Abstract
Salmonella enterica serovar Enteritidis (S. Enteritidis) is a major etiologic agent of nontyphoid salmonellosis in the United States. S. Enteritidis persistently and silently colonizes the intestinal and reproductive tract of laying hens, resulting in contaminated poultry products. The consumption of contaminated poultry products has been identified as a significant risk factor for human salmonellosis. To understand the mechanisms S. Enteritidis utilizes to colonize and persist in laying hens, we used selective capture of transcribed sequences to identify genes overexpressed in the HD11 chicken macrophage cell line and in primary chicken oviduct epithelial cells. From the 15 genes found to be overexpressed in both cell types, we characterized the antimicrobial peptide resistance (AMPR) genes, virK and ybjX, in vitro and in vivo. In vitro, AMPR genes were required for natural morphology, motility, secretion, defense against detergents such as EDTA and bile salts, and resistance to antimicrobial peptides polymyxin B and avian β-defensins. From this, we inferred the AMPR genes play a role in outer membrane stability and/or modulation. In the intestinal tract, AMPR genes were involved in early intestinal colonization and fecal shedding. In the reproductive tract, virK was required in early colonization whereas a deletion of ybjX caused prolonged ovary colonization and egg deposition. Data from the present study indicate that AMPR genes are differentially utilized in various host environments, which may ultimately assist S. Enteritidis in persistent and silent colonization of chickens.
INTRODUCTION
Salmonella enterica serovars are important zoonotic pathogens that cause 1.028 million nontyphoidal salmonellosis cases with approximately 400 deaths annually in the United States (1). Since 1994, Salmonella enterica serovar Enteritidis has been the predominate serovar isolated from nontyphoidal salmonellosis cases in the United States, with a majority of these cases associated with the consumption of contaminated poultry products (2). Since the 1990s, eggs have been a significant source of infection and chicken products have been identified as a significant risk factor for human illness (2–4). In the 1990s, the USDA and FDA implemented regulations on quality control, storage, and transportation and led efforts to improve consumer knowledge of proper storage and cooking of eggs; this resulted in a 50% decrease in S. Enteritidis-induced illnesses by 1999 (4). However, even with these increased control measures in place, S. Enteritidis continues to be a major health risk associated with significant economic losses. For example, in August 2010, the United States had to recall 500 million eggs during an S. Enteritidis outbreak in Iowa that spread to 11 states and caused over 1,939 illnesses (5).
Egg contamination can be a result of horizontal transmission, of transmission from the environment after the egg is laid, or of vertical contamination from S. Enteritidis colonizing the reproductive tract (2). Salmonella can spread in a hen house very quickly through the fecal-oral route. After ingestion, the mildly acidic crop primes Salmonella genes for the acidic environment faced in the intestinal tract, where the organism withstands this pressure due to upregulation of stress response genes such as the rpoS regulon (6). In the intestine, Salmonella evades innate immune responses, such as bile salts and avian β-defensins, to interact with the epithelium (7, 8). Salmonella pathogenicity island 1 (SPI-1)- and SPI-2-encoded type-three secretion system 1 (T3SS-1) and T3SS-2 are used to invade tissue and to establish and maintain Salmonella-containing vacuoles in macrophages, respectively (9, 10).
Macrophages containing Salmonella organisms are the primary vessels for dissemination. To survive inside macrophages, Salmonella has to defend itself against many host killing factors: low Mg2+ and Ca2+, acidic pH, and reactive oxygen species (7). To survive in this environment, Salmonella utilizes the components of T3SS-1 and T3SS-2 and the PhoP/PhoQ regulon (10–13). S. Enteritidis has been shown to colonize the ovary and the reproductive tract within laying hens, with a higher recovery from the ovary than from any other reproductive tissue (14, 15). Successful S. Enteritidis colonization involves inherent characteristics employed to subvert the reproductive innate immune system: phagocytes, antimicrobial peptides, including avian β-defensins, and immunoglobulins (16, 17). Persistent reproductive-tract colonization that leads to egg contamination is confined primarily to S. Enteritidis and is partially due to its ability to survive under these harsh conditions without causing overt clinical signs in the chicken host (18, 19). While much information has been gathered on the significance and mechanisms of T3SS-1 and T3SS-2, the mechanisms of persistence employed by S. Enteritidis remain to be fully understood (20).
Most of our understanding of Salmonella pathogenicity is based on information gathered from S. Typhimurium experiments in mammalian hosts or cell cultures. Translation of data from S. Typhimurium to S. Enteritidis is not direct, especially since they have a 3% genetic difference, accounting for 6.4% of the genome of S. Enteritidis and 9.6% of the genome of S. Typhimurium. One major difference is in the composition of their outer membranes or O-antigens (D1 for S. Enteritidis and B for S. Typhimurium), a key barrier to innate defenses and interaction with the host (21, 22). The studies aimed at understanding the mechanisms of persistence in the chicken host have often involved chicks. The data collected from these experiments do not transpose to infection of mature hens with S. Enteritidis, especially since the immunological landscape changes after the point of lay (23). In the present study, we identified S. Enteritidis genes overexpressed in primary chicken oviduct epithelial cells and in chicken macrophages. From the genes identified, we characterized the antimicrobial peptide resistance (AMPR) genes in vitro and in vivo. The current investigation revealed that AMPR genes used by S. Enteritidis to evade host innate immune defenses also play a role in reproductive-tract colonization and egg deposition.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Spontaneously nalidixic acid-resistant S. Enteritidis, designated strain ZM100, was generated by serial passages in Luria-Bertani (LB) broth with increasing concentrations of nalidixic acid. S. Enteritidis and Escherichia coli strains were cultured aerobically in tryptic soy broth (TSB), Super Optimal broth (SOB) or SOC (SOB plus 20 mM glucose [3.603 g]), or LB broth or on LB agar plates at 37°C. When appropriate, antibiotics were added at the following concentrations: chloramphenicol, 30 μg/ml; ampicillin, 100 μg/ml; nalidixic acid, 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description or relevant genotypea | Reference or source |
|---|---|---|
| Strains | ||
| S. Enteritidis ZM100 | Wild-type S. Enteritidis, Nalr | 24 |
| S. Enteritidis ZM112 | ZM100 Nalr Cmr virK::pEP185.2 | This study |
| S. Enteritidis ZM114 | ZM100 Nalr Cmr ybjX::pEP185.2 | This study |
| S. Enteritidis ZM112C | ZM112 Nalr Cmr Ampr pWSKVirK | This study |
| S. Enteritidis ZM114C | ZM114 Nalr Cmr Ampr pWSKYbjX | This study |
| S. Enteritidis ZM122 | ZM100 Nalr ΔvirK (Δ14–919/930) | This study |
| S. Enteritidis ZM123 | ZM100 Nalr ΔybjX (Δ13–927/969) | This study |
| S. Enteritidis ZM124 | ZM100 Nalr ΔvirK ΔybjX (Δ14–919/930) (Δ13–927/969) | This study |
| S. Enteritidis ZM122C | ZM122 Nalr Ampr pWSKVirK | This study |
| S. Enteritidis ZM123C | ZM123 Nalr Ampr pWSKYbjX | This study |
| E. coli S17-1 | recA Tn7 λpir | SZ collection |
| E. coli Top10F' | F′ lacQQ Tn10 (Tetr) | Invitrogen |
| Plasmids | ||
| pCR2.1 | TA cloning vector, Ampr Kanr lacZα | Invitrogen |
| pEP185.2 | Suicide vector, Cmr | 25 |
| pRDH10 | Cmr, sacB | 26 |
| pWSK29 | Low-copy-number expression vector, Ampr | 27 |
| pZM-16S | pCR2.1 16S rRNA gene | This study |
| pZM-23S | pCR2.1 23S rRNA gene | This study |
| pZM112 | pEP185.2 carrying a fragment of virK | This study |
| pZM114 | pEP185.2 carrying a fragment of ybjX | This study |
| pZM122 | pRDH10 carrying the flanking regions of virK | This study |
| pZM123 | pRDH10 carrying the flanking regions of ybjX | This study |
| pWSKVirK | pWSK29 carrying the virK gene | This study |
| pWSKYbjX | pWSK29 carrying the ybjX gene | This study |
Amp, ampicillin; Cm, chloramphenicol; Kan, kanamycin; Nal, nalidixic acid; Tet, tetracycline.
Cell cultures and culture conditions.
Primary chicken oviduct epithelium cells (COEC) were prepared as described previously (10). Briefly, oviduct tissue (isthmus region) from 20- to 23-week-old Hy-line W36 chickens was obtained from a local poultry producer. After Salmonella-free status was confirmed by PCR, the tissue was washed extensively with Hanks balanced salt solution (HBSS) containing penicillin at 200 U/ml and streptomycin at 200 mg/ml. After treatment with collagenase XI (Sigma) (1 mg/ml), the epithelial cells were released by treatment with 0.25% trypsin–EDTA (Invitrogen), collected via centrifugation at 100 × g for 5 min, and resuspended in minimum essential media (MEM; Invitrogen) supplemented with 15% heat-inactivated fetal bovine serum (FBS), 2% chicken serum (CS), 0.05 mM β-estradiol (Sigma), and 0.01 mg/ml insulin (Sigma). COEC were seeded into 48-well tissue culture plates at a density of 4 × 104 cells per well (for selective capture of transcribed sequences [SCOTS]) or into 96-well plates at a density of 2 × 104 cells per well (for invasion assays) and incubated at 37°C in 5% CO2 for 48 h. COEC were stained with monoclonal antipancytokeratin antibody and fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG and examined with an Olympus IX81 FA microscope. Cultures with more than 80% cytokeratin-positive (epithelial lineage) cells were used in subsequent infections. HD11 chicken macrophage cells (28) were maintained in RPMI 1640 tissue culture medium (Invitrogen) supplemented with 10% FBS and 2% CS at 37°C in 5% CO2. Prior to infections, HD11 cells were seeded into 48-well tissue culture plates at a density of 4 × 105 cells per well (for SCOTS assays) or into 96-well plates at a density of 2 × 105 cells per well (for invasion assays) and incubated for 24 h.
Infection of cell cultures.
Gentamicin protection assays were performed for invasion assays and SCOTS as described previously (29). To prepare the bacterial inoculum, 50 μl of an overnight culture of an S. Enteritidis strain (strain ZM100 only for SCOTS assays) was diluted into 5 ml of fresh TSB or LB broth and incubated aerobically at 37°C for 4 h (logarithmic phase) or 16 h (stationary phase). The S. Enteritidis cultures were harvested by centrifugation for 15 min at 1,500 × g and resuspended in fresh HBSS. S. Enteritidis numbers from each inoculum were determined by measuring their optical density at 600 nm and confirmed by subsequent CFU enumerations by plating 10-fold serial dilutions.
Prior to infections, each cell culture, in experiments performed in triplicate, was washed three times in the appropriate media containing no antibiotics. For SCOTS assays, 200-μl bacterial suspensions containing approximately 8 × 105 CFU of logarithmic-phase ZM100 cells (for COEC) or 8 × 106 CFU stationary-phase ZM100 cells (for HD11) were added into each well to reach a multiplicity of infection (MOI) of 20:1 (bacteria/cells). For invasion and intracellular replication assays, 200-μl bacterial suspensions containing approximately 2 × 107 CFU of either logarithmic- or stationary-phase S. Enteritidis strains (for both COEC and HD11) were added into the triplicate wells to reach an MOI of 20:1 for each well. To synchronize infections, all inoculated cultures were centrifuged at 800 × g for 10 min and then incubated at 37°C in 5% CO2 for 1 h. Extracellular bacteria were removed by treatment with 100 μg/ml gentamicin in MEM (for COEC) or RPMI 1640 (for HD11) at 37°C in 5% CO2 for 1 h. Following gentamicin treatment, infected cells were either lysed or maintained in fresh media containing 50 μg/ml gentamicin for an additional 3 h and 15 h followed by lysis. These time points were designated 1 h postinfection (hpi) (T1), 4 hpi (T4), and 16 hpi (T16). For RNA extraction, infected cells were lysed in TRIzol (200 μl/well). For invasiveness and intracellular replication studies, infected cells were lysed in 0.5% Triton X-100 (100 μl/well). Then, 10-fold serial dilutions of the invaded-cell lysates were plated onto LB agar supplemented with appropriate antibiotics and incubated overnight at 37°C for CFU enumeration. Invasiveness was calculated for each strain as the proportion of inoculum internalized at T1, and intracellular replication, or survival, was calculated as the proportion of S. Enteritidis cells recovered at T4 or T16 compared to the inoculum.
Preparation of bacterial gDNA and rRNA genes.
Genomic DNA (gDNA) was isolated from an overnight ZM100 culture using a Blood & Cell Culture DNA Midi kit according to the manufacturer's instruction (Qiagen). Biotinylation of gDNA was carried out by mixing equal amounts (20 μg) of gDNA and photosensitive biotin (Sigma) in a final volume of 50 μl inside a 0.5-ml tube; the tubes were exposed to strong (200-W) incandescent light for 30 min. The labeled gDNA was extracted with 2-butynal, washed with 70% ethanol, and resuspended in 10 mM Tris-HCl–0.5 mM EDTA (1× TE) buffer.
The 16S and 23S rRNA coding regions of S. Enteritidis were amplified by PCR using primer pair 16S-F1 and 16S-R1 and primer pair 23S-F1 and 23S-R1, respectively (Table 2). The PCR products were cloned into pCR2.1 (Invitrogen). The resulting plasmids, pZM-16S and pZM-23S, were propagated in E. coli TOP10F′ (Invitrogen). Plasmid DNA was extracted using a Wizard Plus Minipreps DNA purification system (Promega) according to the manufacturer's instructions. The DNA concentration was determined based on the A260 spectrophotometer reading.
TABLE 2.
