Abstract
Group B Streptococcus (GBS) causes severe disease in neonates, the elderly, and immunocompromised individuals. GBS species are highly diverse and can be classified by serotype and multilocus sequence typing. Sequence type 17 (ST-17) strains cause invasive neonatal disease more frequently than strains of other STs. Attachment and invasion of host cells are key steps in GBS pathogenesis. We investigated whether four serotype III strains representing ST-17 (two strains), ST-19, and ST-23 differ in their abilities to attach to and invade both decidual cells and lung epithelial cells. Virulence gene expression following host cell association and exposure to amnion cells was also tested. The ST-17 strains differed in their abilities to attach to and invade decidual cells, whereas there were no differences with lung epithelial cells. The ST-19 and ST-23 strains, however, attached to and invaded decidual cells less than both ST-17 strains. Although the ST-23 strain attached to lung epithelial cells better than ST-17 and -19 strains, none of the strains effectively invaded the lung epithelial cells. Notably, the association with host cells resulted in the differential expression of several virulence genes relative to basal expression levels. Similar expression patterns of some genes were observed regardless of cell type used. Collectively, these results show that GBS strains differ in their abilities to attach to distinct host cell types and express key virulence genes that are relevant to the disease process. Enhancing our understanding of pathogenic mechanisms could aid in the identification of novel therapeutic targets or vaccine candidates that could potentially decrease morbidity and mortality associated with neonatal infections.
INTRODUCTION
Group B Streptococcus (GBS) is a leading cause of neonatal sepsis and meningitis and is transferred from mothers to babies in utero or during childbirth (1). Approximately 30% of women are asymptomatically colonized with GBS, and roughly 50% to 70% of babies born to those women become colonized. Neonatal GBS infections are divided in two classes of disease: early-onset disease and late-onset disease. Early-onset disease occurs within the first few days of life, and late-onset disease occurs between 1 week and 3 months of age (2). Current prevention practices rely on antibiotic prophylaxis administered to colonized mothers prior to childbirth. Although these efforts have been successful in preventing early-onset GBS disease, the prevalence of late-onset disease remains the same. In addition, screen-and-treat approaches do not provide a safeguard against premature birth due to invasive GBS infections. Therefore, the identification and development of alternative preventative measures, such as vaccines and drug targets, are greatly needed (3).
GBS strains can be classified into 10 distinct serotypes based on types of capsular polysaccharide (cps) (Ia, Ib, and II to IX), with types Ia, III, and V more often associated with disease than the other types (3, 4). GBS can be further classified using multilocus sequence typing (MLST), which examines the allelic profiles of seven conserved genes and groups the strains into sequence types (STs), providing a classification based on the genetic backbone (5). Serotype III ST-17 GBS strains have been shown to cause a higher frequency of neonatal disease than other STs (6–9).
GBS, like many other pathogens, needs to cross physical barriers within the host to cause disease. Progression of GBS disease involves initial maternal colonization of vaginal epithelial cells, dissemination across extraplacental membranes (causing chorioamnionitis) and across neonatal lung epithelial cells, bloodstream survival, and, in cases of meningitis, penetration of the blood-brain barrier (10). Infection of the newborn is a result of either invasion by a GBS strain(s) that ascends the genital tract to infect through the extraplacental membranes to cause infection in utero or aspiration of infected vaginal fluid as the baby passes through the birth canal (2). In order to cross these anatomical barriers to infection, GBS must be able to adhere to and invade the host cells that comprise these barriers. Previous studies have shown that GBS effectively adheres to and invades epithelial and endothelial cells. Additionally, GBS strains of different serotypes differ in their abilities to associate with host cells (11–16); however, such studies selected strains on the basis of cps type rather than ST. Because cps is horizontally transferred between strains and there is evidence of capsule switching (17, 18), selecting strains based on ST, or genetic backbone, is warranted. Comparing the hypervirulent lineage, ST-17, with other lineages with respect to the ability to attach to and invade host cells will facilitate the identification of factors that play an important role in GBS disease development.
In this study, we determined the level of GBS attachment and invasion of two barriers that are typically encountered during the early stages of an infection. These barriers include decidual cells, which make up the outer layer of the extraplacental membranes, and lung epithelial cells, one site of inoculation in neonates during passage through the birth canal or during aspiration of contaminated amniotic fluid in utero. Four serotype III GBS strains representing ST-17, ST-19, and ST-23 were compared to quantify differences in associations with decidual and lung epithelial cells across and within phylogenetically distinct lineages. Additionally, the expression of known virulence genes was examined in each strain upon association with host cells to better understand the role these factors play in GBS pathogenesis and identify gene targets useful for guiding disease prevention strategies.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Four GBS strains were used in this study, all of them serotype III: GB112, a ST-17 strain isolated from a follow-up vaginal rectal screen of a woman who had recently given birth (19); GB411, a ST-17 strain isolated from a newborn with septicemia (20); GB590, a ST-19 strain isolated from a vaginal rectal screen of a pregnant woman (19); and genome strain NEM316 (ATCC 12403), a ST-23 strain isolated from a newborn with septicemia. Strains were cultured in Todd-Hewitt broth (THB) or agar (THA) or on sheep's blood agar plates (BD) at 37°C with 5% CO2.
Cell culture.
The A549 cell line (ATCC CCL-185), a human alveolar epithelial carcinoma cell line, was maintained by incubation at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) (Gibco) containing 10% fetal bovine serum (FBS) (HyClone) and 2% penicillin-streptomycin (pen/strep) (Gibco). A human endometrial stromal cell (HESC) line, T HESC (ATCC CRL-4003), was cultured in DMEM/Nutrient Mixture Ham's F-12 with l-glutamine (Sigma) supplemented with 1.5 g/liter sodium bicarbonate, 1% BD ITS+ universal culture supplement premix, 10% charcoal-treated FBS (HyClone), and 2% pen/strep (referred to as HESC medium here).
Association assays.
