Abstract
Shiga toxin (Stx)-producing Escherichia coli (STEC) causes hemorrhagic colitis and the hemolytic-uremic syndrome (HUS). STEC strains may produce Stx1a and/or Stx2a or variants of either toxin. A 2006 spinach-associated outbreak of STEC O157:H7 resulted in higher hospitalization and HUS rates than previous STEC outbreaks. The spinach isolate, strain K3995, contains both stx2a and stx2c. We hypothesized that the enhanced virulence of K3995 reflects the combination of stx2 alleles (carried on lysogenic phages) and/or the amount of Stx2 made by that strain. We compared the virulence of K3995 to those of other O157:H7 isolates and an isogenic Stx2 mutant in rabbits and mice. We also measured the relative levels of Stx2 produced from those strains with or without induction of the stx-carrying phage. Some rabbits infected with K3995 exhibited intestinal pathology and succumbed to infection, while none of those infected with O157:H7 strain 2812 (Stx1a+ Stx2a+) died or showed pathological signs. Rabbits infected with the isogenic Stx2a mutant K3995 stx2a::cat were not colonized as well as those infected with K3995 and exhibited no signs of disease. In the streptomycin-treated mouse model, more animals infected with K3995 died than did those infected with O157:H7 strain 86-24 (Stx2a+). Additionally, K3995 produced higher levels of total Stx2 and toxin phage DNA in cultures after phage induction than did 86-24. Our results demonstrate the greater virulence of K3995 compared to other O157:H7 strains in rabbits and mice. We conclude that this enhanced virulence is linked to higher levels of Stx2 expression as a consequence of increased phage induction.
INTRODUCTION
Shiga toxin (Stx)-producing Escherichia coli (STEC) O157:H7 is a food-borne pathogen that causes hemorrhagic colitis (HC) and the hemolytic-uremic syndrome (HUS) (1, 2). E. coli O157:H7 infection results in approximately 63,000 illnesses annually in the United States (3), and the yearly cost of these illnesses is estimated to be $255 million to $405 million (2, 4). The principal reservoir of STEC is the intestinal tracts of bovines and other ruminants. Initial STEC outbreaks were associated with consumption of undercooked hamburger. However, over the last 2 decades a wide variety of food items have been linked with disease due to E. coli O157:H7 infection, such as unpasteurized fruit juices (5), smoked meats (6), and fresh produce (7–10). Moreover, other routes of STEC transmission that are unrelated to ingestion of contaminated foods have been identified, and these include swimming in contaminated water (11–13), contact with infected animals (14, 15), and person-to-person spread (16, 17). These modes of transmission are likely facilitated by the fact that E. coli O157:H7 can cause illness after exposure to <100 organisms (4).
STEC strains are characterized by production of Stx. This toxin is critical to the development of HC and HUS (18, 19), and the genes for Stxs are carried on lambda-like lysogenic converting phages in E. coli (20–23). STEC produces two serologically distinct groups of Stx, i.e., Stx1 and Stx2, and variants of both toxins, all of which are N-glycosidases that inactivate the 60S ribosomal subunits in eukaryotic cells (24). The Stx-mediated injury to the ribosome leads to inhibition of protein synthesis and cell death (24). The Stx2 group of toxins consists of 7 subtypes, of which Stx2a is the prototype (25). The Stx2c variant is 99% identical to Stx2a at the amino acid level and differs by two amino acids in the mature B subunit (26). Epidemiological studies suggest that stx2a+, stx2c+, or stx2a+ stx2c+ E. coli O157:H7 strains are associated with more severe disease in humans than STEC strains with other stx alleles or those that lack the capacity to cause attaching and effacing (AE) lesions (27–30). A 2006 spinach-associated outbreak caused by an stx2a+ stx2c+ E. coli O157:H7 resulted in higher hospitalization and HUS rates than were usually associated with serotype O157:H7 outbreaks. These indicators of serious disease fueled speculation that the implicated spinach E. coli O157:H7 strain had greater virulence potential than did “typical” outbreak isolates (27). Furthermore, the prevalence of E. coli O157:H7 strains that belonged to clade 8 (a specific genotype of serotype O157:H7 based on single nucleotide polymorphisms at 96 loci [15]) and that frequently harbored the stx2a stx2c gene combination increased from 10% to 46% over a 5-year study period that preceded the 2006 spinach outbreak (27).
In this study, we compared the virulence of an E. coli O157:H7 strain that carries both stx2a and stx2c and was isolated from a person during the 2006 spinach outbreak, strain K3995, with different E. coli O157:H7 strains that also carry the stx2a allele either alone or in combination with stx1a. For that purpose we used 3 different animal models. We also evaluated the amount of Stx2 produced in vitro by two of the most pathogenic strains. In addition, we assessed the contribution of Stx2a to the virulence of E. coli O157:H7 strain K3995. We found that E. coli O157:H7 strain K3995 (stx2a+ stx2c+) was more virulent in Dutch belted rabbits than was E. coli O157:H7 isolate 2812 (stx1a+ stx2a+). Further, K3995 was more virulent in streptomycin (Str)-treated mice than was E. coli O157:H7 86-24 (stx2a+), although both strains colonized mice with intact commensal flora (ICF mice) equally. Finally, Dutch belted rabbits infected with K3995 displayed higher colonization and more disease and intestinal pathology and gained less weight than rabbits infected with an stx2a-negative K3995 isogenic mutant.
MATERIALS AND METHODS
Bacterial strains.
E. coli O157:H7 strain K3995 (stx2a+ stx2c+) was isolated from a patient infected during the 2006 spinach-associated outbreak that occurred in the United States (8) and was kindly provided by Nancy Strockbine of the Centers for Disease Control and Prevention (Atlanta, GA). Strain K3995 was isolated from a patient infected in the same outbreak from which a sequenced E. coli O157:H7 strain, TW14359 (31), was isolated. An Stx2a mutant of K3995 was constructed as described below. E. coli O157:H7 strain 2812 (stx1a+ stx2a+) was kindly provided by Darrell Griffin of the Madigan Army Medical Center and was isolated from a patient with hemorrhagic colitis who was infected during a 1993 outbreak associated with contaminated hamburgers (32, 33). E. coli O157:H7 strain 86-24 (stx2a+) was provided by Phil Tarr, then at the Washington University School of Medicine. The nalidixic acid-resistant (Nalr) mutant of 86-24 was provided by Arthur Donohue-Rolfe of Tufts University. Strain 86-24 was isolated in Washington State during a 1986 outbreak associated with contaminated beef products. Additionally, spontaneous Str-resistant (Strr) or Nalr mutants of each strain were isolated and used for animal studies. Throughout this article, Stx2 indicates total toxin (Stx2 and Stx2c) or refers to a study in which the Stx2 toxin subtype was not specified. Stx2a is the prototype Stx2 toxin and is encoded by stx2a. Stx2c indicates that subtype of Stx2 with the associated gene designation stx2c.
Rabbit studies.
Rabbit feces were prescreened for the presence of E. coli by plating on sorbitol MacConkey agar (SMAC) plates. E. coli O157:H7 isolates do not ferment sorbitol (i.e., are sorbitol nonfermenters) and are tan/colorless on SMAC plates. Conversely, most other E. coli isolates ferment sorbitol and are pink on this agar. All rabbits used in this study were free of sorbitol nonfermenters and were found to have <103 CFU/g feces sorbitol fermenters. Additionally, the animals were evaluated for clinical signs of disease, such as diarrhea or hemorrhagic colitis, prior to infection.