Primers used in this studya
| Primer | Sequence (5′–3′) | Amplicon size (bp) |
|---|---|---|
| For SCOTS | ||
| ZM-1 | GACACTCTCGAG ACATCACCGG | NA |
| ZM-2 | TGCTCTAGACGTCGACATGGTT | |
| ZM1-Nw | GACACTCTCGAGACATCACTGG(N9) | NA |
| ZM2-Nw | TGCTCTAGACGTCGACATGGTT(N9) | |
| 16S-F1 | CGGACGGGTGAGTAATGTCT | 1,372 |
| 16S-R1 | ATCACAAAGTGGTAAGCGCC | |
| 23S-F1 | CGGGGGAACTGAAACATCTA | 2,636 |
| 23S-R1 | TCAACGTCGTCGTCTTCAAC | |
| For real-time PCR | ||
| 16srRNA-F | CCTTACGACCAGGGCTACACACG | 94 |
| 16srRNA-R | GGACTACGACGCACTTTATGAGG | |
| hsdS-F | TTGAAAAGACAATCCCACTC | 120 |
| hsdS-R | GGTAACCAACAACTCCCG | |
| orgAa-F | AACGGATAAACTTGTTCCCTGAT | 110 |
| orgAa-R | TCGGTTGCCATAAACTGAG | |
| pgtE-F | AACTGGACTGGAAAATAAAAAATGT | 120 |
| pgtE-R | TATGACCCGATCCCGACG | |
| pipB-F | TCGGTGCAAATTTGTGTTGT | 142 |
| pipB-R | GAGCCGAATAGAATTGCAGC | |
| prgJ-F | GAAAAAGCCTGGAGTAGCC | 95 |
| prgJ-R | GTCCCTGAGAATGCCGTT | |
| prgK-F | ACGCCCTCCATCGTCTGT | 93 |
| prgK-R | TTCGCTGGTATCGTCTCC | |
| sefB-F | CTCCATTTATTGTAACACCACCTAT | 123 |
| sefB-R | TTACACACAACCAATACAAAGACTC | |
| ssaD-F | ATCCAAATAAGCCGCTACCA | 84 |
| ssaD-R | CAAGTTCACAATCCTGTTTACCAA | |
| ssaK-F | CTGTTCCAGCCATTCCACTTCCAT | 113 |
| ssaK-R | TCATCCGAGACGCCTATCGTTATCA | |
| ssaI-F | TGCCTGTAAGCACTCAATCT | 125 |
| ssaI-R | CTGCGGTAATAAAGCACTGG | |
| ssaJ-F | CGTCTCAGGCAAAAATAGC | 118 |
| ssaJ-R | ACGCCAATAAAGGGAAGG | |
| sthC-F | ATTCAGCCCTGACCACCG | 128 |
| sthC-R | ACCTTATGCTTCGCCTTACCA | |
| yifK-F | CGTGGGCGAACTATTTGA | 131 |
| yifK-R | AACTTTGTAGACCAGCGTGA | |
| yjjZ-F | GCGTATTATTGCCTGGAGTGAT | 132 |
| yjjZ-R | AAAATGCCGTAATTGTTTGTGAT | |
| ybjX-F | GACGATGTAGCCCGAATAGG | 81 |
| ybjX-R | TACTGACCAATCTCACCCAAT | |
| virK-ORF1 | GCGAGCTCATGACGATGCAGCAAAG | 138 |
| virK-R | AATAAGGCAACGTAATAC | |
| For mutant construction and complementation | ||
| virK-ORF1 | GCGAGCTCATGACGATGCAGCAAAG | |
| virK-R | AATAAGGCAACGTAATAC | |
| ybjX-ORF1 | GCGAGCTCATGTCGCGGATTACGAT | |
| ybjX-rt-R | CGACGAAAGCTGGCTTTAC | |
| virK-UF1 | CTGGCTTACACAATAGCAG | |
| virK-UR1 | ACTCTAGAGCTGCATCGTCATACTAC | |
| virK-DF1 | ACTCTAGATCTCCCGGTAGAACTATTTC | |
| virK-DR1 | CTGGGTGCATATTGATAC | |
| ybjX-DF2 | TATGGGAATCGAGTGG | |
| ybjX-DR2 | ACTCTAGAGATAGCGTCGTCGAAC | |
| ybjX-UF2 | ACTCTAGAAATCCGCGACATAAGA | |
| ybjX-UR2 | ATCTGGGTCAATCACG | |
| ybjX-ORF1 | GCGAGCTCATGTCGCGGATTACGAT | |
| ybjX-ORF2 | GCTCTAGATTAACGTTTGAATGTGAC | |
| virK-ORF1 | GCGAGCTCATGACGATGCAGCAAAG | |
| virK-ORF2 | GCTCTAGACTACCGGGAGAGGCTGTTA |
The restriction sites integrated into the sequences are underlined. NA, not applicable.
Isolation of RNA.
Total RNA was isolated from strain ZM100-infected cell cultures and ZM100 grown in TSB using TRIzol reagents (Life Technologies). RNA samples were treated with RNase-free DNase I (Ambion), and the RNA concentration was determined by A260 and A280 spectrophotometer readings and agarose gel electrophoresis.
Synthesis of cDNA.
RNA (5 μg) was converted to first-strand cDNA by random priming using a Superscript III system (Invitrogen) and primers ZM1-Nw and ZM2-Nw for intracellular and broth-grown bacteria, respectively. Double-stranded cDNA was generated using Klenow DNA polymerase (Promega) and amplified by PCR using primer ZM1-Nw (for intracellular bacteria) or primer ZM2-Nw (for broth-grown bacteria). PCR was performed using Platinum Taq DNA polymerase (Sigma-Aldrich) under the following conditions: initial denaturation of 3 min at 94°C followed by 25 cycles at 94°C for 45 s, at 58°C for 45 s, and at 72°C for 2 min and a final elongation at 72°C for 10 min. The amplified cDNA was precipitated in 100% ethanol (2.5 volumes of cDNA) with 3 M sodium acetate (NaOac) (0.1 volume of cDNA) and 1 μl glycogen (1 μg/ml) and resuspended in 10 mM N-(2-hydroxyethyl)-piperazine-N′-3-propanesulfonic acid–1 mM EDTA (1× EPPS-EDTA). DNA concentrations were determined based on the A260 spectrophotometer reading.
SCOTS.
To block rRNA coding regions, pZM-16S, pZM-23S (cloned rRNA gene), and biotinylated gDNA were mixed at a ratio of 5:5:1 (μg), fragmented by sonication, and resuspended in 1× EPPS-EDTA. The rRNA gene-blocked gDNA (8 μl) and the amplified cDNA (3 μg plus 8 μl 1× EPPS-EDTA) were denatured separately at 98°C for 3 min. Following addition of 2 μl 5 M NaCl to each reaction tube, the denatured gDNA and cDNA were self-annealed at 67°C for 30 min. The gDNA and cDNA were then mixed and hybridized at 67°C for 20 h. The cDNA molecules hybridized to the biotinylated gDNA were captured by incubation with streptavidin-coated beads and subsequent elution according to the manufacturer's instruction (Dynal). The eluted cDNA was amplified by PCR using specific primer ZM-1 for intracellular bacteria or ZM-2 for extracellular bacteria. For each time point/cell type/growth condition combination, amplified cDNAs from the 10 parallel reactions were combined and subjected to another round of hybridization. Three rounds of hybridizations were carried out to enrich the cDNA species representing S. Enteritidis gene transcripts at a given time under a given set of growth conditions. Following enrichments, competitive hybridizations were performed: rRNA gene-blocked and biotinylated gDNAs were prehybridized with cDNA derived from broth-grown bacteria at 67°C for 4 h and then hybridized with cDNA of intracellular bacteria for an additional 20 h. The hybridized cDNA specific to intracellular bacteria was captured using streptavidin-coated beads and amplified using primer ZM-1. Three rounds of competitive hybridizations with 10 parallel reactions in each round were performed to enrich the transcripts specific to intracellular bacteria. The cDNA specific to intracellular bacteria was cloned into pCR2.1. Following transformation of E. coli TOP10F′, the insertions of all clones were sequenced and compared to the S. enterica genomes using the BLASTN algorithm. Transcripts identified by two independent SCOTS procedures following two independent infections were considered intracellularly expressed.
RT-PCR.
Reverse transcriptase PCR (RT-PCR) was conducted using MultiScribe reverse transcriptase (Invitrogen) and SYBR green PCR master mix (Applied Biosystems). The primer sequences of Salmonella genes were obtained from the Entrez Nucleotide database and are listed in Table 2. Reverse transcription of total RNA (2 μg) in a volume of 100 μl containing 5.5 mM MgCl2, 500 μM deoxynucleoside triphosphate (dNTP), 2.5 μM random hexamers, and 1.25 U of MultiScribe reverse transcriptase was performed at 42°C for 30 min. The resultant cDNA product was used as a template (4 μl/reaction) for subsequent real-time PCR (ABI Prism 7700; Applied Biosystems). PCR was carried out in a volume of 25 μl under the following conditions: 95°C for 10 min followed by 45 amplification cycles of 95°C for 15 s, 58°C for 30 s, and 72°C for 30 s in the presence of 1× SYBR green PCR master mix (Applied Biosystems). The threshold cycle (2−ΔΔCT ± standard deviation [SD]) method was used to qualify transcriptional changes within host cells (30), and the expression of 16S rRNA was used to normalize the cDNA concentrations of different samples.
Construction of mutants.
Mutants and complemented strains were constructed using the primers listed in Table 2. The initial insertion mutants, strains with virK (ZM112) or ybjX (ZM114) inserted, were constructed for inactivation of the target gene as previously described (31). Briefly, DNA fragments carrying the 5′ termini of virK and ybjX were amplified by PCR using primer pair virK-ORF1 and virK-R and primer pair ybjX-ORF1 and ybjX-rt-R, respectively. The PCR products were cloned into pCR2.1TOPO (Invitrogen) and subcloned into the SacI and XbaI sites of pEP185.2, a suicide vector coding for chloramphenicol resistance (25). The resulting plasmids, pZM112 and pZM114, were introduced into E. coli strain S17-1λpir by chemical transformation and transferred into strain ZM100 by conjugation. Exconjugants with a pEP185.2 insertion into the chromosome of strain ZM100 were selected by determination of resistance to chloramphenicol and nalidixic acid. These mutants were used in initial in vitro phenotypic characterizations. For complementation, the open reading frames (ORF) of virK and ybjX were amplified by PCR using primer pair VirK-ORF1 and VirK-ORF2 and primer pair YbjX-ORF1 and YbjX-ORF2. The PCR products were cloned directionally under the control of the lac promoter of pWSK29, a low-copy-number expression vector (27), which generated plasmids pWSKVirK and pWSKYbjX. These plasmids were introduced into the corresponding mutant strains by electroporation and selection for resistance to ampicillin. The resultant strains were designated ZM112C and ZM114C, respectively.
To avoid possible polar effects, unmarked ΔvirK (ZM122) and ΔybjX (ZM123) deletion mutants and a ΔvirK ΔybjX (ZM124) double mutant were constructed using allelic exchange mutagenesis as previously described (31). Briefly, the upstream and downstream regions of the genes were amplified by PCR using primer pair virK-UF1 and virK-UR1, primer pair virK-DF1 and virK-DR1, primer pair ybjX-UF2 and ybjX-UR2, and primer pair ybjX-DF2 and ybjX-DR2, respectively. After ligation of the upstream and downstream products, the fusion was reamplified by PCR and cloned into pCR2.1TOPO. The fusion was then subcloned into vector pRDH10, a suicide vector that carries the sacB and chloramphenicol resistance genes and is λpir dependent (26). The resulting plasmids, pZM122 and pZM123, were chemically transformed into E. coli S17-1λpir, selected by resistance to chloramphenicol, and then transferred to ZM100 by conjugation. Exconjugants were selected by resistance to nalidixic acid and chloramphenicol for the insertion of the plasmid into the genome. Selection for a second recombination leading to unmarked deletion was done in 10% sucrose LB broth followed by growth on 5% sucrose LB plates at 30°C. The colonies sensitive to chloramphenicol were subjected to PCR to screen for the deletion of interest. Each unmarked deletion was complemented with its respective pWSK29 plasmid harboring the ORF of the gene as described above. The unmarked deletion for each gene was confirmed by DNA sequencing. These unmarked deletion mutants were used in further in vitro phenotypic characterizations as well as in vivo phenotypic characterizations. The strains and plasmids are listed in Table 1.
Bacterial cell morphology assay.
Bacterial morphology for ZM100, ΔvirK, ΔybjX, ΔvirK ΔybjX, ΔvirK pWSKVirK (ZM122C), and ΔybjX pWSKYbjX (ZM123C) strains was determined by microscopic examination of logarithmic and stationary cultures at 4 h and 16 h of growth. Three fields were captured at 400× from three separate experiments by a Sony microscope camera and analyzed for the average Feret length using ImageJ morphometric analysis (32). After conversion to micrometers (12.156 pixels/μm), the data are presented as the average length per strain per time point.
Cell motility assay.
To test the ability of the ZM100, ΔvirK, ΔybjX, ΔvirK ΔybjX, ΔvirK pWSKVirK, and ΔybjX pWSKYbjX strains to swim, bacteria were inoculated into 0.3% agar LB plates as previously described (33). Briefly, equal amounts of logarithmic-phase (4-h) cultures were spotted in the middle of 0.3% agar plates and incubated at 37°C for 3 h, at which point their motility diameter was measured. The data are represented as relative motility values (percentage of ZM100 motility).
EDTA and DOC sensitivity assays.
