Abstract
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 causes hemorrhagic diarrhea and potentially fatal renal failure in humans. Ruminants are considered to be the primary reservoir for human infection. Vaccines that reduce shedding in cattle are only partially protective, and their underlying protective mechanisms are unknown. Studies investigating the response of cattle to colonization generally focus on humoral immunity, leaving the role of cellular immunity unclear. To inform future vaccine development, we studied the cellular immune responses of cattle during EHEC O157:H7 colonization. Calves were challenged either with a phage type 21/28 (PT21/28) strain possessing the Shiga toxin 2a (Stx2a) and Stx2c genes or with a PT32 strain possessing the Stx2c gene only. T-helper cell-associated transcripts at the terminal rectum were analyzed by reverse transcription-quantitative PCR (RT-qPCR). Induction of gamma interferon (IFN-γ) and T-bet was observed with peak expression of both genes at 7 days in PT32-challenged calves, while upregulation was delayed, peaking at 21 days, in PT21/28-challenged calves. Cells isolated from gastrointestinal lymph nodes demonstrated antigen-specific proliferation and IFN-γ release in response to type III secreted proteins (T3SPs); however, responsiveness was suppressed in cells isolated from PT32-challenged calves. Lymph node cells showed increased expression of the proliferation marker Ki67 in CD4+ T cells from PT21/28-challenged calves, NK cells from PT32-challenged calves, and CD8+ and γδ T cells from both PT21/28- and PT32-challenged calves following ex vivo restimulation with T3SPs. This study demonstrates that cattle mount cellular immune responses during colonization with EHEC O157:H7, the temporality of which is strain dependent, with further evidence of strain-specific immunomodulation.
INTRODUCTION
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is a bacterial zoonotic disease of global importance (1). Ruminants, particularly cattle, are the predominant reservoir, and humans become infected following fecal contamination of food, the environment, and water (2–6). Many cases are sporadic (7); however, large outbreaks occur periodically. While colonization of cattle is largely asymptomatic (8), humans typically develop painful hemorrhagic diarrhea. A significant minority of patients progress to potentially fatal hemolytic-uremic syndrome (HUS), a form of renal failure resulting from Shiga toxin (Stx; also known as verotoxin)-induced endothelial dysfunction (9).
Significant efforts have been made in abattoirs and meat-processing plants to reduce the contamination of beef produce reaching the consumer. While these efforts have been partially successful, food-borne outbreaks have not been eliminated, and environmental transmission remains unmitigated. A range of on-farm interventions have been proposed to reduce bacterial shedding by cattle, including dietary manipulation, phage therapy, antimicrobial treatment, probiotic administration, and vaccination (10–15). Recent reviews and meta-analyses have identified probiotics and vaccines as the most tractable and efficacious control measures (12, 16, 17). Two commercial vaccines have been developed to date. One involves a method of extraction from bacteria cultured under iron-restricted conditions that enriches for membrane components (Epitopix, Willmar, MN, USA). The other is a supernatant preparation that contains type III secreted proteins (T3SPs) (Econiche; Bioniche Life Sciences, Belleville, Ontario, Canada). Vaccination of cattle with recombinant T3SPs has been demonstrated to reduce colonization rates in cattle, an effect enhanced by the addition of H7 flagellin (18, 19).
To our knowledge, the mechanism of protection of these vaccines has not been demonstrated. EHEC O157:H7 uses its type III secretion system (T3SS) to deliver a range of effector proteins to host epithelial cells (20, 21). The T3SS includes an EspA translocon filament, capped by the pore-forming EspB/EspD (EspB/D) complex. The construction of the translocon and the delivery of effector proteins by the T3SS are carefully regulated (22). Injected effectors include the translocated intimin receptor (Tir), which binds to the bacterial surface protein intimin and is central to the formation of actin pedestals and intimate attaching and effacing (A/E) lesions (23). It is reasonable to hypothesize that antibodies directed against the structural components of the T3SS, such as EspA/B/D, or against adhesion factors, such as intimin and its cognate receptor Tir, may block bacterial binding. In support of this hypothesis, vaccination of sows with intimin has been shown to reduce the level of EHEC O157:H7 colonization of suckling piglets (24), presumably through antibody-mediated blocking of bacterial binding. In addition, bovine colostrum has been shown to reduce T3SS-mediated hemolysis in vitro (25), and passive transfer of EspB, intimin, and neutralizing antibodies against Stx2 to calves has been demonstrated following maternal vaccination (26). H7 flagellin has also been demonstrated to act as an adhesion factor, and anti-H7 antibodies reduce the level of binding of bacteria to primary cell cultures (27).
Despite high antibody titers following vaccination with these antigens, vaccination is only partially protective (13, 14, 28–31), while antibody titers are inconsistently correlated with bacterial shedding (32–34). The serological response of cattle to colonization has also been studied extensively, and antibodies against O157 lipopolysaccharide (LPS), H7, Tir, intimin, EspA, EspB, EspD, EspM2, NleA, TccP, Stx1, and Stx2 have been demonstrated (25, 33–36). Despite this, prior exposure to EHEC O157:H7 results in only partial and transient protection against reinfection (33, 34, 37).
While cattle can shed detectable levels of EHEC O157:H7 for a considerable period, most animals are able to clear the infection successfully (37–39). Given the limitations of antibody-mediated protection, it is likely that other innate and adaptive responses play important roles in bacterial clearance. It is also rational to hypothesize that once a microcolony has formed on an epithelial cell, and its cellular processes are thus being manipulated by secreted bacterial effectors, the best way of effectively clearing the infection is to dispose of the colonized cell. This raises the prospect of an important role for cellular immunity in bacterial clearance. However, to our knowledge, only two studies have considered the role of cellular immunity during EHEC O157:H7 colonization in ruminants; these identified lymphoproliferative cellular responses to heat-killed EHEC O157:H7 (40) and recombinant antigens (41) in bovine peripheral blood mononuclear cells and ovine rectal lymph node cells, respectively. The case for a protective cellular immune response is further strengthened by the observations that colonized bovine epithelial cells are exfoliated in vivo (42) and that some strains are efficiently internalized by bovine epithelial cells both in vivo and in vitro (43).
While colonization of mice by Citrobacter rodentium has been used as a model for study of the interaction of an A/E lesion-forming bacterium with its mammalian host (44), none of the mouse models reported to date have demonstrated convincing colonization and A/E lesion formation with EHEC O157:H7 (45–51). The relevance of studies on mice to the interaction of EHEC O157:H7 with its natural bovine host is therefore unclear. In addition to being the natural reservoir of infection, cattle can also be sampled repeatedly during colonization, allowing one to control for interanimal variation resulting from the use of outbred populations.
We hypothesized that by quantifying T-helper cell-associated gene transcripts at the terminal rectums of cattle during the course of EHEC O157:H7 colonization, it would be possible to identify and determine the direction of the cellular immune response at the primary site of colonization. Our results show that there is a TH type 1 skew to the response at the rectal mucosa, the temporal nature of which differs by strain, and that colonization with both the PT21/28 and the PT32 strain results in an increase in the proportion of CD4+ T cells within the rectal mucosal T-cell population. We also present data analyzing regional lymph node responses to bacterial proteins that suggest a role for both innate and adaptive cellular immunity in the bovine response to colonization.
MATERIALS AND METHODS
Bacterial strains, inocula, and T3SP preparation.
The strains used in this study are listed in Table 1. Glycerol stocks were resuscitated on lysogeny broth (LB) agar and were incubated at 37°C overnight. To generate the inocula for the calf studies, a single colony of strain 9000 (phage type 21/28 [PT21/28]), PT32 strain 10671, Zap1380 (a naturally derived nalidixic acid [Nal]-resistant variant of 9000), or Zap1381 (a naturally derived Nal-resistant variant of 10671) was used to inoculate separate flasks of LB, which were incubated at 37°C overnight. Inocula for each calf were prepared by mixing 5 ml of an overnight LB culture of 9000 with 5 ml of an overnight LB culture of Zap1380 or by mixing 5 ml of an overnight LB culture of 10671 with 5 ml of an overnight LB culture of Zap1381, representing a final dose of ∼1 × 109 CFU per calf.
TABLE 1.
EHEC O157:H7 strains used in this study
| Strain | Shiga toxin gene(s) | Phage type | Origin | Modification(s) | Source or reference |
|---|---|---|---|---|---|
| 9000 (WX009000S01E) | stx2a, stx2c | PT21/28 | Cattle feces | None | 86 |
| Zap1380 | stx2a, stx2c | PT21/28 | Strain 9000 | Nalr | This study |
| 10671 (WX010671S01E) | stx2c | PT32 | Cattle feces | None | 86 |
| Zap1381 | stx2c | PT32 | Strain 10671 | Nalr | This study |
| Zap193 (NCTC 12900) (WT) | None | NAb | NCTCa | None | NCTC |
| Zap1143 (ΔsepL mutant) | None | NA | Strain Zap193 | Nalr, ΔsepL | 52 |
National Collection of Type Cultures (NCTC), Public Health England (https://www.phe-culturecollections.org.uk/collections/nctc.aspx).
NA, not available.
To generate the T3SP preparations for the study of antigen recall responses, a single colony of Zap193 (wild type [WT]) or Zap1143 (ΔsepL) was used to inoculate 10 ml LB, which was incubated at 37°C (200 rpm) overnight. The WT strain secretes predominantly structural components of the T3SS, while the ΔsepL strain instead secretes a wider variety of effector proteins (52). Two milliliters of the overnight culture was used to inoculate 500 ml minimal essential medium (MEM)-HEPES (Sigma-Aldrich, Gillingham, United Kingdom), which was cultured at 37°C (200 rpm) to an optical density at 600 nm (OD600) of 0.8. Bacteria were pelleted, and supernatants were filter sterilized (0.2-μm low protein binding filters; Millipore, Watford, United Kingdom). Proteins were precipitated overnight at 4°C using trichloroacetic acid (VWR International, Lutterworth, United Kingdom) at a final concentration of 10% (vol/vol). Precipitated proteins were pelleted at 5,000 × g for 30 min at 4°C and the supernatant discarded. Pellets were suspended in 1.5 M Tris-HCl (pH 8.8) and were dialyzed in phosphate-buffered saline (PBS) across a regenerated cellulose membrane with a molecular size cutoff rating of 3.5 kDa (Spectrum Labs, Breda, The Netherlands). Protein preparations were checked by separation using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were stained with Coomassie blue (Invitrogen, Paisley, United Kingdom) (see Fig. S1 in the supplemental material). The total-protein concentration was estimated using a bicinchoninic protein assay kit (Pierce Biotechnology, Rockford, IL, USA) and was read using a microplate reader at 562 nm (Dynex Technologies, Worthing, United Kingdom). The lipopolysaccharide (LPS) concentration was estimated using the EndoLISA kit (Hyglos GmbH, Bernried, Germany) and was read using a Synergy HT microplate reader (BioTek, Winooski, VT, USA) fitted with 360/40-nm wavelength excitation and 460/40-nm wavelength emission filters.
Animal experiments.
Oral bacterial challenge was performed at the Moredun Research Institute (MRI) under Home Office license 60/3179. Ethical approval was obtained from the MRI Animal Experiments Committee. Two groups of six conventionally reared male dairy calves were randomly assigned to separate rooms in the MRI High Security Unit (HSU). Two calves were housed in conventional pens. The average age of the calves at the time of challenge was 12 ± 2 weeks. At this age, all the animals were weaned, i.e., they were ruminants yet small enough to be handled safely in the HSU. Fecal samples obtained from each calf prior to challenge were confirmed to be negative for EHEC O157:H7 by immunomagnetic separation (IMS) performed according to the manufacturer's instructions (anti-EHEC O157 Dynabeads; Invitrogen). Five additional age-matched calves that had been used as controls in other studies were used as sources of additional lymph node and rectal mucosal samples. Fecal samples collected postmortem from these calves were screened for the presence of EHEC O157:H7 using IMS. EHEC O157:H7 was not isolated from any of the samples.
Four calves in one HSU room were orally challenged by orogastric intubation with 500 ml PBS containing 10 ml of an overnight LB culture of strain 9000 and its naturally derived Nal-resistant variant. Four calves in the other HSU room were orally challenged in the same way with strain 10671 and its naturally derived Nal-resistant variant. The other two calves (sentinels) in each HSU room were administered 500 ml PBS only.
