Abstract
In Escherichia coli, the small regulatory noncoding RNA (sRNA) RyhB and the global ferric uptake regulator (Fur) mediate iron acquisition and storage control. Iron is both essential and potentially toxic for most living organisms, making the precise maintenance of iron homeostasis necessary for survival. While the roles of these regulators in iron homeostasis have been well studied in a nonpathogenic E. coli strain, their impact on the production of virulence-associated factors is still unknown for a pathogenic E. coli strain. We thus investigated the roles of RyhB and Fur in iron homeostasis and virulence of the uropathogenic E. coli (UPEC) strain CFT073. In a murine model of urinary tract infection (UTI), deletion of fur alone did not attenuate virulence, whereas a ΔryhB mutant and a Δfur ΔryhB double mutant showed significantly reduced bladder colonization. The Δfur mutant was more sensitive to oxidative stress and produced more of the siderophores enterobactin, salmochelins, and aerobactin than the wild-type strain. In contrast, while RyhB was not implicated in oxidative stress resistance, the ΔryhB mutant produced lower levels of siderophores. This decrease was correlated with the downregulation of shiA (encoding a transporter of shikimate, a precursor of enterobactin and salmochelin biosynthesis) and iucD (involved in aerobactin biosynthesis) in this mutant grown in minimal medium or in human urine. iucD was also downregulated in bladders infected with the ΔryhB mutant compared to those infected with the wild-type strain. Our results thus demonstrate that the sRNA RyhB is involved in production of iron acquisition systems and colonization of the urinary tract by pathogenic E. coli.
INTRODUCTION
RyhB is a small regulatory noncoding RNA (sRNA) involved in the regulation of several genes in response to iron availability in Escherichia coli (1). Iron is an essential cofactor for many metabolic enzymes involved in biological reactions, such as respiration, DNA biosynthesis, the tricarboxylic acid (TCA) cycle, and gene regulation. However, iron is toxic under oxygen-rich conditions, as it represents a source of highly reactive hydroxyl radicals, which can generate oxidative stress. Iron is thus both essential and potentially toxic for most living organisms, making the precise maintenance of iron homeostasis necessary for survival (2–4). In E. coli, iron acquisition and storage control is mediated by the global ferric uptake regulator (Fur) and the sRNA RyhB (1, 5–7). Under iron-rich conditions, Fe2+-Fur acts as a negative regulator of ryhB and iron uptake genes. When iron availability is limited, Fur becomes inactive, and subsequently, the production of RyhB and iron acquisition systems is initiated in order to restore iron homeostasis (1, 4, 8–10). In addition, Fur regulates several genes involved in other cellular processes, such as metabolic pathways, acid tolerance, virulence factor production, and protection against oxidative stress, in many pathogens, including E. coli, Pseudomonas aeruginosa, Vibrio cholerae, and Salmonella enterica (11–16).
Many RyhB targets are nonessential iron-using proteins, such as succinate dehydrogenase (sdh operon), superoxide dismutase (sodB), ferritins (ftnA and bfr), and aconitase and fumarase, enzymes of the tricarboxylic acid cycle (acnA and fumA) (1, 5). By inhibiting the synthesis of these proteins under iron-limited conditions, RyhB promotes availability of iron for essential iron-using proteins required for many biological processes (respiration, oxygen transport, and DNA biosynthesis) (17). RyhB has also been shown to promote synthesis of the siderophore enterobactin in nonpathogenic E. coli K-12 (18, 19). Siderophores are high-affinity iron-chelating molecules that contribute to bacterial survival by sequestering iron from the host. RyhB-dependent regulation of enterobactin siderophore production is mediated by (i) repressing the translation of cysE mRNA, which allows more serine to be used for enterobactin production (19); and (ii) activating shiA mRNA translation to increase shikimate (a metabolic precursor of enterobactin biosynthesis) acquisition from the environment (18). RyhB is also required for normal expression of the enterobactin biosynthesis polycistron, entCEBAH (19). More recently, in E. coli K-12, RyhB was also shown to activate the translation of cirA mRNA, encoding an outer membrane receptor involved in the uptake of precursor and breakdown products of enterobactin, namely, 2,3-dihydroxybenzoic acid (DHBA) and 2,3-dihydroxybenzoic serine (DHBS), respectively (20). Thus, RyhB plays an essential role in bacteria that require adaptation to iron starvation. Moreover, this sRNA also contributes to many pathogenicity determinants, such as acid resistance in Shigella flexneri (21); eukaryotic cell invasion and cell-to-cell spread in Shigella dysenteriae (22, 23); motility, chemotaxis, and biofilm formation in Vibrio cholerae (24, 25); biosynthesis of capsular polysaccharide and iron acquisition in Klebsiella pneumoniae (26); and oxidative stress resistance, survival inside human macrophages, and iron acquisition in Salmonella enterica serovar Typhi (27).
Pathogenic E. coli strains are responsible for intestinal or extraintestinal infections in humans and animals (28). Extraintestinal pathogenic E. coli (ExPEC) causes urinary tract infections (UTIs), neonatal meningitis, and septicemia in humans, as well as systemic infections in poultry and livestock (29–32). UTIs affect millions of women annually and result in significant health care costs and morbidity worldwide. UTIs are one of the most common bacterial infections, and uropathogenic E. coli (UPEC) is the predominant causal agent, representing up to 80% of community-acquired UTIs. Despite existing antimicrobial treatments, recurrent episodes of UTI are common, and bacterial strains are becoming increasingly more resistant to many currently used antimicrobial agents (33–35). UPEC strains possess many virulence factors (adhesins, toxins, iron acquisition systems, and capsular antigens) that promote bacterial growth and persistence within the urinary tract in a broad range of hosts (33, 36). UPEC strains have to be able to acquire essential nutrients, such as iron, during an infection of the urinary tract, regardless of its limited bioavailability. To overcome iron starvation within the host, E. coli strains are able to produce up to 4 different siderophores: enterobactin, salmochelins, aerobactin, and yersiniabactin. Whereas most E. coli strains, including commensal strains, produce enterobactin, salmochelins, aerobactin, and yersiniabactin are associated with pathogenic E. coli strains (37, 38). Salmochelins are glycosylated molecules of enterobactin that are synthesized by the iroBCDE gene products, while aerobactin is synthesized by the iucABCD gene products (39–42). Interestingly, it has been reported that salmochelins and aerobactin are often associated with ExPEC strains and contribute to their virulence (38, 43–51).
While the roles of RyhB and Fur in iron homeostasis have been well studied in nonpathogenic E. coli K-12, their impact on the production of virulence-associated factors is still unknown for ExPEC strains. In this study, we thus investigated the individual and combined roles of these regulators in iron acquisition, oxidative stress resistance, and virulence of the UPEC strain CFT073. First, we evaluated the colonization abilities of this strain in a mouse model of urinary tract infection, in the presence/absence of RyhB and/or Fur. We then analyzed the effects of these regulators on oxidative stress resistance and quantified siderophore production by mass spectrometry. Production of siderophores in the presence/absence of RyhB and/or Fur was then correlated with expression of siderophore biosynthesis genes in minimal medium, human urine, and infected bladders of mice.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strain CFT073 (O6:H1:K2) was isolated from the blood of a woman with acute pyelonephritis (52, 53). For bacterial culture, LB-Lennox broth and tryptic soy agar (Difco Laboratories, Detroit, MI) were routinely used. Strains were also grown in human urine, which was collected from healthy female volunteers of 20 to 30 years of age who had no history of UTI or antibiotic use in the prior 2 months. Urine samples were collected from human healthy donors with their consent as required by the ethics committee of INRS. Each urine sample was immediately filter sterilized (0.2-μm pore size), pooled, frozen at −30°C, and used within 1 week. Bacteria were also grown in iron-poor M63-glycerol minimal medium containing 39 mM KH2PO4, 80 mM K2HPO4, and 15 mM (NH4)2SO4. The pH was adjusted to 7.5 with KOH, and the medium was supplemented with 1 mM MgCl2, 0.1 mM CaCl2, 1 mM thiamine, and 0.6% (vol/vol) glycerol. Iron-poor medium was prepared in plastic bottles to reduce trace contamination of iron. When necessary, the following antibiotics and reagents were used at the indicated concentrations: chloramphenicol, 30 μg ml−1; gentamicin, 15 μg ml−1; ampicillin, 100 μg ml−1; kanamycin, 30 μg ml−1; and diaminopimelic acid (DAP), 50 μg ml−1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli K-12 strains | ||
| EM1055 | DJ480, a ΔX74lac derivative of MG1655 | 1 |
| EM1238 | EM1055 ryhB::cat Catr | 1 |
| DH5α | F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG ϕ80lacZΔM15 Δ(lacZYA-argF) U169 hsdR17 (rK− mK+) λ− | Bethesda Laboratories |
| MGN-617 | thi thr leu tonA lacY glnV supE ΔasdA4 recA::RP4 2-Tc::Mu [λ pir] Kanr | 72 |
| S17-1 (λ pir) | λ pir lysogen of S17-1 (Tpr Smr thi pro ΔhsdR hsdM+ recA::RP4 2-Tc::Mu-kan::Tn7) | 73 |
| JW0669 | BW25113 fur::FRT | 74 |
| UPEC strains | ||
| CFT073 | Wild-type pyelonephritis strain | 52 |
| QT1081 | CFT073 ΔlacZYA::FRT | 58 |
| QT2230 | CFT073 ΔryhB::cat Catr | This study |
| QT2469 | QT2230 attTn7::ryhB Catr Genr | This study |
| QT2634 | QT2230 Δfur::FRT Catr | This study |
| QT2637 | CFT073 Δfur::FRT | This study |
| QT2642 | QT2634 attTn7::fur Catr Genr | This study |
| QT2808 | QT2642 Δlac::ryhB Catr Genr Kanr | This study |
| Plasmids | ||
| pKD46 | oriR101 repA101(Ts) araBp-gam-bet-exo Ampr | 54 |
| pCP20 | Flp helper plasmid Ts replicon; Catr Ampr | 54 |
| pGEM-T Easy | Ampr | Promega |
| pIJ319 | pGEM-T::fur | This study |
| pIJ321 | pGEM-T::ryhB | This study |
| pIJ253 | pGP704::Tn7T-Gen; Ampr Genr | 55 |
| pIJ326 | pIJ253::ryhB (from pIJ321); Ampr Genr | This study |
| pIJ336 | pIJ253::fur (from pIJ319); Ampr Genr | This study |
| pIJ258 | pST6K::tnsABCD; Kanr | 55 |
| pIJ266 | pBluescript II SK(+)::lacZ′-kan-′lacA; Kanr Ampr | 46 |
| pIJ377 | pIJ266::ryhB (from pIJ321); Kanr Ampr | This study |
| pMEG-375 | sacRB mobRP4 oriR6K Catr Ampr | 50 |
| pIJ386 | pMEG-375::lacZ′-kan-ryhB-′lacA (from pIJ377); Catr Kanr Ampr | This study |
Construction of mutants and complemented strains.
