Abstract
Acinetobacter baumannii is an extracellular opportunistic human pathogen that is becoming increasingly problematic in hospitals. In the present study, we demonstrate that the A. baumannii Omp 33- to 36-kDa protein (Omp33-36) is a porin that acts as a channel for the passage of water. The protein is found on the cell surface and is released along with other porins in the outer membrane vesicles (OMVs). In immune and connective cell tissue, this protein induced apoptosis by activation of caspases and modulation of autophagy, with the consequent accumulation of p62/SQSTM1 (sequestosome 1) and LC3B-II (confirmed by use of autophagy inhibitors). Blockage of autophagy enables the bacterium to persist intracellularly (inside autophagosomes), with the subsequent development of cytotoxicity. Finally, we used macrophages and a mouse model of systemic infection to confirm that Omp33-36 is a virulence factor in A. baumannii. Overall, the study findings show that Omp33-36 plays an important role in the pathogenesis of A. baumannii infections.
INTRODUCTION
Microbial pathogenesis is defined by factors that favor colonization (adhesion, motility, and biofilm formation), development of infection (cytotoxicity, inflammation, iron acquisition, and serum complement resistance), and persistence on either animate or inanimate surfaces (resistance to antibiotics, disinfectants, and desiccation; broad substrate utilization for growth; and biofilm formation). In relation to the development of infection, two mechanisms have been associated with intrinsic cellular defense: apoptosis and autophagy. Apoptosis is a process of programmed cell death that may be dependent on or independent of caspases. Caspases play a central role in the transduction of regulatory apoptotic signals (1). Autophagy involves cell degradation of the contents of lysosomal compartments associated with several proteins, for example, p62 and LC3B. p62 is an adapter protein that is selectively degraded by autophagy (2). This protein is known to accumulate when autophagy is inhibited (3). Likewise, when autophagy is activated, the levels of this protein decrease. LC3B is a protein that participates in maturation of the autophagosome, and it occurs in two forms: LC3B-I (cytosolic form) and LC3B-II (lipidated form and membrane-bound autophagosome).
Acinetobacter baumannii is an opportunistic, nonfermentative, nonflagellated Gram-negative bacillus that forms part of the normal flora of human skin, the gastrointestinal tract, and the upper respiratory tract. It is also common in soil and freshwater environments (4, 5). The bacterium has recently emerged to be an important nosocomial pathogen that causes pneumonia, septicemia, urinary tract infections, and meningitis (6). Consequently, it has been included in the Infectious Diseases Society of America (IDSA) hit list of the six most dangerous microbes (7). The cytotoxicity of A. baumannii has been ascribed to several proteins (8).These include two outer membrane proteins, the OmpA protein and the Omp 33- to 36-kDa protein (Omp33-36) (9, 10). However, thus far, the mechanism underlying the well-established relationship between the cytotoxicity induced by these outer membrane proteins and the development of apoptosis is not clear, and only OmpA has been studied (11, 12).
Omp33-36 (called Omp33 or Omp34 in some Acinetobacter spp.) expression is associated with resistance to carbapenem antibiotics (imipenem and meropenem). In a previous study in which our group cloned the gene encoding Omp33-36 of A. baumannii, here named the mapA (modulating autophagy protein from Acinetobacter) gene, we noted that the predicted amino acid sequence is typical of Gram-negative bacterial porins, although Omp33-36 is not very similar to other known outer membrane proteins (identity, 25 to 30%) (13).
The aim of the present study was to determine the role of Omp33-36 in the development of cytotoxicity (apoptosis and autophagy mechanisms) in several different cell types, including immune and connective tissue cells, and thus in A. baumannii infections.
MATERIALS AND METHODS
Characterization of Omp33-36 as a porin. (i) Expression of the mapA gene in Xenopus oocytes.
To express mapA (ENA accession number AJ831523) in Xenopus oocytes, the cDNA insert was excised with BamHI and ligated into Xenopus expression vectors. The mRNA was transcribed and capped using an in vitro transcription kit (Ambion T7 mMessage mMachine; AMS Biotechnology Ltd., Oxford, United Kingdom) according to the manufacturer's instructions.
Xenopus oocytes were prepared as previously described (14). Briefly, isolated oocytes were treated with collagenase, and after 24 h, healthy-looking stage V or VI oocytes were injected with either 50 nl mRNA (1 μg μl−1) or 50 nl diethyl pyrocarbonate (DEPC)-treated water. The oocyte swelling rate was determined as follows: after 3 days of incubation in ND96 [15 mM HEPES, pH 7.6, 88 mM NaCl, 1 mM KCl, 2.4 mM NaHCO3, 0.3 mM Ca(NO3)2, 0.41 mM CaCl2, 0.82 mM MgSO4] at 18°C, each oocyte was transferred into a 1:5 dilution of ND96. Changes in the cell volume were photographed with a digital camera, with images being recorded every 20 s to obtain 10 images in 2 min. The cell volume was calculated using the Sigmascan Pro (version 5) image analysis program (SPSS Inc.). The changes in protoplast volume over time were used to determine the exponential rate constant, k. The osmotic water permeability coefficient (Pf) was calculated according to the following equation: (V0/S0){k/[Vw(osmolin − osmolout)]}, where V0 is the initial cell volume, S0 is the initial cell surface area, Vw is the molar volume of water (18 cm3 mol−1), k is the fitted exponential rate constant of the initial phase of the swelling, and osmolin and osmolout are the internal and external osmolarities, respectively.
To study porin functionality, 50 μM mercuric chloride (HgCl2) was added to ND96 prior to Pf measurement, to block porin pore formation (15).
(ii) Omp33-36 purification and analysis.
The mapA gene was PCR amplified from the genomic DNA of clinical strain A. baumannii JC7/04 (13). Primer design omitted the region corresponding to the peptide signal and was based on the sequence information available for A. baumannii mapA (GenBank accession number AJ831523). NcoI and HindIII sites were introduced through the primers at the 5′ and 3′ ends of the amplicon, respectively. A stop codon upstream from the HindIII site was incorporated in the reverse primer. The following primers were used: forward primer 5′-GGAGATTATccatggCTCAATTTGAAGTTCCAAGGA-3′ and reverse primer 5′-CGACGACCCAGaagcttAAGCTTTTATTAATGATGATGATGATGATGGAAACGGAATTTAGCAT-3′, where the lowercase italic sequences represent the NcoI and HindIII restriction endonuclease sites.
Amplified mapA was subsequently cloned into the NcoI and HindIII sites of the pRAT expression vector to yield the construct pRAT-His-mapA (pRAT-His33), which was confirmed by sequencing.
Bacterial cells were grown in Luria-Bertani (LB) broth containing 100 mg ampicillin liter−1 at 37°C. Exponentially growing cultures (optical density at 600 nm [OD600] = 0.6 to 0.8) were induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Sigma, St. Louis, MO) and harvested by centrifugation after 12 h. Escherichia coli DH5α cells were resuspended in lysis buffer (50 mM Tris-HCl, pH 8, 500 mM NaCl, 5 mM EDTA) and lysed by sonication. Inclusion bodies were harvested by centrifugation at 17,000 × g for 20 min in a buffer containing 50 mM Tris-HCl, pH 8, 500 mM KCl, 5 mM EDTA, 1% sodium deoxycholate, 1% lauryl sulfobetaine, and 1% Triton X-100, before being recentrifuged and washed several times with water to remove the detergent. The concentration of total Omp33-36 was measured by UV spectroscopy, and the protein was analyzed by SDS-PAGE.
Inclusion bodies of His-tagged Omp33-36 were solubilized in 5 M guanidinium chloride buffer containing 50 mM Tris-HCl, pH 8. The protein was isolated by Ni-nitrilotriacetic acid (NTA) affinity chromatography by means of an Akta purifier system (GE Healthcare) and Ni-NTA columns (HisTrap; HP Amersham Biosciences). The eluent consisted of a linear gradient of 0 to 500 mM imidazole in solubilization buffer. The resulting 25 ml of dilute protein solution was dialyzed overnight against 5 liters of water and centrifuged at 17,000 × g. The resulting pellet was resuspended in 50 mM Tris-HCl, pH 8, containing 5 M guanidinium chloride. Pooled fractions typically contained 2 mg protein/ml.
Edman degradation analysis of the N-terminal sequence was carried out with a Procise 494 analyzer (Applied Biosystems, Foster City, CA). The amino acid sequence of the N-terminal region of the 33- to 36-kDa polypeptide was as follows: Tyr-Gln-Phe-Glu-Val-Gln-Gly-Gln-Ser-Glu.
Refolding buffer (20 mM Tris hydrochloride [Tris-HCl], pH 8, 1 mM EDTA, 1% N,N-dimethyldodecylamine N-oxide [LDAO; Fluka, St. Louis, MO]) was cooled at 4°C for 30 min with stirring, after which Omp33-36 (2 mg/ml of solution buffer) was added and the mixture was quickly diluted 1:5 in a buffer containing 1% LDAO, 1 mM EDTA, and 20 mM Tris-HCl, pH 8. The protein solution was dialyzed against 5 liters of a buffer containing 20 mM Tris-HCl, pH 8, 1 mM EDTA, and 0.1% LDAO for 48 h at 4°C with two changes of the same buffer. The refolded protein (2 mg/ml) showed no signs of precipitation after 12 h.
Small aggregates and particulate matter were removed from the protein solution by ultracentrifugation at 17,000 × g for 30 min at 4°C. Omp33-36 was then further purified by ion-exchange chromatography and gel filtration chromatography. A fast-flow 5-ml Q-Sepharose column (HiTrap Q FF; Amersham Biosciences) was equilibrated with 3 bed volumes of binding buffer (20 mM Tris-HCl, pH 8, 150 mM NaCl, 0.1% LDAO) and with 1 bed volume of elution buffer (20 mM Tris-HCl, pH 8, 1 M NaCl, 0.06% LDAO). Omp33-36 was loaded onto the column at a flow rate of 2 ml/min and then purified with Q-Sepharose by using a linear gradient of 0 to 1 M NaCl in 20 mM Tris-HCl, pH 8, and 0.06% LDAO. The protein was concentrated by filtration through a Falcon 10-kDa-molecular-mass-cutoff membrane (Roche). For gel filtration chromatography, the Sephacryl S-300 (HiPrep 16/60 Sephacryl S-300 high-resolution) column was equilibrated with 3 volumes of 20 mM Tris-HCl, pH 8, 150 mM NaCl, and 0.06% LDAO. Omp33-36 was loaded and then eluted with the same buffer at a flow rate of 0.5 ml/min.
To study the secondary structure of Omp33-36, circular dichroism spectra were recorded in a Jasco J-810 spectropolarimeter (Jasco, Easton, MD) at ambient temperature. Omp33-36, at a concentration of 0.2 mg/ml in 20 mM Tris-HCl, pH 8, 150 mM NaCl, and 0.06% LDAO, was placed in a quartz cuvette with a 0.2-cm path length.
