Abstract
Streptococcus pneumoniae is a fermentative microorganism and causes serious diseases in humans, including otitis media, bacteremia, meningitis, and pneumonia. However, the mechanisms enabling pneumococcal survival in the host and causing disease in different tissues are incompletely understood. The available evidence indicates a strong link between the central metabolism and pneumococcal virulence. To further our knowledge on pneumococcal virulence, we investigated the role of lactate dehydrogenase (LDH), which converts pyruvate to lactate and is an essential enzyme for redox balance, in the pneumococcal central metabolism and virulence using an isogenic ldh mutant. Loss of LDH led to a dramatic reduction of the growth rate, pinpointing the key role of this enzyme in fermentative metabolism. The pattern of end products was altered, and lactate production was totally blocked. The fermentation profile was confirmed by in vivo nuclear magnetic resonance (NMR) measurements of glucose metabolism in nongrowing cell suspensions of the ldh mutant. In this strain, a bottleneck in the fermentative steps is evident from the accumulation of pyruvate, revealing LDH as the most efficient enzyme in pyruvate conversion. An increase in ethanol production was also observed, indicating that in the absence of LDH the redox balance is maintained through alcohol dehydrogenase activity. We also found that the absence of LDH renders the pneumococci avirulent after intravenous infection and leads to a significant reduction in virulence in a model of pneumonia that develops after intranasal infection, likely due to a decrease in energy generation and virulence gene expression.
INTRODUCTION
Streptococcus pneumoniae is an important cause of bacteremia, meningitis, otitis media, and septicemia. In addition, the bacterium is also a frequent occupant of the human nasopharynx, where it resides without causing symptoms (1). The increasing trend of antibiotic-resistant strains and the high occurrence of genome variability and plasticity among the pneumococcal strains limit the effective use of existing vaccines (1). Hence, a better understanding of the pneumococcal pathogenesis is required to combat this microbe.
One approach to such a better understanding relies on the recognition that microbial virulence and central metabolism are inherently linked (2–4). Indeed, several studies have implicated central metabolic pathways in pneumococcal virulence (3–5). For example, NADH oxidase, which reoxidizes NADH and reduces molecular oxygen to water, was shown be required for the efficient development of competence for genetic transformation and for virulence and persistence in mice (6). The other well-studied protein that links pneumococcal central metabolism to virulence is the glycolytic enzyme α-enolase, which has been shown to be responsible for binding to plasminogen (7).
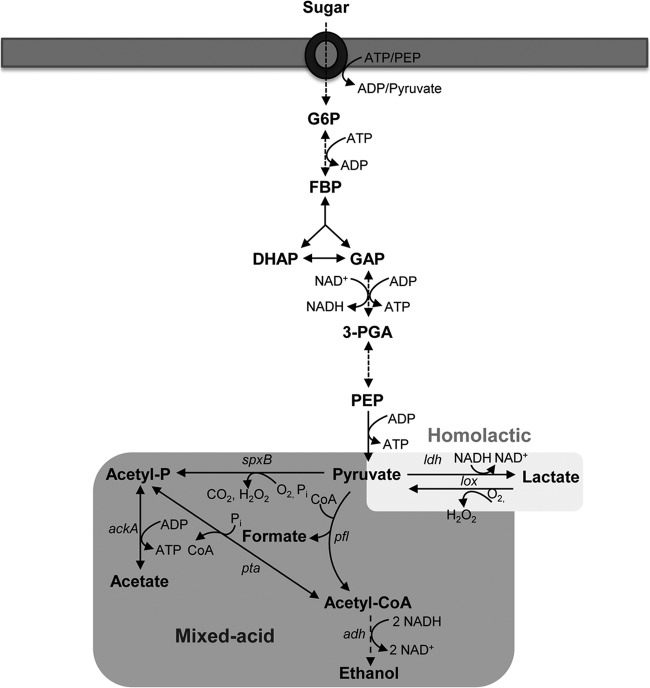
The efficient utilization of available nutrients for generation of metabolic energy is fundamental to pneumococcal survival in vivo (3, 4). The pneumococcus generates its metabolic energy through fermentative breakdown of sugars, as it is devoid of a complete set of genes required for respiration (8). Conversion of carbohydrates by the classical Embden-Meyerhof-Parnas pathway leads to generation of pyruvate and NADH and a net gain of two ATP molecules per mole of glucose (9) (Fig. 1). The further conversion of pyruvate is then achieved through two paths: homolactic and mixed-acid fermentations (Fig. 1) (10). While homolactic fermentation generates mainly lactic acid as the main metabolic end product, under certain conditions, such as aerobiosis, sugar limitation, or in the presence of sugars less preferred than glucose (for example, galactose), products other than lactate are generated through mixed-acid fermentation (10).
FIG 1.
Schematic representation of sugar metabolism in S. pneumoniae. Sugars are internalized and phosphorylated, and the resulting sugar-phosphates are converted through the Embden-Meyerhof-Parnas pathway to pyruvate. This metabolite is further converted to fermentation end products. Abbreviations: ldh, lactate dehydrogenase; pfl, pyruvate formate lyase; spxB, pyruvate oxidase; adh, alcohol dehydrogenase; ack, acetate kinase; pta, phosphotransacetylase; G6P, glucose 6-phosphate; FBP, fructose 1,6-bisphosphate; DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde 3-phosphate; 3-PGA, 3-phosphoglycerate; PEP, phosphoenolpyruvate.
We showed previously that mixed-acid fermentation in pneumococcus is mediated through the activity of pyruvate formate lyase (PFL), which converts pyruvate to acetyl coenzyme A (acetyl-CoA) and formate under microaerobic and anaerobic conditions (4). Further metabolism of acetyl-CoA by phosphotransacetylase/acetate kinase or by aldehyde and alcohol dehydrogenases generates acetate or ethanol, respectively. While acetate formation produces an additional molecule of ATP, ethanol production from acetyl-CoA regenerates two molecules of NAD+ (Fig. 1) (10).
A key enzyme in homolactic fermentation is lactate dehydrogenase (LDH), which converts pyruvate to lactate and regenerates NAD+ for the continuation of glycolysis (10, 11). LDH activity is widely spread in all the domains of life, and in certain bacteria LDH activity is encoded by multiple ldh genes (11, 12). Besides its role in fermentative metabolism, accumulating evidence suggests its involvement in microbial virulence and pathogenesis. In Enterococcus faecalis, it has been shown that resistance to different environmental stresses is determined by the ability of the microbe to maintain redox balance via LDH (13). In the absence of LDH, the bacterium was shown to be susceptible to various stressors, including solvents, oxidative stress, hypochlorite, detergent, acidity, and salt. Furthermore, LDH deficiency led to attenuation of the ability of the microbe to colonize host organs in a murine model of systemic infection (13). In Staphylococcus aureus, nitric oxide-induced LDH activity was shown to be important for metabolic adaptation to nitrosative stress by maintaining redox homeostasis and to be essential for virulence (14). In Bacillus cereus, ldhA mutation reduced fermentative growth and downregulated enterotoxin gene expression under both anaerobiosis and aerobiosis (15).
The pneumococcal genome has a single ldh copy (8). Previous attempts to mutate the pneumococcal ldh, either through introduction of a stop codon or through insertional mutation by single-crossover integration of the nonreplicative plasmid pAM239 carrying an ldh fragment, were unsuccessful unless an ectopic copy of the gene was provided (6). Hence, it was concluded that ldh was essential. Here, we report for the first time the construction of an ldh mutant of S. pneumoniae D39. This strain provided the basis to investigate the contribution of LDH to pneumococcal carbon metabolism and virulence by growth studies, end product analysis, in vivo nuclear magnetic resonance (NMR), and virulence tests using a mouse model of pneumococcal infection. We found that LDH is important for pneumococcal growth, metabolism, and virulence.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Streptococcus pneumoniae serotype 2 strain D39 and its unencapsulated derivative strain R6 were used in this study. Routinely, pneumococci were grown in brain heart infusion (BHI) broth or on blood agar plates supplemented with 5% (vol/vol) defibrinated horse blood (Oxoid, Basingstoke, United Kingdom) under microaerophilic conditions at 37°C. Where appropriate, spectinomycin (100 μg/ml) or kanamycin (500 μg/ml) was added to the culture medium.