Three days after oral challenge, 10 g surface feces taken directly from the rectum was suspended in 90 ml sterile PBS. Samples were collected daily for the first 2 weeks and then every other day. Tenfold serial dilutions were prepared in PBS, and 100 μl from three dilutions across a 1,000-fold range of dilutions was plated out in triplicate onto cefixime-tellurite sorbitol MacConkey agar (CT-SMAC) plates. Resuspended feces were stored at 4°C overnight. Plates were incubated at 37°C overnight and colonies enumerated at the most suitable dilution. Five to 10 colonies from each plate were confirmed as O157 positive by using a latex agglutination kit (Oxoid, Basingstoke, United Kingdom). The CFU per gram of feces was calculated by multiplying the mean colony count of the triplicate plates by the appropriate dilution factor. Where no colonies were observed, broth enrichment was carried out with 1 ml of the resuspended feces added to 9 ml tryptone soya broth (TSB; Oxoid). TSBs were incubated at 37°C overnight and were plated onto CT-SMAC plates. Overnight bacterial growth was tested for O157 by latex agglutination. Feces that were negative by direct plating but positive after broth enrichment were assigned a value of 10 CFU/g.
Rectal biopsy specimens were taken at −5, 7, 14, 21, and 31 days postchallenge. A local anesthetic was applied to the anal sphincter (5% EMLA; Astra Zeneca, Luton, United Kingdom). A rectal speculum (Veterinary Instrumentation, Sheffield, United Kingdom) was used to access the rectal mucosa. Pinch biopsy specimens from the rectal mucosa were taken from two opposing sites approximately 5 cm proximal to the rectoanal junction. The position of each biopsy specimen was recorded, and the site was avoided at subsequent samplings. Biopsy specimens were immediately placed into RNAlater (Ambion, Paisley, United Kingdom) and were stored at 4°C overnight before storage at −80°C.
At the end of the trial (31 to 33 days), calves were killed using 60 ml intravenous pentobarbital (Animalcare, York, United Kingdom). Rectal lymph nodes (RLN), mesenteric lymph nodes (MLN) (100 cm proximal to the ileocecocolic junction) and prescapular lymph nodes (PsLN) were collected and were transported in 35 ml preparation medium (Hanks buffered saline solution [HBSS] without calcium and magnesium, 2% heat-inactivated [56°C, 30 min] fetal calf serum [HI-FCS], 10 mg/ml gentamicin [Sigma-Aldrich], 200 IU/ml penicillin, and 200 μg/ml streptomycin). A 0.5-cm-wide strip of rectal mucosa was dissected and was placed in PBS containing 2 mg/ml polymyxin B (Sigma-Aldrich) for subsequent isolation of rectal mucosal lymphocytes.
Assessment of gene expression.
RNA was extracted from the rectal biopsy specimens using the RNeasy Plus minikit (Qiagen, Hilden, Germany). Biopsy specimens were disrupted in Precellys CK28 tubes using 5 23-s cycles at 6,200 rpm in a tissue homogenizer (Peqlab, Salisbury Green, United Kingdom). Tubes were put on ice for 2 min between cycles. Residual DNA was digested on the column with DNase I (Qiagen). The RNA yield was measured using a NanoDrop ND1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) and RNA quality assessed using an RNA 6000 Nano kit for total RNA and a model 2100 Bioanalyzer (Agilent Technologies, Wokingham, United Kingdom). All RNA samples had an RNA integrity number (RIN) score of >7.0.
cDNA was synthesized as per the manufacturer's instructions, using SuperScript II (Invitrogen) reverse transcriptase (RT), RNAseOUT (Invitrogen), a deoxynucleoside triphosphate (dNTP) mixture (Invitrogen), and oligo(dT)23 primers (Sigma-Aldrich), from 1 μg RNA, where 0.5 μg was pooled from each of the two biopsy specimens taken at each time point. The reaction volume was 20 μl. Cycling conditions were as follows: 42°C for 2 min followed by the addition of reverse transcriptase, 42°C for 50 min, and 70°C for 15 min. cDNA was diluted 3-fold using PCR water and was stored at −20°C prior to use.
Quantitative PCR (qPCR) was conducted in triplicate in 96-well qPCR plates using Precision master mix with ROX (Primer Design, Southampton, United Kingdom) and an ABI Prism 7000 or ABI Prism 7500 (Applied Biosystems, Paisley, United Kingdom) qPCR instrument as per the manufacturer's instructions. The reaction volume was 20 μl. Cycling conditions were 95°C for 15 min and 50 cycles of 95°C for 15 s and 60°C for 60 s, during which time fluorescence was read, followed by melting curve analysis.
Reference genes were selected using the bovine geNorm kit (Primer Design) containing primers for ATP5B, eukaryotic initiation factor 2, subunit beta (EIF2B2), actin, beta (ACTB), succinate dehydrogenase complex, subunit A (SDHA), RPL12, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). qPCRs were prepared using 5 μl cDNA from each time point for three animals. qBasePLUS2 software, version 2.4 (Biogazelle, Zwijnaarde, Belgium), was used to select the GAPDH and ATP5B genes as the most stably expressed genes. Primers and standard curve plasmids for GAPDH and ATP5B were supplied by Primer Design and were used as per the manufacturer's instructions. All other primers in this study were supplied by Eurofins Genomics (Acton, United Kingdom).
Plasmids for standard curve generation for bovine interleukin 10 (IL-10), transforming growth factor β1 (TGF-β1), gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), IL-4, and IL-13 were available in-house. Fragments of bovine FoxP3, RAR-related orphan receptor C (RORC), IL-17, IL-22, T-bet, and GATA3 were cloned using the external primers shown in Table S1 in the supplemental material, which were designed using Primer BLAST (NCBI, Bethesda, MD, USA), and their specificity was checked against the Bos taurus RefSeq mRNA database. Gene fragments were generated using KOD Hot Start (Millipore) DNA polymerase, a dNTP mixture (Invitrogen), and bovine cDNA synthesized from one rectal biopsy sample as per the manufacturer's instructions. The reaction volume was 50 μl. Cycling was carried out according to the following touchdown PCR (53) protocol: 95°C for 3 min; 15 cycles of 95°C for 30 s, 70°C decreasing by 1°C per cycle for 45 s, and 72°C for 60 s; 20 cycles of 95°C for 30 s, 55°C for 45 s, and 72°C for 60 s; and 70°C for 10 min. PCR products were run on a 2% agarose gel and bands of the expected size excised and purified using the QIAquick gel extraction kit (Qiagen). Two microliters of the purified amplicon mixture was A-tailed by incubation with 5 U of GoTaq DNA polymerase (Promega, Southampton, United Kingdom), 2 μl 5× GoTaq DNA master mix (Promega), 1 μl 2 mM dATP (Invitrogen), and 4 μl PCR water for 30 min at 70°C. A-tailed amplicons were ligated into pGEM-T Easy vector plasmids (Promega) as per the manufacturer's instructions.
Plasmids were used to transform JM109 competent cells (Promega) as per the manufacturer's instructions, and the transformed cells were plated onto LB agar containing 100 μg/ml ampicillin, 80 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal), and 500 μM isopropyl-β-d-thiogalactopyranoside (IPTG). Two white colonies for each transformation were checked by colony PCR using the internal primers shown in Table S2 in the supplemental material. Internal primers were taken from the literature or were designed using Primer BLAST (NCBI). Specificity was checked against the Bos taurus RefSeq mRNA database. One PCR-positive colony for each gene was used to inoculate 10 ml LB, and the mixture was incubated at 37°C (200 rpm) overnight. Plasmids were purified using the Wizard Plus SV Miniprep DNA purification system (Promega). Plasmids were sequenced by Eurofins Genomics using the T7 and M13 rev (−29) primers. Sequences were checked against the Bos taurus RefSeq RNA database using BLAST (NCBI), and internal primers (see Table S2) were checked using the Sequence Manipulation Suite, version 2 (Bioinformatics Organization, MA, USA). Plasmid molecular weight was estimated using the Sequence Manipulation Suite by updating the published plasmid sequence with the inserted sequences. One microgram of plasmid DNA was linearized using the restriction enzyme NdeI (Promega) at 37°C for 30 min. Linearization was confirmed by agarose gel electrophoresis. Digests were purified using the QIAquick PCR purification kit (Qiagen); DNA was quantified; and linearized plasmids were diluted using PCR water to 109 copies per μl.
Plasmids were diluted across the dynamic ranges indicated in Table S3 in the supplemental material and were used to generate a standard curve for each gene on each qPCR plate run. qPCR mixtures were prepared using 1 μl cDNA/well. For ATP5B, GAPDH, IFN-γ, T-bet, IL-17, RORC, TNF-α, and IL-10, Precision master mix was used as per the geNorm experiment described above. For the lower-copy-number transcripts GATA3, FoxP3, IL-4, IL-22, IL-13, and TGF-β1, the SYBR GreenER qPCR SuperMix (Invitrogen) was used as per the manufacturer's instructions. Reactions were performed in duplicate, and the reaction volume was 25 μl. Cycling conditions were 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 60 s, during which time fluorescence was read, followed by melting curve analysis.
Standard curves were calculated from the threshold cycle (CT) values using ABI Prism 7000 SDS software, version 1.2.3 (Applied Biosystems), and the calculated gene copies/μl cDNA were exported in comma-separated variable files. An SQL script (MySQL Community Server, version 5.1) was used to calculate the arithmetic mean for the technical repeats and then to normalize the copy number/μl cDNA for each gene to the geometric mean copy number/μl cDNA for the reference genes ATP5B and GAPDH as described previously (54).
Analysis of mucosal T-cell populations.
Rectal mucosal strips collected postmortem were cut into 1-cm by 2-cm pieces, placed in 25 ml digestion medium consisting of Dulbecco's modified Eagle medium (DMEM; Sigma-Aldrich) supplemented with 10% HI-FCS, 75 IU/ml collagenase (Sigma-Aldrich), and 20 μg/ml dispase I neutral protease, grade 1 (Roche, Basel, Switzerland), and incubated at 37°C (200 rpm) for 90 min. The digested samples were shaken vigorously and were centrifuged at 405 × g for 5 min. The supernatant was passed through a 100-μm filter (BD Biosciences, San Jose, CA), made up to 50 ml with lymphocyte medium (RPMI 1640 [Gibco], 10% HI-FCS, 200 mM l-glutamine [Invitrogen], 0.004% β-mercaptoethanol [Sigma-Aldrich], 100 IU/ml penicillin, and 100 μg/ml streptomycin [Invitrogen]), and centrifuged at 405 × g for 5 min. The supernatant was discarded and the pellet resuspended in 50 ml lymphocyte medium. This process was repeated until the supernatant was clear after centrifugation. The pellet was resuspended in 10 ml lymphocyte medium; cells were enumerated; and a V bottom 96-well plate was seeded with 1 × 106 cells/well. Unless otherwise stated, incubations were carried out for 30 min at 4°C, and 2% fluorescence-activated cell sorting (FACS) buffer (2% HI-FCS in PBS) was used for washes and antibody dilutions. The antibodies used are listed in Table 2. Unstained, no-primary-antibody, and single-stained controls were prepared. The cells were washed once, resuspended, and incubated with anti-bovine CD3 and anti-bovine CD4, CD8, or TcR1-N24(δ). The cells were washed three times, resuspended, and incubated with anti-mouse IgG1 and anti-mouse IgG2a or anti-mouse IgG2b for 20 min. The cells were subsequently washed and were resuspended in 2% FACS buffer. Data were acquired using a FACSCalibur flow cytometer (BD Biosciences) with a total of 10,000 events collected in the gated region. Compensation and analysis were conducted using FlowJo, version 7.5 (Tree Star, Ashland, OR, USA).
TABLE 2.
Antibodies used in this study
| Reactivity | Clone/host | Isotype | Conjugate | Supplier |
|---|---|---|---|---|
| Bovine CD3 | MM1A | IgG1 | None | Washington State University, Pullman, WA |
| Bovine CD4 | ILA12 | IgG2a | None | In-house |
| Bovine CD8 | ILA105 | IgG2a | None | In-house |
| Bovine TcR1-N24(δ) | GB21A | IgG2b | None | Washington State University, Pullman, WA |
| Bovine NKp46 | AKS1 | IgG1 | Alexa Fluor 488 | AbD Serotec, Oxford, UK |
| Bovine IFN-γ | CC302 | IgG1 | Alexa Fluor 647 | AbD Serotec, Oxford, UK |
| Mouse IgG | Goat | NAa | Alexa Fluor 488 | Molecular Probes, Paisley, UK |
| Ki67 | Rabbit | NA | None | Abcam, Cambridge, UK |
| Rabbit Ig | Goat | NA | Alexa Fluor 405 | Molecular Probes |
| Mouse IgG1 | Goat | NA | Alexa Fluor 647 | Molecular Probes |
| Mouse IgG2a | Goat | NA | Phycoerythrin | Molecular Probes |
| Mouse IgG2b | Goat | NA | Alexa Fluor 488 | Molecular Probes |
NA, not applicable.
