Abstract
Although primary syphilis lesions heal spontaneously, the infection is chronic, with subsequent clinical stages. Healing of the primary chancre occurs as antibodies against outer membrane antigens facilitate opsonophagocytosis of the bacteria by activated macrophages. TprK is an outer membrane protein that undergoes antigenic variation at 7 variable regions, and variants are selected by immune pressure. We hypothesized that individual TprK variants escape immune clearance and seed new disseminated lesions to cause secondary syphilis. As in human syphilis, infected rabbits may develop disseminated secondary skin lesions. This study explores the nature of secondary syphilis, specifically, the contribution of antigenic variation to the development of secondary lesions. Our data from the rabbit model show that the odds of secondary lesions containing predominately TprK variant treponemes is 3.3 times higher than the odds of finding TprK variants in disseminated primary lesions (odds ratio [OR] = 3.3 [95% confidence interval {CI}, 0.98 to 11.0]; P = 0.055) and that 96% of TprK variant secondary lesions are likely seeded by single treponemes. Analysis of antibody responses demonstrates significantly higher antibody titers to tprK variable region sequences found in the inoculum compared to reactivity to tprK variant sequences found in newly arising secondary lesions. This suggests that tprK variants escape the initial immune response raised against the V regions expressed in the inoculum. These data further support a role for TprK in immune evasion and suggest that the ability of TprK variants to persist despite a robust immune response is instrumental in the development of later stages of syphilis.
INTRODUCTION
Syphilis, caused by Treponema pallidum, is a complex disease that manifests as multiple clinical stages. Infection begins with development of a primary chancre at the site of inoculation. The primary chancre heals as opsonic antibodies against surface-exposed antigens facilitate opsonophagocytosis and clearance by activated macrophages (1). Despite the presence of circulating antibodies that are functionally opsonic (2–7), the second stage of syphilis appears as disseminated treponemes multiply and establish new infectious lesions throughout the body. These new lesions may develop as a macular or papular rash on the skin, as well as mucosal lesions. Approximately 90% of untreated patients develop secondary syphilis (8), and 22.5% of them have recurrent secondary lesions, that is, multiple successive bouts of this disseminated infectious rash (8). The infected individual then enters latency and may, after years, develop tertiary manifestations (8).
The development of secondary syphilis is puzzling, because the immune response has already been quite effective in healing the primary chancre, and high titers of circulating antibodies are present (9). This pattern of successive episodes of clinical disease during infection is reminiscent of other diseases in which antigenic variation of the infectious agent accounts for repeated cycles of pathology. For example, the waves of spirochetemia and fever in tick-borne relapsing fever are due to antigenic variation of the variable membrane proteins (VMPs) of Borrelia hermsii (10). Antigenic variation of outer membrane antigens is a hallmark of many chronic multistage infectious diseases, including Lyme disease (VMP-like sequence E [VlsE] in Borrelia burgdorferi) (11), anaplasmosis (major surface protein 2 [MSP2] in Anaplasma marginale) (12), and trypanosomiasis (variant surface glycoprotein [VSG] in Trypanosoma brucei) (13). We sought to elucidate the mechanisms by which T. pallidum is able to evade the early immune response to cause the second stage of syphilis.
Antigenic variation of TprK is hypothesized to be central to T. pallidum's ability to escape antibody binding and opsonophagocytosis, thus permitting it to persist in the host (14, 15). Multiple lines of evidence support this hypothesis. TprK is predicted to be located in the outer membrane and oriented so that each of its 7 variable (V) regions is exposed and accessible to host antibodies (16). Sequence variation is restricted to these 7 variable regions (14, 17), and variation occurs by segmental gene conversion, with multiple donor sites located distant from the single tprK expression site (18). Infection-induced antibodies are focused against V regions, where sequence variation abrogates binding of antibodies raised to different V regions (17). Lastly, selection of TprK variants is dependent on an intact acquired immune response, as shown in the rabbit model (15).
T. pallidum organisms are routinely passaged and studied in the rabbit model, which closely recapitulates the disease in humans. During or following resolution of primary chancres in rabbits (i.e., after intradermal [i.d.] inoculation), generalized secondary syphilis may appear in infected rabbits, and lesions are readily visible on the skin if the fur is kept clipped (19). These lesions are considered to be true secondary lesions because they appear in the face of an effective immune response, just as in human infection. Rabbits may also develop disseminated skin lesions as their initial clinical manifestation after intravenous (i.v.) infection (20, 21). In this case, the initial immune response is just developing as these disseminated primary lesions appear. Because T. pallidum cannot be grown in vitro, we utilize disseminated primary infection in the rabbit model to “clone” T. pallidum, that is, to obtain isogenic strains of T. pallidum with nearly perfect identity at the tprK locus (22), suggesting that each disseminated primary lesion is seeded by a single treponemal cell. Such isogenic strains have allowed us to monitor the development of antigenic variation and immune selection of variants during the course of infection (18).
In this study, we sought to address the questions of (i) whether secondary lesions are also seeded by single treponemes and (ii) how treponemes are able to persist to cause the second stage of syphilis despite the presence of an immune response that actively clears treponemes from the primary chancre. We tested the hypothesis that, as treponemes are cleared from the healing primary lesion, single escape variants with unique tprK sequences seed skin sites, leading to the new disseminated lesions of secondary syphilis. Here, we demonstrate that treponemes expressing variant TprK proteins escape the immune response and seed new secondary lesions.
MATERIALS AND METHODS
Animal experiments.
Male New Zealand White rabbits were used for propagating strains and for the experiments described in this study. The animals were fed an antibiotic-free diet and housed at 16 to 18°C. All protocols involving animals were approved in advance by the University of Washington (UW) Institutional Animal Care and Use Committee.
Chicago C strain isolation.
The Chicago strain of T. pallidum was isolated from a chancre in 1951 (21). As we described previously, the Chicago C isolate was obtained after i.v. inoculation of a naive rabbit and harvesting of disseminated skin lesions (22) and has been used extensively in studies of TprK antigenic variation (15, 17, 18, 23–25). Because the Chicago C strain is known to have a high rate of tprK variation, special care was taken to ensure each inoculum was as isogenic at tprK as possible. In this study, prior to both i.d. and i.v. infections, the Chicago C isolate was reisolated as described previously (17, 22) with the following modification. After taking biopsy specimens of discrete skin lesions and mincing the tissue in 500 μl of sterile normal rabbit serum (NRS) (collected from healthy rabbits nonreactive in both venereal disease research laboratory [VDRL] and fluorescent treponemal antibody absorption [FTA-ABS] tests), the serum-treponeme suspension was removed from the residual skin material, gently mixed with 500 μl of sterile glycerol, and then immediately frozen in a dry ice-ethanol slurry to maintain the viability of the bacteria. The remaining material from the same skin biopsy specimen was homogenized in lysis buffer (10 mM Tris [pH 8], 0.1 M EDTA, 0.5% SDS) and also frozen in a dry ice-ethanol slurry.