To rule out the presence of potential bacterial contaminants, such as either peptidoglycan (PGC) or lipopolysaccharide (LPS), an assay was carried out using cell lines permanently expressing Toll-like receptor 2 (TLR2) and TLR4, HEK293/TLR2 (human embryonic kidney 293 cells stably transfected with the human TLR2a gene) and HEK293/TLR4/MD2/CD14 (human embryonic kidney 293 cells stably transfected with TRL4a, MD2, and CD14 genes) cells, respectively (16) (Invitrogen, Toulouse, France). The cells were plated at a concentration of 5 × 105 cells/well and exposed to 10 μg Omp33-36/ml for 4 h. The negative control consisted of unstimulated cells. The positive controls were HEK293/TLR2 cells stimulated with 0.01, 1, or 10 μg PGC (from Bacillus subtilis; Sigma-Aldrich)/ml or HEK293/TLR4/MD2/CD14 cells stimulated with 0.01, 1, or 10 μg LPS (from E. coli O111:B4; Sigma-Aldrich)/ml. The interleukin-8 (IL-8) secreted into the medium by these cells in response to either stimulant was detected by a conventional enzyme-linked immunosorbent assay (ELISA; interleukin-8 high-sensitivity human ELISA set; ImmunoTools). The ELISA reaction was coupled to alkaline phosphatase, and the reaction product was detected at a wavelength of 405 nm in an ELISA reader (Lab Systems Multiscan Plus).
Apoptosis in Omp33-36-treated cells. (i) Fluorescence microscopy studies of cell morphology and flow cytometry analysis of DNA fragmentation.
HEp-2 cells were seeded in a chamber slide (Lab-Tek, Rochester, NY) at a concentration of 5 × 104 cells/well and treated with 8 μg Omp33-36/ml for 4 or 12 h. To prevent apoptosis, 50 mM Z-VAD-FMK (BD Biosciences), a general caspase inhibitor, was added to the cells for 1 h. When required, the cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min, washed twice with wash buffer (PBS and 0.05% Tween 20), permeabilized with 0.1% Triton X-100 in PBS for 1 to 5 min, and finally, washed three times. In colocalization experiments, live mitochondria were stained with 10 μg JC-1/ml and the J-9 mitochondrial potential sensor (Molecular Probes, Eugene, OR) for 30 min at 37°C before fixation. Staining with the F-actin probe fluorescein isothiocyanate (FITC)-phalloidin (Sigma) was performed following the manufacturer's recommendations. Phosphatidylserine flipping from the inner to the outer leaflet of the plasma membrane was assessed by in vitro binding of the FITC-coupled serum factor annexin V, as detected by fluorescence microscopy. Annexin V-FITC was added to cells seeded in a chamber slide, and the cells were treated with 8 μg Omp33-36/ml for 4 and 12 h. After incubation for 15 min in the dark and one wash with PBS, the cells were counterstained with 1 μg propidium iodide (PI)/ml to determine the fraction of permeable (necrotic) cells. The coverslips were mounted with Ultracruz mounting medium, and the cells were stained with DAPI (4′,6-diamidino-2-phenylindole; Santa Cruz Biotech, Santa Cruz, CA), which stains bacterial and eukaryotic DNA. The apoptotic index (percentage of apoptotic cells) was determined microscopically at 4 and 12 h by use of an Olympus BX61 fluorescence microscope and DP manager software.
To study DNA fragmentation, HEp-2 and HeLa cells were cultured in 6-well microtiter plates (5 × 105 cells/well) and challenged with 8 μg Omp33-36/ml for 12 h. Untreated cells were used as the negative control. To ensure that Omp33-36 exposure was responsible for the observed fragmentation, the protein (8 μg/ml) was incubated with the same volume of an undiluted rabbit polyclonal antibody against Omp33-36 for 1 h at 4°C. At the end of the incubation period, the protein that had been incubated with the polyclonal antibody was added to the cells. To exclude the possibility that the antibodies themselves had an effect on HEp-2 and HeLa cells, the same amount of antibody alone was added to the cultures; no effect on the cells was observed (data not shown). DNA fragmentation in HEp-2 and HeLa cells exposed to 8 μg Omp33-36/ml for 12 h was determined with a modified terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay (APO-BrdU TUNEL assay kit), as described by the manufacturer (BD Pharmingen). Adherent cells (which were detached with trypsin) and detached cells were collected separately, and 20,000 cells per sample were assessed in a BD FACSCalibur flow cytometer.
(ii) Caspase detection by immunoblotting.
The cells were lysed in lysis buffer (50 mM Tris-HCl, 10% glycerol, 1% Triton X-100, 0.1% SDS, 150 mM NaCl, 5 mM EDTA, 5 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 50 mM NaF, 50 mM phosphate glycerol, protease inhibitor cocktail [Roche]) for 30 min on ice. The lysates were cleared by centrifugation and quantified in a NanoDrop ND-1000 spectrophotometer. Proteins from each sample were separated by SDS-PAGE on 12% acrylamide gels, followed by electrotransfer onto a polyvinylidene difluoride membrane (Roche). The blots were blocked in 5% nonfat skimmed milk and incubated with primary antibodies. Proteins were visualized using horseradish peroxidase-conjugated secondary antibodies, followed by enhanced chemiluminescence (ECL Plus kit; Amersham Pharmacia Biotech), and detected with a LAS3000 chemiluminescence detector (Fujifilm).
HEp-2 cells treated with Omp33-36 were incubated with primary antibodies against the following proteins, which were added to the medium at the concentrations recommended by the manufacturer: procaspase 3/caspase 3, procaspase 9/caspase 9 (BD Pharmingen), and tubulin (Sigma).The studies were conducted in three different experiments.
Autophagy studies. (i) Electron microscopy analysis of alterations in cellular structures.
HEp-2 cells incubated for 4, 12, and 20 h with 8 μg Omp33-36/ml were rinsed, detached, and centrifuged. The resulting pellets were immediately fixed in 2.5% cold glutaraldehyde in 0.2 M sodium cacodylate buffer, pH 7.4, for 4 h at room temperature and postfixed in 1% osmium tetroxide in 0.1 M sodium cacodylate buffer, pH 7.4, for 1 h at 4°C. The pellets were then dehydrated in a graded acetone series and embedded in Spurr medium. Semithin sections (1 μm thick) were cut, stained with toluidine blue sodium tetraborate, and observed under a light microscope. Ultrathin sections (70 nm) obtained from the same specimens were stained with uranyl acetate and lead citrate and were observed by transmission electron microscopy (TEM; JEOL JEM 1010 microscope, 80 kV).
(ii) Real-time PCR studies, immunoblotting, and electron microscopy analysis of the modulation of the autophagy in mapA-transfected HeLa Tet-On cells.
The mapA gene without a signal peptide was PCR amplified from the A. baumannii JC7/04 chromosome, using the primers CRLOmpFW (gcggccgcATGTATCAATTTGAAGTTCA) and CRLOmpRV (gtcgacTTAGAAACGGAATTTAGCATTTAA) (where the lowercase italic sequences represent the NotI and SalI restriction endonuclease sites), and cloned into pTRE2Hyg, a mammalian expression vector (Clontech). The resulting plasmids were purified with a High Pure plasmid isolation kit (Roche). A Savant SpeedVac concentrator (SPD121P; Thermo Scientific) was used to obtain DNA of the appropriate concentration and purity.
A HeLa Tet-On Advanced (HTAC) cell line (Clontech) was transfected with pTRE2Hyg-mapA by using a FuGene HD transfection kit (Roche) following the manufacturer's instructions. Positive clones harboring pTRE2Hyg-mapA (HTAC-pTRE33Hyg cells) were obtained using the method described in the Tet-Off and Tet-On gene expression systems user manual (Clontech). Briefly, at 48 h posttransfection, 200 μg/ml of hygromycin was added as a selective agent for the transfected cells; the medium was replaced every 4 days. After 3 weeks, hygromycin-resistant colonies were isolated and transferred to individual wells. As a negative control, the HeLa Tet-On cells were transfected with pTRE2Hyg as described above.
To produce a stably transfected cell line, all hygromycin-resistant colonies were induced with 1 μg doxycycline/ml for 24, 48, and 72 h, and mapA gene expression was confirmed by real-time reverse transcription-PCR (RT-PCR). DNase-treated RNA was obtained from 106 cells (0.4 to 0.6 absorbance units) by using a High Pure RNA isolation kit (Roche, Germany) and 100 ng of RNA per sample. Controls without reverse transcriptase confirmed the absence of contaminating DNA in the samples. For the RT-PCR studies, a LightCycler 480 RNA master hydrolysis probe (Roche, Germany) and UPL TaqMan probes (Roche, Germany) were used together with the following probes and primers, and the concentrations were adjusted to achieve efficiencies of 90 to 110%: for mapA, foward primer 5′-CAAGATGCTGTAACTGCTCGTACT-3′, reverse primer 5′-CAATAGCCATGTTAGTGCCATC-3′, and probe sequence 5′-TGGCAACA-3′(number 145); for the β-actin gene, foward primer 5′-TCCTCCCTGGAGAAGAGCTAC-3′, reverse primer 5′-GGCTGGAAGAGTGCCTCA-3′, and probe sequence 5′-CAGGCAGC-3′ (number 27).
Gene expression was normalized using the single-copy housekeeping gene β-actin and then calibrated relative to the expression of that gene, which was assigned a value of 1.0, in HTAC-pTRE33Hyg cells without induction. All experiments were performed in triplicate. The clone with the highest level of mapA expression was selected for the experiments. In the case of HeLa cells transfected with pTRE2Hyg, one hygromycin-resistant colony was selected at random for further experiments.
Studies of the expression of p62 were carried out with the following probe and primers: probe sequence 5′-CTGCTGGG-3′ (number 3), forward primer 5′-GGGCTGAAATGGCATGAG-3′, and reverse primer 5′-GGCAAGGTCCTGTGAGTAGG-3′.
As before, gene expression was normalized using the β-actin housekeeping gene and then calibrated relative to the expression of this gene, which was assigned a value of 1.0, in HTAC-pTRE2Hyg cells.
Cell death markers were assessed by immunoblotting mapA-transfected HeLa cells as described above. Anti-LC3B (Sigma) and a polyclonal antibody against Omp33-36 served as primary antibodies.
In TEM studies, HeLa cells transfected with pTRE2Hyg or pTRE2Hyg-mapA (HTAC-pTRE33Hyg cells) were induced with 1 μg doxycycline/ml for 72 and 24 h, respectively. These cells were rinsed, detached, and harvested by centrifugation. The resulting pellets were fixed immediately, as described above, before being dehydrated. Semithin (1-μm) sections were obtained and observed by light microscopy. In addition, ultrathin (70-nm) sections were prepared for TEM and analyzed using a JEOL JEM 1010 transmission electron microscope (80 kV).
(iii) Immunoblot assays with autophagy inhibitors.
The p62, LC3B, and caspase 8 levels were studied in wild-type (wt) murine embryonic fibroblasts (MEFs) incubated with 8 μg Omp33-36/ml. Moreover, p62 and LC3B levels were analyzed in combination with the autophagy inhibitors wortmannin (1 μM; an inhibitor of the class III phosphatidylinositol 3-kinase [PI3K] activity) and bafilomycin (0.1 μM; an inhibitor of the fusion between autophagosomes and lysosomes) for 0, 8, 16, and 24 h.
