium (5 g/liter NaCl, 5 g/liter yeast extract, 10 g/liter tryptone), unless otherwise indicated. For agar plates, LB medium was solidified with 1.4% (wt/vol) agar (LB-agar), unless otherwise stated. For overnight cultures, a single bacterial colony growing on LB-agar plates was inoculated into 10 ml LB medium and incubated for 14 to 15 h at 30°C and 120 rpm.
Phage propagation and bacterial lysogen generation.
Phage GIL01 and GIL16 stocks were obtained by mitomycin C (AppliChem) induction as described previously (18). For lysogeny establishment, susceptible host GBJ002 was grown to an optical density at 595 nm (OD595) of ∼0.5 in 10 ml LB medium at 30°C and 120 rpm. Two hundred microliters of the bacterial culture was infected with equal volumes of 10-fold serial dilutions of GIL01 and GIL16 stocks, and infection proceeded as described elsewhere (18). Final lysogens growing on LB-agar were screened by PCR using the Tect1-Tect2 and Tect3-Tect4 sets of primers described previously (18), and then the PCR products were sequenced to confirm GIL01 and GIL16 lysogeny. A positive colony for each phage was selected and retained for the study. GBJ002 lysogens for GIL01 and GIL16 were named GBJ002/GIL01 and GBJ002/GIL16, respectively.
Cellular and colony morphologies.
The cellular morphology of overnight cultures was observed under phase-contrast using a DMI6000B inverted microscope (Leica). For colony morphology, 10-fold serial dilutions of overnight bacterial cultures were plated on LB-agar plates and incubated for 48 h at 30°C. The resulting bacterial colonies were scanned (HP Scanjet G4010) at 600 dots per inch (dpi) and compared. To facilitate the differentiation of phenotypes, image correction was applied to all the scanned plates as follows: +40% contrast, +20% brightness, and +50% sharpness.
Measurement of bacterial growth kinetics in liquid medium.
Overnight bacterial cultures were diluted 1,000-fold in LB medium, and 250 μl was inoculated into a 96-well polystyrene microtiter plate (Greiner Bio-One). Bacterial growth was measured by following the OD595 with a Multiskan FC microplate photometer (Thermo Fisher Scientific, Inc.) every hour over a period of 27 h of incubation at 30°C with background shaking. For phage induction, mitomycin C was added to the inoculated wells to a final concentration of 1 μg/ml at 3 h of bacterial growth, and the OD595 continued to be monitored. Ten replicate wells were used for each bacterial culture tested. Control wells containing noninoculated medium were located on each plate. LB medium was used as a reference and adjusted to an OD595 of 0 in the data analysis.
To verify the stability of lysogeny, serial 10-fold dilutions of bacterial cultures were taken at 27 h and spread onto LB-agar plates. After overnight incubation at 30°C, 20 individual colonies for each bacterial strain were randomly selected, regrown in LB-agar plates supplemented with Nal (15 μg/ml), and screened by PCR using the sets of primers Tect1-Tect2 and Tect3-Tect4 (18). Bacterial growth in LB-agar plates with Nal was used to confirm the purity of the cultures, since strain GBJ002 is Nal resistant (Nalr).
Measurements of phage plaque production versus bacterial growth.
One hundred fifty microliters of an overnight culture of the nonlysogenic strain GBJ002 was inoculated into 150 ml of LB medium contained in a 500-ml Erlenmeyer flask. Bacterial growth (at 30°C and 120 rpm) was measured by determination of the OD595 with a spectrophotometer (Ultrospec 2100 Pro; Amersham-GE Healthcare) every 15 min during the first 4 h of growth and then every 30 min for up to 10 h of growth. At specific time points, 100 μl of the bacterial culture was infected separately with equal volumes of GIL01 and GIL16 fresh stocks (1 × 105 PFU/ml). Phage stocks were prepared by mitomycin C induction from lysogens GBJ002/GIL01 and GBJ002/GIL16 as described above, and phage titration was done by the double-agar method (see below) using a GBJ002 culture with an OD595 of ∼0.5. The time points selected for measurement of bacterial growth were 2 h, every 30 min between 3 and 9 h, 10 h, and 24 h. Bacterium-phage mixtures were incubated for 30 min, and 10-fold dilutions were plated with molten LB-top agar (0.6% [wt/vol]) onto LB double-agar plates. Bacterium-phage mixtures were prepared in duplicate for each time point. Plates were incubated overnight at 30°C, and the numbers of PFU were counted and analyzed for plaque morphology. Due to the turbidity of the phage plaques, photographs for plaque morphologies were acquired using a scanner (Agfa Snap Scan Touch) at 300 dpi. To work with the same backlight background, image acquisition parameters were the same for all plates scanned. To facilitate the differentiation of plaque phenotypes, image correction was applied as follows: −20% contrast and +40% brightness.
Metabolic profiles.
To compare the bacterial exploitation of a panel of metabolic substrates, the API 50 CH and API 20 E strip systems (bioMérieux) were used. This resulted in 70 biochemical tests that allowed, among other analyses, the measurement of carbon source utilization and carbohydrate fermentation by strain GBJ002 and derivative lysogens. The assays were performed according to the manufacturer's instructions.
Antibiograms.
The antibiotic susceptibility of strains GBJ002, GBJ002/GIL01, and GBJ002/GIL16 was tested using 17 different antibiotic disks (Neo-Sensitabs; Rosco Diagnostica). Overnight cultures were adjusted at an OD595 of ∼1, 100 μl of these suspensions was placed into 5 ml Mueller-Hinton broth (Bio-Rad) containing 0.6% (wt/vol) agar, and the contents were gently mixed and poured into Mueller-Hinton agar (Bio-Rad) plates. The antibiotic disks were placed onto the solidified agar, and the plates were incubated at 30°C for 24 h. Inhibition zones were measured for each antibiotic tested.
Biofilm formation assay by microtiter plate method.
Biofilm formation was evaluated in 96-well polystyrene microtiter plates (Greiner Bio-One) as described elsewhere (24), with the following modifications: 200 μl of a 1:100 dilution in LB medium from an overnight bacterial culture was placed in each well. After 48 h of incubation at 30°C, bacterial cells were removed and the wells were gently washed twice with distilled H2O. Bound cells were stained with 250 μl per well of a 0.1% (wt/vol) aqueous solution of crystal violet. The microtiter plate was incubated at room temperature for 30 min. After three gentle washings with distilled H2O, the microtiter plate was dried upside down for 1 h. Crystal violet dye was solubilized with 225 μl ethanol absolute (VWR). The absorbance was quantified in an automatic plate reader (Multiskan FC microplate photometer; Thermo Fisher Scientific) at 595 nm. Five replicate wells were used for each bacterial culture tested.
Swarming motility assays.