Subsequently, DNA was extracted from the lysis buffer sample, and tprK sequences from multiple lesions were analyzed for isogenicity at tprK by fragment length analysis (FLA) of PCR amplicons of V6 (the most diverse V region in the Chicago strain) and by sequencing of cloned tprK amplicons as described below. Because nearly all variation in tprK involves insertions or deletions, finding a single V6 size by FLA and a single sequence by sequencing strongly supports isogenicity. After identifying lesions that appeared to have the most homogeneous tprK sequences, the corresponding viable treponemes frozen in glycerol were recovered by adding 1 ml of sterile NRS to thaw the sample and injecting the mixture intratesticularly (i.t.) into a naive rabbit. At peak orchitis (3 to 4 weeks postinfection), the isogenic population was expanded once by passaging in naive animals (1 to 2 weeks). Because some tprK sequence variation can occur during these propagation steps, we retained samples of the experimental inocula so that sequence variation that occurred during propagation could be analyzed.
Experimental infection. (i) Intradermal inoculation to cause the primary stage of syphilis and development of secondary syphilis.
Rabbits were infected i.d. with 0.1 ml of a 107-treponeme/ml suspension at 10 marked sites along the back. The rabbits' backs were meticulously kept free of fur by daily clipping throughout the experiment. As primary lesions appeared, progressed, and healed, measurements of erythema, induration, and ulceration were noted. When secondary lesions began to develop, they were biopsied as quickly as possible after appearance to prevent development of additional tprK variation within the lesion. In selecting lesions for biopsy, a distinction was made between true secondary lesions and satellite lesions. The latter develop in an annular pattern at the leading edges of primary lesions; true secondary lesions develop as diffuse single macular and then papular lesions at least 1 cm from the edge of a primary lesion. At this early stage of development of secondary lesions, the full lesion was easily encompassed by a 4-mm-diameter biopsy punch. Animals were monitored for lesion development for 100 days.
(ii) Intravenous inoculation to obtain disseminated primary skin lesions.
For comparison with secondary lesions, we also examined disseminated primary lesions. Rabbits were inoculated with 108 total treponemes intravenously into the marginal ear vein under general anesthesia. Again, the fur on the animals' backs was clipped daily to prevent hair accumulation. As described above for secondary lesions, disseminated primary lesions were biopsied as quickly as possible after they appeared (∼4 weeks postinfection). Unlike secondary lesions, large numbers of disseminated primary lesions developed nearly simultaneously in a given rabbit, so the animals were euthanized to permit biopsy of a large number of lesions.
Biopsy.
Full skin thickness 4-mm biopsy specimens were taken, after euthanasia or with local lidocaine anesthesia, using sterile biopsy punches. Each biopsy sample was transferred to a sterile dish and minced thoroughly, utilizing sterile forceps and scalpel, before homogenizing in a solution of 400 μl of lysis buffer. The entire 400 μl of the biopsy specimen preparation was first incubated with proteinase K (100 mg/ml) for 8 to 16 h at 56°C, and then, DNA was isolated using the QIAamp DNA minikit (Qiagen, Chatsworth, CA), with the addition of a second wash step with 500 μl of AW1 solution (Qiagen). DNA was eluted in 200 μl of nuclease-free water.
DNA analysis.
tprK sequences in secondary and disseminated primary lesions were analyzed with a two-pronged approach. First, single variable region sequences were analyzed by FLA to determine the relative proportions of size variants within a sample (15). FLA is a very sensitive measure of V region sequences that differ in size but cannot distinguish V region amplicons that are the same size but that have different sequences. Second, tprK was amplified directly from lesions, and the amplicon was sequenced. The addition of amplicon sequence analysis addresses the limitations of FLA mentioned above.
From DNA samples, tprK was amplified using primers that encompass the entire open reading frame (ORF) (see Table S1 in the supplemental material) as previously described (16). The tprK amplicons were then sequenced directly. The products of a second tprK amplification from both inocula (i.d. and i.v.) and from DNA extracted from secondary lesions were cloned into a sequencing vector (Invitrogen, Grand Island, NY), and 10 tprK clones containing inserts were sequenced per sample as previously described (26). Sanger sequencing was performed at the UW Biochemistry Core Facility or GeneWiz (South Plainfield, NJ). The sequences obtained were analyzed using BioEdit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html).
Individual V regions were amplified for FLA as previously described (15) with the following modification: PIG-tailed primers (listed in Table S1 in the supplemental material) were used to nullify Taq adenosine addition artifacts (27). Because of low DNA concentrations in a few lesions, a nested PCR was used to obtain some FLA data; this nesting step did not alter the FLA size ratio values compared to nonnested samples (data not shown).
Quantitative real-time PCR (qPCR) was used to probe the inoculum for specific V region sequences to determine whether specific V region sequences found in secondary lesions had also been present in low numbers in the inoculum. Primers and probes for individual V regions are shown in Table S1 in the supplemental material. The probes were 5′ 6-carboxyfluorescein (FAM) labeled and quenched with 3′ 6-carboxytetramethylrhodamine (TAMRA) (Eurofins, Huntsville, AL). Briefly, high-performance liquid chromatography (HPLC)-purified primers and probes were used at 0.4 μM and 0.2 μM concentrations, respectively, with 2.5 μl of template DNA and 2× TaqMan Fast Advance (Applied Biosystems, Grand Island, NY) in a 10-μl reaction mixture. Reactions were performed in triplicate and analyzed on a Viia7 real-time PCR system (Applied Biosystems) with the following cycling conditions: two hold cycles, each at 50°C for 2 min and 95°C for 20 s, and then, 50 cycles of 95°C for 1 s and 62°C for 20 s. PCRs for V4 were run with an annealing temperature of 64°C instead of 62°C. Each primer-probe set was assessed for its specificity for the corresponding target DNA sequence (data not shown). Copy numbers were extrapolated from standard curves constructed using 10-fold serial dilutions of plasmids containing full-length tprK sequences from lesion and inoculum sequences. A primer-probe set complementary to the conserved region between V1 and V2 was used as a positive control (data not shown). The sensitivities of all qPCRs ranged from 3.729 to 3.771 copies detected in 10-μl reaction mixtures. The proportions of any variant sequences found in the inoculum were calculated as the number of copies of variant 1 or 2 divided by the total number of tprK copies detected (defined as variant 1 plus variant 2 plus the inoculum sequence).