T HESCs were first decidualized as previously described (21). Briefly, cells were grown to approximately 50% confluence and treated with 0.5 mM 8-bromo-cyclic amp (cAMP) (Sigma) for 3 to 6 days. Decidualization was confirmed by examining the expression of prolactin and insulin-like growth factor (IGF)-binding protein 1, which are upregulated following decidualization. Assays were not performed until the cells reached 100% confluence such so no part of the bottom of the well was exposed since the bacteria attached well to the plates. Bacterial strains were grown in THB to mid-log phase, washed once with phosphate-buffered saline (PBS), and resuspended in infection medium (HESC medium with 2% charcoal-treated FBS, no ITS+, and no antibiotics). The same infection medium was used for both cell lines for comparisons of associations across cell lines without factoring in effect of the medium used. Prior to infection, host cells were washed three times with PBS. They were then infected with GBS strains in the infection medium at a multiplicity of infection (MOI) of one bacterial cell per host cell. After 2 h of incubation at 37°C with 5% CO2, wells were washed three times with PBS to remove nonadherent bacteria.
To determine the number of associated bacteria (attached and invaded), host cells were lysed with 0.1% Triton X-100 (Sigma) for 30 min at 37°C. Lysates were subjected to gentle vortex mixing to further disrupt the host cells and liberate intracellular bacteria. After serial dilution, lysates were then plated on THA and incubated overnight at 37°C, and CFU were counted. To test invasion, once nonadherent bacteria were washed away as described above, infection medium containing 100 μg/ml of gentamicin (Gibco) and 5 μg/ml of penicillin G (Sigma) was added to each well and incubated at 37°C for 1 h to kill extracellular bacteria. Wells were then washed with PBS two times, and intracellular bacteria were enumerated as described above for associated bacteria. The number of attached bacteria was calculated by subtracting the number invaded from the number associated. All data were expressed as percentages of the total number of bacteria per well after the 2-h infection. Assays were run in triplicate at least three times.
Amnion cell isolation.
Human extraplacental membranes were collected from healthy, nonsmoking, singleton pregnancies undergoing scheduled cesarean delivery prior to onset of labor at the University of Michigan Birth Center as previously described (22). The University of Michigan Institutional Review Board approved this research (IRBMED no. HUM0037054). Immediately following delivery, the membranes were transported to the laboratory in Dulbecco's phosphate-buffered saline (DPBS). Membranes were rinsed with DPBS, and blood clots were removed. Membranes were then subjected to blunt dissection to separate the choriodecidua from the amnion. Amnion tissue was used to isolate amnion cells using methods adapted from three protocols (23–25). Briefly, amnion cells were digested with 0.25% trypsin–EDTA (Gibco) at 37 C for 30 min. Amnion tissue was transferred to fresh trypsin-EDTA, and the digestion described above was repeated. Following each digestion, the trypsin-EDTA was neutralized with medium (DMEM:F12 supplemented with 10% FBS and pen/strep). Cells were pelleted by centrifugation, washed in medium, pelleted again, and resuspended in medium containing epidermal growth factor (EGF) (Peprotech) (DMEM:F12 supplemented with 10% FBS, pen/strep, and 10 ng/ml EGF). Amnion cells were seeded at 500,000 cells/well (12-well plates) in 1 ml medium and grown to 70% to 80% confluence, and the medium was changed on day 2 of culture (DMEM:F12 supplemented with 10% FBS and 100 ng/ml EGF without antibiotics). Viability of amnion cells prior to plating was assessed using trypan blue. After 2 days of culture, cell morphology and growth were assessed. Cells were infected on day 3 after forming a monolayer.
RNA preparation and real-time PCR (qRT-PCR).
Bacterial RNA was isolated from samples in the association assay described above. The four types of samples collected were bacterial cells for establishment of the basal activity (growth in cell culture media with no exposure to host cells), bacterial cells in suspension (in the media, not attached to host cells), bacterial cells associated with host cells, and bacterial cells that invaded host cells. Samples used for RNA extraction were prepared using RNAprotect Bacteria Reagent (Qiagen) and an RNeasy minikit (Qiagen) as described in the “Enzymatic Lysis, Proteinase K Digestion, and Mechanical Disruption of Bacteria” protocol in the RNAprotect handbook with the addition of incubation with mutanolysin during the treatment with proteinase K and lysozyme. RNA samples were then treated with a Turbo DNA-free kit (Ambion) and checked for DNA contamination by PCR without prior reverse transcription (RT). cDNA was synthesized from 1 μg RNA with random primers using an iScript Select cDNA synthesis kit (Bio-Rad). Quantitative RT-PCR (qRT-PCR) was performed in a 15-μl reaction mixture using iQ SYBR Supermix (Bio-Rad) and 10 μM (each) gene-specific primers. The list of primers used can be found in Table 1. Amplification and detection of specific products were performed using a CFX384 Touch Real-Time PCR detection system (Bio-Rad) under the following conditions: 1 cycle of 3 min at 95°C and 39 cycles of 95°C for 10 s and 60°C for 30 s. Relative gene expression levels were calculated using the comparative threshold cycle (CT) method (2−ΔΔCT method) (26) with gyrA as the internal control gene and normalization of expression relative to basal levels of expression. A 2-fold change in the gene expression level was considered significant.
TABLE 1.
Oligonucleotide primers used for qRT-PCR
| Gene | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| gyrA | CGGGACACGTACAGGCTACT | CGATACGAGAAGCTCCCACA |
| srr1 | GTCACTTCCGTTTGTTCTGCC | CTGGTAGGTGGAGCGAGTTT |
| srr2 | GCTGTAGTTGGAGGGACGAC | TTACTTCTGGCGCAACCACT |
| bibA | CTAGCGGAAACTTGGTGGCT | GGCTTCACCCGTTGATGGTA |
| hvgA | ATACAAATTCTGCTGACTACCG | TTAAATCCTTCCTGACCATTCC |
| lrrG | TACGCCAGATTCCTTTGGCA | CTTCTGCCGCTGCATTTCG |
| lmb | GATCCCTTGCCCAAGCTTCT | TCCAATCAGGTGCAGGCATT |
| scpB | GTAACTACGCTCAAGCTATC | CCCAAAGCTACTATCATTAC |
| spb1 | TCGCTGTTAGTGGCGAGTTT | TGTCTCAGCGGCAAAAGCTA |
| fbsB | GCGATTGTGAATAGAATGAGTG | ACAGAAGCGGCGATTTCATT |
| cylE | TGGAAATTGCTAAGTTAGATAACG | AGCCCTCGTTAAGTTTGCCA |
| iagA | CCCCCAAGTTTCGGGAGTTT | ACGTTTGACATTACGGTCGGT |
| sip | CATCGACAATGGCAGCTTCG | GCTGTCCACGTCGTATCTGT |
| ponA | AGGAAGTTTGGCTTGGGCTT | AGCGAGCAAAGCAAGTTGTG |
| cylX | TGGCTTGTATAAAACGCGGCT | CAACGACACTGCCATCAGCA |
Statistical analysis.
