Abstract
Enterohemorrhagic Escherichia coli (EHEC), a subgroup of Shiga toxin (Stx)-producing E. coli (STEC), is a leading cause of diarrhea and hemolytic-uremic syndrome (HUS) in humans. However, urinary tract infections (UTIs) caused by this microorganism but not associated with diarrhea have occasionally been reported. We geno- and phenotypically characterized three EHEC isolates obtained from the urine of hospitalized patients suffering from UTIs. These isolates carried typical EHEC virulence markers and belonged to HUS-associated E. coli (HUSEC) clones, but they lacked virulence markers typical of uropathogenic E. coli. One isolate exhibited a localized adherence (LA)-like pattern on T24 urinary bladder epithelial cells. Since the glycosphingolipids (GSLs) globotriaosylceramide (Gb3Cer) and globotetraosylceramide (Gb4Cer) are well-known receptors for Stx but also for P fimbriae, a major virulence factor of extraintestinal pathogenic E. coli (ExPEC), the expression of Gb3Cer and Gb4Cer by T24 cells and in murine urinary bladder tissue was examined by thin-layer chromatography and mass spectrometry. We provide data indicating that Stxs released by the EHEC isolates bind to Gb3Cer and Gb4Cer isolated from T24 cells, which were susceptible to Stx. All three EHEC isolates expressed stx genes upon growth in urine. Two strains were able to cause UTI in a murine infection model and could not be outcompeted in urine in vitro by typical uropathogenic E. coli isolates. Our results indicate that despite the lack of ExPEC virulence markers, EHEC variants may exhibit in certain suitable hosts, e.g., in hospital patients, a uropathogenic potential. The contribution of EHEC virulence factors to uropathogenesis remains to be further investigated.
INTRODUCTION
Escherichia coli is one of the most versatile microorganisms that rapidly colonizes the gastrointestinal tract of newborns (1). Although E. coli usually represents an important commensal of the normal intestinal microbiota, other variants that are able to cause infections exist (1, 2). On the basis of their virulence properties and the site of infection, these bacteria are classified as intestinal pathogenic E. coli (IPEC), which are associated with diarrhea, and extraintestinal pathogenic E. coli (ExPEC), which cause infections beyond the intestinal tract (3). Enterohemorrhagic E. coli (EHEC) belongs to the IPEC group of bacteria and represents one of the main causative agents of diarrhea, hemorrhagic colitis (HC), and hemolytic-uremic syndrome (HUS) in humans (4). The characteristic main EHEC virulence markers include the locus of enterocyte effacement (LEE) pathogenicity island and Shiga toxin (Stx)-encoding bacteriophages. Additionally, other toxins, including the cytolethal distending toxin (CDT), EHEC hemolysin (Hly), serine protease EspP, subtilase cytotoxin, cyclomodulin Cif, and different siderophore systems, as well as adhesins can also contribute to EHEC pathogenesis (5). These EHEC virulence factors are absent from archetypal ExPEC strains.
Similarly, the main ExPEC virulence factors, such as P fimbriae and α-hemolysin, are usually not expressed in EHEC and other IPEC isolates (1, 6–8). EHEC as well as other diarrheagenic E. coli isolates are generally not considered to cause extraintestinal infection (1, 2, 4). Nevertheless, cases of urinary tract infection (UTI) where EHEC isolates have been isolated from the corresponding urine samples have been reported. The first report of EHEC urine isolates was released in 1994 (9). Although the frequency of UTI caused by EHEC seems to be low (10), many questions regarding the virulence potential and pathogenicity of EHEC isolates in the urinary tract still remain unanswered. In a recent study, we reported on atypical uropathogenic E. coli (UPEC) isolates (11). Some of these atypical UPEC isolates from hospital patients represented IPEC pathotypes, such as EHEC, enteropathogenic E. coli (EPEC), or enteroaggregative E. coli (EAEC), whereas others combined the virulence properties of both IPEC and ExPEC isolates.
To better understand this phenomenon and to further address the question of whether EHEC, despite the absence of typical ExPEC virulence-associated factors, may have a uropathogenic potential, we further geno- and phenotypically characterized three EHEC isolates that were collected from the urine of hospital patients suffering from UTIs. We assessed the ability of the EHEC isolates to adhere to human urinary bladder cells (line T24). Moreover, we determined the ability of Stx produced by these organisms to bind to the neutral glycosphingolipids (GSLs) globotriaosylceramide (Gb3Cer) and globotetraosylceramide (Gb4Cer), known Stx receptors in human endothelial cells (12), which, as we showed, are also present in T24 cells as well as in murine bladder tissue. We analyzed whether Stx of the EHEC urine isolates is cytotoxic toward T24 cells. The transcript levels of the stx genes (the stx1c, stx2a, and stx2b alleles) upon growth of the EHEC strains in pooled human urine, lysogeny broth (LB), RPMI 1640 cell culture medium, or simulating colonic environment medium (SCEM) were compared by quantitative reverse transcription-PCR (qRT-PCR). Finally, the ability of the EHEC urine isolates to cause UTI was monitored in an experimental murine model of UTI, and their general fitness in pooled human urine was analyzed in competitive growth experiments in vitro with model UPEC strain 536.
MATERIALS AND METHODS
Clinical isolates studied.
The three EHEC isolates used in this study are listed in Table 1 and were obtained from the urine of alert hospital patients with E. coli bacteriuria (defined as colony counts of >5 × 105 CFU ml−1) in clean-voided urine (11). These female patients had been admitted to the Department of Urology, University Hospital Würzburg, Würzburg, Germany, and suffered from cystitis or hemorrhagic cystitis (Table 1). None of the patients had been admitted to the hospital because of diarrhea, nor did they exhibit such symptoms during their hospital stay. The EHEC strains were routinely cultivated in LB (13) or on LB agar plates at 37°C, but they also grew on sorbitol MacConkey agar with cefixime and potassium tellurite (CT-SMAC; Oxoid, Wesel, Germany).
TABLE 1.
Characteristics of EHEC isolates from UTI casesa
| Characteristic | Value or result for strain: |
||
|---|---|---|---|
| UR5703/202 | UR5779/201 | UR131 | |
| Age (yr) | 69 | 22 | 44 |
| Disease | Cystitis | Cystitis | Hemorrhagic cystitis |
| Serotype | O145:H− | Ont:H− | O76:H19 |
| ST, CC | ST32, CC32 | ST330, CC10 | ST675 |
| Phylogenetic group | A | E | B1 |
| Presence or type of toxin genes | |||
| cdt I, II, III, IV, V | − | − | − |
| astA | − | − | − |
| α-hlyA | − | − | − |
| cnf1, cnf2 | − | − | − |
| cif | + | − | − |
| vat | − | − | − |
| estIA | − | + | − |
| sat | − | − | − |
| EHEC hlyA | + | + | + |
| espP subtype | α-espP | γ-espP | − |
| stx genotype | stx2a | stx2a | stx1c + stx2b |
| Presence or type of adhesin genes | |||
| fimH | + | − | + |
| papA | − | − | − |
| sfa-foc | − | − | − |
| eae subtype | eae γ | eae ι-2 | − |
| bfpA | − | − | − |
| iha | + | − | − |
| saa | − | − | + |
| efa1 | + | − | − |
| sfpA | − | − | − |
| paa | + | − | − |
| aap | − | − | − |
| pet | − | − | − |
| aggA | − | − | − |
| aafA | − | − | − |
| agg3A | − | − | − |
| hdaA | − | − | − |
| aaf5A | − | − | − |
| Presence or type of other genes | |||
| irp2 | − | + | − |
| fyuA | − | + | − |
| chuA | + | + | − |
| tir subtype | EHEC tir | + (nt) | − |
| cdiAB | − | − | + |
| escV | + | + | − |
| espF | + | + | − |
| espG | + | − | − |
| map | + | + | − |
| terE | + | + | − |
| ureD | + | + | − |
| Presence of plasmid genes | |||
| etpD | − | − | − |
| katP | − | − | − |
| traT | − | + | + |
| Phenotypes | |||
| Fermentation of sorbitol | + | + | + |
| Growth on CT-SMAC | + | + | − |
| EHEC Hly expression | + | + | + |
| Stx Vero cell titer | 128 | 256 | 256 |
| Stx T24 cell titer | 8 | 8 | 32 |
| Production of colicin | − | − | + |
| Production of aerobactin | − | − | − |
| Motility | − | − | + |
| Adherence pattern | |||
| HEp-2 cells | Weak, undefined | Weak, undefined | LA |
| T24 cells | Weak, undefined | Weak, undefined | LA |
All cases occurred in females. ST, sequence type; CC, clonal complex; LA, localized adherence; nt, nontypeable.
Virulence gene detection.
We performed a set of PCR analyses to detect the presence of various IPEC and ExPEC virulence-associated genes as previously described (11, 14–16). The Stx-encoding genes were subtyped according to the recently published sequence-based protocol and designated in agreement with the modified nomenclature for Stx and stx (17). For identification of different eae alleles, we applied different previously published PCR schemes (18, 19). Subtyping of espP genes was performed using allele-specific PCRs (20, 21). All PCRs were carried out with REDTaq ReadyMix PCR mix (Sigma-Aldrich, Deisenhofen, Germany) in a total volume of 25 μl containing 2 μl of template DNA.
Total RNA isolation.
