Abstract
The betaproteobacteria “Aromatoleum aromaticum” pCyN1 and “Thauera” sp. strain pCyN2 anaerobically degrade the plant-derived aromatic hydrocarbon p-cymene (4-isopropyltoluene) under nitrate-reducing conditions. Metabolite analysis of p-cymene-adapted “A. aromaticum” pCyN1 cells demonstrated the specific formation of 4-isopropylbenzyl alcohol and 4-isopropylbenzaldehyde, whereas with “Thauera” sp. pCyN2, exclusively 4-isopropylbenzylsuccinate and tentatively identified (4-isopropylphenyl)itaconate were observed. 4-Isopropylbenzoate in contrast was detected with both strains. Proteogenomic investigation of p-cymene- versus succinate-adapted cells of the two strains revealed distinct protein profiles agreeing with the different metabolites formed from p-cymene. “A. aromaticum” pCyN1 specifically produced (i) a putative p-cymene dehydrogenase (CmdABC) expected to hydroxylate the benzylic methyl group of p-cymene, (ii) two dehydrogenases putatively oxidizing 4-isopropylbenzyl alcohol (Iod) and 4-isopropylbenzaldehyde (Iad), and (iii) the putative 4-isopropylbenzoate-coenzyme A (CoA) ligase (Ibl). The p-cymene-specific protein profile of “Thauera” sp. pCyN2, on the other hand, encompassed proteins homologous to subunits of toluene-activating benzylsuccinate synthase (termed [4-isopropylbenzyl]succinate synthase IbsABCDEF; identified subunits, IbsAE) and protein homologs of the benzylsuccinate β-oxidation (Bbs) pathway (termed BisABCDEFGH; all identified except for BisEF). This study reveals that two related denitrifying bacteria employ fundamentally different peripheral degradation routes for one and the same substrate, p-cymene, with the two pathways apparently converging at the level of 4-isopropylbenzoyl-CoA.
INTRODUCTION
Biological degradation of aromatic hydrocarbons is challenged by their extraordinary chemical stability arising from the exclusive presence of apolar C—C and C—H bonds and the resonance stabilization of the aromatic ring (1). Aerobic bacteria employ a variety of oxygen-dependent enzymes, i.e., oxygenases, for initial substrate activation (2). One such enzyme is the membrane-bound methyl-hydroxylating toluene/xylene monooxygenase (XylM) encoded on the TOL plasmid of Pseudomonas putida mt-2 (3). XylM is a non-heme di-iron monooxygenase, which forms (methyl)benzyl alcohol from (alkyl)toluene. Likewise, a bifunctional monooxygenase (CymAa, hydroxylase subunit; CymAb, reductase subunit) of P. putida F1 was previously demonstrated to convert p-cymene to 4-isopropylbenzyl alcohol (4). Thus, hydroxylation of benzylic methyl groups represents one important mode for the activation of aromatic hydrocarbons under oxic conditions. However, due to the strict O2 dependence of these enzymes, this reaction is not feasible for bacterial oxidation of such substrates under anoxic conditions.
The catabolic capacity of anaerobic bacteria utilizing aromatic hydrocarbons under strictly anoxic conditions (O2 independent) has been studied intensively over the last 2 decades, with particular emphasis on the diversity and physiology of new isolates (e.g., 5–7) and the elucidation of novel biochemical reactions (e.g., 2, 8–10). The addition of toluene to fumarate, yielding benzylsuccinate by an enzymatic radical reaction, was originally discovered in the related denitrifying betaproteobacteria Thauera aromatica K172T (11) and Azoarcus sp. strain T (12). This type of reaction proved to be archetypical among diverse facultatively and obligately anaerobic bacteria for the initial activation of the benzylic methyl group in toluene, all three xylene isomers (13–15), or 2-methylnaphthalene (16, 17), as well as the subterminal methylene group in n-alkanes (18, 19) and the benzylic methylene group in ethylbenzene (20). In the case of the latter, addition to fumarate is only known for energy-limited sulfate-reducing bacteria, while denitrifying “Aromatoleum aromaticum” strains EbN1 and EB1 activate ethylbenzene via hydroxylation to (S)-1-phenylethanol (21–23). Taken together, benzylic methyl groups in aromatic hydrocarbons are activated only via addition to fumarate in all described cases of anaerobic bacterial metabolism, while an O2-independent hydroxylation has not been demonstrated to date. In contrast, both reaction types have been reported for the phenolic compound p-cresol, the benzylic methyl group of which is known to be anaerobically functionalized via hydroxylation in denitrifying P. putida NCIB 9866 (24, 25) and in iron-reducing Geobacter metallireducens GS-15T (26) or via addition to fumarate in sulfate-reducing Desulfobacterium cetonicum 480 (27) and Desulfobacula toluolica Tol2T (28). p-Cresol differs markedly from the aromatic hydrocarbon p-cymene with respect to chemical reactivity and, hence, potential biochemical activation (see Discussion).
Following initial activation of (alkyl)toluenes, further degradation proceeds to the intermediate (alkyl)benzoyl-coenzyme A (CoA). In the case of (alkyl)toluene-derived (alkyl)benzylsuccinate(s), a modified β-oxidation yielding succinyl-CoA and (alkyl)benzoyl-CoA is employed, as originally described for toluene degradation in T. aromatica K172T (29, 30). In contrast, conversion of ethylbenzene-derived (S)-1-phenylethanol to benzoyl-CoA in “A. aromaticum” EbN1 involves further dehydrogenation to acetophenone, carboxylation, activation with coenzyme A, and thiolytic cleavage of the thioester yielding benzoyl-CoA and acetyl-CoA (23, 31, 32). Benzoyl-CoA is further metabolized to acetyl-CoA via a central degradation pathway involving reductive dearomatization and hydrolytic ring cleavage, followed by conventional β-oxidation (8).
Most of the currently known denitrifying bacteria (pure cultures) capable of anaerobic degradation of aromatic hydrocarbons belong to the “Aromatoleum”/Azoarcus/Thauera cluster within the betaproteobacterial Rhodocyclus group (33–36). The bacterial strains investigated in this study, “A. aromaticum” pCyN1 and “Thauera” sp. strain pCyN2, represent the first pure cultures reported to anaerobically oxidize a para-alkylated toluene, namely, the monoterpenoid aromatic hydrocarbon p-cymene (4-isopropyltoluene), completely to CO2 (35). In contrast to strain pCyN2, however, strain pCyN1 also anaerobically grows with 4-ethyltoluene and toluene (Table 1).
TABLE 1.
Anaerobic activation of selected (alkyl)toluenes in denitrifying betaproteobacteria
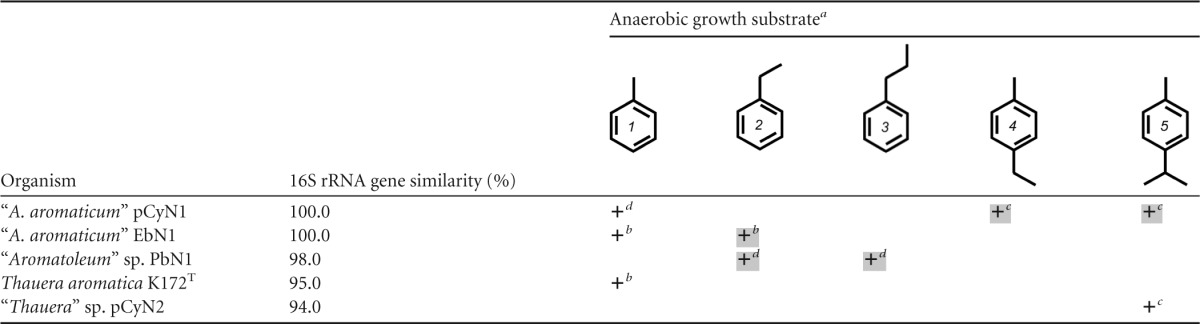
Structure numbers correspond to the following compounds: 1, toluene; 2, ethylbenzene; 3, n-propylbenzene; 4, 4-ethyltoluene; 5, p-cymene. Growth is indicated as follows: empty cell, no growth with the substrate; +, growth with the substrate and arylsuccinate formation (no box) or hydroxylation of a benzylic methyl/methylene group (shaded box).
Based on proteogenomic and metabolite evidence (35; this study).
In this study, the peripheral anaerobic degradation pathways for p-cymene in “A. aromaticum” pCyN1 and “Thauera” sp. pCyN2 were investigated.
MATERIALS AND METHODS
Strains, growth conditions, and cultivation experiments.
The denitrifying p-cymene-degrading bacterial strains “A. aromaticum” pCyN1 and “Thauera” sp. pCyN2 have been maintained in our laboratory since their isolation (35). Both strains were cultivated under nitrate-reducing conditions (7 mM nitrate) and an anoxic atmosphere of N2-CO2 (90:10, vol/vol) in ascorbate-reduced (4 mM) and bicarbonate-buffered defined mineral medium (34). (Alkyl)toluenes were supplied diluted (p-cymene, 5%; 4-ethyltoluene, 2%; toluene, 2%; all vol/vol) in an inert carrier phase of deaerated 2,2,4,4,6,8,8-heptamethylnonane (HMN). 4-Isopropylbenzoate (2 mM), succinate (5 mM), or 2,3-2H2-fumarate (10 mM) was added from sterile aqueous stock solutions. Growth was determined by measuring the optical density at 660 nm (OD660) in a spectrophotometer (UV-1202; Shimadzu, Kyoto, Japan). Prior to cultivation for DNA sequencing or proteomic or metabolite analyses, “A. aromaticum” pCyN1 and “Thauera” sp. pCyN2 were revived from cryopreserved stock cultures (dating back to the year 2000) as previously described (37) and adapted to anaerobic growth with the respective substrate for at least five passages (i.e., five consecutive transfers into fresh medium).