Antibody reactivity.
After initial infection, sera were collected weekly for 10 weeks from each rabbit and stored at −80°C until tested. Prior to use, serum samples were heat inactivated at 56°C for 30 min. Synthetic peptides, designed to reflect individual V region sequences found in the inoculum and in variant T. pallidum isolates from secondary lesions (see Table S3 in the supplemental material), were produced to >70% purity (GenScript Corporation, Piscataway, NJ) and used as antigens. Sera were tested by enzyme-linked immunosorbent assay (ELISA) at a 1/20 dilution, as previously described (14, 17), for the ability to bind V region peptides, with the following modification: washing steps (4 30-s washes with 350 μl phosphate-buffered saline [PBS]-0.05% Tween 20) were carried out on a MultiWash Advantage plate washer (TriContinent, Grass Valley, CA). Each sample was tested in triplicate.
Phylogenetic analysis.
DNA alignments were constructed using ClustalW and adjusted manually in BioEdit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Phylip version 3.695 (http://evolution.genetics.washington.edu/phylip.html) was used to construct bootstrapped (100 times) maximum-parsimony cladogram trees. Consensus cladograms were visualized with FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Statistical analysis.
To compare the presence of variant TprK sequences in secondary versus disseminated primary lesions, we used logistic regression for clustered data (28) to estimate the odds ratio (OR) between case-control status and lesion positivity. A two-tailed, paired Student t test was used to compare antibody reactivity (A405) to predominant inoculum V region peptides versus secondary-lesion variant peptides. A P value of <0.05 was considered significant. Bootstrap support of >70/100 was considered significant for the phylogenetic cladogram analyses (29).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences obtained (KM015522 to KM016072) are listed in Table S2 in the supplemental material.
RESULTS
Secondary lesions are likely seeded by single treponemes.
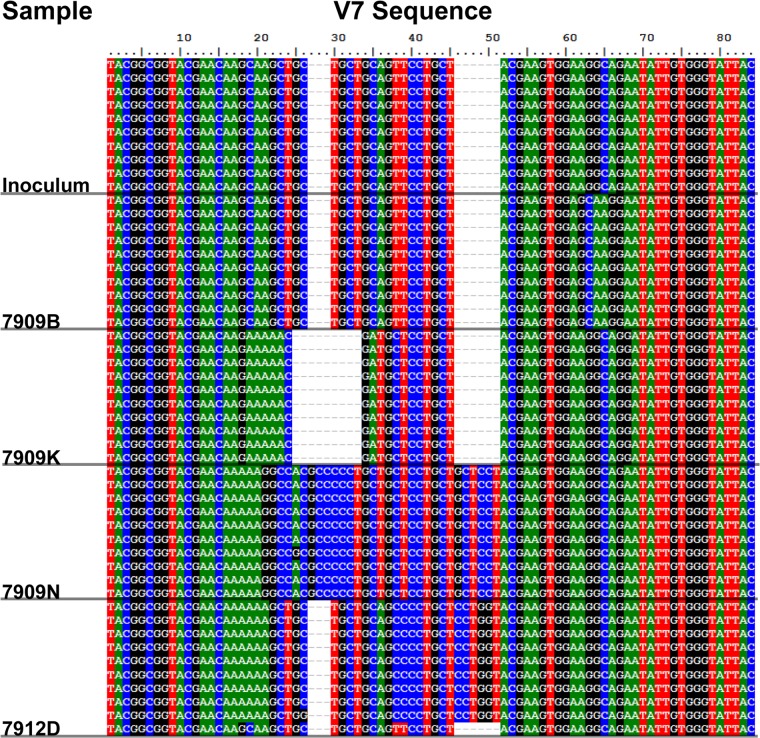
In order to obtain isogenic “clones” of Treponema pallidum for these and many past studies, we collected individual disseminated skin lesions from i.v.-infected animals. Careful analysis of treponemal DNA from individual lesion biopsy specimens led us to conclude that each disseminated skin lesion is likely seeded by a single treponeme, because the lesions contained nearly identical tprK full-ORF sequences. In this study, we sought to determine how secondary lesions arise, i.e., whether they are an expansion of a diverse nest of treponemes or the result of clonal expansion of a single treponemal cell. To address this question, we sequenced 10 clones of the tprK amplicon from each disseminated secondary skin lesion. All sequence changes preserved the tprK expression site open reading frame; there were no early terminations or changes in reading frames. Examples of V7 sequences are shown in Fig. 1. Insertions, deletions, and nonsynonymous substitutions distinguish each of the 4 variants shown in the figure from the inoculum sequences. Homology within each secondary lesion results in the appearance of a “blocking” pattern of sequences shown for each lesion.
FIG 1.
Examples of TprK V7 sequences among secondary lesions compared to the inoculum. For each lesion (identified by number on the left), DNA sequences were obtained from 10 Escherichia coli clones containing the tprK ORF. Note that the sequence variation in V7 is seen as insertions/deletions and base changes (numbers at the top indicate base pair positions). The identity, or near identity, of the 10 sequences within each lesion supports the single-cell origin of the lesion.
We scored a lesion as having arisen from a single tprK variant treponeme if ≥70% of its sequenced clones were identical and different from the inoculum in at least one V region. The 70% cutoff was chosen prior to data analysis because, as T. pallidum divides, some sequence change can occur. Any data-driven selection of a cutoff would introduce a type 1 error, increasing bias in subsequent analysis. Because our inoculum was nearly homogeneous in sequence at tprK, we cannot determine the number of treponemes that may have seeded lesions containing primarily inoculum sequences. By the time a lesion becomes clinically evident, many cell divisions have occurred, and some V region change is inevitable, even without immune selection. Using this definition, we found that 96% (26/27) of tprK variant secondary lesions were likely seeded by single treponemes.