An RNeasy minikit (Qiagen, Hilden, Germany) was used for the extraction of total RNA from EHEC strains grown at 37°C in pooled, sterile-filtered (pore size, 0.2 μm) human urine, LB, RPMI 1640 cell culture medium, or SCEM (22) for incubation periods of 2, 4, and 12 h. Residual coextracted DNA was removed from the purified RNA using an Ambion Turbo DNA-free kit (Life Technologies, Darmstadt, Germany).
qRT-PCR.
qRT-PCR was performed as previously described (23). A 2× SensiMix SYBR one-step kit (Peqlab Biotechnologie, Erlangen, Germany) and 200 nM each primer (17, 24) were used to measure the relative expression of mRNA of stxA1c, stxB2a, and stxB2b. The stx1c and stx2b primers were annealed at 58°C, and the stx2a primers were annealed at 62°C. stx transcript levels were normalized to those of gapA (glyceraldehyde-3-phosphate dehydrogenase) mRNA and compared with those at the 2-h time point. The experiments were performed three times with at least three independent RNA preparations. Comparison of the means of the relative transcript levels and of groups of relative transcript levels was performed using the Student t test and the two-way analysis of variance test, respectively.
Cell culture.
Eukaryotic cells were cultivated at 37°C in a 5% CO2 atmosphere in the presence of 10% fetal calf serum (FCS; PAA Laboratories, Cölbe, Germany). T24 cells (ATCC HTB-4) were grown in McCoy's 5A medium (PAA or Lonza, Cologne, Germany) containing 2 mM l-glutamine. HEp-2 cells (ATCC CCL-23) were cultivated in minimal essential medium with Earle's salts (Biochrom, Berlin, Germany) containing 2 mM l-glutamine, 1× nonessential amino acids (PAA), and 1 mM sodium pyruvate, whereas Vero cells (ATCC CCL-81) were grown in Dulbecco's modification of Eagle medium (DMEM; PAA or Biochrom).
Adhesion assay.
The patterns of adherence of the bacterial isolates to cells of both the human laryngeal HEp-2 and human bladder T24 epithelial cell lines were assayed as described previously (11), with the following modification. HEp-2 cells and T24 cells were grown as semiconfluent monolayers in Eagle's minimum essential medium (EMEM) and McCoy's 5A medium, respectively, in 4-well chamber slides for 24 to 48 h. The bacteria were grown under static conditions overnight in DMEM without FCS. On the next day, approximately 4 × 107 CFU was incubated on the cultured cell lines for 3 h at 37°C in 5% CO2 in the presence of 0.5% d-mannose in DMEM without FCS. Afterwards, the cells were washed three times with Hanks' balanced salt solution (HBSS), fixed with methanol for 1 min, and stained for 30 min with Giemsa stain freshly diluted 1:10 in water. The adherence pattern was evaluated under oil immersion with a bright-field microscope (Axiostar; Zeiss, Jena, Germany). The reference strains EAEC 042 and K-12 C600, as well as bfp-positive and bfp-negative isogenic EPEC variants 1083/87 and E. coli 904/90 (25–27), were included in this assay to validate our results.
Cytotoxicity assay.
Cytotoxicity tests were performed in 96-well plates as described before (14). Stx-mediated cellular damage was determined by using both Vero cells and T24 cells. One hundred microliters of serial dilutions of sterile-filtered bacterial overnight LB culture supernatants was added to confluent cell monolayers, and the cells were incubated for 72 h. Cell detachment was indirectly quantified by extracting crystal violet from remnant adherent cells and determining the optical density of the eluate. The Stx titer was defined as the highest dilution of culture supernatant that caused cytotoxicity in 50% of cells.
Extraction, isolation, and purification of neutral GSLs from T24 and Vero cells and from murine urinary bladders.
Neutral GSLs were extracted and purified from total cells or bladders as previously described (12, 28–30). The neutral GSL-containing fractions were adjusted to defined volumes of chloroform-methanol (2/1, vol/vol), and the amounts of GSLs corresponded to the amounts from 1 × 104 cells/μl (Vero cells) and 2 × 104 cells/μl (T24 cells) or 0.1 mg (wet weight)/μl of murine bladders.
Reference GSLs and antibodies.
Two neutral GSL reference mixtures were used for antibody- and Stx2-mediated overlay assays. Reference mixture 1 included GSLs from human erythrocytes, which contain lactosylceramide (Lc2Cer), globotriaosylceramide (Gb3Cer), and globotetraosylceramide (Gb4Cer) as the major compounds (31). GSL reference mixture 2 consisted of equimolar amounts of Gb3Cer and Gb4Cer (28). GSLs were designated according to the recommendations of IUPAC-IUB (32).
High-performance TLC and overlay assay.
GSL reference mixtures and purified neutral GSLs from the cell lines were separated by thin-layer chromatography (TLC) and either stained with orcinol or submitted to a TLC overlay assay (28). Antibody-mediated immunodetection of Gb3Cer and Gb4Cer and binding of Stx to Gb3Cer and Gb4Cer were visualized as described before (29).
Structural characterization of immunostained GSLs.
Immunostained GSLs were extracted from the silica gel as described before (28, 29). Mass spectrometric (MS) analysis of silica gel extracts from immunopositive GSL bands was performed by use of a quadrupole time of flight (Q-TOF) instrument (Micromass, Manchester, United Kingdom) equipped with a nanoelectrospray manipulator essentially as described before (33). The redissolved GSL silica gel extracts were analyzed in the positive-ion mode. For structural elucidation by tandem MS (MS2) experiments, low-energy collision-induced dissociation (CID) was performed with argon as the collision gas. The fragment ions obtained were assigned according to the nomenclature introduced by Domon and Costello (34).
Bacterial growth competition assay in pooled human urine.
To investigate the general fitness of the atypical UPEC strains in urine, pooled human urine was inoculated with one of the atypical UPEC strains (streptomycin sensitive) and with archetypal UPEC isolate 536 (streptomycin resistant) (35) in a 1:1 ratio. The cultures were incubated under static conditions at 37°C for 24 h. On the next day, a fresh urine culture was inoculated with the 24-h culture starting with an optical density at 600 nm of 0.02. The ratio of the atypical UPEC strain and archetypal UPEC strain 536 in the 24-h culture and the 48-h subculture was calculated after determination of the CFU count on LB agar and on streptomycin selective LB agar. The values represent the means and standard deviations of at least three independent experiments.
Animal experiments.
Groups of 8 to 16 female C57BL/6 mice (Charles River Laboratories, Sulzfeld, Germany) were transurethrally inoculated with 5 × 108 to 5 × 109 CFU of UPEC strain 536 (positive control), E. coli K-12 strain MG1655 (negative control), or the EHEC isolates, as previously described (36, 37). Bladder and kidney infections were evaluated by means of the unpaired, nonparametric one-tailed Mann-Whitney test. For the isolation of neutral GSLs, the urinary bladders of 10 3- to 5-month-old male C57BL/6 mice, which had a total tissue wet weight of 301.5 mg, were further processed according to previously described protocols (30). Animal experiments were performed according to the Guidelines for the Care and Use of Laboratory Animals (38) in compliance with German regulations (Tierschutzgesetz) and with permission of the regional government (AZ 55.2-2531.01–53/09).
RESULTS
Molecular epidemiology of EHEC strains isolated from hospital patients with UTIs.
On the basis of molecular epidemiological data, EHEC isolate UR5703/202 was allocated to sequence type 32 (ST32), clonal complex 32 (CC32), and serotype O145:H−, strain UR5779/201 was assigned to ST330, CC10, and serotype Ont:H− (where nt indicates nontypeable), and UR131 was assigned to ST675 and serotype O76:H19 (11). In this context, the three microorganisms resemble the typical HUS-associated EHEC strains HUSEC022 (ST137, CC32, O145:H−), HUSEC002 (ST330, CC10, Ont:H−), and HUSEC039 (ST675, O76:H19), respectively (39), and could clearly be classified as intestinal pathogens.
IPEC and ExPEC virulence gene content of UTI-associated EHEC isolates.
The three isolates were positive for EHEC hlyA (Table 1) but differed in their overall EHEC virulence gene repertoire. UR5703/202 and UR5779/201 carried stx2a and represented LEE-positive EHEC strains due to the presence of the escV, eae, espF, and map genes (Table 1). These two strains carried additional EHEC virulence markers, i.e., map, terE, and ureD. E. coli UR5703/202 also harbored paa, efa1, espG, tir, and cif. In contrast, strain UR131 harbored the stx1c and stx2b genes and the EHEC autoagglutinating adhesin-encoding gene saa (Table 1). saa has often been reported in LEE-negative EHEC strains (40), which is consistent with the absence of eae and LEE-associated genes in UR131. Strain UR5779/201 carried the estIA gene (Table 1), which codes for the heat-stable enterotoxin (STa) and is usually characteristic for enterotoxigenic E. coli (16). The three urinary EHEC isolates did not contained either the EPEC biomarker bfpA or any of the tested virulence loci of EAEC or aggregative adherence fimbria-encoding determinants (pet, aap, aggA, aafA, agg3A, hdaA, aaf5A), some of which occur in the EHEC O104:H4 hybrid that caused the large outbreak in 2011 (14).
Characteristic ExPEC virulence genes coding for toxins (α-hemolysin, CDT I to IV, CNF), capsules, or protectins were not detected in the three EHEC isolates. Type 1, P, S, and F1C fimbrial adhesins represent important ExPEC virulence factors (41, 42). None of the EHEC isolates, however, carried the P, S, or F1C fimbrial determinants. The type 1 fimbrial adhesin gene fimH was detected in isolates UR5703/202 and UR131. Expression of type 1 fimbriae has been closely associated with the early development of UTI (43, 44). Genes coding for G fimbriae or the M blood group-specific, Dr, or afimbrial adhesins were not detected. Only isolate UR5703/202 was iha positive (Table 1). Screening for iron acquisition determinants, including chuA, fyuA, irp2, iroN, and iutA, revealed that strain UR5779/201 harbors the yersiniabactin and the hemin uptake system. Strain UR5703/202 is chuA positive. E. coli UR131 lacks the yersiniabactin and hemin uptake systems but carries the cdiAB gene cluster, which is required for contact-dependent growth inhibition of other bacteria (Table 1). UR5779/201 and UR131 possess traT, characteristic for large (virulence) plasmids (Table 1). Altogether, our screening for IPEC and ExPEC virulence determinants demonstrated that the three urine isolates represent typical EHEC variants and lack the well-known major virulence determinants of ExPEC which have been described to be involved in uropathogenesis.