Rabbit infection studies were done as previously described (34, 35). Dutch belted rabbits (Covance, Denver, PA) aged 5 to 8 weeks were fasted overnight, and water was removed from the cages 2 h prior to bacterial inoculation. Animals were anesthetized and received 5 or 10 ml of 10% sodium bicarbonate solution by intragastric gavage through an infant feeding tube and then were inoculated by gavage with Strr E. coli O157:H7 strains suspended in 1 ml phosphate-buffered saline (PBS). After inoculation, rabbits were monitored daily for colonization and development of clinical signs of disease for 7 days. Rabbits that showed signs of disease (lethargy, fever, hemorrhagic colitis, or weight loss) and did not respond to interventions (analgesic and rehydration therapy) were euthanized at alternative endpoints: weight loss of more than 20% compared with the preinoculation weight, inability to eat or drink for >12 h, or lack of movement. All other rabbits were euthanized at the conclusion of the study (7 days postinfection [p.i.]). The analgesic was buprenorphine (buprenorphine HCl injection; PharmaForce, Columbus, OH) given at 0.02 mg/kg subcutaneously every 6 to 12 h. Rehydration therapy was administered as a lactate Ringer's solution (Hospira, Inc., Lake Forest, IL) to the rabbits at 25 ml/kg daily until signs resolved. Colonization was monitored by levels of E. coli O157:H7 isolated from the feces. Fecal pellets were collected, weighed, diluted 1:10 by weight into PBS, and then homogenized. The supernatants from the fecal homogenates were serially diluted and plated on MacConkey agar supplemented with Str (40 μg/ml), and plates were incubated overnight at 37°C. Colonies were enumerated to determine CFU/g feces. Organs (including kidneys, small intestine, cecum, and colon) were collected at necropsy for histopathological analyses.
The rabbit studies were conducted at an Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC)-accredited animal facility in the Program of Comparative Medicine at the University of Maryland School of Medicine. All procedures conformed to the guidelines of the Guide for the Care and Use of Laboratory Animals (36) and the policies of the Institutional Animal Care and Use Committee of the University of Maryland School of Medicine. All procedures were in compliance with the publications Biosafety in Microbiological and Biomedical Laboratories (37).
ICF and Str-treated mouse models.
Mouse studies were done as previously described (38, 39). Male Crl:CD-1(ICR, CD-1) or female BALB/cAnNCrl (BALB/c) mice, 10 to 12 g and 3 to 6 weeks old, from Charles River Laboratories (Wilmington, MA) were used for all mouse experiments. Mice were fasted for ∼16 h and deprived of water 1 to 2 h prior to bacterial inoculation. For the Str-treated mouse model, mice were given drinking water with Str (5 g/liter) 18 h before infection. Str treatment continued throughout the course of the experiment. To infect animals in both models, overnight cultures of the E. coli O157:H7 strain were pelleted by centrifugation and resuspended in glucose-PBS (1:200). Mice were fed 100 μl of inoculum by gavage. Intestinal colonization of BALB/c mice with intact commensal flora (ICF mice) was monitored by the determination of the levels of Nalr E. coli O157:H7 isolated from the feces. Fecal pellets were collected, weighed, diluted 1:10 by weight into PBS, and homogenized; the fecal homogenate supernatants were serially diluted and plated on MacConkey agar plates supplemented with nalidixic acid (25 μg/ml) and incubated overnight at 37°C. Colonies were enumerated to determine CFU/g feces. (Note that colonization by E. coli O157:H7 was not routinely monitored in the Str-treated mouse model because historical data indicate that high levels of Strr bacteria are maintained for 25 days [38].) In both models, mice were monitored for survival for up to 15 days p.i. These studies were conducted in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals (36). All mouse studies were approved by the Institutional Animal Care and Use Committee of the Uniformed Services University of the Health Sciences. All procedures were in compliance with the publication Biosafety in Microbiological and Biomedical Laboratories (37).
Immuno-dot blotting. (i) Preparation of samples.
Samples for immuno-dot blotting to measure Stx2 levels in cultures of E. coli O157:H7 were prepared as follows. Bacteria were grown overnight in Luria broth (LB). The overnight cultures were then diluted 1:500 in LB and grown at 37°C with shaking. In some cases, after 2 h of growth of the cultures, lysogenic stx2-carrying phage were induced by addition of 15 ng/ml of ciprofloxacin (1/2 the MIC). Aliquots from induced and uninduced cultures were then obtained at various times of continued incubation, and the optical density at 600 nm (OD600) for each such sample was determined. A portion of each sample was then diluted and plated to enumerate CFU, and the remainder was lysed by sonication (2 to 5 min total; 10 s on and 20 s off) or a series of freeze-thaw cycles and then spun at 20,000 × g for 10 min at 4°C to remove debris. The resultant supernatants were used in immuno-dot blot experiments as described below.
(ii) Immuno-dot blotting.
Dot blots were prepared on a 96-well Minifold I Dot-Blot apparatus (Whatman; Sigma-Aldrich, St. Louis, MO). Culture lysate supernatants were serially diluted 2-fold in PBS, and 400 μl of each dilution was applied to a 0.45-μm nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA) with vacuum. The membrane was removed from the apparatus, air dried briefly, and incubated in PBS that contained 0.05% Tween 20 (PBS-T) and 5% nonfat dry milk for 1 h at room temperature. Primary antiserum (rabbit α-Stx2 polyclonal [40] precleared against an Stx2− E. coli O157:H7 strain) diluted 1:200 in PBS-T with 5% milk was then applied to the membrane in the dot blot apparatus, and the membrane was further incubated overnight at 4°C in the covered dot blot apparatus. The membrane was then removed from the apparatus, washed several times with PBS-T, and then incubated for 1 h at room temperature with goat anti-rabbit IgG(H+L)–horseradish peroxidase (HRP) conjugate (Bio-Rad, Hercules, CA) diluted 1:2,000 in PBS-T and 5% nonfat dry milk, applied to the membrane in the dot blot apparatus. The membrane was removed from the apparatus and washed several times with PBS-T. Signal was visualized with the Opti-4CN substrate kit (Bio-Rad, Hercules, CA) according to the manufacturer's directions.
(iii) Evaluation of integrated pixel density.
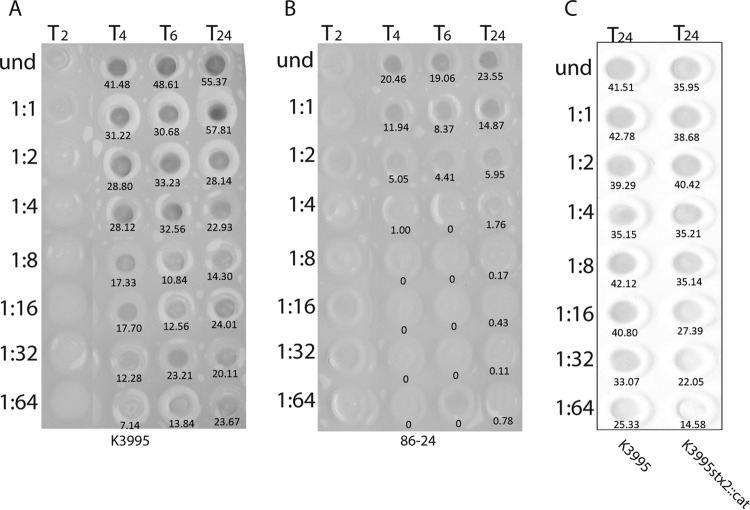
The relative intensities of the spots on the blot were quantified by assessing integrated pixel density using Adobe Photoshop CS5 Extended (version 12.0 ×32). For the purpose of analysis, the color of the blot was inverted so the background was black and the dots were white. A circle was drawn to encompass the spot, and the measurement was recorded. The same circle was used to measure all samples on each blot. To compare the values derived from blots in Fig. 4A and B, the values of the spots corresponding to the final three dilutions at the 2-h time point on both blots were averaged to provide a background value that was subtracted from all other spot intensities. The blot in Fig. 4C was assessed separately from those in Fig. 4A and B.
FIG 4.