Bacterial swarming motility assays were carried out as described previously with some modifications (25). Briefly, 9-cm petri dishes were prepared with 25 ml of LB medium solidified with two agar concentrations: 0.6 and 1.0% (wt/vol). Swarming plates were inoculated with 3 μl of bacterial cultures with an OD595 of ∼0.1, and the cultures were incubated at 30°C. Bacterial swarming diameters were measured at 24, 48, and 72 h after inoculation. Swarming assays were repeated three times with three replicates each.
Sporulation rate.
Overnight bacterial cultures were adjusted with LB to an OD595 of 0.3 in order to have approximately the same number of cells. One hundred microliters of these suspensions was spread on agar-water (1.5% [wt/vol]) plates (26) and allowed to sporulate for 5 days at 30°C. One loopful of bacteria was harvested from sporulating cultures and resuspended in 1.5 ml ice-cold 0.85% (wt/vol) NaCl, and two aliquots of 500 μl of bacterium-spore suspensions were prepared. One aliquot was heated at 80°C for 10 min and then kept on ice for 10 min, while the other aliquot was kept on ice during this process. Serial 10-fold dilutions were prepared and 100 μl of each dilution was plated onto LB-agar plates. Growing colonies were counted after 48 h of incubation at 30°C. The sporulation rate was determined as the ratio between the average colony counts of heated suspensions (spores) and the average colony counts before heat shock (viable cells and spores).
Statistical analysis.
The statistical significance of the colony diameters displayed by nonlysogenic strain GBJ002 and lysogens GBJ002/GIL01 and GBJ002/GIL16 was analyzed by the nonparametric Kruskal-Wallis test, since the data did not show a normal distribution using the Shapiro-Wilks test. The statistical significance of the biofilm formation and swarming motility data for nonlysogenic strain GBJ002 and lysogens GBJ002/GIL01 and GBJ002/GIL16 was determined using the one-way analysis of variance (ANOVA) test. The Tukey multiple-comparison test was used for comparisons of means. Statistical analyses were performed using the software package InfoStat (version 2011; InfoStat Group, College of Agricultural Sciences, National University of Cordoba, Cordoba, Argentina).
RESULTS
Establishment of tectiviral lysogeny and its impact on colony morphology.
In order to work with the same genetic background, strain GBJ002 was subjected to infection with single turbid plaques of phage GIL01 or GIL16. The resulting lysogens (GBJ002/GIL01 and GBJ002/GIL16) were screened by PCR, and amplification products were sequenced to confirm either GIL01 or GIL16 lysogeny establishment (data not shown). It is important to note that strain GBJ002 is a plasmid-cured strain that does not harbor the genetic determinants coding for the insecticidal Cry toxins. This strain was chosen because it is a known host for tectiviruses GIL01 and GIL16 (18). The lack of plasmids allowed assessment of whether or not tectiviral lysogeny has an impact on bacterial life traits not associated with plasmids.
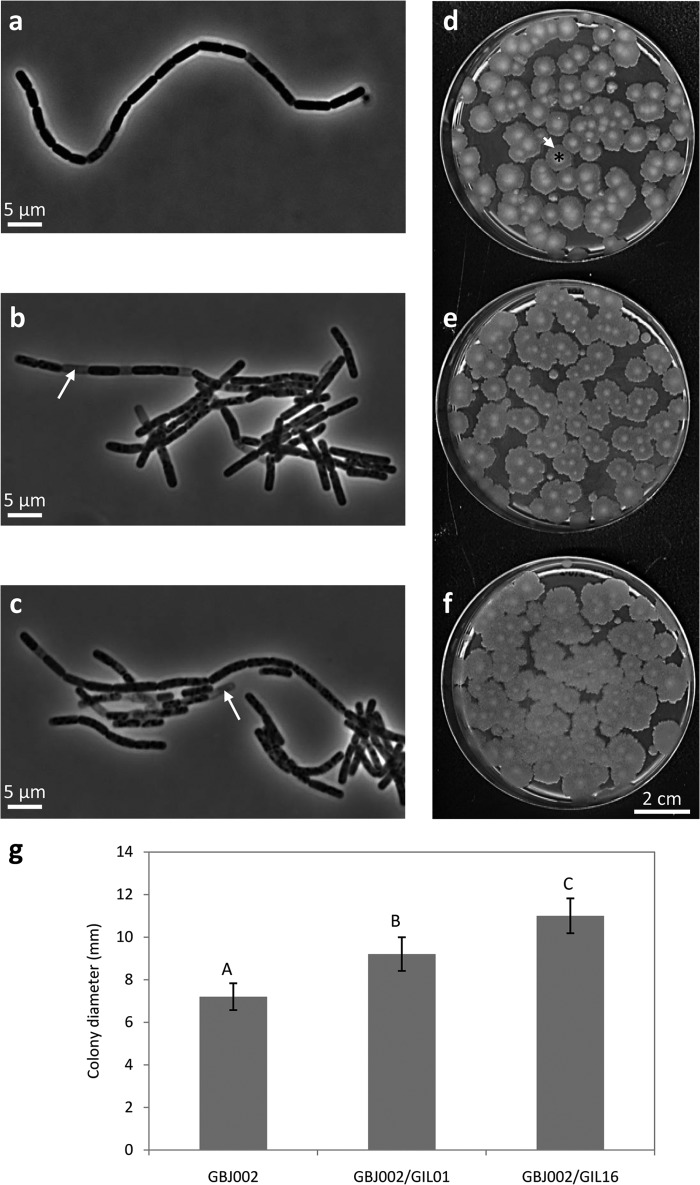
The cellular and colony morphologies of GBJ002 and the two lysogens were evaluated (Fig. 1). Microscopic observation of overnight cultures indicated that there were not outstanding morphological differences among the strains (Fig. 1a to c). Cell dimensions and cell chain formation were similar. In contrast, the proportion of empty cells observed was higher in the two lysogens (∼10%) than in the nonlysogenic strain (∼1%). When colony morphologies were analyzed after 48 h of incubation at 30°C, all strains presented circular, white-bone, matte colonies with the characteristic fried egg phenotype of B. thuringiensis (27) (Fig. 1d to f). However, the lysogens had significantly (Kruskal-Wallis test, P < 0.05) larger colonies than nonlysogenic strain GBJ002 (Fig. 1g). Comparison of colony diameters between lysogenic strains GBJ002/GIL01 and GBJ002/GIL16 also showed that they were statistically significantly (Kruskal-Wallis test, P < 0.05) different, with lysogen GBJ002/GIL16 producing the largest colonies (Fig. 1g). The most important difference pointed to the fried egg phenotype (Fig. 1d to f). Whereas in GBJ002 this phenotype presented very well defined equivalents of the egg white and yolk, in the lysogens GBJ002/GIL01 and GBJ002/GIL16, the yolk was less prominent and bright and the egg white zone was larger than that of GBJ002. This difference was more obvious in the lysogen GBJ002/GIL16 (Fig. 1f), as its egg yolks appeared to be fuzzy and less bright than those of the other two strains. Also, the colony edges of the lysogenic strains were more irregular than those of GBJ002.