Ex vivo restimulation experiments.
Lymph nodes extracted postmortem were first washed twice with 25 ml preparation medium and then placed in a 100-mm by 25-mm petri dish containing 15 ml preparation medium. The nodes were repeatedly incised and were then transferred to a stomacher bag and placed in a stomacher for 30 s. The contents were then filtered through a 70-μm nylon filter (Fisher Scientific, Loughborough, United Kingdom) and were made up to 20 ml with preparation medium. The suspension was underlaid with 10 ml Ficoll-Paque (GE Healthcare, Little Chalfont, United Kingdom) and was centrifuged at 800 × g for 30 min at 4°C. The mononuclear cells were harvested from the top of the Ficoll layer and were washed twice in PBS before being resuspended in lymphocyte medium. Spare cells were resuspended in 10% (vol/vol) dimethyl sulfoxide (DMSO; Sigma-Aldrich) in HI-FCS and were stored in cryovials at 1 × 107 cells/ml in liquid nitrogen. Round-bottom 96-well plates were seeded with 1 × 105 cells/well and were stimulated with 5 μg/ml WT or ΔsepL T3SPs. Controls included concanavalin A (ConA: 5 μg/ml; Sigma-Aldrich), lymphocyte medium, and 38 or 78 endotoxin units (EU)/ml O111:B4 LPS (Sigma-Aldrich), corresponding to the LPS concentration of the WT or ΔsepL T3SPs, respectively. Cultures were incubated for 5 days under a humidified 5% CO2 atmosphere at 37°C. Cultures were pulsed with 1 μCi/well of [3H]thymidine (PerkinElmer, Waltham, MA) for the final 18 h of incubation by removing 50 μl of the supernatant and replacing it with [3H]thymidine-containing lymphocyte medium. The removed supernatants were stored at −20°C prior to use. Cells were harvested onto glass fiber filters (PerkinElmer, Waltham, MA, USA). [3H]thymidine incorporation was quantified using an automated scintillation counter (PerkinElmer) and was expressed as counts per minute, with each test performed in triplicate. Stimulation indices (SI) were calculated by dividing the mean value for ConA by the mean value for the medium control or by dividing the mean value for the T3SPs by the mean value for the relevant LPS control.
IFN-γ release was measured using high-binding 96-well enzyme-linked immunosorbent assay (ELISA) plates (Corning, Amsterdam, The Netherlands) and a commercial bovine IFN-γ ELISA kit (Mabtech AB, Nacka Strand, Sweden) as per the manufacturers' instructions. Results below the limit of detection were assigned a value of 16 pg/ml. The supernatants of triplicate wells harvested as described above were pooled, and IFN-γ was assayed in duplicate. IFN-γ release indices were calculated by dividing the mean value for ConA by the mean value for the medium control or by dividing the mean value for the T3SPs by the mean value for the relevant LPS control.
Proliferation marker expression.
For each animal, one vial of rectal lymph node cells stored in liquid nitrogen was resuscitated in lymphocyte medium and was used to seed round-bottom 96-well plates at 5 × 105 cells/well. Cells were stimulated in duplicate with 5 μg/ml heat-treated (110°C for 30 min) WT T3SPs. Controls and culture conditions were as described above. Brefeldin A (10 μg/ml; Sigma-Aldrich) was added for the final 5 h of incubation by removing 50 μl of the supernatant and replacing it with spiked lymphocyte medium. Duplicate stimulated cultures were first pooled and then split evenly between 6 wells of a 96 well round-bottom plate.
The antibodies used are listed in Table 2. Unless stated otherwise, incubation was carried out for 30 min at 4°C, and 5% FACS buffer (5% HI-FCS, 0.02% sodium azide in PBS) was used for washes and antibody dilutions. Unstained, no-primary-antibody, single-stained, and fluorescence-minus-one (FMO) controls were prepared. Cells were pelleted, resuspended, and incubated with anti-bovine CD4, CD8, TcR1-N24(δ), or NKp46. Cells were washed twice, resuspended, and incubated with anti-mouse IgG. Cells were washed twice with PBS and were incubated with Live/Dead Fixable Near-IR reactive dye (Invitrogen) in 0.1% DMSO in PBS. Cells were washed in PBS and were fixed using 1% paraformaldehyde for 10 min at room temperature. Cells were washed in PBS and were permeabilized overnight in permeabilization/block buffer (5% FACS buffer, 0.2% saponin [Sigma-Aldrich], 20% heat-inactivated mouse serum [Invitrogen]). Cells were pelleted, resuspended, and incubated for 1 h with anti-bovine IFN-γ and anti-Ki67 in permeabilization buffer (5% FACS buffer, 0.2% saponin). Cells were washed twice in permeabilization buffer, resuspended, and incubated for 1 h with anti-rabbit Ig in permeabilization buffer. Cells were washed twice in permeabilization buffer and were resuspended in PBS. The entire sample was analyzed using a MACSQuant flow cytometer (Miltenyi Biotec, Surrey, United Kingdom). Compensation and data analysis were conducted using FlowJo, version 10.0.7 (Tree Star). The gating strategy is shown in Fig. S2 in the supplemental material.
Statistical analysis.
Statistical analysis of RT-qPCR data was performed using SAS, version 9.3 (SAS Institute Inc., Cary, NC, USA). All other statistical analyses and plotting of graphs were performed using R, version 3.1.0 (55), and the ggplot2 (56), lattice (57), nlme (58), lme4 (59), plyr (60), reshape (61), multcomp (62), grid (55), gridExtra (63), and lsmeans (64) packages.
T-helper cell-associated gene copy numbers were analyzed for the four orally challenged calves in each group and the two control calves. The fold change in gene expression from the prechallenge level was calculated for each gene and was log10 transformed. These log values were analyzed using linear mixed models, with log10 prechallenge gene copies, experimental group, days postchallenge, and the interaction of the experimental group with the number of days postchallenge fitted as fixed effects. Animals were fitted as random effects. The use of log values ensured that normality requirements for the mixed model were satisfied.
Flow cytometry counts of surface marker-positive versus surface marker-negative CD3+ T cells isolated from the terminal rectum were analyzed using generalized linear models. These fitted the experimental group as the only explanatory variable and fitted a dispersion parameter to allow for overdispersion in the data. Comparisons between each pair of experimental groups were made using Tukey tests. Flow cytometry counts of Ki67-positive versus Ki67-negative, surface marker-positive rectal lymph node cells were also analyzed using generalized linear models with the experimental group, the ex vivo stimulant, and the interaction of the experimental group with the ex vivo stimulant fitted as explanatory variables, and a dispersion parameter was included to allow for overdispersion in the binomial data. Comparisons between each pair of experimental groups were made using Tukey tests.
Log10-transformed thymidine incorporation, IFN-γ release, stimulation indices, and IFN-γ release indices were analyzed using linear mixed models with restricted maximum likelihood. The models fitted the experimental group, ex vivo stimulant, and lymph node as fixed effects. Additionally, two-way interactions between every two main effects were tested and were retained in the model if significant. The animal, the interaction of the animal with the lymph node, and the interaction of the animal with the stimulant were fitted as random effects. Comparisons between each pair of experimental groups were made using Tukey tests. When there was a significant interaction between the experimental group and the lymph node or stimulant, pairwise comparisons were made within the results for each lymph node or stimulant, again using Tukey tests.
RESULTS
Bacterial shedding.
Individual shedding curves for each calf are shown in Fig. 1. EHEC O157:H7 was detectable at >103 CFU/g feces in all orally challenged calves from the first sampling at 3 days postchallenge until days 8 and 10 for the PT32 and PT21/28 strains, respectively, with peak shedding occurring between days 3 and 7. Bacteria were present at >103 CFU/g in both sentinel calves in the PT21/28 group from the first sampling until day 10, with peak shedding occurring at days 6 and 8. Shedding did not exceed 103 CFU/g until day 4 for one of the two PT32 sentinel calves and exceeded 103 CFU/g only on day 8 for the other, with peak shedding occurring at days 7 and 8. Shedding after day 10 was more heterogeneous in all calves, with some animals exceeding 103 CFU/g at multiple samplings while others remained below 103 CFU/g for the remainder of the study. These results confirm that the subsequent immunological analyses were conducted on successfully colonized animals.
FIG 1.
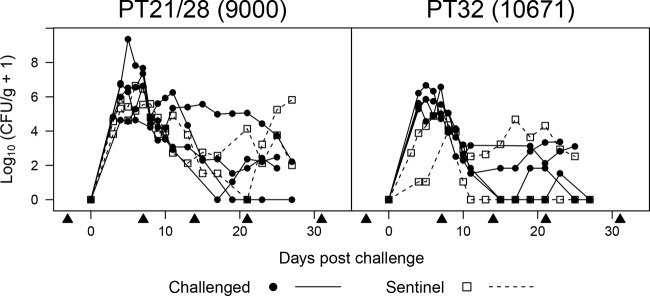
Fecal shedding of EHEC O157:H7 by experimentally infected weaned calves. Six calves were housed in each of two separate HSU rooms. Four calves in each HSU room were dosed with EHEC O157:H7 (strain 9000 in one room and strain 10671 in the other) by orogastric intubation (challenged), while two calves in each room received PBS only (sentinel). Each curve represents an individual animal. Arrowheads indicate rectal biopsy time points.
Gene expression at the terminal rectum during colonization.
The inflammatory transcript TNF-α, TH1-associated transcripts IFN-γ and T-bet, TH2-associated transcripts IL-4, IL-13, and GATA3, TH17-associated transcripts IL-17, IL-22, and RORC, and regulatory T cell (TReg)-associated transcripts IL-10, TGF-β1, and FoxP3 were quantified in rectal biopsy specimens taken from orally challenged calves at −5, 7, 14, 21, and 31 days postchallenge. Since the exact time of challenge was unknown for the sentinel animals, only transcript data from the orally challenged calves were analyzed. Transcripts for IL-4 and IL-13 were below the limit of quantification of 100 and 1,000 copies per μl of cDNA, respectively (data not shown). The log10-transformed relative change in expression from the prechallenge level was calculated for the remaining data and was analyzed using linear mixed models for each transcript of interest. The tests of the fixed effects for each model are shown in Table S4 in the supplemental material. The predicted means and 95% confidence intervals (95% CI) of the model outputs are shown in Fig. 2 and Table S5 in the supplemental material.
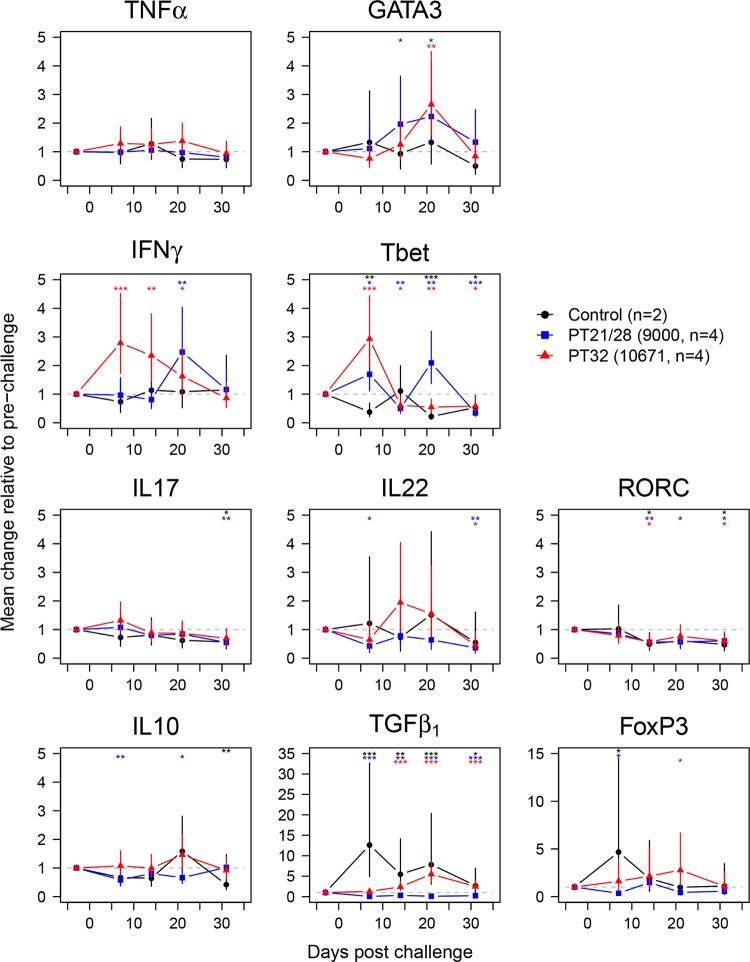
FIG 2.