Bacteria were also grown in chemically defined medium (CDM) containing disodium β-glycerophosphate (21 g/liter), sodium pyruvate (0.01%, wt/vol) and choline (0.001%, wt/vol), at 37°C without pH control (initial pH 6.5) (4, 16). Glucose (1%, wt/vol) or galactose (1%, wt/vol) was used as the carbon source. S. pneumoniae was grown under anaerobic conditions in rubber-stoppered bottles (200 ml). Cultures were inoculated with overnight cultures (incubation time, at least 9 h) grown in CDM with the desired sugar, to bring the initial optical density at 600 nm (OD600) to approximately 0.05. Growth was monitored by measuring the OD600. Growth rates (μ) were calculated through linear regressions of the plots of ln(OD600) versus time during the exponential growth phase.
Construction of mutants.
We mutated ldh in D39 and R6 strains. To construct mutants, an approximately 2-kb genomic region containing the target gene was amplified with the appropriate primers (Table 1). For transposition reactions, approximately 200 ng of PCR fragments was mixed with 200 to 400 ng of donor mariner plasmid pR412, which contains a spectinomycin resistance cassette, and incubated in the presence of purified Himar1 transposase, as described previously (4). Gaps in transposition products were repaired with T4 DNA polymerase (New England BioLabs, Hitchin, United Kingdom) and subsequently by Escherichia coli ligase (New England BioLabs). Repaired transposition products were transformed into S. pneumoniae D39 using a synthetic competence-inducing peptide. Transformants were selected for spectinomycin resistance, and insertion of the resistance cassette was confirmed by PCR using transposon-specific primer MP127 or MP128 with appropriate chromosomal primers. In addition, mutations were also confirmed by sequencing (17). A representative strain designated SPD1078M was selected for further study.
TABLE 1.
Oligonucleotide primers used in this study
| Primer namea | Primer sequence (5′–3′) | Target gene in D39 (reference) |
|---|---|---|
| SPD0049RTF | GAGACGCGAGCCATTAAGG | comA |
| SPD0049RTR | GGGATCTGGATCGGCAATATGA | |
| SPD0065RTF | GGACCTCTTTGTAACAGGAA | bgaC |
| SPD0065RTR | CATCTGCCAATTCCTTAGGA | |
| SPD0265RTF | CCTTGCAAAAGAAGTAGGCG | adh |
| SPD0265RTR | AGCACGAATGGAGTCAACAG | |
| SPD0287RTF | GGATTTTTATTAAACCATTAT | hyl |
| SPD0287RTR | TGCTTCCGTCCTCCGTCTGAC | |
| SPD0323RTF | TCACTTTATCGTTATGGACTGGC | cps2H |
| SPD0323RTR | CTCAAGTAACTATTGACACCACC | |
| SPD0420RTF | TGGTGTTTACGCACGTCTTG | pflB |
| SPD0420RTR | CATCAACCCCGTAAAGGTCAC | |
| SPD0420F | GACAGTTGTTGAAGCACAAG | pflB |
| SPD0420R | CTCAATGCATCCAAGGCATC | |
| SPD0636RTF | CGTCACCTTCACATGACACC | spxB |
| SPD0636RTR | CATGTTGAATGCTCCGTCAC | |
| SPD0709RTF | TCGTGTGGCTGCCAAGCGTG | gyrB |
| SPD0709RTR | GGCTGATCCACCAGCTGAGTC | |
| SPD1126RTF | TCAACTATACTTTGCCAGAAGC | adh |
| SPD1126RTR | CCATCACCACCACAACATTC | |
| SP1078F | CCAAAATTCCCATGACCATC | ldh |
| SP1078R | AAAGAGGCCTTTCCAAATCC | |
| SPD1078RTF | TGCAGCTCAATACTCTGAC | ldh |
| SPD1078RTR | GTTAGCAGCAACAAGGAAG | |
| LDHNcoI | CGCCATGGTGCCTCATTTCACAATTAG | ldh |
| LDHBamHI | ACGGGATCCTTAGTTTTTAGAAGCTTCTTGG | |
| SPD1504RTF | CGTCGTCATCACTGTAGG | nanA |
| SPD1504RTR | CAGACTATCGCGTTGTTG | |
| SPD1726RTF | AACGACAGTCGCCTCTATCC | ply |
| SPD1726RTF | AGAAAGCTATCGCTACTTGCC | |
| SPD1834RTF | TGATCGTTACTGACCATGCC | adh |
| SPD1834RTR | GCACGCATAATCTCAGTACC | |
| SPD1865RTF | AGTTATTCGCAAGCCAACAG | adh |
| SPD1865RTR | CAACAATCCCAATCCCTTCG | |
| MP 127 | CCGGGGACTTATCAGCCAACC | pR412-specific target (36) |
| MP 128 | TACTAGCGACGCCATCTATGTG | pR412-specific target (36) |
| MALF | GCTTGAAAAGGAGTATACTT | NAb |
| PCEPR | AGGAGACATTCCTTCCGTATC | NA |
Primers with an F or R tag were used for amplification of gene targets for mutational work, while those with RTF or RTR tags were utilized for gene expression analysis.
NA, not applicable; the recognition sites for MalF and pCEPR are in pCEP (19).
To construct the LDH mutant in the strain R6 background, the mutated region was amplified from SPD1078M with SPD1078F and SPD1078R, and the amplicons were transformed into S. pneumoniae strain R6 as described above. The mutant in the R6 background was designated SPR1078M.
Analysis of suppressor mutations in SPD1078M.
To determine whether the mutation of ldh introduced suppressor mutations, SPD1078M was transformed with genomic DNA from strain R304, which possesses a point mutation in rpsL41, conferring resistance to streptomycin, as described previously (18). The resulting strain was designated SPD1078MSm, and its genomic DNA was subsequently transformed into wild-type strain D39. The transformants were selected separately in streptomycin and spectinomycin. The transformation frequency for the ldh mutation was compared to that of the reference marker, which was within the same DNA and therefore is directly comparable. Normally, if a cassette is accepted readily or, at worst, at 20% of the efficiency of the reference marker, it is considered that the gene under study is not essential. However, if the ratio is roughly 10 times less than this (i.e., 2% of reference), then cotransfer of 1, if it was less than 100×, cotransfer of 2, and so on, suppressor mutations are assumed (18).
Complementation of SPD1078M.
To eliminate the possibility of polar effects due to ldh mutation, SPD1078M was complemented with an intact copy of the gene using pCEP, which is a nonreplicative plasmid that allows controlled gene expression following ectopic integration into the chromosome (19). The entire ldh sequence, as well as its 194-bp upstream sequence, was amplified with LDHNcoI and LDHBamHI primers (Table 1), which were modified to include NcoI and BamHI sites, respectively. The amplicons were digested with NcoI and BamHI and were ligated into similarly digested pCEP. An aliquot of ligation mixture was transferred into E. coli Stellar competent cells (Clonetech, Saint-Germain-en-Laye, France) as described by the manufacturer. The transformants were selected on kanamycin-containing Luria-Bertani medium agar plates. Colony PCR with MalF and pCEPR primers, whose recognition sites are localized immediately up- and downstream of the cloning site, respectively, and which amplify an ∼263-bp product from the empty vector, produced an ∼1,250-bp product (data not shown), indicating that successful cloning had taken place (the additional 987 bp represents the ldh sequence). The recombinant plasmid was purified using a commercial kit (Qiagen, Crawley, United Kingdom), and a portion was transformed into strain SPD1078M as described above. The transformants were selected on blood agar plates supplemented with spectinomycin and kanamycin. The complemented strain was designated SPD1078comp.