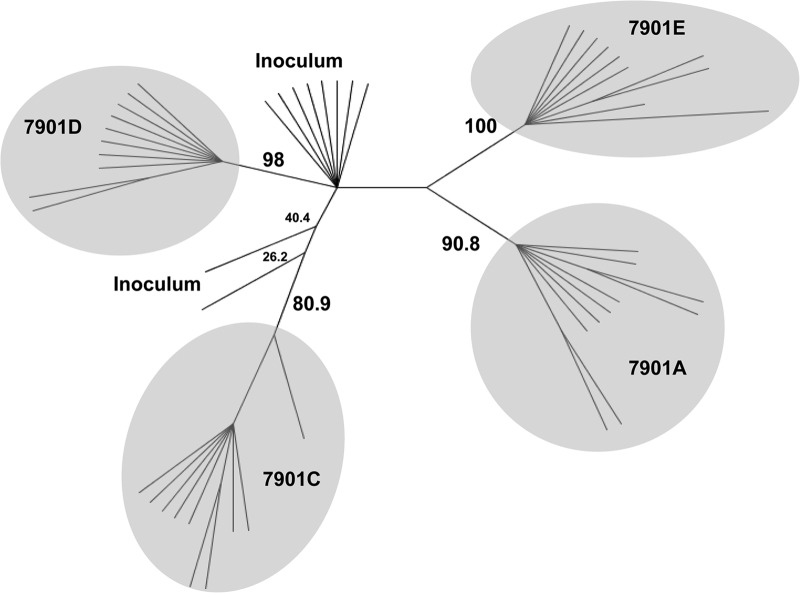
As a more stringent assessment, we used the data obtained from sequencing clones from these lesions to construct a maximum-parsimony cladogram of the full-length tprK ORFs. For all 27 tprK variant secondary lesions studied, representing 8 rabbits, we found a clustering pattern in which the majority of sequences from a single lesion cluster tightly together and each lesion is separate from the others. A representative example of 4 lesions arising in rabbit 7901 is shown in Fig. 2, demonstrating tight clustering of sequences within each secondary lesion; these lesions are separate from each other, with significant bootstrap support. These data further support our hypothesis that secondary lesions are seeded by single treponemes.
FIG 2.
Phylogenetic analysis of full-ORF tprK DNA sequences from secondary lesions. A representative example of phylogenetic relationships (a maximum-parsimony cladogram) among the inoculum and the individual secondary lesions (identified by rabbit number and lesion letter) that developed in rabbit 7901 is shown. Lesion 7901B did not contain amplifiable treponemes. The clusters enclosed by shaded ovals correspond to the sequences from 10 plasmid clones per lesion. Bootstrap values of the major branches are shown, demonstrating separation of the lesion sequences from the inoculum sequence. Note that the sequences for each lesion are in tight clusters, consistent with a single-cell origin for each lesion.
TprK variants cause secondary lesions.
In addition to demonstrating the tight clustering of tprK clone sequences, Fig. 2 also exemplifies the dispersion of variant sequence clusters away from the cluster of inoculum sequences. This pattern of branching is evidence of new secondary lesions being founded by tprK variant treponemes during infection. If tprK sequence variation abrogates antibody binding to variant bacterial cells during the primary stage of syphilis, then we would expect that TprK variant treponemes would escape killing by the host immune response and could seed new lesions (i.e., the secondary stage of syphilis).
We hypothesized that the treponemes multiplying in secondary lesions are doing so in the presence of an active immune response that is raised against the antigens expressed by the inoculum and that is responsible for healing the primary lesions. In contrast, disseminated primary lesions (developing after i.v. infection) in a naive rabbit serve to reflect only the basic variation rate of tprK without immune pressure. We used this method to compare the proportion of disseminated lesions containing variant tprK treponemes versus inoculum-like sequences in the presence (secondary lesions) and absence (disseminated primary lesion) of an effective immune response. Lesions with tprK variants were defined as those in which, by FLA, at least one V region contained <30% inoculum sequence. Sequences obtained by direct amplicon sequencing were used to identify variant sequences when FLA sizes were the same as those found in the inoculum. Careful analysis, by individual V region, of the secondary and disseminated primary lesions revealed the proportions of lesions arising from variant treponemes; they are shown, by rabbit, in Fig. 3A and B, respectively. Of the 10 rabbits in the secondary-syphilis study, two animals did not develop any secondary lesions and one animal (rabbit 7909) produced a very large number of secondary lesions. The numbers of secondary lesions per rabbit varied from 0 to 24 (median = 3; mean = 4.3), and they appeared 17 to 73 days postinfection (median = 39; mean = 40.5). In contrast, the disseminated primary lesions appeared on all rabbits between days 16 and 24 postinfection and totaled more than 100 lesions on 5 rabbits. In rabbit 7909, which had an unusually high number of secondary lesions, only 42% (10/24) of the lesions carried variant sequences. In contrast, in the 7 remaining rabbits that developed secondary lesions, 89% (17/19) of the lesions contained only variant sequences. In 5 of these 7 rabbits, all secondary lesions that developed contained variant sequences. Overall, as defined by FLA and amplicon sequence analysis, tprK variants seeded 63% (27/43) of all secondary lesions, compared to 34% (17/50) for disseminated primary lesions. We used logistic regression for clustered data (28) to estimate the odds ratio for the presence of TprK variant treponemes in secondary versus disseminated primary lesions. The odds of secondary lesions containing predominately TprK variant treponemes are 3.3 times higher than the odds of finding TprK variants in disseminated primary lesions (OR = 3.3 [95% confidence interval {CI}, 0.98 to 11.0]; P = 0.055) (Table 1). Excluding rabbit 7909 from the analysis, the calculated OR is 16.4 (95% CI, 4.44 to 62.5; P < 0.0001).
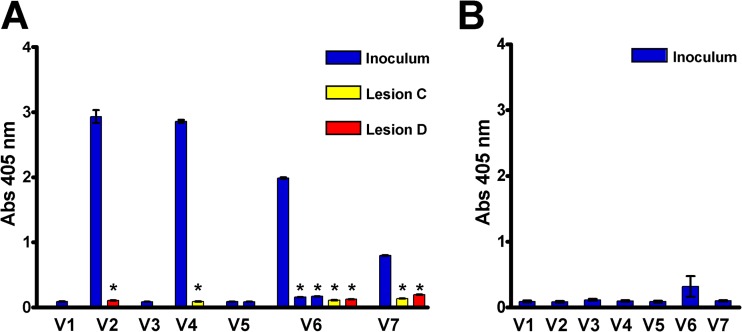
FIG 3.
TprK variants found in secondary lesions escape antibody binding. (A) Antibody binding (ELISA) of antiserum (collected at the time of development of secondary lesions [60 days postinfection]) from rabbit 7912 against synthetic peptides representing V region sequences identified in the inoculum (blue bars) and in two secondary lesions (red and yellow bars) that arose in the rabbit. Antibody reactivity is seen to the predominant sequences of the inoculum V2, V4, V6, and V7 regions, but significantly lower reactivity was detected to the V region sequences found in the secondary lesions. Note also that there is significantly lower antibody reactivity to the two minority V6 sequences that were present in the inoculum (also in blue). Sera from all rabbits with secondary syphilis were similarly tested and showed the same pattern of reactivity to the majority inoculum sequences, but not to the minor inoculum sequences or the variant secondary-lesion sequences. (B) Mean (±standard error [SE]) ELISA results for antisera (collected at the time of lesion appearance [mean, 22 days postinfection]) from rabbits with disseminated primary syphilis (n = 5 rabbits) showing lack of antibody reactivity directed to the V region sequences from the inoculum. *, two-tailed, paired Student t test; P < 0.001 compared to antibody binding to the peptide representing the majority sequences identified in the inoculum. Abs 405 nm, absorbance at 405 nm.