Primers used for cloning and generation of mutants and complemented strains are listed in Table S1 in the supplemental material. Enzymes used for generation of constructs were purchased from Fermentas or New England BioLabs. All mutants were generated by the method described by Datsenko and Wanner (54). Briefly, using a lambda red recombination procedure, ryhB and fur were deleted and replaced by chloramphenicol and kanamycin resistance cassettes, respectively. E. coli strains EM1238 and JW0669 were used to amplify the ryhB::cat allele, using primers CMD1171 and CMD1172, and the fur::kan allele, using primers CMD290 and CMD291, respectively. The kanamycin resistance cassette was then removed by using plasmid pCP20 expressing the FLP recombinase (54). The deletion of the kanamycin resistance cassette was confirmed by PCR using primers CMD290 and CMD291.
The mutant strains were complemented by inserting the ryhB gene or the fur gene at the attTn7 site of the chromosome as described by Crépin et al. (55). Briefly, the fur and ryhB genes and their native promoters were amplified from CFT073 genomic DNA by using primer pairs CMD1269/CMD1270 and CMD1271/CMD1272, respectively. The amplified products were then inserted into the pGEM-T Easy vector (Promega), and recombinant plasmids were introduced into E. coli DH5α by electroporation. The construct containing the fur gene was verified using primer pairs CMD175/CMD176 and CMD1269/CMD1270. The construct containing the ryhB gene was verified using primer pairs CMD175/CMD176 and CMD1271/CMD1272. Plasmids harboring fur or ryhB in the direction opposite from that of the lacZ gene were used for subsequent constructions (pIJ319 and pIJ321, respectively). Plasmid pIJ319 was then digested with KpnI, whereas pIJ321 was digested with XhoI and XmaI. The fragments were inserted into the multiple-cloning site (MCS) of the mini-Tn7-containing vector pGP-Tn7-Gm digested with the corresponding restriction enzymes, generating plasmids pIJ326 (ryhB gene in the same direction as the aacC1 gene) and pIJ336 (fur gene in the direction opposite from that of the aacC1 gene). These vectors were verified by PCR using primer pair CMD1067/CMD1068 or CMD1269/CMD1270 for the fur gene and primer pair CMD1271/CMD1272 for the ryhB gene. Strain MGN-617 containing either pIJ326 or pIJ336 was then conjugated overnight at 30°C on LB agar plates supplemented with DAP, with either the ΔryhB mutant or the Δfur mutant containing the plasmid pIJ258, carrying the tnsABCD transposase genes required for transposition of Tn7 at the attTn7 site. The bacterial lawn was then serially diluted, spread on LB agar plates supplemented with gentamicin, and incubated at 37°C. Colonies were verified for sensitivity to kanamycin and ampicillin, indicating the likelihood of integration at attTn7 and the loss of pIJ258. Insertion into the attTn7 site was then verified by PCR using primer pair CMD1073/CMD1269 for complementation of the fur gene and primer pair CMD1073/CMD1272 for complementation of the ryhB gene.
The ΔryhB Δfur mutant strain was also complemented by inserting the ryhB gene at the lac site of the chromosome. Complementation at the lac site was achieved by allelic exchange. First, pIJ321 was digested with XhoI and XmaI, and the resulting restriction fragment (containing the ryhB gene) was inserted into XhoI/XmaI-digested pIJ266 to generate plasmid pIJ377. The vector pIJ266 is a pBluescript II SK(+) derivative in which a kanamycin resistance cassette was inserted between the lacZ and lacA genes. pIJ377 was then digested with AvrII to generate a fragment containing lacZ′-kan-ryhB-lacA′. This fragment was inserted into the digested pMEG-375 plasmid, a sacB-based allelic exchange plasmid (50), to generate the suicide plasmid pIJ386. This plasmid was then introduced by conjugation into strain CFT073 ΔryhB Δfur attTn7::fur (QT2639) by using MGN-617, and allelic exchange into the lac site was then verified by production of white colonies on MacConkey lactose agar plates and by PCR using primer pair CMD1394/CMD1272.
Bacterial growth experiments.
For bacterial growth experiments, overnight LB cultures were diluted 100-fold in either fresh LB medium, M63-glycerol minimal medium, or human urine, and strains were grown for 3 h at 37°C with agitation. Strains were then diluted 100-fold in the appropriate medium without antibiotics and cultured in quadruplicate at 37°C in 100-well, sterile, covered microplates containing 250 μl medium/well. Plates were then incubated at 37°C with agitation in a Bioscreen C automated microbiology growth curve analysis system (Growth Curves), and the optical density at 600 nm (OD600) was measured every 15 min for 24 h.
For coculture experiments, overnight LB cultures of strains were washed twice and resuspended in the same volume of human urine. The strains were then inoculated in equivalent numbers (each corresponding to an OD600 of 0.025) into human urine and cultured at 37°C without agitation. At different times, serial dilutions were plated on MacConkey agar to determine bacterial counts.
Hydrogen peroxide resistance assays.
Overnight LB cultures were washed with either LB medium or M63-glycerol minimal medium, and the OD600 was adjusted at 0.5 (corresponding to 107 CFU/ml). Fifty-microliter aliquots of these bacterial cultures were added to 900 μl of either LB medium or M63-glycerol minimal medium. Fifty microliters of H2O2 was then added to the growth medium, to a final concentration of 5 mM. Bacterial counts were determined at 0 min. After 30 min of incubation at 37°C, survival analysis was performed by plating serial dilutions on LB agar plates for three independent experiments for each strain.
Analyses of siderophores from culture supernatants.
For production and detection of siderophores, 6-h cultures grown in LB medium were diluted 50-fold in M63-glycerol minimal medium, and strains were cultured at 37°C with agitation. Siderophores from culture supernatants were obtained as described by Caza et al. (47). Briefly, supernatants of 17-h cultures were obtained following centrifugation of bacterial cells at 3,200 × g for 15 min and filtration of the supernatants on 0.2-μm membranes. Aliquots of 1 ml of supernatant were then prepared in 5% (vol/vol) formic acid, and 0.12 ng ml−1 of 5,6,7,8-tetradeutero-4-hydroxy-2-heptylquinoline and 0.25 ng ml−1 of 5,6,7,8-tetradeutero-3,4-dihydroxy-2-heptylquinoline were added as internal controls. Each strain was cultured in triplicate, and a sample of each culture supernatant was analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Liquid chromatography-mass spectrometry analyses.
Multiple-reaction monitoring (MRM) analyses were performed using a Waters 2795 Alliance HT instrument coupled to a Micromass Quattro Premier XE spectrometer (Micromass MS Technologies). Samples were injected onto a Kinetex 2.6-μm C8 4.6- by 100-mm column at a flow rate of 400 μl min−1, with a linear gradient of water-acetonitrile with 1% acetic acid. The analyses were performed in positive electrospray ionization mode with a cone voltage of 30 V as described by Caza et al. (47).
RNA extraction and quantitative reverse transcription-PCR (qRT-PCR).
For gene expression analysis, overnight LB cultures of strains were washed twice and resuspended in the same volume of M63-glycerol minimal medium or human urine. The strains were then diluted 50-fold in fresh M63-glycerol minimal medium and 100-fold in human urine and cultured at 37°C with agitation. RNAs were extracted from bacterial cultures grown in M63-glycerol minimal medium or in human urine from three independent experiments for each strain during the late exponential growth phase (OD600 = 0.9 and 0.3, respectively), using a Nucleospin RNAII kit (Macherey-Nagel) according to the manufacturer's instructions. To provide immediate stabilization of RNA, 2 volumes of RNA Protect (Qiagen) was added to a volume of bacterial cells. After 5 min at room temperature, samples were centrifuged at 5,000 × g for 15 min, and pellets were stored at −80°C for RNA extraction. RNAs were also extracted from infected bladders and kidneys at 6 h and 48 h postinfection (p.i.) by use of TRIzol reagent (Invitrogen) according to the manufacturer's recommendations. All RNA extractions were followed by rigorous DNase treatments with a Turbo DNA-free kit (Ambion). Total RNA concentrations were estimated using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific).
An iScript cDNA synthesis kit and an Ssofast Evagreen Supermix kit (Bio-Rad) were used for qRT-PCR experiments according to the manufacturer's instructions. Control RT-PCRs, omitting reverse transcriptase from the reaction mixture, were performed to check for DNA contamination of the RNA preparations. The tus gene was used as a housekeeping control (56). The calculated threshold cycle (CT) for each reaction was normalized to the CT of the tus gene amplified from the corresponding sample. The fold change compared to the wild-type strain was calculated using the 2−ΔΔCT method (57). Genes with a fold change above or below the defined threshold of 2 were considered to be expressed differentially. Primers used for qRT-PCR analysis are listed in Table S1 in the supplemental material.
Experimental UTIs in mice.
All animal experiments complied with the Canadian Council on Animal Care (CCAC) and the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) guidelines for the care and use of laboratory animals. The experimental protocol for experimental UTI was approved by the Institutional Committee for Protection of Animals (Comité Institutionnel de Protection des Animaux [CIPA], Centre National de Biologie Expérimentale) under protocol number CIPA 1206-03.
Experimental infections were carried out using either competition (coinfection) or single-strain infection models as described previously (56, 58). Prior to inoculation, strains were grown for 16 h at 37°C with shaking (250 rpm) in 55 ml of LB medium. For coinfection experiments, cultures of the wild-type and derivative strains were mixed 1:1. Six-week-old CBA/J female mice were inoculated transurethrally with 20 μl of a 1:1 mixture containing 5 × 108 CFU of the virulent ΔlacZYA derivative of the UPEC CFT073 strain (QT1081) and 5 × 108 CFU of either CFT073 ΔryhB (QT2230), CFT073 Δfur (QT2637), CFT073 ΔryhB Δfur (QT2634), CFT073 ΔryhB complemented with ryhB (QT2469), or CFT073 Δfur complemented with fur (QT2642). Experiments were also performed with 20 μl of a 1:1 mixture containing 5 × 108 CFU of the wild-type CFT073 strain and 5 × 108 CFU of lac-negative CFT073 ΔryhB Δfur complemented with both ryhB and fur (QT2808). At 48 h p.i., mice were euthanized, and bladders and kidneys were aseptically removed, homogenized, diluted, and plated on MacConkey agar to determine bacterial counts.