Quantification of extracellular metabolites.
Samples (2 ml) of cultures growing in CDM containing 1% (wt/vol) glucose were collected at different points during growth, centrifuged (6,700 × g, 10 min, 4°C), and filtered (Millex-GN 0.22-μm filters), and the supernatant solutions were stored at −20°C until analysis by high-performance liquid chromatography (HPLC) or 1H-NMR. Glucose and metabolic products were quantified by HPLC using an HPX-87H anion-exchange column (Bio-Rad Laboratories, Hemel Hempstead, United Kingdom) and a refractive index detector (Shodex RI-101; Showa Denko, Munich, Germany) at 60°C with 5 mM H2SO4 as the elution fluid and a flow rate of 0.5 ml/min. Chromeleon software was used for data treatment as described before (20). Given that galactose and pyruvate have similar retention times under the HPLC conditions used, these two compounds were quantified by 1H-NMR using a Bruker AMX300 spectrometer (Bruker BioSpin GmbH, Karlsruhe, Germany) and a 5-mm inverse detection probe head. Formic acid (sodium salt) was added to the samples and was used as an internal concentration standard.
Batch cultivations in bioreactors with pH control.
The pneumococcal strains were grown in CDM in a 2-liter bioreactor (Biostat B plus; Sartorius, Madrid, Spain), with the pH controlled at 6.5 and under anaerobic (argon atmosphere) conditions. For anaerobic growth, the medium was degassed by flushing argon during the hour preceding inoculation. Glucose was used as a carbon source at a final concentration of about 55 mM. The pH was kept at 6.5 by the automatic addition of 10 M NaOH, and the temperature was set to 37°C. Culture homogenization was achieved by maintaining an agitation speed of 50 rpm.
In vivo 13C-NMR experiments.
S. pneumoniae strains R6 and SPR1078M were grown under anaerobic conditions with pH control as described above, harvested in the late exponential phase of growth (OD600, 1.9), centrifuged (5,750 × g, 7 min, 4°C), washed twice with 50 mM potassium phosphate buffer with 1% (wt/vol) choline (pH 6.5) (5,750 × g, 5 min, 4°C), and suspended to a protein concentration of 9 to 15 mg/ml in 50 mM potassium phosphate with 1% (wt/vol) choline, pH 6.5 (16). In vivo NMR experiments were performed using an online system described earlier (21). Glucose (20 mM), specifically labeled with 13C on carbon one, was added to the cell suspension at time point zero. The time course of glucose consumption, amount of product formation, and changes in the pools of intracellular metabolites were monitored in vivo, which refers to analysis in living cells. When the substrate was exhausted and no changes in the resonances of intracellular metabolites were observed, an NMR sample extract was prepared as described previously (16). Carbon-13 spectra were acquired at 125.77 MHz on a Bruker DRX500 spectrometer (Bruker BioSpin GmbH). All in vivo experiments were run using a quadruple nucleus probe head at 37°C, as described before (16). Lactate and acetate were quantified in the NMR sample extract by 1H-NMR in a Bruker AMX300 (Bruker BioSpin GmbH). The concentration of other metabolites was determined in fully relaxed 13C spectra of the NMR sample extracts as previously described (21). Due to the fast pulsing conditions used for acquiring in vivo 13C spectra, correction factors for resonances due to the C-1 and C-6 of fructose 1,6-bisphosphate (FBP) (0.39 ± 0.04), lactate (0.53 ± 0.01), and acetate (0.93 ± 0.09) were determined to convert peak intensities into concentrations. The concentration of labeled lactate (in R6 cell extracts) or acetate (in SPR1078M cell extracts) determined by 1H-NMR was used as a standard to calculate the concentration of the other metabolites in the sample as described previously (21). Intracellular metabolite concentrations were calculated using a value of 3.0 μl/mg of protein, determined for the intracellular volume of S. pneumoniae (9). Although individual experiments are illustrated in each figure, each type of in vivo NMR experiment was repeated at least twice, and the results were highly reproducible. The values reported are averages from at least two experiments.
Measurement of lactate dehydrogenase activity.
Lactate dehydrogenase activity was determined in cells grown without pH control under semiaerobic conditions. Cells were grown until late exponential phase, centrifuged (5,750 × g, 5 min, 4°C), washed twice with potassium phosphate buffer 10 mM (pH 7.0), and suspended in the same buffer. Crude extracts were prepared by passage through a French press (twice at 120 MPa; SLM Aminco Instruments, Golden Valley, MN, USA) and centrifugation for 30 min (16,100 × g, 4°C) to remove cell debris. LDH activity was assayed at 37°C in a Beckman Coulter DU 800 spectrophotometer as described previously (22). One unit of enzyme activity is defined as the conversion of 1 μmol of substrate per minute under the experimental conditions used. The protein concentration was determined with the bicinchoninic acid (BCA) protein assay kit (Pierce, Rockford, IL, USA). Specific activity was expressed as units (μmol/min) per milligram of protein (U/mg of protein).
RNA extraction from bacterial cells and purification.
The extraction of RNA was done by the TRIzol method using mid-log-phase cultures, as described previously (23, 24). Before use, the RNA was treated with amplification grade DNase I (Qiagen) and subsequently purified with an RNeasy minikit (Qiagen).
Extraction of pneumococcal RNA from infected tissues.
To prepare the bacterial inoculum for infection, an overnight culture of strain D39 was used to inoculate fresh BHI serum broth (80% [vol/vol] BHI and 20% [vol/vol] bovine serum). When the OD500 reached 1.6, bacteria were stored at −80°C in small aliquots until needed. Outbred 9-week-old female MF1 mice (Harlan Olac, Bicester, United Kingdom) were intranasally infected with 50 μl phosphate-buffered saline (PBS) containing 1 × 106 cells of strain D39 (4, 25). As soon as the mice became severely lethargic, blood was collected by cardiac puncture under terminal anesthesia. After the mice were killed by cervical dislocation, the lungs and nasopharynx were removed and homogenized on ice in 10 ml of sterile PBS using a tissue homogenizer. To separate pneumococci from host cells, lung homogenates and clotted blood samples were centrifuged at 900 × g for 6 min at 4°C. Supernatants were subsequently centrifuged at 15,500 × g for 2 min at 4°C, and the bacterial pellet was stored at −80°C until further processing. Prior to isolation of bacteria, 20 μl of homogenate was removed, serially diluted in PBS, and plated onto blood agar in order to enumerate pneumococci and to exclude the presence of a contaminating microflora. RNA extraction and purification were done as described in “RNA Extraction from Bacterial Cells and Purification.”
Quantitative RT-PCR.
First-strand cDNA synthesis was performed on approximately 1 μg of DNase-treated total RNA, immediately after isolation, using random hexamers and 200 U of SuperScript II reverse transcriptase (Invitrogen, Paisley, United Kingdom) at 42°C for 55 min. cDNA (15 ng) was amplified in a 20-μl reaction volume that contained 1× SYBR green PCR master mix (Bioline, London, United Kingdom) and 3 pmol of each primer (indicated with “RTF” or “RTR” tags in Table 1). The transcription level of specific genes was normalized to gyrB transcription, which was amplified in parallel with SPD0709F and SPD0709R primers. The results were analyzed by the comparative threshold cycle (CT) method (2).
In vivo virulence studies.