Sufficient cell material for proteomic studies was generated as described previously (38). Six parallel cultures (400 ml in 500-ml flat glass bottles) of anaerobic p-cymene- or succinate-grown cells were harvested during active growth at the half-maximal OD660s of 0.28 (p-cymene) and 0.33 (succinate) for “A. aromaticum” pCyN1 and at 0.27 (p-cymene) and 0.37 (succinate) for “Thauera” sp. pCyN2.
For metabolite analyses, cultures of anaerobic p-cymene-, 4-isopropylbenzoate-, or succinate-adapted cells of “A. aromaticum” pCyN1 and “Thauera” sp. pCyN2 were grown in duplicates (400 ml in 500-ml Schott flasks) and harvested upon transition into the stationary growth phase due to nitrate/nitrite limitation. In addition, such cultivations were also carried out with 4-ethyltoluene, toluene, or 2,3-2H2-fumarate in the case of “A. aromaticum” pCyN1. The cultures were inactivated by incubation at 85°C in a water bath, chilled on ice, acidified to pH 1.5, and extracted with diethyl ether three times as previously described (18).
Metabolite analyses.
Diethyl ether extracts (see above) were treated with anhydrous Na2SO4 to remove carry-over of water and then methylated prior to gas chromatography-mass spectrometry (GC-MS) analysis using a solution of diazomethane in diethyl ether. To avoid interferences from coeluting compounds, the extract obtained upon growth of “A. aromaticum” pCyN1 with 4-ethyltoluene was subsequently derivatized with N-methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA). GC-MS measurements were performed using a Trace GC-MS (Thermoelectron, Dreieich, Germany). The GC was equipped with a temperature-programmable injection system and a BPX5-fused silica capillary column (length, 50 m; inner diameter, 0.22 mm; film thickness, 0.25 μm [SGE]). Helium was used as the carrier gas at a constant flow rate of 1 ml min−1. The GC oven temperature was programmed from 50°C (1 min isothermal) to 310°C (30 min isothermal) increasing at a rate of 3°C min−1. The MS was operated in electron ionization mode at an ion source temperature of 230°C. Full-scan mass spectra were recorded over the mass range of 50 to 600 Da at a rate of 2.5 scans s−1. Identification of metabolites was based on comparison of GC retention times and mass spectra with those of authentic standards or from interpretation of mass spectra. For stereochemical assignment, arylsuccinates were derivatized with (R)-1-phenylethanamine and analyzed as recently described by Jarling et al. (39).
4-Ethylbenzyl alcohol, 4-ethylbenzaldehyde, 4-ethylbenzoic acid, 4-isopropylbenzyl alcohol, 4-isopropylbenzaldehyde, 4-isopropylbenzoic acid, N-methyl-N-nitroso-p-toluenesulfonamide (Diazald), and sodium sulfate were purchased from Sigma-Aldrich, Germany. (4-Isopropylbenzyl)succinate was synthesized from 4-isopropylbenzylbromide via 1-(iodomethyl)-4-isopropylbenzene and dimethyl 2-(4-isopropylbenzyl)succinate, and its structure and purity were determined by nuclear magnetic resonance (NMR) and high-resolution mass spectrometry (HRMS) analyses as outlined in the supplemental material.
Proteomic analyses.
Soluble proteins for two-dimensional difference gel electrophoresis (2D DIGE) were prepared as described previously (40), and the protein content was determined according to the method developed by Bradford (41). Isoelectric focusing was performed using 24-cm-long immobilized pH gradient (IPG) strips with a nonlinear (NL) pH range from 3 to 11 (GE Healthcare, Munich, Germany) in an IPGphor system (GE Healthcare). Separation according to molecular mass in 12.5% polyacrylamide gels was conducted in an Ettan Dalt II system (GE Healthcare). CyDye DIGE Fluor dyes (200 pmol) were applied for preelectrophoretic labeling of 50 μg of protein sample. Protein extracts from p-cymene-grown cells served as test states and were labeled with Cy3. Protein extracts of succinate-grown cells served as the reference state and were labeled with Cy5. The internal standard comprised equal amounts of extracts of p-cymene- and succinate-grown cells, which were labeled with Cy2. Each individual gel was loaded with equal amounts of the reference state, the test state, and the internal standard. Four gels were prepared per strain, with each gel comprising the protein extract of one biological replicate per substrate. Digital gel images were directly acquired after electrophoresis using a Typhoon 9400 scanner (GE Healthcare) and analyzed with DeCyder software (version 7.0; GE Healthcare). Abundance changes were considered to be significant, when a fold change of ≥1.5 (42) was detected (t test value of <10−3) and when the protein spot was matched in at least three of the four gels.
Separate colloidal Coomassie brilliant blue (cCBB)-stained gels with 300 μg of protein loaded per gel were prepared for protein identification by mass spectrometry. Protein spots specifically formed in p-cymene-adapted cells were excised with an EXQuest spot cutter (Bio-Rad, Munich, Germany), washed, and digested as described previously (43). Tryptic peptides were spotted onto Anchorchip steel targets (Bruker Daltonik GmbH, Bremen, Germany) and analyzed with an UltrafleXtreme matrix-assisted laser desorption ionization–two-stage time of flight (MALDI-TOF/TOF) mass spectrometer (Bruker Daltonik GmbH) (43).
Membrane proteins from two biological replicates per strain and substrate were prepared as described recently and separated in a Mini-Protean Tetra System (Bio-Rad) according to molecular mass in small-sized SDS-PAGE gels (43). Each sample lane was cut in four slices, and each slice was cut into ∼1-mm3 pieces, which were washed, reduced, alkylated, and tryptically digested (43). Generated peptides were separated with an UltiMate 3000 RSLCnano system (Thermo Fisher Scientific, Germering, Germany) in a trap column setup equipped with an analytical column (C18; pore size, 100 Å; bead size, 2 μm; inner diameter, 75 μm; length, 25 cm [Thermo Fisher Scientific]) applying a linear gradient as described previously (44). Continuous analysis of the eluent was performed with an online-coupled ion trap mass spectrometer (amaZon ETD; Bruker Daltonik GmbH) operated as described previously (43).
Protein identification was performed with an in-house Mascot server (version 2.3; Matrix Science, London, United Kingdom) via the ProteinScape platform (version 3.1; Bruker Daltonik GmbH). Acquired mass spectra were searched against the translated genomes of “A. aromaticum” pCyN1 and “Thauera” sp. pCyN2, applying a target-decoy strategy. Identified peptides of all samples per strain and growth condition were compiled, and proteins were accepted only when identified by two or more peptides.
Calculation of standard Gibbs free energy change.
The free energy change under standard conditions (ΔG0′) for the complete oxidation of p-cymene coupled to denitrification (for the stoichiometric equation, see reference 35) was calculated from published free energies of formation from the elements () (45) or, in the case of p-cymene, using a calculated estimation of (150.3 kJ mol−1) (see Table 1 in reference 46).
Assembly of gene clusters, gene function analysis, phylogenetic analysis, and nucleotide sequence accession numbers.
Construction of shotgun libraries, DNA sequencing, and protein sequence-directed assembly of gene clusters was essentially performed as described previously (23). Open reading frame (ORF) finding and automatic functional assignment involved the HTGA (High-Throughput Genome Annotation) system (23) and RAST (Rapid Annotation using Subsystem Technology) (47). ARTEMIS (48) was used to manually refine the start position of predicted ORFs. Refinement of functional predictions of identified proteins was performed using the BLAST function of the UniProt knowledgebase (UniProtKB [http://www.uniprot.org/]) and InterPro (49) (see Table S1 in the supplemental material).
The phylogenetic analysis of the catalytically active α-subunits of the putative p-cymene dehydrogenase (CmdA; “A. aromaticum” pCyN1) and (4-isopropylbenzyl)succinate synthase (IbsA; “Thauera” sp. pCyN2) was performed in two consecutive steps using MegAlign of the Lasergene software package (version 8.0.2; DNASTAR, Madison, WI, USA). First, selected protein sequences were aligned with Clustal W (50). Second, based on the Clustal W-generated alignments, MegAlign constructs rooted trees using 1,000 bootstrap iterations. In doing so, a new artificial node is introduced on which bootstrapping cannot be performed. This is indicated by NA (not applicable) in the trees.
Nucleotide sequence accession numbers.
Nucleotide sequences of relevant proteins (Table 2) were submitted to the GenBank under accession numbers KM105874 to KM105880 (“A. aromaticum” pCyN1) and KM105881 to KM105894 (“Thauera” sp. pCyN2) (see Table S1 in the supplemental material).
TABLE 2.
