Abstract
Uropathogenic Escherichia coli (UPEC) is responsible for the majority of urinary tract infections (UTIs), which are some of the world's most common bacterial infections of humans. Here, we examined the role of FNR (fumarate and nitrate reduction), a well-known global regulator, in the pathogenesis of UPEC infections. We constructed an fnr deletion mutant of UPEC CFT073 and compared it to the wild type for changes in virulence, adherence, invasion, and expression of key virulence factors. Compared to the wild type, the fnr mutant was highly attenuated in the mouse model of human UTI and showed severe defects in adherence to and invasion of bladder and kidney epithelial cells. Our results showed that FNR regulates motility and multiple virulence factors, including expression of type I and P fimbriae, modulation of hemolysin expression, and expression of a novel pathogenicity island involved in α-ketoglutarate metabolism under anaerobic conditions. Our results demonstrate that FNR is a key global regulator of UPEC virulence and controls expression of important virulence factors that contribute to UPEC pathogenicity.
INTRODUCTION
FNR (for fumarate and nitrate reduction) is known to be a global regulator which controls gene expression, facilitating bacterial adaptation to anaerobic conditions. The fnr gene was first identified by Lambden and Guest in the mid-1970s in their pioneering work on the characterization of mutants that could not use fumarate or nitrate (1). FNR is a member of a well-characterized global transcription factor family that has two conserved domains: the N-terminal ligand-binding domain for the O2 signal and the C-terminal DNA-binding domain. The N-terminal sensory domain contains five cysteine residues, four of which (Cys20, Cys23, Cys29, and Cys122) were shown to be required for in vivo binding of either [4Fe-4S]2+ or [2Fe-2S]2+ (2, 3). FNR is activated under anaerobic conditions by the acquisition of one [4Fe-4S]2+ molecule per protein, which promotes dimerization and enhances DNA binding to target promoters. Under aerobic conditions, molecular oxygen triggers the conversion of the [4Fe-4S]2+ into [2Fe-2S]2+. This conversion causes a conformational change within the FNR protein, turning it into a monomeric inactive form, preventing DNA binding and interactions with the transcription machinery (4).
The C-terminal DNA-binding domain recognizes specific FNR-binding sequences within FNR-controlled promoters. FNR-binding sites can be located at variable positions within the promoter region (5) and can have only a partial match to the consensus sequence of TTGATNNNNATCAA. The FNR regulon has been well studied in nonpathogenic Escherichia coli, and up to 125 regulon members have been identified so far (6). These include genes encoding enzymes for the anaerobic oxidation of carbon sources, such as the glycerol and formate dehydrogenases; enzymes for the anaerobic reduction of alternate terminal electron acceptors, such as the nitrate, fumarate, and dimethyl sulfoxide (DMSO) reductases; and proteins for the transport of these carbon sources or electron acceptors. FNR also represses the synthesis of enzymes required for aerobic respiration, such as NADH dehydrogenase II. As a consequence, compounds such as fumarate, nitrate, or other reducible substrates can replace oxygen as the terminal electron acceptors, thus providing alternate electron transport chains for generating energy via oxidative phosphorylation (6).
A role for FNR in bacterial virulence was first indicated by its requirement for the replication of Salmonella enterica serovar Typhi within epithelial cells (7). It has been further studied in Salmonella enterica serovar Typhimurium (ATCC 14028s), where FNR works as a positive regulator of genes involved in motility, flagellar biosynthesis, and pathogenesis (8). This regulation was confirmed by phenotype analysis with an fnr mutant, which was nonmotile, lacking in flagella, unable to survive inside macrophages, and attenuated in a murine model of mucosal and acute infection. The inability of the fnr deletion mutant to survive inside macrophages was likely due to its sensitivity to the reactive oxygen species generated by phagocyte NADPH oxidase. In addition, many of the virulence genes in the Salmonella pathogenicity island 1 (SPI-1), as well as the srfABC operon, were significantly downregulated in the fnr mutant strain (8). More recently, FNR was shown to modulate Shigella virulence (9). Dysentery-causing Shigella flexneri encounters changes in oxygen tension as it progresses along the gastrointestinal tract, and its type III secretion system (T3SS), which is essential for cell invasion and virulence, is impacted by the oxygen concentration. Expression of spa32 and spa33, the virulence genes that regulate secretion through the T3SS, is regulated by FNR, thereby affecting Shigella's entry into cells in response to available oxygen in vivo (9).
On the basis of the importance of FNR for bacterial adaptation to anaerobic conditions and the many phenotypes shared by the fnr mutants in Salmonella, Shigella, and E. coli, it has generally been assumed that FNR is important for the virulence of pathogenic E. coli. However, experimental evidence for a more general role of FNR, i.e., beyond facilitating adaptation to an anaerobic environment, in contributing to intramacrophage survival has so far been lacking. To address these questions, we constructed and characterized an fnr mutant and a complemented strain in a uropathogenic E. coli (UPEC) strain. We found that deletion of fnr resulted in significantly decreased virulence in vivo and in vitro. These phenotypes were associated with a loss of hemolytic activity and motility and the reduced expression of type 1 and P fimbriae, leading to decreased adherence to and invasion of host cells. In addition, FNR was found to target a novel pathogenicity island. The results indicate that FNR in UPEC plays, in addition to its established role as a regulatory switch between aerobic and anaerobic metabolism, a role in regulating virulence gene expression much more extensive than was previously believed.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and plasmids.
The wild-type strain used was UPEC CFT073, isolated from a patient that was admitted to the University of Maryland Hospital with acute pyelonephritis characterized by bacteriuria of 105 CFU/ml, pyuria, fever, and no other identified source of infection (10). The strains and plasmids used in this study are listed in Table S1 in the supplemental material. Aerobic growth was achieved by shaking in air at 180 rpm, and anaerobic growth was achieved by incubating in a Bactron chamber (Sheldon Manufacturing, Inc., OR) filled with a gas mixture (N2, 90%; CO2, 5%; H2, 5%). For genetic manipulations, all E. coli strains were routinely grown in Luria-Bertani (LB) broth medium. Selective antibiotics and IPTG (isopropyl-β-d-thiogalactopyranoside) were added when necessary at the following concentrations: ampicillin (Amp), 100 μg · ml−1; kanamycin (Kan), 50 μg · ml−1; chloramphenicol (Chl), 25 μg · ml−1; and IPTG, 0.1 mM (11).
Recombinant DNA techniques.
PCR, DNA ligation, electroporation, and DNA gel electrophoresis were performed as described by Sambrook and Russell (12), unless otherwise indicated. All oligonucleotide primers were purchased from Integrated DNA Technologies (IDT; Coralville, IA) and are listed in Table S2 in the supplemental material. All restriction and DNA-modifying enzymes were purchased from New England BioLabs (NEB; Ipswich, MA) and used on the basis of the supplier's recommendations. Recombinant plasmids, PCR products, and restriction fragments were purified using QIAquick PCR purification kits or MinElute gel extraction kits (Qiagen, CA) as recommended by the supplier. DNA sequencing was performed at the DNA facility, Iowa State University.
Deletion mutants were constructed using the bacteriophage lambda red recombinase system described by Datsenko and Wanner (13). Chromosomal transcriptional lacZ fusions were performed using a CFT073 strain with the deletion of the original lacZYA genes. The homologous recombination constructions used the suicide plasmid pVIK112 carrying a fragment of the complete 5′ region or 3′ region of the target gene, leaving the target functional (a 3′-region fusion was used only for hlyA and hlyD, and a 5′-region fusion was used for all other genes) (11, 14). For complementation, the coding sequences of genes plus their putative promoter regions were amplified from the CFT073 strain and independently cloned into pGEN-MCS (15) using EcoRI and SalI restriction sites.
Agglutination assays.
Tests for agglutination were performed as described earlier (16). For analysis of the fim operon, suspensions (10%) of yeast (Saccharomyces cerevisiae) cells were used, and for analysis of the Pap operon, type O human red blood cells (RBCs) (10%) that had been washed 3 times in phosphate-buffered saline (PBS) were used. Briefly, yeast cells or human RBCs were mixed on a glass slide with PBS or with bacteria in PBS in the presence and absence of d-mannose. Agglutination was read after 10 min at room temperature. The strength of the agglutination was determined by measurement of the titer of serial 2-fold dilutions of bacterial suspensions in PBS.
Motility assay.
The motility of the wild type (WT), the fnr mutant, and the Δfnr strain complemented with pGEN-fnr was evaluated under anoxic conditions as described in reference 8. Briefly, 10 μl of anaerobically grown (16 h) cells was stabbed onto LB agar (0.25% agar) plates and incubated at 37°C for 16 h. The diameter of the growth halo was used as a measure of motility.
Determination of the switch orientation of fim ON and OFF cells.