Female MFI outbred mice (Harlan Olac) were used for virulence testing. A standardized inoculum of pneumococcal strains was prepared as described above. To determine the virulence of pneumococcal strains, mice were lightly anesthetized with 2.5% (vol/vol) fluothane over oxygen (1.5 to 2 liters/min). A 50-μl sample of PBS containing approximately 1 × 106 S. pneumoniae cells was given into the nostrils gradually. The inoculum dose was confirmed by viable counting on blood agar plates. Mice were monitored for disease signs (hunching, piloerection, or lethargy) for 7 days (26), and those that reached the severely lethargic stage were considered to have reached the endpoint of the assay and were killed humanely. The time to this point was defined as “survival time.” Mice that were alive 7 days after infection were deemed to have survived the infection. To determine the development of bacteremia in each mouse, approximately 20 μl of venous blood was obtained from intranasally infected mice at predetermined time points after infection. Viable counts in blood were determined by serial dilution in sterile PBS and plating onto blood agar plates supplemented with 5% (vol/vol) defibrinated horse blood with the appropriate antibiotic. Median survival times were analyzed using the Mann-Whitney U test.
Mouse experiments at the University of Leicester were performed under appropriate project (no. 60/4327) and personal (no. 80/10279) licenses according to the United Kingdom Home Office guidelines and local ethical approval. Where appropriate, the procedures were carried out under anesthetization with isoflurane. Mice were kept in individually ventilated cages in a controlled environment and were frequently monitored after infection to comply with the requirement of the project license.
For intravenous infections, approximately 5 × 105 CFU/ml S. pneumoniae in 100 μl PBS, pH 7.0, was administered via the dorsal tail vein. The inoculum was confirmed by plating on blood agar as above. Data were analyzed by analysis of variance followed by the Bonferroni posttest. Statistical significance was set at P values of <0.05.
RESULTS
Mutation of ldh and analysis of compensatory mutations.
In order to study the role of pneumococcal LDH, an isogenic ldh mutant was constructed using in vitro mariner mutagenesis. Seven transformants were obtained (Fig. 2). The approximate positions of insertions in these transformants were mapped by PCR using MP127 and SP1078F primers. The sizes of the amplicons indicated potentially 7 insertions in the coding region, representing 6 independent mutational events. Two transformants, designated SPD1078-3 and SPD1078-4 (Fig. 2A, lanes 3 and 4, respectively), were analyzed further by sequencing. The result showed that the minicassette had been inserted in regions 152 and 506 nucleotides from the 5′-end of ldh in SPD1078-3 and SPD-1078-4, respectively. SPD1078-3 was redesignated SPD1078M for further study.
FIG 2.
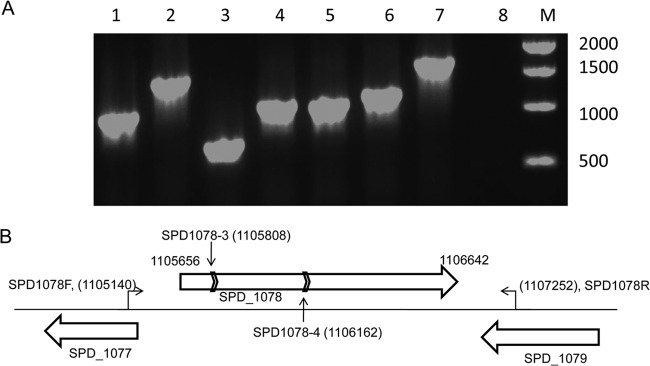
Analysis of transformants (A) and schematic representation of the genomic region containing putative ldh (SPD_1078) (B). (A) The presence of the spectinomycin cassette is analyzed in transformants 1 to 7 using the primers SPD1078F and MP127, which has recognition sites in the spectinomycin resistance cassette. D39 is used as a negative control (lane 8). Lane M shows 1-kb DNA markers (New England Biolab), and the size of each band is given in base pairs. (B) The chromosome is represented with a thin solid line, and genes are shown with a block arrow. The genomic locations of SPD1078F and SPD1078R primers, which amplified the genetic locus for mutation, are shown, and the position of the first nucleotide recognized by each primer and the coordinates of ldh have been indicated. The vertical arrows show the positions of mutations in transformants SPD1078-3 and SPD1078-4, and the chevrons represent the orientation of the spectinomycin cassette. The diagram is not drawn to scale.
As ldh had been designated an essential gene previously (6), the possibility that suppressor mutations would allow the acceptance of ldh mutation in SPD1078M was investigated. However, no evidence for suppressor mutations could be obtained (data not shown).
Impact of ldh mutation on glucose and galactose metabolism.
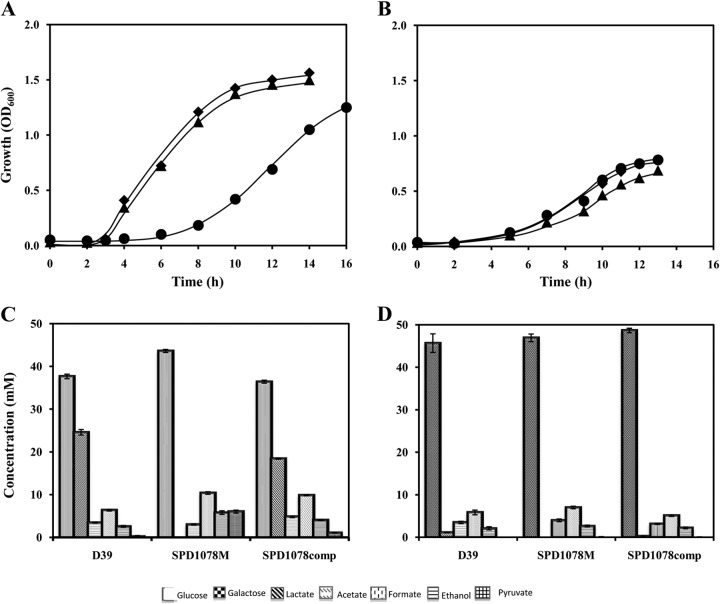
To evaluate the impact of ldh mutation on pneumococcal metabolism, the growth profiles of D39 and SPD1078M were studied in CDM supplemented with 1% glucose or galactose (Fig. 3). The ldh mutant showed a 2.5-fold-lower growth rate than its parent when glucose was used as the sole carbon source. In contrast, with galactose, loss of LDH had no effect on growth rate (Table 2). These results clearly indicate a more prominent role of LDH in the metabolism of glucose than of galactose.
FIG 3.
Growth profiles and end products from glucose and galactose metabolism. D39 and its LDH-deficient mutant were grown in CDM supplemented with 1% glucose (A) or galactose (B) under anaerobic or aerobic conditions at 37°C and initial pH 6.5. Concentrations of substrate in the medium and fermentation end products after growth arrest of strains D39 (◆), SPD1078M (●), and SPD1078comp (▲) on glucose (C) or galactose (D).
TABLE 2.
Growth rate and maximal OD600 of D39, SPD1078M, and SPD1078comp in glucose and galactosea
| Strain | Glucose |
Galactose |
||
|---|---|---|---|---|
| μ (h−1) | Max OD600 | μ (h−1) | Max OD600 | |
| D39 | 0.76 ± 0.02 | 1.53 ± 0.04 | 0.34 ± 0.02 | 0.77 ± 0.05 |
| SPD1078M | 0.31 ± 0.03 | 1.21 ± 0.03 | 0.36 ± 0.03 | 0.78 ± 0.03 |
| SPD1078comp | 0.79 ± 0.03 | 1.46 ± 0.02 | 0.31 ± 0.01 | 0.71 ± 0.02 |
Strains were grown in CDM supplemented with 1% (wt/vol) glucose or galactose at 37°C and initial pH 6.5. Values are averages from 3 independent experiments ± standard deviations. Max, maximal; μ, growth rate.
Analysis of fermentation products revealed that lactate production is completely abolished in strain SPD1078M (Fig. 3). This result is in agreement with the presence of a single copy of LDH-encoding genes in the genome sequence of strains R6 and D39 (27, 28). Deletion of LDH in strain D39 caused a shift to a mixed-acid fermentation profile on glucose with production of pyruvate, acetate, and ethanol. In SPD1078M, the accumulation of pyruvate indicates a metabolic constraint in the alternative fermentative steps downstream of the pyruvate node. Thus, LDH is seemingly the most efficient enzyme in pyruvate reduction and concomitant NAD+ regeneration in S. pneumoniae. The amount of ethanol produced doubles in the LDH-negative strain, which indicates a role for alcohol dehydrogenase in the reduction of NADH resulting from glycolysis.