Identified proteins involved in the anaerobic degradation of p-cymene to 4-isopropylbenzoyl-CoA in “A. aromaticum” pCyN1 and “Thauera” sp. pCyN2

Proteins prepared from membrane protein-enriched fractions of anaerobically grown p-cymene-adapted cells.
See Table S1 in the supplemental material for annotation and accession numbers of the proteins.
Fold change in protein abundance compared to anaerobically grown succinate-adapted cells of strain pCyN1 or pCyN2.
Identified from multiple spots. For details, see Fig. S2 in the supplemental material.
Comigrating protein in the same spot. For details, see Fig. S2 in the supplemental material.
RESULTS
To elucidate the peripheral degradation pathway(s) for p-cymene to 4-isopropylbenzoyl-CoA in the studied organisms “A. aromaticum” pCyN1 and “Thauera” sp. pCyN2, substrate-adapted cells of both strains were analyzed for specifically formed metabolites and proteins. The degradation pathways reconstructed on the basis of these findings are depicted in Fig. 1.
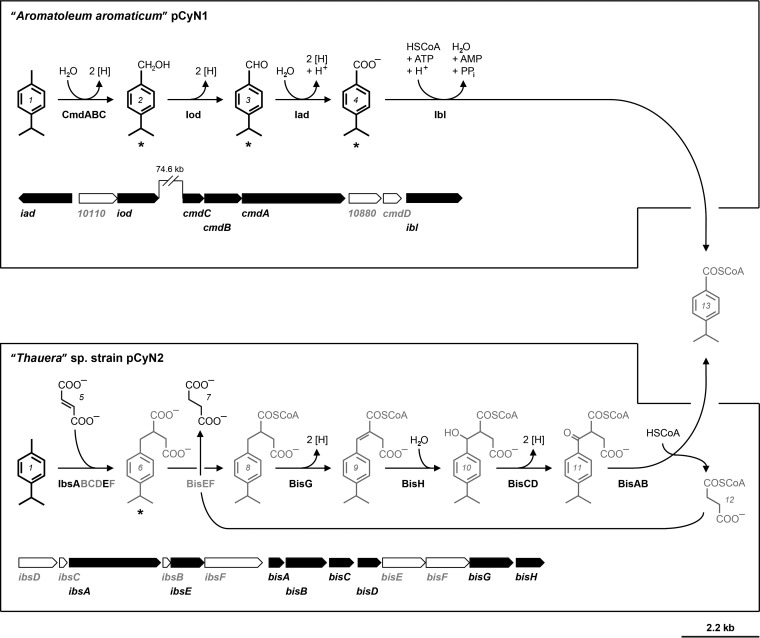
FIG 1.
Peripheral reaction sequences for the conversion of p-cymene to 4-isopropylbenzoyl-CoA in “A. aromaticum” pCyN1 and “Thauera” sp. pCyN2. Compounds shown in black represent proven growth substrates. Protein and gene names are shown in black (identified) and gray (predicted only). Identified proteins are detailed in Table 2 as well as in Table S1 and Fig. S2 in the supplemental material (except for 10880, encoding a predicted short-chain dehydrogenase/reductase not part of the suggested pathway). Numbered structures correspond to the following compounds: 1, p-cymene; 2, 4-isopropylbenzyl alcohol; 3, 4-isopropylbenzaldehyde; 4, 4-isopropylbenzoate; 5, fumarate; 6, (4-isopropylbenzyl)succinate; 7, succinate; 8, (4-isopropylbenzyl)succinyl-CoA; 9, (E)-(4-isopropylphenyl)itaconyl-CoA; 10, 2-[hydroxy(4-isopropylphenyl)methyl]succinyl-CoA; 11, (4-isopropylbenzyl)succinyl-CoA; 12, succinyl-CoA; 13, 4-isopropylbenzoyl-CoA. Identified metabolites are marked with an asterisk (*) and are detailed in Fig. 2 (see also Fig. S1 in the supplemental material); 4-isopropylbenzoate was also identified in the case of “Thauera” sp. pCyN2.
Metabolite formation during anaerobic growth with (alkyl)toluenes.
Upon anaerobic growth of “A. aromaticum” pCyN1 with toluene, (R)-(+)-benzylsuccinate and benzoate were identified by comparison with authentic standards. In addition, a metabolite was detected, the mass spectrum of which was identical with that of dimethyl phenylitaconate as previously reported by Krieger et al. (13). These findings indicate that “A. aromaticum” pCyN1 degrades toluene via addition to fumarate and subsequent modified β-oxidation to the central intermediate benzoyl-CoA, as known from its close relative “A. aromaticum” EbN1 (51, 52). Furthermore, all necessary genes for this degradation pathway (e.g., BssA displaying 100% sequence identity to its ortholog in strain EbN1) were detected in the genome of “A. aromaticum” pCyN1 (K. Trautwein, A. Strijkstra, and R. Rabus, unpublished data).
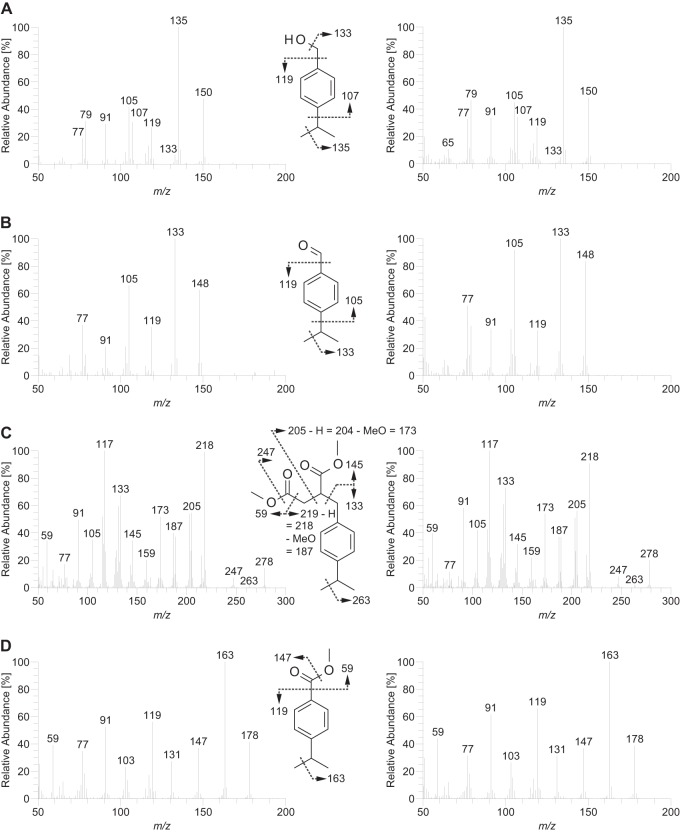
Accordingly, extracts of “A. aromaticum” pCyN1 cultures grown with 4-ethyltoluene and cultures of both strains grown with p-cymene were screened for the presence of expected arylalkylsuccinates and metabolites derived thereof. During growth with p-cymene, “Thauera” sp. pCyN2 formed a metabolite with a molecular ion at m/z 278, i.e., a shift by 42 amu to higher mass in comparison to benzylsuccinate, and with a mass spectrometric fragmentation pattern in agreement with the presence of an isopropyl substituent in the molecule (Fig. 2C). Subsequently, the metabolite was unequivocally identified as dimethyl (4-isopropylbenzyl)succinate by comparison with a standard synthesized using a biomimetic addition of the 4-isopropylbenzyl radical, generated from the corresponding iodide, to fumarate. Derivatization of the free acid with (R)-1-phenylethanamine enabled separation of the racemate. As known for toluene-derived benzylsuccinate (53), only the later-eluting enantiomer is formed, which very likely also has the R-configuration. During anaerobic growth of “Thauera” sp. pCyN2 with p-cymene in the presence of 2,3-2H2-fumarate, a metabolite was formed, the mass spectrum of which was in agreement with the structure of 2,3-2H2-(4-isopropylbenzyl)succinate (see Fig. S1 in the supplemental material). This provided direct evidence for involvement of fumarate in the activation of p-cymene. Furthermore, a metabolite was detected, the mass spectrum of which agrees with the structure of (4-isopropylphenyl)itaconate. In cultures with 2,3-2H2-fumarate, the molecular ion of this metabolite was shifted by 1 amu, thus representing the expected dehydrogenation product of 2,3-2H2-(4-isopropylbenzyl)succinate (see Fig. S1 in the supplemental material). Another metabolite with a molecular ion at m/z 178 was identified as 4-isopropylbenzoate by comparison with a commercially available standard (Fig. 2D). Overall, these results provide evidence that p-cymene is anaerobically degraded in “Thauera” sp. pCyN2 via a pathway analogous to that found for toluene in all previously described toluene-utilizing denitrifying, ferric iron-reducing, and sulfate-reducing bacteria (for an overview, see reference 54).
FIG 2.
Mass spectra of key metabolites of anaerobic p-cymene degradation (left) and of the corresponding authentic standards (right). (A) 4-Isopropylbenzyl alcohol (“A. aromaticum” pCyN1). (B) 4-Isopropylbenzaldehyde (“A. aromaticum” pCyN1). (C) (4-Isopropylbenzyl)succinate as methyl ester upon derivatization with diazomethane (“Thauera” sp. pCyN2). (D) 4-Isopropylbenzoate as methyl ester upon derivatization with diazomethane (both strains).