Amounts of Stx2 produced in vitro by K3995, 86-24, and K3995 stx2a::cat as assessed by immuno-dot blotting. Samples were removed from cultures at the times indicated (in hours) and lysed, and supernatants were applied to the membrane in 2-fold dilutions as specified on the right of each blot. Values below the spots indicate the integrated pixel densities, measured using Adobe Photoshop. Values derived from Photoshop analysis were divided by 1,000 for ease of presentation on the blots. When induced with ciprofloxacin, K3995 (A) produced more toxin than did 86-24 (B), as is evident even at early time points. Similarly, when induced with ciprofloxacin, the amount of toxin produced by K3995 stx2a::cat was increased to levels nearer those made by wt K3995 (C).
Southern and DNA dot blotting.
Southern and DNA dot blot analyses were done as directed in the Roche digoxigenin (DIG) system manual (41). For the Southern blots, total genomic DNA was isolated from the cell pellet (MasterPure DNA purification kit; Epicentre Biotechnologies, Madison, WI) and digested with EcoRI. The resultant restriction fragments were separated on a 0.7% agarose gel, transferred with the iBlot Dry blotting system (Invitrogen, Carlsbad, CA) to a 0.2-μm nylon membrane, fixed by UV, and probed for the stx2a A subunit gene or the chloramphenicol (Chl) acetyltransferase gene (cat). In vitro DNA dot blot samples were prepared as follows. Overnight cultures were diluted 1:500 in LB and grown at 37°C with shaking for 24 h; samples were removed at 2, 4, 6, and 24 h. For cultures that were induced, 15 ng/ml ciprofloxacin (1/2 the MIC) was added at 2 h. Culture samples were collected and the OD determined at 600 nm; a portion of the sample culture was then diluted and plated to determine CFU. Next, total genomic DNA was isolated from the whole-culture samples (MasterPure DNA purification kit; Epicentre Biotechnologies, Madison, WI) and applied to a 0.45-μm nylon membrane (Whatman Nytran SuPerCharge TurboBlotter; Sigma-Aldrich, St. Louis, MO) by vacuum using a 96-well Minifold I Dot-Blot apparatus (Sigma-Aldrich, St. Louis, MO).
The probes for Southern and DNA dot blot analyses were amplified by PCR according to the manufacturer's directions (PCR DIG probe synthesis kit; Roche Applied Sciences). The primers used to amplify the stx2a A subunit gene probe from pJES120 (42) were 5′-GGGTACTGTGCCTGTTACTG-3′ and 5′-TACCACTGAACTCCATTAAC-3′. The primers used to amplify the cat probe from pACYC184 were 5′-CCTATAACCAGACCGTTCAG-3′ and 5′-TAAGCATTCTGCCGACATGG-3′. Detection of the genes encoding phage replication protein O, designated O, was used as a measure of the amount of 933W or 2851 phage present. The primers used to amplify the O probe from the stx2a-bearing phage 933W were 5′-ATACTCAACTTCGGCAGAGG-3′ and 5′-CCAAACATGCCGCCTTGCTG-3′. The primers used to amplify the O probe from the stx2c-bearing phage 2851 were 5′-GAGCATCGCGTGGCAGATAC-3′ and 5′-GTCCTGGCTAATCCACTGAG-3′.
For evaluation of integrated pixel density, the spots on the DNA blot were quantified in Photoshop as described above. Each blot was assessed individually.
Stx2 mutant construction.
K3995 stx2a::cat is a K3995 derivative in which base pairs (bp) 529 to 1012 of stx2a were replaced by cat. This mutant was constructed by the use of the lambda Red recombinase system according to the method of Serra-Moreno et al. (43) as detailed below.
(i) Creation of linear DNA piece stx2a-cat-stx2a.
The linear DNA fragment cat flanked by 230 to 280 bp of stx2a sequence, stx2a-cat-stx2a, was generated as follows. First, a 1,022-bp portion of stx2a was cut from pJES120 (42) with Sau3AI and EcoRI, and that piece was ligated into pUC18 (Thermo Scientific, Pittsburgh, PA) that had been digested with EcoRI and BamHI. The resultant plasmid was designated pTZ101. A SacI site was then created by splicing by overlap extension PCR in the stx2a B subunit gene with primers SFor (5′-GCGGTTTTATTTGCATGAGCTCCTGTTAATGCAATGG-3′) and SRev (5′-CCATTGCATTAACAGGAGCTCATGCAAATAAAACCGC-3′), and the resultant PCR fragment was ligated into pUC18 to generate pTZ105. Since pUC18 had other SacI sites, pTZ105 was then cut with EcoRI to release stx2a, and the stx2a DNA segment with EcoRI overhangs was ligated to EcoRI-cut pUC7 to create pTZ106. cat was amplified from pACYC184 with CatAmpF (5′-CCCGAGCTCCACTTATTCAGGCGTAGCAC-3′) and CatAmpR (5′-CCCATGCATATCGGCACGTAAGAGGTTCC-3′), primers that include SacI (5′-GAGCTC-3′) and NsiI (5′-ATGCAT-3′) restriction sites, respectively, on the 5′ ends. The cat amplicon was cut with SacI and NsiI and inserted into pTZ106 cut with SacI and NsiI to create pTZ109. The stx2a-cat-stx2a fragment for lambda Red recombination was amplified by PCR from pTZ109 with TZ1F (5′-CAGGCGCGTTTTGACCATCTTG-3′) and TZ3R (5′-ACCCACATACCACGAATCAG-3′) primers and then purified with the QIAquick PCR purification kit (Qiagen, Germantown, MD) according to the manufacturer's instructions.
(ii) Transformation of K3995.
K3995 was first transformed with the lambda Red helper plasmid pKD46 by electroporation, as per the method of Serra-Moreno et al. (43). Then, a 50-ml culture of K3995(pKD46) in LB with ampicillin (100 μg/ml) and 0.1 M arabinose was grown at 30°C with shaking to an OD600 of 0.6. The cells were harvested by centrifugation at 3,000 × g for 5 min at 4°C and washed four times with ice-cold distilled water. The cells were then resuspended in 100 μl water, and 500 ng of the stx2a-cat-stx2a linear DNA fragment was added. The cells were electroporated at 2.5 kV, 25 F, and 200 Ω and recovered in 1 ml Super Optimal broth with catabolite repression (SOC) for 1 to 16 h at 37°C without shaking. Recombinants were selected on LB plates supplemented with Chl at 5 μg/ml. Loss of pKD46 was achieved by growth at 42°C and confirmed by sensitivity to ampicillin. Replacement of stx2a with cat was confirmed by Southern blotting hybridization and Vero cell cytotoxicity assay (done as previously described [26, 35, 44]).
Statistical analyses.
All statistical analyses were done through application of the GraphPad Prism v5.04 for Windows software (GraphPad Software, San Diego, CA). The repeated-measures (RM) analysis of variance (ANOVA) was used to assess statistical differences in weight and colonization of infected animals. When infected animals died, the last weight observation was carried forward and used in all subsequent calculations.
RESULTS
Pathogenesis of K3995 in Dutch belted rabbits.
Dutch belted rabbits are susceptible to STEC infection and develop clinical disease that closely resembles human disease (34, 45). Therefore, we compared the virulence of the stx2a+ stx2c+ spinach outbreak isolate K3995 to that of another E. coli O157:H7 outbreak isolate, stx1a+ stx2a+ 2812, in the Dutch belted rabbit model. Rabbits infected with K3995 showed greater morbidity than those infected with 2812: 2/6 K3995-infected rabbits were lethargic, 5/6 K3995-infected rabbits had diarrhea, and 5/6 K3995-infected rabbits lost weight. K3995-infected rabbits also lost more weight than those infected with strain 2812, and this difference was significant on days 5 to 8 p.i. (Fig. 1A). Only one 2812-infected rabbit had diarrhea, and none showed any other signs of morbidity. Finally, 5/6 K3995-infected rabbits succumbed to infection, whereas all of the 2812-infected rabbits survived. Over the course of the study, we assessed the capacities of strains K3995 and 2812 to colonize the rabbit large intestine by enumerating Strr E. coli O157:H7 shed in the feces. Both strains colonized consistently throughout the study (Fig. 1B), although K3995 colonized to a higher level than did 2812.