The metabolism of galactose gave rise to a mixed-acid fermentation profile. As expected, lactate was not produced by the mutant strain (Fig. 3). Complementation of SPD1078M with an intact copy of ldh restored the growth and fermentation profiles on both glucose and galactose (Fig. 3 and Table 2).
Analysis of the effect of ldh mutation on glucose metabolism by in vivo 13C-NMR.
Growth and fermentation profiles showed that glucose catabolism in S. pneumoniae D39 is drastically affected by inactivation of LDH activity. In order to assess the impact of this inactivation on glycolysis and fermentation dynamics, we resorted to in vivo 13C-NMR to follow glucose metabolism in real time. To this end, we transferred the ldh mutation to strain R6, a nonvirulent derivative of D39, in order to prepare dense cell suspensions (16). Of importance, strains D39 (4.0 ± 0.3 U/mg of protein) and R6 (4.3 ± 0.2 U/mg of protein) showed identical levels of LDH activity, while no activity was detected in the LDH-deficient R6 and D39 strains.
The kinetics of [1-13C]glucose (20 mM) consumption, end product formation, and intracellular metabolite pools in R6 and SPR1078M (LDH-derivative mutant in R6) are shown in Fig. 4. Mutation of ldh in R6 caused a 7-fold reduction in the glucose consumption rate (from 0.32 ± 0.01 to 0.05 ± 0.00 μmol/min/mg of protein). The parental strain, R6, displayed a homolactic fermentation profile, with about 89% of the glucose being converted to lactate. In addition to lactate (35.8 ± 0.6 mM), small amounts of acetate (2.4 ± 0.7 mM) were also produced. This result is in agreement with the fermentation profile observed for strain D39 during growth on glucose (Fig. 3). Loss of ldh resulted in a remarkable end product profile change: lactate formation was abolished, and instead glucose was converted to acetate, ethanol, and pyruvate. At the time of glucose exhaustion, maximal concentrations of 9.9 ± 1.0 mM for ethanol, 10.9 ± 2.4 mM for pyruvate, and 3.1 ± 0.0 mM for acetate were observed. Unexpectedly, upon glucose depletion, pyruvate decreased to almost undetectable levels, while acetate increased to concentrations around 20 mM. The accumulation of acetate was characterized by biphasic kinetics: acetate production was slow (0.006 ± 0.002 μmol/min/mg of protein) while glucose was available, and the rate increased about 7-fold at the onset of glucose depletion. This profile indicates inhibition of acetate formation during glucose catabolism, most likely as a result of redox pressure. Under these conditions, NAD+ regeneration at the level of alcohol dehydrogenase seems to be favored over ATP production by acetate kinase (Fig. 1). The glycolytic intermediate FBP accumulated during glucose consumption at concentrations of up to 32 ± 3 mM in R6 and 47 ± 2 mM in SPR1078M and decreased steeply to undetectable levels after substrate depletion. Interestingly, the 3-phosphoglycerate (3-PGA) profiles of the two strains were remarkably different. In strain R6, 3-PGA started to accumulate after glucose exhaustion and remained constant afterwards (5 ± 1 mM), while in strain SPR1078M, 3-PGA became detectable at the onset of glucose exhaustion, peaked (10 mM ± 3 mM) after FBP depletion, and decreased thereafter.
FIG 4.
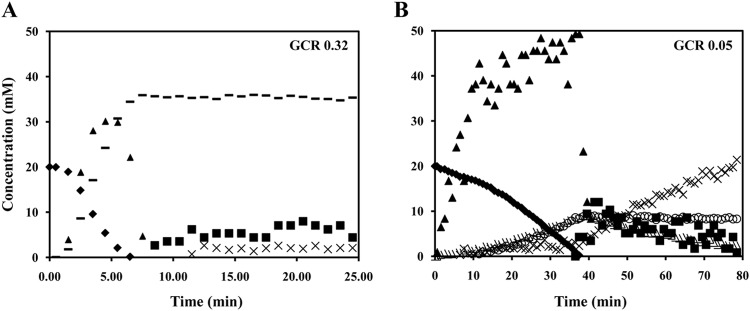
Real-time measurements of metabolites during the metabolism of glucose in S. pneumoniae. Kinetics of [1-13C]glucose (20 mM) consumption, end product formation, and intracellular metabolites in strain R6, an avirulent derivative of D39 (A) and its ldh mutant SPR1078M (B) at 37°C under anaerobic conditions and pH 6.5. Symbols: ◆, glucose; ▲, FBP; ◼, 3-PGA; –, lactate; ×, acetate; ○, ethanol; △, pyruvate. GCR, glucose consumption rate (μmol min−1 mg−1 of protein).
Virulence studies.
LDH's contribution to pneumococcal virulence was determined in a mouse model of pneumococcal infection. The result showed that the median survival time of mice infected intranasally with SPD1078M (99.5 h ± 52.3 h, n = 10) was significantly longer than that of the wild-type-infected group (37 h ± 41 h, n = 10) (P < 0.01). The addition of an intact copy of ldh in SPD1078M reconstituted virulence, because the median survival time of mice infected with the complemented strain SPD1078comp (48 h ± 39.5, n = 10) was similar to that of the wild-type-infected cohort (P > 0.05).
In the wild-type-infected cohort, bacteremia developed 12 h postinfection (log10 1.53 ± 0.52 CFU/ml, n = 10), while in the mutant-infected cohort no bacterial growth in the blood could be detected at this time point (Fig. 5). Moreover, the bacterial load in the blood for the mutant strain was significantly less at 24 and 36 h postinfection [log10 (1.94 ± 0.57) CFU/ml and log10 (2.86 ± 0.80) CFU/ml (n = 10) for 24 and 36 h postinfection, respectively] than for the wild type [log10 (4.3 ± 0.77) CFU/ml and log10 (5.55 ± 0.65) CFU/ml (n = 10) for 24 and 36 h postinfection, respectively] (P < 0.05 for 24 h and P < 0.01 for 36 h). The colony counts in blood for SPD1078comp were similar to those of the wild type at all time points [log10 (0.93 ± 0.5), log10 (3.92 ± 0.63), and log10 (5.10 ± 0.74) (n = 10) for 12, 24, and 36 h postinfection, respectively] (P > 0.05).
FIG 5.
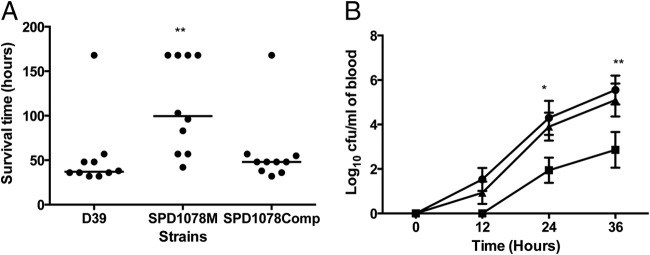
Impaired virulence of pneumococcal strains defective in galactose catabolic pathways following intranasal infection. (A) Survival time of mice after infection with approximately 1 × 106 CFU pneumococci. Symbols show the times at which mice became severely lethargic. The horizontal bars mark the median times to the severely lethargic state. (B) Growth of bacteria in the blood is shown for D39 (●), SPD1078M (●), and SPD1078comp (◆). Each point is the mean of data from 10 mice. Error bars show the standard errors of the means. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 relative to D39.