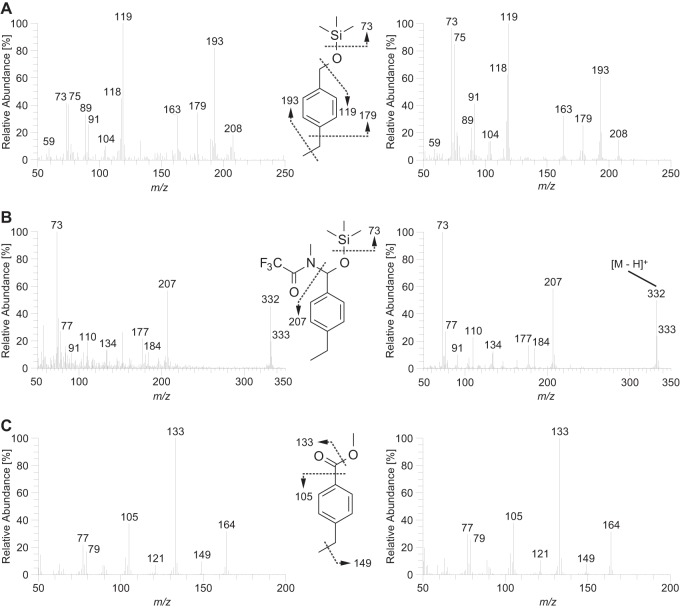
4-Isopropylbenzoate was also formed during anaerobic growth of “A. aromaticum” pCyN1 with p-cymene. Likewise, 4-ethylbenzoate (Fig. 3C) was formed with 4-ethyltoluene as the substrate. Unexpectedly, however, no evidence was obtained for the formation of the corresponding benzylsuccinates or phenylitaconates in these experiments. Instead, the respective benzyl alcohols and benzaldehydes were identified by comparison with commercially available standards (Fig. 2A and B and 3A and B). These findings provide direct metabolic evidence for the O2-independent dehydrogenation/oxidation of a benzylic methyl group as the initial activation reaction during bacterial utilization of alkylated toluenes under anoxic conditions.
FIG 3.
Mass spectra of metabolites of anaerobic 4-ethyltoluene degradation in “A. aromaticum” pCyN1 (left) and of the corresponding authentic standards (right). (A) 4-Ethylbenzyl alcohol as trimethylsilyl ether upon derivatization with MSTFA. (B) 4-Ethylbenzaldehyde as MSTFA adduct (N-[(4-ethylphenyl)(trimethylsiloxy)methyl]-2,2,2-trifluoro-N-methylacetamide) upon derivatization (81). (C) 4-Ethylbenzoate as methyl ester upon derivatization with diazomethane (81).
Proteogenomics of anaerobic p-cymene degradation.
Proteins identified from the soluble (2D DIGE) and membrane protein-enriched fractions and suggested to be involved in anaerobic degradation of p-cymene to 4-isopropylbenzoyl-CoA in “A. aromaticum” pCyN1 and “Thauera” sp. pCyN2, respectively, are presented in Table 2 and Fig. S2 in the supplemental material. The functional predictions of these proteins are compiled in Table S1 in the supplemental material. The proteogenomic findings are summarized in Fig. 1.
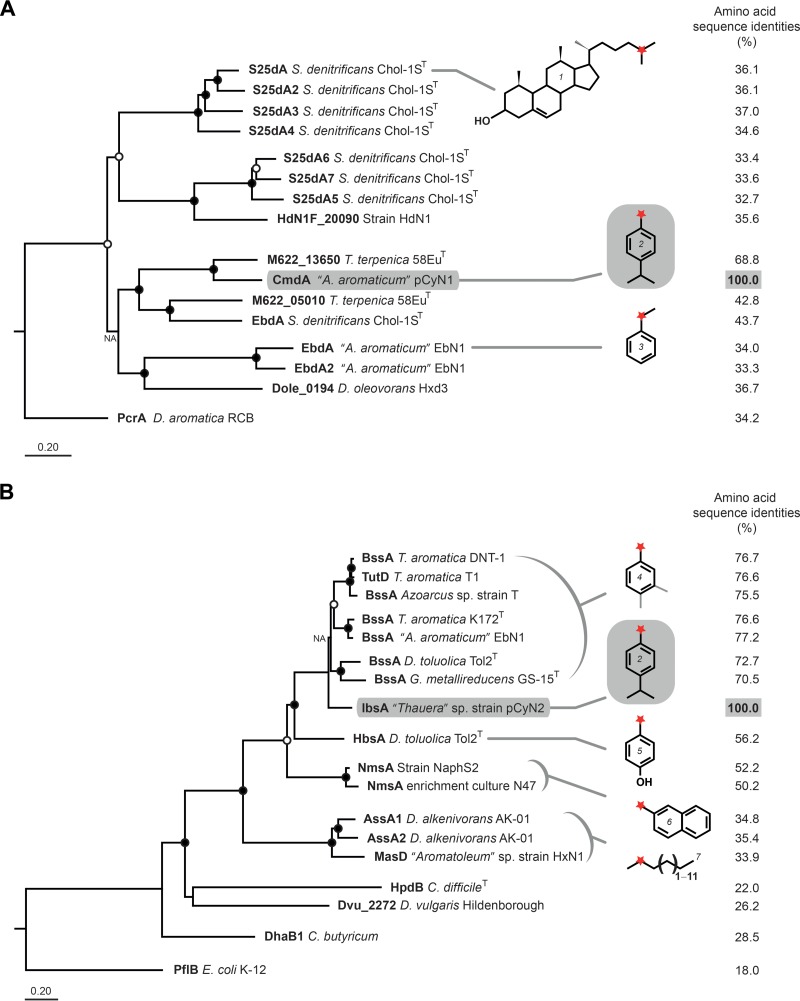
Three specifically and abundantly formed proteins in p-cymene-adapted cells of “A. aromaticum” pCyN1 were identified as homologs of ethylbenzene dehydrogenase (EbdABC; 30 to 55% amino acid sequence identities) from closely related “A. aromaticum” EbN1 (23). Colocalization of genes and homology of protein sequences, as well as key features (see Fig. S3 in the supplemental material) of the presumptive three-subunit p-cymene dehydrogenase CmdABC from “A. aromaticum” pCyN1 closely relate this enzyme to the ethylbenzene dehydrogenase from “A. aromaticum” EbN1 (methylene group hydroxylating) and steroid C-25 dehydrogenase from Sterolibacterium denitrificans Chol-1ST (methanetriyl carbon atom hydroxylating) (Fig. 4A). The large subunit CmdA possesses a twin-arginine motif (R7R8) as part of an N-terminal signal peptide, suggesting translocation of the folded protein via the Tat-secretion pathway (55) into the periplasm. Moreover, CmdA contains the conserved Asp204 and Lys434 residues, which were previously implicated for ethylbenzene dehydrogenase to be involved in coordination of the Mo-bis-molybdopterin guanine dinucleotide (Mo-bisMGD) cofactor in the active site and the conserved binding site for the FS0-[Fe4S4] cluster (56). The CmdB subunit harbors a series of conserved cysteine residues for binding of a total of four Fe-S clusters, and the conserved Met104 and Lys194 residues of the CmdC subunit may function as the two axial ligands of the heme-Fe (56). Based on these findings, the presumptive p-cymene dehydrogenase (CmdABC) is assumed to catalyze the O2-independent hydroxylation of p-cymene to 4-isopropylbenzyl alcohol.
FIG 4.