The orientation of the fim invertible DNA fragment can be determined using a molecular approach described previously (17). In brief, a 669-bp DNA fragment containing the fim invertible element was amplified, subjected to HinfI restriction, and analyzed on a 2% agarose gel. Depending on the orientation of the fim invertible element, this method generates fragments of different sizes (415 and 254 bp when in the ON orientation, 526 and 143 bp when in the OFF orientation).
β-Galactosidase assays.
Overnight LB medium cultures of E. coli containing the gene of interest fused to lacZ were washed once with PBS and were then diluted 1:100 in LB medium or the medium indicated below and grown at 37°C. For analysis of fim, a single colony was taken from an LB agar plate, inoculated into 5 ml of LB medium, and allowed to grow overnight (preinoculum); then, 200 μl was inoculated into a new bottle of 5 ml of LB medium and incubated for 48 h statically at 37°C (18). For analysis of pap, cultures were grown on CFA medium overnight with shaking at 37°C (17). For analysis of hly, cultures were grown on blood agar plates overnight at 37°C under anaerobic conditions (10). For analysis of fli transcriptional fusions, cultures were grown at 37°C in LB medium with shaking until an optical density (OD) of 0.5 was reached (19). For analysis of α-ketoglutarate, cultures were grown anaerobically at 37°C in M9 medium with α-ketoglutarate overnight (11). These cultures were collected when the OD at 600 nm (OD600) was 0.4, diluted 1:10 in Z buffer, and assayed for β-galactosidase activity using ortho-nitrophenyl-β-galactoside (ONPG) as the substrate, as described previously (20). β-Galactosidase activity was reported as the mean from 3 biological replicates and two technical replicates.
EMSAs.
We used the (FnrD154A)2 protein variant because it displayed DNA-binding affinities and transcriptional regulatory activities with various FNR-dependent promoters under aerobic conditions (21). Protein expression was performed as described previously (5). Briefly, E. coli BL21 with pET28a-(FnrD154A)2 was grown in 200 ml of LB medium for 16 h at 25°C with 0.1 mM IPTG. To study the binding of FNR to the DNA probe, electrophoretic mobility shift assays (EMSAs) were performed as described elsewhere (21, 22). Briefly, the FNR-His6 fusion protein was purified to homogeneity using Ni-nitrilotriacetic acid spin columns (5) and dialyzed against binding buffer. DNA probes were amplified using specific primers and purified using a Qiagen MinElute gel extraction kit. EMSAs were performed by adding increasing amounts of purified (FnrD154A)2-His6 fusion protein (0 to 20 ng) to the DNA probe (40 ng) in binding buffer (20 mM Tris [pH 6.8], 10 mM EDTA, 4 mM dithiothreitol, 50 mM NaCl, 10% glycerol, 0.5 mg ml−1 bovine serum albumin [NEB]) for 30 min at 37°C. The reaction mixtures were then subjected to electrophoresis on a 6% polyacrylamide gel in 0.5× TBE buffer (44.5 mM Tris, 44.5 mM boric acid, 1 mM EDTA, pH 8.0) at 200 V for 45 min. The gel was stained in 0.5× TBE buffer containing 1× SYBR Gold nucleic acid staining solution (Life Technologies, Grand Island, NY) for 30 min, and then the image was recorded.
Adherence and invasion assays.
Cell adherence and invasion assays were performed as previously described (23). The cell lines J82 (HTB-1, bladder) and A498 (HTB-44, kidney) (American Type Culture Collection) were cultured in Eagle's minimum essential medium (Gibco) containing 10% fetal bovine serum (FBS; Gibco) at 37°C in 5% CO2.
Cells were plated in sterile 24-well plates with 1 × 105 cells/well 48 h before the experiment. CFT073 wild-type, mutant, and complemented strains were cultured statically in LB medium for 16 h. Before infection, two wells of cultured cells were trypsinized and the cells were counted to estimate the cell number per well. Cells were washed once with 1 ml PBS and were then exposed to bacteria at a multiplicity of infection (MOI) of 10 and washed once with PBS. The 24-well plates were centrifuged (500 × g for 5 min) and incubated for 1 h. For the adherence assays, bacterium-exposed cells were washed four times with 1 ml of sterile PBS and then lysed with 1 ml 0.1% Triton X-100 for 10 min at room temperature. Serial dilutions of cell suspensions were spread onto MacConkey agar plates (Becton Dickinson & Co, Franklin Lakes, NJ), and CFU counts were obtained after overnight growth at 37°C. For the invasion assays, infected cells were washed 4 times with 1 ml of sterile PBS, then reincubated with Eagle's minimum essential medium containing 100 μg/ml gentamicin, and incubated for a further 3 h. At 4 h postinfection (p.i.), cells were washed 4 times with 1 ml of sterile PBS and then lysed with 1 ml of 0.1% Triton X-100 for 10 min at room temperature. Serial dilutions of the cell suspensions were spread onto MacConkey agar plates, and CFU counts were obtained after overnight growth at 37°C. The input dilution of bacteria was also plated to determine the CFU count for each inoculum.
In vivo infection studies.
Mouse infection studies were performed as described previously (11). Female CBA/J mice (age, 6 to 10 weeks) were anesthetized and inoculated via transurethral catheterization with a 20-μl (2 × 109-CFU) challenge inoculum per mouse. Overnight LB medium cultures of UPEC CFT073 and the mutant strain were pelleted and resuspended in sterile PBS, mixed in equal volumes, and adjusted to make the combined challenge inoculum. To determine the initial number of CFU/ml, dilutions of each inoculum were plated onto LB medium plates with and without kanamycin. After 48 h, the mice were euthanized and the bladder and kidneys were aseptically removed, weighed, and homogenized in PBS. Dilutions of the homogenized tissue were then plated on duplicate LB medium plates with and without kanamycin or on plates with different antibiotics to determine the bacterial concentration per unit of tissue (number of CFU/g). After overnight incubation at 37°C, the colonies were counted. The numbers of colonies on kanamycin plates were subtracted from those on LB medium plates to obtain the number of WT bacteria. In the case of the complemented strain, recovered bacteria were plated on LB medium plates with ampicillin and on LB medium plates with ampicillin plus kanamycin. Similarly, the numbers of colonies on plates with ampicillin plus kanamycin were subtracted from those on LB medium plates with ampicillin to obtain the number of complemented bacteria. A group of 10 mice for each dual-strain challenge was used to determine alterations in fitness. The competition assay was performed once. For statistical analysis, a two-tailed Wilcoxon matched-pairs test was used (Prism software; GraphPad, La Jolla, CA), and the threshold for statistical significance was a P value of <0.05.
Ethics statement.
All animal procedures were conducted in accordance with NIH guidelines, the Animal Welfare Act, and U.S. federal law. The experimental protocol for handling animals was approved by the Institutional Animal Care and Use Committee at Iowa State University (protocol number 4-11-7111-Z). All surgery was performed under isoflurane anesthesia, and all efforts were made to minimize suffering. All procedures were done in the presence of a qualified veterinarian.
RESULTS
The fnr mutation attenuates virulence in the mouse model of urinary tract infection (UTI).
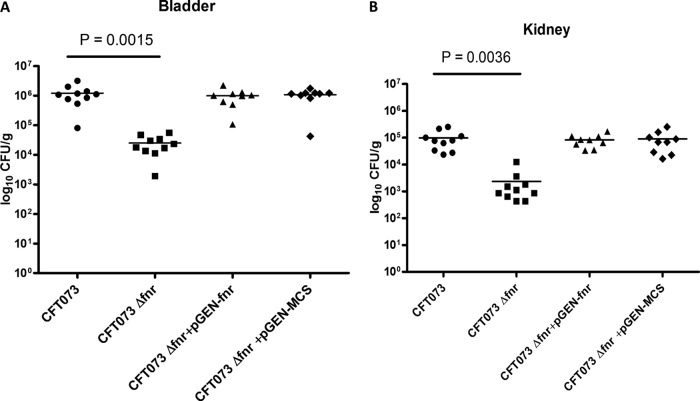
To address the role of FNR in UPEC virulence, we first compared the UPEC CFT073 wild type with its isogenic fnr deletion mutant strain for the ability to colonize the mouse urinary tract at 48 h postinoculation using an in vivo competition assay. Though the fnr mutant grew as well as the WT in LB medium under aerobic conditions, the fnr mutant showed significantly reduced colonization in both the bladder (P < 0.01) (Fig. 1A) and kidneys (P < 0.01) (Fig. 1B) compared to the WT. While the WT strains colonized the bladder at a level of 1.2 × 106 CFU/g, the fnr mutant colonized the bladder at a level of 2.5 × 104 CFU/g, a 48-fold reduction in the median number of CFU/g in the mutant. Similar results were observed for the kidney. The WT colonized the kidneys at a level of 1.1 ×105 CFU/g, while the fnr mutant colonized the kidneys at a level of 2.4 × 103 CFU/g, a 47-fold reduction in the median number of CFU/g in the mutant.