Analysis of the relationship between apoptosis and autophagy. (i) Immunoblotting studies of apoptosis and autophagy.
Fibroblasts in which the bax and bak genes were knocked out (bax−/− bak−/− MEFs) (17) were incubated with 8 μg Omp33-36/ml. The autophagy markers p62 and LC3B were analyzed in immunoblots of the cells prepared after 0, 8, 16, and 24 h of treatment. In addition, wt MEFs were pretreated for 30 min with 5 μM the apoptosis inhibitor Z-VAD-FMK and also incubated with 8 μg Omp33-36/ml for 0, 8, 16, and 24 h.
p62, LC3B, and caspase 8 (apoptosis activity) markers were also analyzed in similarly treated autophagy-defective fibroblasts (atg5−/− MEFs).
(ii) Apoptosis and modulation of autophagy in relation to the ROS response.
Cellular oxidative stress after Omp33-36 treatment was assessed using 10 μM the reactive oxygen species (ROS)-sensitive probe 2′,7′-dichlorofluorescein diacetate (DCFH-DA; Fluka, St. Louis, MO). Briefly, RAW 264.7 cells were treated for 8, 16, and 24 h with 8 μg Omp33-36/ml. Untreated cells and LDAO-treated cells were used as negative controls. Moreover, cells pretreated for 30 min with 20 mM N-acetyl-I-cysteine (NAC1; a chemical inhibitor of ROS; Sigma-Aldrich, Germany) and incubated with 8 μg Omp33-36/ml for 24 h were used as controls. The cells were labeled with 10 μM DCFH-DA in Dulbecco modified Eagle medium (DMEM) for 30 min and then treated again for 15 min either with 8 μg Omp33-36/ml or, for the negative controls, with LDAO and 20 mM NAC1. The cells were then washed once with physiological saline solution, harvested, resuspended in PBS, and finally, visualized by flow cytometry analysis in the FL1 channel of a BD FACSCalibur system (Becton Dickinson, Rutherford, NJ, USA). Fluorescence levels denoted the percentage of cells positive for ROS production. p62, LC3B, caspase 8, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase; Sigma) antibodies were analyzed in RAW 264.7 cells that had been pretreated for 30 min with 20 mM NAC1 and incubated with 8 μg Omp33-36/ml for 0, 8, 16, and 24 h.
Proteomic and immunoblotting analysis of OMVs carrying Omp33-36 in A. baumannii ATCC 17978 and A. baumannii ATCCΔmapA.
A stable mutant carrying the mapA gene (locus tag A1S_3297; ENA accession number AM039631) from the full genome of A. baumannii ATCC 17978 (A. baumannii ATCCΔmapA) and free of foreign plasmid DNA was obtained by gene replacement via double-crossover recombination, confirmed by proteomic and immunoblotting analyses (18).
A. baumannii ATCC 17978 and A. baumannii ATCCΔmapA were cultured in 1 liter of LB broth containing 500 mM NaCl (19), and outer membrane vesicles (OMVs) were extracted following the protocol described by Rumbo et al. (20).
The autophagic markers LC3B and p62 were analyzed in immunoblots of RAW 264.7 cells incubated with 50 μg of OMVs from A. baumannii ATCC 17978 and A. baumannii ATCCΔmapA strains for 0, 8, 16, and 24 h. Moreover, a polyclonal antibody against Omp33-36 was used to confirm the presence of the protein in OMVs from A. baumannii ATCC 17978.
To observe OMV release from both isolates by electron microscopy, the cells were prepared according to the above-described protocol.
Proteomics studies were carried out by two-dimensional gel electrophoresis (2-DE) of the OMVs to detect the presence of Omp33-36. The protein concentration in the extracts was determined by a modified Bradford assay, by using a protein assay kit (Bio-Rad, Germany). For isoelectric focusing, an IPGphor III system (Amersham GE Healthcare, USA) was used with 3-10 nonlinear immobilized pH gradient (IPG) strips (13 cm; GE Healthcare, Uppsala, Sweden). Proteins were solubilized in 8 M urea, 2% (wt/vol) CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 40 mM dithiothreitol (DTT), and 0.5% (vol/vol) IPG buffer (GE Healthcare). Isoelectric focusing was carried out at 30 V for 12 h, followed by 250 V for 1 h, 500 V for 1.5 h, 1,000 V for 1.5 h, a gradient to 8,000 V over 1.5 h, and maintenance at 8,000 V for a further 4 h. All steps were carried out at 20°C. Prior to the second dimension (SDS-PAGE), the focused IPG strips were equilibrated twice for 15 min each time in buffer containing 50 mM Tris-HCl, pH 8.8, 6 M urea, 30% (vol/vol) glycerol, and 2% (wt/vol) SDS. A trace of bromophenol blue-DTT (1%, wt/vol) was added to the mixture for the first equilibration step, and 2.5% (wt/vol) iodoacetamide was added to the mixture for the second equilibration step. SDS-PAGE was performed on 12% or 15% polyacrylamide gels. Analytical 2-DE gels were stained with silver nitrate, and the gels were loaded with 25 μg of total protein. For preparative gels, mass spectrometry-compatible silver staining, in which the gels were loaded with at least 150 μg of total protein, was carried out. The gels were scanned with an image scanner (GE Healthcare, Biosciences), and the data were analyzed with Image Master Platinum software, v.6.0 (GE Healthcare, Biosciences). For matrix-assisted laser desorption ionization–tandem time of flight (MALDI-TOF/TOF) mass spectrometry, the selected spots were excised from the gels and destained for 5 min with a solution containing 20% (wt/vol) sodium thiosulfate and 1% (wt/vol) potassium ferricyanide. The supernatant was removed and the gel spots were washed twice with 25 mM ammonium bicarbonate in 50% (vol/vol) acetonitrile for 20 min and then in acetonitrile, dried in a SpeedVac apparatus (Savant), and digested overnight with 20 μg trypsin (Roche, USA)/ml in 25 mM ammonium bicarbonate at 37°C. When necessary, tryptic peptides were passed through C18 Zip-Tip pipette tips (Millipore, USA), mixed with 3 mg α-cyano-4-hydroxycinnamic acid/ml (as the matrix) in 0.1% trifluoroacetic acid in 50% acetonitrile, and subjected to MALDI-TOF/TOF analysis (4800 proteomics analyzer; Applied Biosystems). For protein identification, databases (NCBInr) were queried with Mascot data files obtained from the MALDI-TOF/TOF mass spectral data. The Mascot scores for all protein identifications were higher than the accepted threshold of significance (P < 0.05). In the case of the Mascot results, monoisotopic masses were used to search the databases, allowing a peptide mass accuracy of 0.6 Da and one partial cleavage. Methionine oxidation and carbamidomethyl modification of cysteine were considered.
Electron microscopy of the bacteria and the autophagosome structure.
For TEM studies, RAW 264.7 macrophages were incubated for 12 h with A. baumannii ATCC 17978 cells and prepared as described above. Ultrathin sections (70 nm) were examined and photographed with a JEOL JEM 1010 transmission electron microscope (80 kV).
Intracellular persistence of A. baumannii ATCC 17978 and ATCCΔmapA strains and virulence in a mouse model of infection.
To assay survival within macrophages, 106 exponentially growing (OD600 = 0.4 to 0.6) bacterial cells from A. baumannii strains ATCC 17978 and ATCCΔmapA were collected and cultured with RAW 264.7 macrophages for 12 and 16 h in 6-well plates at a ratio of 20 to 50 bacteria per cell. The plates were then washed three times with serum, after which DMEM supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and 15 μg colistin/ml was added to eliminate external bacteria. After incubation for 2 h, the macrophages were lysed with 1 ml of 0.5% deoxycholate and the lysates (6 wells/strain) were seeded onto LB plates.
Male BALB/c mice (age, 5 to 8 weeks; weight, approximately 20 g) were housed under specific-pathogen-free conditions with antibiotic-free pelleted food and autoclaved water provided ad libitum. All experiments involving mice were approved by the Animal Ethics Committee from an outside organization made up of institutions affiliated with this work.
A. baumannii strains ATCC 17978 and ATCCΔmapA were analyzed for virulence by use of the model previously described by Smani et al. (10). Two groups of 10 mice each were infected intraperitoneally with 0.5 ml of a bacterial suspension containing 5% porcine mucin and either 3.5 × 104 CFU of A. baumannii strain ATCC 17978 or 4 × 104 CFU of A. baumannii strain ATCCΔmapA and monitored for signs of disease over 7 days. Each strain was grown for 18 h at 37°C in LB broth, and the number of cells was adjusted to the appropriate concentration with physiological saline. Inocula were prepared by mixing the bacterial suspensions 1:1 (vol/vol) with a 10% (wt/vol) solution of porcine mucin (Sigma). The bacterial concentrations of the inocula were determined by plating on LB agar. Survival data were compared using the log-rank test, and differences with a P value of <0.0001 were considered statistically significant.
RESULTS AND DISCUSSION
Omp33-36 is a porin. (i) Expression of Omp33-36 in Xenopus oocytes.
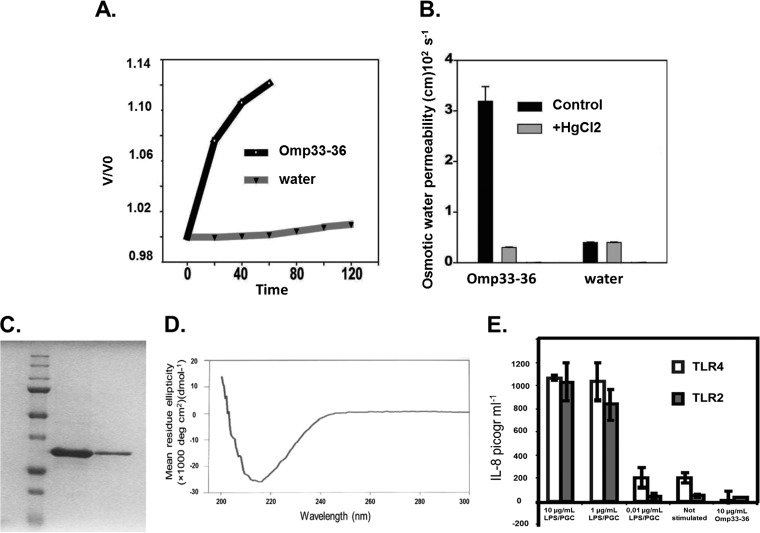
To assess whether Omp33-36 acts as an outer membrane channel, we expressed its cRNA in Xenopus oocytes and then determined the permeability (Pf) of the cells to water (Fig. 1A and B). The values for the Pf of oocytes expressing Omp33-36 to water were higher than those of control oocytes (Fig. 1A), suggesting that Omp33-36 permits the passage of water and is thus similar to a porin. Outer membrane proteins that form water channels are functionally inhibited by mercurial compounds (21, 22). Thus, the addition of 50 μM mercuric chloride strongly inhibited Omp33-36, reducing the Pf of oocytes expressing Omp33-36 to water to that of control oocytes (Fig. 1B). These results confirm that Omp33-36 acts as a porin that allows the passage of water in A. baumannii.