In the single-strain experimental UTI model, mice were infected as described above, but with a single culture of 1 × 109 CFU of each tested strain. Bacterial counts were determined at 6 h or 48 h p.i. Bladders and kidneys were aseptically removed and bisected; one half of each was used to determine bacterial counts, and the other half was resuspended in TRIzol reagent (Invitrogen) for RNA extractions and subsequent analyses of bacterial gene expression.
Statistical analyses.
All data were analyzed with the Prism 5.01 software package (GraphPad Software, San Diego, CA). The Wilcoxon signed-rank test (two-tailed; P ≤ 0.05) was used to determine statistical significance for coinfections. For single-strain infection experiments, the Mann-Whitney test was used. All other statistical analyses were determined by the Student t test.
RESULTS
RyhB is required for optimal colonization of the murine urinary tract.
To characterize the roles of Fur and RyhB in the pathogenesis of the UPEC strain CFT073, experimental UTIs were performed in CBA/J mice. First, coinfection experiments were performed between the wild-type CFT073 ΔlacZYA strain and one of its derivative mutant strains, i.e., the ΔryhB, Δfur, or ΔryhB Δfur strain. The CFT073 ΔlacZYA strain was previously shown to present no statistical difference from the CFT073 wild-type parent in urinary tract colonization (58). At 48 h p.i., the ΔryhB and Δfur mutants were as virulent as the wild-type strain in the bladder but were outcompeted by the latter in the kidneys (15-fold and 17-fold, respectively) (Fig. 1A). The ΔryhB Δfur double mutant showed a significantly reduced competitive index (CI) relative to the wild-type strain in both the bladder and kidneys. This mutant was outcompeted 46-fold in the bladder and 34-fold in the kidneys (Fig. 1A), indicating that the combined loss of RyhB and Fur has a cumulative effect on competitive colonization of the mouse urinary tract. To confirm the role of these regulators, we then performed coinfection experiments either with the wild-type CFT073 ΔlacZYA strain and one of the complemented strains, i.e., the ΔryhB strain complemented with ryhB (ΔryhB compl.) or the Δfur strain complemented with fur (Δfur compl.), or with the wild-type CFT073 strain and the ΔryhB Δfur strain complemented with ryhB and fur (ΔryhB Δfur compl.). As shown in Fig. 1A, all the complementations significantly improved the competitive fitness of the mutant strains in both the bladder and kidneys. The complementation studies confirmed the important roles of RyhB and Fur in the colonization of the mouse urinary tract and for satisfying molecular Koch's postulates (59).
FIG 1.
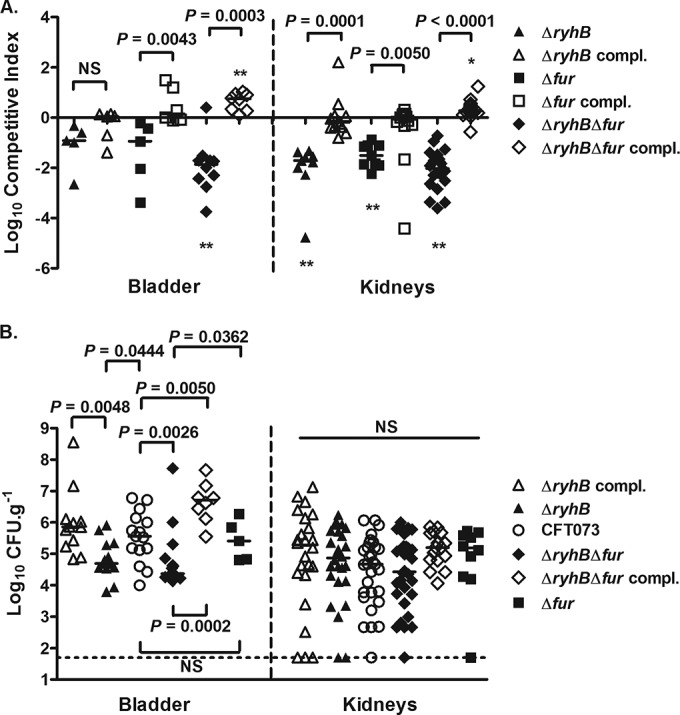
Roles of RyhB and Fur in colonization of the mouse urinary tract. (A) CBA/J mice were coinfected either with a 1:1 ratio of CFT073 ΔlacZYA and one of the mutant or complemented strains (ΔryhB, Δfur, ΔryhB Δfur, ΔryhB compl., or Δfur compl.) or with a 1:1 ratio of the wild-type CFT073 strain and the ΔryhB Δfur complemented strain (with both ryhB and fur). Bladders and kidneys were aseptically removed at 48 h p.i. The proportion of each strain was monitored, and results are represented as log10 competitive indexes (CI). The CI represent the relative numbers of the two tested strains from the tissues sampled compared to the initial numbers of the strains in the inoculum. Negative CI values indicate a decreased capacity of the mutant or complemented strain to compete with the reference strain (CFT073 or CFT073 ΔlacZYA). Horizontal bars indicate the mean log10 CI values. (B) CBA/J mice were infected with either CFT073 or one of its mutant (ΔryhB, Δfur, or ΔryhB Δfur) or complemented (ΔryhB compl. or ΔryhB Δfur compl.) strains. Bladders and kidneys were aseptically removed at 48 h p.i. Results are presented as log10 CFU g−1. Each data point represents a sample from an individual mouse, and horizontal bars indicate the median values. Each kidney was sampled separately. The dashed line represents the limit of detection of bacterial numbers. Statistically significant differences in CI were determined by the Wilcoxon matched-pair test (*, P < 0.05; **, P < 0.005). The Mann-Whitney test was used to determine statistical differences between CI values and to determine the statistical differences in single-strain infection experiments. NS, not significant.
To determine more accurately the role of each of these regulators in the pathogenesis of the UPEC strain CFT073, single-strain infection experiments were performed in CBA/J mice. The fur mutant was as virulent as the wild-type CFT073 strain and colonized the bladder and kidneys as well as the wild-type strain did (Fig. 1B). The ΔryhB and ΔryhB Δfur mutants both demonstrated significantly reduced bacterial numbers in the bladder but not in the kidneys (Fig. 1B). Because the ΔryhB Δfur mutant was significantly attenuated in the bladder compared to the Δfur mutant (Fig. 1B), this might indicate that the observed attenuation of the double mutant was due to the ryhB mutation alone. Complementation of the ΔryhB mutant with the ryhB gene or of the ΔryhB Δfur double mutant with the ryhB and fur genes significantly improved the fitness of these strains in the bladder (Fig. 1B). These results thus demonstrate the important role of iron regulators, particularly RyhB, in the pathogenesis of UPEC strain CFT073.
Since inactivation of ryhB and/or fur could affect the growth of strain CFT073 in iron-poor media, we verified that the attenuated bladder colonization of the ΔryhB mutant was not due to a growth defect, particularly in human urine. This medium may reflect nutrient availability and environmental conditions encountered in the bladder. In an iron-rich medium, all the strains grew at similar rates, except for the Δfur and ΔryhB Δfur mutants, which demonstrated a decreased final OD600 compared to the other strains (Fig. 2A). Moreover, the Δfur and ΔryhB Δfur mutants exhibited a growth lag in iron-poor medium, i.e., M63-glycerol minimal medium, compared to the wild-type strain (Fig. 2B). Growth was restored to the wild-type level in the complemented strains. The ΔryhB mutant exhibited no growth defect in this medium. However, in human urine, all of the mutant strains exhibited the same growth level as the wild-type strain (Fig. 2C). Since the ΔryhB Δfur mutant was outcompeted by the wild-type strain in coinfection experiments (Fig. 1A), we also performed an in vitro competition assay in human urine between the wild-type CFT073 ΔlacZYA strain and one of its derivative mutant strains, i.e., the ΔryhB, Δfur, or ΔryhB Δfur strain. None of the mutants was outcompeted by the wild-type strain at 3, 6, 24, and 48 h postinoculation (see Fig. S1 in the supplemental material). Hence, the attenuation of the ΔryhB and ΔryhB Δfur mutants in the bladder was not a consequence of a defect in growth in urine.
FIG 2.