TABLE 1.
Proportions of lesions arising from TprK variants in secondary and disseminated primary lesionsa
| Lesion type | Rabbit no. | % Variant lesions (no./total) |
|---|---|---|
| Secondary | 7905 | No lesions |
| 7914 | No lesions | |
| 7919 | 100 (1/1) | |
| 7934 | 100 (1/1) | |
| 7898 | 100 (3/3) | |
| 7903 | 100 (3/3) | |
| 7902 | 67 (2/3) | |
| 7901 | 100 (4/4) | |
| 7912 | 75 (3/4) | |
| 7909 | 42 (10/24) | |
| Total | 63 (27/43) | |
| Disseminated primary | 8694 | 40 (4/10) |
| 8696 | 10 (1/10) | |
| 8697 | 40 (4/10) | |
| 8699 | 50 (5/10) | |
| 8700 | 30 (3/10) | |
| Total | 34 (17/50) |
Logistic regression for clustered data yielded an OR of 3.3 (95% CI, 0.98 to 11.0; P = 0.055) for the odds of having predominately TprK variants in secondary lesions versus disseminated primary lesions.
tprK variants found in secondary lesions develop during infection.
tprK variants were defined in this study as those sequences that were not found in our initial sequence analysis (full ORF sequencing of 10 clones) of the inoculum. Our initial sequencing did reveal some minor populations with different V regions in the inoculum, and this information was considered when identifying and calculating the contribution of variants to secondary lesions above. It is quite possible, however, that there were very small populations with different tprK sequences that went undetected by our initial sequence analysis and that some of the sequences that we found in secondary lesions may in fact have been present in the initial inoculum. It would be likely that any minority sequences present in the inoculum might be seen in lesions from multiple rabbits. To examine this possibility, we chose to interrogate our secondary-lesion inoculum by PCR for V region sequences that had been found in secondary lesions from multiple animals.
To interrogate our inoculum, we performed a targeted PCR-based search for 7 individual V3, V4, V5, V6, and V7 sequences (no variants were found at V1, and only single lesions contained V2 variants). DNA extracted from the inoculum was probed with specific TaqMan probes that were complementary to the tprK variant, as well as known inoculum sequences. None of the tested variants for V4, V5, V6, and V7 were detectable in the inoculum. In contrast, two V3 variants that had been seen in secondary lesions from multiple rabbits were found in the inoculum and were estimated to be present in the inoculum at 8.17% and 1.29% of tprK copies detected. These two V3 variants (different from the inoculum only at V3) were solely responsible for identifying 15% (4/27) of secondary lesions as tprK variant lesions, and we acknowledge that they came from the inoculum. Even though these minor variants were present in the inoculum, we examined whether they might have survived to cause secondary lesions because they were able to escape immune detection.
TprK variants escape humoral immunity.
Giacani et al. demonstrated that, in the face of an active immune response, treponemes expressing variant TprK sequences are positively selected (15). To test whether new TprK variants have a selective advantage over the inoculum treponemes in our study, we examined whether antibodies recognizing the secondary-lesion variant sequences were present in rabbits at the time that secondary lesions developed. This was accomplished by ELISA using synthetic peptides based on inoculum and tprK variant sequences. Antibody reactivity was detected against the V2, V4, V5, V6, and V7 peptide sequences of the majority inoculum sequences for all 10 rabbits in the secondary-syphilis group; a representative example is shown in Fig. 3A (blue bars). In contrast, the same sera collected at the time that secondary lesions arose (day 60 postinfection) had significantly less (P < 0.001) detectable antibody reactivity to the minority populations that were present in the inoculum (Fig. 3A, blue bars) or to the variant V2, V4, V6, and V7 sequences found in the secondary lesions (Fig. 3A, red and yellow bars). Similar findings were seen for each of the remaining 9 rabbits in the secondary-syphilis study (data not shown). These data suggest that antibodies to V2, V4, V6, and V7 select against the inoculum majority treponemes by facilitating bacterial clearance from primary lesions. TprK variants, even those present as minority populations in the inoculum, can escape clearance because there is no antibody against the specific TprK variant protein being expressed. These organisms subsequently evade the immune response and are able to cause secondary lesions.
In the rabbits infected i.v. to establish disseminated primary lesions, little to no antibody was detectable, even to majority inoculum TprK V region sequences, at the time that disseminated lesions developed (Fig. 3B). These findings are consistent with the concept that treponemes can survive to cause lesions when specific anti-V region antibodies are not present.
DISCUSSION
There are no known nonhuman reservoirs for venereal syphilis, making the disease a potential candidate for vaccine eradication, but infection-induced immunity is not complete (30), and reinfections occur (31). Efforts to understand the treponeme-host interface have focused on characterizing outer membrane proteins (OMPs) (32) of T. pallidum. This is especially difficult, considering that the treponemes cannot be cultured in vitro and that in vitro manipulations of the spirochete easily damage the fragile outer membrane. It is known that, during infection, a small population of treponemes develop that are resistant to opsonization and thus escape immune clearance, presumably due to alterations in their outer membrane proteins (9). Clinically, this makes sense, as, despite a robust immune response that clears most treponemes and heals primary chancres, later episodes of infectious skin lesions (secondary syphilis) and chronic infection occur.
To understand T. pallidum's ability to evade opsonophagocytosis, an understanding of the spirochetes' reported “paucity of outer membrane proteins” is needed (33). Few T. pallidum candidate OMPs have been studied in detail (3, 6, 34–38), but included among them are the Tpr proteins, specifically TprK. TprK is the only T. pallidum antigen known to undergo sequence variation during the course of infection (15, 17, 39) and is hypothesized to be central to immune escape and persistence (16). Sequence variation is confined to 7 discrete variable regions and occurs by a gene conversion mechanism (4, 18). The TprK variation mechanism is found not only in the syphilis subspecies (Treponema pallidum pallidum), but also in the other subspecies of T. pallidum (Treponema pallidum endemicum, Treponema pallidum pertenue), the Fribourg-Blanc isolate, and the agent of rabbit syphilis Treponema paraluiscuniculi (25). All of these treponemes can cause chronic infection and have intact TprK variation and expression systems, with V region donor sites (25). Additionally, in silico modeling predicts a beta barrel structure for TprK that places each variable region on exposed external loops, where antibody binding may drive selection of new variants. Indeed, TprK variants are selected by the acquired immune response (15), and B cell epitopes are restricted to the variable regions (14) during persistent infection. Although tprK variation itself has been well described, its relationship to the discrete clinical stages of syphilis had not yet been demonstrated. This report demonstrates an association between tprK variation and syphilis disease progression from the primary to the secondary stage in the rabbit model.