FIG 1.
(A) Omp33-36 acts as a channel (porin) for the passage of water. The x axis shows the time course of osmotic swelling of individual Xenopus oocytes injected with water or the cRNA of Omp33-36 from A. baumannii. Oocytes in ND96 were perfused at time zero with a 5-fold dilution of ND96 in distilled water. On the y axis, V0 is the initial cell volume and V is the cell volume after treatment. (B) The porin is an aquaporin. The osmotic permeability (Pf) to water of individual oocytes injected with water or the cRNA of Omp33-36 from A. baumannii (bars labeled Omp33-36) was determined. Oocytes in ND96 were perfused at time zero with a 5-fold dilution of ND96 in distilled water (bars labeled water). The Pf values reflect the volume changes determined in three different oocytes. When indicated, the inhibition assay was performed in the presence of mercury (50 μM HgCl2, 10 min preincubation) dissolved in the same distilled water used for diluting ND96. (C) Purification of Omp33-36. A Coomassie blue-stained gel of purified Omp33-36 from A. baumannii is shown. SDS-PAGE on a 12% acrylamide gel did not reveal any bacterial contaminants in 16 μg of purified Omp33-36. (D) β-Barrels as a secondary structural feature of the porin. The circular dichroism spectrum of Omp33-36 in Tris-HCl, pH 8, 150 mM NaCl, and 0.06% LDAO is shown. The Omp33-36-LDAO spectrum revealed β-barrels as the secondary structure. (E) Absence of contaminants in Omp33-36. The results of the IL-8 ELISA performed with the culture medium of HEK293/TLR4 cells stimulated for 4 h with 10, 1, or 0.01 μg of E. coli LPS/ml, not stimulated, or stimulated with 10 μg pure Omp33-36/ml are shown. HEK293/TLR2 cells were challenged in the same way, except that commercial peptidoglycan was used. Pure Omp33-36 did not contain any bacterial contaminants that triggered TLR2 or TLR4 activation.
(ii) Purification of A. baumannii Omp33-36 and the β-barrel structure of the protein.
Omp33-36 was overproduced in E. coli cells, then purified first on a Ni-NTA affinity column and then by ion-exchange and size exclusion chromatography, and finally, concentrated in Tris-HCl, pH 8, 150 mM NaCl, and 0.06% LDAO. The aim of this three-step purification protocol was to remove any bacterial contaminants that may have coeluted with the protein (Fig. 1C), especially major components of the bacterial membrane.
The circular dichroism spectrum of purified Omp33-36 (Fig. 1D) was consistent with a secondary structure rich in β-barrels, which are also a major feature of other bacterial porins (22).
To confirm that purified Omp33-36 induces cytotoxicity in human cells, it was first necessary to rule out the presence of bacterial contaminants, such as LPS and PGCs, which may have coeluted with the protein during the purification process. The toxic effects of LPS and PGCs are mediated by specific Toll-like receptors, which in turn stimulate interleukin-8 (IL-8) secretion (23, 24). Accordingly, we measured IL-8 release from HEK293 cells permanently transfected with a plasmid encoding either the PGC receptor Toll-like receptor 2 (TLR2) or the LPS receptor Toll-like receptor 4 (TLR4) (16) and stimulated with purified Omp33-36. Omp33-36 did not induce the release of IL-8 into the medium in either cell line, as the results were the same as those obtained with the negative noninduced controls (Fig. 1E). Moreover, when HEK293/TLR2 and HEK293/TLR4 cells were stimulated with LPS and PGCs at concentrations as low as 1 μg/ml, the levels of IL-8 released were higher than the those released by cells in which IL-8 release was induced by the presence of Omp33-36 (P < 0.001). We therefore concluded that the Omp33-36 preparation was free of bacterial contaminants.
Porins are fundamental for bacterial survival, mediating the diffusion of nutrients into the cell and toxic metabolites out of the cell. Our findings are consistent with those of Smani et al. (10), who reported that the loss of Omp33-36 was associated with decreased fitness in A. baumannii, probably because of reduced nutrient intake.
Omp33-36 induces apoptosis in human cells. (i) Omp33-36 produces apoptotic morphological changes in human cells.
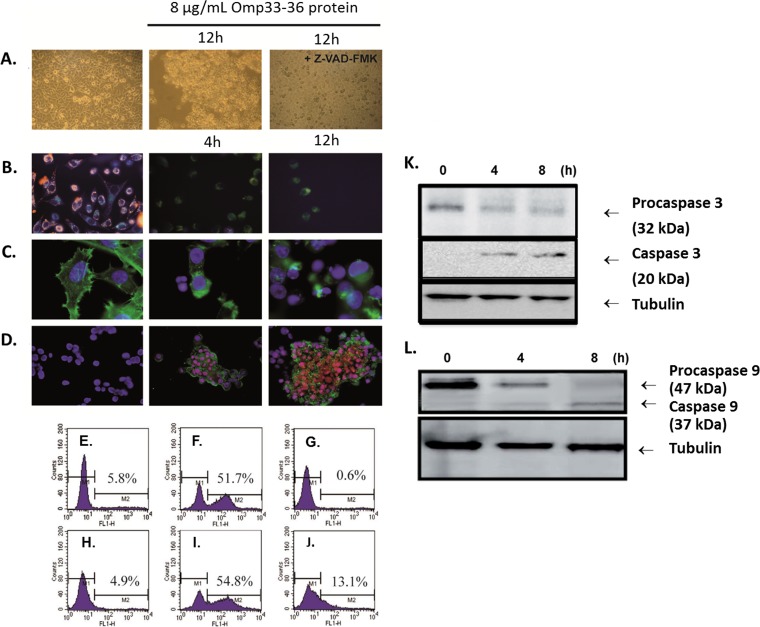
Apoptosis, or programmed cell death, differs from necrosis in that it is associated with the appearance of specific morphological features: cellular shrinkage, blebbing of the cytoplasmic membrane, chromatin condensation, nuclear fragmentation, and DNA cleavage (Fig. 2A). To determine whether Omp33-36 induces apoptosis in human cells, we challenged a healthy population of HEp-2 human epithelial cells for 12 h (Fig. 2A, center) with a concentration of purified Omp33-36 equal to that used by Choi and colleagues in experiments in which apoptosis was induced by the porin OmpA (11).We observed cellular shrinkage, membrane blebbing, and rounded cells that detached from the culture plate, as reported by Choi et al. (11) for A. baumannii OmpA. These morphological changes were reversed by the addition to the culture of 5 μM Z-VAD-FMK, which inhibits apoptosis induced by diverse stimuli (Fig. 2A, right).
FIG 2.
(A) Morphological changes indicative of apoptosis by bright-field microscopy. The changes in HEp-2 cells incubated with 8 μg/ml of Omp33-36 for 12 h (center and right) and in the absence of this protein (left) are shown. The cells were pretreated for 1 h with 5 μM the caspase inhibitor Z-VAD-FMK (right). Magnifications, ×20. (B to D) Apoptotic features analyzed by fluorescence microscopy. (B) Untreated cells accumulated JC-1 (red) in polarized mitochondria (left). In cells incubated with Omp33-36 for 4 h (center) or 12 h (right), the green monomeric form of JC-1 accumulated throughout the cytoplasm (both images, center and right). Magnifications, ×40. (C) Costaining of the cytoskeleton with phalloidin and of nuclear DNA with DAPI. Round nuclei and a structured cytoskeleton are evident in untreated cells (left), whereas cells incubated with Omp33-36 for 4 h (center) or 12 h (right) show a collapsed cytoskeleton and apoptotic bodies. Magnifications, ×100. (D) Treated cells were stained with DAPI, PI, and annexin V, and untreated cells were treated with DAPI only. Cells incubated with Omp33-36 for 4 h (center) and 12 h (right) were stained with annexin V and in some cases with PI, which specifically stains fragmented DNA. Magnifications, ×40. The images are representative of those from three independent experiments, in which at least 50 cells were scored each time. (E to J) Flow cytometric analysis of apoptosis by staining by use of an APO-BrdU kit (TUNEL assay). HEp-2 cells (E to G) and HeLa cells (H to J) were seeded at a density of 5 × 105 cells and were then either challenged with 8 μg Omp33-36/ml (F, G, I, J) shortly thereafter or left untreated (E, H). To block Omp33-36, the cells were incubated for 1 h at 4°C with excess anti-Omp33-36 antibody, which was added to the cultures together with Omp33-36 (G, J). After 12 h, the cells were harvested and analyzed for apoptosis, as described in Materials and Methods. (K, L) Caspase cleavage during Omp33-36-induced apoptosis. HEp-2 cells were challenged at different times with 8 μg Omp33-36/ml. Cell lysates were resolved by SDS-PAGE on 12% gels and immunoblotted with specific antibodies. Tubulin served as a loading control. (K) Lysates prepared from HEp-2 cells exposed to 8 μg/ml of Omp33-36 for 4, 8, and 12 h were blotted against procaspase 3 and caspase 3. (L) Lysates prepared from HEp-2 cells exposed to 8 μg/ml of Omp33-36 for 4 and 8 h were blotted against procaspase 9 and caspase 9.
Changes in mitochondrial permeability (Fig. 2B), induced by loss of the electrochemical gradient across the mitochondrial membrane, are an early feature of apoptosis. In the mitochondria of healthy cells, the dye JC-1 is seen as fluorescent red aggregates (Fig. 2B, left). With the onset of apoptosis, the mitochondrial membrane potential is lost and JC-1 no longer accumulates in the mitochondria, remaining in the cytoplasm as green fluorescent monomers (Fig. 2B, center and right). The cytoskeleton of the studied cells was stained with FITC-phalloidin, and the nuclear DNA was stained with DAPI (Fig. 2C). In HEp-2 cells exposed to Omp33-36, there was a clear loss of cytoskeletal structure, accompanied by cytoplasmic retraction (Fig. 2C, center and right). We also followed Omp33-36-induced apoptosis in HEp-2 cells by staining them with DAPI, PI, and annexin V (Fig. 2D). PI stains fragmented DNA, a characteristic feature of apoptosis, and annexin V binds to phosphatidylserine, present on the surface of apoptotic cells. HEp-2 cells incubated with Omp33-36 were positive (green) for annexin V (Fig. 2D, center and right), and the colocalization of PI (red) with DAPI (blue) within their apoptotic nuclei resulted in a pink coloration. These pink nuclei were also smaller than those that were positive only for DAPI. After incubation of the cells for 8 h with Omp33-36, the apoptotic index increased by 43% (P < 0.001, Mann-Whitney U-test).
All of these results suggest that apoptosis can be induced by A. baumannii Omp33-36 in human cells.
(ii) DNA fragmentation induced by Omp33-36 in HeLa and HEp-2 cells is inhibited by anti-Omp33-36 antibody.