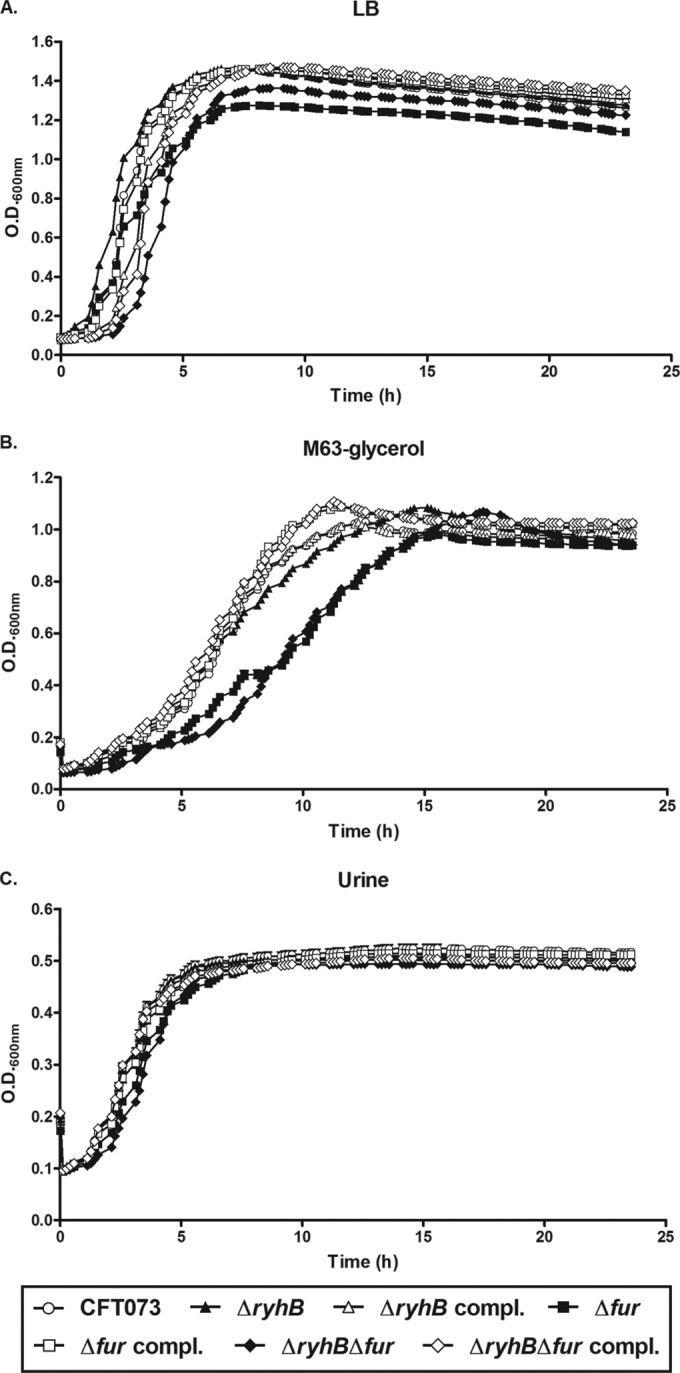
Roles of RyhB and Fur in the growth of UPEC strain CFT073. Growth curves (OD600) are shown for UPEC strain CFT073, mutant strains (ΔryhB, Δfur, and ΔryhB Δfur), and complemented strains (ΔryhB compl., Δfur compl., and ΔryhB Δfur compl.) grown with agitation at 37°C in either iron-rich LB medium (A), iron-poor M63-glycerol minimal medium (B), or human urine (C). Growth curves were obtained using a Bioscreen C apparatus. Results are the mean values and standard deviations of results from three biological experiments.
Fur, but not RyhB, is involved in oxidative stress resistance.
In nonpathogenic E. coli, RyhB regulates genes involved in oxidative stress, such as the superoxide dismutase gene sodB (1). It has also been shown that deregulation of iron homeostasis increases sensitivity to reactive oxygen species, such as H2O2, in Salmonella enterica serovar Typhi and S. Typhimurium (27, 60). Therefore, we determined whether Fur and/or RyhB contributes to H2O2 resistance in UPEC strain CFT073. Wild-type strain CFT073 and the different mutant and complemented strains were exposed to oxidative stress by addition of H2O2 to strains incubated in either iron-rich medium (LB) or iron-poor medium (M63-glycerol minimal medium). As shown in Fig. 3, deletion of ryhB did not significantly alter H2O2 sensitivity when strains were grown in either LB medium or M63-glycerol medium. In LB and M63-glycerol media, 52% and 51% of the bacterial suspension of the wild-type strain, respectively, survived, compared to 62% and 69% survival, respectively, for the ΔryhB mutant. The Δfur mutant was significantly more sensitive to H2O2 than its parental strain in LB medium (13% survival) (Fig. 3A) and in M63-glycerol medium (26% survival) (Fig. 3B). Interestingly, the additional loss of ryhB in the Δfur mutant further increased the sensitivity of the strain to H2O2, particularly in M63-glycerol minimal medium (1.38% survival) (Fig. 3B). H2O2 resistance was restored to the wild-type level in all the complemented strains (Fig. 3), confirming the predominant role of Fur in oxidative stress resistance. These data thus indicate that the role of RyhB alone for optimal colonization of the murine urinary tract is not due to an increase in sensitivity to oxidative stress.
FIG 3.
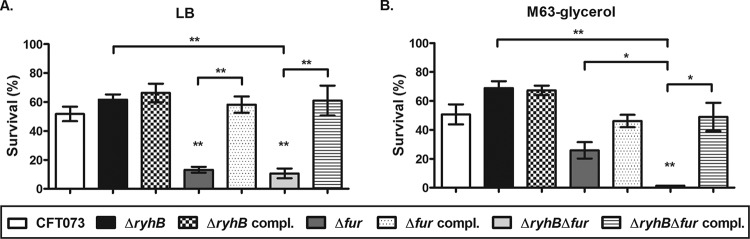
Roles of RyhB and Fur in oxidative stress resistance. UPEC strain CFT073, mutant strains (ΔryhB, Δfur, and ΔryhB Δfur), and complemented strains (ΔryhB compl., Δfur compl., and ΔryhB Δfur compl.) were exposed to 5 mM H2O2 for 30 min in either iron-rich LB medium (A) or iron-poor M63-glycerol minimal medium (B). Results are presented as percentages of surviving bacteria after 30 min of H2O2 exposure and are the mean values and standard deviations of results from three biological experiments. Statistical significance was calculated by the Student t test (*, P < 0.05; **, P < 0.005).
RyhB promotes siderophore production, whereas Fur represses it.
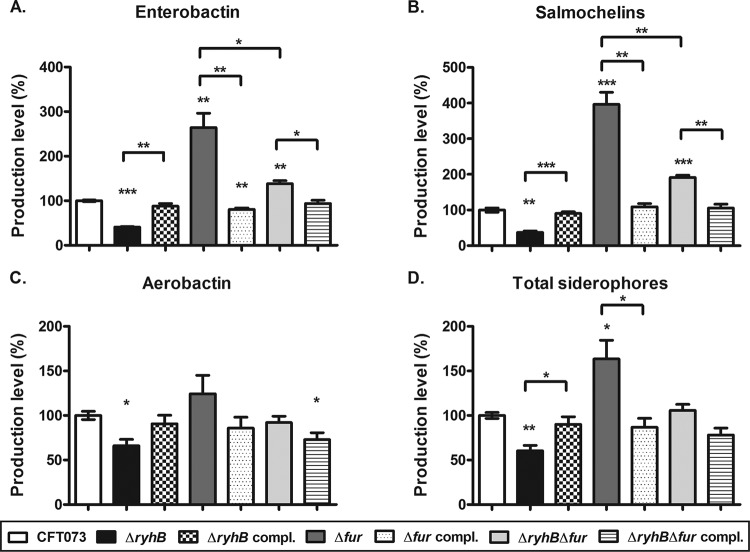
Because several studies on nonpathogenic E. coli and Salmonella enterica previously demonstrated the roles of Fur and RyhB in siderophore production (18, 19, 27), we determined whether these regulators have an impact on siderophore production in the UPEC strain CFT073. We compared production of the three types of siderophores by strain CFT073 (enterobactin, salmochelins, and aerobactin) directly from supernatants of bacterial cultures grown in iron-poor medium. As shown in Fig. 4, the ΔryhB mutant produced lower levels of siderophores than the wild-type strain. This mutant secreted 2.45-fold, 2.7-fold, and 1.51-fold less enterobactin, salmochelins, and aerobactin, respectively, than the wild-type strain (Fig. 4A to C). The ΔryhB mutant produced 1.65-fold less total siderophores than the wild-type strain (Fig. 4D). The Δfur mutant produced 2.64-fold more enterobactin, 3.97-fold more salmochelins, and 1.24-fold more aerobactin than the wild-type strain (Fig. 4A to C). The Δfur mutant produced 1.63-fold more total siderophores than the wild-type strain (Fig. 4D). Deletion of ryhB in the Δfur mutant resulted in a decrease in siderophore production levels to levels more similar to those of the wild-type strain (Fig. 4), even if enterobactin and salmochelin production was still higher in this strain than in the wild-type strain (1.38-fold and 1.91-fold higher, respectively) (Fig. 4A and B). Complementation of the mutant strains with ryhB (ΔryhB compl.), fur (Δfur compl.), or ryhB and fur (ΔryhBΔfur compl.) restored siderophore production to levels similar to those observed in the wild-type strain (Fig. 4). The complemented strains regained wild-type production levels, although enterobactin production in the fur-complemented strain was significantly less than that in the wild-type strain and aerobactin production was significantly less in the ryhB- and fur-complemented strain (1.23-fold and 1- to 39-fold less, respectively) (Fig. 4A and C). Overall, these results demonstrate that RyhB and Fur have opposite effects on siderophore production by UPEC strain CFT073.
FIG 4.
Roles of RyhB and Fur in siderophore production. Production of siderophores secreted into the supernatants of UPEC strain CFT073, mutant strains (ΔryhB, Δfur, and ΔryhB Δfur), and complemented strains (ΔryhB compl., Δfur compl., and ΔryhB Δfur compl.) in iron-poor M63-glycerol minimal medium was quantified by LC-MS analyses. Production levels of enterobactin (A), salmochelins (B), aerobactin (C), and total siderophores (D) are presented as the percentages produced by each strain compared to the wild-type strain. Results are the mean values and standard deviations of results from three biological experiments. Statistical significance was calculated by the Student t test (*, P < 0.05; **, P < 0.005; ***, P < 0.0001).
Since RyhB and Fur regulate the production of the 3 types of siderophores produced by UPEC strain CFT073, we then compared the relative amounts of each of these siderophores produced by this strain. In both the wild-type and all mutant and complemented strains, aerobactin was the predominant siderophore. In terms of total siderophore production, the CFT073 strain secreted 78.9% aerobactin, 13.3% enterobactin, and 7.8% salmochelins, as shown in Fig. 5. ryhB deletion did not significantly alter the proportion of siderophores (86.2% aerobactin, 9% enterobactin, and 4.8% salmochelins) (Fig. 5). However, in the Δfur mutant, the proportion of aerobactin decreased considerably (59.4%), whereas the portion of catecholate siderophores increased correspondingly, with enterobactin representing 21.4% and salmochelins 19.2% of the siderophores (Fig. 5). Alteration of siderophore production by RyhB may explain the attenuation of the ΔryhB mutant in a single-strain murine model of UTI (Fig. 1B).
FIG 5.