Phenotypic characterization of EHEC strains isolated from hospital patients with UTIs.
Whereas all three strains fermented sorbitol, only UR5703/202 and UR5779/201 grew on CT-SMAC, demonstrating their resistance to tellurite, which is a typical feature of EHEC O157:H7 and the major non-O157 EHEC serotypes (45). All three isolates expressed EHEC Hly and Stx, as demonstrated by their ability to produce a typical enterohemolysin phenotype and cause cytotoxicity on Vero cells, respectively (Table 1). In addition, E. coli UR131 produced colicins which inhibit the growth of E. coli DH5α (Table 1). Accordingly, the growth characteristics of the strains and the expression of typical EHEC toxins further corroborated the allocation of these isolates to the EHEC pathotype.
Phenotypes of UTI-associated adherence of EHEC strains to HEp-2 and T24 cells.
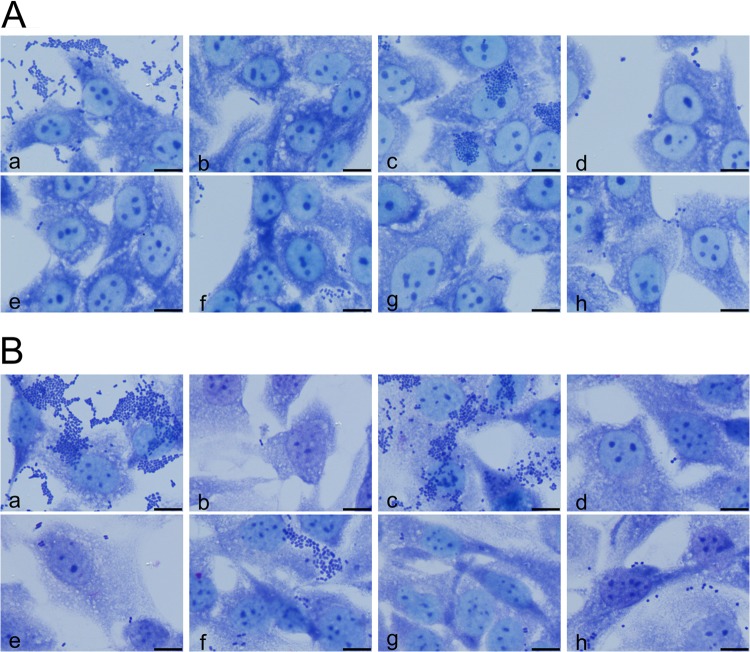
EHEC strains as well as typical EPEC strains adhere to the surface of HEp-2 cells as compact microcolonies, known as localized adherence (LA) (46). Strain UR131 exhibited an LA-like adherence pattern with HEp-2 cells which was weaker than that of the bfp-positive EPEC strain 1083/87, which was used as a positive control for LA adherence. The absence of the bfp gene cluster in this strain background (E. coli 904/90) resulted in the loss of the LA adherence phenotype. E. coli strains UR5779/201 and UR5703/202 did not markedly adhere to this cell line, similar to EHEC O157:H7 strain EDL933 (Fig. 1A). As the three strains UR131, UR5779/201, and UR5703/202 were recovered from patients suffering from UTIs, we also evaluated their pattern of adherence to the T24 human urinary bladder epithelial cell line (Fig. 1B). Here, again, E. coli isolate UR131, but not E. coli UR5703/202 and UR5779/201, clearly showed a characteristic LA-like adherence pattern similar to that of bfp-positive EPEC strain 1083/87, whereas EHEC O157:H7 strain EDL933, the bfp-negative EPEC strain 904/90, and the two isolates UR5779/201 and UR5703/202 did not adhere to T24 cells under the conditions used in this assay.
FIG 1.
Localized adherence pattern of EHEC isolates from hospitalized patients with UTIs. The adherence patterns of the urine isolates were investigated on HEp-2 cells (A) and on T24 urinary bladder epithelial cells (B). (a) EAEC strain 042 (positive control); (b) E. coli K-12 strain C600 (negative control); (c) EPEC O157:H45 strain 1083/97 (bfp positive); (d) EPEC O157:H45 strain 904/90 (bfp negative); (e) EHEC O157:H7 strain EDL933; (f) UR131; (g) UR5703/202; (h) UR5779/201. Bars = 10 μm.
Differential sensitivity of Vero and T24 cells to Stx.
As the atypical UPEC (aUPEC) isolates were able to interact with T24 cells, we investigated their susceptibility to Stxs. The Stx-containing culture supernatants of the three EHEC isolates were toxic for Vero cells, with Stx titers ranging from 128 to 256. T24 cells also responded to Stx but displayed a markedly decreased susceptibility, with titers ranging from 8 to 32 (Table 1). These results confirm that the urine EHEC isolates express Stx, which efficiently impairs the viability of Vero cells and, to a lesser extent, also that of T24 bladder epithelial cells. To demonstrate that the cytotoxic effect observed with the aUPEC strains resulted from Stx expression and not from other undefined factors, we compared the cytotoxicity of isogenic eae- and stx-positive and eae-positive and stx-negative E. coli O145:H28/NM (where NM represents nonmotile) strain pairs, which belong to the same clone as aUPEC isolate UR5703/202 and share the virulence characteristics, except for the presence of the stx gene (45, 47) (see Table S1 in the supplemental material). A cytotoxic effect could be detected in the cytotoxicity assay only for the stx-positive variants and not for the stx-negative variants. This finding supports the idea that the cytotoxicity observed in this assay is due to Stxs released by the aUPEC strains to the culture medium.
Interaction of Stx2 variants with neutral GSLs isolated from Vero and T24 cell lines.
In light of these results, we evaluated the GSL composition of T24 and Vero cells. GSLs are cognate receptors of Stxs and the P fimbrial UPEC adhesin (48, 49). We detected Gb3Cer and Gb4Cer in neutral GSL preparations of both cell lines. Vero cells contained more Gb3Cer and Gb4Cer than T24 cells, as deduced from the orcinol staining (see Fig. S1A, lanes b and c, in the supplemental material). This quantitative difference was stronger for Gb3Cer than for Gb4Cer, as shown in the TLC overlay assays whose results are shown in Fig. S1B and C (lanes b and c) in the supplemental material, using anti-Gb3Cer and anti-Gb4Cer antibodies, respectively. We tested whether Stx2a expressed by isolates UR5703/202 and UR5779/201 and Stx2b expressed by E. coli UR131 interacted with Gb3Cer and Gb4Cer of Vero and T24 cells (Fig. 2A to C). Stx2a and Stx2b generally interacted with Gb3Cer and Gb4Cer from Vero and T24 cells. Stx2 binding to Gb3Cer was much stronger than that to Gb4Cer, as indicated by the strong positive reaction with Gb3Cer and the weak binding to Gb4Cer of reference mixture 2, which contained equimolar quantities of the two GSLs (Fig. 2A to C, lanes d). Less interaction was detectable between the two Stx2 subtypes and the GSL fractions from T24 cells than the GSL fractions from Vero cells (Fig. 2A to C, lanes b and c). This can be explained by the lower content of Gb3Cer and Gb4Cer in the GSL preparation of T24 cells than in that of Vero cells (see Fig. S1 in the supplemental material). In the context of the expression of Gb3Cer and Gb4Cer, it is noteworthy that both Stx receptors were detectable in the neutral GSL fraction of C57BL/6 mouse bladders, suggesting their presence in bladder epithelial cells (see Fig. S2 in the supplemental material). The cognate receptors of Stxs are expressed in immortalized bladder epithelial cells as well as in murine bladders. Accordingly, Stxs released by aUPEC isolates can bind to Gb3Cer and Gb4Cer in the urinary tract and may affect urothelial cells.
FIG 2.
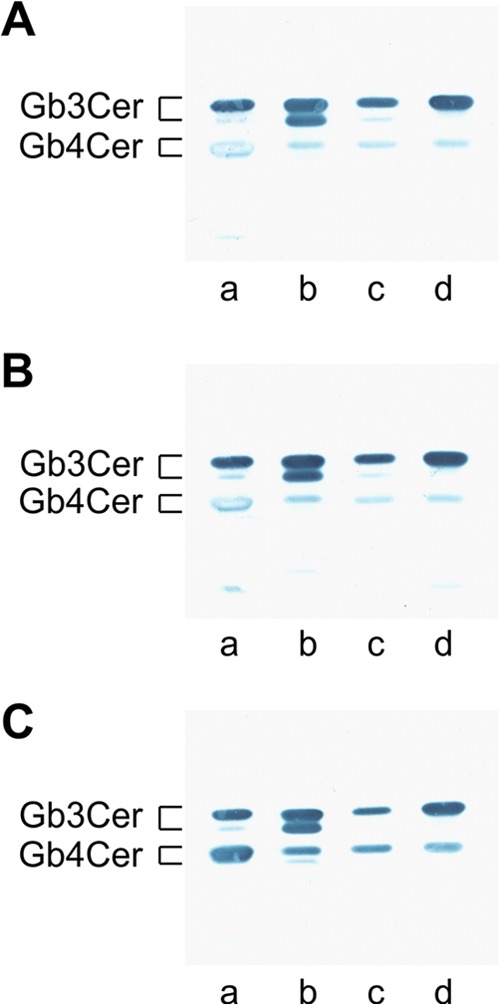
Stx2-mediated detection of Gb3Cer and Gb4Cer in neutral GSL preparations from Vero and T24 cells. (A to C) GSLs in amounts equivalent to those from 5 × 105 cells were applied (lanes b, Vero cells; lanes c, T24 cells). GSL reference mixture 1, composed of Gb4Cer as the predominant GSL and Gb3Cer in a lower abundance (lanes a), and reference mixture 2, containing equimolar amounts of Gb3Cer and Gb4Cer (lanes d), were present at 2 μg and 1 μg, respectively. Supernatants of EHEC isolates UR5703/202 (Stx2a) (A), UR5779/201 (Stx2a) (B), and UR131 (Stx2b) (C) were investigated.