In HEp-2 and HeLa cells exposed to Omp33-36, DNA fragmentation was detected by use of an APO-BrdU kit (as described in Materials and Methods) in 51.7% (Fig. 2F) and 54.8% (Fig. 2I) of the cells, respectively. Preincubation of Omp33-36 with a polyclonal antibody against the protein for 1 h prevented DNA fragmentation (0.6% and 13.1%; Fig. 2G and J, respectively). To discount the possibility that the antibodies themselves have an effect on HEp-2 and HeLa cells, the same amount of antibody was added to the cultures and was found to have no effect on the cells at all (data not shown).
(iii) Omp33-36 activates caspases in HEp-2 cells (Fig. 2K and L).
A group of intracellular proteases called caspases is responsible for the deliberate disassembly of the cell into apoptotic bodies during apoptosis (1). Caspases are present as inactive proenzymes that are activated by proteolytic cleavage. Caspases 9 and 3 are situated at pivotal junctions in apoptosis pathways. HEp-2 cells challenged with 8 μg/ml of Omp33-36 displayed caspase activation. Specifically, we observed a decrease in the amount of procaspase 3 (at 4 and 8 h) and an increase in its active form, caspase 3. The same transformation was observed for procaspase 9 and its active form, caspase 9.
The activation of proapoptotic proteins of the Bcl-2 family causes mitochondrial damage, which leads to the release of cytochrome c into the cytoplasm and, thus, to apoptosome activation, so that caspase 9 is cleaved from its inactive procaspase 9 state (47 kDa) to its active form (37 kDa). Activated caspase 9 (Fig. 2L) in turn activates caspase 3 (Fig. 2K), triggering the final phases of the apoptosis cascade.
The involvement of bacterial porins in the development of apoptosis is known for several pathogens (25), but as noted in the introduction, only the 38- to 40-kDa Omp protein (OmpA) has been implicated in this process in A. baumannii (9, 11). However, OmpA differs from Omp33-36 in that although the former causes cytotoxicity by targeting the host cell through a specific nuclear localization signal region (KTKEGRAMNRR), this motif is lacking in Omp33-36 (26).
Involvement of Omp33-36 in the modulation of autophagy. (i) Structures derived from processes of apoptosis and autophagy are present in HEp-2 cells incubated with Omp33-36 (Fig. 3).
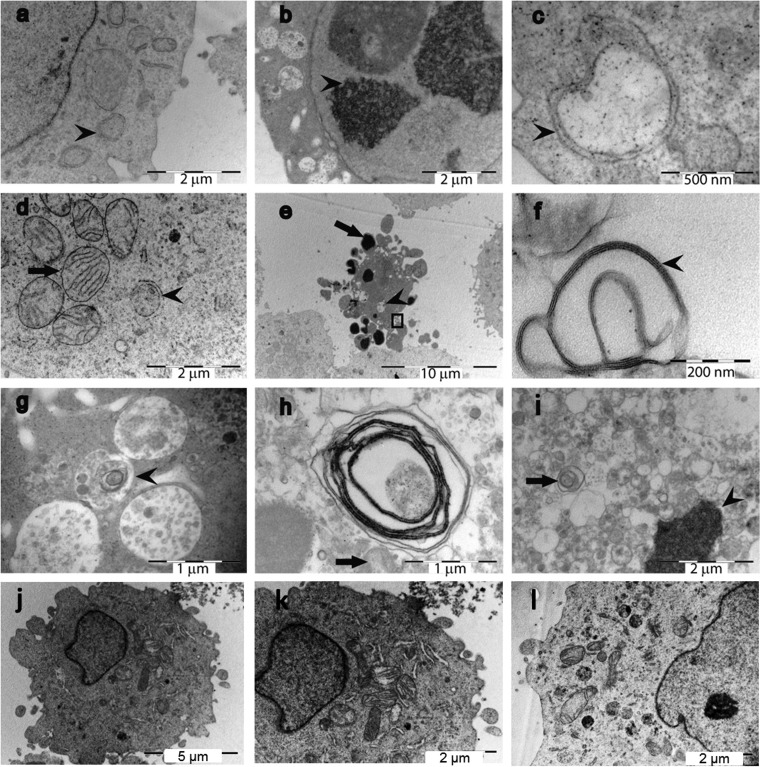
FIG 3.
TEM analysis of Omp33-36-treated HEp-2 cells reveals signs of apoptosis and autophagocytosis. The cells were incubated for 4 h (a, b), 12 h (c to g), and 20 h (h, i) with 8 μg Omp33-36/ml or left untreated (j to l). (a) Double-membrane organelles (arrowhead), a “gold standard” biomarker of autophagy; (b) cytoplasmic content in vacuoles and condensed DNA (arrowhead); (c) double-membrane vacuole (arrowhead); (d) an organelle with a double membrane (arrowhead) surrounded by intact mitochondria with cristae (arrow); (e) a typical apoptotic cell with apoptotic bodies (arrow) and vesicles containing intracellular material (arrowhead); the area inside the square is amplified in panel f; (f) multilamellar organelle composed of a series of closely apposed membranes (arrowhead); (g) presumably damaged organelles (arrowhead) typical of multilamellar structures; (h) vesicular structure with an onion-ring-like appearance (arrow); (i) after 20 h, most of the cytoplasm is filled with cytoplasm-containing vacuoles (arrow) and the DNA is condensed (arrowhead); (j) a healthy HEp-2 cell; its cytoplasm is amplified in panel k; (k) intact mitochondria with cristae; (l) at 20 h, cytoplasm of an untreated HEp-2 cell showing healthy organelles.
Electron microscopic analysis showed that while unstimulated HEp-2 cells showed a healthy cell morphology (the presence of a nucleolus, uncondensed DNA, cytoplasmatic organelles, mitochondria with cristae), cells incubated for 4, 12, or 20 h with 8 μg Omp33-36/ml revealed a significant increase in the number of apoptotic cells (Fig. 3) with a shrunken cytoplasm exhibiting DNA condensation and apoptotic bodies, as well as double-membrane organelles and onion ring-like multilamellar bodies as markers of autophagy (Fig. 3). This indicates that the protein is involved in apoptosis and autophagy.
(ii) Transfection of mapA gene in HeLa Tet-On cells and modulation of autophagy.
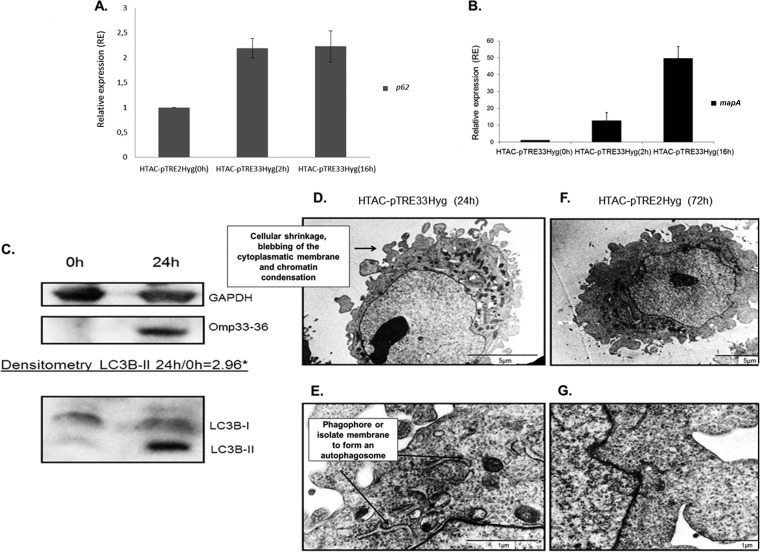
To confirm the results obtained with purified Omp33-36 and to fully rule out the putative effect of bacterial contaminants, the mapA gene under the control of a doxycycline-inducible promoter was cloned into a eukaryotic vector that was then used to transfect HeLa Tet-On Advanced cells (HTAC-pTRE33Hyg cells). The relative expression of the p62 and mapA genes in mapA transfectants induced with doxycycline for 2 and 16 h was determined (Fig. 4A and B). The p62 marker was induced at 2 to 16 h, which indicates blockage of autophagy.
FIG 4.
(A, B) Modulation of autophagy was analyzed by RT-PCR in HeLa Tet-On Advanced (HTAC) cells transfected with mapA cloned in pTRE2Hyg (HTAC-pTRE33Hyg cells). p62 gene expression was studied by RT-PCR at 2 h and 16 h after doxycycline induction of HTAC-pTRE33Hyg cells. Relative expression was based on HTAC-pTRE2Hyg expression (y axis) at 16 h, which was assigned a value of 1. The same samples were analyzed for mapA expression by HTAC-pTRE33Hyg cells at 0 h, 2 h, and 16 h after doxycycline induction. (C) Detection of Omp33-36 and LC3B accumulation in HTAC-pTRE33Hyg cells by immunoblotting for the detection of Omp33-36 and LC3B-I/LC3B-II by polyclonal antibodies in HTAC-pTRE33Hyg cells under the control of a doxycycline-inducible promoter. *, densitometry of LC3B-II 0 and 24 h after doxycycline induction of HTAC-pTRE33Hyg cells. (D, E) Presence of the features of apoptosis and probable phagophore structures (isolate membrane to form an autophagosome) determined by TEM of HTAC-pTRE33Hyg cells expressing mapA after 24 h of doxycycline induction. (F, G) Absence of characteristics of the apoptosis and autophagy mechanisms determined by TEM of HTAC cells transfected with pTRE2Hyg (empty plasmid) after 72 h of doxycycline induction (negative control).
Induction of mapA expression by incubation of the transfectants with doxycycline for 24 h was followed by monitoring of the increase in the autophagy marker LC3B-II (Fig. 4C), as revealed by Western blotting and densitometry analysis. Electron microscopy revealed a membranous cistern from the endoplasmic reticulum (a probable phagophore or isolation membrane) that increases in length (27). Phagophore membranes grow and eventually join ends to form a closed compartment called an autophagosome (Fig. 4E). These structures were not found in the negative control (Fig. 4G).
(iii) Studies with autophagy inhibitors.
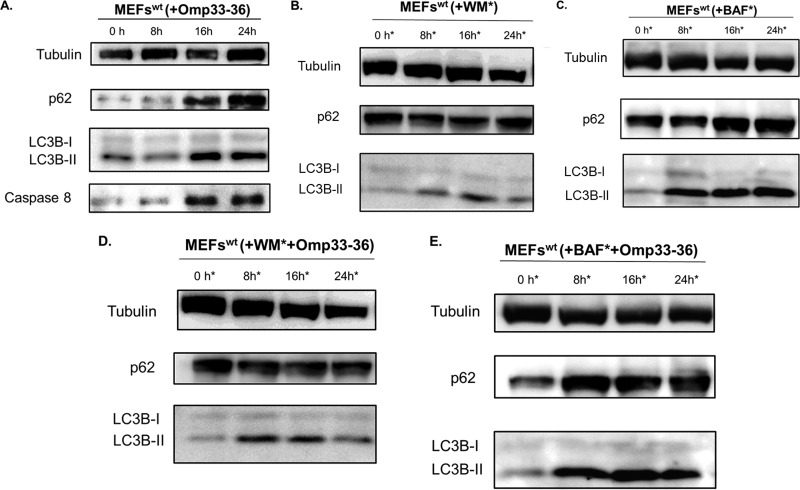
As shown Fig. 5A, in wt MEFs there was an increase in the levels of caspase 8 and in two autophagic markers (p62 and LC3B-II), indicating an activation of the apoptosis and a modulation of autophagy, respectively, by Omp 33-36. These markers were studied in both the presence and absence of Omp33-36 and with wortmannin (an inhibitor of PI3K, an enzyme needed for autophagosome formation) and bafilomycin (a specific inhibitor of the H+-ATPase [V-ATPase] vacuolar type in cells which inhibits the acidification of organelles containing this enzyme, such as lysosomes and endosomes) (Fig. 5B to E). Omp33-36 showed behavior similar to that of the inhibitors: with wortmannin, no autophagosomes were formed (p62 accumulation but no LC3B-II accumulation), while autophagosomes were formed and accumulated in the presence of bafilomycin (p62 and LC3B-II accumulation), as was also observed with the protein Omp33-36 alone (Fig. 5A).