FIG 1.
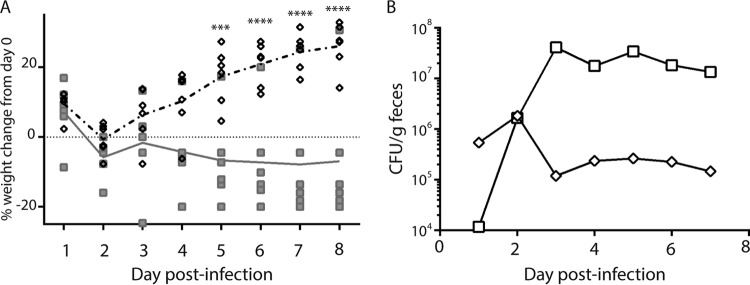
E. coli O157:H7 K3995 is more virulent in Dutch belted rabbits than is E. coli O157:H7 2812. (A) Percent weight change of rabbits infected with K3995 (□) or 2812 (◇). Each symbol represents one animal; the line indicates mean percent weight change. K3995-infected rabbits lost more weight than did rabbits infected with 2812, and that difference was significant on days 5, 6, 7, and 8 postinfection (***, P < 0.001; ****, P < 0.0001). (B) K3995-infected rabbits (□) were more highly colonized than rabbits infected with 2812 (◇), beginning on day 3 postinfection and continuing through the remainder of the study. Limit of detection, 102 CFU/g feces.
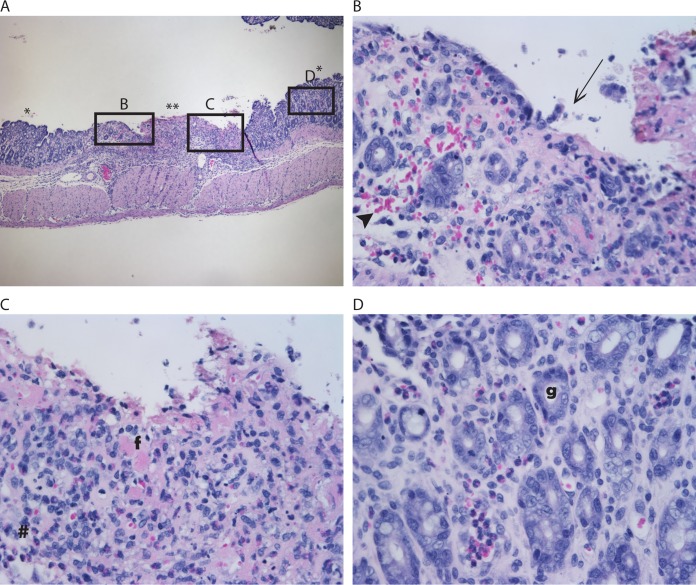
Gastrointestinal (GI) organs and kidneys were collected after euthanasia and assessed for histopathology. The organs of the 2812-infected rabbits did not show any pathology that differed significantly from that of the organs of the control rabbits. However, various degrees of damage to the GI tract were seen in all K3995-infected rabbits, especially in the cecum, a finding that has previously been reported for E. coli O157:H7-infected rabbits (46). Additionally, we noted extensive damage in the colons of two rabbits that succumbed to infection with K3995; the histological sections from the colon of one of those rabbits are shown in Fig. 2 (compare to a histological section from the colon of an uninfected rabbit, shown in Fig. S1 in the supplemental material).
FIG 2.
Histopathological analysis of colon from a K3995-infected rabbit that succumbed to disease. (A) Section of colon. Note contrast of less-affected (*) and ulcerated (**) tissue. Magnification, ×40. (B to D) Enlargements of the boxed areas in panel A. Note hemorrhage as evidenced by extravasated erythrocytes (arrowhead) and the gradual loss of epithelium (arrow) (B) and the transition to total effacement of the mucosa (C), characterized by loss of glands with replacement by necrotic debris, neutrophils (#), and fibrin (f). Magnification, ×400. (D) Area of less-affected colon, displaying typical architecture; note the presence of glands (g). Magnification, ×400.
Colonization and virulence capacity of K3995 in mice.
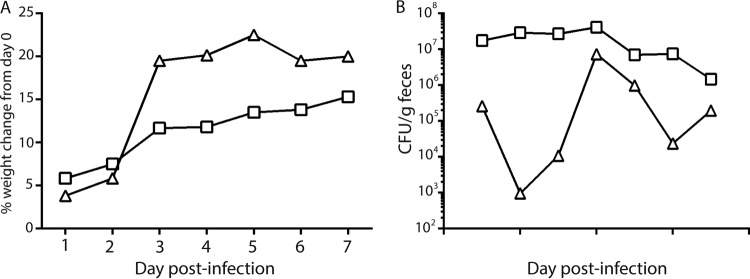
Previous work from our lab showed that Stx2a plays a role in colonization and virulence of mice by E. coli O157:H7 strain 86-24 (39, 47) (strain 2812 is not virulent in ICF mice [data not shown]). Here we sought to assess the contribution of Stx2c to the colonization of a strain that also makes Stx2a compared to that of a strain that makes only Stx2a. Therefore, we compared the colonization capacities of strains 86-24 and K3995 in a mouse with an intact commensal flora (ICF). For that purpose, ICF BALB/c mice were infected with 86-24 or K3995, and intestinal colonization by the E. coli O157:H7 strains was monitored by enumeration of the Nalr bacteria shed in the feces. We found that strains 86-24 and K3995 colonized ICF BALB/c mice to similar levels (Fig. 3A). Next, to compare the pathogenicities of strains 86-24 and K3995, we used the Str-treated CD-1 mouse model. The Str-treated mouse model permits high levels of colonization by E. coli over time and is therefore a more consistent model in which to assess virulence of E. coli O157:H7 than is the ICF model. The Str-treated, infected mice were monitored for survival over 15 days. We found that 37% of the mice succumbed to K3995 infection, while only 3% of mice died after infection with 86-24 (Fig. 3B).
FIG 3.
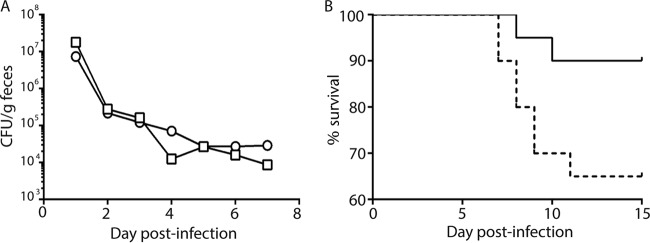
Colonization and virulence of K3995 compared to 86-24 in mice. (A) CFU/g feces of K3995 (□) and 86-24 (○). K3995 and 86-24 colonize ICF BALB/c mice similarly. (B) However, K3995 (dashed line) is more virulent than 86-24 (solid line) in Str-treated CD-1 mice (n = 25).
Total Stx2 production by K3995 and 86-24.
We next compared the relative amounts of total Stx2 (includes Stx2c for K3995) produced by the E. coli O157:H7 strains by immuno-dot blotting with a polyclonal anti-Stx2 antibody probe. That antiserum detects Stx2a and Stx2c (48). Although K3995 made levels of Stx2 similar to those made by 86-24 (data not shown), when the cultures were induced with ciprofloxacin, K3995 produced at least 8-fold more toxin than did 86-24 (Fig. 4A and B).