Phylogenetic relationship of the catalytic α-subunits of p-cymene dehydrogenase (CmdA) from “A. aromaticum” pCyN1 (A) and (4-isopropylbenzyl)succinate synthase (IbsA) from “Thauera” sp. pCyN2 (B). CmdA and IbsA are marked in bold and highlighted in gray. Sequence identities (percentages) are based on alignment of amino acid sequences using Clustal W. Open circles indicate >70% and closed circles indicate >90% bootstrap support. NA indicates nodes where bootstrapping is not applicable (see Materials and Methods). The bar below each tree corresponds to 20 inferred amino acid substitutions per 100 amino acids. The red star marks carbon atoms targeted during the initial activation reactions of cholesterol (structure 1), p-cymene (2), ethylbenzene (3), toluene, m- or p-xylene (4), p-cresol (5), 2-methylnaphthalene (6), and n-alkanes of chain lengths C6 to C16 (7). Abbreviations of enzyme catalytic subunits in panel A are as follows: S25dA, steroid C-25 dehydrogenase; S25dA2-7, steroid C-25 dehydrogenase-like; HdN1F_20090, annotated as dimethylsulfide dehydrogenase; M622_13650, uncharacterized; M622_05010, uncharacterized; EbdA and EbdA2, ethylbenzene dehydrogenase(-like); Dole_0194, molybdopterin oxidoreductase; PcrA, perchlorate reductase. Abbreviations of enzymes (catalytic subunits) in panel B are as follows: BssA and TutD, benzylsuccinate synthase; HbsA, (hydroxybenzyl)succinate synthase; NmsA, (2-naphthylmethyl)succinate synthase; AssA and MasD, (1-methylalkyl)succinate synthase; HpdB, 4-hydroxyphenylacetate decarboxylase; Dvu_2272, putative formate acetyltransferase; DhaB1, glycerol dehydratase; PflB, pyruvate formate lyase. Genus abbreviations are as follows: S. denitrificans, Sterolibacterium denitrificans; T. terpenica, Thauera terpenica; “A. aromaticum,” “Aromatoleum aromaticum”; D. oleovorans, Desulfococcus oleovorans; D. aromatica, Dechloromonas aromatica; T. aromatica, Thauera aromatica; D. toluolica, Desulfobacula toluolica; G. metallireducens, Geobacter metallireducens; D. alkenivorans, Desulfatibacillum alkenivorans; C. difficile, Clostridium difficile; D. vulgaris, Desulfovibrio vulgaris; C. butyricum, Clostridium butyricum; E. coli, Escherichia coli. UniProtKB accession numbers for proteins in panel A, from top to bottom (omitting CmdA), are as follows: UniProtKB H9NN89, H9NN92, H9NN93, H9NN94, H9NNA1, H9NNA4, H9NN98, E1VLA0, T0B0B4, S9ZMT3, H9NNA7, Q5P5I0, Q5NZV2, A8ZSU1, and Q47CW6. UniProtKB accession numbers for proteins in panel B, from top to bottom (omitting IbsA), are as follows: Q8L1A3, O68395, Q8VPT7, O87943, Q5P6A2, K0NCD2, Q39VF1, K0ND30, D8FBK7, D2XBH8, B8FEM4, B8FF75, A9J4K4, Q84F16, Q729S7, A9LAS8, and P09373.
Two further specifically formed proteins in p-cymene-adapted cells of “A. aromaticum” pCyN1 are homologs (>70% sequence identities) of recently described geraniol (GeoA) and geranial dehydrogenase (GeoB) from anaerobically monoterpene-degrading Castellaniella defragrans 65Phen (57) and are suggested to oxidize 4-isopropylbenzyl alcohol via the aldehyde intermediate to 4-isopropylbenzoate (enzymes from “A. aromaticum” pCyN1 termed Iod and Iad for 4-isopropylbenzyl alcohol and 4-isopropylbenzaldehyde dehydrogenase, respectively). Notably, GeoA of C. defragrans 65Phen also converts 4-isopropylbenzyl alcohol (57). Finally, the identified putative CoA ligase (termed Ibl) could catalyze formation of CoA-activated 4-isopropylbenzoate prior to further degradation.
In contrast, differential proteomic analysis of p-cymene- versus succinate-adapted “Thauera” sp. pCyN2 cells allowed p-cymene-specific identification of the catalytic α-subunit of a tentative (4-isopropylbenzyl)succinate synthase (IbsA; 77% amino acid sequence identity with the homologous BssA of benzylsuccinate synthase from T. aromatica K172T). Moreover, most of the enzymes required for the modified β-oxidation of the succinate moiety of (4-isopropylbenzyl)succinate yielding 4-isopropylbenzoyl-CoA were specifically identified in p-cymene-adapted cells (BisABCDGH with 71 to 81% amino acid sequence identities compared to the homologous Bbs enzymes of T. aromatica K172T). All genes coding for initial (4-isopropylbenzyl)succinate synthase and the enzymes of β-oxidation colocalize in the genome of “Thauera” sp. pCyN2 in an operon-like structure (Fig. 1). Encoded proteins not identified by proteomics (IbsDCBF and BisEF) also revealed high sequence identities to the respective proteins in T. aromatica K172T (see Table S1 in the supplemental material).
As summarized in Table 1, a notable property of “A. aromaticum” pCyN1 is fumarate-dependent activation of toluene and methyl group hydroxylation of p-cymene and 4-ethyltoluene. A possible explanation for the incapacity of “A. aromaticum” pCyN1 to degrade the latter two compounds via addition to fumarate and subsequent β-oxidation is the absence of ibs and bis gene clusters (see above) from the genome of “A. aromaticum” pCyN1 (K. Trautwein and R. Rabus, unpublished data). Moreover, the distinct phylogenetic branching of IbsA from toluene/xylene-converting BssA homologues (Fig. 4B) (see the next section of the text) as well as the bulkiness of the 4-isopropyl and 4-ethyl groups suggests that bss and bbs gene products from “A. aromaticum” pCyN1 cannot accommodate and/or convert p-cymene and 4-ethyltoluene.
Phylogenetic relations of key enzymes for anaerobic p-cymene degradation in “A. aromaticum” pCyN1 and “Thauera” sp. pCyN2.
The CmdA protein of “A. aromaticum” pCyN1 (catalytic α-subunit of predicted p-cymene dehydrogenase) phylogenetically belongs to the DdhA/SerA/EbdA clade (58) of the prokaryotic complex iron-sulfur molybdoenzyme (CISM) family (59). CmdA of “A. aromaticum” pCyN1 together with uncharacterized molybdoenzymes from Thauera terpenica 58EuT and S. denitrificans Chol-1ST (substrates currently unknown) constitutes a distinct cluster clearly separated from the catalytic subunits of ethylbenzene dehydrogenase (EbdA) from “A. aromaticum” EbN1 and steroid C-25 dehydrogenase (S25dA) from S. denitrificans Chol-1ST as closest characterized relatives (Fig. 4A). In conjunction, the molybdenum-containing subunits of these three types of hydroxylating enzymes cluster separately from the α-subunit of perchlorate reductase (PcrA) of Dechloromonas aromatica RCB as representative for molybdoenzymes involved in anaerobic respiration (60).
The phylogenetic analysis of the IbsA protein of “Thauera” sp. pCyN2 (Fig. 4B) revealed this predicted catalytic α-subunit of (4-isopropylbenzyl)succinate synthase to affiliate with the cluster of known toluene- and/or xylene-activating benzylsuccinate synthases from denitrifying, ferric iron-reducing, and sulfate-reducing bacteria (61, 62) and to be more distantly related to p-cresol-activating (hydroxybenzyl)succinate synthase (HbsA) (28), 2-methylnaphthalene-activating (2-naphthylmethyl)succinate synthases (NmsA) (16, 17), or n-alkane-activating (1-methylalkyl)succinate synthases (MasD and AssA) (63, 64). Within the cluster of toluene/xylene-activating benzylsuccinate synthases, the IbsA protein apparently forms a distinct branch, which may reflect mechanistic and steric constraints exerted by the 4-isopropyl group.
DISCUSSION
Known initial anaerobic hydroxylation reactions at benzylic methyl groups and tertiary carbon atoms.
The reaction mechanism of ethylbenzene dehydrogenase from “A. aromaticum” EbN1 has already been intensively studied based on its crystal structure (56) and ab initio and quantum chemical modeling (65, 66), as well as by substrate and inhibitor studies (67). Essentially, methylene group hydroxylation proceeds via two one-electron transfer steps in the catalytic α-subunit (EbdA), where the initial radical-type transition state/intermediate results from C—H bond cleavage and where the subsequent carbocation-type transition state/intermediate precedes the actual hydroxyl-group transfer. The electron transfers in the α-subunit are associated with stepwise reduction of the Mo cofactor from Mo(VI) to Mo(IV). Reoxidation of the latter by the FS0-[Fe4S4] cluster additionally present in the α-subunit further requires transfer of electrons to cytochrome c via the four Fe-S clusters in the β-subunit (EbdB) and the heme group in the γ-subunit (EbdC).
The only other known enzyme catalyzing an O2-independent hydroxylation is the long-studied p-cresol methylhydroxylase (Pch) from anaerobically grown P. putida NCIB 9866, targeting the benzylic methyl group of the phenolic compound p-cresol (24, 25). Pch is a flavocytochrome protein, with the large subunit (PchF) bearing a covalently bound flavin adenine dinucleotide (FAD) and the small subunit (PchC) having a covalently bound heme. An initial one-step transfer of two electrons from p-cresol to FAD yields a quinone-methide intermediate (4-methylidenecyclohexa-2,5-dien-1-one). This is subsequently transformed to 4-hydroxybenzyl alcohol via a nucleophilic attack of a water molecule at the methide carbon atom, i.e., a formal 1,6-addition of water to the quinone-methide intermediate (25, 68). Based on homology, a similar mechanism for p-cresol hydroxylation may be envisioned for Pch from G. metallireducens GS-15T (69). It may be assumed that the hydroxyl group plays an integral role in the reaction to allow formation of the quinone-methide intermediate based on electron donation either from one of the lone pairs of the hydroxyl group or from the corresponding oxyanion. Because of the lack of free electron pairs at alkyl carbon atoms and the implausibility of creating an intermediate carbanion, a similar mechanism of enzymatic transformation may be excluded for p-cymene, requiring that hydroxylation of its methyl group differs mechanistically from that of p-cresol.