Mass spectrometric identification of Gb3Cer and Gb4Cer lipoforms.
Structural characterization of the Gb3Cer (Fig. 3) and Gb4Cer (Fig. 4) variants present in T24 cells was performed by electrospray ionization (ESI)–Q-TOF MS from the silica gel extract of antibody-positive bands (cf. Fig. S1B and C, lanes c, in the supplemental material). The overview mass spectrum (MS1) derived from the Gb3Cer-positive double band is marked by a dotted rectangle in the inset in Fig. 3A. The mass spectrum indicated the presence of highly abundant Gb3Cer species with d18:1 sphingosine and long-chain fatty acid acyl residues (C24:0 at m/z 1,158.80, C24:1 at m/z 1,156.79, and C22:0 at m/z 1.130.79) in the ceramide moieties, in accordance with an intensively stained upper band, which is visible in the inset. Additionally, a Gb3Cer variant with a C16:0 fatty acid acyl residue allocated to the lower band was detected at m/z 1,046.69. To give evidence of the tentatively assigned structures, MS2 experiments were performed, and the low-energy CID spectrum obtained from Gb3Cer (d18:1, C24:0) is shown as a representative example in Fig. 3B, together with the corresponding fragmentation scheme and the molecular structure (Fig. 3C). The spectrum reveals a complete series of Y-type ions, i.e., Y2 ions at m/z 996.74, Y1 ions at m/z 834.72, and Y0 ions at m/z 672.65, which denote, together with amendatory Z-type ions, the loss of three hexose moieties. The nonreducing part of the sugar chain is represented by a full series of B-type ions, i.e., B1 ions at m/z 185.07, B2 ions at m/z 347.13, and B3 ions at m/z 509.18; the corresponding C-type ions; as well as internal fragment ions assigned as 0.2A2 at m/z 305.11 and 0.2A3 at m/z 467.19.
FIG 3.
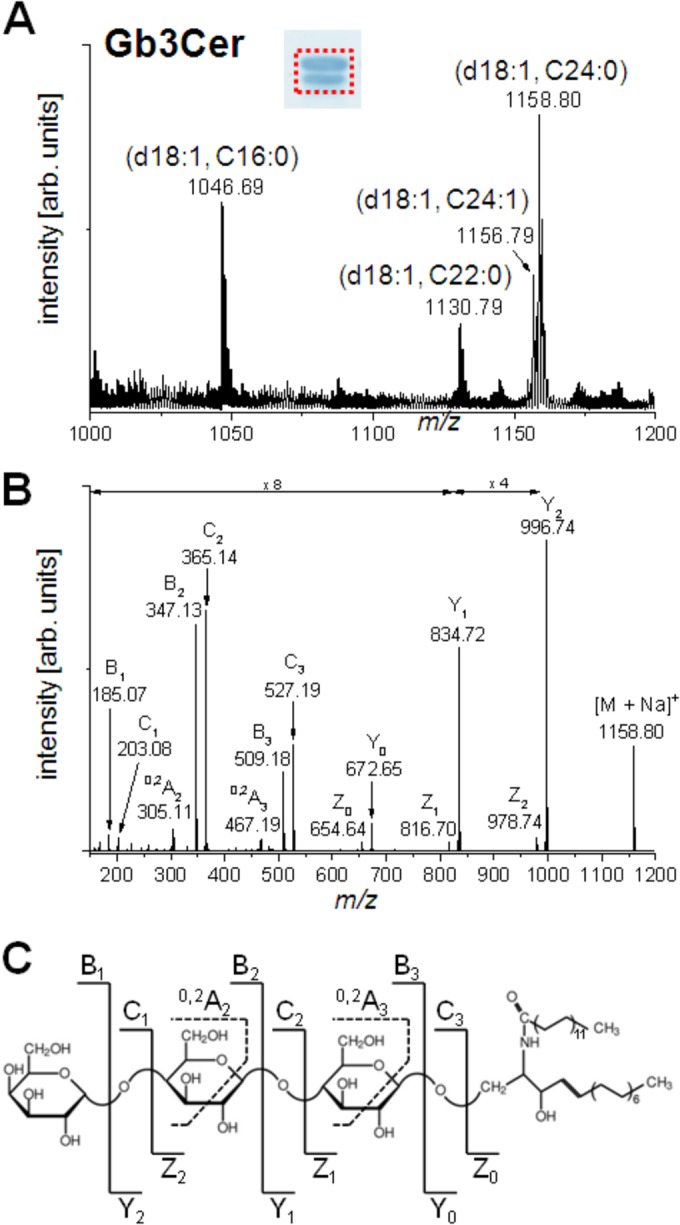
Positive-ion-mode ESI–Q-TOF mass spectra of antibody-detected Gb3Cer species from T24 cells. (A) Overview of the MS1 spectrum obtained from the silica gel extract of the TLC overlay assay (shown in the inset) performed with an aliquot of GSL at an amount corresponding to the amount from 1 × 106 cells. The area comprising the immunostained double band, from which the silica gel was scraped off, is indicated by a dotted rectangle. (B) Low-energy CID MS2 spectrum of monosodiated Gb3Cer (d18:1, C24:0) with m/z 1,158.80. (C) Molecular structure and explanatory fragmentation scheme of Gb3Cer (d18:1, C24:0). arb., arbitrary.
FIG 4.
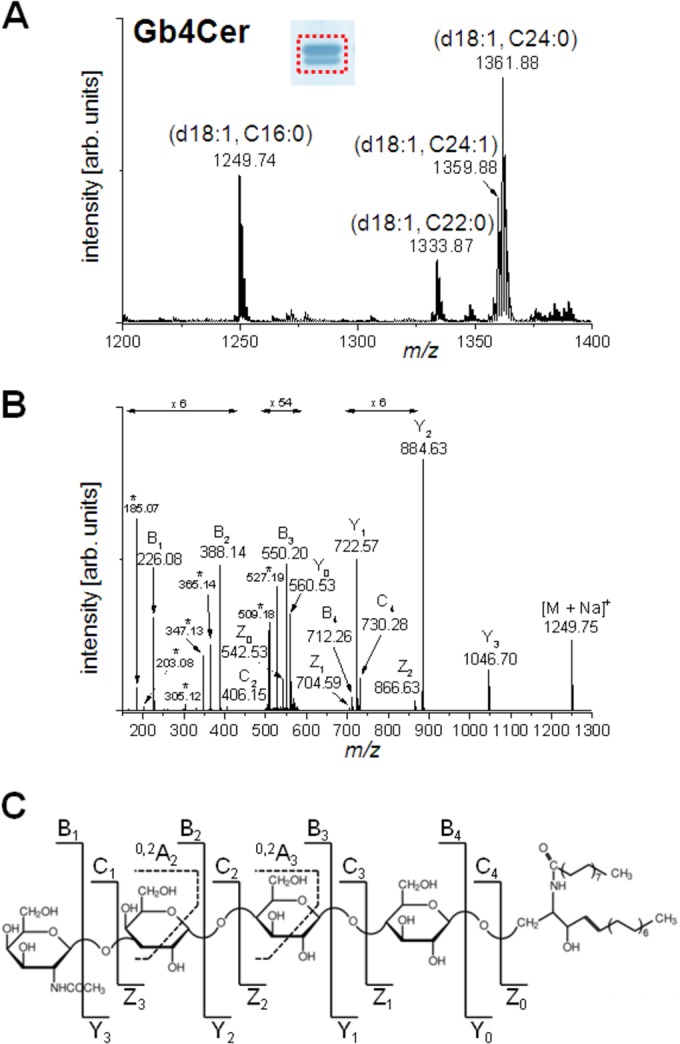
Positive-ion-mode ESI–Q-TOF mass spectra of antibody-detected Gb4Cer species from T24 cells. (A) Overview of the MS1 spectrum obtained from the silica gel extract of the TLC overlay assay (shown in the inset) performed with an aliquot of GSL at an amount corresponding to the amount from 6 × 105 cells. The area comprising the immunostained double band, from which the silica gel was scraped off, is indicated by a dotted rectangle. (B) Low-energy CID MS2 spectrum of monosodiated Gb4Cer (d18:1, C16:0) with m/z 1,249.75. Fragment ions generated by internal cleavages are marked by asterisks. (C) Molecular structure and explanatory fragmentation scheme of Gb4Cer (d18:1, C16:0).