We then assessed the amounts of Stx2a- and Stx2c-converting phage DNA in the K3995 and 86-24 cultures. Since stx2a is found on a 933W-type phage in both strains and stx2c is on the 2851-type phage (49), we assessed the amount of phage DNA with a probe for the gene of the bacteriophage replication protein O (the gene for replication protein O is not homologous between 933W and 2851). More Stx2a-converting phage DNA was present in K3995 cultures than was found in 86-24 cultures (Fig. 5A), a finding that suggests that more spontaneous phage induction takes place in cultures of K3995 than in cultures of 86-24. As expected, Stx2c-converting phage DNA was detected only in cultures of strain K3995 (Fig. 5B).
FIG 5.
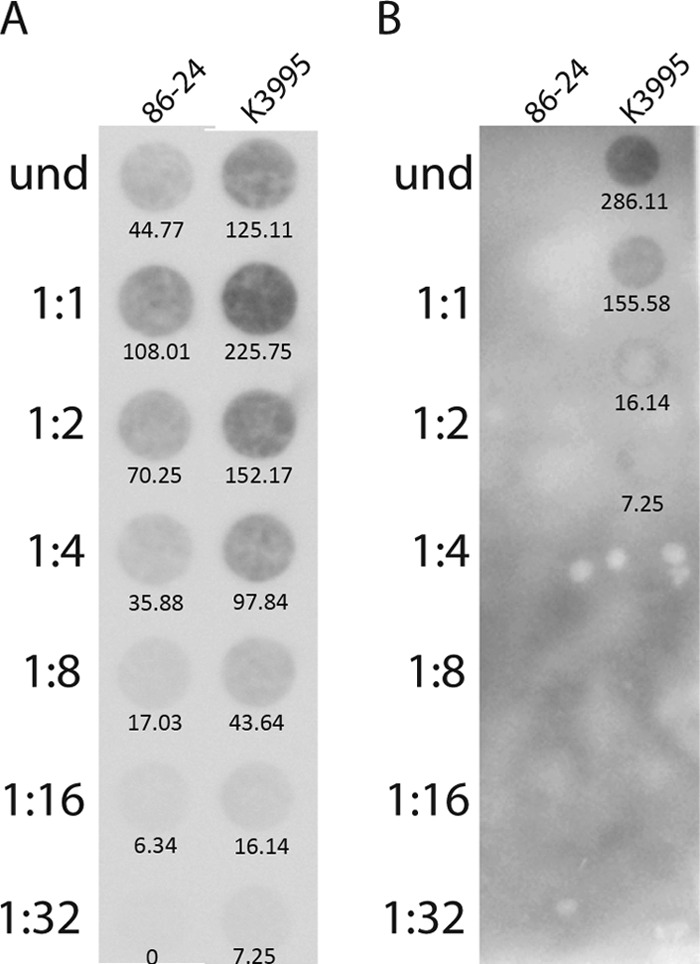
Dot blot of stx2a-converting (933W) or stx2c-converting (2851) phage DNA. Samples from an in vitro culture were applied to a nylon membrane in 2-fold dilutions as indicated and probed for the gene for the 933W or 2851 replication protein O. Values below the dots indicate the integrated pixel densities, measured using Adobe Photoshop. Values derived from Photoshop analysis were divided by 1,000 for ease of presentation on the blots. (A) More 933W phage DNA was detected in the K3995 culture than in the culture of 86-24. (B) 2851 phage DNA was found only in K3995.
Contribution of Stx2 to the virulence of K3995.
Collectively our data suggest that the increased virulence of K3995 compared to 86-24 is linked to higher levels of total Stx2 production in K3995 after induction, a finding that may reflect the presence of two toxin genes carried on separate inducible phages. Therefore, we constructed an Stx2a− K3995 strain by replacing stx2a with cat. We assessed wild-type (wt) K3995 and the Stx2a mutant for toxin production and found that K3995 stx2a::cat produced less toxin than did K3995 (data not shown), an expected result since the mutant strain has only one toxin gene (stx2c) and the parent strain has two. However, when K3995 stx2a::cat cultures were induced with ciprofloxacin, Stx2c production was increased over basal levels (Fig. 4C), a result that indirectly shows that the phage that harbors the gene for Stx2c can be induced in K3995.
We next assessed the virulence of K3995 stx2a::cat compared to that of wt K3995 in Dutch belted rabbits and ICF mice. Dutch belted rabbits were infected with wt K3995 or K3995 stx2a::cat and assessed for morbidity and colonization over 7 days. K3995-infected rabbits gained less weight (Fig. 6A) and displayed more disease than did rabbits infected with K3995 stx2a::cat: 2/6 K3995-infected rabbits had diarrhea and 4/6 were lethargic, while none of the K3995 stx2a::cat-infected rabbits showed any signs of disease. Additionally, K3995 colonized rabbits to a higher level and more consistently than did K3995 stx2a::cat (Fig. 6B). Similarly, K3995 colonized ICF mice more highly than K3995 stx2a::cat, and this difference was significant on day 2 p.i. (Fig. 7).
FIG 6.
The stx2a mutant K3995 stx2a::cat, is less virulent in Dutch belted rabbits than wt K3995. (A) Percent weight change of rabbits infected with K3995 (□) or K3995 stx2a::cat (△). The line indicates mean percent weight change. K3995-infected rabbits gained less weight than rabbits infected with K3995 stx2a::cat. (B) Mean CFU/g feces of K3995 (□) or K3995 stx2a::cat (△). K3995 colonized rabbits to a higher level and more consistently than K3995 stx2a::cat. Limit of detection, 102 CFU/g feces.
FIG 7.
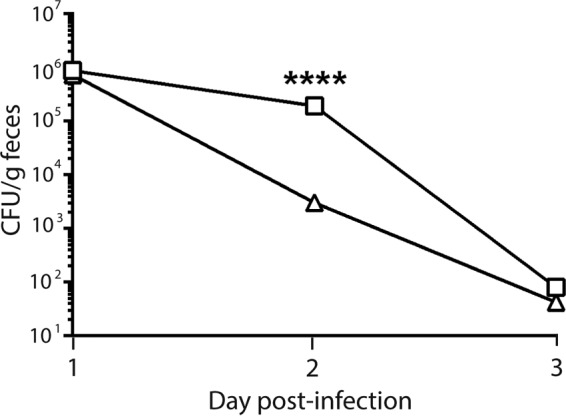
Mean CFU/g feces of K3995 (□) or K3995 stx2a::cat (△). K3995 colonizes ICF mice more highly than the stx2a mutant K3995 stx2a::cat. ICF mice infected with K3995 or K3995 stx2a::cat did not colonize well or remain colonized for longer than 3 days p.i.; however, on day 2 p.i. colonization levels of K3995 were higher than those of K3995 stx2a::cat, and this difference was significant on this day. Limit of detection, 102 CFU/g feces. ****, P < 0.0001.
DISCUSSION
While epidemiological data suggest that the Stx2a+ Stx2c+ E. coli O157:H7 strain implicated in the 2006 spinach outbreak was more virulent than previous E. coli O157:H7 outbreak strains (27), that hypothesis has not been specifically addressed in an animal model of E. coli O157:H7 pathogenesis. In this study, we found that K3995, isolated from a patient infected in that 2006 spinach-associated outbreak, was more pathogenic than two other E. coli O157:H7 strains in Dutch belted rabbits and in Str-treated CD-1 mice (strain 2812 was tested in this model previously [32]). The virulence of K3995 was associated with an increase in the Stx2a (933W)- and Stx2c (2851)-encoding phage DNA and total Stx2 production after induction with ciprofloxacin compared to strain 86-24. We deem induction with ciprofloxacin to be a proxy for stress-related stx2-encoding phage induction in vivo.
In the Dutch belted rabbit model, K3995 caused more morbidity and mortality than 2812, an stx1a+ stx2a+ E. coli O157:H7 isolate from a fast-food chain outbreak. Additionally, histopathological damage was evident in the colon of a Dutch belted rabbit that succumbed to K3995 infection. Pathology in the colon of an E. coli O157:H7-infected Dutch belted rabbit has not been previously shown. Garcia et al. did note some small spots of blood, edema, and thickening on “serosal surfaces of the cecum and/or proximal colon” in one E. coli O157-infected rabbit, although no image was shown (45).