The recently described molybdenum-containing steroid C-25 dehydrogenase (S25d) from S. denitrificans Chol-1ST (70, 71) is the first O2-independent enzyme capable of hydroxylating a tertiary carbon atom in the side chain of cholesterol. The subunits of heterotrimeric S25d show modest amino acid sequence identities (36, 56, or 32% for the α-, β-, or γ-subunit) to the respective subunits of ethylbenzene dehydrogenase and contain all functionally relevant conserved amino acids (see Fig. S3 in the supplemental material) (71). S25d is assumed to employ a similar catalytic mechanism for hydroxylation as elaborated for ethylbenzene dehydrogenase with the required tertiary carbocation at C-25 of cholesterol being much more stable than a primary carbocation derived from either of the two terminal carbon atoms (C-26 or C-27) (71).
Reactivity of C—H bonds at benzylic methyl groups and tertiary carbon atoms.
Based on the above-described mechanism for ethylbenzene dehydrogenase via a “carbocation-type transition state” (56, 65–67), it is useful to consider heterolytic C—H bond dissociation energies in the context of the hydroxylation of p-cymene. Values for formation of a carbenium ion and a hydride have been reported to decrease in the following order: toluene (992 kJ mol−1 [72] or 994 kJ mol−1 [73]) > isobutane (978 kJ mol−1 [72] or 979 kJ mol−1 [73]) > p-xylene (966 kJ mol−1 [72]) > ethylbenzene (947 kJ mol−1 [73]). The lower values for p-xylene and ethylbenzene than for toluene can be ascribed to hyperconjugative stabilization of the carbenium ion center by the p- and α-methyl groups, respectively (74, 75). Values for 4-ethyltoluene and p-cymene are not available in the literature, but the known similarity of the hyperconjugative effect of methyl (Me), ethyl (Et), and isopropyl (iPr) groups (albeit iPr < Et < Me) (74, 76) suggests comparable stabilization. Given that the heterolytic C—H bond dissociation energy of p-xylene is ∼12 kJ mol−1 lower than that of isobutane and assuming that the tertiary carbon atom in the sterol side chain is oxidized by a mechanism similar to that of ethylbenzene in “A. aromaticum” EbN1, this should also be true for the methyl groups in p-xylene, p-cymene, and 4-ethyltoluene. The p-alkyl group thus stabilizes the resonance structures of the intermediate carbenium based on its hyperconjugative effect. We suggest that o-alkyltoluenes might also be substrates for the enzyme(s) described, but not the m-isomers. This is supported by the heterolytic C—H bond dissociation energies of the three xylene isomers (72). Notably, “A. aromaticum” pCyN1 does not utilize o-cymene (35), which could indicate that its transformation may be hindered at a downstream step of the degradation pathway.
Formation of (4-isopropylbenzyl)succinate in anaerobically p-cymene-grown “Thauera” sp. pCyN2 is consistent with a homolytic mechanism for this hydrocarbon substrate. The homolytic C—H bond dissociation energies of toluene (369 [72] or 368 kJ mol−1 [77]) and p-xylene (367 kJ mol−1 [72]) are nearly identical but are slightly higher than the value for ethylbenzene (358 kJ mol−1 [77]). A value for p-cymene is not available in the literature, but it is reasonable to assume comparable stabilization of a presumptive intermediate 4-alkylbenzyl radical independent of the type of alkyl substituent, in this case isopropyl, following the arguments provided above for the heterolytic C—H bond dissociation.
Concluding remarks.
The related betaproteobacterial strains pCyN1 and pCyN2 employ two fundamentally different reaction sequences to anaerobically convert p-cymene to 4-isopropylbenzoyl-CoA. This difference should not be due to energetic constraints, as both strains can yield the same high energy from p-cymene degradation (ΔG0′ = −5,354 kJ mol−1 p-cymene). The distinctiveness of the herein reported enzymes from other known anaerobic hydrocarbon-activating enzymes with respect to substrate type (p-cymene) and phylogenetic branching could add to (i) the compound-specific resolution power of biogeographic studies on genes encoding these enzymes (78) and (ii) the prospects of biomimetic approaches for the industrially relevant C—H bond activation (79). Further degradation of 4-isopropylbenzoyl-CoA is currently being investigated by pursuing possibilities of degradation via a route similar to the recently described 4-methylbenzoyl-CoA pathway (80) or the classical anaerobic benzoyl-CoA pathway (8).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Daniela Thies (Bremen, Germany) and Christina Hinrichs (Oldenburg, Germany) for technical assistance. We thank Fritz Widdel (Bremen) for continuous support of proteomic work in our group. B. T. Golding and M. Drozdowska also thank the EPSRC Mass Spectrometry Service at the University of Wales (Swansea) for mass spectrometric analyses.
This study was supported by the Deutsche Forschungsgemeinschaft within the framework of Priority Program 1319.
Footnotes
Published ahead of print 26 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02385-14.
REFERENCES
- 1.Wilkes H, Schwarzbauer J. 2010. Hydrocarbons: an introduction to structure, physico-chemical properties and natural occurrence, p 1–48 In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, Germany. [Google Scholar]
- 2.Widdel F, Musat F. 2010. Diversity and common principles in enzymatic activation of hydrocarbons, p 981–1009 In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, Germany. [Google Scholar]
- 3.Austin RN, Buzzi K, Kim E, Zylstra GJ, Groves JT. 2003. Xylene monooxygenase, a membrane-spanning nonheme diiron enzyme that hydroxylates hydrocarbons via a substrate radical intermediate. J. Biol. Inorg. Chem. 8:733–740. [DOI] [PubMed] [Google Scholar]
- 4.Eaton RW. 1997. p-Cymene catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. J. Bacteriol. 179:3171–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Widdel F, Rabus R. 2001. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr. Opin. Biotechnol. 12:259–276. 10.1016/S0958-1669(00)00209-3. [DOI] [PubMed] [Google Scholar]
- 6.Widdel F, Knittel K, Galushko A. 2010. Anaerobic hydrocarbon-degrading microorganisms: an overview, p 1997–2021 In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, Germany. [Google Scholar]
- 7.Kaser FM, Coates JD. 2010. Nitrate, perchlorate and metal respirers, p 2033–2047 In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, Germany. [Google Scholar]
- 8.Fuchs G, Boll M, Heider J. 2011. Microbial degradation of aromatic compounds—from one strategy to four. Nat. Rev. Microbiol. 9:803–816. 10.1038/nrmicro2652. [DOI] [PubMed] [Google Scholar]
- 9.Philipp B, Schink B. 2012. Different strategies in anaerobic biodegradation of aromatic compounds: nitrate reducers versus strict anaerobes. Environ. Microbiol. Rep. 4:469–478. 10.1111/j.1758-2229.2011.00304.x. [DOI] [PubMed] [Google Scholar]
- 10.Boll M, Löffler C, Morris BEL, Kung JW. 2014. Anaerobic degradation of homocyclic aromatic compounds via arylcarboxyl-coenzyme A esters: organisms, strategies and key enzymes. Environ. Microbiol. 16:612–627. 10.1111/1462-2920.12328. [DOI] [PubMed] [Google Scholar]
- 11.Biegert T, Fuchs G, Heider J. 1996. Evidence that anaerobic oxidation of toluene in the denitrifying bacterium Thauera aromatica is initiated by formation of benzylsuccinate from toluene and fumarate. Eur. J. Biochem. 238:661–668. 10.1111/j.1432-1033.1996.0661w.x. [DOI] [PubMed] [Google Scholar]
- 12.Krieger CJ, Roseboom W, Albracht SP, Spormann AM. 2001. A stable organic free radical in anaerobic benzylsuccinate synthase of Azoarcus sp. strain T. J. Biol. Chem. 276:12924–12927. 10.1074/jbc.M009453200. [DOI] [PubMed] [Google Scholar]
- 13.Krieger CJ, Beller HR, Reinhard M, Spormann AM. 1999. Initial reactions in anaerobic oxidation of m-xylene by the denitrifying bacterium Azoarcus sp. strain T. J. Bacteriol. 181:6403–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morasch B, Schink B, Tebbe CC, Meckenstock RU. 2004. Degradation of o-xylene and m-xylene by a novel sulfate-reducer belonging to the genus Desulfotomaculum. Arch. Microbiol. 181:407–417. 10.1007/s00203-004-0672-6. [DOI] [PubMed] [Google Scholar]