Data are reported as means ± standard deviations (SD). GraphPad Prism 5.0 was used for statistical analysis. For the GBS growth rates and association assays, statistical analysis was performed using one-way analysis of variance (ANOVA) and Tukey's posttest. For gene expression analysis, statistical analysis was performed using two-way ANOVA and Bonferroni's posttest. Differences with P values of <0.05 were considered statistically significant.
RESULTS
Bacterial growth rates did not differ among GBS isolates.
The bacterial growth rate has previously been shown to influence the invasiveness of GBS in respiratory epithelial cells (27). Therefore, it was important that the strains used in this study exhibited similar growth rates to reduce the effect of this parameter on the experiments. To determine the growth characteristics of the four strains used in this study, each strain was grown in the infection medium used for the association assays over a 6-h period and the optical density at 600 nm (OD600) and CFU/ml were determined every hour (Fig. 1). No significant differences in growth rates were observed for all four strains. Similar results were observed when the strains were grown in THB (not shown).
FIG 1.
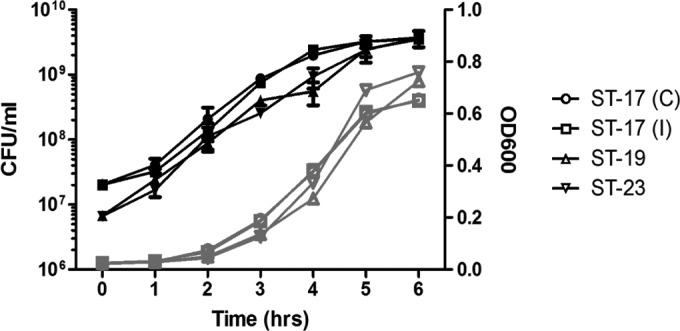
Bacterial growth curves. Bacterial cultures of the four strains used in this study were incubated at 37°C for a 6-h period during which the cultures were sampled every hour to determine CFU/ml (black lines) and OD600 (gray lines). C, colonizing; I, invasive.
Association with host cells varies among serotype III GBS strains.
Previous studies have shown that GBS strains of different serotypes vary in attachment and invasion of host cells, but few have compared GBS strains of the same serotype. Association assays were performed to determine if serotype III strains differed in their abilities to attach to and invade host cells. Decidualized human endometrial stromal cells (T HESCs) were used as an extraplacental membrane invasion model, and A549 lung epithelial cells were used as a model for infection through the lungs.
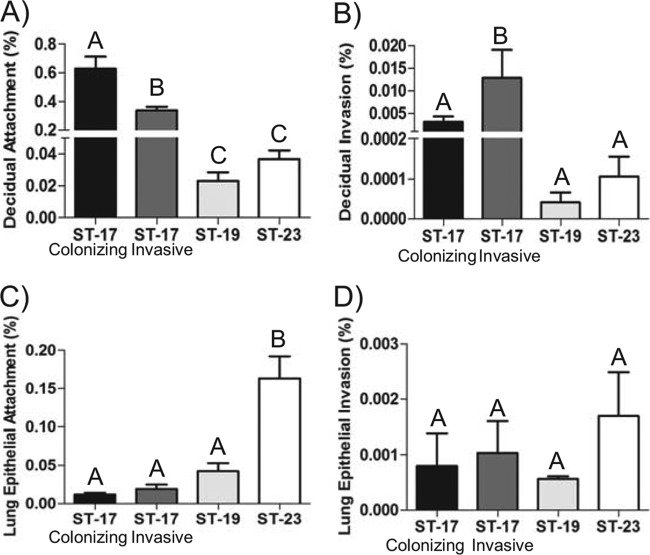
The hypervirulent ST-17 lineage is most often associated with neonatal sepsis and meningitis. It is known that ST-17 strains have a greater propensity to cause severe disease than strains of other lineages (6–9); therefore, we first compared strains within this lineage. Two ST-17 GBS isolates were selected: one colonizing strain isolated from a vaginal rectal swab of a healthy pregnant woman and one invasive strain isolated from a newborn baby with septicemia. Interestingly, significantly more bacteria of the colonizing strain attached to decidual cells than bacteria of the invasive strain (P < 0.01), whereas the invasive strain invaded the decidual cells significantly more extensively than the colonizing strain (P < 0.05) (Fig. 2A and B). No difference was seen in the abilities of the two strains to attach to lung epithelial cells (Fig. 2C), and neither strain effectively invaded the lung epithelial cells (Fig. 2D). Overall, the levels of attachment and invasion of decidual cells were much higher than those seen with lung epithelial cells.
FIG 2.
GBS association with host cells. Host cell lines were infected with GBS at a multiplicity of infection (MOI) of 1 for 2 h. The number of bacteria is expressed relative to the total number of bacteria in the well after the infection. (A and B) Attachment to (A) and invasion of (B) decidual cells. (C and D) Attachment to (C) and invasion of (D) lung epithelial cells. Bars represent means ± SD of the results. Bars labeled with different letters are significantly different from each other. Experiments were repeated in triplicate at least three times.
In a previous study, it was reported that both ST-17 and ST-19 strains were more often associated with invasive disease but that ST-23 strains were linked to asymptomatic colonization (9). To determine if variations exist in the ability to associate with host cells across these diverse GBS genotypes of the same serotype, the same association assays were used to compare two strains representing ST-19 and ST-23 to the colonizing and invasive ST-17 strains. The ST-23 strain attached to lung epithelial cells significantly more extensively than the ST-19 strain (P < 0.001). However, there was no difference with respect to attachment to decidual cells, and neither strain effectively invaded either cell type (Fig. 2). Both strains had an enhanced ability to attach to the lung epithelial cells compared to the decidual cells. Additionally, both ST-17 strains attached to decidual cells significantly more than either the ST-23 strain or the ST-19 strain (P < 0.01) (Fig. 2A), whereas the ST-23 strain attached to the lung epithelial cells more than strains ST-19 and ST-17 (P < 0.001) (Fig. 2C).
Virulence gene expression levels did not differ among ST-17 strains.