In our study of secondary syphilis, there was some clinical variability among the rabbits. Eight of the 10 rabbits infected intradermally subsequently developed secondary lesions during the 100-day observation period in our studies. Seven of these 8 rabbits developed a small number of lesions (1 to 4), and the majority of the lesions (17/19; 89%) were likely seeded by single TprK variants. The two animals that failed to develop secondary lesions had comparable primary-lesion progression, as well as typical serological responses as measured by VDRL titers and ELISA reactivity to the conserved lipoprotein p47 recombinant antigen (data not shown). Interestingly, these two animals had 2- to 5-fold-greater antibody reactivity to a TprC epitope than the other 8 rabbits in this group (P < 0.001) (data not shown), which is noteworthy in light of the recent characterization of TprC as an outer membrane protein (6).
Rabbit 7909 was unusual in that he developed many lesions at once, similar to what was observed in the rabbits with disseminated primary lesions after i.v. infection. Further, the ratio of variant TprK sequences to the inoculum sequence in lesions in rabbit 7909 approached the ratio observed after i.v. infection. We therefore sought, but did not uncover, an explanation for the unusual course in this single rabbit. Antibody titers (e.g., VDRL and anti-V region) for 7909 were not different from those for the remaining 9 rabbits. There was slight variation in primary-lesion progression, including the time to ulceration and the diameters of lesions among the 10 animals, but 7909 was not an outlier in this regard. Thus, there was no obvious evidence of an inadequate immune response in the rabbit. We were unable to monitor the time to lesion healing in rabbit 7909, however, because he was euthanized early for collection of many lesion biopsy specimens.
In analyzing the sequence data from each secondary lesion, it was apparent that certain tprK escape variant sequences (especially for V3) were found in multiple lesions and in different rabbits. This led us to ask whether these “TprK escape variants” developed de novo during the experimental infection or whether they were represented in the inoculum at a low frequency that was not detected in the initial sequencing of the inoculum (10 clones). Our careful examination of the inoculum by qPCR demonstrates, however, that the vast majority of the tprK variants found in secondary lesions developed during infection as opposed to being small subpopulations already present in the inoculum.
In our examination of antibody responses to TprK, antibody reactivity was detected in all rabbits to the V2, V4, V6, and V7 sequences expressed by the majority population in the inoculum. Based upon our ELISA data, however, no antibody reactivity was detected to V3 peptides, even to the inoculum V3 sequence. It was therefore puzzling that four secondary lesions had tprK sequences differing from the inoculum only at V3, perhaps suggesting that variation at V3 may be sufficient for immune escape. It is possible that V3 may be part of an important conformational epitope and that anti-V3 antibodies fail to bind the short linear peptides in our ELISA studies.
Our studies require that we use as the inoculum a T. pallidum clone that has nearly identical sequences in the tprK expression site so that we can clearly identify variants that develop. We have shown that, despite our efforts to maintain a tprK clonal population, some low-level diversity inevitably accumulates during expansion of the treponemes prior to inoculation. Examples of this diversity are found at V3 (minority populations detected by qPCR), as well as at V5 and V6 (minority populations detectable by sequencing 10 clones). In our studies, we demonstrate that a negligible antibody response develops to minority TprK sequences expressed in the inoculum. LaFond et al. showed that, following infection with a T. pallidum suspension that contains diverse tprK sequences, antibodies are developed to TprK V region sequences from multiple inoculum subpopulations, but the frequencies of these minority populations in the inoculum were not provided (17). From our data, it appears that minority populations that account for less than 10% of the inoculum (calculated from qPCR data or 1/10 inoculum clones) do not elicit antibodies to their V regions (Fig. 3). This suggests that the antibodies formed to the majority population in the inoculum (and not the infrequent minority population) positively select for the minority inoculum populations, as well as new variants that develop during infection.
In natural human infection, people are infected with populations of treponemes that are heterogeneous at tprK (26). This higher TprK diversity of the inoculum in human infections is important in the pathogenesis of syphilis because a diverse inoculum with multiple minority populations may serve as a source of scattered and early disseminating (e.g., to the central nervous system and other organs) tprK variants that are not recognized by the antibodies induced by the growth of the majority populations remaining at the site of the developing primary chancre. Our demonstration that secondary lesions are seeded by immune escape variants, regardless of whether they develop during primary infection or are present as minority populations in the inoculum, supports a critical role for TprK in the pathogenesis of syphilis and in the progression of the infection through multiple clinical stages.
The clinical variability described above for our outbred rabbits replicates the wide range in clinical presentations of syphilis infection in humans. For example, the rash of secondary syphilis is variable in appearance: it may be quite florid in some patients, while symptoms go unnoticed in others (40, 41). In addition to heterogeneity in host responses, part of the variability in human clinical presentation may be associated with T. pallidum strain-specific differences and with inoculum size (30, 42), both of which are controlled in our experimental model. We know that some strains of T. pallidum vary the sequence in the tprK expression site more readily than other strains (39). Such strains might be expected to cause more apparent clinical disease, such as more florid or recurrent secondary lesions, but this has not yet been explored in the experimental model. It is also possible that treponemes can actively modulate the amount of TprK protein on cell surfaces, potentially eliminating the targets of some opsonic antibodies. Strain-specific differences in the quantity of tprK mRNA may suggest a mechanism of tprK transcriptional regulation (25, 43), although there is evidence that tprK is expressed throughout the course of primary intradermal infection (15). There are no data on tprK expression during the latent stage, in which downregulation of opsonic targets might be most advantageous to the persistent organisms.
During early infection, T. pallidum infection elicits a robust humoral and cellular immune response (14), clearing the primary chancre, and then a small subpopulation effectively evades host opsonophagocytosis (9). A similar mechanism of clearance and immune evasion likely occurs during healing of the secondary stage, leaving persistent organisms that may remain quiescent during latency or escape to proliferate at sites of tertiary manifestations.
TprK's clear role in facilitating immune evasion and persistence has important implications for understanding not only the natural history, but also the epidemiology, of syphilis infection. The basic reproductive rate (R0) of an infectious disease predicts whether that disease can spread through a population and is directly proportional to the duration of infectiousness (44, 45). Evading host opsonophagocytosis may greatly increase the duration of the infectious period of syphilis in at least two possible ways: (i) by increasing the duration of the primary stage, as TprK variants locally maintain the chancre as they replace the initial infecting phenotype to which the initial immune response was generated, and (ii) by facilitating the development of the widespread infectious lesions of the secondary stage of syphilis and, potentially, the recurrence of secondary lesions in some infected persons (8). We propose that TprK antigenic variation increases T. pallidum's infectious period by facilitating immune escape and persistence to cause later stages in vivo, thus increasing the basic reproductive rate of syphilis in a population and contributing to T. pallidum's success as a pathogen.