The MS1 overview spectrum obtained from the silica gel extract of the immunopositive Gb4Cer double band is depicted in Fig. 4A, together with the results of the corresponding overlay assay in the inset. The variation within the ceramide part due to the substitution with different fatty acid acyl chains is similar to that observed for Gb3Cer. The prominent constituents detected corresponded to Gb4Cer with d18:1 and C16:0, C22:0, and C24:1/C24:0 fatty acid acyl chains at m/z 1,249.74, 1,333.87, and 1,359.88/1,361.88, respectively. The structural elucidation by low-energy CID is shown for the ions with m/z 1,249.75 in Fig. 4B together with the auxiliary fragmentation scheme of Gb4Cer (d18:1, C16:0) (Fig. 4C). The MS2 spectrum shows a full series of Y-type ions starting with Y3 ions at m/z 1,046.70, indicative of the loss of the terminal N-acetylhexosamine moiety, and ending with Y0 ions at m/z 560.53, representing the ceramide part. A complete set of complementary B-type ions, i.e., B1 to B4, comprising one to four sugar units, was detected, as were a few C-type, Z-type, and internal fragment ions (marked by asterisks in Fig. 4B), giving rise to the structural assignment Gb4Cer (d18:1, C16:0).
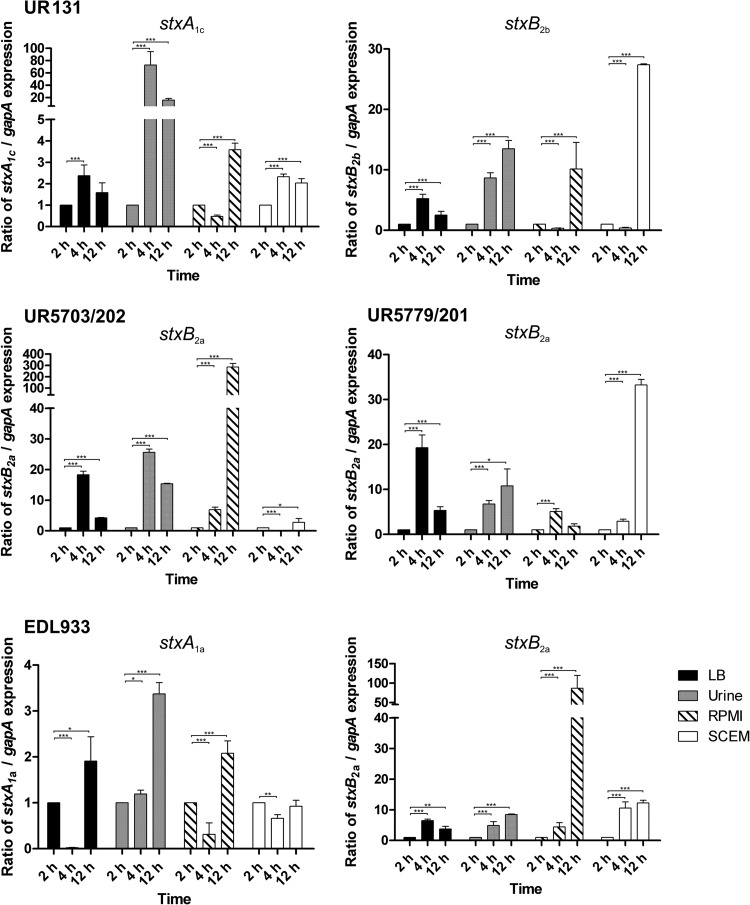
Transcription of stx1c, stx2a, and stx2b subtypes of the three UTI-associated EHEC isolates.
To analyze the impact of different growth media on the transcription of the stx genes in the different atypical UPEC isolates, we compared the relative levels of expression of the individual stx alleles upon growth in pooled human urine, LB, RPMI 1640 cell culture medium, and SCEM, which mimics the colonic environment. EHEC O157:H7 model isolate EDL933 was included as a reference (Fig. 5). Interestingly, we observed marked differences in the relative transcription of the individual stx alleles in response to different growth media and growth phases. Generally, the relative transcription of the stx1 alleles (stxA1c of E. coli UR131 as well as stxA1a of E. coli EDL933) was markedly lower than the relative transcription of the stx2 alleles (stxB2a, stxB2b). The levels of relative transcription of the stx alleles were often, but not always, higher in the stationary phase than in the early or late exponential growth phase (Fig. 5).
FIG 5.
Real time-PCR-based quantification of transcript levels of different stx alleles in EHEC O157:H7 isolate EDL933 and urine isolates UR5703/202, UR5779/201, and UR131. Data represent the normalized fold expression levels of the different stx alleles upon growth in pooled human urine, LB, RPMI 1640, or SCEM after 2 h, 4 h, and 12 h of growth. Error bars show the standard errors of the means of three independent experiments and technical replicates. The individual stx transcript levels were normalized to the transcript level of the housekeeping gene gapA and compared with the level at the starting point (2 h). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Whereas the transcription of stxA1c of E. coli UR131 strongly responded only to growth in urine, growth in RPMI 1640 cell culture medium or SCEM did not drastically affect its relative transcription. In this strain, the last two growth media had a significant positive effect on stxB2b transcription in the stationary phase. Significantly increased stxA1c and stxB2b transcript levels in pooled human urine relative to those in LB were observed for E. coli UR131 (P < 0.0001). Except for the transcription of stxB2a of E. coli UR5779/201 at the 4-h time point, the relative stxB2a transcription of isolates UR5703/202 and UR5779/201 was significantly higher in urine than in LB (P < 0.0001). The relative transcription of stxB2a of strains UR5703/202 and EDL933 in response to the different growth conditions was similar. Here, a significantly increased relative transcription of stxB2a was observed upon prolonged growth in RPMI 1640 medium. Interestingly, the relative transcription of stxB2a of E. coli UR5779/201 did not resemble that of stxB2a of strains UR5703/202 and EDL933 and was not markedly increased in RPMI 1640 medium. Instead, growth in SCEM resulted in significantly increased stxB2a transcription.
These data show an allele-specific stx transcription pattern depending on the stx subtype, growth phase, and growth medium. Our data demonstrate that the stx genes are also transcribed upon growth in pooled human urine. This may support a virulence-associated function of Stx during uropathogenesis.
Some UTI-associated EHEC strains are able to cause ascending UTI.
An experimental murine model of ascending UTI was used to assess and compare the uropathogenic potential of the three EHEC isolates. Groups of 8 to 16 mice were transurethrally inoculated with 5 × 108 to 5 × 109 CFU of strain 536, nonpathogenic E. coli K-12 strain MG1655, or the atypical UTI isolates. According to the bacterial counts in the bladders and in the kidneys at 72 h postinfection (Fig. 6A and B, respectively), EHEC strains UR131 and UR5703/202 were recovered in nearly equal numbers from the bladder tissue (1 × 105 to 5 × 105 CFU/g bladder tissue), like archetypal UPEC strain 536 (Fig. 6A). However, only infection with UPEC strain 536 resulted in bacterial loads in the bladder tissue significantly higher than those achieved with infection with the negative control, nonpathogenic E. coli K-12 strain MG1655. EHEC strains UR131 and UR5703/202 also colonized the kidneys (Fig. 6B) at least as efficiently as UPEC strain 536 did and reached significantly higher numbers of CFU than the nonpathogenic strain MG1655, which was unable to ascend to the kidneys. Infection with strain UR131 even resulted in significantly higher bacterial loads in the kidney than infection with UPEC strain 536 did (Fig. 6B). Accordingly, these two EHEC strains exhibit a potential to cause UTI which is comparable to that of archetypal UPEC strain 536. Interestingly, the type 1 fimbrial adhesin gene fimH could be amplified from both isolates. EHEC isolate UR5779/201, however, was not able to cause UTIs in mice and neither colonized the bladder nor ascended to the kidneys (Fig. 6A and B). Curiously, fimH could not be detected by PCR in this strain. Our results suggest that under certain circumstances members of the EHEC pathotype are able to cause UTIs, despite the fact that such organisms are usually not considered uropathogens.
FIG 6.
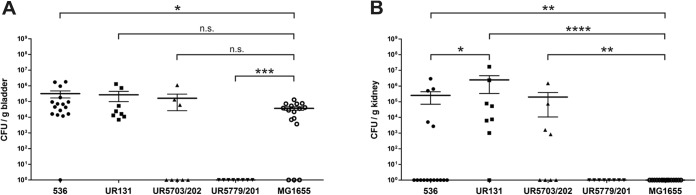
Urovirulence of EHEC urine isolates. Bladder (A) and kidney (B) colonization levels were determined 72 h after transurethral inoculation of female C57BL/6 mice. The mice were independently challenged with UPEC strain 536 (positive control), EHEC urine isolate UR131, UR5703/202, or UR5779/201, or nonpathogenic E. coli K-12 strain MG1655 (negative control). Horizontal bars represent the mean number of CFU/g tissue for each population; the whiskers display the respective standard errors of the means. Significant differences in the bacterial organ load compared to that for the negative controls are indicated: n.s., no significant difference; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Competitive growth experiments with model UPEC isolate 536 demonstrated that aUPEC strains UR131 and UR5703/202 could not be outcompeted by strain 536. The typical UPEC strain 536 represented more than 90% of the entire culture population after 48 h of static growth in pooled human urine in vitro only when it was in coculture with aUPEC isolate UR5779/201 (Fig. S3 in the supplemental material). This observation corroborates the results of the experimental ascending UTI model, where only E. coli UR131 and UR5703/202 and not strain UR5779/201 were able to efficiently colonize the murine bladder and to ascend to the kidneys (Fig. 6).
DISCUSSION
Certain EHEC strains have the potential to cause UTIs.