FIG 1.
Cell and colony morphologies of B. thuringiensis serovar israelensis strain GBJ002 (a, d) and derivative lysogenic strains GBJ002/GIL01 (b, e) and GBJ002/GIL16 (c, f). (a to c) Phase-contrast micrographs of overnight bacterial cultures in LB medium. White arrows, empty cells. (d to f) Colonies growing in LB-agar (1.5%). Images were acquired after 48 h of incubation. The fried egg phenotype is highlighted in panel d by an asterisk (yolk) and a short white arrow (egg white). (g) Colony diameters of bacteria growing on LB-agar plates after 48 h of incubation. Each datum plotted represents the average of 10 colonies. The error bars indicate the standard deviation values. Means with different letters (A, B, C) indicate significant differences (Kruskal-Wallis test, P < 0.05). Similar observations were made in 3 independent experiments.
Tectiviral lysogeny influence on bacterial growth.
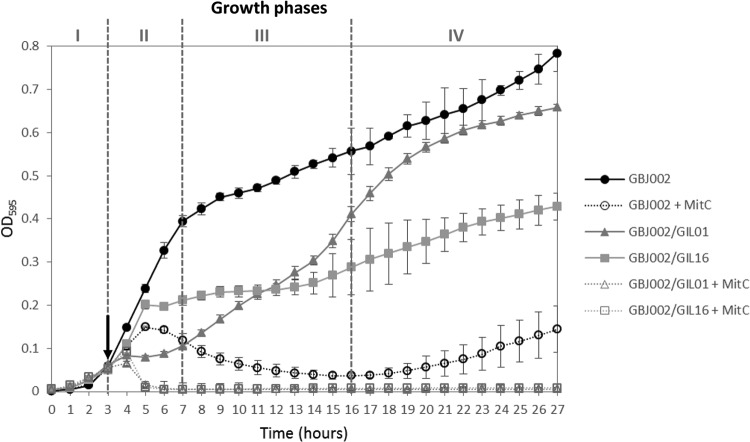
Since lysogeny might affect other bacterial phenotypes, the growth curves of strains GBJ002, GBJ002/GIL01, and GBJ002/GIL16 were compared with and without early-phase induction by mitomycin C. The growth of nonlysogenic strain GBJ002 occurred in four phases during the analyzed time (27 h): lag phase (phase I; during the first 3 h), exponential phase (phase II), phase III, and phase IV (Fig. 2). When mitomycin C was added at the end of the lag phase, GBJ002 continued growing for approximately 1 h before the optical density started to decrease due to the strong toxic effects of this DNA-damaging compound. Interestingly, at the end of phase IV the cell density started to rise again. Lysogens GBJ002/GIL01 and GBJ002/GIL16 displayed growth curves different from each other and from that of GBJ002 (Fig. 2). When lysogenic cultures were noninduced, GBJ002/GIL01 presented a lower growth rate (change in OD595/change in time in a growth interval [ΔOD595/Δt]) than GBJ002/GIL16 at phase II. However, during early phase III, GBJ002/GIL01 increased its growth rate, which, at the end of phase III, was higher than that of GBJ002/GIL16 yet lower than that of GBJ002 (ΔOD595/Δt for late phase III, GBJ002/GIL16 < GBJ002/GIL01 < GBJ002). Indeed, GBJ002/GIL16 phase III was equivalent to a stationary phase, where bacterial growth was strongly reduced, i.e., the optical density of GBJ002/GIL16 was much less than the optical densities of GBJ002 and GBJ002/GIL01. Similar to the optical density of GBJ002 treated with mitomycin C, lysogens' optical densities started to increase during phase IV, but GBJ002/GIL01 proliferated in a more pronounced way than GBJ002/GIL16. When lysogenic cultures were induced by mitomycin C after the lag phase, the OD595 decreased below the limit of detection after 2 h of treatment (Fig. 2). No increment in the lysogens' optical densities was observed at any of the remaining phases, indicating bacterial death. These results indicate that a high bacterial mortality rate occurs when tectiviruses are induced by DNA-damaging treatments like mitomycin C during an early bacterial growth phase of the bacterial lysogens.
FIG 2.
Comparison of growth kinetics of B. thuringiensis serovar israelensis strain GBJ002 and derivative lysogenic strains GBJ002/GIL01 and GBJ002/GIL16. Bacterial growth was measured by determination of the OD595 for 27 h at 30°C. Black arrow, the time when mitomycin C (MitC; final concentration, 1 μg/ml) was added to the bacterial cultures. Gray dashed lines separate bacterial growth phases I to IV. Each datum point represents the mean value from 10 replicate wells. The error bars indicate the standard deviation values.
To evaluate the stability of lysogeny, serial 10-fold dilutions of bacterial cultures were plated into LB-agar medium at 27 h. After overnight incubation, 20 colonies were randomly selected from different plates, regrown in fresh LB-agar plates supplemented with Nal, and screened by PCR. GBJ002/GIL01 and GBJ002/GIL16 lysogenic colonies were 100% positive for lysogeny. No bacterial colonies grew from lysogenic cultures treated with mitomycin C, confirming bacterial death.
Phage particle production and plaque morphology depend on the bacterial growth phase.
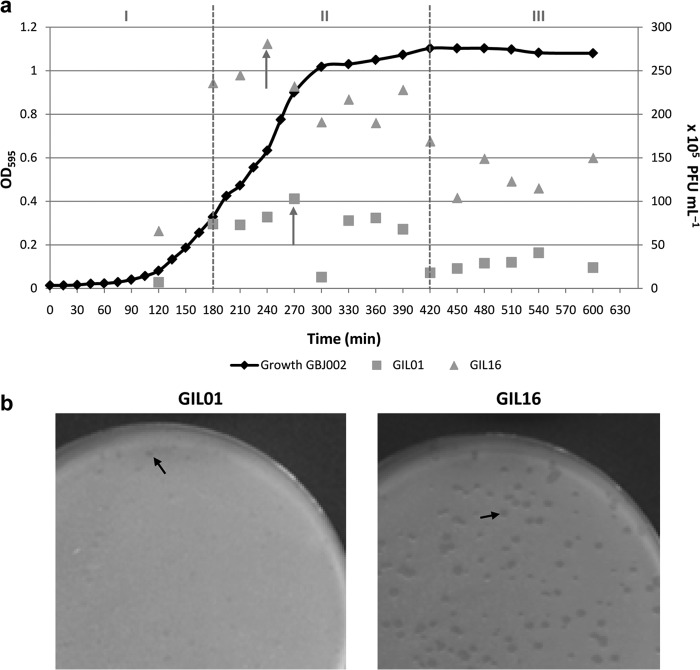
In order to compare the lytic actions of phages GIL01 and GIL16, nonlysogenic strain GBJ002 was infected with either GIL01 or GIL16 at different phases of bacterial growth. For this purpose, the growth kinetics for strain GBJ002 was followed by measurement of the OD595 for up to 10 h, and then bacterial growth was allowed to reach 24 h. Several samples were taken during this interval of time, infected separately with approximately equal titers of GIL01 or GIL16 phage stocks (GIL01, 1.3 × 107 PFU/ml; GIL16, 1 × 107 PFU/ml), and plated. After overnight incubation, the numbers of PFU were analyzed (Fig. 3) at the different growth phases of strain GBJ002 described above.