Gene expression at the terminal rectums of calves orally challenged with EHEC O157:H7. Data represent predicted means and 95% confidence intervals from linear-mixed-model analyses of changes in log10-transformed gene expression from prechallenge levels. Statistically significant differences from prechallenge levels are indicated for control (black symbols), strain 9000 (PT21/28)-challenged (blue symbols), and strain 10671 (PT32)-challenged (red symbols) calves. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Analysis of TNF-α as a general inflammatory indicator was included. There was no statistically significant change in expression at any time point in either control or challenge groups, indicating that the biopsy procedure and challenge did not induce a detectable inflammatory response. Levels of GATA3, the only TH2-associated transcript that could be quantified, increased gradually and peaked with 2.23-fold (95% CI, 1.20- to 4.14-fold; P = 0.0134) and 2.65-fold (95% CI, 1.56- to 4.50-fold; P = 0.001) increases at 21 days postchallenge in the PT21/28 and PT32 groups, respectively. There was no statistically significant change in the control group.
The patterns of expression of the TH1-associated transcripts were noticeably different for the two strains. IFN-γ expression in the PT32-challenged calves increased 2.79-fold (95% CI, 1.72- to 4.52-fold; P = 0.0002) by 7 days postchallenge and declined gradually, remaining significantly above prechallenge levels until 21 days postchallenge. This was in stark contrast to induction in the PT21/28-challenged calves, where IFN-γ expression remained unchanged until its peak at 21 days of 2.47 times prechallenge levels (95% CI, 1.51 to 4.04 times prechallenge levels; P = 0.001). There was no change in IFN-γ expression in the control animals. Changes in T-bet expression in PT32-challenged calves also peaked at 7 days (2.93-fold; 95% CI, 1.93- to 4.44-fold; P < 0.0001), but T-bet expression declined more rapidly than IFN-γ expression, to below prechallenge levels by 14 days (0.61-fold; 95% CI, 0.4- to 0.92-fold; P = 0.0214), where it remained until the end of the trial. T-bet expression in the PT21/28-challenged group was noticeably different, with biphasic induction at 7 days (1.69-fold; 95% CI, 1.11- to 2.59-fold; P = 0.0169) and 21 days (2.09-fold; 95% CI, 1.37- to 3.19-fold; P = 0.0016). In the control group, T-bet expression was significantly lower than prechallenge levels at 7 (0.38-fold; 95% CI, 0.21- to 0.69-fold; P = 0.0026), 21 (0.22-fold; 95% CI, 0.12- to 0.39-fold; P < 0.0001), and 31 (0.53-fold; 95% CI, 0.29- to 0.95-fold; P = 0.0346) days postchallenge. For all three groups, T-bet expression was significantly below prechallenge levels by day 31. Taken together, these results suggest a strong TH type 1 skew in the response to colonization, the temporal dynamics of which differed by strain.
None of the three TH17-associated transcripts IL-17, IL-22, and RORC demonstrated a statistically significant increase in expression following challenge. IL-22 expression was somewhat variable within the control and PT32-challenged groups, as evidenced by the wide 95% CI. IL-17 expression remained unchanged until day 31, when it was significantly lower than prechallenge levels in the control group (0.57-fold; 95% CI, 0.32- to 0.99-fold; P = 0.0467) and in PT21/28-challenged calves (0.55-fold; 95% CI, 0.37- to 0.81-fold; P = 0.0047). RORC expression followed a similar pattern, with statistically significant downregulation in PT21/28-challenged calves (0.56-fold; 95% CI, 0.37- to 0.85-fold; P = 0.0089), PT32-challenged calves (0.57-fold; 95% CI, 0.38- to 0.87-fold; P = 0.0111), and control calves (0.50-fold; 95% CI, 0.28- to 0.89-fold; P = 0.0218) by 14 days. IL-22 expression in the PT21/28 group was down at day 7 (0.43-fold; 95% CI, 0.20- to 0.89-fold; P = 0.0257), with no other statistically significant changes in expression until day 31, when it was reduced in the PT21/28 (0.36-fold; 95% CI, 0.17- to 0.76-fold; P = 0.0094) and PT32 (0.44-fold; 95% CI, 0.21- to 0.92-fold; P = 0.0314) groups.
The TReg-associated transcripts TGF-β1 and FoxP3 demonstrated strong induction in control calves at 7 days, with changes of 12.64-fold (95% CI, 4.89- to 32.70-fold; P < 0.0001) and 4.67-fold (95% CI, 1.50- to 14.54-fold; P = 0.0102), respectively. Unlike FoxP3 expression, which returned to baseline, TGF-β1 expression remained elevated until the end of the study. This was in contrast to IL-10 expression, which was unchanged in the control calves until it was downregulated on day 31 (0.42-fold; 95% CI, 0.24- to 0.74-fold; P = 0.0047). All three genes were downregulated at various time points relative to prechallenge levels in PT21/28-challenged calves: IL-10 was downregulated on days 7 (0.58-fold; 95% CI, 0.40- to 0.84-fold; P = 0.0062) and 21 (0.66-fold; 95% CI, 0.46- to 0.96-fold; P = 0.0295), TGF-β1 was downregulated from day 7 (0.05-fold; 95% CI, 0.02- to 0.11-fold; P < 0.0001) until the end of the trial, and FoxP3 was downregulated on day 7 only (0.35-fold; 95% CI, 0.13- to 0.96-fold; P = 0.0423). In contrast, IL-10 expression remained unchanged in PT32-challenged calves; however, TGF-β1 expression was upregulated from day 14 (2.38-fold; 95% CI, 1.53- to 3.71-fold; P = 0.0005) until the end of the trial, and FoxP3 was upregulated on day 21 only (2.77-fold; 95% CI, 1.15- to 6.67-fold; P = 0.0249) in this group.
T cell subsets at the terminal rectum after colonization.
Lymphocytes were isolated from the terminal rectum postmortem and were analyzed by flow cytometry. The numbers of CD4+, CD8+, and γδ T-cell receptor-positive (γδTCR+) T cells as proportions of CD3+ cells are presented in Fig. 3. The experimental group was a statistically significant variable for the proportions of CD4+ (degrees of freedom [df] = 2; deviance = 7,852.1; P = 0.0009) and CD8+ (df = 2; deviance = 1,209.1; P = 0.0335) T cells but not for the proportions of γδTCR+ T cells (df = 2; deviance = 1,290.1; P = 0.1006). Tukey comparisons confirmed that the proportions of CD4+ cells were significantly higher at 58.8% (95% CI, 48.4 to 68.5%; Z-score [z] = 3.602; P = 0.0009) in the PT21/28-challenged calves and 63.5% (95% CI, 54.8 to 71.4%; z = 4.496; P < 0.0001) in the PT32-challenged calves than in the control group (31.4%; 95% CI, 22.5 to 41.9%), suggesting a CD4+ T-cell infiltrate into the rectal mucosa in challenged calves. The proportion of CD8+ cells was significantly lower, at 15.8% (95% CI, 11.5 to 21.2%; z = −2.892; P = 0.0106), in the PT21/28-challenged calves than in the control group (27.5%; 95% CI, 21.8 to 34.1%).
FIG 3.
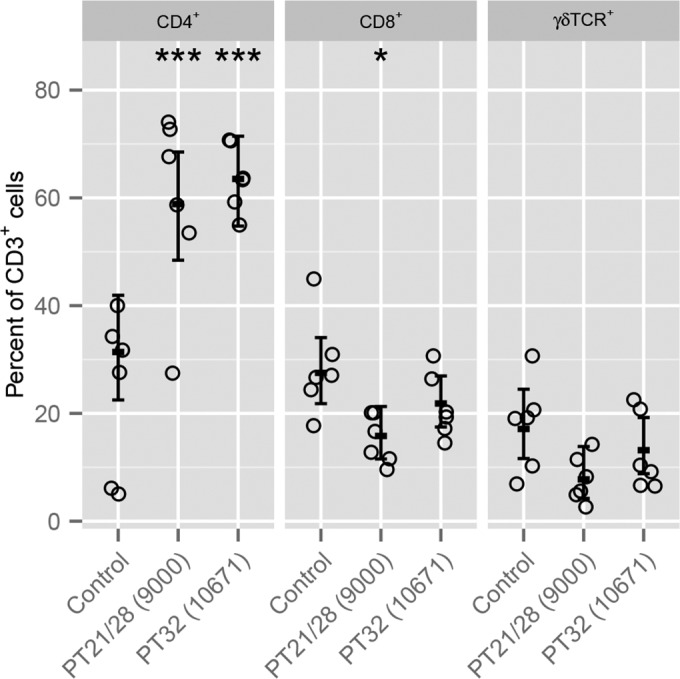
Flow cytometry analysis of CD3+ T-cell subsets at the terminal rectums of EHEC O157:H7-colonized calves postmortem. Circles represent results for individual animals. Error bars represent the predicted means and 95% confidence intervals from generalized linear model analysis for each T-cell subset. The significance of differences found by Tukey comparisons between challenged and age-matched control calves is indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Ex vivo restimulation of regional lymph node cells.
Fresh lymph node cells were extracted postmortem and were restimulated for 5 days with concentrated bacterial secreted protein preparations that include primarily T3SPs. Responses to a wild-type (WT) T3SP preparation consisting predominantly of translocon proteins were compared to responses to a T3SP preparation from an isogenic ΔsepL mutant containing minimal translocon-associated proteins but increased amounts of secreted effector proteins (52). To distinguish between antigen-specific responses and nonspecific proliferation in response to LPS, control wells were spiked with EHEC O111:B4 LPS, so that the final concentration of LPS in control wells was matched to that in the T3SP preparations (WT LPS and ΔsepL LPS). Levels of thymidine incorporation and IFN-γ release are presented in Fig. 4 as the fold change from the level for the respective LPS control (Fig. 4A and B) and as absolute thymidine incorporation (Fig. 4C) and IFN-γ concentration (Fig. 4D).
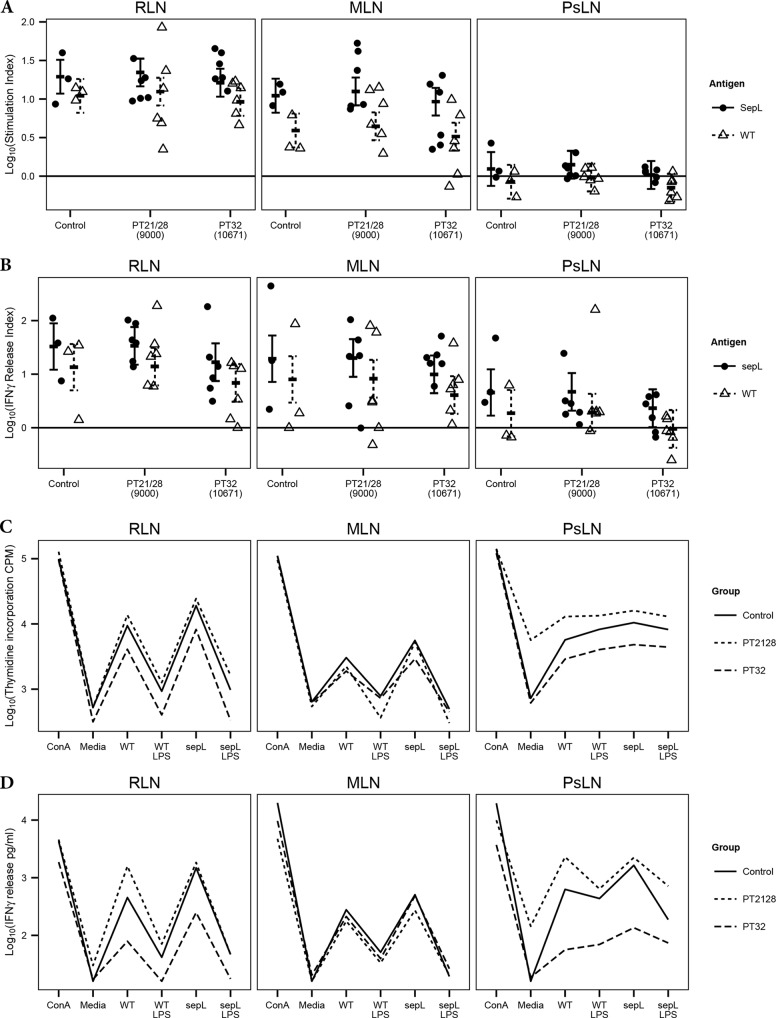
FIG 4.