FIG 1.
Deletion of fnr attenuates virulence in the mouse model of UTI by UPEC CFT073. The WT and fnr mutant strains, the wild-type strain containing the empty vector (pGEN-MCS), and the mutant strain containing the complementation plasmid (pGEN-fnr) were mixed in a 1:1 ratio and approximately 2 × 109 CFU was transurethrally inoculated into female mice. Two days after infection, the mice were sacrificed and their bladders (A) and kidneys (B) were aseptically removed. WT and fnr mutant bacteria were recovered by plating homogenized tissue samples on LB medium or LB medium containing kanamycin, and CFU counts were determined. The wild-type strain containing the empty vector (pGEN-MCS) and the mutant strain containing the complementation plasmid (pGEN-fnr) were recovered by plating homogenized tissue samples on LB medium with ampicillin or LB medium with both ampicillin and kanamycin. Each dot represents the log10 number of CFU/g in the bladder or kidney from an individual animal, and the detection limit was 1,000 CFU/g. Bars indicate the median log10 number of CFU/g. A two-tailed Wilcoxon matched-pairs test was performed, and the difference in the colonization levels of the WT and mutants was considered statistically significant if P was <0.05.
To verify that the impact on colonization is not due to a secondary mutation, in vivo complementation experiments were performed. A stable low-copy-number plasmid, pGEN-MCS (11), was used to clone the coding region of fnr plus its predicted promoter region, and the obtained plasmid was transformed into the mutant to construct the complemented strain. As shown in Fig. 1, reintroduction of fnr back into the mutants restored colonization in both the bladder and kidney to WT levels. The fnr mutant containing the empty vector (pGEN-MCS) demonstrated an expected colonization defect in bladder and kidney colonization compared to the ability of the WT with the empty vector to colonize the bladder and kidney (not shown). These results demonstrate that the fnr mutation attenuated virulence in the mouse model of UTI.
The fnr mutant was impaired in adherence and invasion.
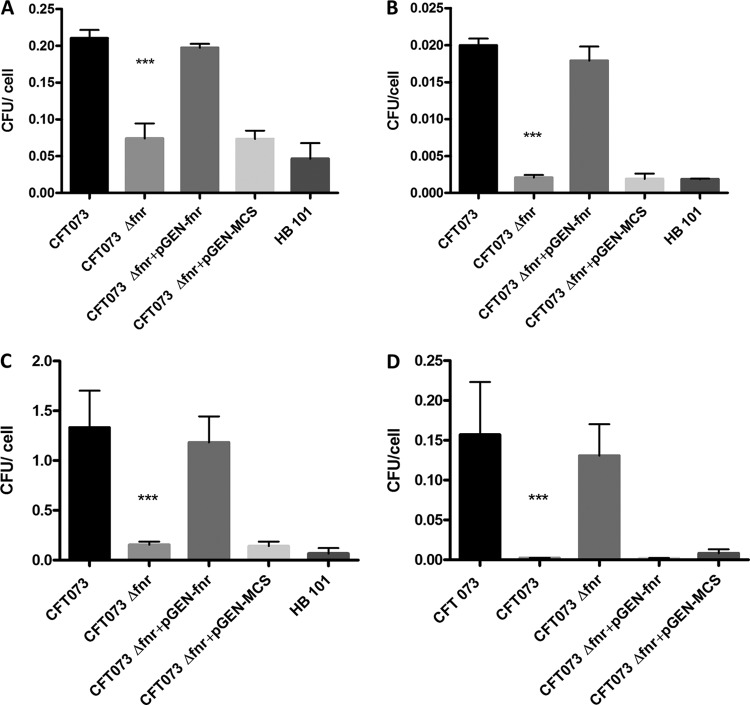
Successful colonization of the urinary tract relies on UPEC's ability to adhere to and invade host cells and tissues. Cell models of UPEC adherence to and invasion of bladder and kidney epithelial cells have been established (23). To determine the effect of the fnr mutation on the rates of adherence to and invasion of epithelial cells in vitro, cultured bladder epithelial (J82) cells were infected with the WT, fnr mutant, and complemented strains. The results showed that deletion of fnr significantly decreased UPEC's adherence to (P < 0.01) and invasion of (P < 0.01) bladder cells (Fig. 2A and B); whereas the WT strain displayed an association of 0.21 bacterium per J82 cell, the fnr mutant displayed an association of 0.07 bacterium per cell, a 3-fold reduction in host cell adherence. Similarly, we observed that while the WT strain showed an invasion of 0.02 bacterium per cell, the fnr mutant showed an invasion of 0.002 bacterium per cell. When we expressed these values as the percentage of bacterial invasion, which is calculated by dividing the number of intracellular bacteria at 4 h p.i. by the number of the corresponding adherent bacteria at 1 h p.i. and taking the amount of associated bacteria to be 100%, we found that 9.5% of the adherent WT cells actually invaded J82 cells but that only 2.8% of the adherent fnr mutant strain was able to invade the cells. Thus, the mutant strain showed a 3-fold reduction in the level of invasion of bladder epithelial cells compared to that for the WT. Complementation of the mutant by reintroduction of fnr restored the levels of adherence and invasion to WT levels (Fig. 2A and B). These results suggest that mutation of fnr reduces the levels of adherence and invasion of UPEC to bladder cells.
FIG 2.
Adherence to and invasion of the bladder J82 cell line and A498 kidney cell line by CFT073 and its mutants. J82 cells (A and B) and A498 cells (C and D) were infected at an MOI of 10 CFU/cell, as described in Materials and Methods. For the association assays (A and C), cells were lysed at 1 h postinfection, and the extracts were plated onto LB agar for enumeration, For the invasion assays (B and D), at 1 h postinfection, cells were washed with PBS and incubated for a further 3 h in the presence of gentamicin. The cells were then lysed, and the extracts were plated onto LB agar for counting of the CFU. E. coli HB101 was used as a negative control. The values shown are means plus standard deviations for quadruplicate samples from four independent experiments. Significant differences are indicated by asterisks (***, P < 0.0001 compared to the WT and mutant).
We next determined the impact of the fnr mutation on adherence to and invasion of kidney epithelial (A498) cells. Deletion of fnr significantly decreased UPEC's levels of adherence to and invasion of kidney cells (P < 0.01) (Fig. 2C and D), with the fold changes being even higher than those seen with bladder epithelial cells. Whereas the WT strain displayed an adherence of 1.33 bacteria per A498 cell, the fnr mutant displayed an adherence of 0.15 bacterium per cell, a 9-fold reduction compared to the level for the WT. Similarly, the WT invaded at a rate of 0.16 bacterium per cell, while the fnr mutant showed an invasion rate of 0.002 bacterium per cell. When we express these values as the percentage of bacterial invasion, we found that 11.8% of the adherent WT bacteria actually invaded A498 cells but only 1.6% of the adherent fnr mutant cells were able to invade cultured kidney epithelial cells. Thus, the mutant strain showed a 7-fold reduction in the level of invasion of kidney epithelial cells compared to that of the WT. Also, reintroduction of fnr back into the mutant restored WT levels of adherence and invasion (Fig. 2C and D). All of these results suggest that the fnr mutation impairs the ability of UPEC to adhere to and invade bladder and kidney cells.
Deletion of fnr reduced the expression of type 1 fimbriae in UPEC.
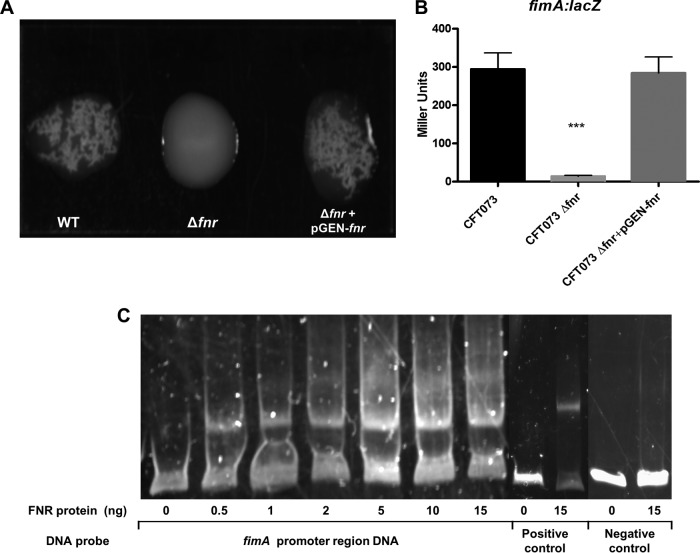
We have shown that deletion of fnr significantly decreased UPEC's colonization ability in vivo and also significantly reduced its level of attachment to and invasion of human bladder and kidney epithelial cell culture lines. These pathogenic mechanisms require the expression of diverse virulence factors, including fimbrial and afimbrial adhesins. Among them, type 1 fimbriae are prominent for their role in the establishment of a UTI (24, 25). We therefore analyzed whether fnr deletion affects the expression of type 1 fimbriae, as detected by a mannose-sensitive yeast agglutination (MSYA) assay, which tests the ability of bacteria to bind mannoside-containing receptors on the surface of yeast cells. The growth of the WT and fnr mutant was analyzed in various culture media (LB medium, tryptone bile agar [TBA], CFA, and tryptic soy agar [TSA]; data not shown) under different culture conditions (data not shown), and the medium and conditions in which they showed the same growth rates were chosen for the semiquantitative MSYA assay. Using serially diluted bacteria suspended in PBS, we observed agglutination of yeast cells at a dilution of WT cell suspensions higher (16-fold, i.e., a dilution containing 16 times more bacterial cells) than that for the fnr mutant; the complemented strain recovered the agglutination ability to the WT level (Fig. 3A). These results indicate that the fnr mutation caused a substantial decrease in the expression of type 1 fimbriae on the cell surface.