The ZM100, ΔvirK, ΔybjX, ΔvirK ΔybjX, ΔvirK pWSKVirK, and ΔybjX pWSKYbjX strains were tested for their sensitivity to EDTA and deoxycholic acid (DOC) as previously described (34, 35). Briefly, approximately 1 × 108 CFU bacteria were added to warm 0.5% LB agar and the mixture was poured over 1.5% LB agar plates. Once the plates had dried, filter disks containing 0.5 M EDTA were placed in the center of the agar and incubated without inversion at 37°C for 16 h. The zones of inhibition were measured in millimeters, and the data are presented as relative sensitivity values (percent difference from ZM100). For the determinations of DOC sensitivity, LB agar plates containing 1% DOC and plain LB agar plates were inoculated with 10-fold serial dilutions of bacterial culture and incubated overnight at 37°C for enumeration. Percent inhibition of growth was calculated using the following formula: [(CFU on plain LB agar − CFU on 1% DOC LB agar)/(CFU on plain LB agar)] × 100.
Cell supernatant 2D SDS-PAGE assay.
Cell supernatant proteins were obtained from the ZM100, ΔvirK, ΔybjX, and ΔvirK ΔybjX strains as previously described (36). Briefly, bacterial cultures were grown in triplicate overnight at 37°C and 250 rpm, subcultured at 1:50× into fresh LB broth, and incubated 4 h at 37°C and 250 rpm. Culture supernatants from approximately 12 × 109 CFU of each strain were recovered by centrifugation at 5,000 × g at 4°C for 15 min and filtration through a 0.45-μm-pore-size sterile filter. The supernatant was concentrated using centrifugal filters (molecular weight cutoff, 3,000), and the proteins in the supernatant were obtained by methanol-chloroform protein precipitation. A portion of the total supernatant proteins recovered was used to measure the concentration using a Bradford assay and was also visualized on a 12.5% SDS-PAGE gel stained with 0.1% silver stain to check for purity. One-fourth of the total supernatant proteins (representing 3 × 109 CFU per strain) were analyzed by two-dimensional (2D) SDS-PAGE. Nonlinear isoelectric focusing strips (pH 3 to 10, 7 cm in length) were rehydrated overnight with proteins dissolved in UT Chaps buffer consisting of 7 M urea, 2 M thiourea, and 4% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} with DeStreak, pharmalytes, amylytes, and bromophenol blue (GE Healthcare), subjected to 770 V-h, and then separated on a 12.5% SDS-PAGE gel and stained with 0.2% silver stain. After three independent experiments were completed, densitometric analysis with ImageJ determined the differences between the ΔvirK, ΔybjX, and ΔvirK ΔybjX deletion mutants and wild-type ZM100 in spot densities (32). Spots whose density was greater or lesser than 30% of the density of ZM100 were excised, reduced, and alkylated with iodoacetamide, digested with trypsin, and then processed on a ThermoFisher LTQ or OrbiTrap linear ion trap mass spectrometer using nano-liquid chromatography (nano-LC) peptide separations. Proteins were analyzed with Scaffold 4.1 for specific protein identities by using BLAST analysis and NCBI and UniProt databases (37).
Polymyxin B sensitivity assay.
The S. Enteritidis ZM100, virK, ybjX, virK pWSKVirK, and ybjX pWSKYbjX strains were grown at 37°C in N-minimal media containing 10 mM MgCl as described previously (38). The pH of the medium was buffered with 100 mM Tris-HCl (pH 7.4). Approximately 5 × 105 CFU were inoculated into 1 ml LB containing 2.5 μg polymyxin B (Sigma) and incubated at 37°C for 1 h. Serial dilutions of bacterial suspensions were prepared in phosphate-buffered saline (PBS) and plated on LB agar plates for CFU enumeration. Polymyxin B sensitivity was expressed using the following equation: log reduction = [log input (CFU/ml) − log viability (CFU/ml)].
Avian beta-defensin sensitivity assay.
The mature peptide of avian beta-defensin-6 (AvBD6) (SPIHACRYQRGVCIPGPCRWPYYRVGSCGSGLKSCCVRNRWA) with three disulfide bridges (Cys1-Cys5, Cys2-Cys4, and Cys3-Cys6) was synthesized by LifeTein LLC (Hillsborough, NJ). The sensitivity of S. Enteritidis strains to AvBD-6 was determined using a microbroth dilution method (39). In brief, overnight cultures were diluted in fresh Mueller-Hinton broth to achieve a 0.5 McFarland turbidity standard (approximately 5 ×107 CFU/ml). Equal volumes (50 μl of 5 × 105 CFU) of bacterial suspension and AvBD-6 (32 μg/ml) were mixed and incubated at 37°C for 1 h. For controls, PBS was used to replace AvBD-6. Following incubation, serial dilutions of bacterial suspensions were prepared in PBS and plated on LB agar plates for CFU enumeration. AvBD sensitivity was expressed as the percentage of growth inhibition relative to the growth of the AvBD-free control: [100 × (CFU of PBS-treated culture − CFU of AvBD-treated culture)/CFU of PBS-treated culture].
Ethics statement.
Animal rearing, maintenance, and euthanasia were performed according the recommendations by the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and our Texas A&M University Institutional Animal Care and Use Committee SACC-approved Animal Use Protocol (permit number 2011-143).
Animal experiments.
Female Hy-Line W36 chicks, obtained at 1 day of age from a local hatchery without vaccination for S. Enteritidis, were maintained at 30°C until they were 7 weeks old and were then moved to floor pens maintained at room temperature until they were 21 weeks old. They were given feed and water at libitum. Three hens were sacrificed prior to the experiments, and their spleen, oviduct, ovary, and cecum were tested by PCR to rule out Salmonella-positive status.
Prior to oral inoculations, the hens were housed in an animal biosafety level 2 (ABSL-2) housing facility with 2 hens per cage and grouped into six cage stacks so that seven groups of 12 hens were isolated from each other. While on a schedule of 16 h of light and 8 h of dark, the hens were given water and standard layer diet ad libitum for the duration of the infection. The overnight cultures of S. Enteritidis strains were diluted 1:50× with sterile LB broth and incubated for 16 h at 37°C and 250 rpm. The resultant cultures were harvested by centrifugation at 3,000 rpm at 4°C for 20 min and resuspended in sterile saline solution at a final concentration of 5 × 109 CFU/ml. Each group of 12 hens was orally inoculated with 1 ml of one bacterial strain per hen; one group of hens was inoculated with 1 ml of sterile saline solution to serve as a control. At the same time each day postinfection (dpi), all viable eggs and a fecal sample were collected from each cage. From 1 dpi to 5 dpi, the fecal and egg samples represented two hens; from 6 dpi to 10 dpi, the fecal and egg samples represented one hen. At 5 dpi and 10 dpi, six hens from each group were euthanized and the spleen, oviduct (isthmus region), ovary, cecum, and ileum were collected and stored at −80°C.
For bacterial enumeration, approximately 1 g of feces was resuspended in 10 ml of buffered peptone water (BPW) and 10-fold serial dilutions were plated onto XLT4 agar followed by incubation at 37°C for 16 h. The BPW reaction mixture was then enriched overnight at 37°C and plated on XLT4 agar, and any growth seen was arbitrarily assigned a value of 5 CFU/g, the detection threshold for the procedure. The eggs were washed three times in 70% ethanol, placed in 50 ml of BPW, and homogenized for 5 min. A portion of the homogenized egg-BPW mixture was plated onto selective LB agar for enumeration, and the remainder was enriched for 2 days at 37°C, with plating of 24-h and 48-h enrichments performed on selective LB agar. Tissue samples were sterilely collected, weighed, homogenized for 30 s in 5 ml PBS, and plated on selective LB for CFU enumeration. The cecal and ileal contents were weighed, resuspended in 5 ml BPW, and then plated on XLT4 and selective LB for enumeration. The cecal and ileal tissues were washed three times in PBS, homogenized for 30 s in 5 ml PBS, and plated on selective LB for enumeration. Homogenates were enriched overnight at 37°C and plated on selective LB agar. Cultures that were negative in the initial plating but had growth after enrichment were arbitrarily assigned values of 2.5 CFU/homogenate for the spleen, ovary, and oviduct tissue and 12.5 CFU/homogenate for the cecum contents. The detection thresholds for the different types of samples were arithmetically determined using the following formulas: arbitrary tissue CFU/homogenate = [(1 CFU/0.5 ml) × 5 ml]/4 and arbitrary cecum content CFU/homogenate = [(1 CFU/0.1 ml) × 5 ml]/4. When possible, three colonies from each culture with a positive result were tested by PCR to validate the accuracy of the visual counts. The data are expressed as log CFU/g for the feces and tissue and as log CFU/ovary for the ovary.
Statistics.
Statistical analyses using one-way analysis of variance (ANOVA) were performed to determine significant differences among the groups in the different experiments, and Student's t test was used to determine any significant differences between the individual strains tested in each experiment (P < 0.05).
RESULTS
Detection of S. Enteritidis genes overexpressed in infected chicken cells by selective capture of transcribed sequences (SCOTS).
To better understand the mechanism of S. Enteritidis colonization in chickens, SCOTS procedures were performed to identify the genes preferentially expressed in a macrophage cell line (HD11) and primary chicken oviduct epithelial cells (COEC), two main cell types utilized by S. Enteritidis for systemic infection and reproductive-tract colonization. Of the 48 genes identified, 37 were overexpressed in COEC, 26 in HD11, and 15 in both types of cells (Table 3). The intracellular expression of selected genes was further confirmed by quantitative real-time PCR using 16S rRNA as a reference gene (Fig. 1). The genes specific to COEC consisted of those encoding SPI-1 T3SS components, restriction modification enzymes, oxidative stress resistance, proteins involved in fimbrial biogenesis, and outer membrane assembly. The elevated transcription of COEC-specific genes occurred mainly at 1 hpi. The genes expressed within HD11 cells at 1 hpi or 4 hpi included those involved in nitrate reduction, cell wall synthesis, and O-antigen conversion as well as the anaerobically induced tdc operon. S. Enteritidis genes overexpressed in both cell types comprised those encoding the SPI-2 T3SS apparatus, the SPI-5 pipB gene, and genes responsible for antimicrobial peptide resistance (AMPR) (virK and ybjX). Increased expression of the genes was detected in both cell types at 4 hpi, suggesting the significance of these genes in intracellular survival or replication inside COEC and HD11. In addition, several genes of unknown functions (such as yifK and yjjZ, hypothetically involved in transport, with the latter being overexpressed around 30-fold) were overexpressed in both types of cells (Fig. 1).
TABLE 3.
Genes overexpressed by S. Enteritidis in chicken oviduct epithelial cells and macrophagesa
| Category | Gene | Description and possible function | COEC result |
HD11 result |
||
|---|---|---|---|---|---|---|
| 1 h | 4 h | 1 h | 4 h | |||
| Virulence, SPI-1 | orgA | Type III secretion, host cell invasion | + | |||
| prgJ | Type III secretion, host cell invasion | + | ||||
| prgK | Type III secretion, host cell invasion | + | ||||
| Virulence, SPI-2 | ssaD | Type III secretion, intracellular survival | + | + | ||
| ssaI | Type III secretion, intracellular survival | + | + | |||
| ssaJ | Type III secretion, intracellular survival | + | + | |||
| ssaM | Type III secretion, intracellular survival | + | + | |||
| ssaK | Type III secretion, intracellular survival | + | + | |||
| Virulence, SPI-5 | pipB | T3SS-2 secreted protein | + | + | ||
| Antimicrobial peptide resistance | pgtE | Outer membrane protease E | + | + | ||
| virK | Intracellular survival | + | + | |||
| ybjX | Putative virK homologue | + | + | |||
| fumA | Fumarate hydratase | + | ||||
| rpiA | Ribose 5-phosphate isomerase | + | ||||
| yliG | Putative Fe-S oxidoreductase | + | ||||
| ahpC | Alkyl hydroperoxide reductase, oxidative stress resistance | + | ||||
| acs | Acetyl-coenzyme A synthetase | + | ||||
| gip | Glyoxylate-induced protein | + | ||||
| hemX | Uroporphyrinogen III methylase | + | ||||
| hyi | Hydroxypyruvate isomerase | + | ||||
| gilnD | Uridylyltransferase | + | ||||
| Membrane transport, metabolism, stress response | narU-narY | Respiratory nitrate reductase, nitrite extrusion protein | + | |||
| pheA | Prephenate dehydratase | + | ||||
| ybbP | Putative inner membrane ABC transporter | + | ||||
| yiaH | Putative inner membrane protein | + | ||||
| sb35 | Hydrolase of HD superfamily | + | ||||
| yifK | Putative ABC transporter | + | + | |||
| yfdZ | Putative aminotransferase | + | + | |||
| yin-cysE | Putative mandelate racemase/muconate-lactonizing enzyme, serine acetyltransferase | + | + | |||
| yraO-yraP | Putative phosphoheptose isomerase | + | ||||
| trpS | Tryptophanyl-tRNA synthetase | + | + | |||
| tdcB | l-Threonine/l-serine permease, anaerobically inducible | + | ||||
| tdcC-tdcD | Catabolic threonine dehydratase, anaerobic metabolism | + | ||||
| Restriction modification | spa1514 | Putative DNA/RNA-nonspecific endonuclease | + | |||
| hsdM | DNA methylase, protecting DNA against endonuclease | + | ||||
| hsdS | DNA methylase, protecting DNA against endonuclease | + | ||||
| hsdR | DNA methylase, protecting DNA against endonuclease | + | ||||
| Cell wall and surface structure | sefB | Fimbrial periplasmic chaperone, pilus assembly, adherence | + | |||
| sthC | Fimbrial usher protein, adherence | + | ||||
| murC | UDP-N-acetylmuramate:alanine ligase, cell wall synthesis | + | ||||
| mpl | Murein peptide ligase, cell wall synthesis | + | ||||
| gtrB | Glucosyl transferase, O-antigen conversion | + | ||||
| yjjZ | Inner membrane protein, function unknown | + | + | |||
| yfiO | Lipoprotein, outer membrane assembly | + | ||||
| Transcription | arcB | Aerobic respiration control sensor, global regulation | + | |||
| nusB | Transcription antitermination | + | + | + | ||
| rpoN | RNA polymerase sigma-54, nitrogen assimilation | + | ||||
COEC, chicken oviduct epithelial cells; HD11, macrophages.