FIG 5.
(A) Accumulation of autophagy markers (modulation of the autophagy) in the presence of Omp33-36. The accumulation of p62, LC3B, and caspase 8 was analyzed by Western blotting in wt MEFs. Cells incubated with 8 μg/ml of Omp33-36 were tested at 0, 8, 16, and 24 h. (B, D) Inhibition of autophagy before formation of autophagosomes. Immunoblotting was performed to determine the levels of p62 and LC3B in wt MEFs in the presence of 1 μM wortmannin (WM), which was added at 0, 8, 16, and 24 h (*), in the presence and absence of 8 μg/ml of Omp33-36. (C, E) Inhibition of autophagy and accumulation of autophagosomes. Western blotting was used to study the autophagy process by determining p62 and LC3B levels in the presence of 0.1 μM bafilomycin (BAF), which was added at 0, 8, 16, and 24 h (*), in the presence and absence of 8 μg/ml of Omp33-36.
The results obtained by expression studies, immunoblotting, and electron microscopy suggested that in HeLa cells transfected with mapA, expression of the protein arrested the process of autophagy. These results support those obtained with the purified protein in the absence and presence of autophagy inhibitors, confirming that the protein blocks autophagy, with the consequent accumulation of autophagosomes (28, 29).
Autophagy enables eukaryotic cells to capture cytoplasmic components for degradation within lysosomes and has classically been studied as a response to several processes, including infectious diseases (30). In this pathway, bacteria that invade the cytoplasm are engulfed and degraded. Pathogens have developed diverse strategies to evade host cell autophagy, so that not only do they escape lysosomal degradation but they also exploit this pathway to their advantage (31). For example, the intracellular pathogens Coxiella burnetii (32), Brucella (33), and Legionella (34) modulate both autophagy and apoptosis to establish an intracellular niche in the autophagosomes for cellular replication. Moreover, even Staphylococcus aureus (29) and Serratia marcescens (35), best known as extracellular pathogens, can induce cell death by intracellular mechanisms. Recently, Acinetobacter calcoaceticus-A. baumannii complex strains associated with apoptosis in human epithelial cells were shown to induce the formation of autophagic vacuoles, although neither the mechanism nor the bacterial factor involved in this process was explored (36).
The bacterial proteins shown to be involved in the modulation of autophagy by the accumulation of autophagosomes (preventing bacterial degradation) include the virulence factors VirG, VacA, and SipB from, respectively, Shigella flexneri, Helicobacter pylori (32), and Salmonella enterica serovar Typhimurium (37).
Relationship between apoptosis and autophagy induced by Omp33-36.
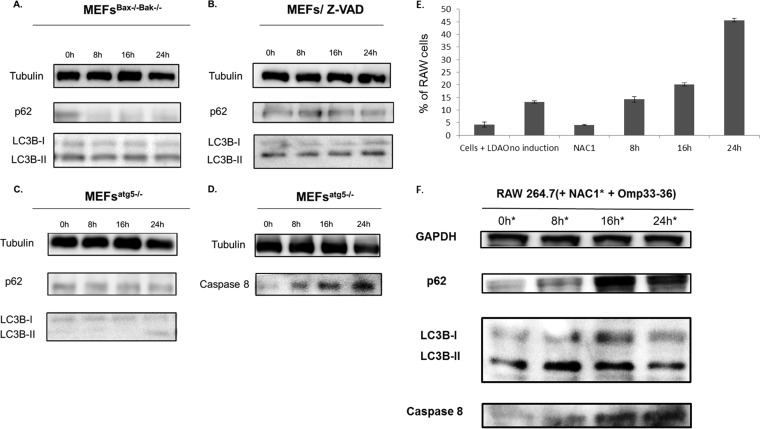
The relationship between apoptosis and autophagy induced by Omp33-36 was studied in bax−/− bak−/− MEFs or wt MEFs treated with the caspase inhibitor Z-VAD-FMK in the presence of Omp33-36. Both the conversion of LC3B to LC3B-II and p62 degradation occurred (Fig. 6A and B), indicative of the development of autophagy. Finally, when the experiments were performed in autophagy-defective atg5−/− MEFs (in which no significant conversion of LC3B to LC3B-II or degradation of p62 occurs), incubation with 8 μg Omp33-36/ml still resulted in apoptosis, determined on the basis of the presence of caspase 8 levels similar to those in wild-type cells (Fig. 6C and D).
FIG 6.
(A, B) Activation of the autophagy mechanism occurs without development of apoptosis. Western blotting was used to study the autophagy markers p62 and LC3B in cells defective for apoptosis (bax−/− bak−/− MEFs [MEFsBax−/−Bak−/−] and wt MEFs pretreated with 5 μM Z-VAD [MEFs/Z-VAD]). The cells were incubated with 8 μg/ml of Omp33-36 for 0, 8, 16, and 24 h. (C, D) Apoptosis occurs without autophagy. Western blotting was used to analyze the autophagy markers (p62 and LC3B) and apoptosis antibodies (caspase 8) in defective autophagic cells (atg5−/− MEFs). Cells were also incubated with 8 μg/ml of Omp33-36 for 0, 8, 16, and 24 h. (E) Time course of ROS production by cells subjected to oxidative stress (indicated as the percentage of RAW 264.7 cells) after incubation with Omp33-36 (8, 16, and 24 h). Negative controls: cells + LDAO, Omp33-36 buffer solution; no induction, cells alone; and NAC1, cells plus 20 mM NAC1. All negative-control experiments were done at 24 h. There was a significant increase (P < 0.001, Mann-Whitney U test) in ROS production over time in the presence of Omp33-36. (F) Inhibition of ROS response as consequence of apoptosis and blockage of autophagy. An immunoblot with anti-LC3B, anti-p62, and anti-caspase 8 antibodies in RAW 264.7 cells treated for 30 min with 20 mM NAC1 and 8 μg Omp33-36/ml at 0, 8, 16, and 24 h is shown.
ROS production and apoptosis/modulation of autophagy are mediated by Omp33-36.
Challenge of RAW 264.7 macrophages with pure Omp33-36 for 0, 8, 16, and 24 h resulted in a significant (P < 0.001) increase in ROS production (Fig. 6E) in negative-control cells incubated with NAC1 (an inhibitor of ROS production) and LDAO buffer.
Finally, caspase 8 levels increased and p62 and LC3B accumulated in the presence of Omp33-36 and ROS inhibitor (NAC1) (Fig. 6F).
Overall, our results show that the Omp33-36 induces apoptosis (by a caspase-dependent mechanism) and modulates autophagy (an increase in LC3BII and the accumulation of p62). When caspase-dependent apoptosis is blocked by chemical inhibitors or in fibroblasts defective in apoptotic activity, the induction of autophagy by Omp33-36 is nonetheless preserved (an increase in LC3B-II and degradation of p62). However, in autophagy-defective fibroblasts, Omp33-36 continues to activate apoptosis, as measured by the maintenance of caspase 8 cleavage.
The relationship between autophagy and apoptosis is complex, as the processes can act synergistically or antagonistically, depending on the cell type, stimulus, and cellular environment (38). For instance, Xi et al. (39) studied the interplay between autophagy and apoptosis in human rhabdomyosarcoma cells infected with enterovirus 71. These authors observed that caspase 3, a member of the apoptosis cascade, was an important regulator in autophagosome maturation (LC3B conversion) and, thus, in the relationship between autophagy and apoptosis. In rhabdomyosarcoma cells in which the autophagy gene atg5 was silenced (no LC3B conversion), caspase 3 activation and, thus, apoptosis was also inhibited. However, when caspase 3 was inhibited, ATG5 activity (LC3B conversion) and p62 degradation remained intact, allowing autophagy to proceed.
Finally, evidence from several studies suggests that macrophages produce ROS during microbial attack (40, 41). Our results show that the ROS response is a consequence rather than a cause of activation of apoptosis and modulation of autophagy.
Omp33-36 as a surface protein on A. baumannii cells that is released in OMVs.
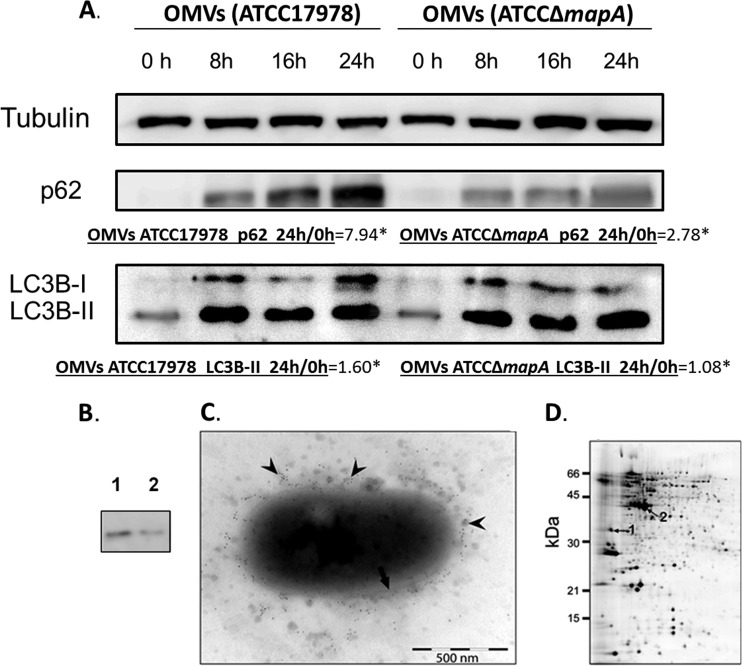
OMVs were purified from late-exponential-phase cultures of A. baumannii ATCC 17978 and A. baumannii ATCCΔmapA, as described in Materials and Methods. The levels of p62 were higher (by almost 8 times, as shown by densitometry studies) in the presence the OMVs from A. baumannii ATCC 17978 than in the presence of OMVs from the mutant A. baumannii ATCCΔmapA (Fig. 7A). The level of LC3B-II increased over time in the presence of OMVs from both strains, indicating that the OMVs with Omp33-36 strongly block autophagy. However, there was not an important blockage with the OMV without Omp33-36, but we cannot rule out the possible involvement of other porins.