FIG 3.
UPEC type I fimbria regulation. (A) Yeast agglutination assay with dilutions of the WT (1:16); Δfnr (1:1), and Δfnr/pGEN-fnr (1:16) strains. Bacteria were grown statically for 48 h, agglutination was read after 10 min at room temperature, and the strength of the agglutination was determined by measurement of the titer of serial 2-fold dilutions of the bacterial suspensions in PBS. The experiments were performed four times in quadruplicate. (B) β-Galactosidase activity assay for expression of fimA. fimA-lacZ transcriptional fusion strains were grown statically in LB medium for 48 h at 37°C. β-Galactosidase activity was measured, and the values shown are means plus standard deviations for triplicate samples from three independent experiments. Significant differences are indicated by asterisks (***, P < 0.0001 compared to the WT and mutant). (C) Nonradioactive EMSA of binding of (FnrD154A)2-His6 to the promoter regions. The PCR product of the fimA promoter region was used as the probe at 300 ng per reaction mixture. Purified (FnrD154A)2-His6 fusion protein was added to each reaction mixture at different concentrations, as indicated; ydfZ promoter region DNA probes with and without the FNR protein were used as positive controls, and fimA coding region DNA probes with and without the FNR protein were used as negative controls. DNA fragments were stained with SYBR green.
The fimA gene encodes the major subunit of type 1 fimbriae. Transcriptional studies were first performed to compare the expression of the fim operon in the phase-variation-proficient WT and the fnr mutant, which carry the same fim-lacZ genes in the chromosome. A significant downregulation of fimA was observed in the fnr deletion background compared to the level of regulation in the WT (Fig. 3B), suggesting that FNR enhances type 1 fimbriation at the transcriptional level, consistent with our agglutination data.
To determine if FNR directly regulates fimA expression, an electrophoretic mobility shift assay (EMSA) was performed. The promoter region of fimA was predicted by the BProm program (SoftBerry). The potential binding site of FNR was identified with Patser software (version 3d) (8). DNA fragments containing the potential binding site were then PCR amplified for use as probes (202 nucleotides in size, starting from position −158 to position +44 relative to the position of the translational start codon). Fragments amplified from the coding region of fimA were used as negative controls. As shown in Fig. 3C, the FNR fusion protein was able to shift the promoter fragment of fimA but not the control fragment. These results suggest that FNR directly regulates the expression of fimA.
FNR affected phase variation of type 1 fimbriae.
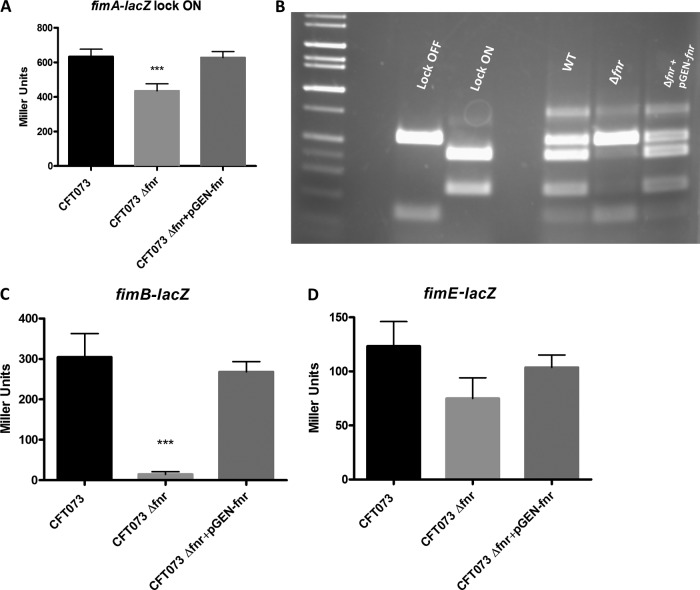
Deletion of fnr resulted in a higher fold change in the level of fimA expression in phase-variation-proficient strains (Fig. 3). To rule out the possibility of interference of the phase variation on fimA expression, we next performed expression studies in phase variation-deficient wild-type (CFT073 ON and CFT073 OFF) and fnr mutant (CFT073 Δfnr ON and CFT073 Δfnr OFF) strains; we observed that fimA expression in CFT073 Δfnr ON was not downregulated as much as it was in the phase-variation-proficient strain (Fig. 4A), indicating that FNR also affects the phase variation switch of type 1 fimbriae. No differences in fimA expression were observed between CFT073 OFF and CFT073 Δfnr OFF (data not shown). We then tested the effect of the fnr mutation on the phase ON/OFF switch. The fnr mutation blocked the phase switch in the lock OFF phase, in contrast to the findings for the wild type. Furthermore, complementation of the fnr mutant restored the ON orientation to the level for the parent strain (Fig. 4B). These results demonstrate that deletion of fnr affected the phase variation of type 1 fimbriae.
FIG 4.
UPEC type 1 fimbria phase variation. (A) β-Galactosidase activity assay for expression of fimA in lock ON strains. fimA-lacZ transcriptional fusion strains were grown statically in LB medium for 48 h at 37°C. β-Galactosidase activity was measured, and the values shown are means plus standard deviations for triplicate samples from three independent experiments. (B) Phase variation electrophoresis. PCR products were purified, digested with HinfI for 4 h, and analyzed on a 2% agarose gel. fimB-lacZ and fimE-lacZ transcriptional fusion strains were grown statically in LB medium for 48 h at 37°C. (C, D) β-Galactosidase activity assay for expression of fimB (C) and fimE (D). β-Galactosidase activity was measured, and the values shown are means plus standard deviations for triplicate samples from three independent experiments. Significant differences are indicated by asterisks (***, P < 0.0001 compared to the WT and mutant).
The recombinases FimB and FimE have been previously associated with the phase variation of type 1 fimbriae in E. coli (27). We analyzed the expression of fimB and fimE using fimB-lacZ and fimE-lacZ fusions in the chromosome. Our results showed that fimB transcription was downregulated 20-fold in the fnr mutant (Fig. 4C) and fimE transcription was downregulated 2-fold compared to the levels of transcription in the wild type (Fig. 4D). Expression of fimB and fimE was restored to the wild-type level in the complemented strain (Fig. 4C and D). According to these results, FNR affects the phase variation of type 1 fimbriae, probably by upregulating the transcription of the fimB recombinase. However, the regulatory mechanisms of fimB by FNR are still unknown.
Deletion of fnr decreased expression of type P fimbriae in UPEC.
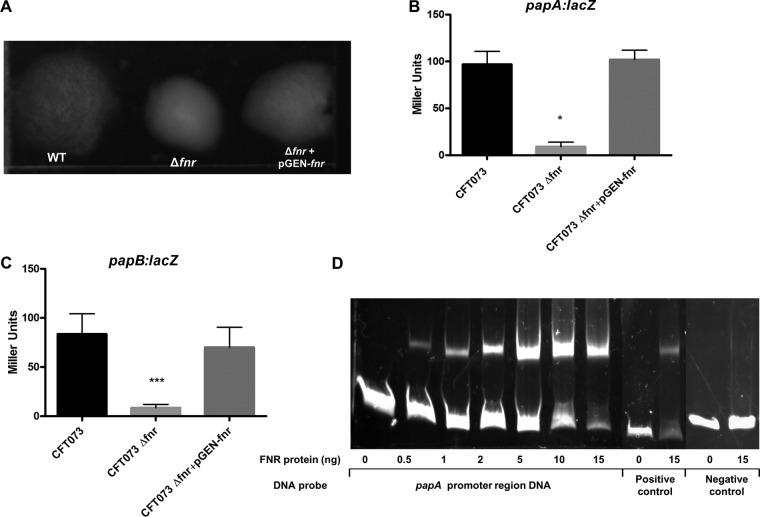
P fimbriae appeared to be especially important in E. coli pyelonephritis, and their subtle roles in uroepithelial cell culture models have been previously described (17). We thus analyzed whether deletion of fnr affects the expression of P fimbriae. P-fimbriated E. coli specifically binds to receptors on the surface of human red blood cells, and so the expression of P fimbriae can be detected by the human red blood cell agglutination assay (16). The agglutination assay showed that the UPEC wild-type CFT073 strain agglutinated human red blood cells (serotype O) in a mannose-independent way, and the agglutination titers obtained for the wild type were 32-fold higher than those obtained for the fnr mutant (Fig. 5A). Reintroducing the fnr gene into the mutant allowed the agglutination ability of the fnr mutant to recover to the wild-type level. These results suggest that the fnr mutation significantly decreases the expression of P fimbriae.