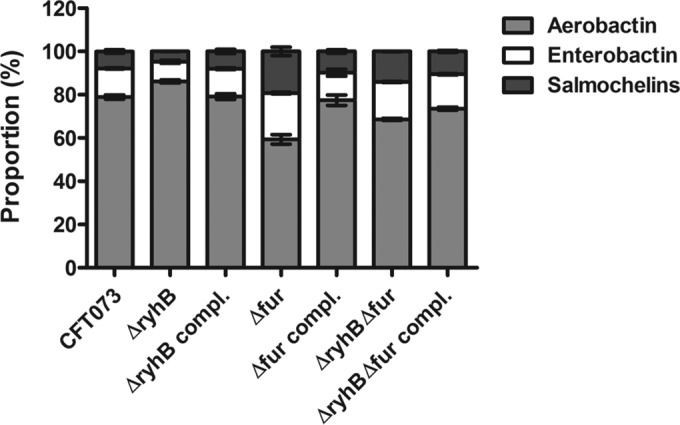
Proportions of siderophores produced by UPEC strain CFT073 and derivatives. Production of siderophores secreted into the supernatants of UPEC strain CFT073, mutant strains (ΔryhB, Δfur, and ΔryhB Δfur), and complemented strains (ΔryhB compl., Δfur compl., and ΔryhB Δfur compl.) in iron-poor M63-glycerol minimal medium was quantified by LC-MS analyses. For each strain, results are presented as the percentage of each siderophore produced compared to the total siderophore production level. Results are the mean values and standard deviations of results from three biological experiments.
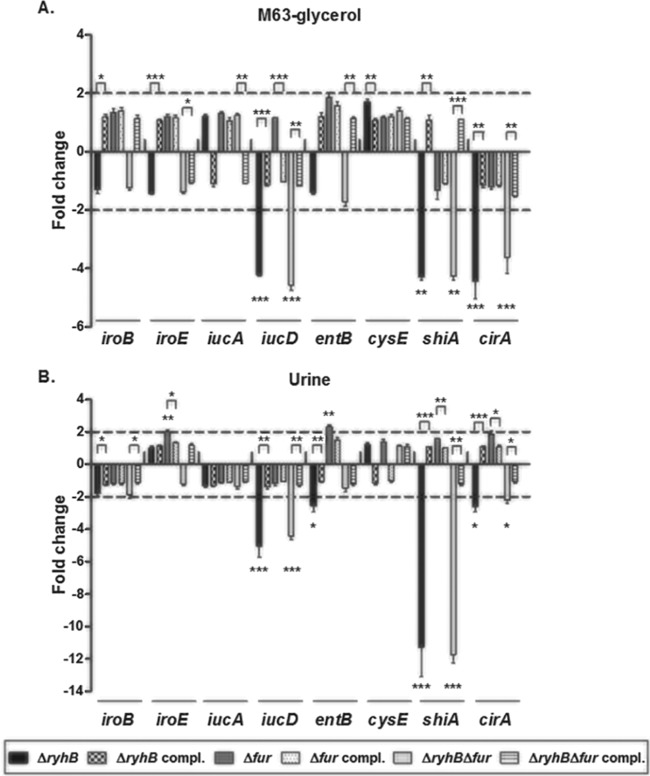
RyhB activates siderophore gene expression in vitro and in human urine.
Previous results tend to demonstrate that Fur may be able to repress genes involved in siderophore biosynthesis, whereas RyhB may activate these genes, in the UPEC strain CFT073. To verify this hypothesis, qRT-PCR experiments were performed to measure expression of siderophore biosynthesis genes. We thus performed these experiments on RNAs extracted from strains cultured in M63-glycerol minimal medium. RyhB has already been shown to regulate the cysE and shiA genes, encoding a serine acetyltransferase and a shikimate permease, respectively, in nonpathogenic E. coli (18, 19). These proteins promote serine and shikimate availability, and both substrates are required for enterobactin synthesis. Therefore, we determined the expression of these genes as well as a gene directly involved in enterobactin biosynthesis, entB, and another known RyhB target, cirA, a DHBS receptor-encoding gene (20). Because UPEC CFT073 produces both salmochelins and aerobactin in addition to enterobactin, we also investigated the expression of genes involved in production of these siderophores, i.e., iroB and iroE for salmochelins and iucA and iucD for aerobactin. During the late exponential phase of growth, expression of iucD was downregulated 4.2-fold in the ΔryhB mutant and 4.6-fold in the ΔryhB Δfur mutant compared to the level in the wild-type strain (Fig. 6A). Moreover, expression of shiA was downregulated 4.3-fold in both the ΔryhB mutant and the ΔryhB Δfur mutant compared to level in the wild-type strain. Expression of cirA was downregulated 4.4-fold and 3.6-fold in the ΔryhB and ΔryhB Δfur mutants, respectively (Fig. 6A). None of the other genes tested were differentially expressed between the ΔryhB mutants and the wild-type strain. Since M63-glycerol minimal medium is an iron-poor medium, it is not surprising that expression of all siderophore genes was not significantly different in the Δfur mutant compared to the wild-type strain (Fig. 6A). Complementation with ryhB or ryhB and fur restored expression of differentially expressed genes (iucD, shiA, and cirA) to the wild-type level (Fig. 6A). Moreover, even though iroB, iroE, and cysE were not differentially expressed between the ΔryhB mutant and the wild-type strain, complementation of this mutant with ryhB resulted in minor changes in gene expression, as the fold changes of expression were significantly different between the ΔryhB mutant and its complemented strain. Differential expression was also observed between the ΔryhB Δfur mutant and its complemented strain for iroE, iucA, and entB (Fig. 6A). This suggests that RyhB may have an impact on iroB, iroE, iucA, entB, and cysE expression, but this effect was minor in the medium used, as no differences were observed between the ΔryhB mutant and the wild-type strain. Finally, all of these results correlate with the decrease in siderophore production levels observed in the ΔryhB strain (Fig. 4).
FIG 6.
Roles of RyhB and Fur in expression of siderophore genes in M63-glycerol minimal medium and human urine. Expression of genes for the mutant and complemented strains in M63-glycerol minimal medium (A) or in human urine (B) was compared to that of the wild-type CFT073 strain. The dashed lines correspond to the cutoffs for a significant difference in expression. Results are the mean values and standard deviations of results from three biological experiments. Statistical significance was calculated by the Student t test (*, P < 0.05; **, P < 0.005; ***, P < 0.0001).
Since we observed an attenuation of the ΔryhB mutant in colonization of the bladder in the murine UTI model, we investigated whether RyhB also plays a role in siderophore gene expression in human urine. In human urine, ryhB was expressed the same as in M63-glycerol medium in the wild-type strain (data not shown). As shown in Fig. 6B, expression of iucD, shiA, and cirA was also downregulated in human urine for the ΔryhB and ΔryhB Δfur mutants (4.9-fold and 4.4-fold less than the wild-type level, respectively, for iucD, 10.7-fold and 11.7-fold less than the wild-type level, respectively, for shiA, and 2.6-fold and 2.2-fold less than the wild-type level, respectively, for cirA). Moreover, expression of entB was downregulated 2.5-fold in the ΔryhB mutant in human urine (Fig. 6B). Once again, expression of iucD, entB, shiA, and cirA was also restored in the corresponding complemented strains (Fig. 6B). Differential fold changes were observed between the ΔryhB and ΔryhB Δfur mutants and their respective complemented strains for iroB (Fig. 6B), indicating once again that RyhB may have a slight effect on gene expression. In contrast to the case in minimal medium, no difference was observed for iroE, iucA, and cysE expression between the ΔryhB and ΔryhB Δfur mutants and their respective complemented strains (Fig. 6B). This suggests that differences in iron concentration or other components in urine compared to minimal medium may result in differences in regulation by RyhB. We also observed that iroE and entB expression was upregulated 2-fold and 2.3-fold, respectively, in the Δfur mutant compared to the wild-type strain grown in human urine and that this expression was restored to wild-type levels in the complemented fur strain (Fig. 6B). As no differences were observed between the Δfur mutant and the wild-type strain in M63-glycerol medium (Fig. 6A), this suggests that human urine contains somewhat more iron than minimal medium.
RyhB regulates iucD expression in infected bladders of mice.
Finally, to understand the role of RyhB in the virulence of the UPEC strain CFT073, we determined if expression of genes implicated in siderophore production was also regulated by RyhB and/or Fur in vivo. We quantified siderophore gene expression in the bladders and kidneys of infected mice at 48 h p.i., i.e., when the ΔryhB and ΔryhB Δfur mutants were attenuated in the bladders of mice (Fig. 1B). First, we quantified ryhB expression in bladders and kidneys of mice infected with the wild-type strain and found that ryhB was not expressed in infected kidneys but was detected in all infected bladders (data not shown). This suggests that RyhB may not be as important in kidneys during colonization at 48 h p.i. At this time of infection, we were also unable to detect iroE, shiA, and cirA expression in either bladders or kidneys of infected mice for any of the strains. Interestingly, iucD expression was downregulated 10.7-fold in the ΔryhB mutant and 14.5-fold in the ΔryhB Δfur mutant compared to the wild-type strain in infected bladders (Fig. 7A). However, we were able to quantify iucD expression for only 3/5 mice infected with the ΔryhB and ΔryhB Δfur mutants, consistent with the effect of ryhB on downregulation of iucD. Furthermore, iucD was also expressed 2.6-fold less in the Δfur mutant than in the wild-type strain, though this was not a significant difference (Fig. 7A). This unexpected impact of fur mutation on iucD expression might be due to an indirect effect, given the large number of genes that are regulated by Fur. A Fur-regulated gene differentially expressed in the Δfur mutant may affect iucD expression. Expression of iroB, iucA, entB, and cysE was not significantly different from that in the wild-type strain in the ΔryhB and ΔryhB Δfur mutants, but iroB was expressed 2.2-fold more in the Δfur mutant than in the wild-type strain (Fig. 7A). Finally, these results indicate that the ΔryhB mutant demonstrates a reduced expression of iucD in vivo, which may result in lower production of aerobactin during infection of the bladder.
FIG 7.