FIG 1.
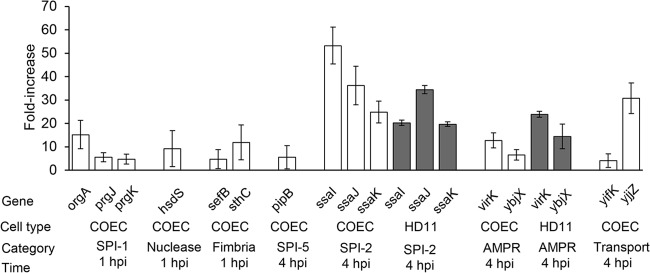
Quantitative analysis of genes overexpressed by S. Enteritidis upon infection in COEC and HD11 cells. Relevant genes found to be overexpressed in COEC or HD11 cells from the SCOTS experiment were quantified for their intracellular expression in these cell types using reverse transcriptase real-time PCR; the fold increase is shown (2−ΔΔCT ± SD, where SD is the standard deviation). Data shown are from three independent experiments performed with duplicate runs per assay. For each gene, the cell type, category, and time postinfection at which the expression occurred are shown under the graph.
Sensitivity of the S. Enteritidis ΔybjX mutant to EDTA and bile acids.
To determine the sensitivity of wild-type (ZM100), ΔvirK (ZM122), ΔybjX (ZM123), and ΔvirK ΔybjX (ZM124) S. Enteritidis to EDTA, each strain was seeded into LB agar and tested for its ability to grow in the presence of a filter disc containing 0.5 M EDTA. The zones of inhibition were measured and are shown as representative of relative sensitivity levels (Fig. 2A). The ΔybjX mutant, but not the ΔvirK and ΔvirK ΔybjX mutants, was significantly more sensitive to 0.5 M EDTA (9.1% more sensitive than the wild type; P < 0.05). Growth in 0.5 M EDTA was restored to the wild-type phenotype when pWSKYbjX was introduced into the corresponding mutant strain. To test the sensitivity of strains to bile acid, bacteria were grown in LB agar containing 1% deoxycholic acid (1% DOC). As shown in Fig. 2B, the ΔvirK ΔybjX mutant was significantly more susceptible (99.6% killed; P < 0.05) than any other strain to 1% DOC. These data indicate that AMPR genes play distinct roles in the susceptibility of S. Enteritidis to EDTA and 1% DOC, with ybjX having a greater role in this defense.
FIG 2.
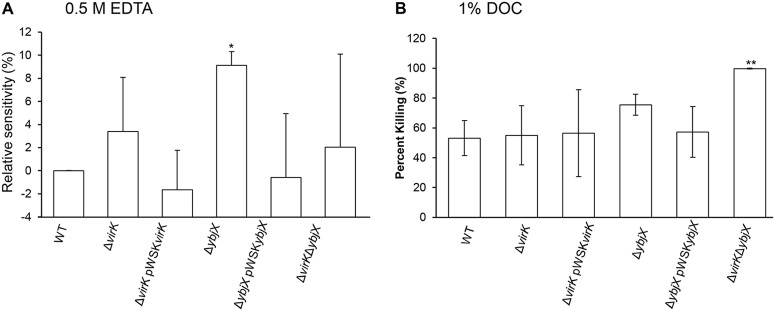
Sensitivity of AMPR mutants to EDTA and bile acid. Wild-type S. Enteritidis (WT; ZM100) and S. Enteritidis ΔvirK (ZM122), ΔybjX (ZM123), ΔvirK ΔybjX (ZM124), ΔvirK pWSKVirK (ZM122C), and ΔybjX pWSKYbjX (ZM123C) were tested for their sensitivity to EDTA and bile acid. (A) Sensitivity to 0.5 M EDTA was measured as zones of inhibition, and data are presented as percent WT inhibition. The ΔybjX mutant exhibited significantly increased sensitivity to 0.5 M EDTA. (B) Sensitivity to bile acid was measured by comparing bacterial growth in LB to that in LB containing 1% DOC, and data are presented as percent killing by DOC. The ΔvirK ΔybjX mutant was significantly more sensitive than the WT strain to 1% DOC. Data shown are from three independent experiments. A single asterisk denotes statistical significance at P < 0.05, and double asterisks denote statistical significance at P < 0.01.
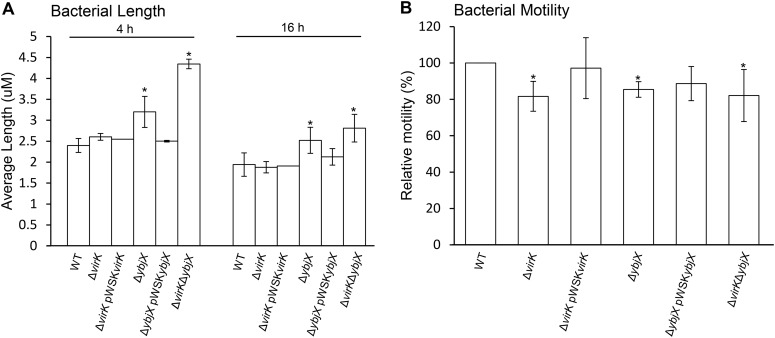
Altered cell morphology and motility of S. Enteritidis ΔybjX and ΔvirK ΔybjX mutants.
To characterize the functional contributions of AMPR genes to S. Enteritidis pathogenicity, we examined the differences between the wild-type and mutant S. Enteritidis strains in morphology and motility. During the logarithmic phase of growth and the stationary phase of growth (4 h and 16 h, respectively), the ΔybjX and ΔvirK ΔybjX mutants, but not the ΔvirK mutant, formed long filaments with average lengths (μm) and chain-forming tendencies exceeding those of the wild-type strain (Fig. 3A; P < 0.05). At logarithmic phase, the ΔvirK ΔybjX mutant exhibited a greater length (4.3 μm) than any of the other strains. Wild-type morphology was restored to the ΔybjX mutant when a cloned ybjX gene (pWSKYbjX) was introduced into the strain (Fig. 3A). Regardless of the variations in cell length, all three mutants showed significantly decreased motility (81.7% compared to 85.4% ZM100 motility; P < 0.05) (Fig. 3B). The motility defects of the ΔvirK and ΔybjX mutants were partially restored when the mutants were complemented with pWSKVirK and pWSKYbjX, respectively (Fig. 3B).
FIG 3.
Average length and motility of AMPR mutants. (A) Wild-type S. Enteritidis (WT; ZM100) and S. Enteritidis ΔvirK (ZM122), ΔybjX (ZM123), ΔvirK ΔybjX (ZM124), ΔvirK pWSKVirK (ZM122C), and ΔybjX pWSKYbjX (ZM123C) were grown to log phase (4 h) and stationary phase (16 h), photographed at ×400 magnification, and analyzed for average Feret length using ImageJ morphometric analysis. The ΔybjX and ΔvirK ΔybjX strains exhibited increased average lengths in both phases of growth. (B) Log-phase S. Enteritidis strains were inoculated onto 0.3% agar, and the swimming diameter (mm) of each strain was determined. Relative motility levels were determined following 3 h of incubation and are presented as percentages of WT motility. All three AMPR mutants showed decreased motility compared to WT strain ZM100. Data shown are from three independent experiments with duplicates in each assay. A single asterisk denotes statistical significance (P < 0.05).
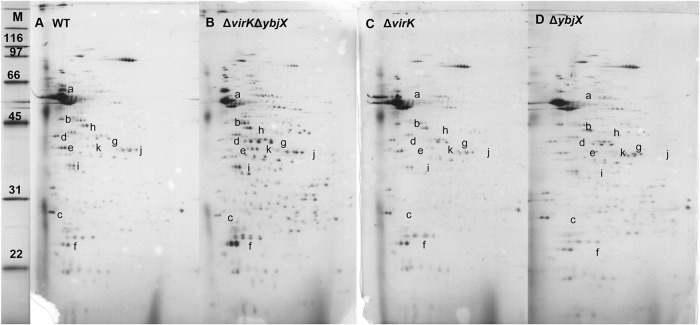
Altered profiles of secreted proteins of S. Enteritidis AMPR mutants.
The increased susceptibility of AMPR mutants to EDTA and deoxycholic acid along with the increased bacterial cell length suggested an altered outer membrane. To evaluate the integrity of the outer membrane of the AMPR mutants, we chose to assess their ability to appropriately secrete proteins. We extracted total proteins from the supernatants of the following log-phase S. Enteritidis strains: the wild type (ZM100), the ΔvirK mutant (ZM122), the ΔybjX mutant (ZM123), and the ΔvirK ΔybjX mutant (ZM124). We separated the supernatant proteins using 2D SDS-PAGE and analyzed the differences in protein spot densities. The 2D SDS-PAGE analysis revealed differences in the protein quantities between the wild-type strain and the mutant strains, especially the ΔvirK ΔybjX mutant (Fig. 4). We initially chose spots that exhibited at least a 30% difference between the wild-type and mutant strain results in densitometric analysis. Selected spots were excised and sequenced by LC-tandem mass spectrometry (LC-MS/MS) to determine their identities. The flagellum-associated proteins (FliC, FliK, FlgD, and FlgE) and Salmonella T3SS-1 invasion protein (SipD) were most abundant in the wild-type supernatant, decreased in abundance the supernatants of the ΔvirK and ΔybjX mutants, and most strikingly decreased in abundance in the supernatant of the ΔvirK ΔybjX mutant. The FliC protein, phase 1 flagellin, was not detectable in any of the 2D SDS-PAGE gels of the ΔybjX and ΔvirK ΔybjX mutants. The supernatants of the ΔvirK ΔybjX and ΔybjX mutants contained more bacterial cell wall proteins or plasma proteins, including EF Tu, a cell membrane-associated elongation factor Tu, OsmY, a periplasmic osmotically inducible protein, MalE, a periplasmic maltose transporter protein, GlpQ, a periplasmic glycerophosphoryl diester phosphodiesterase, EF Ts, a cytosolic elongation factor Ts, and GapA, a cytosolic glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Fig. 4 and Table 4).
FIG 4.
Differences in in vitro protein secretion. The proteins secreted into the culture supernatant from the log-phase wild-type strain and the AMPR mutants were extracted, separated using 2D SDS-PAGE, and analyzed densitometrically. The ΔvirK ΔybjX mutant displayed the greatest differences in spot densities (indicated by lowercase letters) compared to the wild-type strain. Representative 2D SDS-PAGE gels are shown as follows: lane M, protein ladder (kDa); lane A, wild-type S. Enteritidis (ZM100); lane B, S. Enteritidis ΔvirK ΔybjX (ZM124); lane C, S. Enteritidis ΔvirK (ZM122); lane D, S. Enteritidis ΔybjX (ZM123). Experiments were repeated three times with triplicate cultures in each run.
TABLE 4.
S. Enteritidis proteins that differ in their levels of in vitro-secreted abundance between the wild-type strain and AMPR mutants
| Spot IDa (see Fig. 5) | Molecular mass (kDa) | Gene symbol | Description | Cellular location | Protein coverage (%) | GenBank ID | Order of secreted protein abundanceb |
|---|---|---|---|---|---|---|---|
| a | 52 | fliC | Phase-1 flagellin | Secreted | 28.0 | AAA27085.1 | WT > Δvc |
| b | 42 | fliK | Flagellar hook length control protein | Secreted | 22.0 | P26416.2 | WT > Δv > Δy > ΔvΔy |
| c | 24 | flgD | Flagellar basal-body rod modification protein | Secreted | 10.0 | P0A1J0.1 | WT > Δv = Δy > ΔvΔy |
| d | 42 | flgE | Flagellar hook protein | Secreted | 40.0 | P0A1J2.2 | WT > Δv = Δy > ΔvΔy |
| e | 37 | sipD | T3SS-1 cell invasion protein | Secreted | 34.0 | Q56026.1 | WT > Δv > Δy > ΔvΔy |
| f | 21 | osmY | Osmotically inducible protein | Periplasm | 27.0 | P0AFH9.1 | ΔvΔy > Δv = Δy > WT |
| g | 43 | malE | Maltose ABC transporter substrate binding protein | Periplasm | 44.0 | P19576.2 | ΔvΔy > Δy > Δv > WT |
| h | 43 | tuf | Elongation factor Tu | Cell membrane | 5.3 | A7ZSL4.1 | ΔvΔy > Δv = Δy > WT |
| i | 30 | tsf | Elongation factor Ts | Cytosol | 13.0 | A8ALC0.2 | ΔvΔy > Δv = Δy > WT |
| j | 36 | gapA | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH-A) | Cytosol | 8.8 | P0A9B4.2 | ΔvΔy > Δy > Δv > WT |
| k | 41 | glpQ | Glycerophosphoryl diester phosphodiesterase | Periplasm | 6.7 | P09394.2 | ΔvΔy > Δy > Δv > WT |
ID, identifier.
The order of protein abundance was determined densitometrically from three 2D SDS-PAGE gels for the following S. Enteritidis strains: the wild type (WT), ΔvirK mutant (Δv), ΔybjX mutant (Δy), and ΔvirK ΔybjX mutant (ΔvΔy).
FliC protein secretion was not detectable in the ΔybjX (Δy) and ΔvirK ΔybjX (ΔvΔy) mutants with our methods.