FIG 7.
(A) Blockage of autophagy by OMVs carrying porins (with and without Omp33-36). p62 and LC3B levels of OMVs from A. baumannii ATCC 17978 and A. baumannii ATCCΔmapA analyzed by Western blotting. *, densitometry of p62 and LC3B-II at 0 and 24 h from OMVs of A. baumannii ATCC 17978 and A. baumannii ATCCΔmapA. (B) Confirmation of the presence of the Omp33-36 in OMVs. Pure Omp33-36 (0.3 μg/ml) (lane 1) and 20 μl of OMVs prepared from A. baumannii ATCC 17978 (lane 2) were resolved by SDS-PAGE on 12% gels, immunoblotted with a polyclonal antibody against Omp33-36, and visualized with a secondary antibody linked to horseradish peroxidase. A unique band between 43 and 34 kDa (lane 1) was detected. (C) Examination of OMVs by TEM analysis of an A. baumannii ATCC 17978 cell. Arrow, bacterial membrane; arrowheads, OMVs released from A. baumannii. Omp33-36 was detected with a polyclonal antibody against the protein and a gold-labeled anti-rabbit antibody. (D) Presence of the porins detected by proteomic analysis in the OMVs. The 2-DE gel (IEF) shows A. baumannii ATCC 17978 OMV proteins. The numbered spots in the gel indicate the Omp33-36 (spot 1) and OmpA (spot 2) proteins identified by MALDI-TOF/TOF analysis. All gels (12% acrylamide) were silver stained and loaded with 25 μg total protein.
The Western blotting assays used to detect the protein revealed a unique band corresponding to a 33- to 36-kDa protein both in the pure protein and in the OMVs (Fig. 7B, lanes 1 and 2, respectively), which are thus a vehicle for transmission of this protein.
The location of Omp33-36 on the outer membrane suggested that the protein also occurred within the OMVs released by A. baumannii and many other Gram-negative bacteria (42, 43). Both the bacterial membrane and the OMVs surrounding the cell were visualized by TEM analysis of A. baumannii cells (Fig. 7C). The presence of Omp33-36 in the OMVs was confirmed immunohistochemically with a polyclonal antibody against Omp33-36 and a gold-labeled antirabbit antibody, as described in Materials and Methods.
Proteomic analysis detected Omp33-36 as well as OmpA, a major A. baumannii porin that also triggers apoptosis in mammalian cells (44). Purified OMVs were analyzed on a 2-DE gel, and both OmpA and Omp33-36 were identified by MALDI-TOF/TOF analysis (Fig. 7D). These results are consistent with the release of A. baumannii outer membrane porins embedded in OMVs, including Omp33-36 and OmpA.
OMVs are natural vehicles for directing bacterial factors into host cells and tissues, as has been described for different bacterial pathogens (45). In addition to secreting a virulence factor that induces cellular apoptosis, Gram-negative bacteria release active toxins associated with OMVs (20, 43, 46). The role of OMVs in the interaction between A. baumannii and eukaryotic cells is unclear, although the delivery of bacterial proteins and other bacterial components present in OMVs to eukaryotic cells has been demonstrated (47). The findings of the present study suggest that in A. baumannii, OMVs provide a vehicle for the transport of Omp33-36 as well as other porins and virulence factors (44, 48).
A. baumannii ATCC 17978 inside autophagosomes.
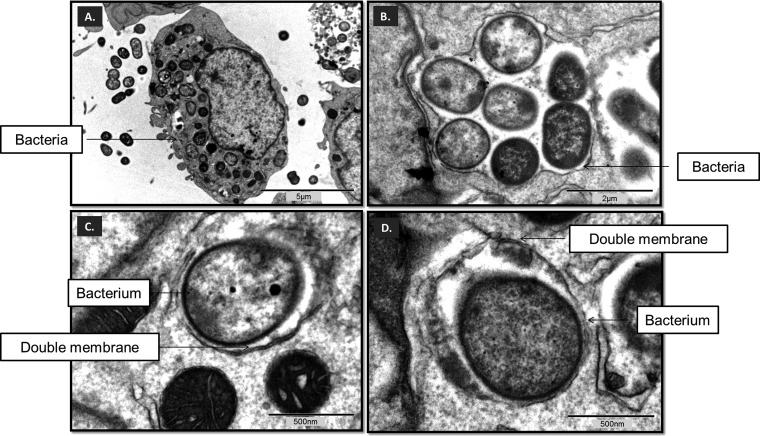
The interaction between microbes and macrophages was visualized in a TEM study, which showed A. baumannii cells persisting within double-membrane structures (i.e., autophagosomes), which is key to the development of infection (Fig. 8).
FIG 8.
(A to D) Autophagosomes (double-membrane structures) with A. baumannii cells replicating within them. (A) TEM of RAW 264.7 macrophages incubated for 12 h with A. baumannii ATCC 17978; (B) group of intact bacteria enclosed by double membranes in RAW 264.7 macrophages; (C and D) individual bacterium surrounded by double membranes (arrows) in RAW 264.7 macrophages.
The efficacy of autophagy in eliminating pathogenic organisms or providing a niche for their replication depends on the nature of the pathogen. A group of intracellular microbes is targeted by autophagy. Avoidance of phagosome maturation is one of the strategies used by a pathogen to enable its replication and intracellular survival within the host cell. Indeed, one of the most remarkable characteristics of Mycobacterium tuberculosis is its ability to control the fate of the bacterium-containing phagosome by blocking its maturation into a degradative compartment (49).
Impact of A. baumannii Omp33-36 on cytotoxicity in macrophages and virulence in a mouse model of systemic infection.
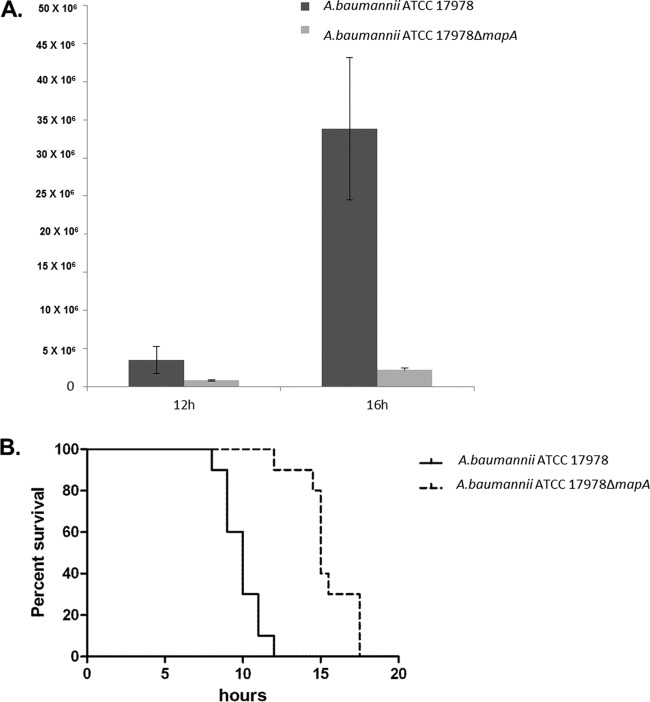
Survival assays were performed to study the persistence and survival of A. baumannii in macrophages as well as the role of Omp33-36 in the process. In RAW 264.7 macrophages incubated for 16 h with A. baumannii ATCC 17978 and A. baumannii ATCCΔmapA, significantly higher numbers of CFU/ml were recovered from wild-type cells than from the mutants (35 × 106 versus 3 × 106 CFU) (Fig. 9A). The experiment could not be carried out for longer than 20 h after infection because the percentage of dead macrophages was much higher in the A. baumannii ATCC 17978 infection (62.31%) than in the A. baumannii ATCCΔmapA infection (12.65%). These findings suggest the higher cytotoxicity of A. baumannii ATCC 17978 expressing Omp33-36 in the macrophage model of infection.
FIG 9.
(A) Greater intracellular persistence of A. baumannii ATCC 17978 (carrying Omp33-36). The numbers of CFU (indicated on the y axis) recovered after infection with A. baumannii ATCC 17978 and A. baumannii ATCCΔmapA for 12 h and 16 h are shown. (B) Omp33-36 is a virulence factor in animal models. The survival of BALB/c mice (n = 10 per group) following intraperitoneal infection of 3.5 × 104 CFU of A. baumannii ATCC 17978 or 4 × 104 CFU of A. baumannii ATCCΔmapA is shown. Survival was significantly higher (P < 0.0001) in mice infected with the wild-type strain.
To assess the impact of our findings in a mammalian animal model (Fig. 9B), we determined the virulence of the wild-type (A. baumannii ATCC 17978) and mutant (A. baumannii ATCCΔmapA) strains in a mouse model of systemic infection. All 10 mice infected with 3.5 × 104 CFU of the wild-type strain and 1 of the 10 mice infected with 4 × 104 CFU of the ATCCΔmapA mutant strain died within the first 12 h. The remaining mice died within the following 6 h. These data suggest that Omp33-36 participates in the virulence of A. baumannii and are consistent with the findings of Smani et al. (10), who concluded that Omp33-36 plays an important role in the fitness and virulence of A. baumannii. However, in that study the molecular mechanisms of cytotoxicity were not further explored.
In summary, in this study we showed the following: (i) Omp33-36 is a porin released within OMVs; (ii) Omp33-36 induces apoptosis through caspase activation and subsequently modulates autophagy, which does not proceed in the absence of apoptosis; (iii) ROS is produced as a consequence of these processes; (iv) A. baumannii ATCC 17978 strains (carrying this protein) in macrophage cells persisted intracellularly (inside autophagosomes), causing cytotoxicity; and (v) Omp33-36 is a virulence factor in a systemic model of mouse infection.
Finally, in light of our findings, together with those of Islam and colleagues (50), on the strong immunogenic capacity of Omp33-36, we can recommend this protein as a therapeutic target in the fight against infections caused by A. baumannii.
ACKNOWLEDGMENTS
C. Rumbo was supported by a doctoral grant (PFIS) from the Instituto de Salud Carlos III. M. Tomás is currently the recipient of financial support from the Miguel Servet Programme CHU A Coruña and the Instituto de Salud Carlos III (SERGAS/Ministerio de Sanidad). M. Carvajal was supported by a grant from the Fundación Séneca (Comunidad Autónoma de la Región de Murcia). This work was supported by Magic Bullet (to G. Bou), a project funded by the European Union's Directorate General for Research and Innovation, through the Seventh Framework Program for Research and Development (grant agreement 278232); the Spanish Network for Research in Infectious Diseases (REIPI-RD12/0015); and Spanish Ministry of Health/FEDER funding (ISCIII-FIS PI12/00552 to G. Bou, FIS PI10/00056 and PI13/02390 to M. Tomás).
We thank Catalina Sueiro and Ada Castro for their kind help with microscopic techniques and Bruce Patrick Dos Santos for help in preparing the manuscript.