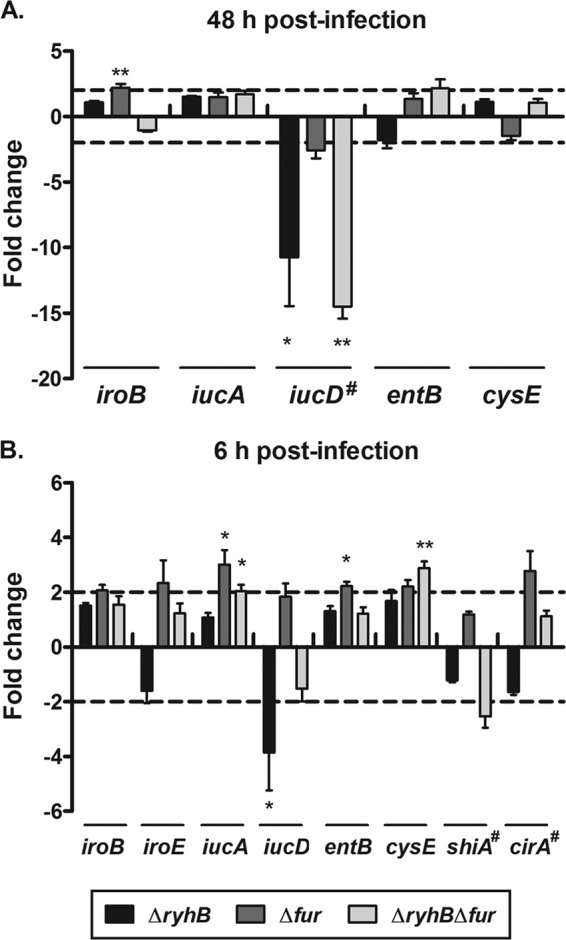
Roles of RyhB and Fur in expression of siderophore genes in infected bladders of mice. Expression of genes for the mutant strains in infected bladders at 48 h p.i. (A) and 6 h p.i. (B) was compared to that of the wild-type CFT073 strain. The dashed lines correspond to the cutoffs for a significant difference in expression. Results are the mean values and standard deviations of results for five bladders infected with each strain. Statistical significance was calculated by the Student t test (*, P < 0.05; **, P < 0.005). #, these genes were detected in 3/5 bladders infected with the ΔryhB and ΔryhB Δfur mutants.
Because several genes implicated in siderophore production were not expressed in infected bladders at 48 h p.i., we investigated their expression in mice infected by the wild-type CFT073 strain or one of its derivative mutants at an early stage postinfection. We thus performed single-strain infection experiments in CBA/J mice and analyzed bacterial numbers and gene expression at 6 h p.i. As shown in Fig. 8, the ΔryhB mutant colonized the bladder and kidneys as well as the wild-type strain did. In contrast, the Δfur and ΔryhB Δfur mutants both demonstrated significantly reduced bacterial numbers in the bladder, and the ΔryhB Δfur mutant was also attenuated in the kidneys (Fig. 8). This suggests that Fur but not RyhB is important for establishment of infection and that RyhB but not Fur is important for persistence within the urinary tract, as only the ΔryhB mutant was attenuated at 48 h p.i. (Fig. 1B). We then quantified siderophore gene expression in the bladders of infected mice at 6 h p.i. At this stage, ryhB was also expressed in all bladders of mice infected with the wild-type strain (data not shown). In contrast to the case at 48 h p.i., we were able to detect iroE, shiA, and cirA expression in any of the strains. Expression of iroB, iroE, entB, shiA, and cirA was not significantly different from that in the wild-type strain in the ΔryhB and ΔryhB Δfur mutants, but iucA and cysE were expressed 2-fold and 2.9-fold more, respectively, in the ΔryhB Δfur mutant than in the wild-type strain (Fig. 7B). iucA and entB were also expressed 3-fold and 2.2-fold more in the Δfur mutant than in the wild-type strain (Fig. 7B). Finally, iucD expression was also downregulated 3.9-fold at this stage in the ΔryhB mutant compared to the wild-type strain (Fig. 7B). As the ΔryhB mutant was not attenuated in bladders at 6 h p.i., these results indicate that aerobactin may not be important initially for the establishment of infection, but it might be required for bacterial persistence in the bladder.
FIG 8.
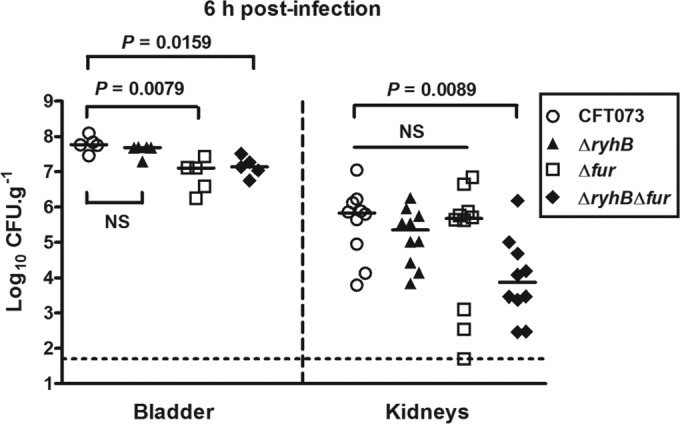
Roles of RyhB and Fur in colonization of the mouse urinary tract at 6 h postinfection. CBA/J mice were infected with either CFT073 or one of its mutants (ΔryhB, Δfur, or ΔryhB Δfur), and bladders and kidneys were aseptically removed at 6 h p.i. Results are presented as log10 CFU g−1. Each data point represents a sample from an individual mouse, and horizontal bars indicate the median values. Each kidney was sampled separately. The Mann-Whitney test was used to determine the statistical differences. The dashed line represents the limit of detection of bacterial numbers. NS, not significant.
DISCUSSION
In this study, we examined the importance of the sRNA RyhB for iron acquisition and virulence of UPEC strain CFT073. The ΔryhB mutant was attenuated for infection of the bladder, indicating that strain CFT073 requires RyhB in vivo. The ΔryhB mutant also produced lower levels of siderophores (enterobactin, salmochelins, and aerobactin) than the wild-type strain, demonstrating the importance of RyhB for iron acquisition in a pathogenic E. coli strain. Moreover, the iucD gene, involved in aerobactin synthesis, was downregulated in the ΔryhB mutant grown in minimal medium, human urine, and infected bladders. These findings suggest that, in vivo, RyhB mediates iron uptake via the regulation of siderophore gene expression, especially that of aerobactin, to allow colonization and multiplication within the urinary tract.
Whereas this is the first demonstration of the impact of RyhB in the virulence of an ExPEC strain, several studies have demonstrated that this sRNA is implicated in virulence-associated processes of other pathogenic bacteria. In S. flexneri, RyhB represses the expression of ydeP, a gene required for the virulence-associated phenotype of extreme acid resistance (21). In S. dysenteriae, effector protein secretion, plaque formation, and invasion of eukaryotic epithelial cells are suppressed by RyhB through regulation of virB (23). In V. cholerae, RyhB modulates the expression of several genes controlling motility, chemotaxis, and biofilm formation. However, a ΔryhB mutant is still able to infect mice (24, 25). Production of capsular polysaccharide and iron acquisition systems, two factors required for survival within the human host, are regulated by RyhB in K. pneumoniae (26). S. enterica encodes two homologs of RyhB, RyhB-1 and RyhB-2 (also termed RfrA and RfrB). They are involved in protection against oxidative stress, bactericidal antibiotic and acid resistance, and survival within epithelial cells and macrophages (7, 61). RyhB-2 is also implicated in the regulation of motility in S. Typhimurium, and the highest expression of both sRNAs in S. Typhi was obtained during interaction with host cells, which correlates with their role in virulence (27, 62).
The role of RyhB in the virulence of the UPEC strain CFT073 can be linked to its effect on siderophore production, as the levels of all siderophores secreted by this strain, i.e., enterobactin, salmochelins, and aerobactin, were decreased in the ΔryhB mutant. The enterobactin decrease was correlated with the role of RyhB in the expression of genes involved in production of this siderophore. shiA and cirA were both repressed in the ΔryhB and ΔryhB Δfur mutants, which corresponds to what was previously observed in a nonpathogenic E. coli K-12 strain (18, 20). For that strain, the authors presented evidence that under conditions of iron limitation, RyhB base pairs with shiA and cirA mRNAs, thereby favoring translation and transcript stabilization. Since ryhB, shiA, and cirA are conserved between nonpathogenic strains and CFT073, we can hypothesize that RyhB regulation of cirA and shiA in CFT073 depends on the same regulatory mechanism. However, whereas RyhB expression allows normal expression of entCEBAH in E. coli K-12 grown in M63 minimal medium (19), this sRNA had no significant effect on entB expression in UPEC CFT073 cells grown in the same medium. In contrast, RyhB was important for normal expression of entB in human urine, indicating that, depending on the medium, RyhB may also influence ent expression in UPEC strains, such as CFT073. RyhB also represses cysE expression in nonpathogenic E. coli and reorients the amino acid metabolism toward enterobactin siderophore production (19). Despite a difference in cysE expression between the CFT073 ΔryhB mutant and its complemented strain in M63-glycerol minimal medium, cysE was not significantly differentially expressed in the ΔryhB mutant compared to the wild-type strain in M63-glycerol minimal medium, human urine, or infected bladders. In nonpathogenic E. coli, RyhB pairs at the ribosome-binding site of cysE mRNA, thus resulting in the inhibition of translation initiation (19). Since the cysE gene is identical between nonpathogenic K-12 strains and CFT073, RyhB should be able to pair with cysE mRNA in CFT073. In strain CFT073, other environmental cues might be necessary to allow RyhB pairing with cysE mRNA. Even if RyhB had no effect on cysE expression in CFT073 under the tested conditions, enterobactin production was diminished in the ΔryhB mutant as well as in the nonpathogenic strain (19). Since salmochelins are glycosylated forms of enterobactin, it is not surprising that salmochelin production was also reduced in a ΔryhB mutant. Moreover, shiA encodes a permease of shikimate, an intermediary metabolite of aromatic amino acid, folic acid, ubiquinone, and enterobactin biosynthesis. Since RyhB regulates shiA expression in human urine, we speculate that downregulation of shiA would impair shikimate transport and negatively affect enterobactin and salmochelin production in this medium.
The CFT073 ΔryhB mutant also secreted less aerobactin, a patho-specific siderophore, than that seen with the wild-type strain, and iucD was downregulated in the ΔryhB and ΔryhB Δfur mutants compared to the wild-type strain in M63-glycerol minimal medium, human urine, and infected bladders. Aerobactin is commonly produced by E. coli strains isolated from patients with UTI, bacteremia, or other extraintestinal infections (63). More recently, it was shown that 52% of UPEC strains isolated from hospitalized and ambulatory patients possess aerobactin genes (43). The role of aerobactin in the virulence of ExPEC is well documented for UPEC and avian-pathogenic E. coli (APEC) strains. Indeed, aerobactin is involved in the virulence of APEC strains χ7122 and E058, as different iuc mutants presented decreased colonization of several organs during systemic infection of poultry (45, 46, 50, 51). An iucD mutant of the UPEC strain U17 was also attenuated in a chicken model of colibacillosis (45). In a murine model of infection, a mutant of the UPEC strain CFT073 deficient in enterobactin and aerobactin production was also attenuated in the colonization of bladders and kidneys (48). Moreover, transcriptomic analyses of CFT073 during UTI in mice demonstrated that iucD was upregulated in vivo (64). Altogether, these studies demonstrate the important role of this siderophore in virulence of ExPEC strains. Because iucD was downregulated in the CFT073 ΔryhB mutant both in vitro and in vivo, attenuation of this mutant in the murine UTI model can be correlated with a decreased production of aerobactin. Moreover, in Shigella flexneri, combined deletion of iucD and ferrous iron acquisition systems (feoB and/or sitA) leads to smaller plaque formation on Henle cell monolayers (65). A recent study also demonstrated that deletion of iucA in hypervirulent Klebsiella pneumoniae leads to less growth in human ascites fluid and to less virulence in mice (66).