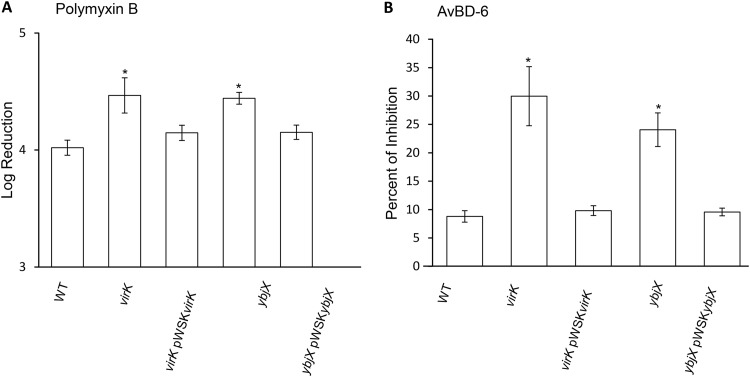
Increased susceptibility of S. Enteritidis AMPR mutants to antimicrobial peptides.
To assess the contributions of AMPR genes to antimicrobial peptide resistance, a series of bacterial growth inhibition or killing assays were performed. Treatment of S. Enteritidis strains with polymyxin B, a potent antimicrobial peptide, resulted in a 4-log reduction of the wild-type strain level and about 4.4-log reductions of the levels of the ΔvirK and ΔybjX mutants (Fig. 5A). Subsequently, S. Enteritidis strains were treated with AvBD-6, which resulted in growth inhibitions of 24.1% and 30% for virK and ybjX mutant strains, respectively, compared to 8.8% for the wild-type S. Enteritidis strain (Fig. 5B). Introduction of pWSKVirK and pWSKYbjX into the corresponding mutant strains complemented the increased sensitivity to polymyxin B and AvBD-6. These results indicate that both virK and ybjX contribute to the resistance of S. Enteritidis to antimicrobial peptides, an important arm of the host innate immune system.
FIG 5.
Sensitivity of AMPR mutants to antimicrobial peptides. Wild-type S. Enteritidis (ZM100) and S. Enteritidis virK (ZM112), ybjX (ZM114), virK pWSKVirK (ZM112C), and ybjX pWSKYbjX (ZM114C) were tested for their sensitivity to antimicrobial peptides. (A) S. Enteritidis strains were exposed to a general, potent antimicrobial peptide, polymyxin B, for 1 h and then plated for CFU enumerations. Results are presented as the log reduction in CFU. virK and ybjX mutants showed significantly increased susceptibility. (B) S. Enteritidis strains were exposed to a host-specific antimicrobial peptide, AvBD-6, using a microbroth dilution method. Sensitivity to AvBD-6 is expressed as percent growth inhibition. Both the virK mutant and the ybjX mutant had increased susceptibility to AvBD-6. Data shown are from three independent experiments, and the average values are shown in parentheses. A single asterisk denotes significant differences (P < 0.05).
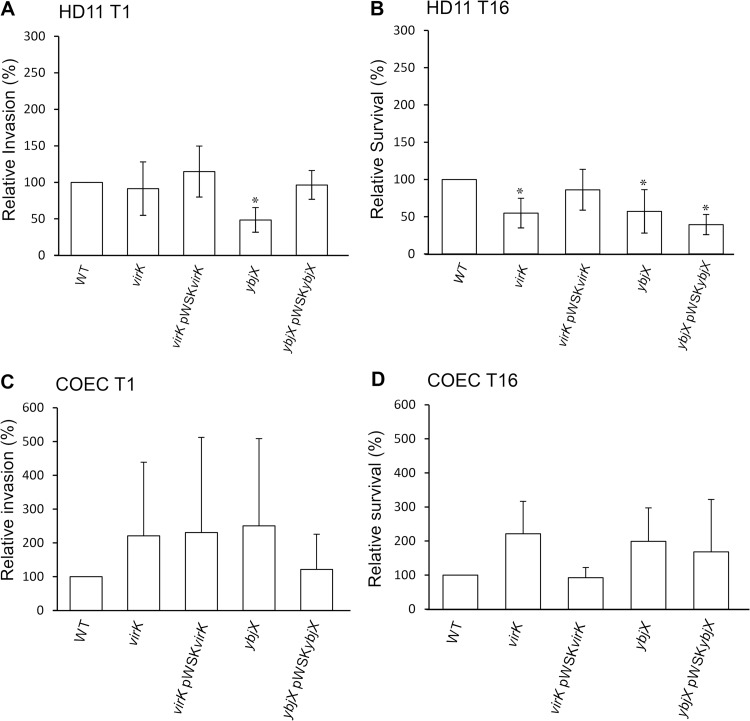
Altered interactions of S. Enteritidis AMPR mutants with macrophage HD11 and COEC.
To assess the interaction between the AMPR mutants and chicken cells, we used a gentamicin protection assay to test the contribution of virK and ybjX to the entry and survival of S. Enteritidis in avian macrophages (HD11) and primary chicken oviduct epithelial cells (COEC). Our results showed that virK was not involved in the entry of S. Enteritidis into HD11 macrophages (Fig. 6A). In contrast, disruption of ybjX resulted in significantly decreased entry into HD11 cells (45% of wild type; P < 0.05). The defect demonstrated by the ybjX mutant was fully restored by introducing pWSKYbjX into the ybjX mutant (Fig. 6A). At 16 hpi, lower numbers of intracellular bacteria were recovered from HD11 cells infected with the virK and ybjX mutants than from those infected with the wild type (Fig. 6B; P < 0.05). The survival defect of the virK mutant, but not that of the ybjX mutant, was partially complemented by the cloned gene. The ybjX and virK mutants did not show any significant defect in entry of and intracellular survival in COEC (Fig. 6C and D).
FIG 6.
Relative invasion (or entry) and survival of AMPR mutants in chicken macrophage and reproductive epithelial cells. HD11 chicken macrophages and COEC were infected with stationary-phase wild-type S. Enteritidis (WT; ZM100) and S. Enteritidis virK (ZM112), ybjX (ZM114), virK pWSKVirK (ZM112C), and ybjX pWSKYbjX (ZM114C), and a gentamicin protection assay was performed. S. Enteritidis bacteria recovered after 1 hpi were analyzed to determine their ability to enter macrophages (A) or COEC (C); the results are shown as percentages of the WT CFU index counts. S. Enteritidis bacteria recovered after 16 hpi were analyzed to determine their ability to survive inside macrophages (B) or COEC (D); the results are shown as averages of the percentages of the WT recovery levels. Each assay was run in triplicate and repeated three times. A single asterisk denotes statistical significance (P < 0.05).
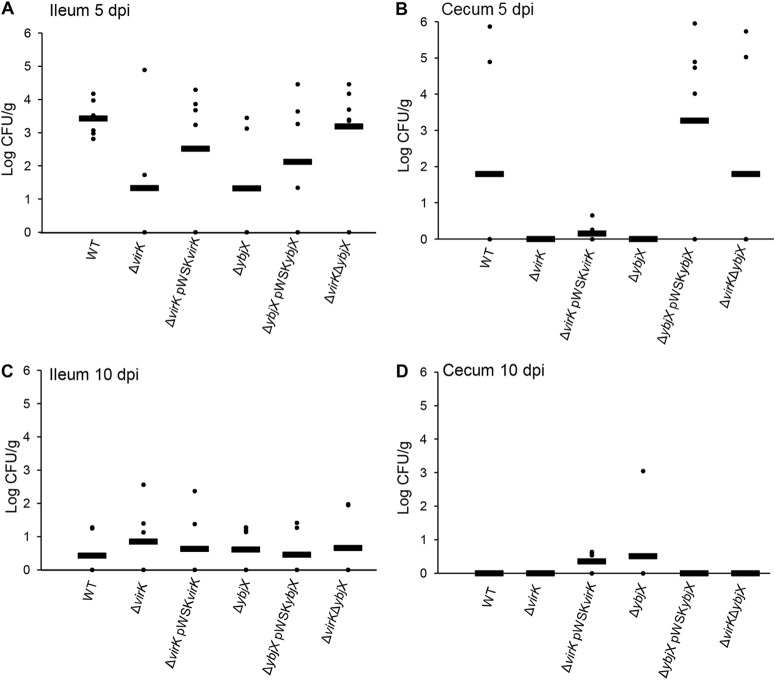
Reduced ability of S. Enteritidis AMPR mutants to survive in the intestinal lumen and to shed in feces.
To investigate the contribution of virK and ybjX genes to intestinal colonization and environmental spread during S. Enteritidis infection in chickens, we collected fecal samples at the same time every day following oral inoculation of 21-week-old laying hens with 5 × 109 CFU of wild-type or mutant strains. At 5 and 10 dpi, we cultured ileal and cecal tissue samples and the contents of the euthanized hens. We did not observe any significant differences in tissue bacterial loads among experimental groups (Fig. 7). In contrast, the bacterial loads of the ΔvirK ΔybjX mutant in cecal content were significantly lower than those of the wild-type strain (Fig. 8B; P < 0.05). Infection with the ΔvirK ΔybjX mutant also resulted in a significant decrease in fecal S. Enteritidis loads (Fig. 9A; P < 0.05) from 1 dpi to 5 dpi and significantly reduced number of shedders throughout the course of infection (Table 5). The compromised ability of the ΔvirK ΔybjX mutant to survive in cecal content from 1 dpi to 5 dpi coincided with reduced fecal bacterial loads and reduced numbers of shedders. Decreased fecal Salmonella load was also detected following infection with the ΔybjX pWSKYbjX mutant (Fig. 9A), suggesting a possibility of compromised survival of this strain in the fecal environment.
FIG 7.
S. Enteritidis bacterial loads in intestinal tissue. Wild-type (ZM100) and ΔvirK (ZM122), ΔybjX (ZM123), ΔvirK ΔybjX (ZM124), ΔvirK pWSKVirK (ZM122C), and ΔybjX pWSKYbjX (ZM123C) bacteria were used to orally infect 21-week-old laying hens (12 hens per strain) at a dose of 5 × 109 CFU per hen. S. Enteritidis intestinal loads were determined at 5 and 10 dpi. No statistical significance of the results of comparisons of WT and mutant strains was observed. (A) Ileum at 5 dpi (PvirK = 0.0546 and PybjX = 0.0556). (B) Cecum at 5 dpi (PvirK = 0.1186 and PybjX = 0.1186). (C) Ileum at 10 dpi (P > 1). (D) Cecum at 10 dpi (PybjX = 0.1022). Dots represent individual bird bacterial loads, and bars represent the averages.
FIG 8.
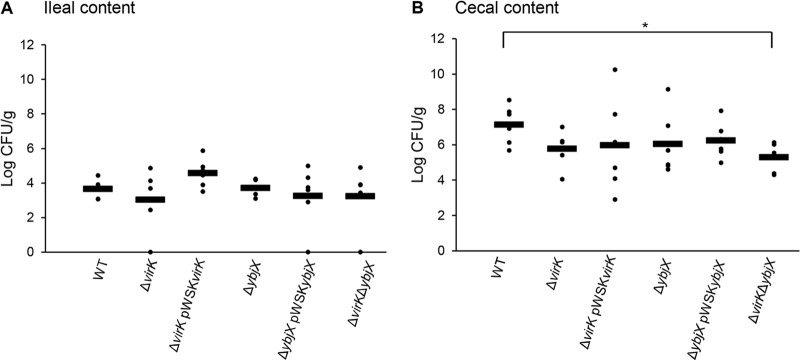
S. Enteritidis bacterial load in intestinal content. Wild-type (ZM100), ΔvirK (ZM122), ΔybjX (ZM123), ΔvirK ΔybjX (ZM124), ΔvirK pWSKVirK (ZM122C), and ΔybjX pWSKYbjX (ZM123C) bacteria were used to orally infect 21-week-old laying hens (12 hens per strain) with 5 × 109 CFU per hen. The number of S. Enteritidis bacteria in intestinal content was determined at 5 dpi. A significant difference between the WT strain and the ΔvirK ΔybjX double-mutation mutant in bacterial loads in the cecal content (B) was detected (* denotes P < 0.05). Dots represent individual bird loads, and bars represent the averages.
FIG 9.
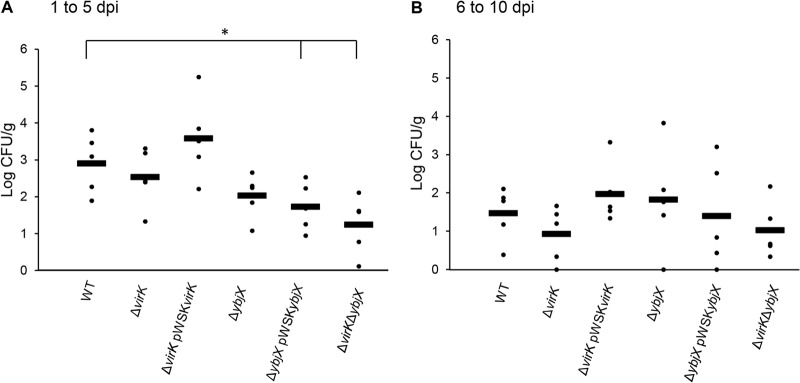
Fecal bacterial load of AMPR mutants during the early and late stages of infection. Wild-type S. Enteritidis (ZM100) and S. Enteritidis ΔvirK (ZM122), ΔybjX (ZM123), ΔvirK ΔybjX (ZM124), ΔvirK pWSKVirK (ZM122C), and ΔybjX pWSKYbjX (ZM123C) were used to orally infect 21-week-old laying hens (12 hens per strain) at a dose of 5 × 109 CFU per hen. A sample of feces was collected daily from each bird. Tenfold dilutions of samples in peptone-buffered water were plated to determine fecal bacterial loads. (A) Fecal bacterial loads from 1 to 5 dpi. (B) Fecal loads from 6 to 10 dpi. Significant differences in fecal loads collected from 1 to 5 dpi were detected (a single asterisk denotes P < 0.05). Each dot represents the average bacterial load of each strain per dpi, and each bar represents the overall average from 5 days.