We also tested SPD1078M through the intravenous route (Fig. 6). The results showed that the mutant was almost avirulent, as 16 of 17 mice survived to the end of experiment at 7 days whereas all of the wild-type-infected group failed to survive beyond 48 h (44 h ± 6.7 h, n = 17) (P < 0.0001). Furthermore, the median survival time of the cohort infected with the complemented strain (54 h ± 49 h, n = 10) was not significantly different from that of the cohort infected with the wild type (P > 0.05) (Fig. 6A). The bacterial load was also determined after intravenous infection. As shown in Fig. 6B, SPD1078M numbers progressively decreased after infection, from log10 (4.26 ± 0.18) CFU/ml (n = 10) at 1 h postinfection to log10 (1.76 ± 0.42) CFU/ml (n = 10) at 48 h postinfection, while D39 numbers increased progressively from log10 (4.4 ± 0.23) CFU/ml at 1 h postinfection to log10 (6.17 ± 0.37) CFU/ml (n = 10) at 36 h postinfection. The growth pattern of SPD1078comp was similar (P > 0.05) to that of the wild type [log10 (4.27 ± 0.11) CFU/ml, log10 (3.65 ± 0.08) CFU/ml, log10 (4.80 ± 0.31) CFU/ml, and log10 (5.70 ± 0.60) CFU/ml (n = 10) for 1, 12, 24, and 36 h postinfection, respectively].
FIG 6.
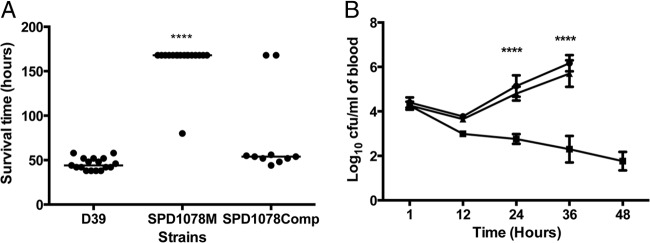
(A) Survival of mice after intravenous infection with pneumococcal strains. Each point represents one animal. The median survival time is given with a horizontal bar. ****, P < 0.0001 relative to D39. (B) Numbers of D39 (●), SPD1078M (●), and SPD1078comp (◆) in the blood after intravenous infection. Each point is the mean of data from 10 mice. Error bars show the standard errors of the means.
Gene expression analysis.
To investigate the reasons for attenuated virulence in SPD1078M, we measured the expression of a group of genes whose products have been suggested to be involved either in fermentative metabolism (adh, encoding alcohol dehydrogenase; pflB, pyruvate formate lyase; and spxB, pyruvate oxidase) or in virulence and competence (bgaC, encoding β-galactosidase; comA, competence protein; cps2H, capsule synthesis; hyl, hyaluronidase; ply, pneumolysin; and zmpC, zinc metalloproteinase) in the ldh mutant relative to the wild-type bacteria recovered from nasopharynx, lungs, and blood (Table 3). The results showed that the expression levels of pflB and adh (SPD_1834) were significantly higher for SPD1078M in all tissues than for D39 (P < 0.05), in line with our observations that in the absence of LDH, pyruvate is dissimilated through the PFL route. In addition to adh, carried by SPD_1834, the D39 genome has three additional genes annotated as alcohol dehydrogenase; these are SPD_0265, SPD_1126, and SPD_1865. Of these, SPD_1126 expression in the mutant did not change significantly relative to the wild type in all tissues, while SPD_0265 and SPD_1865 expression increased in nasopharynx (P < 0.05), which may imply that their activity also contributes to compensate for the lack of LDH. Moreover, comA expression in SPD1078M, responsible for export and maturation of competence stimulating peptide and shown to be important for competence regulation, decreased significantly relative to that in D39 for all tissues (P < 0.05).
TABLE 3.
Fold changes in gene expression in SPD1078M relative to D39 in different tissues
| Type of gene and gene namea (locus tag) | Mean fold change in gene expression (SD) in: |
||
|---|---|---|---|
| Nasopharynx | Lungs | Blood | |
| Metabolic genes | |||
| adh (SPD_0265) | 2.03 (0.41) | 1.5 (0.22) | 2.38 (0.83) |
| adh (SPD_1126) | 0.52 (0.23) | 0.94 (0.06) | 0.77 (0.08) |
| adh (SPD_1834) | 4.19 (0.4) | 3.71 (0.34) | 5.15 (1.24) |
| adh (SPD_1865) | 2.3 (0.47) | 1.38 (0.32) | 1.55 (0.31) |
| ldh (SPD_1078) | 0.42 (0.02) | 0.01 (0.15) | 0.43 (0.02) |
| spxB (SPD_0636) | 1.85 (0.79) | 4.46 (0.63) | 1.33 (0.18) |
| pflB (SPD_0420) | 4.25 (0.52) | 2.72 (0.3) | 6.52 (0.6) |
| Virulence genes | |||
| bgaC (SPD_0065) | 0.66 (0.29) | 1.27 (0.42) | 1.4 (0.45) |
| cps2H (SPD_0323) | 0.94 (0.44) | 0.75 (0.19) | 0.32 (0.09) |
| comA (SPD_0049) | 0.38 (0.18) | 0.07 (0.02) | 0.05 (0.01) |
| hyl (SPD_0287) | 1.45 (0.19) | 0.78 (0.22) | 1.24 (0.11) |
| nanA (SPD_1504) | 1.23 (0.36) | 1.97 (0.48) | 1.89 (0.68) |
| ply (SPD_1726) | 1 (0.38) | 0.72 (0.21) | 0.94 (0.27) |
Gene names: adh, alcohol dehydrogenase; ldh, lactate dehydrogenase; spxB, pyruvate oxidase; pflB, pyruvate formate lyase; bgaC, β-galactosidase C; cps2H, polysaccharide polymerase; comA, competence factor transporting ATP-binding protein; hyl, hyaluronate lyase; nanA, neuraminidase A; ply, pneumolysin.
The expression analysis of virulence-associated genes showed that in the mutant the expression of a capsule synthesis gene, cps2H, decreased 3-fold (± 0.09, n = 6) in the blood, whereas SPD1078M was unable to grow, especially after intravenous infection, relative to D39 (P < 0.05) (Table 3). Expression of nanA in the mutant recovered from the lungs (1.97-fold ± 0.48-fold, n = 6) and blood (1.89-fold ± 0.68-fold, n = 6) increased relative to the wild type. However, there was no significant change in the expression of bgaC, hyl, and ply (coding for β-galactosidase C, hyaluronidase, and pneumolysin, respectively) between the two strains among the tissues (P > 0.05).
DISCUSSION
We investigated LDH's role in the pneumococcal metabolism and virulence, using an isogenic ldh mutant. Unlike what occurred in previous work (6), we were able to obtain viable ldh mutants. The likely reason for successful inactivation of ldh in this study could be due to the use of mariner mutagenesis rather than the plasmid-based mutation system, which was the method of choice for Chapuy-Regaud et al. (6). In fact, the successful mutation of pneumococcal ldh by mariner mutagenesis was also reported in another study (29), where the number of mutations in each gene, determined by barcode sequencing, was used to assess the gene essentiality, and ldh was classified as “likely essential” rather than as an “essential” gene.
We found that the inactivation of ldh reduces the pneumococcal growth rate, particularly on glucose, decreases glucose consumption, leads to mixed-acid fermentation and pyruvate accumulation, and leads to a significant increase in ethanol production. These results emphasize the importance of LDH in the reduction of pyruvate and redox balance maintenance. The loss of this key enzyme impaired substantially the regeneration of NAD+, as evidenced by the high accumulation of pyruvate and the reduced formation of acetate in favor of ethanol. The ultimate consequence of this metabolic constraint is the remarkable reduction (7-fold) in glucose consumption by the LDH-deficient strain. In line with this view is the accentuated accumulation of acetate upon depletion of glucose. In the absence of substrate, the pressure to regenerate NAD+ is relieved, and consequently 3-PGA and pyruvate are converted via the more energetically favorable pathway to acetate (Fig. 1 and 4). The maximal concentration of the glycolytic intermediate FBP was elevated in the ldh mutant compared to the wild type. A similar pattern has been reported for an LDH-deficient strain of Lactococcus lactis (11), an organism for which an inverse relationship between FBP accumulation and glucose consumption was established (30). This association holds true for S. pneumoniae (Fig. 4), since the wild-type strain with the highest glucose consumption showed the lowest accumulation of FBP (30 mM, 17 mM less than the ldh mutant).