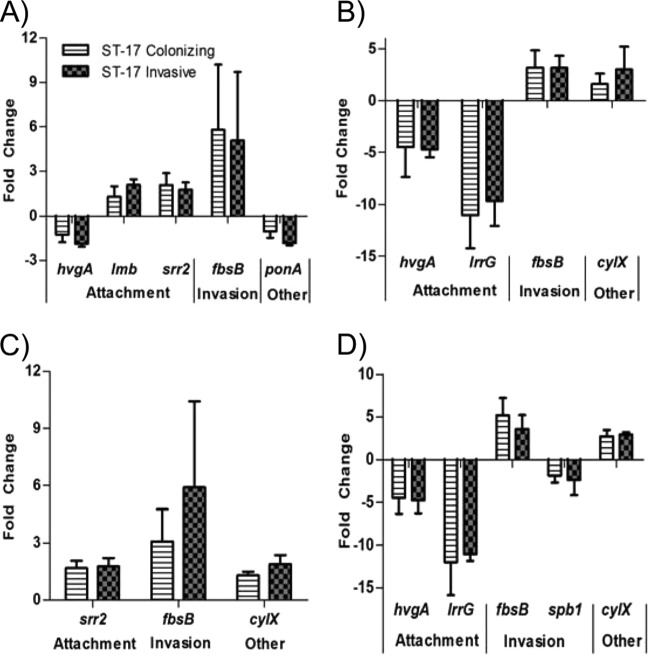
To examine the effect host cells have on bacterial virulence gene expression and to determine if differential expression of bacterial virulence genes could help explain some of the differences seen in host cell association, we tested the expression of several known virulence genes upon either attachment to or invasion of host cells relative to basal levels of expression. The complete list of genes tested and their functions is shown in Table 2. Although no significant differences between the two ST-17 strains in gene expression were detected, 8 of the 12 genes tested were differentially expressed upon associating with host cells relative to the basal gene expression (Fig. 3). Of note, fbsB was highly upregulated under all conditions tested and both hvgA and lrrG were highly downregulated upon invasion of both cell types.
TABLE 2.
Virulence factors tested in this study
| Category | Gene | Product | Function/characteristic(s) | Reference |
|---|---|---|---|---|
| Attachment | srr | Serine-rich repeat protein | Surface adhesins; two genetically distinct variants, srr1 and srr2; srr2 is specific to ST-17 strains | 38, 39 |
| bibA | Immunogenic bacterial adhesin | Binds human C4-binding protein to resist opsonophagocytosis; promotes adherence to epithelial cells | 40 | |
| hvgA | Hypervirulent GBS adhesin | Homologous to BibA; occurs on the same genetic locus; specific to ST-17 strains | 28 | |
| lrrG | Leucine-rich repeat protein | Binds epithelial cells and elicits protective immunity | 34 | |
| lmb | Laminin-binding protein | Promotes GBS colonization and translocation into bloodstream | 31, 32 | |
| scpB | C5a peptidase | Dual function: cleaves and inactivates complement and promotes binding to epithelial cells and fibronectin | 33 | |
| Invasion | spb1 | Surface protein of GBS | Pilus backbone of pilus island (PI)-2b; promotes invasion of epithelial cells; specific to ST-17 strains | 41, 42 |
| fbsB | Fibrinogen-binding protein B | Promotes invasion into epithelial cells | 36 | |
| cylE | CylE protein | Predicted to function as an N-acyltransferase in the biosynthesis of the GBS pigment granadaene required for hemolytic/cytolytic activity of GBS | 43 | |
| iagA | Invasion-associated gene | Anchors lipoteichoic acid to the cell membrane; involved in invasion of brain microvascular endothelial cells | 44 | |
| Other | sip | Surface immunogenic protein | Surface protein that elicits cross-protective immunity | 35 |
| ponA | Penicillin-binding protein 1a | Promotes resistance to antimicrobial peptides | 31 | |
| cylX | CylX protein | Homologous to component of acetyl-CoA carboxylase; predicted to function in biosynthesis of GBS pigment granadaene | 43 |
FIG 3.
Virulence gene expression in ST-17 strains. The expression levels of 12 virulence genes were determined upon attachment to and invasion of host cells in a ST-17 colonizing strain and ST-17 invasive strain. The genes were categorized as “attachment,” “invasion,” or “other” according to their function (see Table 2). Only the genes with at least a 2-fold change in expression relative to basal expression are represented here. (A and B) Gene expression upon attachment to (A) and invasion of (B) decidual cells. (C and D) Gene expression upon attachment to (C) and invasion of (D) lung epithelial cells. Bars represent means ± SD of the results of three independent experiments.
To determine whether the ST-17 strains behave similarly in a more physiologically relevant cell type than immortalized cell lines, both strains were exposed to amnion cells isolated from the extraplacental membranes of three women after childbirth. Following bacterial RNA extraction and gene expression profiling, we observed upregulation of four genes (srr2, lmb, fbsB, and cylX) that were also upregulated in response to decidual or lung epithelial cell exposure in one or both of the strains (Fig. 3A and C). Expression levels of each gene were then compared across exposures. No significant differences in gene expression were observed for the invasive ST-17 strain across all three cell types (Fig. 4A); however, expression levels of two genes differed for the colonizing ST-17 strain (Fig. 4B). fbsB was upregulated by at least 2-fold when exposed to all cell types, but the magnitude of the change was significantly lower with amnion cells than with decidual cells. Additionally, cylX was significantly downregulated with amnion cells, whereas expression levels were unchanged with decidual and lung epithelial cell attachment.
FIG 4.

GBS ST-17 virulence gene expression levels compared across cell exposure categories. The expression levels of four virulence genes upregulated upon host cell attachment (Fig. 3) were examined after exposure to amnion cells isolated from human placental tissues and compared to the expression levels seen after decidual and lung epithelial cell exposure. The genes were categorized as “attachment,” “invasion,” or “other” according to their function (see Table 2). (A) Virulence gene expression of the invasive ST-17 strain. (B) Virulence gene expression of the colonizing ST-17 strain. Dashed lines mark a 2-fold change in expression. Bars represent means ± SD of the results of three independent experiments. Asterisks indicate significant differences in gene expression levels between exposures for each gene (*, P < 0.05; **, P < 0.01).
Differential expression of virulence genes across GBS STs.