TABLE 5.
Hens shedding S. Enteritidis strains in their feces each day postinfection (dpi)a
| Strain | No. of hens shedding S. Enteritidis strains on indicated dpi/total no. of hens (% positive) |
% avg | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| S. Enteritidis WT | 4/6 (66.7) | 4/6 (66.7) | 4/6 (66.7) | 4/6 (66.7) | 4/6 (66.7) | 4/6 (66.7) | 3/6 (50.0) | 1/6 (16.7) | 4/6 (66.7) | 3/6 (50.0) | 58.3 |
| S. Enteritidis ΔvirK | 1/6 (16.7) | 4/6 (66.7) | 5/6 (83.3) | 5/6 (83.3) | 4/6 (66.7) | 3/6 (50.0) | 2/6 (33.3) | 0/6 (0.0) | 3/6 (50.0) | 1/6 (16.7) | 46.7 |
| S. Enteritidis ΔvirK pWSKVirK | 5/6 (83.3) | 5/6 (83.3) | 3/6 (50.0) | 6/6 (100.0) | 3/6 (50.0) | 5/6 (83.3) | 3/6 (50.0) | 2/6 (33.3) | 2/6 (33.3) | 3/6 (50.0) | 61.7 |
| S. Enteritidis ΔybjX | 3/6 (50.0) | 3/6 (50.0) | 4/6 (66.7) | 2/6 (33.3) | 4/6 (66.7) | 3/6 (50.0) | 5/6 (83.3) | 2/6 (33.3) | 0/6 (0.0) | 3/6 (50.0) | 48.3 |
| S. Enteritidis ΔybjX pWSKYbjX | 3/6 (50.0) | 3/6 (50.0) | 3/6 (50.0) | 2/6 (33.3) | 3/6 (50.0) | 4/6 (66.7) | 4/6 (66.7) | 2/6 (33.3) | 1/6 (16.7) | 1/6 (16.7) | 41.7 |
| S. Enteritidis ΔvirK ΔybjX | 2/6 (33.3) | 2/6 (33.3) | 3/6 (50.0) | 1/6 (16.7) | 1/6 (16.7) | 1/6 (16.7) | 3/6 (50.0) | 2/6 (33.3) | 4/6 (66.7) | 1/6 (16.7) | 33.3* |
Fecal samples were collected daily during the infection challenge of 21-week-old laying hens with the S. Enteritidis strains. One gram of feces was cultured to determine fecal shedding. The fecal samples from 1 dpi to 5 dpi represented two hens, and the samples from 6 dpi to 10 dpi represented one hen. A single asterisk denotes a significant difference in the average number of shedding hens inoculated with the double mutant versus the WT (P < 0.05).
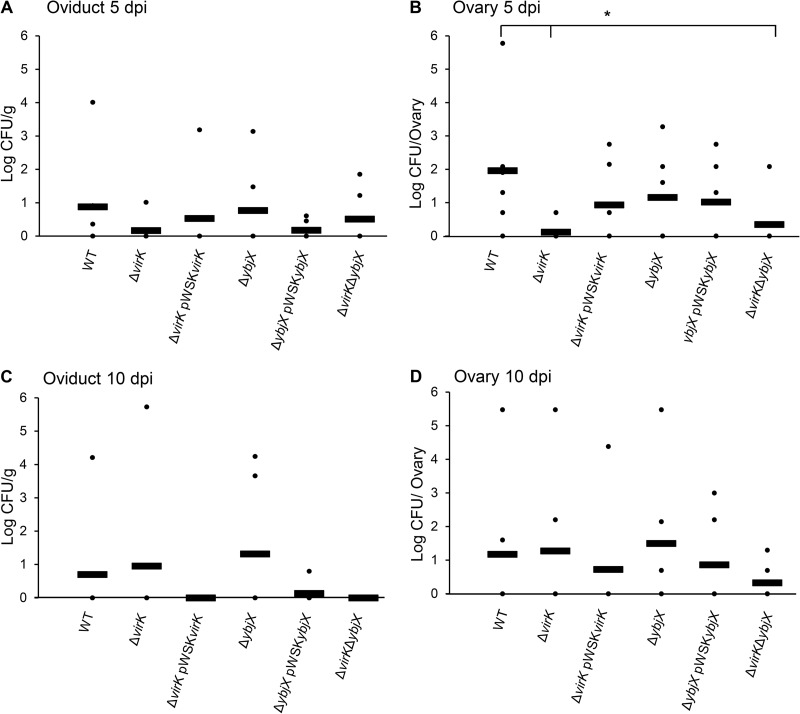
S. Enteritidis AMPR genes involved in reproductive-tract colonization and egg contamination.
To investigate the roles played by AMPR genes during S. Enteritidis colonization of the reproductive tract of laying hens and, ultimately, contamination of eggs, we collected eggs at the same time every day and ovaries and oviduct (isthmus) tissues at 5 dpi and 10 dpi from orally inoculated hens. The egg and tissue samples were tested for the presence and bacterial load of S. Enteritidis.
At 5 dpi, significantly reduced numbers of S. Enteritidis were recovered from the ovaries of the hens infected with the ΔvirK and ΔvirK ΔybjX mutants compared to those infected with the wild-type strain (Fig. 10B; P < 0.05). In contrast, the ΔybjX mutant did not show a defect in ovary colonization at 5 dpi. At 10 dpi, no significant difference in ovary bacterial load was observed among treatment groups. No significant difference in oviduct bacterial load was observed among treatment groups at 5 dpi and 10 dpi.
FIG 10.
The bacterial loads of AMPR mutants in reproductive tissues. Wild-type S. Enteritidis (ZM100) and S. Enteritidis ΔvirK (ZM122), ΔybjX (ZM123), ΔvirK ΔybjX (ZM124), ΔvirK pWSKVirK (ZM122C), and ΔybjX pWSKYbjX (ZM123C) were used to orally infect 21-week-old laying hens (12 hens per strain) at a dose of 5 × 109 CFU per hen. Oviducts (isthmus) and ovaries were collected at 5 and 10 dpi, homogenized, and plated to determine S. Enteritidis loads. (A) Oviduct (isthmus) at 5 dpi. (B) Ovary at 5 dpi. (C) Oviduct (isthmus) at 10 dpi. (D) Ovary at 10 dpi. Each dot represents the bacterial load in an individual bird, each bar represents the average bacterial load in an individual group, and a single asterisk denotes statistical significance (P < 0.05).
As shown by analysis of S. Enteritidis-positive ovaries at 5 dpi (Table 6), the wild-type strain infected the greatest proportion of ovaries (83.3%) and the ΔvirK and ΔvirK ΔybjX mutants infected the lowest proportion of ovaries (16.7%). At 10 dpi, the ΔybjX mutant infected the greatest proportion of ovaries (66.7%) whereas the ΔvirK and ΔvirK ΔybjX mutants infected as many as the wild type (33.3%). Comparing these data to the percentages of contaminated eggs, we noticed that the ΔybjX mutant was the only strain isolated from egg contents from 6 dpi to 9 dpi. The complemented ΔybjX strain (ZM123C) was less able to contaminate eggs and to colonize ovaries (Table 6). The data suggest that AMPR genes play distinct roles in reproductive-tract colonization, with virK contributing to reproductive-tract colonization and egg deposition during the early stages of infection and ybjX playing a counteractive role in reproductive persistence and egg deposition.
TABLE 6.
S. Enteritidis deposition in eggs and colonization in ovariesa
| Strain | No. of ovaries with positive results/total no. of ovaries (% positive) on indicated dpi |
No. of eggs with positive results/total no. of eggs (% positive) on indicated dpi |
Avg % positive | Total no. of eggs with positive results/total no. of eggs (% positive) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 10 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |||
| WT | 5/6 (83.3) | 2/6 (33.3) | 1/5 (20.0) | 1/5 (20.0) | 1/5 (20.0) | 1/3 (33.3) | 0/6 (0.0) | 0/2 (0.0) | 0/2 (0.0) | 0/1 (0.0) | 0/2 (0.0) | 0/4 (0.0) | 9.33 | 4/35 (11.4) |
| ΔvirK | 1/6 (16.7) | 2/6 (33.3) | 4/8 (50.0) | 1/7 (14.3) | 1/6 (16.7) | 0/4 (0.0) | 0/6 (0.0) | 0/4 (0.0) | 0/3 (0.0) | 0/4 (0.0) | 0/4 (0.0) | 0/6 (0.0) | 8.1 | 6/52 (11.5) |
| ΔvirK pWSKVirK | 3/6 (50.0) | 1/6 (16.7) | 1/6 (16.7) | 2/8 (25.0) | 0/3 (0.0) | 2/5 (40.0) | 2/8 (25.0) | 0/6 (0.0) | 0/3 (0.0) | 0/3 (0.0) | 0/2 (0.0) | 0/7 (0.0) | 10.7 | 7/51 (13.7) |
| ΔybjX | 3/6 (50.0) | 4/6 (66.7) | 2/6 (33.3) | 1/7 (14.3) | 1/2 (50.0) | 2/4 (50.0) | 2/4 (50.0) | 1/4 (25.0) | 1/1 (100) | 0/2 (0.0) | 1/3 (33.3) | 0/4 (0.0) | 35.5* | 11/37 (29.7) |
| ΔybjX pWSKYbjX | 3/6 (50.0) | 2/6 (33.3) | 2/3 (66.7) | 0/2 (0.0) | 0/2 (0.0) | 0/3 (0.0) | 0/2 (0.0) | 0/4 (0.0) | 0/2 (0.0) | 0/1 (0.0) | 0/2 (0.0) | 0/4 (0.0) | 6.7 | 2/25 (8.0) |
| ΔvirK ΔybjX | 1/6 (16.7) | 2/6 (33.3) | 2/5 (40.0) | 1/3 (33.3) | 2/3 (66.7) | 1/2 (50.0) | 0/3 (0.0) | 0/2 (0.0) | 0/1 (0.0) | 0/1 (0.0) | 0/1 (0.0) | 0/2 (0.0) | 19.0 | 6/23 (26.1) |
Eggs were collected at the same time daily during the infection challenge of 21-week-old laying hens with 5 × 109 CFU of S. Enteritidis strains. Data for eggs with positive results are in boldface. The number of hens with positive ovary colonization results is shown as the fraction of the total number of hens tested at each of two tissue collection time points. A single asterisk denotes a significant difference from the results determined for the WT strain in the average numbers of eggs with positive results on the indicated dpi (%) and in the amounts of eggs with positive results on the indicated dpi (P < 0.05).
DISCUSSION
The main vehicle of S. Enteritidis infection in humans is thought to be contaminated poultry products (3). The prevalence of S. Enteritidis-associated infections has declined since 1999, but outbreaks associated with this organism seem to persist in our society (4, 5). Compared to other serovars, persistent outbreaks have been hypothesized to occur because S. Enteritidis is more suitable to persistent colonization of the reproductive tract of laying hens without inducing overt clinical symptoms (17). While many studies have confirmed that S. Enteritidis exploits inherent differences to purposefully colonize the reproductive tract and contaminate eggs better than other serovars, the exact mechanisms for this action have yet to be discovered (15, 17, 18, 40).
Many experiments performed with S. Enteritidis focus on the mechanisms of the T3SS-1 and T3SS-2 during invasion and intracellular replication within various chicken tissues (10, 41–43). Recent studies have focused on survival strategies of S. Enteritidis under the stressful conditions encountered by pathogens of the chicken such as those present in egg white. Those studies have identified mutations in rpoS, SPI-14 genes, and ksgA that cause specific attenuation in S. Enteritidis virulence and specific attenuation in chicken liver invasion and macrophage survival (44, 45). In the present study, we used a selective capture of transcribed genes (SCOTS) assay to identify the genes overexpressed by S. Enteritidis upon entry and survival in chicken macrophages (HD11) and chicken oviduct epithelial cells (COEC) and characterized those genes that were identified as antimicrobial peptide resistance genes (AMPR genes).
Salmonella enterica utilizes macrophages as a transport vessel to invade systemic sites within the host (46). Once S. Enteritidis invades the reproductive tract, successful egg contamination by S. Enteritidis most likely happens during egg development, with S. Enteritidis colonization predominantly found in the isthmus of the oviduct (15, 47). Therefore, in our study, selection of chicken macrophage HD11 cells and COEC was essential to identify those genes employed by S. Enteritidis during successful and persistent reproductive-tract colonization. The genes found to be overexpressed in HD11 and COEC consisted of a cohort of stress response, transport, cell wall and DNA modification, fimbrial, AMPR, and virulence genes. The identification of SPI-1 genes at 1 hpi and an SPI-2 gene at 4 hpi validated the authenticity of the results of the SCOTS experiment, as these genes are known to be involved in invasion and intracellular replication, respectively (11, 20, 48). The overexpression of SPI-2, SPI-5, and AMPR genes was confirmed by real-time PCR, indicating the utilization of these genes for replication and survival within these chicken cells. SPI-2 genes and the SPI-5 pipB gene have been studied for their effects during S. Enteritidis infection in hens, but there is a lack of knowledge of the role AMPR genes virK and ybjX play during S. Enteritidis infection in chickens (10, 24, 42, 43).