Str-treated CD-1 mice are susceptible to infection with certain strains of E. coli O157:H7. Such mice remain consistently colonized to a high level with the infecting Strr strain, and the infected, Str-treated mice develop acute tubular necrosis and sometimes die from the effects of Stx2 (subtypes 2a, 2c, and 2dact) (38, 40, 42, 50). In this investigation, we showed that K3995 is more virulent than 86-24, an stx2a+ E. coli O157:H7 outbreak strain, in such Str-treated mice. We hypothesize that the reason that K3995 was more pathogenic than 86-24 in Str-treated mice is because the 933W phage DNA and Stx2a levels do not increase after induction with ciprofloxacin to the same degree in 86-24 as they do in K3995.
There is one description of TW14359 (another clinical isolate from the 2006 spinach-associated outbreak) in an animal model (51). In that study, researchers assessed the susceptibility of infant germfree mice to a number of E. coli O157:H7 strains, including TW14359, 86-24, and 93-111 (from the same outbreak in which 2812 was isolated [27, 32, 33]) and found that the prevalence of disease in the germfree mice infected with TW14359 was similar to the occurrence of disease in mice infected with 93-111 or 86-24. These results differ from ours, as we found that K3995 is more virulent than 2812 and 86-24 in rabbits and Str-treated mice, respectively. The discrepancies between the study by Eaton et al. (51) and ours is most likely due to the differences in the models used to assess virulence. For example, Dutch belted rabbits and Str-treated mice do not remain persistently colonized with greater than 108 CFU E. coli O157:H7/g feces (34, 38), whereas germfree mice shed high levels (109 CFU) of E. coli O157:H7 over the course of the experiment (51). This higher level of E. coli O157:H7 colonization in germfree mice may reflect the fact that by definition, germfree animals do not have intestinal flora, whereas rabbits have intact commensal flora to compete with the incoming E. coli O157:H7 infecting strain, and even the Str-treated mice have some gut flora. The capacity of the germfree mice to remain highly colonized over an extended period most likely allows an accumulation of systemic toxin that leads to kidney disease and death in that model. In contrast, when animals are colonized for only a short period or at a lower level, the amount of toxin delivered is limited and differences in toxin production by the infecting strains may be more readily apparent.
We compared the amounts of total Stx2 (including Stx2c) produced by K3995 and 86-24 (we chose 86-24 for our comparison as it was more virulent than 2812 in this study). K3995 produced levels of total toxin similar to those produced by 86-24 under noninducing conditions. However, when induced with ciprofloxacin, Stx2 was found in greater amounts in K3995 cultures than in 86-24 cultures. Additionally, we showed that the higher level of Stx2 production was associated with an increased level of stx2a-bearing phage (933W) DNA in K3995 compared to 86-24. As 933W phage induction has been shown to occur in the mouse intestine (52), it is therefore reasonable to hypothesize that in vivo, K3995 would also produce more Stx2 than 86-24; in fact, Eaton et al. quantitated Stx2 in the cecal contents of infected germfree mice and found that TW14359 produced about six times more toxin (5,843 ± 1,518 ng/ml) than did 86-24 (921 ± 111 ng/ml) (51). Since infection with TW14359 and 86-24 resulted in similar amounts of disease in the germfree mice, it may be that the amount of Stx2a produced by 86-24 is sufficient for maximal disease in that model and no further disease would be observed with a strain that produces more toxin. Our finding that total Stx2 levels are higher in K3995 than in 86-24 after induction are supported by in vitro studies in which Abu-Ali et al. showed an increase in transcript levels of total stx2 in TW14359 compared to Sakai (an stx1a+ stx2a+ E. coli O157:H7 outbreak strain) as well as increased stx2 transcript from clade 8 strains (to which K3995 belongs) compared to clade 2 strains (53). However, it is important to measure toxin protein amounts as well, because stx2 transcript levels do not necessarily correlate with Stx2 levels as measured by Western blotting (54).
Our results that showed that K3995 produces more toxin than 86-24 are in contrast to a study that found that Stx2 levels were decreased in an E. coli K-12 strain that carried two Stx2-bearing phages compared to an isogenic strain which had only one such phage (55). Our findings may differ from that study for several reasons. First, our comparison is between two wt, nonisogenic E. coli O157:H7 strains lysogenized with one (86-24) or two different (K3995) Stx2 phages. Second, in the previous study the stx2 gene on the second phage was replaced with an antibiotic resistance cassette. Therefore, there were two phages present but only one stx gene in the K-12 strains used in that investigation. In contrast, K3995 harbors two toxin-converting phages and two stx2 genes. Third, Serra-Moreno et al. (55) used stx2 phages isolated from cattle, while K3995 is a human isolate. Our results that suggest that total Stx2 production is higher after induction in an E. coli O157:H7 strain that carries two toxin-encoding phages are in agreement with a later report in which stx transcription was upregulated in a double lysogen compared to an isogenic single lysogen of E. coli K-12 (56). However, Neupane et al. (54) found that levels of stx2 transcript varied when they examined different wt E. coli O157:H7 strains lysogenized with two stx2-bearing phages. These investigators concluded that differences in toxin production by such strains appears to be strain specific (54). Thus, toxin levels in wt E. coli O157:H7 strains that carry more than one toxin-encoding phage will have to be evaluated on an individual basis.
To assess the contributions of Stx2 and Stx2c to the pathogenesis of K3995, we created an Stx2a− derivative of K3995 by replacing stx2a with cat. K3995 stx2a::cat-infected Dutch belted rabbits gained more weight than did wt-infected rabbits, and K3995 stx2a::cat colonized both rabbits and ICF BALB/c mice at a lower level than did wt K3995. That the Stx2a− strain colonizes to a lesser extent than the wt strain is not surprising given that Stx2a enhances the colonization of E. coli O157:H7 in ICF BALB/c mice (47). Although we found that the Stx2c+ strain K3995 stx2a::cat is not sufficient to cause disease in Dutch belted rabbits, there may be a role for Stx2c in pathogenesis, as wt K3995 was more virulent than 86-24 (which produces Stx2a only). We did find that K3995 stx2a::cat produces Stx2c, albeit at a low level, and that the amount of Stx2c produced could be increased by induction with ciprofloxacin. Moreover, previous studies showed that an E. coli O157:H7 strain that makes Stx2c only (40) and an E. coli K-12 producing plasmid-borne stx2c (42) are virulent in Str-treated CD-1 mice. While we made numerous attempts to generate a specific stx2c mutation in K3995, we were unsuccessful and so we could not directly answer the question of the role for Stx2c in the virulence of K3995 in Dutch belted rabbits or Str-treated mice.
High hospitalization and HUS rates were associated with the 2006 spinach-associated E. coli O157:H7 outbreak (27). Few studies have set out to specifically address the increased virulence of the strain responsible for that outbreak (27, 53, 57), and those studies were done in vitro. Indeed, to our knowledge, there is only one other description of pathogenesis of this strain in an animal model of E. coli O157:H7 infection (51). Here we showed that the E. coli O157:H7 spinach outbreak isolate K3995 is more virulent in two animal models than other E. coli O157:H7 outbreak isolates, and we correlated the virulence of K3995 to the presence of stx2a and to total Stx2 production.
Supplementary Material
ACKNOWLEDGMENTS
We thank Farhang Alem, Steven Darnell, and James Vergis for technical assistance and Cara Olsen for facilitation of statistical analyses. We also thank Theresa Nolan, Dawn Engel, Kayla Yi, Kathryn Kopanke, and Michelle Izuka for providing excellent veterinary technical support.