Proliferation of, and IFN-γ release by, bovine lymph node cells isolated postmortem and restimulated ex vivo for 5 days with either 5 μg/ml ConA, medium only, 5 μg/ml EHEC O157:H7 T3SPs from the sepL deletion strain Zap1143 (ΔsepL) or its isogenic wild-type (WT) strain Zap193, or medium containing a matched concentration of commercial EHEC O111:B4 LPS. ΔsepL T3SP preparations contain mainly secreted effector proteins, while WT T3SP preparations consist mainly of structural translocon components. The use of these two different T3SP preparations allows the relative contributions of structural and effector proteins to antigen recall responses to be compared. Cells were isolated from rectal lymph nodes (RLN), mesenteric lymph nodes (MLN), and prescapular lymph nodes (PsLN). (A and B) Proliferation (A) and IFN-γ release (B) are expressed as indices, representing fold changes in the response to T3SPs from levels with the relevant LPS controls. Circles represent individual animals. (C and D) Interaction plots illustrate mean thymidine incorporation (C) and IFN-γ release (D) by lymph node cells from each experimental group in response to different stimuli. Error bars represent predicted means and 95% confidence intervals from linear-mixed-model analysis.
Antigen-specific responses.
The stimulation index (fold change in thymidine incorporation from that for the relevant LPS control) provides an assessment of antigen-specific proliferation in response to the two T3SP preparations. Model selection showed that the interaction between the ex vivo stimulant and the lymph node was significant (P = 0.0078), demonstrating that the antigen-specific responses were different within each lymph node. Tukey comparisons (see Table S6 in the supplemental material) show that the stimulation index for the ΔsepL T3SPs was 1.77-fold higher (standard error [SE] = 1.16; t ratio [TR] = 3.8; P = 0.0058) than that for the WT T3SPs at the RLN and 2.83-fold higher (SE = 1.16; TR = 6.9; P < 0.0001) at the MLN, but there was no difference at the control PsLN (SE = 1.16; TR = 2.5; P = 0.1391). The stimulation index for the ΔsepL T3SP was 15.7-fold higher (SE = 1.28; TR = 11.0; P < 0.0001) at the RLN and 8.9-fold higher (SE = 1.28; TR = 8.7; P < 0.0001) at the MLN than at the PsLN, but there was no difference between the RLN and MLN (SE = 1.28; TR = 2.3; P = 0.2342). The stimulation index values for the WT T3SP, however, differed for the three different types of lymph nodes, with 2.8-fold (SE = 1.28; TR = 4.1; P = 0.0024) and 13.0-fold (SE = 1.28; TR = 10.2; P < 0.0001) higher values at the RLN than at the MLN and PsLN, respectively, and a 4.6-fold higher value (SE = 1.28; TR = 6.1; P < 0.0001) at the MLN than at the PsLN.
Given the TH type 1 skew identified by RT-qPCR in response to colonization, IFN-γ release was used as a second readout for lymphocyte activation and was again expressed as the fold increase over the level for the relevant LPS control (IFN-γ release index). None of the mixed-model interactions were significant, indicating that the difference between the T3SP preparations was the same across all the lymph nodes. Tukey comparisons (see Table S7 in the supplemental material) demonstrate that the IFN-γ release index was 2.4-fold higher (SE = 1.21; z = 4.5; P < 0.0001) for the ΔsepL T3SP than for the WT T3SP. The index was also 7.2-fold higher (SE = 1.58; z = 4.3; P < 0.0001) at the RLN and 4.3-fold higher (SE = 1.58; z = 3.2; P = 0.00578) at the MLN than at the PsLN. There was no difference between the RLN and MLN (SE = 1.58; z = 1.1; P = 0.61659).
Overall responsiveness of regional lymph node cells.
The interaction plots (Fig. 4C and D) indicate that both total thymidine incorporation and IFN-γ release (i.e., values not normalized to the LPS control) in response to the T3SP preparations and LPS were lower at the RLN and PsLN in PT32-challenged calves than in PT21/28-challenged calves. This observation was confirmed by formal statistical testing. For thymidine incorporation, the interactions between the experimental group and the lymph node (P = 0.0079) and between the ex vivo stimulant and the lymph node (P < 0.0001) were significant. Tukey comparisons between experimental groups within each type of lymph node (see Table S8 in the supplemental material) show lower levels of thymidine incorporation—2.64-fold lower (SE = 1.44; TR = 2.6; P = 0.0355) at the RLN and 3.4-fold lower (SE = 1.44; TR = 3.3; P = 0.0071) at the PsLN—across all ex vivo stimulant groups (ConA, medium only, WT T3SP, WT LPS, ΔsepL T3SP, and ΔsepL LPS) for PT32-challenged calves than for PT21/28-challenged calves.
For IFN-γ release, the interactions between the experimental group and the ex vivo stimulant (P = 0.0061), between the experimental group and the lymph node (P = 0.0043), and between the ex vivo stimulant and the lymph node (P < 0.0001) were all significant. Tukey comparisons of IFN-γ release between experimental groups within each node–ex vivo stimulant combination (see Table S9 in the supplemental material) showed that at the RLN, only the antigen-specific responses varied by experimental group. The WT T3SPs resulted in a 12.2-fold (SE = 2.06; TR = 3.5; P = 0.0035) reduction in IFN-γ release in PT32-challenged calves from that in PT21/28-challenged calves. As with thymidine incorporation, there were no significant differences at the MLN, while at the PsLN, IFN-γ release was 7.1-fold (SE = 2.06; TR = 2.7; P = 0.0251), 28.7-fold (SE = 2.06; TR = 4.7; P = 0.0001), 10.6-fold (SE = 2.06; TR = 3.3; P = 0.0061), 13.4-fold (SE = 2.06; TR = 3.6; P = 0.0025), and 8.8-fold (SE = 2.06; TR = 3.0; P = 0.0119) higher in PT21/28-challenged calves than in PT32-challenged calves in response to medium only, WT T3SPs, WT LPS, ΔsepL T3SPs, and ΔsepL LPS, respectively.
Immunophenotyping of proliferating rectal lymph node cells in response to WT T3SP.
Resuscitated RLN cells were incubated for 5 days with ConA, medium only, treated WT T3SPs, or EHEC O111:B4 LPS. The proportions within each subset of cells expressing the proliferation marker Ki67 are presented in Fig. 5. Data were analyzed using generalized linear models, and analysis-of-deviance tables are shown in Table S10 in the supplemental material. Retaining the interaction between the experimental group and the ex vivo stimulant in the models caused comparisons between experimental groups to be based on unweighted averages across the stimulants.
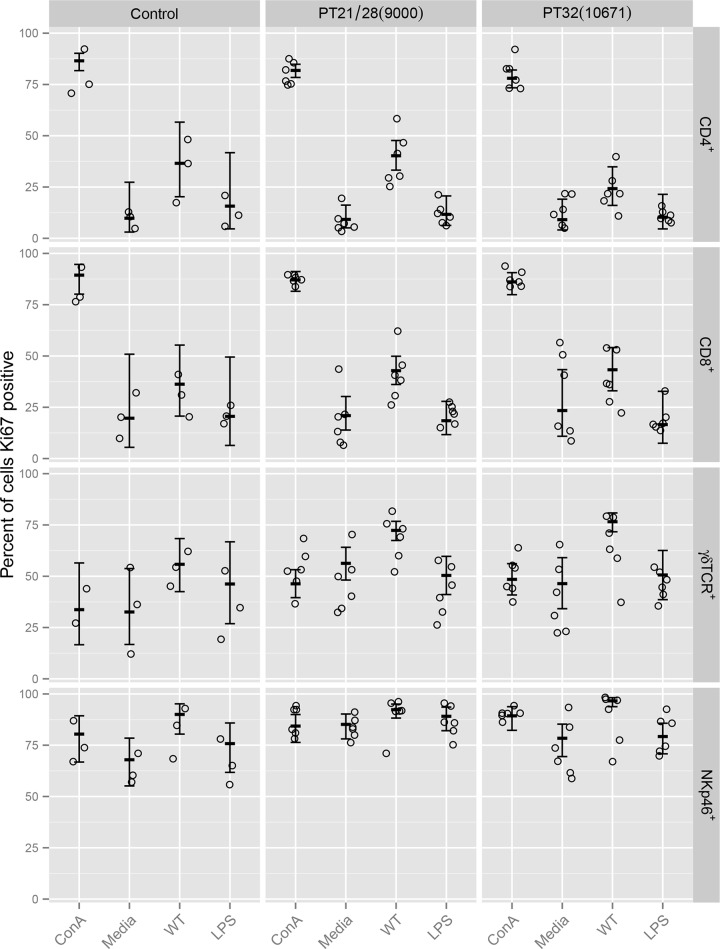
FIG 5.
Flow cytometry analysis of proliferation marker (Ki67) expression by different lymphocyte subsets isolated postmortem from the rectal lymph nodes of control and EHEC O157:H7-challenged calves. Frozen cells were resuscitated and were restimulated ex vivo for 5 days with 5 μg/ml ConA, medium only, 5 μg/ml treated EHEC O157:H7 T3SPs (WT), or medium containing a matched concentration of commercial EHEC O111:B4 LPS (LPS). Circles represent individual animals. Error bars represent predicted means and 95% confidence intervals from generalized-linear-model analysis for each lymphocyte subset.
Tukey comparisons for each model are shown in Table S11 in the supplemental material. For all four cell types, there was no significant change in the proportion of cells staining positive for Ki67 in response to LPS compared to the medium control, while the proportions of CD4+ and CD8+ cells, but not γδTCR+ or NKp46+ cells, staining Ki67 positive increased in response to ConA. These results confirm that none of the cell types proliferated in response to LPS, while only the CD4+ and CD8+ T cells proliferated in response to ConA.
In PT21/28- but not PT32-challenged animals, a significantly higher proportion of CD4+ T cells stained Ki67 positive in response to WT T3SPs than in response to the LPS control. Conversely, in PT32- but not PT21/28-challenged animals, there was a significantly higher proportion of NKp46+ cells staining Ki67 positive in response to WT T3SPs than in response to the LPS control. In both groups of challenged calves, a higher proportion of CD8+ and γδTCR+ T cells stained Ki67 positive in response to WT T3SPs than in response to the LPS control.
DISCUSSION
The objective of this study was to characterize the temporal pattern of expression of a panel of transcripts associated with different T-helper cell polarizations at the terminal rectum, which is the principal site of EHEC O157:H7 colonization in calves (65). These results were then used to inform experiments in which regional lymph node cells were extracted and restimulated ex vivo with T3SP antigens.
Since the results of this study will be used to guide future vaccine development, the selection of clinically relevant strains was an important consideration. In the United Kingdom, PT21/28 strains are associated with higher levels of shedding in cattle than PT32 strains (66) and represent the predominant isolates from human HUS cases (67). In addition, PT21/28 strains are more likely to possess both Stx2a and Stx2c genes, whereas bovine PT32 strains are more likely to possess the Stx2c genes only (68). While cattle do not suffer from the same severe endothelial sequelae of Stx exposure as humans, there is a significant body of evidence that Stx can affect bovine leukocyte function (40, 69–74). Given the important epidemiological differences between these phage types and their propensity to possess different Stx types, we wanted to compare components of the cellular immune response in cattle following colonization with representative PT21/28 and PT32 strains. The strains used in this study were selected because they had common characteristics of these phage types in cattle. PT21/28 strain 9000 contains both Stx2a- and Stx2c-encoding genes and was isolated from a bovine fecal pat with relatively high EHEC O157:H7 levels, while PT32 strain 10671 contains Stx2c only and was isolated from a bovine fecal pat with relatively low EHEC O157:H7 levels.
Both these strains successfully colonized all the orally challenged cattle and were efficiently transmitted to the two sentinel calves. This study does not have the power to make comparisons between the shedding profiles of orally challenged and those of sentinel calves or between the transmissibilities of the two strains; however, these results will be used to inform power calculations leading to larger trials that will specifically address these questions.