Aerobactin is produced sequentially by the proteins IucD, IucB, IucA, and IucC, which are expressed from the iucABCD operon. A surprising result is that RyhB is involved in iucD expression but not iucA expression, although both genes are localized on the same operon. Expression of this operon is controlled by a Fur-regulated promoter upstream of iucA (67). During a transcriptomic analysis of CFT073 during UTI, Snyder et al. showed that the relative expression of iucD in vivo was higher than the relative expression of the other 3 genes of the operon (64). These data, together with the results obtained in our study, suggest that within the operon, only the iucD gene is specifically subjected to a direct or indirect effect of RyhB. To confirm this hypothesis, we quantified iucB and iucC gene expression in the wild-type CFT073 strain and its derivative mutant strains grown in M63-glycerol minimal medium. These genes were not differentially expressed in either the ΔryhB, Δfur, or ΔryhB Δfur mutant compared to the wild-type strain (see Fig. S2 in the supplemental material), confirming that iucD is the only gene within the operon that is subjected to RyhB regulation. In silico analyses using RNA Hybrid (68) allowed the identification of putative RyhB targets within the iucCD genes (data not shown). While it has already been shown that RyhB is able to stabilize the polycistronic mRNA of the vibrioferrin siderophore in Vibrio parahaemolyticus (69), the mechanism allowing aerobactin regulation by RyhB does not involve stabilization of all of the polycistronic mRNA. Moreover, iucD gene expression may depend on an as yet unidentified promoter upstream of this gene that RyhB may affect directly or indirectly. A more in-depth analysis will be needed to determine exactly how RyhB contributes to regulation of expression of iucD without affecting expression of other genes within the same operon.
We present evidence that the Fur protein does not have an important impact on the virulence of the UPEC strain CFT073, as a Δfur mutant was not attenuated in a single-strain infection model of UTI in mice. Previous reports have shown that several Fur-regulated genes involved in iron acquisition are upregulated during urinary tract infection, indicating iron limitation, and thus Fur inactivation, in the urinary tract (64). This is in accordance with the lack of impact of the fur mutation observed in this study for the single-strain infection model. The Δfur mutant was, however, outcompeted by the wild-type strain in kidneys during coinfection experiments, suggesting that the Fur regulon is nevertheless important for competing efficiently during UTI in the murine model. The Fur protein contributes to virulence of many bacterial pathogens, but no studies have yet demonstrated the role of this regulator in ExPEC virulence in animal models (16). This is also somewhat surprising, as many enzymes involved in oxidative stress resistance and protection against reactive oxygen species, a virulence-associated determinant of pathogenic bacteria, are regulated by Fur (16). For CFT073, we demonstrated that the Δfur mutant was more sensitive to H2O2 than the wild-type parental strain in iron-poor and iron-rich media. Similarly, for S. Typhi, it has been shown that the sRNAs RyhB-1 and RyhB-2 are responsible for sensitivity of the Δfur mutant to H2O2, as enzymes required in response to oxidative stress resistance, such as superoxide dismutases, are repressed by RyhB (5, 27). However, for CFT073, the ΔryhB Δfur double mutant was more sensitive to H2O2 than the Δfur mutant in iron-rich medium and iron-poor medium, suggesting that other, unknown mechanisms are involved in the oxidative stress response in this strain. This also highlights that deregulation of iron homeostasis, in addition to oxidative stress, is detrimental for the ΔryhB Δfur double mutant.
During the course of infection, pathogenic bacteria encounter various environments that differ in iron availability. Their survival is mediated in part by their capacity to respond precisely and rapidly to these changes in metal availability in order to efficiently control its acquisition, storage, and utilization. sRNAs are an efficient way to respond to environmental cues, as they are expressed rapidly and require only a fraction of the energy necessary to produce proteins. More and more studies highlight the role of the iron-regulated sRNA RyhB in iron homeostasis and virulence of pathogenic bacteria, demonstrating the importance of such a regulator. Several factors are crucial for UPEC fitness and virulence, such as iron and heme acquisition or metabolism (48, 70, 71). For example, some enzymes of the TCA cycle are known targets of RyhB in E. coli K-12 (5). It would thus be interesting to identify new RyhB targets in UPEC strains, as this sRNA may be a common regulator between different pathways of nutrient acquisition and metabolism. The role of RyhB in iron homeostasis and virulence of pathogenic E. coli strains and other bacterial pathogens merits further attention to more fully elucidate its mechanism of action and to discover targets which, in addition to conserved iron homeostasis and metabolic functions, may also include pathogen-specific systems that contribute to colonization and survival during infection of the host. Based on the importance of this regulatory RNA for iron homeostasis and virulence, this sRNA may represent a target of choice to counter pathogenic E. coli and other bacterial pathogens.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Fondation Armand-Frappier (G.P.), a Natural Sciences and Engineering Research Council Canada (NSERC) discovery grant (RGPIN 250129-07) (C.M.D.), the Centre de Recherche en Infectiologie Porcine et Aviaire (C.M.D.), the Canada Research Chairs program (C.M.D.), and a team grant from the Fonds Québécois de la Recherche sur la Nature et les Technologies (C.M.D., F.D., and E.M.).
Footnotes
Published ahead of print 22 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02287-14.
REFERENCES
- 1.Massé E, Gottesman S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:4620–4625. 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parrow NL, Fleming RE, Minnick MF. 2013. Sequestration and scavenging of iron in infection. Infect. Immun. 81:3503–3514. 10.1128/IAI.00602-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cassat JE, Skaar EP. 2013. Iron in infection and immunity. Cell Host Microbe 13:509–519. 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews SC, Robinson AK, Rodriguez-Quinones F. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215–237. 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 5.Massé E, Vanderpool CK, Gottesman S. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187:6962–6971. 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salvail H, Massé E. 2012. Regulating iron storage and metabolism with RNA: an overview of posttranscriptional controls of intracellular iron homeostasis. Wiley Interdiscip. Rev. RNA 3:26–36. 10.1002/wrna.102. [DOI] [PubMed] [Google Scholar]
- 7.Oglesby-Sherrouse AG, Murphy ER. 2013. Iron-responsive bacterial small RNAs: variations on a theme. Metallomics 5:276–286. 10.1039/c3mt20224k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hantke K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172–177. 10.1016/S1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- 9.McHugh JP, Rodriguez-Quinones F, Abdul-Tehrani H, Svistunenko DA, Poole RK, Cooper CE, Andrews SC. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478–29486. 10.1074/jbc.M303381200. [DOI] [PubMed] [Google Scholar]
- 10.Braun V. 2001. Iron uptake mechanisms and their regulation in pathogenic bacteria. Int. J. Med. Microbiol. 291:67–79. 10.1078/1438-4221-00103. [DOI] [PubMed] [Google Scholar]
- 11.Carpenter BM, Whitmire JM, Merrell DS. 2009. This is not your mother's repressor: the complex role of fur in pathogenesis. Infect. Immun. 77:2590–2601. 10.1128/IAI.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calderwood SB, Mekalanos JJ. 1987. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J. Bacteriol. 169:4759–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellermeier JR, Slauch JM. 2008. Fur regulates expression of the Salmonella pathogenicity island 1 type III secretion system through HilD. J. Bacteriol. 190:476–486. 10.1128/JB.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ochsner UA, Vasil AI, Vasil ML. 1995. Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin A expression: purification and activity on iron-regulated promoters. J. Bacteriol. 177:7194–7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. 1995. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J. Bacteriol. 177:2305–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Troxell B, Hassan HM. 2013. Transcriptional regulation by ferric uptake regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 3:59. 10.3389/fcimb.2013.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massé E, Salvail H, Desnoyers G, Arguin M. 2007. Small RNAs controlling iron metabolism. Curr. Opin. Microbiol. 10:140–145. 10.1016/j.mib.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Prevost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Massé E. 2007. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol. Microbiol. 64:1260–1273. 10.1111/j.1365-2958.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- 19.Salvail H, Lanthier-Bourbonnais P, Sobota JM, Caza M, Benjamin JA, Mendieta ME, Lepine F, Dozois CM, Imlay J, Massé E. 2010. A small RNA promotes siderophore production through transcriptional and metabolic remodeling. Proc. Natl. Acad. Sci. U. S. A. 107:15223–15228. 10.1073/pnas.1007805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salvail H, Caron MP, Belanger J, Massé E. 2013. Antagonistic functions between the RNA chaperone Hfq and an sRNA regulate sensitivity to the antibiotic colicin. EMBO J. 32:2764–2778. 10.1038/emboj.2013.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oglesby AG, Murphy ER, Iyer VR, Payne SM. 2005. Fur regulates acid resistance in Shigella flexneri via RyhB and ydeP. Mol. Microbiol. 58:1354–1367. 10.1111/j.1365-2958.2005.04920.x. [DOI] [PubMed] [Google Scholar]