FIG 3.
Tectivirus plaque production compared against the host growth curve. (a) The growth of B. thuringiensis serovar israelensis strain GBJ002 (black line) was measured by determination of the OD595 (left y axis) for 10 h at 30°C. Gray dashed lines, equivalent time frames for bacterial growth phases I to III indicated in Fig. 2. At each time point on the GBJ002 growth curve, the host strain was infected by either GIL01 (squares) or GIL16 (triangles) purified phage stocks. Squares and triangles, the number of PFU/ml (right y axis) obtained at each time point for the respective phage; gray arrows, the highest titers obtained for each phage. (b) Plaque morphology after overnight incubation for phages GIL01 and GIL16 on lawns of host strain GBJ002, when it was infected at 4.5 h of growth. Black arrows, very turbid plaques produced by GIL01 and turbid large plaques produced by GIL16.
When GBJ002 was infected at the end of the lag phase (phase I), the PFU titers were low and plaques were very turbid, regardless of the phage tested. At mid-phase II of GBJ002 growth, there was a notable difference between the number of PFU (Fig. 3a) and the plaque morphology (Fig. 3b) produced by the phages tested. Phage GIL16 made turbid plaques larger (1.5 to 2 mm) than those made by phage GIL01 (<1 mm; very turbid and almost imperceptible). The PFU counts for GIL16 were always higher than those for GIL01. Also, during GBJ002 growth phase II, the highest phage titers and larger plaque sizes were obtained for both phages, but GIL16 produced more PFU with a larger plaque morphology than GIL01. The highest titer of phage GIL16 (2.8 × 107 PFU/ml) was obtained when GBJ002 was infected at 4 h of growth, while the highest titer of GIL01 (1 × 107 PFU/ml) was obtained when the host was infected at 4.5 h of growth. The PFU counts decreased when GBJ002 was infected at phase III of growth (phage titers obtained at 10 h of host growth, 2.4 × 106 PFU/ml for GIL01 and 1.5 × 107 PFU/ml for GIL16). Altogether, these results indicate that the spontaneous induction of the GIL01 and GIL16 lytic cycle largely depends on the host growth phase, and even though GIL16 is a temperate phage, it seems to be more virulent for strain GBJ002 than phage GIL01.
Tectiviral lysogeny does not affect host metabolic profile and antibiotic susceptibility.
To compare the possible impact of lysogeny on bacterial metabolism, 70 biochemical tests were used to evaluate the performance of carbohydrate metabolism and other biochemical characteristics of strain GBJ002 and lysogens GBJ002/GIL01 and GBJ002/GIL16. As shown in Table 1, all three strains displayed the same metabolic profiles, with one minor exception: the metabolism of glycogen seemed to be faster in the two lysogenic strains than in strain GBJ002. Also, the antibiotic susceptibility of the studied strains was evaluated using 17 antibiotics. The inhibition zones obtained for each antibiotic did not result in significant differences between the three strains tested (Table 2).
TABLE 1.
Metabolic profiles and biochemical characteristics of B. thuringiensis strain GBJ002 and derivative lysogens GBJ002/GIL01 and GBJ002/GIL16 determined using API 50 CH and API 20 E systemsa
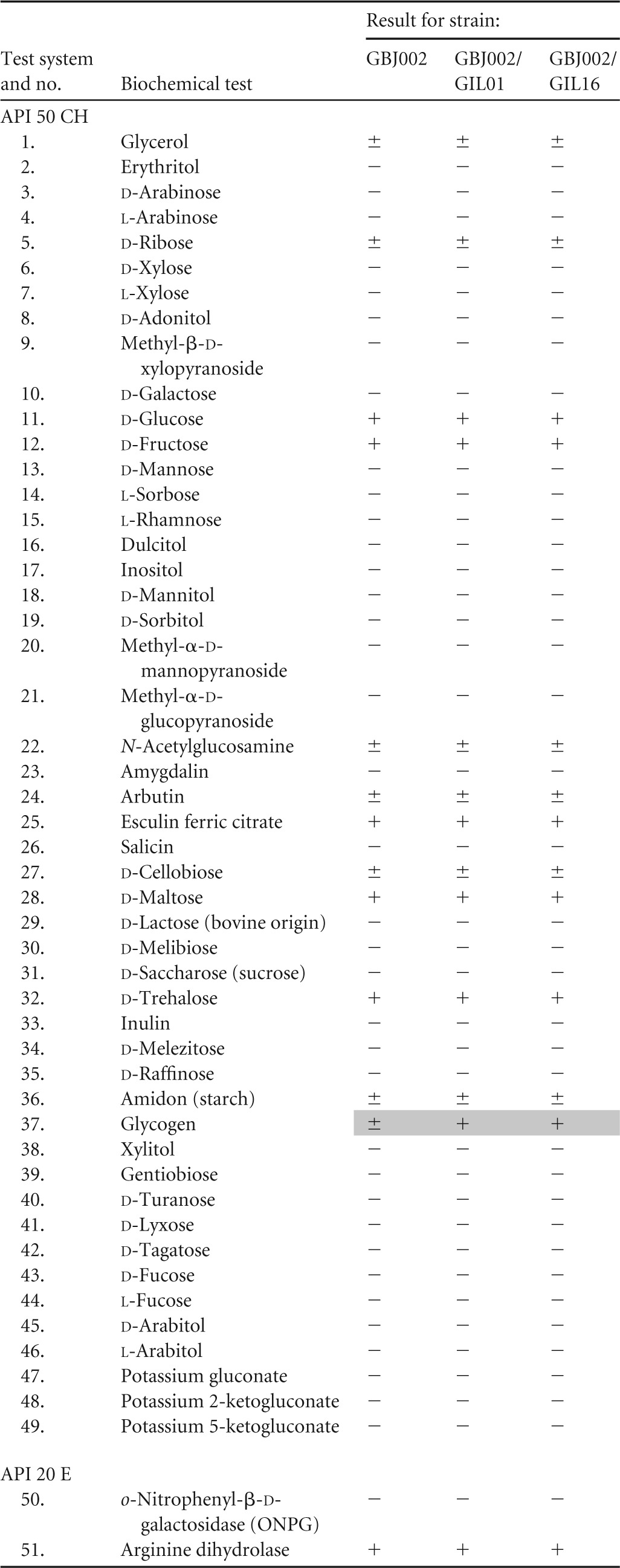
API 50 CH and API 20 E strip systems (bioMérieux) were used. Key to symbols: +, positive test result; −, negative test result; ±, slow activity (after 48 h). Differences between the reference strain and its two isogenic lysogenic derivatives are highlighted in gray. Tests 62 to 70 are for fermentation of the indicated sugar.