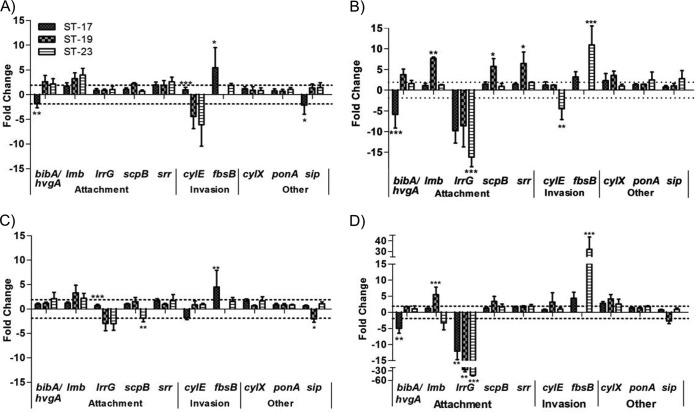
To determine if gene expression levels differ across diverse STs, virulence gene expression levels of strains of ST-17, ST-19, and ST-23 were compared. Since no significant differences between the two ST-17 strains in gene expression were observed, the data were combined for this comparison. Upon attachment to decidual cells, seven of the genes tested were differentially expressed by at least 2-fold in one or more of the STs (Fig. 5A). No significant differences between the ST-19 and ST-23 strains in expression levels were detected for all genes, whereas the expression levels of four genes significantly differed in the ST-17 strains from those seen with ST-19 and ST-23. Although the ST-17 strains attached to decidual cells more than the ST-19 and ST-23 strains in the association assays, none of the attachment genes examined were upregulated in the ST-17 strains, and the ST-19 and ST-23 strains upregulated expression of bibA whereas both ST-17 strains downregulated expression of hvgA, a homologue of bibA unique to ST-17 (28). Upon invasion of decidual cells, 10 of the virulence genes tested were differentially expressed (Fig. 5B). No significant difference between the three STs in expression levels was detected for cylX, ponA, and sip, whereas one strain had significantly different expression levels for the remaining genes.
FIG 5.
Virulence gene expression across GBS STs. The expression levels of known virulence genes were determined upon attachment to and invasion of host cells using ST-17, ST-19, and ST-23 strains. The genes were categorized as “attachment,” “invasion,” or “other” according to their function (see Table 2). (A and B) Gene expression upon attachment to (A) and invasion of (B) decidual cells. (C and D) Gene expression upon attachment to (C) and invasion of (D) lung epithelial cells. Dashed lines mark 2-fold change in expression. Expression of iagA was examined but is not included because it was not differentially expressed under any condition. Bars represent means ± SD of the results of three independent experiments. Asterisks indicate significant differences in gene expression levels between the STs for each gene (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Note that the y axis in panel D differs slightly from those in the other panels.
Attachment to lung epithelial cells resulted in differential regulation of seven virulence genes tested in this study (Fig. 5C); of these, no significant difference in expression was detected among the strains for three genes. Additionally, the expression patterns for lmb, srr, and fbsB were the same upon attachment to both cell types. Although there was no difference in lung epithelial cell invasion levels among the strains in the association assays (Fig. 2D), invasion resulted in differential regulation of the same 10 genes that were differentially regulated following decidual-cell invasion (Fig. 5D). Specifically, expression of bibA and hvgA, lrrG, fbsB, cylX, and ponA in response to lung epithelial cells showed a pattern similar to that observed upon decidual-cell invasion. Of the 10 genes differentially regulated, six of them showed no significant difference in expression levels among the strains.
DISCUSSION
Several studies have previously reported that unique GBS strains differ in their abilities to attach to and invade host cells (11–16). However, the strains tested in the prior studies were chosen based on serotype, a phenotypic characterization dictated by capsular polysaccharide genes, rather than on a genotypic characterization, such as ST. The present study newly examined the ability of diverse GBS STs of the same serotype (serotype III) to associate with host cell types representing different anatomical barriers GBS would encounter early during infection. The three STs used in this study were ST-17, ST-19, and ST-23, which represent the most common genotypes worldwide (5). ST-17 has been suggested to be a hypervirulent lineage because of its association with neonatal meningitis compared to other lineages. Because ST-19 strains are also more often associated with disease than with asymptomatic colonization, comparing two invasive and distinct lineages is important. ST-23 strains are more often associated with asymptomatic colonization than other strains and therefore provide a useful comparison between invasive and colonizing STs.
The two ST-17 strains differed in their abilities to associate with decidual cells. The colonizing strain attached more than the invasive strain, while the invasive strain invaded more than the colonizing strain. This shows that two strains of the same ST can differ in their abilities to associate with host cells. However, levels of attachment and invasion of lung epithelial cells were the same for both strains and were much lower than those seen with decidual cells. Additionally, the ST-17 strains attached to decidual cells more than the ST-19 and ST-23 strains, but the ST-23 strain attached to lung epithelial cells more than the ST-17 and ST-19 strains. Overall, few differences in invasive ability were detected across the three STs. Collectively, these data show that there is variation in association with host cells within the same serotype and suggest that strains of the same phylogenetic lineage also differ in their abilities to associate with host cells. Further studies using larger numbers of strains of each ST are needed to further characterize the ability to associate with host cells of each ST. In addition, it is difficult to draw conclusions about invasive versus colonizing strains because all invasive strains begin the disease process as asymptomatic colonizers.
One limitation of this study was that the invasion assay used accounted only for the bacteria that pass through the cell layer using the transcellular route even though there are other routes that could be used by GBS to cross anatomical barriers (29). In a previous study using electron microscopy, Soriani et al. found that GBS strains use a paracellular route to cross cervical epithelial cells. The majority of GBS were found in the spaces between the cells, with very few passing through the cells themselves (30). Therefore, variation in invasive ability among GBS strains needs to be further tested using other assays that account for the paracellular route of invasion as well. Additionally, the use of polarized cells may more accurately represent the epithelial cell barriers in vivo.
Differential regulation of virulence genes in response to host cell association was also examined in this study. Interestingly, even though differences between the colonizing and invasive ST-17 strains in associations with decidual cells were detected, there were no differences in the expression levels of the virulence genes tested, showing that GBS strains belonging to the same phylogenetic lineage have similar transcriptional responses upon attachment to and invasion of decidual and lung epithelial cells. This suggests that the differences in association characteristics could be due to genetic variation in the genes themselves rather than to differences in gene regulation or could result from other unknown virulence factors. Additionally, we examined the expression levels of four genes in the ST-17 strains following exposure to primary amnion cells and compared them to the levels seen following lung epithelial- and decidual-cell exposure. Overall, gene expression levels were similar across all cell types, showing that using immortalized cell lines to assess GBS gene expression in response to host cells accurately represents the response to primary cells from human tissues.