Despite the limited diversity of means of energy generation due to the absence of respiration, there is a certain degree of flexibility in the fermentative steps, in terms of availability of different enzymes that can convert pyruvate to different end products of sugar fermentation. This flexibility is surmised to be a determinant of in vivo fitness, by allowing the microbe to generate energy from different sugars under different environmental conditions during infection. For example, in the respiratory tract the main sugar is known to be galactose, found within the structure of mucin (4, 25), whereas blood is rich in glucose (31). In addition, the factors that are important for the outcome of bacterial sugar metabolism, such as oxygen concentration, also vary in different tissue sites (32). We demonstrated the essential role of PFL activity in galactose metabolism. In the absence of PFL activity, the pneumococcus was less able than the wild type to grow on galactose, and no mixed-acid products could be formed (4). Our results also showed that the lack of PFL diminished pneumococcal virulence both after intranasal and after intravenous infection of mice, very likely to be due to decreased synthesis of ATP and changes in membrane lipid composition (4). In addition, POX's role in pneumococcal biology has been studied (3). The lack of this enzyme attenuates pneumococcal virulence in both pneumonia and systemic infection models, which can be attributed to a decrease in acetyl phosphate levels, downregulation of adhesive proteins, and altered capsule production (2, 3). In this study, we determined LDH's contribution to pneumococcal virulence. The mutant strain was avirulent when administered directly into blood through the intravenous route. Given that blood is rich in glucose (31), impaired utilization of this sugar and growth defects in environments containing it (Fig. 3 and 4) are seemingly the reason for the progressive decrease in the pneumococcal numbers in blood. Our observation is supported by a recent study that showed that synthesis of LDH in pneumococcus cultured in blood-containing medium increases relative to the synthesis in the absence of blood (33), demonstrating the importance of LDH in pneumococcal survival in blood. On the other hand, although after intranasal infection, SPD1078M was attenuated in virulence, it could still maintain its invasiveness. This is very likely due to the presence of sugars other than glucose that can facilitate growth in the respiratory tract, combined with the metabolic flexibility at the pyruvate node. Alcohol dehydrogenase can, to a certain extent, replace LDH in redox balance maintenance (10). Furthermore, it is known that sugars such as galactose, which is plentiful within the structure of host glycoproteins in the respiratory tract, induce a shift to mixed-acid metabolism in strain D39 (4). Our gene expression data support this assumption, because the expression of pflB, spxB, and adh, whose products are important for mixed-acid fermentation, was higher in SPD1078M than in the wild-type strain. It is intriguing that while the increased expression of these genes in SPD1078M compensates for the lack of LDH to a certain degree in the respiratory tract, it does not do so in blood. It is plausible that the environmental conditions in the respiratory tract allow SPD1078M to undergo metabolic reprogramming, whereas in blood this is not possible. In addition, we determined downregulation of a capsule synthesis gene in SPD1078M, which may also contribute to reduction of virulence in SPD1078M.
The essentiality of LDH in systemic infection was also reported in previous studies with other bacteria (13, 14). In E. faecalis, which has two ldh genes (13), mutation of either ldh did not affect the microbe's virulence in a mouse systemic-infection model, but the mutation of both ldh genes resulted in significantly lower bacterial counts in both the kidneys and the liver for the double mutants than for the wild type. In S. aureus, in which LDH activity is encoded by two ldh genes, ldh1 and ldh2, mutation of ldh1, but not ldh2, reduced the microbe's virulence after intravenous infection relative to the wild type. Moreover, inactivation of both genes rendered the microbe avirulent. Because of its role in bacterial virulence, LDH was suggested as an attractive drug target (13).
Glycolysis and pyruvate metabolism are integrated metabolic processes, and their regulation in LAB has been shown to occur both at the transcriptional and at the metabolic levels (9, 21). Most of our knowledge on regulation of glycolysis and pyruvate metabolism has been deduced from the studies of dairy organisms, such as L. lactis and lactobacilli (LAB) (10, 34). The pneumococcus is a pathogen that occupies a very different ecological niche from the dairy LAB; hence, it is highly plausible that the regulation of these processes may differ. This view is supported not only by our results but also by those from studies with other related human pathogens, such as Streptococcus mutans and Streptococcus salivarius (35). A detailed understanding of the central metabolism and its subtle connections to pneumococcal virulence and pathogenesis is still missing. This important research area will be a major focus of our studies in the near future.
ACKNOWLEDGMENTS
Firas A. Y. Al-Bayati is in receipt of a scholarship from the Government of Iraq. This work was partially supported by the Fundação para a Ciência e a Tecnologia (FCT), PTDC/SAU-MII/100964/2008. P. Gaspar acknowledges FCT for the award of a postdoc grant (SFRH/BPD/31251/2006).
The NMR spectrometers are part of The National NMR Network (REDE/1517/RMN/2005), supported by “Programa Operacional Ciência e Inovação (POCTI) 2010” and FCT.
We also thank Calum Johnston and Jean-Pierre Claverys, Université de Toulouse, for kindly sharing their plasmids.
Footnotes
Published ahead of print 22 September 2014
REFERENCES
- 1.Kadioglu A, Weiser JN, Paton JC, Andrew PW. 2008. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6:288–301. 10.1038/nrmicro1871. [DOI] [PubMed] [Google Scholar]
- 2.Carvalho SM, Farshchi Andisi V, Gradstedt H, Neef J, Kuipers OP, Neves AR, Bijlsma JJ. 2013. Pyruvate oxidase influences the sugar utilization pattern and capsule production in Streptococcus pneumoniae. PLoS One 8:e68277. 10.1371/journal.pone.0068277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spellerberg B, Cundell DR, Sandros J, Pearce BJ, Idanpaan-Heikkila I, Rosenow C, Masure HR. 1996. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol. Microbiol. 19:803–813. 10.1046/j.1365-2958.1996.425954.x. [DOI] [PubMed] [Google Scholar]
- 4.Yesilkaya H, Spissu F, Carvalho SM, Terra VS, Homer KA, Benisty R, Porat N, Neves AR, Andrew PW. 2009. Pyruvate formate lyase is required for pneumococcal fermentative metabolism and virulence. Infect. Immun. 77:5418–5427. 10.1128/IAI.00178-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auzat I, Chapuy-Regaud S, Le Bras G, Dos Santos D, Ogunniyi AD, Le Thomas I, Garel JR, Paton JC, Trombe MC. 1999. The NADH oxidase of Streptococcus pneumoniae: its involvement in competence and virulence. Mol. Microbiol. 34:1018–1028. 10.1046/j.1365-2958.1999.01663.x. [DOI] [PubMed] [Google Scholar]
- 6.Chapuy-Regaud S, Duthoit F, Malfroy-Mastrorillo L, Gourdon P, Lindley ND, Trombe MC. 2001. Competence regulation by oxygen availability and by Nox is not related to specific adjustment of central metabolism in Streptococcus pneumoniae. J. Bacteriol. 183:2957–2962. 10.1128/JB.183.9.2957-2962.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergmann S, Rohde M, Chhatwal GS, Hammerschmidt S. 2001. alpha-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40:1273–1287. 10.1046/j.1365-2958.2001.02448.x. [DOI] [PubMed] [Google Scholar]