EHEC strains have occasionally been documented to be the causative agents of UTIs in humans (9, 50–55). Many questions regarding the urovirulence mechanisms and the uropathogenicity of EHEC strains remain unanswered. We characterized three EHEC isolates recovered from hospital patients with symptomatic UTIs (Table 1). These three strains belong to typical HUS-associated EHEC clones (11), i.e., HUSEC002 (ST330, CC10, Ont:H−), HUSEC022 (ST137, CC32, O145:H−), and HUSEC039 (ST675, O76:H19) (http://www.ehec.org/index.php?id=5562&L=1). Bacteria with the same traits have frequently been reported to be highly pathogenic and, frequently, HUS-associated diarrheagenic E. coli strains (56–58). Nevertheless, none of the UTI patients had been admitted to the hospital due to diarrhea or HUS. The three UTI-associated EHEC isolates did not carry characteristic ExPEC virulence genes, so how can they cause UTIs?
E. coli UTI isolates have been described to grow significantly faster than commensal E. coli isolates in vitro in urine but not in LB. The increased growth capacity of UPEC strains in urine has been considered a fitness trait (59). In our study, however, the EHEC strains grew in pooled human urine as well as typical UPEC strains did. In another study, the growth rates of commensal E. coli, IPEC, and ExPEC strains in pooled human urine and LB were similar. The authors' conclusion that efficient growth in urine should not be considered a marker of urovirulence (60) corroborates our findings that fast growth in urine is not confined to typical UPEC strains (11, 61). In growth competition assays, where archetypal UPEC isolate 536 and one of the aUPEC isolates were grown together in pooled human urine for 48 h, aUPEC isolates UR131 and UR5703/202 could not be outcompeted by UPEC strain 536 and remained present in large numbers in the mixed culture population, whereas aUPEC strain UR5779/201 was almost completely outcompeted by UPEC strain 536. This indicates that the fitness of aUPEC strains UR131 and UR5703/202 in urine was not markedly reduced relative to that of model UPEC isolate 536, although these two strains represent HUS-associated E. coli bacteria, i.e., typical members of the IPEC pathotype. Strain UR5779/201, which was isolated from a case of symptomatic cystitis, displayed drastically reduced competitiveness compared to that of UPEC strain 536 only in pooled human urine. Accordingly, this strain's fitness in urine seemed to be impaired. This may explain why E. coli UR5779/201 was not able to colonize the bladder and to cause ascending UTI. The molecular basis for the reduced urovirulence in the infection model used in this study remains to be further investigated. The absence of typical adhesins of EHEC, EPEC, EAEC, and ExPEC strains determined in this study suggests that other adhesins may contribute to the adherence of the aUPEC strains in the urinary tract. In the case of saa-harboring strain UR131, we demonstrated LA-like adherence to HEp-2 cells and even more pronounced adherence to T24 cells, even though this strain lacks bundle-forming pili. stx1c-harboring EHEC strains often lack the eae gene but express the adhesin Saa (62). Saa has been suggested to be an alternative adhesin of LEE-negative strains responsible for the so-called semilocalized adherence pattern (62, 63). Consequently, Saa may contribute to the semilocalized adherence to urothelial cells.
Stxs interact with and damage T24 human bladder epithelial cells.
Bacterial adherence to the urothelium represents the first step of UTI. The cognate Stx receptors, GSLs of the globo series, are present in tissues of the human urinary tract, serving as important receptors required for P fimbria-mediated adhesion and ascension of UPEC (64–66).
To assess the uropathogenic potential of the EHEC urine isolates, we investigated the susceptibility of human bladder epithelial cells to Stxs. Vero cells were highly susceptible to Stx (Table 1). Although T24 cells were less susceptible to Stx than Vero cells, they were clearly damaged by Stx (Table 1), and the higher content of Gb3Cer and Gb4Cer in Vero cells correlated with increased Stx susceptibility relative to that of T24 cells (Fig. 2; see also Fig. S1 in the supplemental material). The stx determinants were transcribed in urine, and the high level of relative stx expression compared to the level of growth in LB (Fig. 5) suggests that this EHEC virulence factor may also contribute to uropathogenesis. These data support the hypothesis that Stxs expressed by EHEC strains colonizing the urinary tract may have a virulence-associated function and could injure the human urothelium. The cytotoxicity assays with culture supernatants of isogenic eae-positive and stx-positive E. coli O145:H28/NM and eae-positive stx-negative E. coli O145:H28/NM strain pairs, which belong to the same sequence type as aUPEC strain UR5703/201, corroborated the finding that the Stx content in the supernatant is responsible for the cytotoxicity, whereas the contribution of other undefined factors expressed by aUPEC can be ignored for this phenotype (see Table S1 in the supplemental material).
A considerable structural homology was found for the Gb3Cer and Gb4Cer lipoforms carrying ceramides composed of sphingosine (d18:1) and acyl chains with various chain lengths (C16, C22, and C24) (Fig. 3 and 4). These receptor variants represent the canonical Stx receptors in the human vasculature and are targets for Stxs in EHEC-mediated diseases (12).
Urine EHEC isolates cause ascending UTI in a murine model.
Two of the three EHEC strains colonized the urinary bladder and the kidneys as efficiently as prototypic UPEC strain 536. E. coli strain UR5779/201, however, was not able to colonize either the urinary bladder or the kidneys (Fig. 6). The ability of EHEC to infect the urinary tract was independent of the presence of the LEE pathogenicity island. Binding of the Stx types released by the three urine EHEC isolates to globo-series GSLs, which are abundantly present in mouse bladders, ureters, and kidneys (67, 68), suggests involvement in the pathogenesis of urinary tract infections. Interestingly, only the two fimH-positive isolates, isolates UR131 and UR5703/202, caused ascending UTIs. In the murine UTI model, type 1 fimbriae play an important role in the initial adherence to uroepithelial cells (43, 44). Hung and coworkers reported on amino acid sequence variation in the FimH mannose-binding pocket which was conserved in UPEC strains but contained a sequence variation in EHEC strains (69). They interpreted their findings in light of the attenuation of EHEC virulence in the urinary tract, thus preventing them from colonizing this niche. The role of type 1 fimbria expression of the EHEC strains investigated in our study will require further analysis regarding their contribution to adhesion to urothelial cells. Additionally, the impact of other adhesins and additional IPEC virulence factors expressed by the aUPEC isolates on urovirulence or on fitness and competitiveness in urine should be analyzed in the future.
Our results corroborate the finding that E. coli strains with an IPEC genetic background but without characteristic UPEC virulence genes can have uropathogenic potential. Interestingly, Stxs and P fimbriae recognize Gb3Cer and Gb4cer as cellular receptors, and these GSLs are frequently expressed in the urinary tract. We demonstrate that EHEC can interact with T24 bladder epithelial cells. The findings that (i) stx alleles are transcribed in urine, (ii) Stx binds to Gb3Cer and Gb4Cer expressed in T24 cells as well as in murine bladder tissue, and (iii) T24 cells respond to Stx released by EHEC suggest that Stx, among other factors, may contribute to the uropathogenicity of EHEC. Clearly, Stx may be one of only several virulence factors expressed by IPEC which can promote colonization and infection of the urinary tract. Our real-time RT-PCR analysis indicated that stx transcription is not exclusively induced upon growth in urine (Fig. 5). Transcription of the individual stx alleles seems to be regulated in a strain-specific manner. Other growth media, such as RPMI 1640 cell culture medium or SCEM, which mimics the growth conditions in the colon, may also result in increased relative stx transcription compared to that in LB (Fig. 5).
Although EHEC is clearly not a major cause of UTIs, the isolation of such strains from health care-associated UTI cases, together with the results of our experimental UTI model, corroborate the findings that at least some EHEC isolates might be able to cause UTIs. The underlying molecular mechanisms and bacterial determinants involved require further in-depth investigations. Taking into account the fact that the host's own fecal flora is the primary source of UPEC (70), it is tempting to speculate that the patients from this study could have been asymptomatically colonized with EHEC as the result of a previous intestinal infection.
Supplementary Material
ACKNOWLEDGMENTS
F.T. received a Ph.D. fellowship from the German Academic Exchange Service (DAAD). The study was carried out within the UroGenOmics consortium (Medizinische Infektionsgenomik Program, Federal Ministry of Education and Research [BMBF], grant no. 0315833B). U.D. was supported by the German Research Foundation (DO789/4-1). This work has been further supported by the Interdisciplinary Center for Clinical Research (IZKF) Münster, project no. Müth2/028/10 (to J.M. and I.M.), and the project MU845/4-2 (to J.M. and I.U.K.) of the German Research Foundation.
We thank O. Mantel (Münster) for technical assistance.
Footnotes
Published ahead of print 25 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01701-14.