TABLE 2.
Antibiograms of B. thuringiensis strain GBJ002 and derivative lysogens GBJ002/GIL01 and GBJ002/GIL16a
| Antibiotic family | Antibiotic | Disk concnb | Inhibition halo diam (mm) |
||
|---|---|---|---|---|---|
| GBJ002 | GBJ002/GIL01 | GBJ002/GIL16 | |||
| Penicillins | Oxacillin | 1 | 6 | 6 | 6 |
| Ampicillin | 33 | 14 | 14 | 14 | |
| Penicillin | 5 | 12 | 12 | 12 | |
| Aminoglycosides | Spectinomycin | 200 | 6 | 6 | 6 |
| Amikacin | 40 | 10 | 9 | 10 | |
| Streptomycin | 100 | 11 | 11 | 11 | |
| Kanamycin | 100 | 9 | 9 | 9 | |
| Neomycin | 120 | 9 | 10 | 9 | |
| Gentamicin | 40 | 9 | 8 | 9 | |
| Ansamycin | Rifampin | 30 | 5 | 4 | 4 |
| Glycopeptide | Vancomycin | 5 | 4 | 4 | 5 |
| Macrolide | Erythromycin | 78 | 10 | 10 | 11 |
| Bacitracin | Bacitracin | 40 | 3 | 3 | 3 |
| Tetracycline | Tetracycline | 30 | 6 | 6 | 5 |
| Quinolone | Nalidixic acid | 30 | − | − | − |
| Chloramphenicol | 30 | 7 | 7 | 7 | |
| Fosfomycin | 200 | 5 | 5 | 5 | |
Evaluated by use of Neo-Sensitab antibiotic disks (Rosco Diagnostica). −, negative (the strains are resistant to nalidixic acid).
Concentrations are in micrograms for all antibiotics except bacitracin, for which the concentration is in units.
Tectiviral lysogeny has a negative impact on biofilm formation.
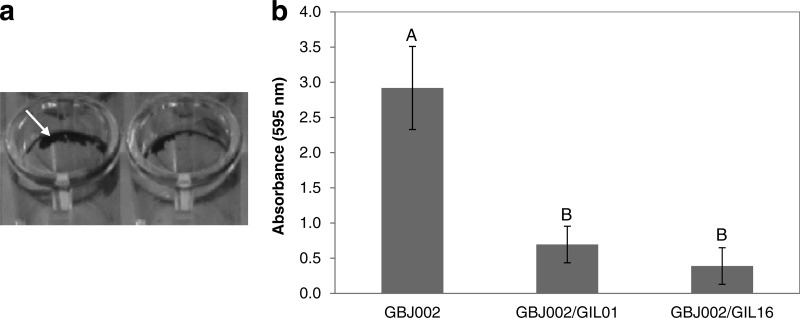
In order to evaluate the impact of lysogeny on biofilm development, strain GBJ002 and lysogens GBJ002/GIL01 and GBJ002/GIL16 were tested for their ability to adhere to an abiotic surface (polystyrene microtiter plates) after incubation for 48 h under static conditions. Biofilms were quantified by crystal violet staining. The biofilms produced by the strains were of the type of a biofilm ring at the side of the well surface at the liquid-air interface (Fig. 4a). On the basis of the OD595 of the solubilized dye, strong differences were found between the nonlysogenic and lysogenic strains (Fig. 4b). Statistical analysis indicated that nonlysogenic strain GBJ002 produced a significantly (Tukey's test, P < 0.05) larger mass of biofilm than the two lysogenic strains. Comparison of the biofilm formation reduction between lysogenic strains GBJ002/GIL01 and GBJ002/GIL16 showed that it was not statistically significant (Tukey's test, P > 0.05). Consequently, tectiviral lysogeny negatively affects the biofilm formation ability in B. thuringiensis, regardless of the tectiviruses tested.
FIG 4.
Biofilm formation of B. thuringiensis serovar israelensis strain GBJ002 and derivative lysogenic strains GBJ002/GIL01 and GBJ002/GIL16. (a) Biofilm formation at the air-liquid interface by nonlysogenic strain GBJ002. Arrow, the pellicle ring stained by crystal violet. (b) Biofilm quantification determined by the crystal violet staining method using measurement of the absorbance at 595 nm. Each plotted datum represents the average of five replicate wells. The error bars indicate the standard deviation values. Means with different letters (A, B) indicate significant differences (Tukey's test, P < 0.05).
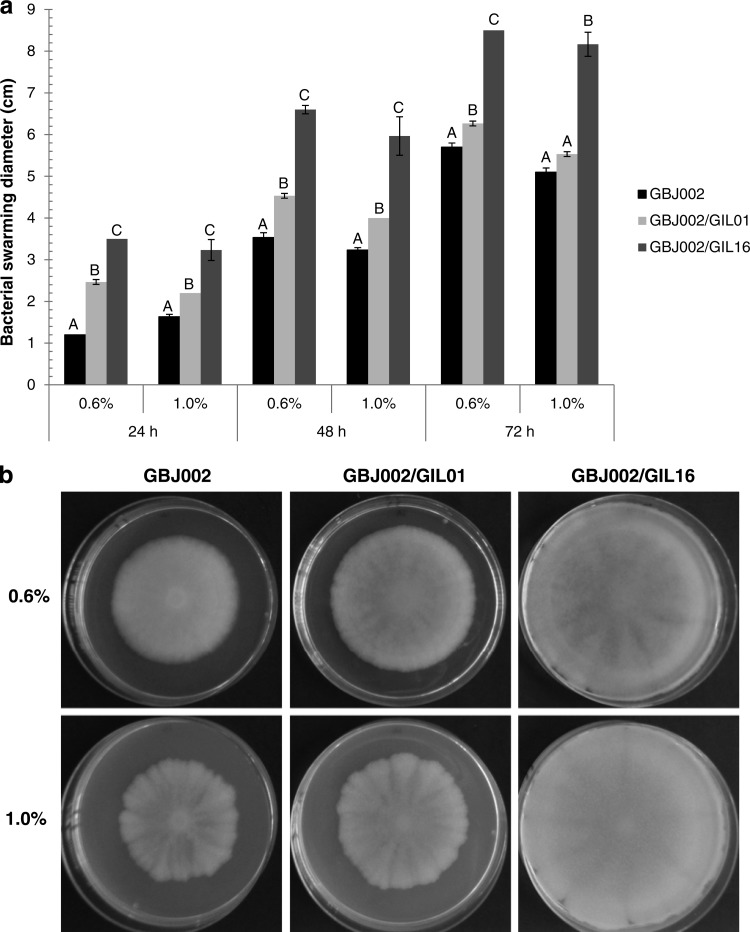
Tectiviral lysogeny enhances swarming motility.