FIG 5.
UPEC type P fimbria regulation. (A) RBC agglutination assay with dilutions of the WT (1:32); Δfnr (1:1), and Δfnr/pGEN-fnr (1:32) strains. Bacteria were grown statically for 48 h, agglutination was read after 10 min at room temperature, and the strength of the agglutination was determined by measurement of the titer of serial 2-fold dilutions of the bacterial suspensions in PBS. The experiments were performed four times in quadruplicate. (B, C) β-Galactosidase activity assay for expression of papA (B) and papB (C). papA-lacZ and papB-lacZ transcriptional fusion strains were grown in CFA medium overnight without shaking at 37°C. β-Galactosidase activity was measured, and the values shown are means plus standard deviations for triplicate samples from three independent experiments. Significant differences are indicated by asterisks (*, P < 0.05 compared to the WT and mutant; ***, P < 0.0001 compared to the WT and mutant). (D) Nonradioactive EMSA of binding of (FnrD154A)2-His6 to the promoter regions. PCR products of the papA promoter region were used as probes at 300 ng per each reaction mixture. Purified (FnrD154A)2-His6 fusion protein was added at different concentrations to each reaction mixture, as indicated; ydfZ promoter region DNA probes with and without the FNR protein were used as positive controls, and papA coding region DNA probes with and without the FNR protein were used as negative controls. DNA fragments were stained with SYBR green.
papA encodes the major subunit of P fimbriae. Using CFT073 wild-type and fnr mutant strains carrying the papA-lacZ fusion in the chromosome, we showed that papA gene expression was significantly downregulated in the fnr mutant at the transcriptional level and the reintroduction of the fnr gene back into the fnr mutant restored papA expression (Fig. 5B). We also evaluated papB expression and also observed that papB gene expression was significantly downregulated in the fnr mutant, indicating that the FNR mutation affects not just papA in the P-fimbria operon. A potential binding site (positions −44 to −23) of FNR was identified in the promoter region of papA using Patser software (version 3d) (8). To determine if FNR directly regulates papA expression, an EMSA was performed. DNA fragments containing a potential binding site of FNR were then PCR amplified for use as probes, and fragments amplified from the coding region of papA were used as negative controls. As shown in Fig. 4D, the FNR fusion protein was able to shift the promoter fragment of papA but not the control fragment. These results demonstrate that FNR directly regulates the expression of papA.
FNR affects motility in UPEC.
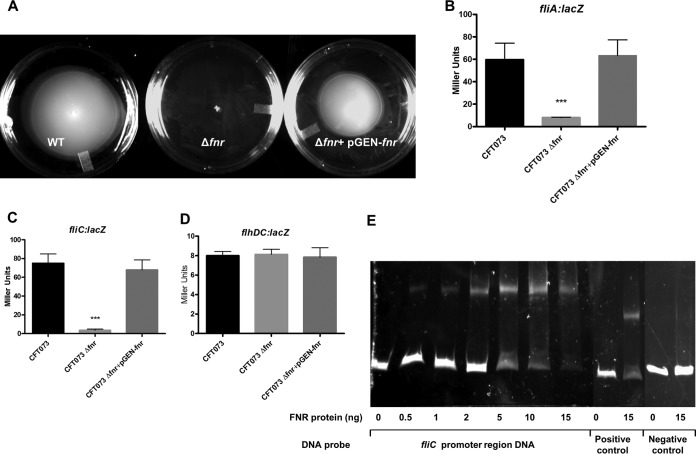
Bacterial flagella likely facilitate the establishment and spread of infection by microbial pathogens within the host, and expression of flagella was coincident with UPEC ascension to the upper urinary tract (15). To correlate FNR with UPEC flagella expression, we compared the motility of the wild type, the fnr mutant, and the fnr complemented strain using soft-agar plates. We observed that deletion of fnr resulted in a nonmotile phenotype in UPEC CFT073 and reintroduction of fnr into the mutant restored its motility capability (Fig. 6A). To determine which gene(s) in the fli and flh operons was downregulated by FNR, we constructed lacZ fusions with fliA, fliC, and flhDC. We observed that the expression of fliA and fliC was significantly downregulated in the fnr mutant strain compared to their levels of expression in the wild type (Fig. 6B and C, respectively), but no significant difference in the expression of flhDC (Fig. 6D) was found between the wild-type and mutant strains. A potential binding site (positions −84 to −105) for the global regulator FNR was found in the promoter region shared by both the fliA and fliC genes, and the direct regulation of a flagellar gene(s) by FNR was also confirmed by EMSA (Fig. 6E).
FIG 6.
Motility regulation. (A) Soft-agar motility assay. Bacterial cultures were stabbed in the middle of each soft-agar plate and incubated at 37°C for 16 h. The experiments were performed four times in quadruplicate. (B to D) β-Galactosidase activity assay for expression of fliA (B), fliC (C), and flhDC (D). fliA-lacZ, fliC-lacZ, and flhDC-lacZ transcriptional fusion strains were grown at 37°C in LB medium with shaking until the OD reached 0.5. β-Galactosidase activity was measured. The values shown are means plus standard deviations for triplicate samples from three independent experiments. Significant differences are indicated by asterisks (***, P < 0.0001 compared to the WT and mutant). (E) Nonradioactive EMSA of binding of (FnrD154A)2-His6 to the promoter regions. PCR products of the fliC promoter region were used as probes at 300 ng per reaction mixture. Purified (FnrD154A)2-His6 fusion protein was added to each reaction mixture at different concentrations, as indicated; ydfZ promoter region DNA probes with and without the FNR protein were used as positive controls, and fliC coding region DNA probes with and without the FNR protein were used as negative controls. DNA fragments were stained with SYBR green.
FNR regulates expression of a novel pathogenicity island.
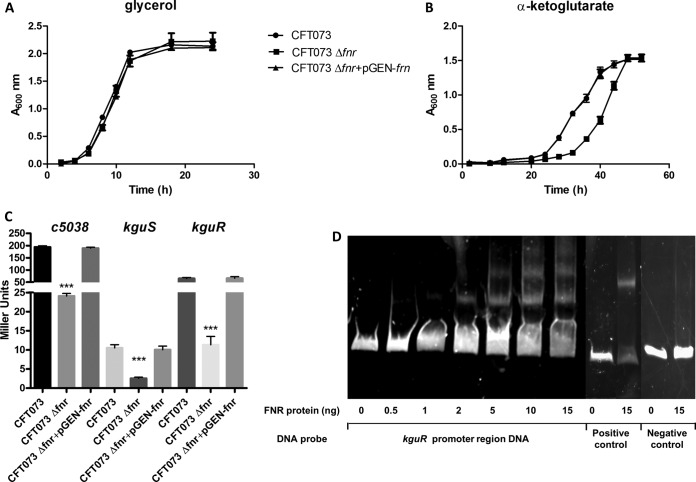
Utilization of α-ketoglutarate mediated by a pathogenicity island under anaerobic conditions has been described to be a novel metabolic trait which increases the adaptability and competitiveness of UPEC in vivo (11). Since FNR is a well-known global regulator mediating bacterial adaptation to the anaerobic environment, we analyzed whether FNR affects the utilization of α-ketoglutarate under anaerobic conditions. Our growth curves showed that the fnr deletion significantly reduced the anaerobic growth of UPEC in M9 minimum medium with α-ketoglutarate as the sole carbon source; in contrast, the fnr mutant did not show a significant growth defect in M9 minimum medium supplemented with glycerol as the sole carbon source compared to the growth of the wild type (Fig. 7A and B). Therefore, we compared the expression of the pathogenicity island genes by the wild type, the fnr mutant, and the complemented mutant strain. We observed that fnr deletion resulted in a significantly decreased expression of pathogenicity island genes, including the α-ketoglutarate transporter gene (c5038), the kinase gene, and the response regulator gene of the novel two-component signal transduction system (kguS-kguR). The complementation assay demonstrated that reintroduction of fnr into the mutant restored the expression of these targeted genes (Fig. 7C).
FIG 7.