- 15.Rotaru A-E, Probian C, Wilkes H, Harder J. 2010. Highly enriched Betaproteobacteria growing anaerobically with p-xylene and nitrate. FEMS Microbiol. Ecol. 71:460–468. 10.1111/j.1574-6941.2009.00814.x. [DOI] [PubMed] [Google Scholar]
- 16.Musat F, Galushko A, Jacob J, Widdel F, Kube M, Reinhardt R, Wilkes H, Schink B, Rabus R. 2009. Anaerobic degradation of naphthalene and 2-methylnaphthalene by strains of marine sulfate-reducing bacteria. Environ. Microbiol. 11:209–219. 10.1111/j.1462-2920.2008.01756.x. [DOI] [PubMed] [Google Scholar]
- 17.Selesi D, Jehmlich N, Bergen von M, Schmidt F, Rattei T, Tischler P, Lueders T, Meckenstock RU. 2010. Combined genomic and proteomic approaches identify gene clusters involved in anaerobic 2-methylnaphthalene degradation in the sulfate-reducing enrichment culture N47. J. Bacteriol. 192:295–306. 10.1128/JB.00874-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rabus R, Wilkes H, Behrends A, Armstroff A, Fischer T, Pierik AJ, Widdel F. 2001. Anaerobic initial reaction of n-alkanes in a denitrifying bacterium: evidence for (1-methylpentyl)succinate as initial product and for involvement of an organic radical in n-hexane metabolism. J. Bacteriol. 183:1707–1715. 10.1128/JB.183.5.1707-1715.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Callaghan AV, Gieg LM, Kropp KG, Suflita JM, Young LY. 2006. Comparison of mechanisms of alkane metabolism under sulfate-reducing conditions among two bacterial isolates and a bacterial consortium. Appl. Environ. Microbiol. 72:4274–4282. 10.1128/AEM.02896-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kniemeyer O, Fischer T, Wilkes H, Glöckner FO, Widdel F. 2003. Anaerobic degradation of ethylbenzene by a new type of marine sulfate-reducing bacterium. Appl. Environ. Microbiol. 69:760–768. 10.1128/AEM.69.2.760-768.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson HA, Pelletier DA, Spormann AM. 2001. Isolation and characterization of anaerobic ethylbenzene dehydrogenase, a novel Mo-Fe-S enzyme. J. Bacteriol. 183:4536–4542. 10.1128/JB.183.15.4536-4542.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kniemeyer O, Heider J. 2001. Ethylbenzene dehydrogenase, a novel hydrocarbon-oxidizing molybdenum/iron-sulfur/heme enzyme. J. Biol. Chem. 276:21381–21386. 10.1074/jbc.M101679200. [DOI] [PubMed] [Google Scholar]
- 23.Rabus R, Kube M, Beck A, Widdel F, Reinhardt R. 2002. Genes involved in the anaerobic degradation of ethylbenzene in a denitrifying bacterium, strain EbN1. Arch. Microbiol. 178:506–516. 10.1007/s00203-002-0487-2. [DOI] [PubMed] [Google Scholar]
- 24.Hopper DJ. 1976. The hydroxylation of p-cresol and its conversion to p-hydroxybenzaldehyde in Pseudomonas putida. Biochem. Biophys. Res. Commun. 69:462–468. 10.1016/0006-291X(76)90544-1. [DOI] [PubMed] [Google Scholar]
- 25.Hopper DJ. 1978. Incorporation of [18O]water in the formation of p-hydroxybenzyl alcohol by the p-cresol methylhydroxylase from Pseudomonas putida. Biochem. J. 175:345–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters F, Heintz D, Johannes J, van Dorsselaer A, Boll M. 2007. Genes, enzymes, and regulation of para-cresol metabolism in Geobacter metallireducens. J. Bacteriol. 189:4729–4738. 10.1128/JB.00260-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Müller JA, Galushko AS, Kappler A, Schink B. 2001. Initiation of anaerobic degradation of p-cresol by formation of 4-hydroxybenzylsuccinate in Desulfobacterium cetonicum. J. Bacteriol. 183:752–757. 10.1128/JB.183.2.752-757.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wöhlbrand L, Jacob JH, Kube M, Mussmann M, Jarling R, Beck A, Amann R, Wilkes H, Reinhardt R, Rabus R. 2013. Complete genome, catabolic sub-proteomes and key-metabolites of Desulfobacula toluolica Tol2, a marine, aromatic compound-degrading, sulfate-reducing bacterium. Environ. Microbiol. 15:1334–1355. 10.1111/j.1462-2920.2012.02885.x. [DOI] [PubMed] [Google Scholar]
- 29.Leutwein C, Heider J. 1999. Anaerobic toluene-catabolic pathway in denitrifying Thauera aromatica: activation an β-oxidation of the first intermediate, (R)-(+)-benzylsuccinate. Microbiology 145:3265–3271. [DOI] [PubMed] [Google Scholar]
- 30.Leuthner B, Heider J. 2000. Anaerobic toluene catabolism of Thauera aromatica: the bbs operon codes for enzymes of β-oxidation of the intermediate benzylsuccinate. J. Bacteriol. 182:272–277. 10.1128/JB.182.2.272-277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kniemeyer O, Heider J. 2001. (S)-1-Phenylethanol dehydrogenase of Azoarcus sp. strain EbN1, an enzyme of anaerobic ethylbenzene catabolism. Arch. Microbiol. 176:129–135. 10.1007/s002030100303. [DOI] [PubMed] [Google Scholar]
- 32.Jobst B, Schühle K, Linne U, Heider J. 2010. ATP-dependent carboxylation of acetophenone by a novel type of carboxylase. J. Bacteriol. 192:1387–1394. 10.1128/JB.01423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders HJ, Kaetzke A, Kämpfer P, Ludwig W, Fuchs G. 1995. Taxonomic position of aromatic-degrading denitrifying pseudomonad strains K 172 and KB 740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the beta subclass of the Proteobacteria. Int. J. Syst. Bacteriol. 45:327–333. 10.1099/00207713-45-2-327. [DOI] [PubMed] [Google Scholar]
- 34.Rabus R, Widdel F. 1995. Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch. Microbiol. 163:96–103. 10.1007/BF00381782. [DOI] [PubMed] [Google Scholar]
- 35.Harms G, Rabus R, Widdel F. 1999. Anaerobic oxidation of the aromatic plant hydrocarbon p-cymene by newly isolated denitrifying bacteria. Arch. Microbiol. 172:303–312. 10.1007/s002030050784. [DOI] [PubMed] [Google Scholar]
- 36.Song B, Häggblom MM, Zhou J, Tiedje JM, Palleroni NJ. 1999. Taxonomic characterization of denitrifying bacteria that degrade aromatic compounds and description of Azoarcus toluvorans sp. nov. and Azoarcus toluclasticus sp. nov. Int. J. Syst. Bacteriol. 49:1129–1140. 10.1099/00207713-49-3-1129. [DOI] [PubMed] [Google Scholar]
- 37.Trautwein K, Lahme S, Wöhlbrand L, Feenders C, Mangelsdorf K, Harder J, Steinbüchel A, Blasius B, Reinhardt R, Rabus R. 2012. Physiological and proteomic adaptation of “Aromatoleum aromaticum” EbN1 to low growth rates in benzoate-limited, anoxic chemostats. J. Bacteriol. 194:2165–2180. 10.1128/JB.06519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wöhlbrand L, Kallerhoff B, Lange D, Hufnagel P, Thiermann J, Reinhardt R, Rabus R. 2007. Functional proteomic view of metabolic regulation in “Aromatoleum aromaticum” strain EbN1. Proteomics 7:2222–2239. 10.1002/pmic.200600987. [DOI] [PubMed] [Google Scholar]
- 39.Jarling R, Sadeghi M, Drozdowska M, Lahme S, Buckel W, Rabus R, Widdel F, Golding BT, Wilkes H. 2012. Stereochemical investigations reveal the mechanism of the bacterial activation of n-alkanes without oxygen. Angew. Chem. Int. Ed. Engl. 51:1334–1338. 10.1002/anie.201106055. [DOI] [PubMed] [Google Scholar]
- 40.Gade D, Thiermann J, Markowsky D, Rabus R. 2003. Evaluation of two-dimensional difference gel electrophoresis for protein profiling. J. Mol. Microbiol. Biotechnol. 5:240–251. 10.1159/000071076. [DOI] [PubMed] [Google Scholar]
- 41.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 42.Zech H, Echtermeyer C, Wöhlbrand L, Blasius B, Rabus R. 2011. Biological versus technical variability in 2-D DIGE experiments with environmental bacteria. Proteomics 11:3380–3389. 10.1002/pmic.201100071. [DOI] [PubMed] [Google Scholar]
- 43.Zech H, Hensler M, Koßmehl S, Drüppel K, Wöhlbrand L, Trautwein K, Hulsch R, Maschmann U, Colby T, Schmidt J, Reinhardt R, Schmidt-Hohagen K, Schomburg D, Rabus R. 2013. Adaptation of Phaeobacter inhibens DSM 17395 to growth with complex nutrients. Proteomics 13:2851–2868. 10.1002/pmic.201200513. [DOI] [PubMed] [Google Scholar]
- 44.Wiegmann K, Hensler M, Wöhlbrand L, Ulbrich M, Schomburg D, Rabus R. 2014. Carbohydrate catabolism in Phaeobacter inhibens DSM 17395, member of the marine Roseobacter clade. Appl. Environ. Microbiol. 80:4725–4737. 10.1128/AEM.00719-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thauer RK, Jungermann K, Decker K. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41:100–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mavrovouniotis ML. 1991. Estimation of standard Gibbs energy changes of biotransformations. J. Biol. Chem. 266:14440–14445. [PubMed] [Google Scholar]