While EHEC O157:H7 can be isolated from multiple sites within the bovine gastrointestinal tract, efficient colonization with A/E lesion formation has been demonstrated at the terminal rectum (65). This offers the opportunity to use minimally invasive rectal biopsy specimens to study the interaction between the bacteria and the mucosal surface at different stages during colonization. It was our hypothesis that by studying the expression of genes associated with different T-helper cell polarizations, it would be possible to determine whether a T-helper response is induced at the site of infection, as well as the type of this response within the TH1, TH2, TH17, and TReg paradigm. Since mathematical modeling of bacterial shedding dynamics suggests that bacterial replication on the mucosal surface declines precipitously 5 to 7 days following oral challenge (75), biopsy specimens were taken at 7 days after challenge and weekly until the end of the study. Previous work using a bovine-specific cDNA microarray to analyze rectal biopsy specimens taken up to 7 days postchallenge demonstrated differential expression of 49 genes in response to colonization across a range of pathways (76). None of the genes analyzed in our study were identified as differentially regulated by the previous microarray approach. Because a subset of 1,676 genes was present on the microarray and the list of these genes is no longer available (Robert Collier, University of Arizona, personal communication), we are unable to determine whether the T-helper cell-associated transcripts we have analyzed were included in that previous study.
Rectal mucosal biopsy specimens were taken at different positions at each sampling time point to avoid resampling the same sites. The stability of TNF-α expression in the control animals indicates that the experimental protocol did not result in a general inflammatory response within the mucosa. In addition, the lack of induction of TNF-α in the challenge groups would also indicate that this cytokine is not an important component of the bovine response to colonization at the terminal rectum. This is significant, because TNF-α is upregulated during C. rodentium colonization in mice (77), while pretreatment of bovine colonic explants with TNF-α has been shown to upregulate mucin secretion and reduce EHEC O157:H7 colonization (78). The observation that TNF-α is not induced at the terminal rectum would imply that this pathway is not active in vivo and is likely to contribute to the difference between the pathogenicity of C. rodentium in mice, which results in weight loss, moderate mortality, and colonic hyperplasia (79, 80), and that of EHEC O157:H7 in cattle, where the infection is asymptomatic and demonstrates only a mild neutrophilic infiltrate, suggesting that while immunogenic, colonization is not a major inflammatory event (42).
The induction of both IFN-γ and T-bet during colonization is indicative of a TH type 1 polarized response at the terminal rectum, which is further supported by the inability to detect either IL-4 or IL-13 transcripts. This is in keeping with studies of C. rodentium in mice, which has been shown to induce IFN-γ but not IL-4 (77), while experiments using knockout mice have confirmed the importance of IFN-γ in bacterial clearance (81).
The lack of a detectable TH17 response in this study, in conjunction with the significant downregulation of RORC, suggests that there are additional important differences between the bovine response to EHEC O157:H7 and the response to C. rodentium in mice, where IL-22, IL17A, and IL-23 p19 are upregulated during colonization (82), while IL-23 (p19−/−) (83), IL-22 (82), IL17A, and IL17F (84) knockout mice demonstrate increased pathology and deficiencies in controlling bacterial replication.
The strain differences in this study were striking. In PT32-challenged calves, as might be anticipated, both IFN-γ and T-bet demonstrated maximal induction around the time of peak shedding at 7 days postchallenge. In PT21/28-challenged calves, however, maximal induction for both genes occurred at 21 days postchallenge, with moderate induction of T-bet but not IFN-γ at 7 days. This strain-specific uncoupling of the IFN-γ and T-bet responses in early infection and the delayed induction of a convincing TH type 1 response were surprising. IFN-γ can be produced by a wide variety of leukocytes, and T-bet can be expressed by both innate lymphoid cells and lymphocytes (85). It is therefore not possible to infer whether the differences in the dynamics of gene expression observed in the PT21/28-challenged animals are due to innate versus adaptive mechanisms or to a change in the kinetics of the response. While research to determine the complete genome sequences of the two challenge strains is ongoing, it is already known that the PT21/28 strain contains both Stx2a and Stx2c lysogens, whereas the PT32 strain contains Stx2c only. The toxin type and level of production may, in part, account for differences, but we also know that there are complex regulatory interactions between prophages and bacterial host gene expression, including colonization factors (86) and core genome regulatory elements (87). Additional in vitro and in vivo work with isogenic Stx and Stx-encoding bateriophage knockout strains is planned to explore these concepts in more detail.
TGF-β1 and FoxP3 were both upregulated in the control animals, suggesting a TReg skew. It is unclear whether this is incidental. Interestingly, the induction of both genes is dampened or absent in the challenged calves compared to unchallenged controls, with only moderate induction in PT32-challenged animals and downregulation in the PT21/28-challenged animals. This provides further evidence supporting a convincing TH1 skew and the reciprocal downregulation of TReg responses.
The induction of GATA3 at 21 days postinfection suggests that there may be a role for TH type 2 immunity later during the course of infection. B-cell-deficient mice have demonstrated impaired clearance of C. rodentium and reduced bacterial shedding following systemic administration of immune sera (88), despite a strong TH type 1 response during early stages of infection, suggesting that while the initial response is biased toward cellular immunity, the role of humoral immunity and/or the presence of a healing response (89) may be important as colonization progresses.
In an attempt to understand the impact of colonization on the T-cell subsets at the terminal rectum, lymphocytes were extracted postmortem and were double stained for CD3 and either CD4, CD8, or γδTCR. The increase in the proportion of CD4+ T cells is indicative of a T-helper cell infiltrate and is strongly suggestive of an adaptive component in the response to colonization. To investigate this further, regional lymph node cells were restimulated ex vivo with two different T3SP preparations. Since we had identified a strong TH type 1 response, IFN-γ release was used alongside thymidine incorporation as a readout of lymphocyte activation. Previous studies using heat-killed bacteria to restimulate ruminant lymphocytes have not controlled for the presence of LPS (40), and our results show that the responsiveness of cells to LPS differs by lymph node, with PsLN cells being more responsive than intestinal lymph node cells. Once the effects of LPS had been controlled for, antigen-specific proliferation and IFN-γ release in response to both T3SP preparations were significantly higher in cells isolated from the two intestinal lymph nodes than in those from the control PsLN, confirming enhanced recognition of EHEC O157:H7 antigens at sites anatomically relevant to colonization. There was no difference between the experimental groups, suggesting that the experimental animals had previously been exposed to some of the T3SP antigens and/or epitopes used in this study, although the animals were screened for EHEC O157 colonization at multiple times prior to the challenges. An alternative but less likely explanation for the response is that these protein preparations could contain effectors that drive lymphocyte proliferation, as is the case with the staphylococcal superantigen TSST (toxic shock syndrome toxin) (90), although one would predict that proliferation would be equivalent throughout all lymph nodes if this were the case. Of interest was the significantly stronger response from all the lymph nodes to the ΔsepL T3SP. Since LPS was controlled for and these preparations were matched by protein concentration, it is reasonable to hypothesize that the secreted effectors are potentially more immunogenic than the structural components. While vaccines containing native secreted and recombinant T3SP structural proteins have been shown to reduce shedding in cattle (13, 28, 29), our current results lend further weight to the consideration of secreted effector proteins in EHEC O157:H7 cattle vaccines and work to define the most immunogenic of these in terms of a TH1 response.
The observation that cells from the RLN and PsLN of PT32-challenged animals are less responsive than those from control and PT21/28-challenged animals is suggestive of systemic immunomodulation by this strain. Paradoxically, however, this effect was not observed at the MLN. This effect was missed when we were examining the stimulation and IFN-γ release indices due to the reduction in responsiveness also affecting the medium and LPS controls. Currently, it is unclear what may be responsible for this immunomodulation. While Stx activity is a prime candidate, it is interesting that it was the Stx2c-positive PT32 strain, and not the PT21/28 strain, which contains both Stx2a and Stx2c, that demonstrated this repression, suggesting that factors other than Stx may be involved.
In an effort to understand which cell types were involved in responding to T3SPs at the RLN and consequently which cell types might be attenuated by PT32 challenge, the proliferation of the different lymphocyte subsets was characterized by flow cytometry. Heat-inactivated protein preparations were used to exclude the possibility of effector bioactivity driving proliferative responses. CD8+ and γδTCR+ T cells from colonized animals demonstrated statistically significant proliferation in response to the T3SPs, independently of LPS and any potential biological activity of the preparations. It is interesting that these antigens are able to drive both γδTCR+ and CD8+ T-cell proliferation, suggesting a role for γδ T cells in the response to colonization and the selection of CD8 T-cell clones that are responsive to major histocompatibility complex (MHC) class I-presented EHEC O157:H7 peptides. While CD8+ T-cell-depleted mice do not demonstrate a deficit in C. rodentium clearance (91), the injection of bacterial effector proteins into host cells via the T3SS means that bacterially derived peptides may be presented on MHC class I molecules. Mathematical modeling of proteasome degradation, TAP processing, and MHC class I loading in humans suggests that secreted EHEC O157:H7 effectors have an altered amino acid composition like that of viral proteins, enabling them to reduce the efficiency by which they are presented by MHC class I molecules (92). The lack of a protective CD8+ T-cell response in mice to an A/E lesion-forming organism does not necessarily mean that these cells do not play an important role in host immunity to EHEC O157:H7 in cattle.
The CD4+ T-cell proliferative response to WT T3SPs seen in PT21/28-challenged calves was absent in PT32-challenged calves, while the NK cell proliferative response seen in PT32-challenged calves was absent in PT21/28-challenged calves. This may indicate that the PT32 strain is effectively cleared via innate mechanisms in contrast to the PT21/28 strain, which may be able to evade components of the innate response, resulting in a stronger adaptive response. Alternatively, the PT32 strain may specifically target adaptive immunity by suppressing CD4+ T-cell function. NK cells have recently been demonstrated to play an important role in C. rodentium clearance (93). If innate immunity is an important component of the immune response in cattle, it may explain why rechallenge experiments demonstrate only a partial and short-term protective effect following bacterial clearance (33). Further work is required to better understand the interaction of these strains with bovine leukocytes.
This study is the first to definitively demonstrate a TH type 1 immune response to EHEC O157:H7 in cattle. The temporal differences in this response between the two strains studied raise important questions about strain-dependent strategies used by these pathogens to evade host immunity and prolong persistence in the gastrointestinal tract. The roles of innate versus adaptive cellular immunity have not been dissected in this study; however, both are likely to be important, and the organism appears to be directly targeting CD4+ T-cell and/or NK cell function through as yet unidentified mechanisms. The serological responses to EHEC O157:H7 vaccination have been extensively studied; however, future vaccine development will need to measure the impact of vaccination on cellular immunity and to test the hypothesis that while high antibody titers may be important in blocking bacterial adhesion, cellular immune responses are required for bacterial clearance. In addition, the targeting of immunomodulatory virulence factors to improve vaccine efficacy should be considered.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by DEFRA project 020714. The MRI and SRUC receive funding from the Scottish Government. D.L.G. and S.P.M. are supported by a BBSRC Institute strategic program at the Roslin Institute. A.C. is supported by a Bioniche Life Sciences, Inc., Ph.D. studentship and BBSRC funding. N.I.A. is supported by the Malaysian Ministry of Education.
We thank Dragan Rogan for his contribution to initial discussions on the role of cellular immunity in the control of E. coli O157 colonization in cattle. We thank the Moredun Research Institute Bioservices Division for excellent care of experimental animals.
Footnotes
Published ahead of print 29 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02462-14.