The AMPR virK and ybjX genes discovered in the SCOTS experiment are a part of the PhoP/PhoQ regulon, which consists of over 40 genes speculated to modulate the bacterial outer membrane to contribute to antimicrobial resistance, virulence, and survival under low-Mg2+ conditions (13, 49–52). In Shigella flexneri, virK is hypothesized to modulate the outer membrane to alter the interaction between IcsP (an actin-modulating protein) and lipid A (52). In S. Typhimurium, a mutation in ybjX (initially termed somA) was found as a suppressor mutation corresponding to a mutation in the MsbB lipid A assembly protein, functionally linking ybjX to outer membrane modification (53). The outer membrane serves several bacterial functions, including stabilization for various functions, such as movement and secretion, and defense against host killing tactics. Mutations that affect outer membrane stability make the bacteria sensitive to detergents, to which Salmonella is naturally resistant (8, 24). We have shown that AMPR genes in S. Enteritidis contribute to outer membrane stability for resistance to EDTA and bile acid deoxycholate (Fig. 4). EDTA is capable of chelating the divalent Mg2+ and Ca2+ cations that link the outer membrane lipopolysaccharide (LPS) molecules. To overcome the reduction in connective cation levels caused by EDTA, Salmonella enterica must actively employ membrane-stabilizing mechanisms, such as addition of myristic acid to the lipid A portion of LPS, to maintain a strong barrier (53). The inability to add more stabilizing fatty acids to anchor LPS molecules in the outer membrane results in increased susceptibility to EDTA (53). Mutations that abolish the synthesis of lipid A (and thus LPS) are lethal to enteric bacteria because LPS makes up a majority of the outer membrane and is responsible for most of its characteristics (54). Deoxycholate is a bile acid, an anionic detergent, which can insert into the bacterial cell membrane, resulting in bacterial death. Salmonella organisms possess an outer membrane that is naturally resistant to the insertion of this detergent (8). Therefore, any alterations in the outer membrane, possibly caused by ΔybjX and ΔvirK ΔybjX, would lead to increased susceptibility to deoxycholate.
In S. Typhimurium, a mutation in msbB caused formation of elongated cells and a mutation in tatB or tatC (encoding twin arginine transport proteins required for transporting outer membrane components) caused long aggregate filaments to form (34, 53). Although a mutation in somA (ybjX) suppressed many phenotypes associated with the msbB mutant, the phenotypes related to a single mutation in somA (ybjX) were not characterized in the previous study (53). In the current investigation, we observed an increase in cell length and filamentous formations for the ΔybjX and ΔvirK ΔybjX mutants (Fig. 3). Therefore, the similar morphologies seen with our mutants and the mutants in previous studies that altered an outer membrane component collectively suggest that AMPR genes in S. Enteritidis play a role in outer membrane modulation.
Flagella contribute to bacterial virulence, and proper flagellum formation and motorization require a stable outer membrane (33, 55, 56). The present study showed that mutations in AMPR genes virK and ybjX have an impact on S. Enteritidis motility. It is likely that these two genes disrupt flagellar function by the same mechanism, because the double-deletion (ΔvirK ΔybjX) mutant displayed motility defects similar to those of the individual (ΔvirK and ΔybjX) mutants (Fig. 3). In S. Typhimurium, a mutation in msbB (waaN) resulted in an inability to secrete Salmonella effector proteins (36, 53). The current study showed that mutations in AMPR genes affect the ability to secrete proteins required for flagellar function (Fig. 5 and Table 4). FliC, FliK, FlgD, and FlgE proteins are secreted through a secretion system similar to the type three and type five secretion systems (56, 57). The inability to secrete these flagellar proteins may explain the decreased motility seen in the three AMPR mutants (58). The decreased secretion of SipD, a T3SS-1 invasion protein, in the AMPR mutants may alter the ability of these cells to invade host tissue (20). Mutations in AMPR genes also caused an increase in the levels of proteins in the supernatant that are naturally found in the cell membrane (Tuf [59]), in the periplasm (OsmY, MalE, and GlpQ [60–62]), or in the cytosol (Tsf and GapA [63, 64]). The decreased secretion of flagellar proteins and the increased presence of membrane proteins in the supernatants of the ΔvirK ΔybjX mutant and the ΔybjX mutant indicate an unstable outer membrane or membrane shearing.
A study in S. Typhimurium has shown that AMPR genes, through direct or indirect modification of LPS, contribute to resistance to antimicrobial peptide polymyxin B (49). We have shown in the present study not only that virK and ybjX are involved in resistance to polymyxin B but also that they are involved in resistance to AvBD-6, a host antimicrobial peptide crucial in the innate immune system (Fig. 5). Therefore, it is obvious that these AMPR genes are required for the stability of the outer membrane of S. Enteritidis, which in turn affects the ability of the organism to coordinate virulence functions and defend against innate antimicrobial peptides.
PhoP/PhoQ-regulated genes have been shown to aid in virulence and play a role in late stages of S. Typhimurium infection in mice (13, 49, 51). The mechanism behind this phenomenon is that genes in the PhoP/PhoQ regulon, known to be upregulated within macrophages, alter the outer membrane structure or composition for defense against the various host killing factors within these phagocytes (7, 13). We have seen that a mutation in ybjX is associated with decreased macrophage entry, due either to a suboptimal interaction with the macrophage or to the inability of macrophages to phagocytize the larger bacteria (Fig. 3 and 6). We have observed a survival defect of AMPR mutants inside macrophages (Fig. 6). However, we did not detect any significant defects in the ability of the mutants to colonize spleen at 5 dpi and 10 dpi (data not shown). Our data are in disagreement with findings of a previous study of S. Typhimurium in mice that suggested that these AMPR genes play a role in late stages of infection (49). At this time, it is not known whether the difference is serovar specific or host specific or both.
To determine the contribution of AMPR genes to intestinal colonization, we analyzed the intestinal and fecal bacterial loads. In early stages of infection, the ΔvirK ΔybjX double mutant was excreted at the lowest level (Fig. 9A), which corresponded to its inability to survive in cecal contents and its inability to withstand deoxycholic acid (Fig. 3 and 8). Complementation of ybjX also caused a reduced fecal bacterial load which could not be explained by the strain's ability to survive in cecal lumen or to resist deoxycholic acid. It has been previously observed that the outer core of LPS is required for the entry of S. Typhimurium into intestinal epithelial cells (65). However, due to experimental variations, we did not observe significant intestinal tissue colonization defects associated with virK and ybjX mutations.
S. Enteritidis is known to preferentially colonize the reproductive tract of laying hens and contaminate eggs without inducing overt clinical signs (16, 17). S. Enteritidis strains recovered from the field have a higher degree of heterogeneity, especially in the glucosylation in the O-chain of the LPS, than S. Typhimurium strains (19, 66). This heterogeneity may be caused by alternative implementations of outer membrane modification stress mechanisms in the various host environments for defense against S. Enteritidis (14, 17). We have seen extensive heterogeneity associated with ybjX. Both deletion and complementation of ybjX could alter the phenotypes of S. Enteritidis in terms of cell length, colony size, and tissue colonization. While a mutation in virK renders S. Enteritidis incapable of colonizing the ovary (Fig. 10B and Table 6), deletion of ybjX resulted in prolonged colonization of ovaries (Table 6) and egg contamination (Fig. 10 and Table 6). It has been previously shown that an S. Enteritidis (wzz) mutant lacking high-molecular-mass LPS (HMM-LPS) was more effective than the wild-type strain with respect to reproductive-tract colonization and egg deposition (67). It was suggested that the presence of HMM-LPS allows S. Enteritidis to silently colonize the chicken host by mitigating reproductive-tract responses to infection but does so without altering the incidence of egg contamination (67). A recent study identified S. Enteritidis LPS biosynthesis as a critical factor involved in egg white persistence (68). Our data suggest that virK promotes ovary colonization at the early stage of infection and that ybjX negatively controls late-stage ovary colonization and egg deposition. The counteractive balance may be achieved through a suppressor role of ybjX, as suggested by a previous study (53). This hypothesis is supported by the observation that introduction of (more than one copy of) a cloned ybjX gene exacerbated phenotypes.
S. Enteritidis and S. Typhimurium are anywhere from 6.4% to 9.6% variant in their genes, with the most obvious difference being the chemical structures of their outer membrane, one of the key components in the interaction between Salmonella and its host (21, 22). Many investigators have argued that the main difference between S. Typhimurium and S. Enteritidis with respect to survival in the chicken host is in the use of stress-induced mechanisms, including outer membrane modulation, and that this accounts for the differences seen in pandemics associated with contaminated poultry products (14, 17, 19, 46). Silent colonization of S. Enteritidis in chicken requires intrinsic abilities to defend against the innate immune system without inducing overt inflammation and damage (16). This report shows that AMPR genes virK and ybjX not only aid in resistance to innate antimicrobial peptides and detergents but are also involved in replication and persistence in chicken tissues and eggs. Also, the effects of AMPR genes seen in S. Enteritidis do not correspond to the effects seen in an infection of mice with S. Typhimurium. Future experiments will be aimed at elucidating the exact mechanistic actions of outer membrane modulation by AMPR genes and looking at how these outer membrane modulations reshape the interaction with the chicken immune system to aid in silent colonization.
ACKNOWLEDGMENTS
We are grateful to Larry Dangott and the staff members of the Texas A&M University Protein Chemistry Laboratory for their assistance in running the experiments using 2D SDS-PAGE gels, tandem mass spectrometry, and protein analysis, Dale Hyatt and the staff members of the Poultry Science Center for raising the hens, and the staff members of the Veterinary Medical Park and the Comparative Medicine Program and Amber Nava at Texas A&M University for all their assistance with the in vivo trials.
Footnotes
Published ahead of print 29 September 2014
REFERENCES
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 17:7–15. 10.3201/eid1701.09-1101p1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braden CR. 2006. Salmonella enterica serotype Enteritidis and eggs: a national epidemic in the United States. Clin. Infect. Dis. 43:512–517. 10.1086/505973. [DOI] [PubMed] [Google Scholar]
- 3.Kimura AC, Reddy V, Marcus R, Cieslak PR, Mohle-Boetani JC, Kassenborg HD, Segler SD, Hardnett FP, Barrett T, Swerdlow DL. 2004. Chicken consumption is a newly identified risk factor for sporadic Salmonella enterica serotype Enteritidis infections in the United States: a case-control study in FoodNet sites. Clin. Infect. Dis. 38(Suppl 3):S244–S252. 10.1086/381576. [DOI] [PubMed] [Google Scholar]
- 4.Patrick ME, Adcock PM, Gomez TM, Altekruse SF, Holland BH, Tauxe RV, Swerdlow DL. 2004. Salmonella enteritidis infections, United States, 1985–1999. Emerg. Infect. Dis. 10:1–7. 10.3201/eid1001.020572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control. 2010. Multistate outbreak of human Salmonella Enteritidis infections associated wtih shell eggs (final update: 8 November 2013). Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/salmonella/enteritidis/. [Google Scholar]
- 6.Fang FC, Libby SJ, Buchmeier NA, Loewen PC, Switala J, Harwood J, Guiney DG. 1992. The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc. Natl. Acad. Sci. U. S. A. 89:11978–11982. 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy K, Travers P, Walport M. 2011. Janeway's immunobiology. Garland Science, Taylor & Francis Group, LLC, New York, NY. [Google Scholar]
- 8.van Velkinburgh JC, Gunn JS. 1999. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 67:1614–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finlay BB, Brumell JH. 2000. Salmonella interactions with host cells: in vitro to in vivo. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:623–631. 10.1098/rstb.2000.0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S, Zhang Z, Pace L, Lillehoj H, Zhang S. 2009. Functions exerted by the virulence-associated type-three secretion systems during Salmonella enterica serovar Enteritidis invasion into and survival within chicken oviduct epithelial cells and macrophages. Avian Pathol. 38:97–106. 10.1080/03079450902737771. [DOI] [PubMed] [Google Scholar]
- 11.Figueira R, Watson KG, Holden DW, Helaine S. 2013. Identification of salmonella pathogenicity island-2 type III secretion system effectors involved in intramacrophage replication of S. enterica serovar Typhimurium: implications for rational vaccine design. mBio 4:e00065. 10.1128/mBio.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giacomodonato MN, Uzzau S, Bacciu D, Caccuri R, Sarnacki SH, Rubino S, Cerquetti MC. 2007. SipA, SopA, SopB, SopD and SopE2 effector proteins of Salmonella enterica serovar Typhimurium are synthesized at late stages of infection in mice. Microbiology 153:1221–1228. 10.1099/mic.0.2006/002758-0. [DOI] [PubMed] [Google Scholar]
- 13.Groisman EA. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835–1842. 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Gast R, Humphrey TJ, Van Immerseel F. 2009. Mechanisms of egg contamination by Salmonella Enteritidis. FEMS Microbiol. Rev. 33:718–738. 10.1111/j.1574-6976.2008.00161.x. [DOI] [PubMed] [Google Scholar]