B. thuringiensis swarming motility and bacterial phenotypes associated with this type of motility (e.g., cell shape and flagellum expression) are tightly controlled by the softness of the medium (25). Therefore, to determine if B. thuringiensis swarming motility is affected by lysogenic tectiviruses, two agar concentrations (0.6 and 1.0% [wt/vol]) were tested. The average swarming diameters for strain GBJ002 and lysogens GBJ002/GIL01 and GBJ002/GIL16 are shown in Fig. 5a. In general, the swarming diameters of the lysogens were larger than those of nonlysogenic strain GBJ002. With 0.6% agar at 24 h of incubation, GBJ002/GIL01 and GBJ002/GIL16 displayed swarming diameters that were double and nearly triple, respectively, the swarming diameter of strain GBJ002. The same effect was observed with 1.0% agar at 24 h, but the swarming diameters were smaller due to the agar concentration, yet the differences from the swarming diameter of strain GBJ002 were statistically significant (Tukey's test, P < 0.05). At 48 h, the same tendency was observed for both agar concentrations: statistically significantly larger swarming diameters of the two lysogens compared to the swarming diameter of strain GBJ002. The swarming diameters of GBJ002/GIL16 were statistically significantly larger than those of GBJ002/GIL01 with both agar concentrations at 24 and 48 h of incubation. At 72 h, the swarming motility differences observed between strain GBJ002 and lysogen GBJ002/GIL01 were less pronounced, yet they were significantly larger for the lysogen growing in 0.6% agar. Remarkably, the swarming diameters of GBJ002/GIL16 were still significant larger than those of GBJ002 and GBJ002/GIL01 with both agar concentrations at 72 h of incubation (Fig. 5b). GBJ002/GIL16 was able to colonize the entire available surface regardless of the agar concentration used. These results indicate that tectiviral lysogeny enhances swarming motility in B. thuringiensis, strongly depending on the tectivirus tested.
FIG 5.
Swarming motility assays performed in LB medium solidified with 0.6 and 1.0% agar. Swarming motility plates were inoculated with B. thuringiensis serovar israelensis strain GBJ002 and derivative lysogenic strains GBJ002/GIL01 and GBJ002/GIL16. (a) Mean values for swarming motility diameters in 0.6 and 1.0% agar at 24, 48, and 72 h. Error bars indicate standard deviation values from 9 measurements from 3 independent experiments. Means with different letters (A, B, C) indicate significant differences among the data for each experimental setup (Tukey's test, P < 0.05). (b) Bacterial swarming motility at 72 h of incubation. Similar observations were made in 3 independent experiments.
Tectiviral lysogeny decreases sporulation rates.
Since bacterial growth was affected by tectiviral lysogeny, the sporulation capacity of the strains was also evaluated by heat treatment. The sporulation efficiency of nonlysogenic strain GBJ002 was ∼50%. The sporulation efficiency of lysogens GBJ002/GIL01 and GBJ002/GIL16 was ∼28 and ∼15%, respectively. This decrease in lysogen sporulation rates compared to the rate for strain GBJ002 represents an approximately 2-fold reduction for GBJ002/GIL01 and an almost 3-fold reduction for GBJ002/GIL16. The fact that phage GIL16 lysogeny has a greater impact than phage GIL01 lysogeny on decreasing the GBJ002 population might be related to the reduction in sporulation capacity detected.
DISCUSSION
Tectiviruses infecting the B. cereus group are, so far, the only known prophages that can reside as autonomously replicating linear plasmids undergoing a lysogenic state in members of this bacterial group (16). Recently, it was shown that two of the most important bioinsecticide serovars of B. thuringiensis used worldwide (i.e., B. thuringiensis serovar kurstaki and B. thuringiensis serovar israelensis) are infected by tectiviruses (18). Phage GIL01 was found in all strains of B. thuringiensis serovar israelensis, whereas GIL16-like phages were found to infect strains belonging to B. thuringiensis serovar kurstaki (18). However, little is known about the interactions taking place between tectiviruses and their respective Gram-positive hosts. Therefore, in this study, different life traits of nonlysogenic B. thuringiensis serovar israelensis plasmid-cured strain GBJ002 and two isogenic derivative tectiviral lysogens infected by either phage GIL01 or phage GIL16 were compared, revealing evidence for a potential contribution of tectiviral lysogeny in the development of different phenotypes in B. thuringiensis.
It has been established that during lysogeny, phages are in a dormant state and their genomes are copied when their host genomes are copied during replication (21). However, some spontaneous prophage induction can still occur in a small percentage of the host population (28, 29). This spontaneous induction is likely responsible for the decrease in the bacterial growth rate observed in lysogens GBJ002/GIL01 and GBJ002/GIL16 compared to that observed in the nonlysogenic strain GBJ002, yet when the tectivirus infection became stable, the dormant phages were passed to the daughter cells in a very efficient manner, since 100% of the colonies recovered had positive PCR signals for tectivirus. Also, these lysogens presented somewhat changed colony morphologies, being more evident in the lysogens infected by phage GIL16. Also, when phages were induced from lysogens GBJ002/GIL01 and GBJ002/GIL16 growing in liquid medium by DNA-damaging (i.e., mitomycin C) treatment, there was a dramatic reduction in the optical densities of the bacterial cultures (below the limit of detection; Fig. 2), indicating that bacterial lysis had occurred in the majority of the cells. Therefore, the induction of tectiviruses in natural environments by specific signals other than mitomycin C (e.g., UV induction) may have important ecological consequences, since they can cause mortality in the majority of B. thuringiensis cells.
Moreover, in order to compare the infection yield of phages GIL01 and GIL16, strain GBJ002 was infected separately with equal titers of purified stocks of phage GIL01 or GIL16 at different phases of the bacterial growth curve. It was found that the highest PFU titers for both phages were obtained when host strain GBJ002 was infected during its exponential phase (phase II; Fig. 3). Also, the plaque morphologies were larger during this growth phase. These results indicate that at phase II the host state favors the induction of the lytic cycle of the phage. After 24 h of growth, the PFU titers dropped and plaques were almost imperceptible in the bacterial host lawns (data not shown), yet lysogeny was established for both phages. As other evolutionary and ecological studies have shown, lysogeny becomes the preferred strategy for phages, particularly when the cell density or nutrient cell resources fall below the lower limit necessary for maintenance of the phage lytic cycle (29, 30).