The opinions or assertions contained herein are the private ones of the authors and are not to be construed as official or reflecting the views of the Department of Defense, the Uniformed Services University of the Health Sciences, or the National Institutes of Health.
This work was supported by National Institutes of Health grants R37 AI020148 to ADO.
Footnotes
Published ahead of print 15 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02361-14.
REFERENCES
- 1.Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA. 1988. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann. Intern. Med. 109:705–712. [DOI] [PubMed] [Google Scholar]
- 2.Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg. Infect. Dis. 11:603–609. 10.3201/eid1104.040739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15. 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann S, Batz MB, Morris JG., Jr 2012. Annual cost of illness and quality-adjusted life year losses in the United States due to 14 foodborne pathogens. J. Food Prot. 75:1292–1302. 10.4315/0362-028X.JFP-11-417. [DOI] [PubMed] [Google Scholar]
- 5.Cody SH, Glynn MK, Farrar JA, Cairns KL, Griffin PM, Kobayashi J, Fyfe M, Hoffman R, King AS, Lewis JH, Swaminathan B, Bryant RG, Vugia DJ. 1999. An outbreak of Escherichia coli O157:H7 infection from unpasteurized commercial apple juice. Ann. Intern. Med. 130:202–209. 10.7326/0003-4819-130-3-199902020-00005. [DOI] [PubMed] [Google Scholar]
- 6.Tilden J, Jr, Young W, McNamara AM, Custer C, Boesel B, Lambert-Fair MA, Majkowski J, Vugia D, Werner SB, Hollingsworth J, Morris JG., Jr 1996. A new route of transmission for Escherichia coli: infection from dry fermented salami. Am. J. Public Health 86:1142–1145. 10.2105/AJPH.86.8_Pt_1.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michino H, Araki K, Minami S, Takaya S, Sakai N, Miyazaki M, Ono A, Yanagawa H. 1999. Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am. J. Epidemiol. 150:787–796. 10.1093/oxfordjournals.aje.a010082. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2006. Ongoing multistate outbreak of Escherichia coli serotype O157:H7 infections associated with consumption of fresh spinach—United States, September 2006. MMWR Morb. Mortal. Wkly. Rep. 55:1045–1046. [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2012. Multistate outbreak of Shiga toxin-producing Escherichia coli O157:H7 infections linked to organic spinach and spring mix blend (final update). Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 10.Centers for Disease Control and Prevention. 2013. Multistate outbreak of Shiga toxin-producing Escherichia coli O157:H7 infections linked to ready-to-eat salads (final update). Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 11.Centers for Disease Control and Prevention. 1996. Lake-associated outbreak of Escherichia coli O157:H7—Illinois, 1995. JAMA 275:1872–1873. 10.1001/jama.1996.03530480016015. [DOI] [PubMed] [Google Scholar]
- 12.Keene WE, McAnulty JM, Hoesly FC, Williams LP, Jr, Hedberg K, Oxman GL, Barrett TJ, Pfaller MA, Fleming DW. 1994. A swimming-associated outbreak of hemorrhagic colitis caused by Escherichia coli O157:H7 and Shigella sonnei. N. Engl. J. Med. 331:579–584. 10.1056/NEJM199409013310904. [DOI] [PubMed] [Google Scholar]
- 13.Friedman MS, Roels T, Koehler JE, Feldman L, Bibb WF, Blake P. 1999. Escherichia coli O157:H7 outbreak associated with an improperly chlorinated swimming pool. Clin. Infect. Dis. 29:298–303. 10.1086/520204. [DOI] [PubMed] [Google Scholar]
- 14.Renwick SA, Wilson JB, Clarke RC, Lior H, Borczyk AA, Spika J, Rahn K, McFadden K, Brouwer A, Copps A, Neil Anderson G, Alves D, Karmali MA. 1993. Evidence of direct transmission of Escherichia coli O157:H7 infection between calves and a human. J. Infect. Dis. 168:792–793. 10.1093/infdis/168.3.792. [DOI] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. 2005. Outbreaks of Escherichia coli O157:H7 associated with petting zoos—North Carolina, Florida, and Arizona, 2004 and 2005. MMWR Morb. Mortal. Wkly. Rep. 54:1277–1280. [PubMed] [Google Scholar]
- 16.Seto EY, Soller JA, Colford JM., Jr 2007. Strategies to reduce person-to-person transmission during widespread Escherichia coli O157:H7 outbreak. Emerg. Infect. Dis. 13:860–866. 10.3201/eid1306.061264. [DOI] [PubMed] [Google Scholar]
- 17.Parry SM, Salmon RL. 1998. Sporadic STEC O157 infection: secondary household transmission in Wales. Emerg. Infect. Dis. 4:657–661. 10.3201/eid0404.980419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffin PM, Tauxe RV. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60–98. [DOI] [PubMed] [Google Scholar]
- 19.Tarr PI, Gordon CA, Chandler WL. 2005. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365:1073–1086. 10.1016/S0140-6736(05)71144-2. [DOI] [PubMed] [Google Scholar]
- 20.Scotland SM, Smith HR, Willshaw GA, Rowe B. 1983. Vero cytotoxin production in strain of Escherichia coli is determined by genes carried on bacteriophage. Lancet ii:216. [DOI] [PubMed] [Google Scholar]
- 21.Huang A, de Grandis S, Friesen J, Karmali M, Petric M, Congi R, Brunton JL. 1986. Cloning and expression of the genes specifying Shiga-like toxin production in Escherichia coli H19. J. Bacteriol. 166:375–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newland JW, Strockbine NA, Miller SF, O'Brien AD, Holmes RK. 1985. Cloning of Shiga-like toxin structural genes from a toxin converting phage of Escherichia coli. Science 230:179–181. 10.1126/science.2994228. [DOI] [PubMed] [Google Scholar]
- 23.Willshaw GA, Smith HR, Scotland SM, Rowe B. 1985. Cloning of genes determining the production of Vero cytotoxin by Escherichia coli. J. Gen. Microbiol. 131:3047–3053. [DOI] [PubMed] [Google Scholar]
- 24.Endo Y, Tsurugi K, Yutsudo T, Takeda Y, Ogasawara T, Igarashi K. 1988. Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. RNA N-glycosidase activity of the toxins. Eur. J. Biochem. 171:45–50. [DOI] [PubMed] [Google Scholar]
- 25.Scheutz F, Teel LD, Beutin L, Pierard D, Buvens G, Karch H, Mellmann A, Caprioli A, Tozzoli R, Morabito S, Strockbine NA, Melton-Celsa AR, Sanchez M, Persson S, O'Brien AD. 2012. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 50:2951–2963. 10.1128/JCM.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitt CK, McKee ML, O'Brien AD. 1991. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H− strain E32511. Infect. Immun. 59:1065–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manning SD, Motiwala AS, Springman AC, Qi W, Lacher DW, Ouellette LM, Mladonicky JM, Somsel P, Rudrik JT, Dietrich SE, Zhang W, Swaminathan B, Alland D, Whittam TS. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. U. S. A. 105:4868–4873. 10.1073/pnas.0710834105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Persson S, Olsen KE, Ethelberg S, Scheutz F. 2007. Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J. Clin. Microbiol. 45:2020–2024. 10.1128/JCM.02591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jelacic JK, Damrow T, Chen GS, Jelacic S, Bielaszewska M, Ciol M, Carvalho HM, Melton-Celsa AR, O'Brien AD, Tarr PI. 2003. Shiga toxin-producing Escherichia coli in Montana: bacterial genotypes and clinical profiles. J. Infect. Dis. 188:719–729. 10.1086/376999. [DOI] [PubMed] [Google Scholar]