REFERENCES
- 1.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140. 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 2.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. 2013. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 26:822–880. 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russo TA, Johnson JR. 2000. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 181:1753–1754. 10.1086/315418. [DOI] [PubMed] [Google Scholar]
- 4.Karch H, Tarr PI, Bielaszewska M. 2005. Enterohaemorrhagic Escherichia coli in human medicine. Int. J. Med. Microbiol. 295:405–418. 10.1016/j.ijmm.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Melton-Celsa A, Mohawk K, Teel L, O'Brien A. 2012. Pathogenesis of Shiga-toxin producing Escherichia coli. Curr. Top. Microbiol. Immunol. 357:67–103. 10.1007/82_2011_176. [DOI] [PubMed] [Google Scholar]
- 6.Johnson JR, Russo TA. 2002. Extraintestinal pathogenic Escherichia coli: “the other bad E. coli. ” J. Lab. Clin. Med. 139:155–162. 10.1067/mlc.2002.121550. [DOI] [PubMed] [Google Scholar]
- 7.Johnson JR, Russo TA. 2005. Molecular epidemiology of extraintestinal pathogenic (uropathogenic) Escherichia coli. Int. J. Med. Microbiol. 295:383–404. 10.1016/j.ijmm.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Köhler CD, Dobrindt U. 2011. What defines extraintestinal pathogenic Escherichia coli? Int. J. Med. Microbiol. 301:642–647. 10.1016/j.ijmm.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Beutin L, Karch H, Aleksic S, Spencker FB, Rosenbaum U. 1994. Occurrence of verotoxin (Shiga-like toxin) producing Escherichia coli in human urinary tract infection. Infection 22:425. 10.1007/BF01715504. [DOI] [PubMed] [Google Scholar]
- 10.Johnson JR, Jerome C, Boster DR, Stapleton AE, Tarr PI. 2002. Analysis of urinary Escherichia coli isolates for ability to produce Shiga toxin. J. Clin. Microbiol. 40:2247–2248. 10.1128/JCM.40.6.2247-2248.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toval F, Köhler CD, Vogel U, Wagenlehner F, Mellmann A, Fruth A, Schmidt MA, Karch H, Bielaszewska M, Dobrindt U. 2014. Characterization of Escherichia coli isolates from hospital inpatients or outpatients with urinary tract infection. J. Clin. Microbiol. 52:407–418. 10.1128/JCM.02069-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betz J, Bielaszewska M, Thies A, Humpf HU, Dreisewerd K, Karch H, Kim KS, Friedrich AW, Müthing J. 2011. Shiga toxin glycosphingolipid receptors in microvascular and macrovascular endothelial cells: differential association with membrane lipid raft microdomains. J. Lipid Res. 52:618–634. 10.1194/jlr.M010819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 14.Bielaszewska M, Mellmann A, Zhang W, Köck R, Fruth A, Bauwens A, Peters G, Karch H. 2011. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. 11:671–676. 10.1016/S1473-3099(11)70165-7. [DOI] [PubMed] [Google Scholar]
- 15.Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261–272. 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 16.Müller D, Greune L, Heusipp G, Karch H, Fruth A, Tschäpe H, Schmidt MA. 2007. Identification of unconventional intestinal pathogenic Escherichia coli isolates expressing intermediate virulence factor profiles by using a novel single-step multiplex PCR. Appl. Environ. Microbiol. 73:3380–3390. 10.1128/AEM.02855-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheutz F, Teel LD, Beutin L, Piérard D, Buvens G, Karch H, Mellmann A, Caprioli A, Tozzoli R, Morabito S, Strockbine NA, Melton-Celsa AR, Sanchez M, Persson S, O'Brien AD. 2012. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 50:2951–2963. 10.1128/JCM.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oswald E, Schmidt H, Morabito S, Karch H, Marches O, Caprioli A. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect. Immun. 68:64–71. 10.1128/IAI.68.1.64-71.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang WL, Köhler B, Oswald E, Beutin L, Karch H, Morabito S, Caprioli A, Suerbaum S, Schmidt H. 2002. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J. Clin. Microbiol. 40:4486–4492. 10.1128/JCM.40.12.4486-4492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bielaszewska M, Stoewe F, Fruth A, Zhang W, Prager R, Brockmeyer J, Mellmann A, Karch H, Friedrich AW. 2009. Shiga toxin, cytolethal distending toxin, and hemolysin repertoires in clinical Escherichia coli O91 isolates. J. Clin. Microbiol. 47:2061–2066. 10.1128/JCM.00201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brockmeyer J, Bielaszewska M, Fruth A, Bonn ML, Mellmann A, Humpf HU, Karch H. 2007. Subtypes of the plasmid-encoded serine protease EspP in Shiga toxin-producing Escherichia coli: distribution, secretion, and proteolytic activity. Appl. Environ. Microbiol. 73:6351–6359. 10.1128/AEM.00920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polzin S, Huber C, Eylert E, Elsenhans I, Eisenreich W, Schmidt H. 2013. Growth media simulating ileal and colonic environments affect the intracellular proteome and carbon fluxes of enterohemorrhagic Escherichia coli O157:H7 strain EDL933. Appl. Environ. Microbiol. 79:3703–3715. 10.1128/AEM.00062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brockmeyer J, Aldick T, Soltwisch J, Zhang W, Tarr PI, Weiss A, Dreisewerd K, Müthing J, Bielaszewska M, Karch H. 2011. Enterohaemorrhagic Escherichia coli haemolysin is cleaved and inactivated by serine protease EspPα. Environ. Microbiol. 13:1327–1341. 10.1111/j.1462-2920.2011.02431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W, Bielaszewska M, Bauwens A, Fruth A, Mellmann A, Karch H. 2012. Real-time multiplex PCR for detecting Shiga toxin 2-producing Escherichia coli O104:H4 in human stools. J. Clin. Microbiol. 50:1752–1754. 10.1128/JCM.06817-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Appleyard RK. 1954. Segregation of new lysogenic types during growth of a doubly lysogenic strain derived from Escherichia coli K12. Genetics 39:440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaudhuri RR, Sebaihia M, Hobman JL, Webber MA, Leyton DL, Goldberg MD, Cunningham AF, Scott-Tucker A, Ferguson PR, Thomas CM, Frankel G, Tang CM, Dudley EG, Roberts IS, Rasko DA, Pallen MJ, Parkhill J, Nataro JP, Thomson NR, Henderson IR. 2010. Complete genome sequence and comparative metabolic profiling of the prototypical enteroaggregative Escherichia coli strain 042. PLoS One 5:e8801. 10.1371/journal.pone.0008801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt H, Rüssmann H, Karch H. 1993. Virulence determinants in nontoxinogenic Escherichia coli O157 strains that cause infantile diarrhea. Infect. Immun. 61:4894–4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Storck W, Meisen I, Gianmoena K, Pläger I, Kouzel IU, Bielaszewska M, Haier J, Mormann M, Humpf HU, Karch H, Müthing J. 2012. Shiga toxin glycosphingolipid receptor expression and toxin susceptibility of human pancreatic ductal adenocarcinomas of differing origin and differentiation. Biol. Chem. 393:785–799. 10.1515/hsz-2012-0165. [DOI] [PubMed] [Google Scholar]
- 29.Kouzel IU, Pohlentz G, Storck W, Radamm L, Hoffmann P, Bielaszewska M, Bauwens A, Cichon C, Schmidt MA, Mormann M, Karch H, Müthing J. 2013. Association of Shiga toxin glycosphingolipid receptors with membrane microdomains of toxin-sensitive lymphoid and myeloid cells. J. Lipid Res. 54:692–710. 10.1194/jlr.M031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Müthing J, Maurer U, Šoštarić K, Neumann U, Brandt H, Duvar S, Peter-Katalinić J, Weber-Schürholz S. 1994. Different distributions of glycosphingolipids in mouse and rabbit skeletal muscle demonstrated by biochemical and immunohistological analyses. J. Biochem. 115:248–256. [DOI] [PubMed] [Google Scholar]
- 31.Souady J, Soltwisch J, Dreisewerd K, Haier J, Peter-Katalinić J, Müthing J. 2009. Structural profiling of individual glycosphingolipids in a single thin-layer chromatogram by multiple sequential immunodetection matched with direct IR-MALDI-o-TOF mass spectrometry. Anal. Chem. 81:9481–9492. 10.1021/ac901948h. [DOI] [PubMed] [Google Scholar]
- 32.Chester MA. 1998. IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature of glyolipids—recommendations 1997. Eur. J. Biochem. 257:293–298. [DOI] [PubMed] [Google Scholar]
- 33.Meisen I, Rosenbrück R, Galla HJ, Hüwel S, Kouzel IU, Mormann M, Karch H, Müthing J. 2013. Expression of Shiga toxin 2e glycosphingolipid receptors of primary porcine brain endothelial cells and toxin-mediated breakdown of the blood-brain barrier. Glycobiology 23:745–759. 10.1093/glycob/cwt013. [DOI] [PubMed] [Google Scholar]
- 34.Domon B, Costello CE. 1988. Structure elucidation of glycosphingolipids and gangliosides using high-performance tandem mass spectrometry. Biochemistry 27:1534–1543. 10.1021/bi00405a021. [DOI] [PubMed] [Google Scholar]