Footnotes
Published ahead of print 25 August 2014
REFERENCES
- 1.Thornberry NA, Lazebnik Y. 1998. Caspases: enemies within. Science 281:1312–1316. 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 2.Komatsu M, Ichimura Y. 2010. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett. 584:1374–1378. 10.1016/j.febslet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. 2009. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc. Natl. Acad. Sci. U. S. A. 106:19860–19865. 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumann P. 1968. Isolation of Acinetobacter from soil and water. J. Bacteriol. 96:39–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong TH, Tan BH, Ling ML, Song C. 2002. Multi-resistant Acinetobacter baumannii on a burns unit—clinical risk factors and prognosis. Burns 28:349–357. 10.1016/S0305-4179(02)00012-8. [DOI] [PubMed] [Google Scholar]
- 6.Bergogne-Bérézin E, Towner KJ. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Talbot GH, Bradley J, Edwards JE, Gilbert D, Scheld M, Bartlett JG, Antimicrobial Availability Task Force of the Infectious Diseases Society of America 2006. Bad bugs need drugs: an update on the development pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin. Infect. Dis. 42:657–668. 10.1086/499819. [DOI] [PubMed] [Google Scholar]
- 8.Smani Y, Docobo-Perez F, McConnell MJ, Pachon J. 2011. Acinetobacter baumannii-induced lung cell death: role of inflammation, oxidative stress and cytosolic calcium. Microb. Pathog. 50:224–232. 10.1016/j.micpath.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Choi CH, Hyun SH, Lee JY, Lee JS, Lee YS, Kim SA, Chae JP, Yoo SM, Lee JC. 2008. Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cell. Microbiol. 10:309–319. 10.1111/j.1462-5822.2007.01041.x. [DOI] [PubMed] [Google Scholar]
- 10.Smani Y, Dominguez-Herrera J, Pachón J. 2013. Association of the outer membrane protein Omp33 with fitness and virulence of Acinetobacter baumannii. J. Infect. Dis. 208:1561–1570. 10.1093/infdis/jit386. [DOI] [PubMed] [Google Scholar]
- 11.Choi CH, Lee EY, Lee YC, Park TI, Kim HJ, Hyun SH, Kim SA, Lee SK, Lee JC. 2005. Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell. Microbiol. 7:1127–1138. 10.1111/j.1462-5822.2005.00538.x. [DOI] [PubMed] [Google Scholar]
- 12.Lee JS, Choi CH, Kim JW, Lee JC. 2010. Acinetobacter baumannii outer membrane protein A induces dendritic cell death through mitochondrial targeting. J. Microbiol. 48:387–392. 10.1007/s12275-010-0155-1. [DOI] [PubMed] [Google Scholar]
- 13.del Mar Tomas M, Beceiro A, Perez A, Velasco D, Moure R, Villanueva R, Martinez-Beltran J, Bou G. 2005. Cloning and functional analysis of the gene encoding the 33- to 36-kilodalton outer membrane protein associated with carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:5172–5175. 10.1128/AAC.49.12.5172-5175.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orsel M, Chopin F, Leleu O, Smith SJ, Krapp A, Daniel-Vedele F, Miller AJ. 2006. Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein-protein interaction. Plant Physiol. 142:1304–1317. 10.1104/pp.106.085209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preston GM, Carroll TP, Guggino WB, Agre P. 1992. Appearance of water channels in Xenopus oocytes expressing red cell CHIP28 protein. Science 256:385–387. 10.1126/science.256.5055.385. [DOI] [PubMed] [Google Scholar]
- 16.Rallabhandi P, Awomoyi A, Thomas KE, Phalipon A, Fujimoto Y, Fukase K, Kusumoto S, Qureshi N, Sztein MB, Vogel SN. 2008. Differential activation of human TLR4 by Escherichia coli and Shigella flexneri 2a lipopolysaccharide: combined effects of lipid A acylation state and TLR4 polymorphisms on signaling. J. Immunol. 180:1139–1147. 10.4049/jimmunol.180.2.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnoult D, Bartle LM, Skaletskaya A, Poncet D, Zamzami N, Park PU, Sharpe J, Youle RJ, Goldmacher VS. 2004. Cytomegalovirus cell death suppressor vMIA blocks Bax- but not Bak-mediated apoptosis by binding and sequestering Bax at mitochondria. Proc. Natl. Acad. Sci. U. S. A. 101:7988–7993. 10.1073/pnas.0401897101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aranda J, Poza M, Pardo BG, Rumbo S, Rumbo C, Parreira JR, Rodríguez-Velo P, Bou G. 2010. A rapid and simple method for constructing stable mutants of Acinetobacter baumannii. BMC Microbiol. 10:279. 10.1186/1471-2180-10-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hood MI, Jacobs AC, Sayood K, Dunman PM, Skaar EP. 2010. Acinetobacter baumannii increases tolerance to antibiotics in response to monovalent cations. Antimicrob. Agents Chemother. 54:1029–1041. 10.1128/AAC.00963-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rumbo C, Fernandez-Moreira E, Merino M, Poza M, Mendez JA, Soares NC, Mosquera A, Chaves F, Bou G. 2011. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55:3084–3090. 10.1128/AAC.00929-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maurel C, Reizer J, Schroeder JI, Chrispeels MJ, Saier MH. 1994. Functional characterization of the Escherichia coli glycerol facilitator, GlpF, in Xenopus oocytes. J. Biol. Chem. 269:11869–11872. [PubMed] [Google Scholar]
- 22.Bay DC, Court DA. 2002. Origami in the outer membrane: the transmembrane arrangement of mitochondrial porins. Biochem. Cell Biol. 80:551–562. 10.1139/o02-149. [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi O, Akira S. 2007. Signaling pathways activated by microorganisms. Curr. Opin. Cell Biol. 19:185–191. 10.1016/j.ceb.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Hornef MW, Henriques-Normark B, Normark S. 2008. The function and biological role of Toll-like receptors in infectious diseases: an update. Curr. Opin. Infect. Dis. 21:304–312. 10.1097/QCO.0b013e3282f88ba3. [DOI] [PubMed] [Google Scholar]
- 25.Boya P, Roques B, Kroemer G. 2001. New EMBO members' review: viral and bacterial proteins regulating apoptosis at the mitochondrial level. EMBO J. 20:4325–4331. 10.1093/emboj/20.16.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon DC, Gurung M, Lee JH, Lee YS, Choi CW, Kim SI, Lee JC. 2012. Screening of nuclear targeting proteins in Acinetobacter baumannii based on nuclear localization signals. Res. Microbiol. 163:279–285. 10.1016/j.resmic.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Tooze SA, Yoshimori T. 2010. The origin of the autophagosomal membrane. Nat. Cell Biol. 12:831–835. 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 28.Mizushima N, Yoshimori T, Levine B. 2010. Methods in mammalian autophagy research. Cell 140:313–326. 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klionsky DJ, Codogno P. 2013. The mechanism and physiological function of macroautophagy. J. Innate Immun. 5:427–433. 10.1159/000351979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swanson MS. 2006. Autophagy: eating for good health. J. Immunol. 177:4945–4951. 10.4049/jimmunol.177.8.4945. [DOI] [PubMed] [Google Scholar]
- 31.Campoy E, Colombo MI. 2009. Autophagy in intracellular bacterial infection. Biochim. Biophys. Acta 1793:1465–1477. 10.1016/j.bbamcr.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Lerena MC, Vázquez CL, Colombo MI. 2010. Bacterial pathogens and the autophagic response. Cell. Microbiol. 12:10–18. 10.1111/j.1462-5822.2009.01403.x. [DOI] [PubMed] [Google Scholar]
- 33.Starr T, Child R, Wehrly TD, Hansen B, Hwang S, López-Otin C, Virgin HW, Celli J. 2012. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe 11:33–45. 10.1016/j.chom.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colombo MI. 2005. Pathogens and autophagy: subverting to survive. Cell Death Differ. 12(Suppl 2):S1481–S1483. 10.1038/sj.cdd.4401767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fedrigo GV, Campoy EM, Di Venanzio G, Colombo MI, García Véscovi E. 2011. Serratia marcescens is able to survive and proliferate in autophagic-like vacuoles inside non-phagocytic cells. PLoS One 6:e24054. 10.1371/journal.pone.0024054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krzymińska S, Frąckowiak H, Kaznowski A. 2012. Acinetobacter calcoaceticus-baumannii complex strains induce caspase-dependent and caspase-independent death of human epithelial cells. Curr. Microbiol. 65:319–329. 10.1007/s00284-012-0159-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hernandez LD, Pypaert M, Flavell RA, Galán JE. 2003. A Salmonella protein causes macrophage cell death by inducing autophagy. J. Cell Biol. 163:1123–1131. 10.1083/jcb.200309161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y. 2004. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 6:1221–1228. 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 39.Xi X, Zhang X, Wang B, Wang T, Wang J, Huang H, Jin Q, Zhao Z. 2013. The interplays between autophagy and apoptosis induced by enterovirus 71. PLoS One 8:e56966. 10.1371/journal.pone.0056966. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Cosgrove K, Coutts G, Jonsson IM, Tarkowski A, Kokai-Kun JF, Mond JJ, Foster SJ. 2007. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J. Bacteriol. 189:1025–1035. 10.1128/JB.01524-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nunoshiba T, DeRojas-Walker T, Wishnok JS, Tannenbaum SR, Demple B. 1993. Activation by nitric oxide of an oxidative-stress response that defends Escherichia coli against activated macrophages. Proc. Natl. Acad. Sci. U. S. A. 90:9993–9997. 10.1073/pnas.90.21.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horstman AL, Kuehn MJ. 2000. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J. Biol. Chem. 275:12489–12496. 10.1074/jbc.275.17.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kesty NC, Mason KM, Reedy M, Miller SE, Kuehn MJ. 2004. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 23:4538–4549. 10.1038/sj.emboj.7600471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin JS, Kwon SO, Moon DC, Gurung M, Lee JH, Kim SI, Lee JC. 2011. Acinetobacter baumannii secretes cytotoxic outer membrane protein A via outer membrane vesicles. PLoS One 6:e17027. 10.1371/journal.pone.0017027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuehn MJ, Kesty NC. 2005. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 19:2645–2655. 10.1101/gad.1299905. [DOI] [PubMed] [Google Scholar]
- 46.Mendez JA, Soares NC, Mateos J, Gayoso C, Rumbo C, Aranda J, Tomas M, Bou G. 2012. Extracellular proteome of a highly invasive multidrug-resistant clinical strain of Acinetobacter baumannii. J. Proteome Res. 11:5678–5694. 10.1021/pr300496c. [DOI] [PubMed] [Google Scholar]
- 47.Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, Van Nhieu GT, van der Goot FG, Sansonetti PJ, Lafont F. 2009. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe 6:137–149. 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 48.Beceiro A, Tomas M, Bou G. 2013. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 26:185–230. 10.1128/CMR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. 2004. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119:753–766. 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 50.Islam AH, Singh KK, Ismail A. 2011. Demonstration of an outer membrane protein that is antigenically specific for Acinetobacter baumannii. Diagn. Microbiol. Infect. Dis. 69:38–44. 10.1016/j.diagmicrobio.2010.09.008. [DOI] [PubMed] [Google Scholar]