- 8.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498–506. 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 9.Ramos-Montanez S, Kazmierczak KM, Hentchel KL, Winkler ME. 2010. Instability of ackA (acetate kinase) mutations and their effects on acetyl phosphate and ATP amounts in Streptococcus pneumoniae D39. J. Bacteriol. 192:6390–6400. 10.1128/JB.00995-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neves AR, Pool WA, Kok J, Kuipers OP, Santos H. 2005. Overview on sugar metabolism and its control in Lactococcus lactis—the input from in vivo NMR. FEMS Microbiol. Rev. 29:531–554. 10.1016/j.fmrre.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Neves AR, Ramos A, Shearman C, Gasson MJ, Almeida JS, Santos H. 2000. Metabolic characterization of Lactococcus lactis deficient in lactate dehydrogenase using in vivo 13C-NMR. Eur. J. Biochem. 267:3859–3868. 10.1046/j.1432-1327.2000.01424.x. [DOI] [PubMed] [Google Scholar]
- 12.Gaspar P, Neves AR, Shearman CA, Gasson MJ, Baptista AM, Turner DL, Soares CM, Santos H. 2007. The lactate dehydrogenases encoded by the ldh and ldhB genes in Lactococcus lactis exhibit distinct regulation and catalytic properties—comparative modeling to probe the molecular basis. FEBS J. 274:5924–5936. 10.1111/j.1742-4658.2007.06115.x. [DOI] [PubMed] [Google Scholar]
- 13.Rana NF, Sauvageot N, Laplace JM, Bao Y, Nes I, Rince A, Posteraro B, Sanguinetti M, Hartke A. 2013. Redox balance via lactate dehydrogenase is important for multiple stress resistance and virulence in Enterococcus faecalis. Infect. Immun. 81:2662–2668. 10.1128/IAI.01299-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richardson AR, Libby SJ, Fang FC. 2008. A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science 319:1672–1676. 10.1126/science.1155207. [DOI] [PubMed] [Google Scholar]
- 15.Laouami S, Messaoudi K, Alberto F, Clavel T, Duport C. 2011. Lactate dehydrogenase A promotes communication between carbohydrate catabolism and virulence in Bacillus cereus. J. Bacteriol. 193:1757–1766. 10.1128/JB.00024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvalho SM, Kuipers OP, Neves AR. 2013. Environmental and nutritional factors that affect growth and metabolism of the pneumococcal serotype 2 strain D39 and its nonencapsulated derivative strain R6. PLoS One 8:e58492. 10.1371/journal.pone.0058492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karlyshev AV, Pallen MJ, Wren BW. 2000. Single-primer PCR procedure for rapid identification of transposon insertion sites. Biotechniques 28:1078, 1080, 1082. [DOI] [PubMed] [Google Scholar]
- 18.Caymaris S, Bootsma HJ, Martin B, Hermans PW, Prudhomme M, Claverys JP. 2010. The global nutritional regulator CodY is an essential protein in the human pathogen Streptococcus pneumoniae. Mol. Microbiol. 78:344–360. 10.1111/j.1365-2958.2010.07339.x. [DOI] [PubMed] [Google Scholar]
- 19.Guiral S, Henard V, Laaberki MH, Granadel C, Prudhomme M, Martin B, Claverys JP. 2006. Construction and evaluation of a chromosomal expression platform (CEP) for ectopic, maltose-driven gene expression in Streptococcus pneumoniae. Microbiology 152:343–349. 10.1099/mic.0.28433-0. [DOI] [PubMed] [Google Scholar]
- 20.Gaspar P, Neves AR, Ramos A, Gasson MJ, Shearman CA, Santos H. 2004. Engineering Lactococcus lactis for production of mannitol: high yields from food-grade strains deficient in lactate dehydrogenase and the mannitol transport system. Appl. Environ. Microbiol. 70:1466–1474. 10.1128/AEM.70.3.1466-1474.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neves AR, Ramos A, Nunes MC, Kleerebezem M, Hugenholtz J, de Vos WM, Almeida J, Santos H. 1999. In vivo nuclear magnetic resonance studies of glycolytic kinetics in Lactococcus lactis. Biotechnol. Bioeng. 64:200–212. . [DOI] [PubMed] [Google Scholar]
- 22.Garrigues C, Loubiere P, Lindley ND, Cocaign-Bousquet M. 1997. Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J. Bacteriol. 179:5282–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hajaj B, Yesilkaya H, Benisty R, David M, Andrew PW, Porat N. 2012. Thiol peroxidase is an important component of Streptococcus pneumoniae in oxygenated environments. Infect. Immun. 80:4333–4343. 10.1128/IAI.00126-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shafeeq S, Yesilkaya H, Kloosterman TG, Narayanan G, Wandel M, Andrew PW, Kuipers OP, Morrissey JA. 2011. The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol. Microbiol. 81:1255–1270. 10.1111/j.1365-2958.2011.07758.x. [DOI] [PubMed] [Google Scholar]
- 25.Terra VS, Homer KA, Rao SG, Andrew PW, Yesilkaya H. 2010. Characterization of novel beta-galactosidase activity that contributes to glycoprotein degradation and virulence in Streptococcus pneumoniae. Infect. Immun. 78:348–357. 10.1128/IAI.00721-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morton DB. 1985. Pain and laboratory animals. Nature 317:106. [DOI] [PubMed] [Google Scholar]
- 27.Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189:38–51. 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoskins J, Alborn WE, Jr, Arnold J, Blaszczak LC, Burgett S, DeHoff BS, Estrem ST, Fritz L, Fu DJ, Fuller W, Geringer C, Gilmour R, Glass JS, Khoja H, Kraft AR, Lagace RE, LeBlanc DJ, Lee LN, Lefkowitz EJ, Lu J, Matsushima P, McAhren SM, McHenney M, McLeaster K, Mundy CW, Nicas TI, Norris FH, O'Gara M, Peery RB, Robertson GT, Rockey P, Sun PM, Winkler ME, Yang Y, Young-Bellido M, Zhao G, Zook CA, Baltz RH, Jaskunas SR, Rosteck PR, Jr, Skatrud PL, Glass JI. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709–5717. 10.1128/JB.183.19.5709-5717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 6:767–772. 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castro R, Neves AR, Fonseca LL, Pool WA, Kok J, Kuipers OP, Santos H. 2009. Characterization of the individual glucose uptake systems of Lactococcus lactis: mannose-PTS, cellobiose-PTS and the novel GlcU permease. Mol. Microbiol. 71:795–806. 10.1111/j.1365-2958.2008.06564.x. [DOI] [PubMed] [Google Scholar]
- 31.Philips BJ, Meguer JX, Redman J, Baker EH. 2003. Factors determining the appearance of glucose in upper and lower respiratory tract secretions. Intensive Care Med. 29:2204–2210. 10.1007/s00134-003-1961-2. [DOI] [PubMed] [Google Scholar]
- 32.Yesilkaya H, Andisi VF, Andrew PW, Bijlsma JJ. 2013. Streptococcus pneumoniae and reactive oxygen species: an unusual approach to living with radicals. Trends Microbiol. 21:187–195. 10.1016/j.tim.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 33.Bae SM, Yeon SM, Kim TS, Lee KJ. 2006. The effect of protein expression of Streptococcus pneumoniae by blood. J. Biochem. Mol. Biol. 39:703–708. 10.5483/BMBRep.2006.39.6.703. [DOI] [PubMed] [Google Scholar]
- 34.Teusink B, Bachmann H, Molenaar D. 2011. Systems biology of lactic acid bacteria: a critical review. Microb. Cell Fact 10(Suppl 1):S11. 10.1186/1475-2859-10-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price CE, Zeyniyev A, Kuipers OP, Kok J. 2011. From meadows to milk to mucosa—adaptation of Streptococcus and Lactococcus species to their nutritional environments. FEMS Microbiol. Rev. 36:949–971. 10.1111/j.1574-6976.2011.00323.x. [DOI] [PubMed] [Google Scholar]
- 36.Martin B, Prudhomme M, Alloing G, Granadel C, Claverys JP. 2000. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol. Microbiol. 38:867–878. 10.1046/j.1365-2958.2000.02187.x. [DOI] [PubMed] [Google Scholar]