- 35.Brzuszkiewicz E, Brüggemann H, Liesegang H, Emmerth M, Ölschläger T, Nagy G, Albermann K, Wagner C, Buchrieser C, Emödy L, Gottschalk G, Hacker J, Dobrindt U. 2006. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl. Acad. Sci. U. S. A. 103:12879–12884. 10.1073/pnas.0603038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagberg L, Engberg I, Freter R, Lam J, Olling S, Svanborg Eden C. 1983. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect. Immun. 40:273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bielaszewska M, Schiller R, Lammers L, Bauwens A, Fruth A, Middendorf B, Schmidt MA, Tarr PI, Dobrindt U, Karch H, Mellmann A. 2014. Heteropathogenic virulence and phylogeny reveal phased pathogenic metamorphosis in Escherichia coli O2:H6. EMBO Mol. Med. 6:347–357. 10.1002/emmm.201303133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 39.Mellmann A, Bielaszewska M, Köck R, Friedrich AW, Fruth A, Middendorf B, Harmsen D, Schmidt MA, Karch H. 2008. Analysis of collection of hemolytic uremic syndrome-associated enterohemorrhagic Escherichia coli. Emerg. Infect. Dis. 14:1287–1290. 10.3201/eid1408.071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jenkins C, Perry NT, Cheasty T, Shaw DJ, Frankel G, Dougan G, Gunn GJ, Smith HR, Paton AW, Paton JC. 2003. Distribution of the saa gene in strains of Shiga toxin-producing Escherichia coli of human and bovine origins. J. Clin. Microbiol. 41:1775–1778. 10.1128/JCM.41.4.1775-1778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klemm P, Hancock V, Schembri MA. 2010. Fimbrial adhesins from extraintestinal Escherichia coli. Environ. Microbiol. Rep. 2:628–640. 10.1111/j.1758-2229.2010.00166.x. [DOI] [PubMed] [Google Scholar]
- 42.Oelschlaeger TA, Dobrindt U, Hacker J. 2002. Virulence factors of uropathogens. Curr. Opin. Urol. 12:33–38. 10.1097/00042307-200201000-00007. [DOI] [PubMed] [Google Scholar]
- 43.Connell I, Agace W, Klemm P, Schembri M, Marild S, Svanborg C. 1996. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. U. S. A. 93:9827–9832. 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gunther NW, IV, Snyder JA, Lockatell V, Blomfield I, Johnson DE, Mobley HL. 2002. Assessment of virulence of uropathogenic Escherichia coli type 1 fimbrial mutants in which the invertible element is phase-locked on or off. Infect. Immun. 70:3344–3354. 10.1128/IAI.70.7.3344-3354.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bielaszewska M, Middendorf B, Köck R, Friedrich AW, Fruth A, Karch H, Schmidt MA, Mellmann A. 2008. Shiga toxin-negative attaching and effacing Escherichia coli: distinct clinical associations with bacterial phylogeny and virulence traits and inferred in-host pathogen evolution. Clin. Infect. Dis. 47:208–217. 10.1086/589245. [DOI] [PubMed] [Google Scholar]
- 46.Liebchen A, Benz I, Mellmann A, Karch H, Gomes TA, Yamamoto D, Hernandes RT, Sampaio J, Sampaio SC, Fruth A, Schmidt MA. 2011. Characterization of Escherichia coli strains isolated from patients with diarrhea in Sao Paulo, Brazil: identification of intermediate virulence factor profiles by multiplex PCR. J. Clin. Microbiol. 49:2274–2278. 10.1128/JCM.00386-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bielaszewska M, Köck R, Friedrich AW, von Eiff C, Zimmerhackl LB, Karch H, Mellmann A. 2007. Shiga toxin-mediated hemolytic uremic syndrome: time to change the diagnostic paradigm? PLoS One 2:e1024. 10.1371/journal.pone.0001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lindberg AA, Brown JE, Stromberg N, Westling-Ryd M, Schultz JE, Karlsson KA. 1987. Identification of the carbohydrate receptor for Shiga toxin produced by Shigella dysenteriae type 1. J. Biol. Chem. 262:1779–1785. [PubMed] [Google Scholar]
- 49.Müsken A, Souady J, Dreisewerd K, Zhang W, Distler U, Peter-Katalinić J, Miller-Podraza H, Karch H, Müthing J. 2010. Application of thin-layer chromatography/infrared matrix-assisted laser desorption/ionization orthogonal time-of-flight mass spectrometry to structural analysis of bacteria-binding glycosphingolipids selected by affinity detection. Rapid Commun. Mass Spectrom. 24:1032–1038. 10.1002/rcm.4480. [DOI] [PubMed] [Google Scholar]
- 50.Chiurchiu C, Firrincieli A, Santostefano M, Fusaroli M, Remuzzi G, Ruggenenti P. 2003. Adult nondiarrhea hemolytic uremic syndrome associated with Shiga toxin Escherichia coli O157:H7 bacteremia and urinary tract infection. Am. J. Kidney Dis. 41:E4. 10.1053/ajkd.2003.50022. [DOI] [PubMed] [Google Scholar]
- 51.Gadea Mdel P, Deza N, Mota MI, Carbonari C, Robatto M, D'Astek B, Balseiro V, Bazet C, Rugnitz E, Livrelli V, Schelotto F, Rivas M, Varela G. 2012. Two cases of urinary tract infection caused by Shiga toxin-producing Escherichia coli O157:H7 strains. Rev. Argent. Microbiol. 44:94–96. [PubMed] [Google Scholar]
- 52.Hogan MC, Gloor JM, Uhl JR, Cockerill FR, Milliner DS. 2001. Two cases of non-O157:H7 Escherichia coli hemolytic uremic syndrome caused by urinary tract infection. Am. J. Kidney Dis. 38:E22. [DOI] [PubMed] [Google Scholar]
- 53.Nguyen QV, Hochstrasser L, Chuard C, Hachler H, Regamey C, Descombes E. 2007. Adult hemolytic-uremic syndrome associated with urosepsis due to Shigatoxin-producing Escherichia coli O138:H. Ren. Fail. 29:747–750. 10.1080/08860220701460418. [DOI] [PubMed] [Google Scholar]
- 54.Starr M, Bennett-Wood V, Bigham AK, de Koning-Ward TF, Bordun AM, Lightfoot D, Bettelheim KA, Jones CL, Robins-Browne RM. 1998. Hemolytic-uremic syndrome following urinary tract infection with enterohemorrhagic Escherichia coli: case report and review. Clin. Infect. Dis. 27:310–315. 10.1086/514656. [DOI] [PubMed] [Google Scholar]
- 55.Tarr PI, Fouser LS, Stapleton AE, Wilson RA, Kim HH, Vary JC, Jr, Clausen CR. 1996. Hemolytic-uremic syndrome in a six-year-old girl after a urinary tract infection with Shiga-toxin-producing Escherichia coli O103:H2. N. Engl. J. Med. 335:635–638. 10.1056/NEJM199608293350905. [DOI] [PubMed] [Google Scholar]
- 56.Beutin L, Zimmermann S, Gleier K. 1998. Human infections with Shiga toxin-producing Escherichia coli other than serogroup O157 in Germany. Emerg. Infect. Dis. 4:635–639. 10.3201/eid0404.980415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elliott EJ, Robins-Browne RM, O'Loughlin EV, Bennett-Wood V, Bourke J, Henning P, Hogg GG, Knight J, Powell H, Redmond D. 2001. Nationwide study of haemolytic uraemic syndrome: clinical, microbiological, and epidemiological features. Arch. Dis. Child. 85:125–131. 10.1136/adc.85.2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sánchez S, Cenoz MG, Martin C, Beristain X, Llorente MT, Herrera-Leon S. 2014. Cluster investigation of mixed O76:H19 Shiga toxin-producing Escherichia coli and atypical enteropathogenic E. coli infection in a Spanish household. Epidemiol. Infect. 142:1029–1033. 10.1017/S0950268813001842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gordon DM, Riley MA. 1992. A theoretical and experimental analysis of bacterial growth in the bladder. Mol. Microbiol. 6:555–562. 10.1111/j.1365-2958.1992.tb01500.x. [DOI] [PubMed] [Google Scholar]
- 60.Alteri CJ, Mobley HL. 2007. Quantitative profile of the uropathogenic Escherichia coli outer membrane proteome during growth in human urine. Infect. Immun. 75:2679–2688. 10.1128/IAI.00076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roos V, Nielsen EM, Klemm P. 2006. Asymptomatic bacteriuria Escherichia coli strains: adhesins, growth and competition. FEMS Microbiol. Lett. 262:22–30. 10.1111/j.1574-6968.2006.00355.x. [DOI] [PubMed] [Google Scholar]
- 62.Friedrich AW, Borell J, Bielaszewska M, Fruth A, Tschäpe H, Karch H. 2003. Shiga toxin 1c-producing Escherichia coli strains: phenotypic and genetic characterization and association with human disease. J. Clin. Microbiol. 41:2448–2453. 10.1128/JCM.41.6.2448-2453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paton AW, Srimanote P, Woodrow MC, Paton JC. 2001. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 69:6999–7009. 10.1128/IAI.69.11.6999-7009.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dodson KW, Pinkner JS, Rose T, Magnusson G, Hultgren SJ, Waksman G. 2001. Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor. Cell 105:733–743. 10.1016/S0092-8674(01)00388-9. [DOI] [PubMed] [Google Scholar]
- 65.Stapleton AE, Stroud MR, Hakomori SI, Stamm WE. 1998. The globoseries glycosphingolipid sialosyl galactosyl globoside is found in urinary tract tissues and is a preferred binding receptor in vitro for uropathogenic Escherichia coli expressing pap-encoded adhesins. Infect. Immun. 66:3856–3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Striker R, Nilsson U, Stonecipher A, Magnusson G, Hultgren SJ. 1995. Structural requirements for the glycolipid receptor of human uropathogenic Escherichia coli. Mol. Microbiol. 16:1021–1029. 10.1111/j.1365-2958.1995.tb02327.x. [DOI] [PubMed] [Google Scholar]
- 67.Lanne B, Olsson BM, Jovall PA, Angström J, Linder H, Marklund BI, Bergström J, Karlsson KA. 1995. Glycoconjugate receptors for P-fimbriated Escherichia coli in the mouse. An animal model of urinary tract infection. J. Biol. Chem. 270:9017–9025. [DOI] [PubMed] [Google Scholar]
- 68.Olsson BM, Karlsson H, Larsson T, Lanne B. 1999. Mass spectrometric analysis of ceramide composition in mono-, di-, tri-, and tetraglycosylceramides from mouse kidney: an experimental model for uropathogenic Escherichia coli. J. Mass Spectrom. 34:942–951. . [DOI] [PubMed] [Google Scholar]
- 69.Hung CS, Bouckaert J, Hung D, Pinkner J, Widberg C, DeFusco A, Auguste CG, Strouse R, Langermann S, Waksman G, Hultgren SJ. 2002. Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection. Mol. Microbiol. 44:903–915. [DOI] [PubMed] [Google Scholar]
- 70.Moreno E, Andreu A, Pigrau C, Kuskowski MA, Johnson JR, Prats G. 2008. Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. J. Clin. Microbiol. 46:2529–2534. 10.1128/JCM.00813-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.