Comparing gene expression levels across all strains tested in this study showed that the expression levels of several virulence genes, upon association with distinct host cells, differed among phylogenetically distinct strains of the same serotype. Three of the five attachment genes examined in this study, lmb, scpB, and lrrG, along with one other gene, sip, have been considered to be potential vaccine targets (3). These four genes are highly conserved and therefore have the potential to elicit protective immunity across GBS serotypes.
The laminin-binding protein, encoded by lmb, assists colonization by adhering to the laminin extracellular matrix protein and has been shown to play a role in adherence to brain microvascular endothelial cells (31, 32). The present study showed that expression of lmb is induced in response to host cell attachment and that there are no significant differences in response across the three STs tested, suggesting that lmb could be a potential vaccine candidate. scpB encodes a dual-function serine protease that promotes adherence to epithelial cells and helps evade the host immune system by cleaving the C5a human complement component (33). Interestingly, even though ScpB appears to be a good vaccine candidate, expression of scpB is upregulated in response to host cell association in only the ST-19 strain and is actually significantly downregulated in the ST-23 strain under conditions of lung epithelial cell attachment. lrrG encodes a highly conserved leucine-rich-repeat surface protein that has been shown to elicit protective immunity in mice, and recombinant LrrG protein adheres to epithelial cells in vitro, thus making it a strong vaccine candidate (34). However, expression of lrrG was highly downregulated upon host cell invasion in the present study, suggesting that lrrG is not expressed early on during GBS infections. LrrG is likely recognized by host cells; therefore, in order to avoid immune system detection, GBS strains downregulate the expression of lrrG. The surface immunogenic protein (Sip) encoded by sip is a protein of unknown function but is conserved across GBS serotypes and elicits protective immunity in mice (35). In the present study, expression of sip upon host cell attachment either was not significantly changed or was downregulated by 2-fold, suggesting that it does not play an important role in host cell attachment. Further studies need to be done to better understand its role in pathogenesis. Upon invasion of decidual cells, expression of lmb, scpB, and srr increased in the ST-19 strain though expression either decreased or remained the same in the ST-17 and 23 strains relative to expression upon attachment to decidual cells. This suggests that the ST-17 and 23 strains no longer need these genes for invasion and turn off their expression whereas the ST-19 strain maintains high expression.
Interestingly, decidual-cell attachment resulted in upregulation of bibA expression in the ST-19 and 23 strains although expression of hvgA, the bibA homologue, was downregulated in both ST-17 strains. This expression pattern was also observed upon invasion of both decidual and lung epithelial cells. bibA and hvgA are located at the same genomic locus, and the regulatory regions between the two genes are highly conserved, with >90% sequence identity (28). Therefore, the observed differences between these two genes in expression levels are not due to variations in the genes themselves; rather, it would seem likely that they are due to differential levels of gene regulation by the different STs.
Expression of fbsB was upregulated upon attachment to host cells and was even higher upon invasion of host cells for both ST-17 strains and for the ST-23 strain. The high upregulation of fbsB upon invasion of host cells is consistent with a previous study that showed that FbsB is important in invasion of epithelial cells but not in attachment (36). The ST-19 strain showed no expression of fbsB for any of the samples tested in this study. Upon further investigation of the fbsB gene sequence in this strain, we found that the fbsB gene sequence contains an inversion of one segment of the gene and an insertion compared to the NEM316 sequence. Further investigation is required to determine if a functional protein is produced and if this sequence is conserved among other ST-19 strains.
The presence of host cells induced differential gene expression in all strains of GBS tested in this study. Although a number of genes were differentially expressed among the three STs, there was no clear connection between differences in the ability to adhere to and invade host cells and expression of virulence genes. Future studies using whole-transcriptome analysis would be helpful in explaining the differences in the ability to associate with host cells and identifying other pathways that are important for adherence to and invasion of host cells among GBS isolates.
One limitation of examining differential gene expression in response to host cells is that gene expression is very dynamic, and the time at which the RNA is sampled could affect which genes we see expressed. In this study, we sampled bacterial RNA at the same time point (after 2 h of infection) for each experiment; it is therefore possible that some of the genes that we did not see differentially expressed could have been expressed at a different time during the infection. For a more complete examination of the transcriptional response to host cells, a full time course may be more appropriate. In addition, there could be differences in protein expression that are not shown by mRNA expression due to posttranslation regulation, which could help explain some of the differences shown in the association assays. Future studies assessing the patterns of protein expression would therefore be beneficial.
The current report provides a comparison of GBS strains based on genetic backbone rather than on serotype, a phenotypic characteristic. The results show that genetically distinct GBS strains of the same serotype differ in their abilities to attach to and invade host cells and differentially express key virulence genes during host cell association. In addition, two strains of the same genotype differ in their abilities to attach to and invade host cells but do not differentially express key virulence genes, suggesting that other, unknown virulence genes are involved in this process. Because these strains were classified by MLST, it is quite possible that additional genetic characteristics are partly responsible for the differences observed. Indeed, Tettelin et al. demonstrated that 20% of the sequence from each strain is partially shared or strain specific (37). Further studies with larger numbers of strains for a full representation of genotypes and whole-genome transcriptome analysis will aid in identifying additional candidate genes important for attachment to and invasion of host cells.
ACKNOWLEDGMENTS
This work was supported by the Global Alliance to Prevent Prematurity and Stillbirth (GAPPS) in collaboration with the Bill and Melinda Gates Foundation (N015615) as well as the National Institutes of Health (AI100903-01).
We thank Maria Tikhonenko and Natalia Porcek for T HESC decidualization and primer design, Rim Al Safadi for help with the association assay, Robert Parker for assistance with qRT-PCR protocols, and Chuanwu Xi for sharing of laboratory resources. We also thank H. Dele Davies for providing GBS strains used in this study and Mark Chames for his assistance with acquiring the placental tissue for amnion cell isolation.