UPEC α-ketoglutarate metabolism regulation. (A, B) In vitro growth of fnr mutants in M9 medium containing α-ketoglutarate as the sole carbon source (A) or M9 medium containing glycerol as the sole carbon source (B). The optical density of the UPEC CFT073 WT and mutants during growth in M9 medium containing α-ketoglutarate or glycerol as the sole carbon source under anaerobic conditions was determined. Growth curves represent the average measurement at each time point in duplicate from three independent experiments. (C) β-Galactosidase activity assay for expression of c5038, kguS, and kguR (C). c5038-lacZ, kguS-lacZ, and kguR-lacZ transcriptional fusion strains were grown anaerobically at 37°C in M9 medium with α-ketoglutarate overnight. β-Galactosidase activity was measured, and the values shown are means plus standard deviations for triplicate samples from three independent experiments. Significant differences are indicated by asterisks (***, P < 0.0001 compared to the WT and mutant). (D) Nonradioactive EMSA of binding of (FnrD154A)2-His6 to the promoter regions. PCR products of the kguR promoter region were used as probes at 300 ng per reaction mixture. Purified (FnrD154A)2-His6 fusion protein was added to each reaction mixture at different concentrations, as indicated; ydfZ promoter region DNA probes with and without the FNR protein were used as positive controls, and fliC coding region DNA probes with and without the FNR protein were used as negative controls. DNA fragments were stained with SYBR green.
By bioinformatics analysis, we found an FNR-binding motif in the promoter region of the KguS/KguR signal transduction system. EMSA was therefore performed using DNA fragments containing the promoter region of the kguS and kguR genes. As shown in Fig. 7D, the FNR fusion protein was able to shift the promoter fragment of kguS-kguR but not the control fragment. These results confirmed that FNR directly regulates the expression of the KguS/KguR two-component signal transduction system. It has been reported that KguS/KguR directly regulates the transport gene c5038 (11); therefore, it is very likely that FNR indirectly regulates other pathogenicity island genes, such as c5038, through KguS/KguR.
Deletion of fnr results in downregulation of hemolysin-encoding genes.
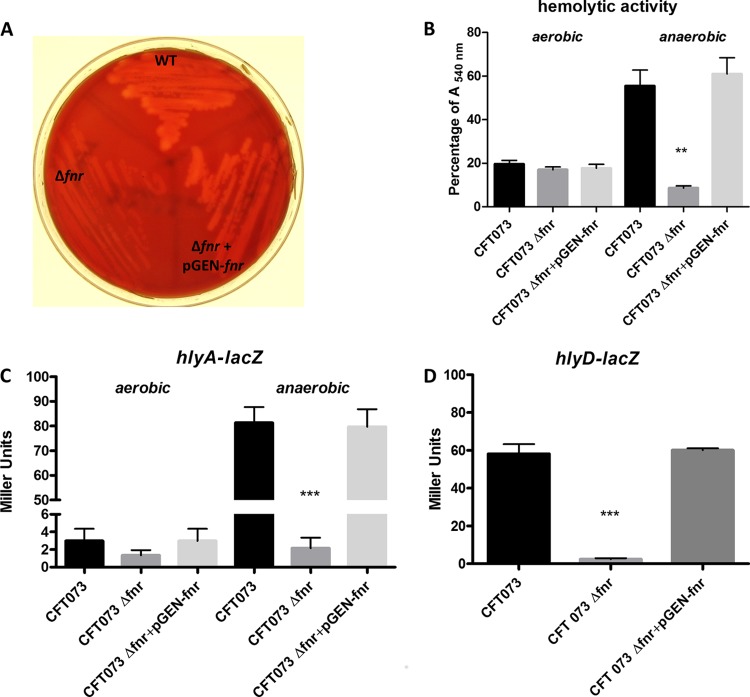
Hemolysin is a very important virulence factor of UPEC (28, 29). We found that UPEC CFT073 showed much stronger hemolysis on blood agar under anaerobic conditions than under aerobic conditions, and the upregulation of hlyCABD was responsible for this hemolytic phenotype (Fig. 8). We reasoned that FNR might be involved in the regulation of hlyA expression. To verify this hypothesis, we compared the hemolytic activity of the wild type with that of the fnr mutant grown on blood agar under both aerobic and anaerobic conditions. As expected, the fnr mutation reduced the hemolytic activity, and complementation rescued the hemolytic activity and restored it to the wild-type level under anaerobic conditions, while no significant difference between the wild type and the fnr mutant was found under aerobic conditions (Fig. 8A and B). A quantitative assay for hemolytic activity further confirmed that the fnr mutant showed significantly weaker hemolysis than the wild-type strain under anaerobic conditions (Fig. 8B).
FIG 8.
UPEC hemolysin regulation. (A) Blood agar hemolytic assay. Bacteria were inoculated into blood agar plates and grown overnight at 37°C under anaerobic conditions. (B) For the hemolytic activity quantification, bacterial strains were grown in the presence of RBCs overnight at 37°C under aerobic and anaerobic conditions, and the OD540 was recorded; a value of 100% was assigned to RBCs lysed with 1% Triton X-100. The values shown are means plus standard deviations for quadruplicate samples from four independent experiments. (C, D) β-Galactosidase activity assay for expression of hlyA (C) and hlyD (D). hlyA-lacZ strains were grown on blood agar plates overnight at 37°C under aerobic and anaerobic conditions, and hlyD-lacZ transcriptional fusion strains were grown on blood agar plates overnight at 37°C under anaerobic conditions. β-Galactosidase activity was measured, and the values shown are means plus standard deviations for triplicate samples from three independent experiments. Significant differences are indicated by asterisks (**, P < 0.001 compared to the WT and mutant; ***, P < 0.0001 compared to the WT and mutant).
We then checked if FNR affected the expression of hlyCABD. Since it was reported that hlyC, hlyA, hlyB, and hlyD share only one promoter (30) and there is a rho-independent terminator localized between hlyA and hlyB in E. coli (31), we constructed hlyA-lacZ and a hlyD-lacZ fusions in the chromosome of CFT073. The fnr mutation significantly downregulated the expression of both hlyA (Fig. 8C) and hlyD (Fig. 8D) under anaerobic conditions, but no significant difference was found under aerobic conditions. The molecular regulatory mechanisms of hlyCABD regulation by FNR in UPEC CFT073 are currently being examined in our laboratories.
DISCUSSION
The global regulator FNR has been recognized to be a major transcription factor involved in bacterial gene expression which participates in numerous regulatory pathways to allow facultative anaerobes to adjust to oxygen deprivation (32). FNR is likely to be important for the virulence of bacterial pathogens that encounter hypoxic and anoxic niches within their host. It serves as the O2 sensor to reprogram bacterial metabolism by activating the genes required for anaerobic respiration and fermentation and inhibiting the genes necessary for aerobic respiration. Accordingly, previous studies demonstrated that FNR of Bordetella pertussis (33), Neisseria meningitidis (34), Pseudomonas aeruginosa (35), and Salmonella enterica serovar Typhimurium (8) is required for optimal growth and survival in vivo. Proteomic analyses also showed that Shigella dysenteriae type 1 switched from aerobic respiration in vitro to anaerobic catabolism in vivo (36). In addition to facilitating adaptation to hypoxic and anoxic metabolism during host colonization and infection, FNR also triggers virulence gene expression and coordinates the function of a type III secretion system, thus being involved in bacterial pathogenesis (36). In this study, we have explored the role of the global regulator FNR in the virulence of UPEC beyond facilitating adaptation to anaerobic metabolism.
The virulence-associated phenotype observed for the fnr mutant was significantly reduced adherence to and invasion of bladder and kidney cells. The ability of UPEC to adhere to and invade cultured bladder and kidney epithelial cells is dependent on the expression of fimbrial and afimbrial adhesins. Type 1 fimbriae are among the most important adhesins of UPEC and have been shown to mediate bacterial adherence to and invasion of cells in vitro and to play a major role in the in vivo colonization process associated with urinary tract infection. In the presence of oxygen, FNR was thought to be inactive. Interestingly, our study showed that FNR could activate the expression of type 1 fimbriae under both aerobic and anaerobic conditions (data not shown) and, notably, played a dual role in the upregulation of type 1 fimbriae by promoting both promoter activity and phase variation. The FNR protein could bind to the promoter region of the fimA gene, which encodes the main subunit of the fimbriae, and directly upregulated the expression of fimA. Also, FNR could activate FimB- and FimE-mediated recombination, thus promoting phase OFF-to-phase ON switching. At this time, the mechanisms by which FNR affects the expression of fimB and fimE are still unknown, and further studies are required to fully understand the underlying mechanisms by which FNR affects both levels of regulation of type 1 fimbriae.
P fimbriae play an important role in the pathogenesis of ascending UTIs and pyelonephritis in humans (37, 38) by promoting epithelial attachment and resistance to filtrate flow, facilitating early bacterial multiplication prior to the onset of ischemia and the infiltration of immune cells (39). Using the human blood red cell agglutination assay, we could demonstrate that deletion of fnr significantly reduced the expression of P fimbriae; in addition, decreased transcription of the main structural gene, papA, was detected in the fnr mutant, suggesting that FNR activates the expression of P fimbriae. The regulation of papA by FNR appears to be direct, since the FNR protein could bind to the promoter region of the papA gene, as shown by the EMSA. Finally, we conclude that the reduced adherence and invasion ability of the fnr deletion mutant was presumably caused by the decreased expression of fimbriae, such as type 1 and P fimbriae.