Analyses of metabolic profiles did not reveal any significant differences between the nonlysogenic strain and the two lysogens tested. Nonetheless, there was a slight difference in the glycogen metabolism of the nonlysogenic strain GBJ002 (in which it was slower than that in the two lysogens). Therefore, further studies that involve the expression of genes coding for glycogen-associated enzymes are necessary to evaluate the genuine contribution of tectiviral lysogeny in the metabolism of this large polymer of glucose. Likewise, no noteworthy differences in the antibiograms displayed by the nonlysogenic strain and the two lysogens were found. Interestingly, the B. cereus ATCC 14579 strain cured of plasmid pBClin15 was more tolerant to antibiotics interfering with DNA integrity (i.e., Nal, norfloxacin, and ciprofloxacin) than the wild-type strain (31). Since the nonlysogenic strain GBJ002 is Nalr, the influence of tectiviral lysogeny on the quinolone response might be masked by the chromosome-encoded antibiotic resistance.
The present study revealed that tectiviral lysogeny can reduce biofilm formation, promote swarming motility, and decrease the sporulation of B. thuringiensis serovar israelensis, all three life traits involved in multicellular bacterial behaviors. In B. subtilis these life traits are associated with the bacterium's ability to differentiate into distinct coexisting cell types (i.e., competent, cannibal, motile, matrix-producing, surfactin-producing, and sporulating cells) (32, 33). Moreover, while studying swarm cell nanoscale characteristics in B. thuringiensis serovar israelensis, it was demonstrated that this bacterium displays different cell types after growing in semisolid medium (25), sustaining previous observations that B. thuringiensis flagellum expression is a prerequisite for biofilm formation in early stages of bacterial development (25, 34, 35). The interesting result that swarming motility is enhanced in the lysogenic strains GBJ002/GIL01 and GBJ002/GIL16 (Fig. 5) suggests that, indeed, tectiviral lysogeny can bestow other ecological advantages, rather than simply conferring immunity to the same type of phage. Swarming is a collective bacterial phenomenon that allows bacteria to migrate rapidly on top of surfaces, allowing them to colonize nutrient-rich environments and facilitating colony spread (36). The fact that phage GIL16 provokes a swarming phenotype in the lysogen significantly larger than that provoked by GIL01 might be related to the more virulent nature of the phage when it infects B. thuringiensis serovar israelensis. The effect of a significantly larger swarming phenotype in the lysogens might also explain the differences in colony morphologies observed. The negative impact on pellicle (biofilm) formation by the two lysogenic strains can be explained by two possibilities. The first is correlated to the induced swarming motility observed in the two lysogens. Cell motility often promotes initial biofilm formation, but ultimately, motility is inhibited during the transition to sessile cell aggregates (37). Therefore, since motility was induced in the two lysogens, it can be expected that surface adhesion and, consequently, biofilm formation would not be stimulated. The second possibility is related to the spontaneous phage induction observed in liquid cultures. Bacterial lysis decreases the bacterial densities and, thus, the number of adhering cells necessary during the first stages of biofilm formation. Also, the low growth rate displayed by the lysogens might be translated into slower biofilm formation. In the same vein, the decrease in bacterial density due to phage induction in the lysogens might result in a downregulation of the sporulation process.
Taken together, the fact that tectiviral lysogeny can affect life traits such as motility and biofilm formation in B. thuringiensis serovar israelensis indicated that tectiviruses may play a role, directly or indirectly, in these complicated and highly regulated behaviors.
Concluding remarks.
Here, the impact of lysogeny by phages GIL01 and GIL16 on diverse life traits of B. thuringiensis serovar israelensis was studied. The results demonstrated that, at least under laboratory conditions, tectiviral lysogeny enhanced swarming motility and decreased biofilm and sporulation capacities, all of which are cell behaviors related to the collective phenotypes and survival traits of B. thuringiensis. Therefore, these findings indicate that lysogeny should be considered in the already complicated and poorly understood life cycle of B. thuringiensis in nature. It is important to note that the experimental setup of this work used full rich medium (LB medium), which might not reflect the real environmental conditions of the different habitats of B. thuringiensis. Also, B. thuringiensis strain GBJ002 is a plasmid-cured strain that does not have the genetic determinants (cry genes) coding for the insecticidal Cry toxins. Therefore, in the environment this strain might behave as a pure saprophyte and not as an insect pathogen. In future experiments, it would be interesting to test the effect of tectiviral lysogeny using B. thuringiensis strains that do harbor the cry genes to analyze the contribution of this type of lysogeny to the pathogenicity of the bacterium in insects. Additional studies that focus on comparing bacterial gene expression in nonlysogenic and tectiviral lysogenic strains, along with analyses of tectivirus genome-encoded factors, will expand the view of how tectiviruses can contribute to the ecology and lifestyles of B. thuringiensis.
ACKNOWLEDGMENTS
We gratefully acknowledge Gustavo Romay for his statistical advice.
This work was supported by the Foundation for Training in Industrial and Agricultural Research (FRIA; a grant to A.G.) and by the National Fund for Scientific Research (FNRS), the Université Catholique de Louvain, and the Research Department of the Communauté Française de Belgique (Concerted Research Action) (grants to J.M.).