Supplementary Material
ACKNOWLEDGMENTS
We thank James Hughes for his expertise in statistical methods and for statistical analyses. We thank Arturo Centurion-Lara and Lorenzo Giacani for their advice and Charmie Godornes and Lauren Tantalo for helpful discussions. We thank Kalli Plummer for assistance in manuscript submission.
This work was supported by the National Institute for Allergy and Infectious Diseases of the National Institutes of Health under award numbers R01AI063940 and R01AI042143. T.B.R. was supported by a supplement to R01AI063940 and by T32AI007140 and T32GM007266.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published ahead of print 15 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02236-14.
REFERENCES
- 1.Lukehart SA, Miller JN. 1978. Demonstration of the in vitro phagocytosis of Treponema pallidum by rabbit peritoneal macrophages. J. Immunol. 121:2014–2024. [PubMed] [Google Scholar]
- 2.Baker-Zander SA, Lukehart SA. 1992. Macrophage-mediated killing of opsonized Treponema pallidum. J. Infect. Dis. 165:69–74. 10.1093/infdis/165.1.69. [DOI] [PubMed] [Google Scholar]
- 3.Cameron CE, Castro C, Lukehart SA, Van Voorhis WC. 1998. Function and protective capacity of Treponema pallidum subsp. pallidum glycerophosphodiester phosphodiesterase. Infect. Immun. 66:5763–5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centurion-Lara A, Castro C, Barrett L, Cameron C, Mostowfi M, Van Voorhis WC, Lukehart SA. 1999. Treponema pallidum major sheath protein homologue Tpr K is a target of opsonic antibody and the protective immune response. J. Exp. Med. 189:647–656. 10.1084/jem.189.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cameron CE, Lukehart SA, Castro C, Molini B, Godornes C, Van Voorhis WC. 2000. Opsonic potential, protective capacity, and sequence conservation of the Treponema pallidum subspecies pallidum Tp92. J. Infect. Dis. 181:1401–1413. 10.1086/315399. [DOI] [PubMed] [Google Scholar]
- 6.Anand A, Luthra A, Dunham-Ems S, Caimano MJ, Karanian C, LeDoyt M, Cruz AR, Salazar JC, Radolf JD. 2012. TprC/D (tp0117/131), a trimeric, pore-forming rare outer membrane protein of Treponema pallidum, has a bipartite domain structure. J. Bacteriol. 194:2321–2333. 10.1128/JB.00101-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruz AR, Ramirez LG, Zuluaga AV, Pillay A, Abreu C, Valencia CA, La Vake C, Cervantes JL, Dunham-Ems S, Cartun R, Mavilio D, Radolf JD, Salazar JC. 2012. Immune evasion and recognition of the syphilis spirochete in blood and skin of secondary syphilis patients: two immunologically distinct compartments. PLoS Negl. Trop. Dis. 6:e1717. 10.1371/journal.pntd.0001717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gjestland T. 1955. The Oslo study of untreated syphilis; an epidemiologic investigation of the natural course of the syphilitic infection based upon a re-study of the Boeck-Bruusgaard material. Acta Derm. Venereol. Suppl. (Stockholm) 35(Suppl 34):3–368,Annex I–LVI. [DOI] [PubMed] [Google Scholar]
- 9.Lukehart SA, Shaffer JM, Baker-Zander SA. 1992. A subpopulation of Treponema pallidum is resistant to phagocytosis: possible mechanism of persistence. J. Infect. Dis. 166:1449–1453. 10.1093/infdis/166.6.1449. [DOI] [PubMed] [Google Scholar]
- 10.Barbour AG, Burman N, Carter CJ, Kitten T, Bergstrom S. 1991. Variable antigen genes of the relapsing fever agent Borrelia hermsii are activated by promoter addition. Mol. Microbiol. 5:489–493. 10.1111/j.1365-2958.1991.tb02132.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhang JR, Hardham JM, Barbour AG, Norris SJ. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:275–285. 10.1016/S0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]
- 12.Brayton KA, Palmer GH, Lundgren A, Yi J, Barbet AF. 2002. Antigenic variation of Anaplasma marginale msp2 occurs by combinatorial gene conversion. Mol. Microbiol. 43:1151–1159. 10.1046/j.1365-2958.2002.02792.x. [DOI] [PubMed] [Google Scholar]
- 13.Vanhamme LPE, McCulloch R, Barry JD. 2001. An update on antigenic variation in African trypanosomes. Trends Parasitol. 17:338–343. 10.1016/S1471-4922(01)01922-5. [DOI] [PubMed] [Google Scholar]
- 14.Morgan CA, Molini BJ, Lukehart SA, Van Voorhis WC. 2002. Segregation of B and T cell epitopes of Treponema pallidum repeat protein K to variable and conserved regions during experimental syphilis infection. J. Immunol. 169:952–957. 10.4049/jimmunol.169.2.952. [DOI] [PubMed] [Google Scholar]
- 15.Giacani L, Molini BJ, Kim EY, Godornes BC, Leader BT, Tantalo LC, Centurion-Lara A, Lukehart SA. 2010. Antigenic variation in Treponema pallidum: TprK sequence diversity accumulates in response to immune pressure during experimental syphilis. J. Immunol. 184:3822–3829. 10.4049/jimmunol.0902788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Centurion-Lara A, Godornes C, Castro C, Van Voorhis WC, Lukehart SA. 2000. The tprK gene is heterogeneous among Treponema pallidum strains and has multiple alleles. Infect. Immun. 68:824–831. 10.1128/IAI.68.2.824-831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LaFond RE, Molini BJ, Van Voorhis WC, Lukehart SA. 2006. Antigenic variation of TprK V regions abrogates specific antibody binding in syphilis. Infect. Immun. 74:6244–6251. 10.1128/IAI.00827-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Centurion-Lara A, LaFond RE, Hevner K, Godornes C, Molini BJ, Van Voorhis WC, Lukehart SA. 2004. Gene conversion: a mechanism for generation of heterogeneity in the tprK gene of Treponema pallidum during infection. Mol. Microbiol. 52:1579–1596. 10.1111/j.1365-2958.2004.04086.x. [DOI] [PubMed] [Google Scholar]
- 19.Strugnell RA, Drummond L, Faine S. 1986. Secondary lesions in rabbits experimentally infected with Treponema pallidum. Genitourin. Med. 62:4–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tantalo LC, Lukehart SA, Marra CM. 2005. Treponema pallidum strain-specific differences in neuroinvasion and clinical phenotype in a rabbit model. J. Infect. Dis. 191:75–80. 10.1086/426510. [DOI] [PubMed] [Google Scholar]