REFERENCES
- 1.Chase-Topping M, Gally D, Low C, Matthews L, Woolhouse M. 2008. Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157. Nat. Rev. Microbiol. 6:904–912. 10.1038/nrmicro2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olsen SJ, Miller G, Breuer T, Kennedy M, Higgins C, Walford J, McKee G, Fox K, Bibb W, Mead P. 2002. A waterborne outbreak of Escherichia coli O157: H7 infections and hemolytic uremic syndrome: implications for rural water systems. Emerg. Infect. Dis. 8:370–375. 10.3201/eid0804.000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Health Canada. 2000. Waterborne outbreak of gastroenteritis associated with a contaminated municipal water supply, Walkerton, Ontario, May–June 2000. Can. Commun. Dis. Rep. 26:170–173. [PubMed] [Google Scholar]
- 4.Armstrong GL, Hollingsworth J, Morris JG., Jr 1996. Emerging foodborne pathogens: Escherichia coli O157:H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol. Rev. 18:29–51. 10.1093/oxfordjournals.epirev.a017914. [DOI] [PubMed] [Google Scholar]
- 5.Ostroff SM, Griffin PM, Tauxe RV, Shipman LD, Greene KD, Wells JG, Lewis JH, Blake PA, Kobayashi JM. 1990. A statewide outbreak of Escherichia coli O157:H7 infections in Washington State. Am. J. Epidemiol. 132:239–247. [DOI] [PubMed] [Google Scholar]
- 6.Ihekweazu C, Carroll K, Adak B, Smith G, Pritchard GC, Gillespie IA, Verlander NQ, Harvey-Vince L, Reacher M, Edeghere O, Sultan B, Cooper R, Morgan G, Kinross PTN, Boxall NS, Iversen A, Bickler G. 2012. Large outbreak of verocytotoxin-producing Escherichia coli O157 infection in visitors to a petting farm in South East England, 2009. Epidemiol. Infect. 140:1400–1413. 10.1017/S0950268811002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Locking ME, O'Brien SJ, Reilly WJ, Wright EM, Campbell DM, Coia JE, Browning LM, Ramsay CN. 2001. Risk factors for sporadic cases of Escherichia coli O157 infection: the importance of contact with animal excreta. Epidemiol. Infect. 127:215–220. 10.1017/S0950268801006045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean-Nystrom EA, Bosworth BT, Cray WC, Jr, Moon HW. 1997. Pathogenicity of Escherichia coli O157:H7 in the intestines of neonatal calves. Infect. Immun. 65:1842–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karmali MA. 2004. Infection by Shiga toxin-producing Escherichia coli: an overview. Mol. Biotechnol. 26:117–122. 10.1385/MB:26:2:117. [DOI] [PubMed] [Google Scholar]
- 10.Rozema EA, Stephens TP, Bach SJ, Okine EK, Johnson RP, Stanford K, McAllister TA. 2009. Oral and rectal administration of bacteriophages for control of Escherichia coli O157:H7 in feedlot cattle. J. Food Prot. 72:241–250. 10.1016/j.jprot.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Callaway TR, Carr MA, Edrington TS, Anderson RC, Nisbet DJ. 2009. Diet, Escherichia coli O157:H7, and cattle: a review after 10 years. Curr. Issues Mol. Biol. 11:67–79. [PubMed] [Google Scholar]
- 12.Sargeant JM, Amezcua MR, Rajic A, Waddell L. 2007. Pre-harvest interventions to reduce the shedding of E. coli O157 in the faeces of weaned domestic ruminants: a systematic review. Zoonoses Public Health 54:260–277. 10.1111/j.1863-2378.2007.01059.x. [DOI] [PubMed] [Google Scholar]
- 13.Potter AA, Klashinsky S, Li Y, Frey E, Townsend H, Rogan D, Erickson G, Hinkley S, Klopfenstein T, Moxley RA, Smith DR, Finlay BB. 2004. Decreased shedding of Escherichia coli O157:H7 by cattle following vaccination with type III secreted proteins. Vaccine 22:362–369. 10.1016/j.vaccine.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Fox JT, Thomson DU, Drouillard JS, Thornton AB, Burkhardt DT, Emery DA, Nagaraja TG. 2009. Efficacy of Escherichia coli O157:H7 siderophore receptor/porin proteins-based vaccine in feedlot cattle naturally shedding E. coli O157. Foodborne Pathog. Dis. 6:893–899. 10.1089/fpd.2009.0336. [DOI] [PubMed] [Google Scholar]
- 15.Stevens MP, van Diemen PM, Dziva F, Jones PW, Wallis TS. 2002. Options for the control of enterohaemorrhagic Escherichia coli in ruminants. Microbiology 148:3767–3778. [DOI] [PubMed] [Google Scholar]
- 16.Varela NP, Dick P, Wilson J. 2013. Assessing the existing information on the efficacy of bovine vaccination against Escherichia coli O157:H7: a systematic review and meta-analysis. Zoonoses Public Health 60:253–268. 10.1111/j.1863-2378.2012.01523.x. [DOI] [PubMed] [Google Scholar]
- 17.Snedeker KG, Campbell M, Sargeant JM. 2012. A systematic review of vaccinations to reduce the shedding of Escherichia coli O157 in the faeces of domestic ruminants. Zoonoses Public Health 59:126–138. 10.1111/j.1863-2378.2011.01426.x. [DOI] [PubMed] [Google Scholar]
- 18.McNeilly TN, Naylor SW, Mahajan A, Mitchell MC, McAteer S, Deane D, Smith DG, Low JC, Gally DL, Huntley JF. 2008. Escherichia coli O157:H7 colonization in cattle following systemic and mucosal immunization with purified H7 flagellin. Infect. Immun. 76:2594–2602. 10.1128/IAI.01452-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNeilly TN, Mitchell MC, Rosser T, McAteer S, Low JC, Smith DG, Huntley JF, Mahajan A, Gally DL. 2010. Immunization of cattle with a combination of purified intimin-531, EspA and Tir significantly reduces shedding of Escherichia coli O157:H7 following oral challenge. Vaccine 28:1422–1428. 10.1016/j.vaccine.2009.10.076. [DOI] [PubMed] [Google Scholar]
- 20.Wong AR, Pearson JS, Bright MD, Munera D, Robinson KS, Lee SF, Frankel G, Hartland EL. 2011. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol. Microbiol. 80:1420–1438. 10.1111/j.1365-2958.2011.07661.x. [DOI] [PubMed] [Google Scholar]
- 21.Jarvis KG, Kaper JB. 1996. Secretion of extracellular proteins by enterohemorrhagic Escherichia coli via a putative type III secretion system. Infect. Immun. 64:4826–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tree JJ, Wolfson EB, Wang D, Roe AJ, Gally DL. 2009. Controlling injection: regulation of type III secretion in enterohaemorrhagic Escherichia coli. Trends Microbiol. 17:361–370. 10.1016/j.tim.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 23.DeVinney R, Stein M, Reinscheid D, Abe A, Ruschkowski S, Finlay BB. 1999. Enterohemorrhagic Escherichia coli O157: H7 produces Tir, which is translocated to the host cell membrane but is not tyrosine phosphorylated. Infect. Immun. 67:2389–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dean-Nystrom EA, Gansheroff LJ, Mills M, Moon HW, O'Brien AD. 2002. Vaccination of pregnant dams with intiminO157 protects suckling piglets from Escherichia coli O157:H7 infection. Infect. Immun. 70:2414–2418. 10.1128/IAI.70.5.2414-2418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vilte DA, Larzabal M, Cataldi AA, Mercado EC. 2008. Bovine colostrum contains immunoglobulin G antibodies against intimin, EspA, and EspB and inhibits hemolytic activity mediated by the type three secretion system of attaching and effacing Escherichia coli. Clin. Vaccine Immunol. 15:1208–1213. 10.1128/CVI.00027-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rabinovitz BC, Gerhardt E, Tironi Farinati C, Abdala A, Galarza R, Vilte DA, Ibarra C, Cataldi A, Mercado EC. 2012. Vaccination of pregnant cows with EspA, EspB, γ-intimin, and Shiga toxin 2 proteins from Escherichia coli O157:H7 induces high levels of specific colostral antibodies that are transferred to newborn calves. J. Dairy Sci. 95:3318–3326. 10.3168/jds.2011-5093. [DOI] [PubMed] [Google Scholar]
- 27.Mahajan A, Currie CG, Mackie S, Tree J, McAteer S, McKendrick I, McNeilly TN, Roe A, La Ragione RM, Woodward MJ, Gally DL, Smith DG. 2009. An investigation of the expression and adhesin function of H7 flagella in the interaction of Escherichia coli O157: H7 with bovine intestinal epithelium. Cell. Microbiol. 11:121–137. 10.1111/j.1462-5822.2008.01244.x. [DOI] [PubMed] [Google Scholar]
- 28.Allen KJ, Rogan D, Finlay BB, Potter AA, Asper DJ. 2011. Vaccination with type III secreted proteins leads to decreased shedding in calves after experimental infection with Escherichia coli O157. Can. J. Vet. Res. 75:98–105. [PMC free article] [PubMed] [Google Scholar]
- 29.Van Donkersgoed J, Hancock D, Rogan D, Potter AA. 2005. Escherichia coli O157:H7 vaccine field trial in 9 feedlots in Alberta and Saskatchewan. Can. Vet. J. 46:724–728. [PMC free article] [PubMed] [Google Scholar]
- 30.Thomson DU, Loneragan GH, Thornton AB, Lechtenberg KF, Emery DA, Burkhardt DT, Nagaraja TG. 2009. Use of a siderophore receptor and porin proteins-based vaccine to control the burden of Escherichia coli O157:H7 in feedlot cattle. Foodborne Pathog. Dis. 6:871–877. 10.1089/fpd.2009.0290. [DOI] [PubMed] [Google Scholar]
- 31.Thornton AB, Thomson DU, Loneragan GH, Fox JT, Burkhardt DT, Emery DA, Nagaraja TG. 2009. Effects of a siderophore receptor and porin proteins-based vaccination on fecal shedding of Escherichia coli O157:H7 in experimentally inoculated cattle. J. Food Prot. 72:866–869. [DOI] [PubMed] [Google Scholar]
- 32.Bretschneider G, Berberov EM, Moxley RA. 2007. Isotype-specific antibody responses against Escherichia coli O157:H7 locus of enterocyte effacement proteins in adult beef cattle following experimental infection. Vet. Immunol. Immunopathol. 118:229–238. 10.1016/j.vetimm.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Naylor SW, Flockhart A, Nart P, Smith DG, Huntley J, Gally DL, Low JC. 2007. Shedding of Escherichia coli O157:H7 in calves is reduced by prior colonization with the homologous strain. Appl. Environ. Microbiol. 73:3765–3767. 10.1128/AEM.02670-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson RP, Cray WC, Jr, Johnson ST. 1996. Serum antibody responses of cattle following experimental infection with Escherichia coli O157:H7. Infect. Immun. 64:1879–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pirro F, Wieler LH, Failing K, Bauerfeind R, Baljer G. 1995. Neutralizing antibodies against Shiga-like toxins from Escherichia coli in colostra and sera of cattle. Vet. Microbiol. 43:131–141. 10.1016/0378-1135(94)00089-F. [DOI] [PubMed] [Google Scholar]
- 36.Asper DJ, Karmali MA, Townsend H, Rogan D, Potter AA. 2011. Serological response of Shiga toxin-producing Escherichia coli type III secreted proteins in sera from vaccinated rabbits, naturally infected cattle, and humans. Clin. Vaccine Immunol. 18:1052–1057. 10.1128/CVI.00068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanderson MW, Besser TE, Gay JM, Gay CC, Hancock DD. 1999. Fecal Escherichia coli O157:H7 shedding patterns of orally inoculated calves. Vet. Microbiol. 69:199–205. 10.1016/S0378-1135(99)00106-6. [DOI] [PubMed] [Google Scholar]
- 38.Carlson BA, Nightingale KK, Mason GL, Ruby JR, Choat WT, Loneragan GH, Smith GC, Sofos JN, Belk KE. 2009. Escherichia coli O157:H7 strains that persist in feedlot cattle are genetically related and demonstrate an enhanced ability to adhere to intestinal epithelial cells. Appl. Environ. Microbiol. 75:5927–5937. 10.1128/AEM.00972-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cray WC, Jr, Moon HW. 1995. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffman MA, Menge C, Casey TA, Laegreid W, Bosworth BT, Dean-Nystrom EA. 2006. Bovine immune response to Shiga-toxigenic Escherichia coli O157:H7. Clin. Vaccine Immunol. 13:1322–1327. 10.1128/CVI.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vande Walle K, De Zutter L, Cox E. 2011. Oral infection with a Shiga toxin-negative Escherichia coli O157:H7 strain elicits humoral and cellular responses but does not protect sheep from colonisation with the homologous strain. Vet. Microbiol. 148:317–322. 10.1016/j.vetmic.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 42.Nart P, Naylor SW, Huntley JF, McKendrick IJ, Gally DL, Low JC. 2008. Responses of cattle to gastrointestinal colonization by Escherichia coli O157:H7. Infect. Immun. 76:5366–5372. 10.1128/IAI.01223-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheng H, Wang J, Lim JY, Davitt C, Minnich SA, Hovde CJ. 2011. Internalization of Escherichia coli O157:H7 by bovine rectal epithelial cells. Front. Microbiol. 2:32. 10.3389/fmicb.2011.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frankel G, Phillips AD, Novakova M, Field H, Candy DC, Schauer DB, Douce G, Dougan G. 1996. Intimin from enteropathogenic Escherichia coli restores murine virulence to a Citrobacter rodentium eaeA mutant: induction of an immunoglobulin A response to intimin and EspB. Infect. Immun. 64:5315–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wadolkowski EA, Burris JA, O'Brien AD. 1990. Mouse model for colonization and disease caused by enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 58:2438–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conlan JW, Perry MB. 1998. Susceptibility of three strains of conventional adult mice to intestinal colonization by an isolate of Escherichia coli O157:H7. Can. J. Microbiol. 44:800–805. 10.1139/w98-056. [DOI] [PubMed] [Google Scholar]
- 47.Isogai E, Isogai H, Kimura K, Hayashi S, Kubota T, Fujii N, Takeshi K. 1998. Role of tumor necrosis factor alpha in gnotobiotic mice infected with an Escherichia coli O157:H7 strain. Infect. Immun. 66:197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagano K, Taguchi K, Hara T, Yokoyama S, Kawada K, Mori H. 2003. Adhesion and colonization of enterohemorrhagic Escherichia coli O157:H7 in cecum of mice. Microbiol. Immunol. 47:125–132. 10.1111/j.1348-0421.2003.tb02795.x. [DOI] [PubMed] [Google Scholar]
- 49.Mundy R, Girard F, FitzGerald AJ, Frankel G. 2006. Comparison of colonization dynamics and pathology of mice infected with enteropathogenic Escherichia coli, enterohaemorrhagic E. coli and Citrobacter rodentium. FEMS Microbiol. Lett. 265:126–132. 10.1111/j.1574-6968.2006.00481.x. [DOI] [PubMed] [Google Scholar]
- 50.Fernandez-Brando RJ, Miliwebsky E, Mejias MP, Baschkier A, Panek CA, Abrey-Recalde MJ, Cabrera G, Ramos MV, Rivas M, Palermo MS. 2012. Shiga toxin-producing Escherichia coli O157: H7 shows an increased pathogenicity in mice after the passage through the gastrointestinal tract of the same host. J. Med. Microbiol. 61:852–859. 10.1099/jmm.0.041251-0. [DOI] [PubMed] [Google Scholar]
- 51.Mohawk KL, Melton-Celsa AR, Zangari T, Carroll EE, O'Brien AD. 2010. Pathogenesis of Escherichia coli O157:H7 strain 86-24 following oral infection of BALB/c mice with an intact commensal flora. Microb. Pathog. 48:131–142. 10.1016/j.micpath.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D, Roe AJ, McAteer S, Shipston MJ, Gally DL. 2008. Hierarchal type III secretion of translocators and effectors from Escherichia coli O157:H7 requires the carboxy terminus of SepL that binds to Tir. Mol. Microbiol. 69:1499–1512. 10.1111/j.1365-2958.2008.06377.x. [DOI] [PubMed] [Google Scholar]
- 53.Korbie DJ, Mattick JS. 2008. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat. Protoc. 3:1452–1456. 10.1038/nprot.2008.133. [DOI] [PubMed] [Google Scholar]
- 54.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.R Core Team. 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 56.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer, New York, NY. [Google Scholar]
- 57.Sarkar D. 2008. Lattice: multivariate data visualization with R. Springer, New York, NY. [Google Scholar]
- 58.Pinheiro J, Bates D, DebRoy S, Sarkar D. R Core Team. 2014. nlme: linear and nonlinear mixed effects models. R package, version 3.1-117 http://CRAN.R-project.org/package=nlme.