- 15.Gast RK, Guraya R, Guard-Bouldin J, Holt PS, Moore RW. 2007. Colonization of specific regions of the reproductive tract and deposition at different locations inside eggs laid by hens infected with Salmonella enteritidis or Salmonella heidelberg. Avian Dis. 51:40–44. 10.1637/0005-2086(2007)051[0040:COSROT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 16.Bedrani L, Helloin E, Guyot N, Rehault-Godbert S, Nys Y. 2013. Passive maternal exposure to environmental microbes selectively modulates the innate defences of chicken egg white by increasing some of its antibacterial activities. BMC Microbiol. 13:128. 10.1186/1471-2180-13-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Immerseel F. 2010. Stress-induced survival strategies enable Salmonella Enteritidis to persistently colonize the chicken oviduct tissue and cope with antimicrobial factors in egg white: a hypothesis to explain a pandemic. Gut Pathog. 2:23. 10.1186/1757-4749-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Vylder J, Raspoet R, Dewulf J, Haesebrouck F, Ducatelle R, Van Immerseel F. 2013. Salmonella Enteritidis is superior in egg white survival compared with other Salmonella serotypes. Poult. Sci. 92:842–845. 10.3382/ps.2012-02668. [DOI] [PubMed] [Google Scholar]
- 19.Guard-Petter J. 2001. The chicken, the egg and Salmonella enteritidis. Environ. Microbiol. 3:421–430. 10.1046/j.1462-2920.2001.00213.x. [DOI] [PubMed] [Google Scholar]
- 20.Ramos-Morales F. 2012. Impact of Salmonella enterica type III secretion system effectors on the eukaryotic host cell. ISRN Cell Biol. 10.5402/2012/787934. [DOI] [Google Scholar]
- 21.Porwollik S, Santiviago CA, Cheng P, Florea L, Jackson S, McClelland M. 2005. Differences in gene content between Salmonella enterica serovar Enteritidis isolates and comparison to closely related serovars Gallinarum and Dublin. J. Bacteriol. 187:6545–6555. 10.1128/JB.187.18.6545-6555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomson NR, Clayton DJ, Windhorst D, Vernikos G, Davidson S, Churcher C, Quail MA, Stevens M, Jones MA, Watson M, Barron A, Layton A, Pickard D, Kingsley RA, Bignell A, Clark L, Harris B, Ormond D, Abdellah Z, Brooks K, Cherevach I, Chillingworth T, Woodward J, Norberczak H, Lord A, Arrowsmith C, Jagels K, Moule S, Mungall K, Sanders M, Whitehead S, Chabalgoity JA, Maskell D, Humphrey T, Roberts M, Barrow PA, Dougan G, Parkhill J. 2008. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 18:1624–1637. 10.1101/gr.077404.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston CE, Hartley C, Salisbury AM, Wigley P. 2012. Immunological changes at point-of-lay increase susceptibility to Salmonella enterica Serovar enteritidis infection in vaccinated chickens. PLoS One 7:e48195. 10.1371/journal.pone.0048195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Zhang MZ, Yan L, Lillehoj H, Pace LW, Zhang S. 2009. Induction of CXC chemokine messenger-RNA expression in chicken oviduct epithelial cells by Salmonella enterica serovar enteritidis via the type three secretion system-1. Avian Dis. 53:396–404. 10.1637/8642-020309-Reg.1. [DOI] [PubMed] [Google Scholar]
- 25.Kinder SA, Badger JL, Bryant GO, Pepe JC, Miller VL. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O8 and construction of a transformable R-M+ mutant. Gene 136:271–275. 10.1016/0378-1119(93)90478-L. [DOI] [PubMed] [Google Scholar]
- 26.Kingsley RA, Reissbrodt R, Rabsch W, Ketley JM, Tsolis RM, Everest P, Dougan G, Bäumler AJ, Roberts M, Williams PH. 1999. Ferrioxamine-mediated iron(III) utilization by Salmonella enterica. Appl. Environ. Microbiol. 65:1610–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199. 10.1016/0378-1119(91)90366-J. [DOI] [PubMed] [Google Scholar]
- 28.Beug H, von Kirchbach A, Doderlein G, Conscience JF, Graf T. 1979. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell 18:375–390. 10.1016/0092-8674(79)90057-6. [DOI] [PubMed] [Google Scholar]
- 29.Hensel M, Shea JE, Raupach B, Monack D, Falkow S, Gleeson C, Kubo T, Holden DW. 1997. Functional analysis of ssaJ and the ssaK/U operon, 13 genes encoding components of the type III secretion apparatus of Salmonella pathogenicity island 2. Mol. Microbiol. 24:155–167. 10.1046/j.1365-2958.1997.3271699.x. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Zhang S, Santos RL, Tsolis RM, Stender S, Hardt WD, Bäumler AJ, Adams LG. 2002. The Salmonella enterica serotype typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves. Infect. Immun. 70:3843–3855. 10.1128/IAI.70.7.3843-3855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abramoff MD, Magalhaes Paulo J, Ram Sunanda J. 2004. Image processing with ImageJ. Biophotonics Int. 11:7. [Google Scholar]
- 33.Toguchi A, Siano M, Burkart M, Harshey RM. 2000. Genetics of swarming motility in Salmonella enterica serovar typhimurium: critical role for lipopolysaccharide. J. Bacteriol. 182:6308–6321. 10.1128/JB.182.22.6308-6321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mickael CS, Lam PK, Berberov EM, Allan B, Potter AA, Köster W. 2010. Salmonella enterica serovar Enteritidis tatB and tatC mutants are impaired in Caco-2 cell invasion in vitro and show reduced systemic spread in chickens. Infect. Immun. 78:3493–3505. 10.1128/IAI.00090-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynolds MM, Bogomolnaya L, Guo J, Aldrich L, Bokhari D, Santiviago CA, McClelland M, Andrews-Polymenis H. 2011. Abrogation of the twin arginine transport system in Salmonella enterica serovar Typhimurium leads to colonization defects during infection. PLoS One 6:e15800. 10.1371/journal.pone.0015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watson PR, Benmore A, Khan SA, Jones PW, Maskell DJ, Wallis TS. 2000. Mutation of waaN reduces Salmonella enterica serovar Typhimurium-induced enteritis and net secretion of type III secretion system 1-dependent proteins. Infect. Immun. 68:3768–3771. 10.1128/IAI.68.6.3768-3771.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646–4658. 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 38.Brodsky IE, Ernst RK, Miller SI, Falkow S. 2002. mig-14 is a Salmonella gene that plays a role in bacterial resistance to antimicrobial peptides. J. Bacteriol. 184:3203–3213. 10.1128/JB.184.12.3203-3213.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wikler MA, Cockerill FR, Craig WA, Dudley MN, Eliopoulos GM, Hecht DW, Hindler JF, Low DE, Sheehan DJ, Tenover FC, Turnidge JD, Weinstein MP, Zimmer BL, Ferraro MJ, Swenson JM. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard—7th ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 40.Berchieri A, Jr, Wigley P, Page K, Murphy CK, Barrow PA. 2001. Further studies on vertical transmission and persistence of Salmonella enterica serovar Enteritidis phage type 4 in chickens. Avian Pathol. 30:297–310. 10.1080/03079450120066304. [DOI] [PubMed] [Google Scholar]
- 41.Rychlik I, Karasova D, Sebkova A, Volf J, Sisak F, Havlickova H, Kummer V, Imre A, Szmolka A, Nagy B. 2009. Virulence potential of five major pathogenicity islands (SPI-1 to SPI-5) of Salmonella enterica serovar Enteritidis for chickens. BMC Microbiol. 9:268. 10.1186/1471-2180-9-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wisner AL, Desin TS, Koch B, Lam PK, Berberov EM, Mickael CS, Potter AA, Köster W. 2010. Salmonella enterica subspecies enterica serovar Enteritidis Salmonella pathogenicity island 2 type III secretion system: role in intestinal colonization of chickens and systemic spread. Microbiology 156:2770–2781. 10.1099/mic.0.038018-0. [DOI] [PubMed] [Google Scholar]
- 43.Wisner AL, Potter AA, Koster W. 2011. Effect of the Salmonella pathogenicity island 2 type III secretion system on Salmonella survival in activated chicken macrophage-like HD11 cells. PLoS One 6:e29787. 10.1371/journal.pone.0029787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah DH, Casavant C, Hawley Q, Addwebi T, Call DR, Guard J. 2012. Salmonella Enteritidis strains from poultry exhibit differential responses to acid stress, oxidative stress, and survival in the egg albumen. Foodborne Pathog. Dis. 9:258–264. 10.1089/fpd.2011.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah DH, Zhou X, Kim HY, Call DR, Guard J. 2012. Transposon mutagenesis of Salmonella enterica serovar Enteritidis identifies genes that contribute to invasiveness in human and chicken cells and survival in egg albumen. Infect. Immun. 80:4203–4215. 10.1128/IAI.00790-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chappell L, Kaiser P, Barrow P, Jones MA, Johnston C, Wigley P. 2009. The immunobiology of avian systemic salmonellosis. Vet. Immunol. Immunopathol. 128:53–59. 10.1016/j.vetimm.2008.10.295. [DOI] [PubMed] [Google Scholar]
- 47.De Buck J, Pasmans F, Van Immerseel F, Haesebrouck F, Ducatelle R. 2004. Tubular glands of the isthmus are the predominant colonization site of Salmonella enteritidis in the upper oviduct of laying hens. Poult. Sci. 83:352–358. 10.1093/ps/83.3.352. [DOI] [PubMed] [Google Scholar]
- 48.García-del Portillo F. 2001. Salmonella intracellular proliferation: where, when and how? Microbes Infect. 3:1305–1311. 10.1016/S1286-4579(01)01491-5. [DOI] [PubMed] [Google Scholar]
- 49.Detweiler CS, Monack DM, Brodsky IE, Mathew H, Falkow S. 2003. virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol. Microbiol. 48:385–400. 10.1046/j.1365-2958.2003.03455.x. [DOI] [PubMed] [Google Scholar]
- 50.Guina T, Yi EC, Wang H, Hackett M, Miller SI. 2000. A PhoP-regulated outer membrane protease of Salmonella enterica serovar typhimurium promotes resistance to alpha-helical antimicrobial peptides. J. Bacteriol. 182:4077–4086. 10.1128/JB.182.14.4077-4086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Navarre WW, Halsey TA, Walthers D, Frye J, McClelland M, Potter JL, Kenney LJ, Gunn JS, Fang FC, Libby SJ. 2005. Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol. Microbiol. 56:492–508. 10.1111/j.1365-2958.2005.04553.x. [DOI] [PubMed] [Google Scholar]
- 52.Wing HJ, Goldman SR, Ally S, Goldberg MB. 2005. Modulation of an outer membrane protease contributes to the virulence defect of Shigella flexneri strains carrying a mutation in the virK locus. Infect. Immun. 73:1217–1220. 10.1128/IAI.73.2.1217-1220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murray SR, Bermudes D, de Felipe KS, Low KB. 2001. Extragenic suppressors of growth defects in msbB Salmonella. J. Bacteriol. 183:5554–5561. 10.1128/JB.183.19.5554-5561.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593–656. 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Josenhans C, Suerbaum S. 2002. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291:605–614. 10.1078/1438-4221-00173. [DOI] [PubMed] [Google Scholar]
- 56.Liu R, Ochman H. 2007. Stepwise formation of the bacterial flagellar system. Proc. Natl. Acad. Sci. U. S. A. 104:7116–7121. 10.1073/pnas.0700266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Badea L, Beatson SA, Kaparakis M, Ferrero RL, Hartland EL. 2009. Secretion of flagellin by the LEE-encoded type III secretion system of enteropathogenic Escherichia coli. BMC Microbiol. 9:30. 10.1186/1471-2180-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crhanova M, Malcova M, Mazgajova M, Karasova D, Sebkova A, Fucikova A, Bortlicek Z, Pilousova L, Kyrova K, Dekanova M, Rychlik I. 2011. LPS structure influences protein secretion in Salmonella enterica. Vet. Microbiol. 152:131–137. 10.1016/j.vetmic.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 59.Defeu Soufo HJ, Reimold C, Linne U, Knust T, Gescher J, Graumann PL. 2010. Bacterial translation elongation factor EF-Tu interacts and colocalizes with actin-like MreB protein. Proc. Natl. Acad. Sci. U. S. A. 107:3163–3168. 10.1073/pnas.0911979107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davidson AL, Shuman HA, Nikaido H. 1992. Mechanism of maltose transport in Escherichia coli: transmembrane signaling by periplasmic binding proteins. Proc. Natl. Acad. Sci. U. S. A. 89:2360–2364. 10.1073/pnas.89.6.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tommassen J, Eiglmeier K, Cole ST, Overduin P, Larson TJ, Boos W. 1991. Characterization of two genes, glpQ and ugpQ, encoding glycerophosphoryl diester phosphodiesterases of Escherichia coli. Mol. Gen. Genet. 226:321–327. [DOI] [PubMed] [Google Scholar]
- 62.Yim HH, Villarejo M. 1992. osmY, a new hyperosmotically inducible gene, encodes a periplasmic protein in Escherichia coli. J. Bacteriol. 174:3637–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karring H, Mathu SG, van Duin J, Clark BF, Kraal B, Knudsen CR. 2004. Qbeta-phage resistance by deletion of the coiled-coil motif in elongation factor Ts. J. Biol. Chem. 279:1878–1884. 10.1074/jbc.M306605200. [DOI] [PubMed] [Google Scholar]
- 64.Tunio SA, Oldfield NJ, Ala'Aldeen DA, Wooldridge KG, Turner DP. 2010. The role of glyceraldehyde 3-phosphate dehydrogenase (GapA-1) in Neisseria meningitidis adherence to human cells. BMC Microbiol. 10:280. 10.1186/1471-2180-10-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoare A, Bittner M, Carter J, Alvarez S, Zaldivar M, Bravo D, Valvano MA, Contreras I. 2006. The outer core lipopolysaccharide of Salmonella enterica serovar Typhi is required for bacterial entry into epithelial cells. Infect. Immun. 74:1555–1564. 10.1128/IAI.74.3.1555-1564.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guard-Petter J, Lakshmi B, Carlson R, Ingram K. 1995. Characterization of lipopolysaccharide heterogeneity in Salmonella enteritidis by an improved gel electrophoresis method. Appl. Environ. Microbiol. 61:2845–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parker CT, Harmon B, Guard-Petter J. 2002. Mitigation of avian reproductive tract function by Salmonella enteritidis producing high-molecular-mass lipopolysaccharide. Environ. Microbiol. 4:538–545. 10.1046/j.1462-2920.2002.00333.x. [DOI] [PubMed] [Google Scholar]
- 68.Raspoet R, Shearer N, Appia-Ayme C, Haesebrouck F, Ducatelle R, Thompson A, Van Immerseel F. 2014. A genome-wide screen identifies Salmonella Enteritidis lipopolysaccharide biosynthesis and the HtrA heat shock protein as crucial factors involved in egg white persistence at chicken body temperature. Poult. Sci. 93:1263–1269. 10.3382/ps.2013-03711. [DOI] [PubMed] [Google Scholar]