The kidney is a critical site in serious urinary tract infections and is characterized by metabolites not found elsewhere in UPEC life cycle. For example, the proximal tubule cells have 10- to 40-fold higher levels of α-ketoglutarate than any other cells (40). We identified the first metabolic trait, which is encoded by a pathogenicity island, to increase UPEC's adaptability and competitiveness during kidney infection (11). The pathogenicity island encodes a novel two-component signal transduction system, KguS/KguR, and its target genes are involved in the anaerobic utilization of α-ketoglutarate. The oxygen tension was shown to modulate the expression of pathogenicity island genes directly via the KguS/KguR system, and oxygen deficiency upregulated the expression of KguS and KguR, suggesting that oxygen modulates the expression of pathogenicity island genes by controlling KguS/KguR expression. However, no sensory domain of oxygen was found in either KguS or KguR, and the mechanisms by which oxygen tension regulates the expression of KguS and KguR remain unknown. Here, we demonstrated that deletion of fnr, which mediates the regulation of oxygen in E. coli (6), substantially reduced the growth of UPEC in M9 medium with α-ketoglutarate as the sole carbon source under anaerobic conditions and deletion of fnr significantly decreased the expression of pathogenicity island genes, including kguS and kguR. Our evidence also showed that FNR directly regulates the expression of the two-component signal transduction system KguS/KguR, thus likely indirectly regulating other pathogenicity island genes, such as c5038, through KguS/KguR. It has been well-known that the global regulator FNR in E. coli K-12 controls 122 operons, which are organized into seven regulatory categories (41). To the best of our knowledge, this is the first report showing that FNR controls the expression of a metabolism island in pathogenic E. coli and contributes to bacterial fitness in vivo.
α-Hemolysin has been shown to be an important virulence factor of UPEC (42). This toxin can lyse eukaryotic cells, providing an opportunity for deeper invasion of UPEC and supplying bacteria with nutrients released from host cells (43). In addition, it has recently been demonstrated that α-hemolysin not only is a pore-forming toxin but also triggers the proteolysis of host proteins to disrupt cell adhesion, inflammatory, and survival pathways (44). The hly operon contains four genes, hlyCABD, encoding the toxin precursor HlyA and other components necessary for toxin activation and secretion (26, 52). Several environmental stimuli, for example, anaerobiosis, have been reported to induce hly expression. However, the underlying mechanisms are not completely understood (45, 46). UPEC CFT073 displayed much higher hemolytic activity under anaerobic conditions than aerobic conditions (Fig. 8), while deletion of fnr reduced the hemolytic activity of CFT073 under anaerobic conditions. The restoration of fnr with its native promoter in the fnr mutant and comparison of hemolytic activity in the resulting complemented strain and in the wild-type strain revealed that the reduced hemolytic activity is indeed due to deletion of the fnr gene. The comparative transcription studies under both aerobic and anaerobic conditions further confirmed that deletion of fnr significantly reduced the transcription of the hly operon under anaerobic conditions. These results indicate that the global regulator FNR, a well-known oxygen sensor, mediates the anaerobiosis induction of the hly operon.
Deletion of fnr also reduced the expression of the UPEC flagellum, whose expression was coincident with UPEC ascension to the upper urinary tract (15), and significantly enhanced the pathogenesis of urinary tract infection caused by UPEC (47).
In summary, our results indicate that, in addition to mediating bacterial adaptation to anaerobic metabolism in vivo, the global regulator FNR contributes to the virulence of UPEC by regulation of the expression of important virulence genes of UPEC. Several known global regulators, such as H-NS (48), RpoS (49), RfaH (50), Hfq (51), and cyclic AMP receptor protein-cyclic AMP (17), have been reported to regulate the virulence of UPEC, and now FNR can be included in the growing list of regulatory networks controlling UPEC virulence. The mechanisms by which these global regulators interplay among each other, thus orchestrating virulence gene regulation cascades in response to the host environment, are still unknown and require further studies.
Supplementary Material
ACKNOWLEDGMENTS
Nicolle L. Barbieri received a scholarship from the Brazilian sponsor agency CNPq (process number 202479/2011-0).
We gratefully acknowledge Michael J. Wannemuehler and Glenn J. Songer for providing access to an anaerobic chamber. We appreciate the kind gift of strains CFT073 lock ON and CFT073 lock OFF from Harry L. T. Mobley and plasmid pET28a-(fnrD154A)2 from Aixin Yan.
Footnotes
Published ahead of print 22 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02315-14.
REFERENCES
- 1.Lambden PR, Guest JR. 1976. Mutants of Escherichia coli K12 unable to use fumarate as an anaerobic electron acceptor. J. Gen. Microbiol. 97:145–160. 10.1099/00221287-97-2-145. [DOI] [PubMed] [Google Scholar]
- 2.Guest JR, Green J, Irvine AS, Spiro S. 1996. The FNR modulon and FNR-regulated gene expression, p 317–342 In Lin ECC, Lynch AS. (ed), Regulation of gene expression in Escherichia coli. R. G. Landes & Co, Austin, TX. [Google Scholar]
- 3.Kiley PJ, Beinert H. 1999. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22:341–352. [DOI] [PubMed] [Google Scholar]
- 4.Crack JC, Green J, Cheesman MR, Le Brun NE, Thomson AJ. 2007. Superoxide-mediated amplification of the oxygen-induced switch from [4Fe-4S] to [2Fe-2S] clusters in the transcriptional regulator FNR. Proc. Natl. Acad. Sci. U. S. A. 104:2092–2097. 10.1073/pnas.0609514104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lazazzera BA, Bates DM, Kiley PJ. 1993. The activity of the Escherichia coli transcription factor Fnr is regulated by a change in oligomeric state. Genes Dev. 7:1993–2005. 10.1101/gad.7.10.1993. [DOI] [PubMed] [Google Scholar]
- 6.Kang YS, Weber KD, Yu Q, Kiley PJ, Blattner FR. 2005. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J. Bacteriol. 187:1135–1160. 10.1128/JB.187.3.1135-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Contreras I, Toro CS, Troncoso G, Mora GC. 1997. Salmonella typhi mutants defective in anaerobic respiration are impaired in their ability to replicate within epithelial cells. Microbiology 143:2665–2672. 10.1099/00221287-143-8-2665. [DOI] [PubMed] [Google Scholar]
- 8.Fink RC, Evans MR, Porwollik S, Vazquez-Torres A, Jones-Carson J, Troxell B, Libby SJ, McClelland M, Hassan HM. 2007. FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028s). J. Bacteriol. 189:2262–2273. 10.1128/JB.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marteyn B, West NP, Browning DF, Cole JA, Shaw JG, Palm F, Mounier J, Prevost MC, Sansonetti P, Tang CM. 2010. Modulation of Shigella virulence in response to available oxygen in vivo. Nature 465:355–358. 10.1038/nature08970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mobley HLT, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai W, Wannemuehler Y, Dell'anna G, Nicholson B, Barbieri NL, Kariyawasam S, Feng Y, Logue CM, Nolan LK, Li G. 2013. A novel two-component signaling system facilitates uropathogenic Escherichia coli's ability to exploit abundant host metabolites. PLoS Pathog. 9:e1003428. 10.1371/journal.ppat.1003428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 13.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalogeraki VS, Winans SC. 1997. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene 188:69–75. 10.1016/S0378-1119(96)00778-0. [DOI] [PubMed] [Google Scholar]
- 15.Lane MC, Alteri CJ, Smith SN, Mobley HL. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc. Natl. Acad. Sci. U. S. A. 104:16669–16674. 10.1073/pnas.0607898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagberg L, Jodal U, Korhonen TK, Lidin-Janson G, Lindberg U, Svanborg Eden C. 1981. Adhesion, hemagglutination, and virulence of Escherichia coli causing urinary tract infections. Infect. Immun. 31:564–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller CM, Aberg A, Straseviciene J, Emody L, Uhlin BE, Balsalobre C. 2009. Type 1 fimbriae, a colonization factor of uropathogenic Escherichia coli, are controlled by the metabolic sensor CRP-cAMP. PLoS Pathog. 5:e1000303. 10.1371/journal.ppat.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bahrani-Mougeot FK, Buckles EL, Lockatell CV, Hebel JR, Johnson DE, Tang CM, Donnenberg MS. 2002. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol. Microbiol. 45:1079–1093. 10.1046/j.1365-2958.2002.03078.x. [DOI] [PubMed] [Google Scholar]