- 22.Africa LA, Murphy ER, Egan NR, Wigley AF, Wing HJ. 2011. The iron-responsive Fur/RyhB regulatory cascade modulates the Shigella outer membrane protease IcsP. Infect. Immun. 79:4543–4549. 10.1128/IAI.05340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy ER, Payne SM. 2007. RyhB, an iron-responsive small RNA molecule, regulates Shigella dysenteriae virulence. Infect. Immun. 75:3470–3477. 10.1128/IAI.00112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis BM, Quinones M, Pratt J, Ding Y, Waldor MK. 2005. Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. J. Bacteriol. 187:4005–4014. 10.1128/JB.187.12.4005-4014.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mey AR, Craig SA, Payne SM. 2005. Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect. Immun. 73:5706–5719. 10.1128/IAI.73.9.5706-5719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang SH, Wang CK, Peng HL, Wu CC, Chen YT, Hong YM, Lin CT. 2012. Role of the small RNA RyhB in the Fur regulon in mediating the capsular polysaccharide biosynthesis and iron acquisition systems in Klebsiella pneumoniae. BMC Microbiol. 12:148. 10.1186/1471-2180-12-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leclerc JM, Dozois CM, Daigle F. 2013. Role of the Salmonella enterica serovar Typhi Fur regulator and small RNAs RfrA and RfrB in iron homeostasis and interaction with host cells. Microbiology 159:591–602. 10.1099/mic.0.064329-0. [DOI] [PubMed] [Google Scholar]
- 28.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140. 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 29.Ron EZ. 2006. Host specificity of septicemic Escherichia coli: human and avian pathogens. Curr. Opin. Microbiol. 9:28–32. 10.1016/j.mib.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Russo TA, Johnson JR. 2000. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 181:1753–1754. 10.1086/315418. [DOI] [PubMed] [Google Scholar]
- 31.Smith JL, Fratamico PM, Gunther NW. 2007. Extraintestinal pathogenic Escherichia coli. Foodborne Pathog. Dis. 4:134–163. 10.1089/fpd.2007.0087. [DOI] [PubMed] [Google Scholar]
- 32.Johnson JR, Russo TA. 2002. Extraintestinal pathogenic Escherichia coli: “the other bad E coli.” J. Lab. Clin. Med. 139:155–162. 10.1067/mlc.2002.121550. [DOI] [PubMed] [Google Scholar]
- 33.Ulett GC, Totsika M, Schaale K, Carey AJ, Sweet MJ, Schembri MA. 2013. Uropathogenic Escherichia coli virulence and innate immune responses during urinary tract infection. Curr. Opin. Microbiol. 16:100–107. 10.1016/j.mib.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Brumbaugh AR, Mobley HL. 2012. Preventing urinary tract infection: progress toward an effective Escherichia coli vaccine. Expert Rev. Vaccines 11:663–676. 10.1586/erv.12.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foxman B. 2003. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Dis. Mon. 49:53–70. 10.1067/mda.2003.7. [DOI] [PubMed] [Google Scholar]
- 36.Nielubowicz GR, Mobley HL. 2010. Host-pathogen interactions in urinary tract infection. Nat. Rev. Urol. 7:430–441. 10.1038/nrurol.2010.101. [DOI] [PubMed] [Google Scholar]
- 37.Garenaux A, Caza M, Dozois CM. 2011. The ins and outs of siderophore mediated iron uptake by extra-intestinal pathogenic Escherichia coli. Vet. Microbiol. 153:89–98. 10.1016/j.vetmic.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 38.Porcheron G, Garenaux A, Proulx J, Sabri M, Dozois CM. 2013. Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence. Front. Cell. Infect. Microbiol. 3:90. 10.3389/fcimb.2013.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carbonetti NH, Williams PH. 1984. A cluster of five genes specifying the aerobactin iron uptake system of plasmid ColV-K30. Infect. Immun. 46:7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Lorenzo V, Bindereif A, Paw BH, Neilands JB. 1986. Aerobactin biosynthesis and transport genes of plasmid ColV-K30 in Escherichia coli K-12. J. Bacteriol. 165:570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fischbach MA, Lin H, Liu DR, Walsh CT. 2005. In vitro characterization of IroB, a pathogen-associated C-glycosyltransferase. Proc. Natl. Acad. Sci. U. S. A. 102:571–576. 10.1073/pnas.0408463102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorsa LJ, Dufke S, Heesemann J, Schubert S. 2003. Characterization of an iroBCDEN gene cluster on a transmissible plasmid of uropathogenic Escherichia coli: evidence for horizontal transfer of a chromosomal virulence factor. Infect. Immun. 71:3285–3293. 10.1128/IAI.71.6.3285-3293.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tarchouna M, Ferjani A, Ben-Selma W, Boukadida J. 2013. Distribution of uropathogenic virulence genes in Escherichia coli isolated from patients with urinary tract infection. Int. J. Infect. Dis. 17:e450–e453. 10.1016/j.ijid.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 44.Ling J, Pan H, Gao Q, Xiong L, Zhou Y, Zhang D, Gao S, Liu X. 2013. Aerobactin synthesis genes iucA and iucC contribute to the pathogenicity of avian pathogenic Escherichia coli O2 strain E058. PLoS One 8:e57794. 10.1371/journal.pone.0057794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao Q, Wang X, Xu H, Xu Y, Ling J, Zhang D, Gao S, Liu X. 2012. Roles of iron acquisition systems in virulence of extraintestinal pathogenic Escherichia coli: salmochelin and aerobactin contribute more to virulence than heme in a chicken infection model. BMC Microbiol. 12:143. 10.1186/1471-2180-12-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caza M, Lepine F, Dozois CM. 2011. Secretion, but not overall synthesis, of catecholate siderophores contributes to virulence of extraintestinal pathogenic Escherichia coli. Mol. Microbiol. 80:266–282. 10.1111/j.1365-2958.2011.07570.x. [DOI] [PubMed] [Google Scholar]
- 47.Caza M, Lepine F, Milot S, Dozois CM. 2008. Specific roles of the iroBCDEN genes in virulence of an avian pathogenic Escherichia coli O78 strain and in production of salmochelins. Infect. Immun. 76:3539–3549. 10.1128/IAI.00455-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torres AG, Redford P, Welch RA, Payne SM. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69:6179–6185. 10.1128/IAI.69.10.6179-6185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ewers C, Li G, Wilking H, Kiessling S, Alt K, Antao EM, Laturnus C, Diehl I, Glodde S, Homeier T, Bohnke U, Steinruck H, Philipp HC, Wieler LH. 2007. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int. J. Med. Microbiol. 297:163–176. 10.1016/j.ijmm.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 50.Dozois CM, Daigle F, Curtiss R., III 2003. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. U. S. A. 100:247–252. 10.1073/pnas.232686799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiong L, Ling J, Gao Q, Zhou Y, Li T, Gao S, Liu X. 2012. Construction of iucB and iucBiutA mutants of avian pathogenic Escherichia coli and evaluation of their pathogenicity. Vet. Microbiol. 159:420–431. 10.1016/j.vetmic.2012.04.024. [DOI] [PubMed] [Google Scholar]
- 52.Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welch RA, Burland V, Plunkett G, III, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HL, Donnenberg MS, Blattner FR. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:17020–17024. 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crépin S, Harel J, Dozois CM. 2012. Chromosomal complementation using Tn7 transposon vectors in Enterobacteriaceae. Appl. Environ. Microbiol. 78:6001–6008. 10.1128/AEM.00986-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crépin S, Houle S, Charbonneau ME, Mourez M, Harel J, Dozois CM. 2012. Decreased expression of type 1 fimbriae by a pst mutant of uropathogenic Escherichia coli reduces urinary tract infection. Infect. Immun. 80:2802–2815. 10.1128/IAI.00162-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 58.Sabri M, Houle S, Dozois CM. 2009. Roles of the extraintestinal pathogenic Escherichia coli ZnuACB and ZupT zinc transporters during urinary tract infection. Infect. Immun. 77:1155–1164. 10.1128/IAI.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Falkow S. 1988. Molecular Koch's postulates applied to microbial pathogenicity. Rev. Infect. Dis. 10:S274–S276. 10.1093/cid/10.Supplement_2.S274. [DOI] [PubMed] [Google Scholar]
- 60.Calderon IL, Morales EH, Collao B, Calderon PF, Chahuan CA, Acuna LG, Gil F, Saavedra CP. 2014. Role of Salmonella Typhimurium small RNAs RyhB-1 and RyhB-2 in the oxidative stress response. Res. Microbiol. 165:30–40. 10.1016/j.resmic.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 61.Kim JN, Kwon YM. 2013. Genetic and phenotypic characterization of the RyhB regulon in Salmonella Typhimurium. Microbiol. Res. 168:41–49. 10.1016/j.micres.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 62.Kim JN, Kwon YM. 2013. Identification of target transcripts regulated by small RNA RyhB homologs in Salmonella: RyhB-2 regulates motility phenotype. Microbiol. Res. 168:621–629. 10.1016/j.micres.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 63.Johnson JR. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 4:80–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Snyder JA, Haugen BJ, Buckles EL, Lockatell CV, Johnson DE, Donnenberg MS, Welch RA, Mobley HL. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 72:6373–6381. 10.1128/IAI.72.11.6373-6381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Runyen-Janecky LJ, Reeves SA, Gonzales EG, Payne SM. 2003. Contribution of the Shigella flexneri Sit, Iuc, and Feo iron acquisition systems to iron acquisition in vitro and in cultured cells. Infect. Immun. 71:1919–1928. 10.1128/IAI.71.4.1919-1928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Russo TA, Olson R, Macdonald U, Metzger D, Maltese LM, Drake EJ, Gulick AM. 2014. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect. Immun. 82:2356–2367. 10.1128/IAI.01667-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Escolar L, Perez-Martin J, de Lorenzo V. 2000. Evidence of an unusually long operator for the fur repressor in the aerobactin promoter of Escherichia coli. J. Biol. Chem. 275:24709–24714. 10.1074/jbc.M002839200. [DOI] [PubMed] [Google Scholar]
- 68.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10:1507–1517. 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tanabe T, Funahashi T, Nakao H, Maki J, Yamamoto S. 2013. The Vibrio parahaemolyticus small RNA RyhB promotes production of the siderophore vibrioferrin by stabilizing the polycistronic mRNA. J. Bacteriol. 195:3692–3703. 10.1128/JB.00162-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hagan EC, Mobley HL. 2009. Haem acquisition is facilitated by a novel receptor Hma and required by uropathogenic Escherichia coli for kidney infection. Mol. Microbiol. 71:79–91. 10.1111/j.1365-2958.2008.06509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alteri CJ, Smith SN, Mobley HL. 2009. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog. 5:e1000448. 10.1371/journal.ppat.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dozois CM, Dho-Moulin M, Bree A, Fairbrother JM, Desautels C, Curtiss R., III 2000. Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the Tsh genetic region. Infect. Immun. 68:4145–4154. 10.1128/IAI.68.7.4145-4154.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Simon R. 1984. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-Mob transposon. Mol. Gen. Genet. 196:413–420. 10.1007/BF00436188. [DOI] [PubMed] [Google Scholar]
- 74.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.