- 59.Bates D, Maechler M, Bolker B, Walker S. 2014. lme4: linear mixed-effects models using Eigen and S4. R package, version 1.1-6 http://CRAN.R-project.org/package=lme4.
- 60.Wickham H. 2011. The split-apply-combine strategy for data analysis. J. Stat. Software 40(1). http://www.jstatsoft.org/v40/i01/. [Google Scholar]
- 61.Wickham H. 2007. 2007. Reshaping data with the reshape package. J. Stat. Software 21(12). http://www.jstatsoft.org/v21/i12/paper. [Google Scholar]
- 62.Hothorn T, Bretz F, Westfall P. 2008. Simultaneous inference in general parametric models. Biom. J. 50:346–363. 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 63.Auguie B. 2012. gridExtra: functions in Grid graphics. R package, version 0.9.1 http://CRAN.R-project.org/package=gridExtra.
- 64.Lenth RV. 2014. lsmeans: least-squares means. R package, version 2.00-5. http://CRAN.R-project.org/package=lsmeans.
- 65.Naylor SW, Low JC, Besser TE, Mahajan A, Gunn GJ, Pearce MC, McKendrick IJ, Smith DG, Gally DL. 2003. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 71:1505–1512. 10.1128/IAI.71.3.1505-1512.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chase-Topping ME, McKendrick IJ, Pearce MC, MacDonald P, Matthews L, Halliday J, Allison L, Fenlon D, Low JC, Gunn G, Woolhouse ME. 2007. Risk factors for the presence of high-level shedders of Escherichia coli O157 on Scottish farms. J. Clin. Microbiol. 45:1594–1603. 10.1128/JCM.01690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lynn RM, O'Brien SJ, Taylor CM, Adak GK, Chart H, Cheasty T, Coia JE, Gillespie IA, Locking ME, Reilly WJ, Smith HR, Waters A, Willshaw GA. 2005. Childhood hemolytic uremic syndrome, United Kingdom and Ireland. Emerg. Infect. Dis. 11:590–596. 10.3201/eid1104.040833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matthews L, Reeve R, Gally DL, Low JC, Woolhouse MEJ, McAteer SP, Locking ME, Chase-Topping ME, Haydon DT, Allison LJ, Hanson MF, Gunn GJ, Reid SWJ. 2013. Predicting the public health benefit of vaccinating cattle against Escherichia coli O157. Proc. Natl. Acad. Sci. U. S. A. 110:16265–16270. 10.1073/pnas.1304978110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moussay E, Stamm I, Taubert A, Baljer G, Menge C. 2006. Escherichia coli Shiga toxin 1 enhances IL-4 transcripts in bovine ileal intraepithelial lymphocytes. Vet. Immunol. Immunopathol. 113:367–382. 10.1016/j.vetimm.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 70.Menge C, Eisenberg T, Stamm I, Baljer G. 2006. Comparison of binding and effects of Escherichia coli Shiga toxin 1 on bovine and ovine granulocytes. Vet. Immunol. Immunopathol. 113:392–403. 10.1016/j.vetimm.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 71.Menge C, Stamm I, Van Diemen PM, Sopp P, Baljer G, Wallis TS, Stevens MP. 2004. Phenotypic and functional characterization of intraepithelial lymphocytes in a bovine ligated intestinal loop model of enterohaemorrhagic Escherichia coli infection. J. Med. Microbiol. 53:573–579. 10.1099/jmm.0.45530-0. [DOI] [PubMed] [Google Scholar]
- 72.Menge C, Blessenohl M, Eisenberg T, Stamm I, Baljer G. 2004. Bovine ileal intraepithelial lymphocytes represent target cells for Shiga toxin 1 from Escherichia coli. Infect. Immun. 72:1896–1905. 10.1128/IAI.72.4.1896-1905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Menge C, Stamm I, Blessenohl M, Wieler LH, Baljer G. 2003. Verotoxin 1 from Escherichia coli affects Gb3/CD77+ bovine lymphocytes independent of interleukin-2, tumor necrosis factor-alpha, and interferon-alpha. Exp. Biol. Med. (Maywood) 228:377–386. [DOI] [PubMed] [Google Scholar]
- 74.Menge C, Wieler LH, Schlapp T, Baljer G. 1999. Shiga toxin 1 from Escherichia coli blocks activation and proliferation of bovine lymphocyte subpopulations in vitro. Infect. Immun. 67:2209–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tildesley MJ, Gally DL, McNeilly TN, Low JC, Mahajan A, Savill NJ. 2012. Insights into mucosal innate responses to Escherichia coli O157: H7 colonization of cattle by mathematical modelling of excretion dynamics. J. R. Soc. Interface 9:518–527. 10.1098/rsif.2011.0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li J, Hovde CJ. 2007. Expression profiles of bovine genes in the rectoanal junction mucosa during colonization with Escherichia coli O157:H7. Appl. Environ. Microbiol. 73:2380–2385. 10.1128/AEM.02262-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Higgins LM, Frankel G, Douce G, Dougan G, MacDonald TT. 1999. Citrobacter rodentium infection in mice elicits a mucosal Th1 cytokine response and lesions similar to those in murine inflammatory bowel disease. Infect. Immun. 67:3031–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xue Y, Zhang H, Wang H, Hu J, Du M, Zhu M-J. 2014. Host inflammatory response inhibits Escherichia coli O157:H7 adhesion to gut epithelium through augmentation of mucin expression. Infect. Immun. 82:1921–1930. 10.1128/IAI.01589-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schauer DB, Zabel BA, Pedraza IF, O'Hara CM, Steigerwalt AG, Brenner DJ. 1995. Genetic and biochemical characterization of Citrobacter rodentium sp. nov. J. Clin. Microbiol. 33:2064–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barthold SW, Coleman GL, Jacoby RO, Livestone EM, Jonas AM. 1978. Transmissible murine colonic hyperplasia. Vet. Pathol. 15:223–236. 10.1177/030098587801500209. [DOI] [PubMed] [Google Scholar]
- 81.Simmons CP, Goncalves NS, Ghaem-Maghami M, Bajaj-Elliott M, Clare S, Neves B, Frankel G, Dougan G, MacDonald TT. 2002. Impaired resistance and enhanced pathology during infection with a noninvasive, attaching-effacing enteric bacterial pathogen, Citrobacter rodentium, in mice lacking IL-12 or IFN-γ. J. Immunol. 168:1804–1812. 10.4049/jimmunol.168.4.1804. [DOI] [PubMed] [Google Scholar]
- 82.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. 2008. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14:282–289. 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 83.Mangan PR, Harrington LE, O'Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. 2006. Transforming growth factor-beta induces development of the TH17 lineage. Nature 441:231–234. 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 84.Ishigame H, Kakuta S, Nagai T, Kadoki M, Nambu A, Komiyama Y, Fujikado N, Tanahashi Y, Akitsu A, Kotaki H, Sudo K, Nakae S, Sasakawa C, Iwakura Y. 2009. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity 30:108–119. 10.1016/j.immuni.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 85.Sciume G, Hirahara K, Takahashi H, Laurence A, Villarino AV, Singleton KL, Spencer SP, Wilhelm C, Poholek AC, Vahedi G, Kanno Y, Belkaid Y, O'Shea JJ. 2012. Distinct requirements for T-bet in gut innate lymphoid cells. J. Exp. Med. 209:2331–2338. 10.1084/jem.20122097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu X, McAteer SP, Tree JJ, Shaw DJ, Wolfson EB, Beatson SA, Roe AJ, Allison LJ, Chase-Topping ME, Mahajan A, Tozzoli R, Woolhouse ME, Morabito S, Gally DL. 2012. Lysogeny with Shiga toxin 2-encoding bacteriophages represses type III secretion in enterohemorrhagic Escherichia coli. PLoS Pathog. 8:e1002672. 10.1371/journal.ppat.1002672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tree JJ, Granneman S, McAteer SP, Tollervey D, Gally DL. 2014. Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol. Cell 55:199–213. 10.1016/j.molcel.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maaser C, Housley MP, Iimura M, Smith JR, Vallance BA, Finlay BB, Schreiber JR, Varki NM, Kagnoff MF, Eckmann L. 2004. Clearance of Citrobacter rodentium requires B cells but not secretory immunoglobulin A (IgA) or IgM antibodies. Infect. Immun. 72:3315–3324. 10.1128/IAI.72.6.3315-3324.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Allen JE, Wynn TA. 2011. Evolution of Th2 immunity: a rapid repair response to tissue destructive pathogens. PLoS Pathog. 7:e1002003. 10.1371/journal.ppat.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Poindexter NJ, Schlievert PM. 1985. Toxic-shock-syndrome toxin 1-induced proliferation of lymphocytes: comparison of the mitogenic response of human, murine, and rabbit lymphocytes. J. Infect. Dis. 151:65–72. 10.1093/infdis/151.1.65. [DOI] [PubMed] [Google Scholar]
- 91.Simmons CP, Clare S, Ghaem-Maghami M, Uren TK, Rankin J, Huett A, Goldin R, Lewis DJ, MacDonald TT, Strugnell RA, Frankel G, Dougan G. 2003. Central role for B lymphocytes and CD4+ T cells in immunity to infection by the attaching and effacing pathogen Citrobacter rodentium. Infect. Immun. 71:5077–5086. 10.1128/IAI.71.9.5077-5086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maman Y, Nir-Paz R, Louzoun Y. 2011. Bacteria modulate the CD8+ T cell epitope repertoire of host cytosol-exposed proteins to manipulate the host immune response. PLoS Comput. Biol. 7:e1002220. 10.1371/journal.pcbi.1002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reid-Yu SA, Small CL, Coombes BK. 2013. CD3− NK1.1+ cells aid in the early induction of a Th1 response to an attaching and effacing enteric pathogen. Eur. J. Immunol. 43:2638–2649. 10.1002/eji.201343435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.