Footnotes
Published ahead of print 18 August 2014
REFERENCES
- 1.Tudela CM, Stewart RD, Roberts SW, Wendel GD, Jr, Stafford IA, McIntire DD, Sheffield JS. 2012. Intrapartum evidence of early-onset group B Streptococcus. Obstet. Gynecol. 119:626–629. 10.1097/AOG.0b013e31824532f6. [DOI] [PubMed] [Google Scholar]
- 2.Doran KS, Nizet V. 2004. Molecular pathogenesis of neonatal group B streptococcal infection: no longer in its infancy. Mol. Microbiol. 54:23–31. 10.1111/j.1365-2958.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- 3.Johri AK, Paoletti LC, Glaser P, Dua M, Sharma PK, Grandi G, Rappuoli R. 2006. Group B Streptococcus: global incidence and vaccine development. Nat. Rev. Microbiol. 4:932–942. 10.1038/nrmicro1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slotved H-C, Kong F, Lambertsen L, Sauer S, Gilbert GL. 2007. Serotype IX, a proposed new Streptococcus agalactiae serotype. J. Clin. Microbiol. 45:2929–2936. 10.1128/JCM.00117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones N, Bohnsack JF, Takahashi S, Oliver KA, Chan MS, Kunst F, Glaser P, Rusniok C, Crook DW, Harding RM, Bisharat N, Spratt BG. 2003. Multilocus sequence typing system for group B Streptococcus. J. Clin. Microbiol. 41:2530–2536. 10.1128/JCM.41.6.2530-2536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohnsack JF, Whiting A, Gottschalk M, Dunn DM, Weiss R, Azimi PH, Philips JB, III, Weisman LE, Rhoads GG, Lin F-YC. 2008. Population structure of invasive and colonizing strains of Streptococcus agalactiae from neonates of six U.S. Academic Centers from 1995 to 1999. J. Clin. Microbiol. 46:1285–1291. 10.1128/JCM.02105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin FC, Whiting A, Adderson E, Takahashi S, Dunn DM, Weiss R, Azimi PH, Philips JB, III, Weisman LE, Regan J, Clark P, Rhoads GG, Frasch CE, Troendle J, Moyer P, Bohnsack JF. 2006. Phylogenetic lineages of invasive and colonizing strains of serotype III group B Streptococci from neonates: a multicenter prospective study. J. Clin. Microbiol. 44:1257–1261. 10.1128/JCM.44.4.1257-1261.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luan S, Granlund M, Sellin M, Lagergård T, Spratt BG, Norgren M. 2005. Multilocus sequence typing of Swedish invasive group B Streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J. Clin. Microbiol. 43:3727–3733. 10.1128/JCM.43.8.3727-3733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manning SD, Springman AC, Lehotzky E, Lewis MA, Whittam TS, Davies HD. 2009. Multilocus sequence types associated with neonatal group B streptococcal sepsis and meningitis in Canada. J. Clin. Microbiol. 47:1143–1148. 10.1128/JCM.01424-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maisey HC, Doran KS, Nizet V. 2008. Recent advances in understanding the molecular basis of group B Streptococcus virulence. Expert Rev. Mol. Med. 10:e27. 10.1017/S1462399408000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma P, Lata H, Arya DK, Kashyap AK, Kumar H, Dua M, Ali A, Johri AK. 2013. Role of pilus proteins in adherence and invasion of Streptococcus agalactiae to the lung and cervical epithelial cells. J. Biol. Chem. 288:4023–4034. 10.1074/jbc.M112.425728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubens CE, Smith S, Hulse M, Chi EY, van Belle G. 1992. Respiratory epithelial cell invasion by group B streptococci. Infect. Immun. 60:5157–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schubert A, Zakikhany K, Pietrocola G, Meinke A, Speziale P, Eikmanns BJ, Reinscheid DJ. 2004. The fibrinogen receptor FbsA promotes adherence of Streptococcus agalactiae to human epithelial cells. Infect. Immun. 72:6197–6205. 10.1128/IAI.72.11.6197-6205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winram SB, Jonas M, Chi E, Rubens CE. 1998. Characterization of group B Streptococcal invasion of human chorion and amnion epithelial cells in vitro. Infect. Immun. 66:4932–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson RL, Lee MK, Soderland C, Chi EY, Rubens CE. 1993. Group B streptococci invade endothelial cells: type III capsular polysaccharide attenuates invasion. Infect. Immun. 61:478–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nizet V, Kim KS, Stins M, Jonas M, Chi EY, Nguyen D, Rubens CE. 1997. Invasion of brain microvascular endothelial cells by group B streptococci. Infect. Immun. 65:5074–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cieslewicz MJ, Chaffin D, Glusman G, Kasper D, Madan A, Rodrigues S, Fahey J, Wessels MR, Rubens CE. 2005. Structural and genetic diversity of group B Streptococcus capsular polysaccharides. Infect. Immun. 73:3096–3103. 10.1128/IAI.73.5.3096-3103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies HD, Jones N, Whittam TS, Elsayed S, Bisharat N, Baker CJ. 2004. Multilocus sequence typing of serotype III group B Streptococcus and correlation with pathogenic potential. J. Infect. Dis. 189:1097–1102. 10.1086/382087. [DOI] [PubMed] [Google Scholar]
- 19.Spaetgens R, DeBella K, Ma D, Robertson S, Mucenski M, Davies HD. 2002. Perinatal antibiotic usage and changes in colonization and resistance rates of group B Streptococcus and other pathogens. Obstet. Gynecol. 100:525–533. 10.1016/S0029-7844(02)02068-9. [DOI] [PubMed] [Google Scholar]
- 20.Davies HD, Raj S, Adair C, Robinson J, McGeer A. 2001. Population-based active surveillance for neonatal group B streptococcal infections in Alberta, Canada: implications for vaccine formulation. Pediatr. Infect. Dis. J. 20:879–884. 10.1097/00006454-200109000-00011. [DOI] [PubMed] [Google Scholar]
- 21.Brosens JJ, Takeda S, Acevedo C, Lewis MP, Kirby PL, Symes EK, Krausz T, Purohit A, Gellersen B, White JO. 1996. Human endometrial fibroblasts immortalized by simian virus 40 large T antigen differentiate in response to a decidualization stimulus. Endocrinology 137:2225–2231. [DOI] [PubMed] [Google Scholar]