- 21.Turner TB, Hollander DH. 1957. Biology of the treponematoses. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 22.Lukehart SA, Marra CM. 2007. Isolation and laboratory maintenance of Treponema pallidum. Curr. Protoc. Microbiol. Chapter 12:Unit 12A 1. 10.1002/9780471729259.mc12a01s7. [DOI] [PubMed] [Google Scholar]
- 23.Morgan CA, Lukehart SA, Van Voorhis WC. 2003. Protection against syphilis correlates with specificity of antibodies to the variable regions of Treponema pallidum repeat protein K. Infect. Immun. 71:5605–5612. 10.1128/IAI.71.10.5605-5612.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giacani L, Jeffrey BM, Molini BJ, Le HT, Lukehart SA, Centurion-Lara A, Rockey DD. 2010. Complete genome sequence and annotation of the Treponema pallidum subsp. pallidum Chicago strain. J. Bacteriol. 192:2645–2646. 10.1128/JB.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giacani L, Brandt SL, Puray-Chavez M, Reid TB, Godornes C, Molini BJ, Benzler M, Hartig JS, Lukehart SA, Centurion-Lara A. 2012. Comparative investigation of the genomic regions involved in antigenic variation of the TprK antigen among treponemal species, subspecies, and strains. J. Bacteriol. 194:4208–4225. 10.1128/JB.00863-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaFond RE, Centurion-Lara A, Godornes C, Rompalo AM, Van Voorhis WC, Lukehart SA. 2003. Sequence diversity of Treponema pallidum subsp. pallidum tprK in human syphilis lesions and rabbit-propagated isolates. J. Bacteriol. 185:6262–6268. 10.1128/JB.185.21.6262-6268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brownstein MJ, Carpten JD, Smith RJ. 1996. Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. Biotechniques 20:1004–1010. [DOI] [PubMed] [Google Scholar]
- 28.Zeger SL, Liang KY. 1986. Longitudinal data analysis for discrete and continuous outcomes. Biometrics 42:121–130. 10.2307/2531248. [DOI] [PubMed] [Google Scholar]
- 29.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 30.Magnuson HJ, Thomas EW, Olansky S, Kaplan BI, DeMello L, Cutler JC. 1956. Inoculation syphilis in human volunteers. Medicine 35:33–82. 10.1097/00005792-195602000-00002. [DOI] [PubMed] [Google Scholar]
- 31.Brewer TH, Peterman TA, Newman DR, Schmitt K. 2011. Reinfections during the Florida syphilis epidemic, 2000–2008. Sex. Transm. Dis. 38:12–17. 10.1097/OLQ.0b013e3181e9afc7. [DOI] [PubMed] [Google Scholar]
- 32.Cameron CE, Lukehart SA. 2014. Current status of syphilis vaccine development: need, challenges, prospects. Vaccine 32:1602–1609. 10.1016/j.vaccine.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radolf JD, Norgard MV, Schulz WW. 1989. Outer membrane ultrastructure explains the limited antigenicity of virulent Treponema pallidum. Proc. Natl. Acad. Sci. U. S. A. 86:2051–2055. 10.1073/pnas.86.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanco DR, Miller JN, Lovett MA. 1997. Surface antigens of the syphilis spirochete and their potential as virulence determinants. Emerg. Infect. Dis. 3:11–20. 10.3201/eid0301.970102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shevchenko DV, Sellati TJ, Cox DL, Shevchenko OV, Robinson EJ, Radolf JD. 1999. Membrane topology and cellular location of the Treponema pallidum glycerophosphodiester phosphodiesterase (GlpQ) ortholog. Infect. Immun. 67:2266–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cameron CE, Brown EL, Kuroiwa JM, Schnapp LM, Brouwer NL. 2004. Treponema pallidum fibronectin-binding proteins. J. Bacteriol. 186:7019–7022. 10.1128/JB.186.20.7019-7022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desrosiers DC, Anand A, Luthra A, Dunham-Ems SM, LeDoyt M, Cummings MA, Eshghi A, Cameron CE, Cruz AR, Salazar JC, Caimano MJ, Radolf JD. 2011. TP0326, a Treponema pallidum beta-barrel assembly machinery A (BamA) orthologue and rare outer membrane protein. Mol. Microbiol. 80:1496–1515. 10.1111/j.1365-2958.2011.07662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houston S, Hof R, Francescutti T, Hawkes A, Boulanger MJ, Cameron CE. 2011. Bifunctional role of the Treponema pallidum extracellular matrix binding adhesin Tp0751. Infect. Immun. 79:1386–1398. 10.1128/IAI.01083-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LaFond RE, Centurion-Lara A, Godornes C, Van Voorhis WC, Lukehart SA. 2006. TprK sequence diversity accumulates during infection of rabbits with Treponema pallidum subsp. pallidum Nichols strain. Infect. Immun. 74:1896–1906. 10.1128/IAI.74.3.1896-1906.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chapel TA. 1980. The signs and symptoms of secondary syphilis. Sex. Transm. Dis. 7:161–164. 10.1097/00007435-198010000-00002. [DOI] [PubMed] [Google Scholar]
- 41.Mindel A, Tovey SJ, Timmins DJ, Williams P. 1989. Primary and secondary syphilis, 20 years' experience. 2. Clinical features. Genitourin. Med. 65:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magnuson HJ, Rosenau BJ. 1948. The rate of development and degree of acquired immunity in experimental syphilis. Am. J. Syph. Gonorrhea Vener. Dis. 32:418–436. [PubMed] [Google Scholar]
- 43.Giacani L, Molini B, Godornes C, Barrett L, Van Voorhis W, Centurion-Lara A, Lukehart SA. 2007. Quantitative analysis of tpr gene expression in Treponema pallidum isolates: differences among isolates and correlation with T-cell responsiveness in experimental syphilis. Infect. Immun. 75:104–112. 10.1128/IAI.01124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brunham RC. 2005. Insights into the epidemiology of sexually transmitted diseases from Ro = ΒcD. Sex. Transm. Dis. 32:722–724. 10.1097/01.olq.0000190093.59071.4d. [DOI] [PubMed] [Google Scholar]
- 45.May RM, Anderson RM. 1987. Transmission dynamics of HIV infection. Nature 326:137–142. 10.1038/326137a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.