- 30.Eklund M, Leino K, Siitonen A. 2002. Clinical Escherichia coli strains carrying stx genes: stx variants and stx-positive virulence profiles. J. Clin. Microbiol. 40:4585–4593. 10.1128/JCM.40.12.4585-4593.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulasekara BR, Jacobs M, Zhou Y, Wu Z, Sims E, Saenphimmachak C, Rohmer L, Ritchie JM, Radey M, McKevitt M, Freeman TL, Hayden H, Haugen E, Gillett W, Fong C, Chang J, Beskhlebnaya V, Waldor MK, Samadpour M, Whittam TS, Kaul R, Brittnacher M, Miller SI. 2009. Analysis of the genome of the Escherichia coli O157:H7 2006 spinach-associated outbreak isolate indicates candidate genes that may enhance virulence. Infect. Immun. 77:3713–3721. 10.1128/IAI.00198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Brien AD, Melton AR, Schmitt CK, McKee ML, Batts ML, Griffin DE. 1993. Profile of Escherichia coli O157:H7 pathogen responsible for hamburger-borne outbreak of hemorrhagic colitis and hemolytic uremic syndrome in Washington. J. Clin. Microbiol. 31:2799–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Centers for Disease Control and Prevention. 1993. Update: multistate outbreak of Escherichia coli O157:H7 infections from hamburgers—western United States, 1992-1993. MMWR Morb. Mortal. Wkly. Rep. 42:258–263. [PubMed] [Google Scholar]
- 34.Panda A, Tatarov I, Melton-Celsa AR, Kolappaswamy K, Kriel EH, Petkov D, Coksaygan T, Livio S, McLeod CG, Nataro JP, O'Brien AD, DeTolla LJ. 2010. Escherichia coli O157:H7 infection in Dutch belted and New Zealand white rabbits. Comp. Med. 60:31–37. [PMC free article] [PubMed] [Google Scholar]
- 35.Zangari T, Melton-Celsa AR, Panda A, Boisen N, Smith MA, Tatarov I, De Tolla LJ, Nataro JP, O'Brien AD. 2013. Virulence of the Shiga toxin type 2-expressing Escherichia coli O104:H4 German outbreak isolate in two animal models. Infect. Immun. 81:1562–1574. 10.1128/IAI.01310-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 37.U.S. Department of Health and Human Services. 2007. Biosafety in microbiological and biomedical laboratories. U.S. Government Printing Office, Washington, DC. [Google Scholar]
- 38.Wadolkowski EA, Burris JA, O'Brien AD. 1990. Mouse model for colonization and disease caused by enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 58:2438–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohawk KL, Melton-Celsa AR, Zangari T, Carroll EE, O'Brien AD. 2010. Pathogenesis of Escherichia coli O157:H7 strain 86-24 following oral infection of BALB/c mice with an intact commensal flora. Microb. Pathog. 48:131–142. 10.1016/j.micpath.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindgren SW, Samuel JE, Schmitt CK, O'Brien AD. 1994. The specific activities of Shiga-like toxin type II (SLT-II) and SLT-II-related toxins of enterohemorrhagic Escherichia coli differ when measured by Vero cell cytotoxicity but not by mouse lethality. Infect. Immun. 62:623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roche Diagnostics. 2008. DIG application manual for filter hybridization. Roche Diagnostics, Indianapolis, IN. [Google Scholar]
- 42.Lindgren SW, Melton AR, O'Brien AD. 1993. Virulence of enterohemorrhagic Escherichia coli O91:H21 clinical isolates in an orally infected mouse model. Infect. Immun. 61:3832–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Serra-Moreno R, Acosta S, Hernalsteens JP, Jofre J, Muniesa M. 2006. Use of the lambda Red recombinase system to produce recombinant prophages carrying antibiotic resistance genes. BMC Mol. Biol. 7:31. 10.1186/1471-2199-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gentry MK, Dalrymple JM. 1980. Quantitative microtiter cytotoxicity assay for Shigella toxin. J. Clin. Microbiol. 12:361–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia A, Bosques CJ, Wishnok JS, Feng Y, Karalius BJ, Butterton JR, Schauer DB, Rogers AB, Fox JG. 2006. Renal injury is a consistent finding in Dutch Belted rabbits experimentally infected with enterohemorrhagic Escherichia coli. J. Infect. Dis. 193:1125–1134. 10.1086/501364. [DOI] [PubMed] [Google Scholar]
- 46.Garcia A, Marini RP, Feng Y, Vitsky A, Knox KA, Taylor NS, Schauer DB, Fox JG. 2002. A naturally occurring rabbit model of enterohemorrhagic Escherichia coli-induced disease. J. Infect. Dis. 186:1682–1686. 10.1086/345371. [DOI] [PubMed] [Google Scholar]
- 47.Robinson CM, Sinclair JF, Smith MJ, O'Brien AD. 2006. Shiga toxin of enterohemorrhagic Escherichia coli type O157:H7 promotes intestinal colonization. Proc. Natl. Acad. Sci. U. S. A. 103:9667–9672. 10.1073/pnas.0602359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith MJ, Melton-Celsa AR, Sinclair JF, Carvalho HM, Robinson CM, O'Brien AD. 2009. Monoclonal antibody 11E10, which neutralizes Shiga toxin type 2 (Stx2), recognizes three regions on the Stx2 A subunit, blocks the enzymatic action of the toxin in vitro, and alters the overall cellular distribution of the toxin. Infect. Immun. 77:2730–2740. 10.1128/IAI.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strauch E, Schaudinn C, Beutin L. 2004. First-time isolation and characterization of a bacteriophage encoding the Shiga toxin 2c variant, which is globally spread in strains of Escherichia coli O157. Infect. Immun. 72:7030–7039. 10.1128/IAI.72.12.7030-7039.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wadolkowski EA, Sung LM, Burris JA, Samuel JE, O'Brien AD. 1990. Acute renal tubular necrosis and death of mice orally infected with Escherichia coli strains that produce Shiga-like toxin type II. Infect. Immun. 58:3959–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eaton KA, Friedman DI, Francis GJ, Tyler JS, Young VB, Haeger J, Abu-Ali G, Whittam TS. 2008. Pathogenesis of renal disease due to enterohemorrhagic Escherichia coli in germ-free mice. Infect. Immun. 76:3054–3063. 10.1128/IAI.01626-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tyler JS, Beeri K, Reynolds JL, Alteri CJ, Skinner KG, Friedman JH, Eaton KA, Friedman DI. 2013. Prophage induction is enhanced and required for renal disease and lethality in an EHEC mouse model. PLoS Pathog. 9:e1003236. 10.1371/journal.ppat.1003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abu-Ali GS, Ouellette LM, Henderson ST, Lacher DW, Riordan JT, Whittam TS, Manning SD. 2010. Increased adherence and expression of virulence genes in a lineage of Escherichia coli O157:H7 commonly associated with human infections. PLoS One 5:e10167. 10.1371/journal.pone.0010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neupane M, Abu-Ali GS, Mitra A, Lacher DW, Manning SD, Riordan JT. 2011. Shiga toxin 2 overexpression in Escherichia coli O157:H7 strains associated with severe human disease. Microb. Pathog. 51:466–470. 10.1016/j.micpath.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Serra-Moreno R, Jofre J, Muniesa M. 2008. The CI repressors of Shiga toxin-converting prophages are involved in coinfection of Escherichia coli strains, which causes a down regulation in the production of Shiga toxin 2. J. Bacteriol. 190:4722–4735. 10.1128/JB.00069-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fogg PC, Saunders JR, McCarthy AJ, Allison HE. 2012. Cumulative effect of prophage burden on Shiga toxin production in Escherichia coli. Microbiology 158:488–497. 10.1099/mic.0.054981-0. [DOI] [PubMed] [Google Scholar]
- 57.Abu-Ali GS, Ouellette LM, Henderson ST, Whittam TS, Manning SD. 2010. Differences in adherence and virulence gene expression between two outbreak strains of enterohaemorrhagic Escherichia coli O157:H7. Microbiology 156:408–419. 10.1099/mic.0.033126-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.