- 47.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. 2012. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28:464–469. 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, de Castro E, Coggill P, Corbett M, Das U, Daugherty L, Duquenne L, Finn RD, Fraser M, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, McMenamin C, Mi H, Mutowo-Muellenet P, Mulder N, Natale D, Orengo C, Pesseat S, Punta M, Quinn AF, Rivoire C, Sangrador-Vegas A, Selengut JD, Sigrist CJA, Scheremetjew M, Tate J, Thimmajanarthanan M, Thomas PD, Wu CH, Yeats C, Yong S-Y. 2012. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40:D306–D312. 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 51.Rabus R, Heider J. 1998. Initial reactions of anaerobic metabolism of alkylbenzenes in denitrifying and sulfate-reducing bacteria. Arch. Microbiol. 170:377–384. 10.1007/s002030050656. [DOI] [Google Scholar]
- 52.Kube M, Heider J, Amann J, Hufnagel P, Kühner S, Beck A, Reinhardt R, Rabus R. 2004. Genes involved in the anaerobic degradation of toluene in a denitrifying bacterium, strain EbN1. Arch. Microbiol. 181:182–194. 10.1007/s00203-003-0627-3. [DOI] [PubMed] [Google Scholar]
- 53.Beller HR, Spormann AM. 1998. Analysis of the novel benzylsuccinate synthase reaction for anaerobic toluene activation based on structural studies of the product. J. Bacteriol. 180:5454–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rabus R. 2005. Biodegradation of hydrocarbons under anoxic conditions, p 277–299 In Ollivier B, Margot M. (ed), Petroleum microbiology. ASM Press, Washington, DC. [Google Scholar]
- 55.Palmer T, Berks BC. 2012. The twin-arginine translocation (Tat) protein export pathway. Nat. Rev. Microbiol. 10:483–496. 10.1038/nrmicro2814. [DOI] [PubMed] [Google Scholar]
- 56.Kloer DP, Hagel C, Heider J, Schulz GE. 2006. Crystal structure of ethylbenzene dehydrogenase from Aromatoleum aromaticum. Structure 14:1377–1388. 10.1016/j.str.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 57.Lüddeke F, Wülfing A, Timke M, Germer F, Weber J, Dikfidan A, Rahnfeld T, Linder D, Meyerdierks A, Harder J. 2012. Geraniol and geranial dehydrogenases induced in anaerobic monoterpene degradation by Castellaniella defragrans. Appl. Environ. Microbiol. 78:2128–2136. 10.1128/AEM.07226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McDevitt CA, Hugenholtz P, Hanson GR, McEwan AG. 2002. Molecular analysis of dimethyl sulphide dehydrogenase from Rhodovulum sulfidophilum: its place in the dimethyl sulphoxide reductase family of microbial molybdopterin-containing enzymes. Mol. Microbiol. 44:1575–1587. 10.1046/j.1365-2958.2002.02978.x. [DOI] [PubMed] [Google Scholar]
- 59.Rothery RA, Workun GJ, Weiner JH. 2008. The prokaryotic complex iron-sulfur molybdoenzyme family. Biochim. Biophys. Acta 1778:1897–1929. 10.1016/j.bbamem.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 60.Bender KS, Shang C, Chakraborty R, Belchik SM, Coates JD, Achenbach LA. 2005. Identification, characterization, and classification of genes encoding perchlorate reductase. J. Bacteriol. 187:5090–5096. 10.1128/JB.187.15.5090-5096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leuthner B, Leutwein C, Schulz H, Hörth P, Haehnel W, Schiltz E, Schägger H, Heider J. 1998. Biochemical and genetic characterization of benzylsuccinate synthase from Thauera aromatica: a new glycyl radical enzyme catalysing the first step in anaerobic toluene metabolism. Mol. Microbiol. 28:615–628. 10.1046/j.1365-2958.1998.00826.x. [DOI] [PubMed] [Google Scholar]
- 62.Winderl C, Schaefer S, Lueders T. 2007. Detection of anaerobic toluene and hydrocarbon degraders in contaminated aquifers using benzylsuccinate synthase (bssA) genes as a functional marker. Environ. Microbiol. 9:1035–1046. 10.1111/j.1462-2920.2006.01230.x. [DOI] [PubMed] [Google Scholar]
- 63.Grundmann O, Behrends A, Rabus R, Amann J, Halder T, Heider J, Widdel F. 2008. Genes encoding the candidate enzyme for anaerobic activation of n-alkanes in the denitrifying bacterium, strain HxN1. Environ. Microbiol. 10:376–385. 10.1111/j.1462-2920.2007.01458.x. [DOI] [PubMed] [Google Scholar]
- 64.Callaghan AV, Wawrik B, Ní Chadhain SM, Young LY, Zylstra GJ. 2008. Anaerobic alkane-degrading strain AK-01 contains two alkylsuccinate synthase genes. Biochem. Biophys. Res. Commun. 366:142–148. 10.1016/j.bbrc.2007.11.094. [DOI] [PubMed] [Google Scholar]
- 65.Szaleniec M, Borowski T, Schühle K, Witko M, Heider J. 2010. Ab initio modeling of ethylbenzene dehydrogenase reaction mechanism. J. Am. Chem. Soc. 132:6014–6024. 10.1021/ja907208k. [DOI] [PubMed] [Google Scholar]
- 66.Szaleniec M, Salwinski A, Borowski T, Heider J, Witko M. 2012. Quantum chemical modeling studies of ethylbenzene dehydrogenase activity. Int. J. Quantum Chem. 112:1990–1999. 10.1002/qua.23143. [DOI] [Google Scholar]
- 67.Knack D, Hagel C, Szaleniec M, Dudzik A, Salwinski A, Heider J. 2012. Substrate and inhibitor spectra of ethylbenzene dehydrogenase: perspectives on application potential and catalytic mechanism. Appl. Environ. Microbiol. 78:6475–6482. 10.1128/AEM.01551-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cunane LM, Chen Z-W, Shamala N, Mathews FS, Cronin CN, McIntire WS. 2000. Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism. J. Mol. Biol. 295:357–374. 10.1006/jmbi.1999.3290. [DOI] [PubMed] [Google Scholar]
- 69.Johannes J, Bluschke A, Jehmlich N, von Bergen M, Boll M. 2008. Purification and characterization of active-site components of the putative p-cresol methylhydroxylase membrane complex from Geobacter metallireducens. J. Bacteriol. 190:6493–6500. 10.1128/JB.00790-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chiang Y-R, Ismail W, Müller M, Fuchs G. 2007. Initial steps in the anoxic metabolism of cholesterol by the denitrifying Sterolibacterium denitrificans. J. Biol. Chem. 282:13240–13249. 10.1074/jbc.M610963200. [DOI] [PubMed] [Google Scholar]
- 71.Dermer J, Fuchs G. 2012. Molybdoenzyme that catalyzes the anaerobic hydroxylation of a tertiary carbon atom in the side chain of cholesterol. J. Biol. Chem. 287:36905–36916. 10.1074/jbc.M112.407304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hayashibara K, Kruppa GH, Beauchamp JL. 2002. Photoelectron spectroscopy of the o-, m-, and p-methylbenzyl radicals. Implications for the thermochemistry of the radicals and ions. J. Am. Chem. Soc. 108:5441–5443. 10.1021/ja00278a011. [DOI] [Google Scholar]
- 73.Sharma RB, Sharma D, Hiraoka K. 1985. Kinetics and equilibria of chloride transfer reactions. Stabilities of carbocations based on chloride and hydride transfer equilibria measurements. J. Am. Chem. Soc. 107:3747–3757. [Google Scholar]
- 74.Cooney BT, Happer DAR. 1987. The Baker-Nathan order: hyperconjugation or a solvent effect? Aust. J. Chem. 40:1537–1544. 10.1071/CH9871537. [DOI] [Google Scholar]
- 75.Alabugin IV, Gilmore KM, Peterson PW. 2011. Hyperconjugation. Wiley Interdiscip. Rev. Comput. Mol. Sci. 1:109–141. 10.1002/wcms.6. [DOI] [Google Scholar]
- 76.DiLabio GA, Ingold KU. 2004. Solvolysis of para-substituted cumyl chlorides. Brown and Okamoto's electrophilic substituent constants revisited using continuum solvent models. J. Org. Chem. 69:1620–1624. 10.1021/jo035693r. [DOI] [PubMed] [Google Scholar]
- 77.McMillen DF, Golden DM. 1982. Hydrocarbon bond dissociation energies. Annu. Rev. Phys. Chem. 33:493–532. [Google Scholar]
- 78.von Netzer F, Pilloni G, Kleindienst S, Krüger M, Knittel K, Gründger F, Lueders T. 2013. Enhanced gene detection assays for fumarate-adding enzymes allow uncovering of anaerobic hydrocarbon degraders in terrestrial and marine systems. Appl. Environ. Microbiol. 79:543–552. 10.1128/AEM.02362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wencel-Delord J, Dröge T, Liu F, Glorius F. 2011. Towards mild metal-catalyzed C—H bond activation. Chem. Soc. Rev. 40:4740–4761. 10.1039/c1cs15083a. [DOI] [PubMed] [Google Scholar]
- 80.Lahme S, Eberlein C, Jarling R, Kube M, Boll M, Wilkes H, Reinhardt R, Rabus R. 2012. Anaerobic degradation of 4-methylbenzoate via a specific 4-methylbenzoyl-CoA pathway. Environ. Microbiol. 14:1118–1132. 10.1111/j.1462-2920.2011.02693.x. [DOI] [PubMed] [Google Scholar]
- 81.Ende M, Luftmann H. 1984. Unerwartete Reaktionsprodukte von N-methyl-N-Trimethylsilyltrifluoracetamid (MSTFA) mit Aldehyden. Tetrahedron 40:5167–5170. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.