- 19.Maes A, Gracia C, Brechemier D, Hamman P, Chatre E, Lemelle L, Bertin PN, Hajnsdorf E. 2013. Role of polyadenylation in regulation of the flagella cascade and motility in Escherichia coli. Biochimie 95:410–418. 10.1016/j.biochi.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 20.Miller J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 21.Shan Y, Pan Q, Liu J, Huang F, Sun H, Nishino K, Yan A. 2012. Covalently linking the Escherichia coli global anaerobic regulator FNR in tandem allows it to function as an oxygen stable dimer. Biochem. Biophys. Res. Commun. 419:43–48. 10.1016/j.bbrc.2012.01.121. [DOI] [PubMed] [Google Scholar]
- 22.Shen Z, Pu XY, Zhang Q. 2011. Salicylate functions as an efflux pump inducer and promotes the emergence of fluoroquinolone-resistant Campylobacter jejuni mutants. Appl. Environ. Microbiol. 77:7128–7133. 10.1128/AEM.00763-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vigil PD, Wiles TJ, Engstrom MD, Prasov L, Mulvey MA, Mobley HL. 2012. The repeat-in-toxin family member TosA mediates adherence of uropathogenic Escherichia coli and survival during bacteremia. Infect. Immun. 80:493–505. 10.1128/IAI.05713-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergsten G, Wullt B, Svanborg C. 2005. Escherichia coli, fimbriae, bacterial persistence and host response induction in the human urinary tract. Int. J. Med. Microbiol. 295:487–502. 10.1016/j.ijmm.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 25.Eto DS, Jones TA, Sundsbak JL, Mulvey MA. 2007. Integrin-mediated host cell invasion by type 1-piliated uropathogenic Escherichia coli. PLoS Pathog. 3:e100. 10.1371/journal.ppat.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia TA, Ventura CL, Smith MA, Merrell DS, O'Brien AD. 2013. Cytotoxic necrotizing factor 1 and hemolysin from uropathogenic Escherichia coli elicit different host responses in the murine bladder. Infect. Immun. 81:99–109. 10.1128/IAI.00605-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bryan A, Roesch P, Davis L, Moritz R, Pellett S, Welch RA. 2006. Regulation of type 1 fimbriae by unlinked FimB- and FimE-like recombinases in uropathogenic Escherichia coli strain CFT073. Infect. Immun. 74:1072–1083. 10.1128/IAI.74.2.1072-1083.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eberspacher B, Hugo F, Bhakdi S. 1989. Quantitative study of the binding and hemolytic efficiency of Escherichia coli hemolysin. Infect. Immun. 57:983–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson JR. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 4:80–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Felmlee T, Pellett S, Welch RA. 1985. Nucleotide sequence of an Escherichia coli chromosomal hemolysin. J. Bacteriol. 163:94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welch RA, Pellett S. 1988. Transcriptional organization of the Escherichia coli hemolysin genes. J. Bacteriol. 170:1622–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiley PJ, Beinert H. 2003. The role of Fe-S proteins in sensing and regulation in bacteria. Curr. Opin. Microbiol. 6:181–185. 10.1016/S1369-5274(03)00039-0. [DOI] [PubMed] [Google Scholar]
- 33.Wood GE, Khelef N, Guiso N, Friedman RL. 1998. Identification of Btr-regulated genes using a titration assay. Search for a role for this transcriptional regulator in the growth and virulence of Bordetella pertussis. Gene 209:51–58. [DOI] [PubMed] [Google Scholar]
- 34.Bartolini E, Frigimelica E, Giovinazzi S, Galli G, Shaik Y, Genco C, Welsch JA, Granoff DM, Grandi G, Grifantini R. 2006. Role of FNR and FNR-regulated, sugar fermentation genes in Neisseria meningitidis infection. Mol. Microbiol. 60:963–972. 10.1111/j.1365-2958.2006.05163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trunk K, Benkert B, Quack N, Munch R, Scheer M, Garbe J, Jansch L, Trost M, Wehland J, Buer J, Jahn M, Schobert M, Jahn D. 2010. Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environ. Microbiol. 12:1719–1733. 10.1111/j.1462-2920.2010.02252.x. [DOI] [PubMed] [Google Scholar]
- 36.Kuntumalla S, Zhang Q, Braisted JC, Fleischmann RD, Peterson SN, Donohue-Rolfe A, Tzipori S, Pieper R. 2011. In vivo versus in vitro protein abundance analysis of Shigella dysenteriae type 1 reveals changes in the expression of proteins involved in virulence, stress and energy metabolism. BMC Microbiol. 11:147. 10.1186/1471-2180-11-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leffler H, Svanborg-Eden C. 1981. Glycolipid receptors for uropathogenic Escherichia coli on human erythrocytes and uroepithelial cells. Infect. Immun. 34:920–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaisanen V, Elo J, Tallgren LG, Siitonen A, Makela PH, Svanborg-Eden C, Kallenius G, Svenson SB, Hultberg H, Korhonen T. 1981. Mannose-resistant haemagglutination and P antigen recognition are characteristic of Escherichia coli causing primary pyelonephritis. Lancet ii:1366–1369. [DOI] [PubMed] [Google Scholar]
- 39.Melican K, Sandoval RM, Kader A, Josefsson L, Tanner GA, Molitoris BA, Richter-Dahlfors A. 2011. Uropathogenic Escherichia coli P and type 1 fimbriae act in synergy in a living host to facilitate renal colonization leading to nephron obstruction. PLoS Pathog. 7:e1001298. 10.1371/journal.ppat.1001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pritchard JB. 1995. Intracellular alpha-ketoglutarate controls the efficacy of renal organic anion transport. J. Pharmacol. Exp. Ther. 274:1278–1284. [PubMed] [Google Scholar]
- 41.Myers KS, Yan H, Ong IM, Chung D, Liang K, Tran F, Keles S, Landick R, Kiley PJ. 2013. Genome-scale analysis of Escherichia coli FNR reveals complex features of transcription factor binding. PLoS Genet. 9:e1003565. 10.1371/journal.pgen.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyd TA, Goldstein L. 1979. Kidney metabolite levels and ammonia production in acute acid-base alterations in the rat. Am. J. Physiol. 236:E289–E295. [DOI] [PubMed] [Google Scholar]
- 43.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140. 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 44.Dhakal BK, Mulvey MA. 2012. The UPEC pore-forming toxin α-hemolysin triggers proteolysis of host proteins to disrupt cell adhesion, inflammatory, and survival pathways. Cell Host Microbe 11:58–69. 10.1016/j.chom.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dobrindt U, Emödy L, Gentschev I, Goebel W, Hacker J. 2002. Efficient expression of the α-haemolysin determinant in the uropathogenic Escherichia coli strain 536 requires the leuX-encoded tRNA5 Leu. Mol. Genet. Genomics 267:370–379. 10.1007/s00438-002-0668-3. [DOI] [PubMed] [Google Scholar]
- 46.Nieto JM, Carmona M, Bolland S, Jubete Y, de la Cruz F, Juárez A. 1991. The hha gene modulates haemolysin expression in Escherichia coli. Mol. Microbiol. 5:1285–1293. 10.1111/j.1365-2958.1991.tb01902.x. [DOI] [PubMed] [Google Scholar]
- 47.Lane MC, Lockatell V, Monterosso G, Lamphier D, Weinert J, Hebel JR, Johnson DE, Mobley HLT. 2005. Role of motility in the colonization of uropathogenic Escherichia coli in the urinary tract. Infect. Immun. 73:7644–7656. 10.1128/IAI.73.11.7644-7656.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Müller CM, Dobrindt U, Nagy G, Emödy L, Uhlin BE, Hacker J. 2006. Role of histone-like proteins H-NS and StpA in expression of virulence determinants of uropathogenic Escherichia coli. J. Bacteriol. 188:5428–5438. 10.1128/JB.01956-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hryckowian AJ, Welch RA. 2013. RpoS contributes to phagocyte oxidase-mediated stress resistance during urinary tract infection by Escherichia coli CFT073. mBio 4(1):e00023-13. 10.1128/mBio.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beloin C, Michaelis K, Lindner K, Landini P, Hacker J, Ghigo J-M, Dobrindt U. 2006. The transcriptional antiterminator RfaH represses biofilm formation in Escherichia coli. J. Bacteriol. 188:1316–1331. 10.1128/JB.188.4.1316-1331.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kulesus RR, Diaz-Perez K, Slechta ES, Eto DS, Mulvey MA. 2008. Impact of the RNA chaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect. Immun. 76:3019–3026. 10.1128/IAI.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagy G, Altenhoefer A, Knapp O, Maier E, Dobrindt U, Blum-Oehler G, Benz R, Emődy L, Hacker J. 2006. Both α-haemolysin determinants contribute to full virulence of uropathogenic Escherichia coli strain 536. Microbes Infect. 8:2006–2012. 10.1016/j.micinf.2006.02.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.