Footnotes
Published ahead of print 26 September 2014
REFERENCES
- 1.Aronson A. 2002. Sporulation and delta-endotoxin synthesis by Bacillus thuringiensis. Cell. Mol. Life Sci. 59:417–425. 10.1007/s00018-002-8434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jensen GB, Hansen BM, Eilenberg J, Mahillon J. 2003. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 5:631–640. 10.1046/j.1462-2920.2003.00461.x. [DOI] [PubMed] [Google Scholar]
- 3.Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Argôlo-Filho R, Loguercio L. 2013. Bacillus thuringiensis is an environmental pathogen and host-specificity has developed as an adaptation to human-generated ecological niches. Insects 5:62–91. 10.3390/insects5010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raymond B, Johnston PR, Nielsen-LeRoux C, Lereclus D, Crickmore N. 2010. Bacillus thuringiensis: an impotent pathogen? Trends Microbiol. 18:189–194. 10.1016/j.tim.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Reyes-Ramirez A, Ibarra JE. 2008. Plasmid patterns of Bacillus thuringiensis type strains. Appl. Environ. Microbiol. 74:125–129. 10.1128/AEM.02133-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aronson AI. 1993. The two faces of Bacillus thuringiensis: insecticidal proteins and post-exponential survival. Mol. Microbiol. 7:489–496. 10.1111/j.1365-2958.1993.tb01139.x. [DOI] [PubMed] [Google Scholar]
- 8.Andrup L, Barfod KK, Jensen GB, Smidt L. 2008. Detection of large plasmids from the Bacillus cereus group. Plasmid 59:139–143. 10.1016/j.plasmid.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Madsen SM, Andrup L, Boe L. 1993. Fine mapping and DNA sequence of replication functions of Bacillus thuringiensis plasmid pTX14-3. Plasmid 30:119–130. 10.1006/plas.1993.1039. [DOI] [PubMed] [Google Scholar]
- 10.Andrup L, Bendixen HH, Jensen GB. 1995. Mobilization of Bacillus thuringiensis plasmid pTX14-3. Plasmid 33:159–167. 10.1006/plas.1995.1017. [DOI] [PubMed] [Google Scholar]
- 11.Andrup L, Jensen GB, Wilcks A, Smidt L, Hoflack L, Mahillon J. 2003. The patchwork nature of rolling-circle plasmids: comparison of six plasmids from two distinct Bacillus thuringiensis serotypes. Plasmid 49:205–232. 10.1016/S0147-619X(03)00015-5. [DOI] [PubMed] [Google Scholar]
- 12.González J, Jr, Carlton BC. 1984. A large transmissible plasmid is required for crystal toxin production in Bacillus thuringiensis variety israelensis. Plasmid 11:28–38. 10.1016/0147-619X(84)90004-0. [DOI] [PubMed] [Google Scholar]
- 13.Jensen G, Andrup L, Wilcks A, Smidt L, Poulsen O. 1996. The aggregation-mediated conjugation system of Bacillus thuringiensis subsp. israelensis: host range and kinetics of transfer. Curr. Microbiol. 33:228–236. 10.1007/s002849900105. [DOI] [PubMed] [Google Scholar]
- 14.Verheust C, Jensen G, Mahillon J. 2003. pGIL01, a linear tectiviral plasmid prophage originating from Bacillus thuringiensis serovar israelensis. Microbiology 149:2083–2092. 10.1099/mic.0.26307-0. [DOI] [PubMed] [Google Scholar]
- 15.Oksanen HM, Bamford DH. 2012. Family Tectiviridae, p 317–321 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy. Ninth report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA. [Google Scholar]
- 16.Gillis A, Mahillon J. 2014. Phages preying on Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: past, present and future. Viruses 6:2623–2672. 10.3390/v6072623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jalasvuori M, Palmu S, Gillis A, Kokko H, Mahillon J, Bamford JKH, Fornelos N. 2013. Identification of five novel tectiviruses in Bacillus strains: analysis of a highly variable region generating genetic diversity. Res. Microbiol. 164:118–126. 10.1016/j.resmic.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 18.Gillis A, Mahillon J. 2014. Prevalence, genetic diversity and host range of tectiviruses among members of the Bacillus cereus group. Appl. Environ. Microbiol. 80:4138–4152. 10.1128/AEM.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verheust C, Fornelos N, Mahillon J. 2005. GIL16, a new Gram-positive tectiviral phage related to the Bacillus thuringiensis GIL01 and the Bacillus cereus pBClin15 elements. J. Bacteriol. 187:1966–1973. 10.1128/JB.187.6.1966-1973.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strömsten NJ, Benson SD, Burnett RM, Bamford DH, Bamford JK. 2003. The Bacillus thuringiensis linear double-stranded DNA phage Bam35, which is highly similar to the Bacillus cereus linear plasmid pBClin15, has a prophage state. J. Bacteriol. 185:6985–6989. 10.1128/JB.185.23.6985-6989.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casjens S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49:277–300. 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- 22.Barksdale L, Arden SB. 1974. Persisting bacteriophage infections, lysogeny, and phage conversions. Annu. Rev. Microbiol. 28:265–299. 10.1146/annurev.mi.28.100174.001405. [DOI] [PubMed] [Google Scholar]
- 23.Brüssow H, Canchaya C, Hardt WD. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560–602. 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Toole GA. 2011. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011:2437. 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillis A, Dupres V, Mahillon J, Dufrene YF. 2012. Atomic force microscopy: a powerful tool for studying bacterial swarming motility. Micron 43:1304–1311. 10.1016/j.micron.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Santana MA, Moccia VCC, Gillis AE. 2008. Bacillus thuringiensis improved isolation methodology from soil samples. J. Microbiol. Methods 75:357–358. 10.1016/j.mimet.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Travers RS, Martin PA, Reichelderfer CF. 1987. Selective process for efficient isolation of soil Bacillus spp. Appl. Environ. Microbiol. 53:1263–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaidelytė A, Jaatinen ST, Daugelavicius R, Bamford JK, Bamford DH. 2005. The linear double-stranded DNA of phage Bam35 enters lysogenic host cells, but the late phage functions are suppressed. J. Bacteriol. 187:3521–3527. 10.1128/JB.187.10.3521-3527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chibani-Chennoufi S, Bruttin A, Dillmann M-L, Brüssow H. 2004. Phage-host interaction: an ecological perspective. J. Bacteriol. 186:3677–3686. 10.1128/JB.186.12.3677-3686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinbauer MG, Suttle CA. 1996. Potential significance of lysogeny to bacteriophage production and bacterial mortality in coastal waters of the Gulf of Mexico. Appl. Environ. Microbiol. 62:4374–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vörös A, Simm R, Kroeger JK, Kolstø AB. 2013. Gene transcription from the linear plasmid pBClin15 leads to cell lysis and extracellular DNA-dependent aggregation of Bacillus cereus ATCC 14579 in response to quinolone-induced stress. Microbiology 159:2283–2293. 10.1099/mic.0.069674-0. [DOI] [PubMed] [Google Scholar]
- 32.Lopez D, Vlamakis H, Kolter R. 2009. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol. Rev. 33:152–163. 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- 33.Lopez D, Kolter R. 2010. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol. Rev. 34:134–149. 10.1111/j.1574-6976.2009.00199.x. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi K. 2007. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol. Microbiol. 66:395–409. 10.1111/j.1365-2958.2007.05923.x. [DOI] [PubMed] [Google Scholar]
- 35.Houry A, Briandet R, Aymerich S, Gohar M. 2010. Involvement of motility and flagella in Bacillus cereus biofilm formation. Microbiology 156:1009–1018. 10.1099/mic.0.034827-0. [DOI] [PubMed] [Google Scholar]
- 36.Verstraeten N, Braeken K, Debkumari B, Fauvart M, Fransaer J, Vermant J, Michiels J. 2008. Living on a surface: swarming and biofilm formation. Trends Microbiol. 16:496–506. 10.1016/j.tim.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 37.Guttenplan SB, Blair KM, Kearns DB. 2010. The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet. 6:e1001243. 10.1371/journal.pgen.1001243. [DOI] [PMC free article] [PubMed] [Google Scholar]