- 22.Boldenow E, Jones S, Lieberman RW, Chames MC, Aronoff DM, Xi C, Loch-Caruso R. 2013. Antimicrobial peptide response to group B Streptococcus in human extraplacental membranes in culture. Placenta 34:480–485. 10.1016/j.placenta.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ilancheran S, Michalska A, Peh G, Wallace EM, Pera M, Manuelpillai U. 2007. Stem cells derived from human fetal membranes display multilineage differentiation potential. Biol. Reprod. 77:577–588. 10.1095/biolreprod.106.055244. [DOI] [PubMed] [Google Scholar]
- 24.Pratama G, Vaghjiani V, Tee JY, Liu YH, Chan J, Tan C, Murthi P, Gargett C, Manuelpillai U. 2011. Changes in culture expanded human amniotic epithelial cells: implications for potential therapeutic applications. PLoS One 6:e26136. 10.1371/journal.pone.0026136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu T, Cheng W, Liu T, Guo L, Huang Q, Jiang L, Du X, Xu F, Liu Z, Lai D. 2010. Human amniotic epithelial cell feeder layers maintain mouse embryonic stem cell pluripotency via epigenetic regulation of the c-Myc promoter. Acta Biochim. Biophys. Sin. (Shanghai) 42:109–115. 10.1093/abbs/gmp115. [DOI] [PubMed] [Google Scholar]
- 26.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3:1101–1108. 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 27.Malin G, Paoletti LC. 2001. Use of a dynamic in vitro attachment and invasion system (DIVAS) to determine influence of growth rate on invasion of respiratory epithelial cells by group B Streptococcus. Proc. Natl. Acad. Sci. U. S. A. 98:13335–13340. 10.1073/pnas.241079098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tazi A, Disson O, Bellais S, Bouaboud A, Dmytruk N, Dramsi S, Mistou M-Y, Khun H, Mechler C, Tardieux I, Trieu-Cuot P, Lecuit M, Poyart C. 2010. The surface protein HvgA mediates group B Streptococcus hypervirulence and meningeal tropism in neonates. J. Exp. Med. 207:2313–2322. 10.1084/jem.20092594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim KS. 2008. Mechanisms of microbial traversal of the blood-brain barrier. Nat. Rev. Microbiol. 6:625–634. 10.1038/nrmicro1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soriani M, Santi I, Taddei A, Rappuoli R, Grandi G, Telford JL. 2006. Group B Streptococcus crosses human epithelial cells by a paracellular route. J. Infect. Dis. 193:241–250. 10.1086/498982. [DOI] [PubMed] [Google Scholar]
- 31.Rajagopal L. 2009. Understanding the regulation of group B streptococcal virulence factors. Future Microbiol. 4:201–221. 10.2217/17460913.4.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spellerberg B, Rozdzinski E, Martin S, Weber-Heynemann J, Schnitzler N, Lütticken R, Podbielski A. 1999. Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin. Infect. Immun. 67:871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng Q, Stafslien D, Purushothaman SS, Cleary P. 2002. The group B streptococcal C5a peptidase is both a specific protease and an invasin. Infect. Immun. 70:2408–2413. 10.1128/IAI.70.5.2408-2413.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seepersaud R, Hanniffy SB, Mayne P, Sizer P, Le Page R, Wells JM. 2005. Characterization of a novel leucine-rich repeat protein antigen from group B Streptococci that elicits protective immunity. Infect. Immun. 73:1671–1683. 10.1128/IAI.73.3.1671-1683.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brodeur BR, Boyer M, Charlebois I, Hamel J, Couture F, Martin D. 2000. Identification of group B streptococcal Sip protein, which elicits cross-protective immunity. Infect. Immun. 68:5610–5618. 10.1128/IAI.68.10.5610-5618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gutekunst H, Eikmanns BJ, Reinscheid DJ. 2004. The novel fibrinogen-binding protein FbsB promotes Streptococcus agalactiae invasion into epithelial cells. Infect. Immun. 72:3495–3504. 10.1128/IAI.72.6.3495-3504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O'Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U. S. A. 102:13950–13955. 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seifert KN, Adderson EE, Whiting AA, Bohnsack JF, Crowley PJ, Brady LJ. 2006. A unique serine-rich repeat protein (Srr-2) and novel surface antigen (epsilon) associated with a virulent lineage of serotype III Streptococcus agalactiae. Microbiology 152:1029–1040. 10.1099/mic.0.28516-0. [DOI] [PubMed] [Google Scholar]
- 39.Samen U, Eikmanns BJ, Reinscheid DJ, Borges F. 2007. The surface protein Srr-1 of Streptococcus agalactiae binds human keratin 4 and promotes adherence to epithelial HEp-2 cells. Infect. Immun. 75:5405–5414. 10.1128/IAI.00717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santi I, Scarselli M, Mariani M, Pezzicoli A, Masignani V, Taddei A, Grandi G, Telford JL, Soriani M. 2007. BibA: a novel immunogenic bacterial adhesin contributing to group B Streptococcus survival in human blood. Mol. Microbiol. 63:754–767. 10.1111/j.1365-2958.2006.05555.x. [DOI] [PubMed] [Google Scholar]
- 41.Chattopadhyay D, Carey AJ, Caliot E, Webb RI, Layton JR, Wang Y, Bohnsack JF, Adderson EE, Ulett GC. 2011. Phylogenetic lineage and pilus protein Spb1/SAN1518 affect opsonin-independent phagocytosis and intracellular survival of group B Streptococcus. Microbes Infect. 13:369–382. 10.1016/j.micinf.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adderson EE, Takahashi S, Wang Y, Armstrong J, Miller DV, Bohnsack JF. 2003. Subtractive hybridization identifies a novel predicted protein mediating epithelial cell invasion by virulent serotype III group B Streptococcus agalactiae. Infect. Immun. 71:6857–6863. 10.1128/IAI.71.12.6857-6863.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whidbey C, Harrell MI, Burnside K, Ngo L, Becraft AK, Iyer LM, Aravind L, Hitti J, Waldorf KMA, Rajagopal L. 2013. A hemolytic pigment of group B Streptococcus allows bacterial penetration of human placenta. J. Exp. Med. 210:1265–1281. 10.1084/jem.20122753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doran KS, Engelson EJ, Khosravi A, Maisey HC, Fedtke I, Equils O, Michelsen KS, Arditi M, Peschel A, Nizet V. 2005. Blood-brain barrier invasion by group B Streptococcus depends upon proper cell-surface anchoring of lipoteichoic acid. J. Clin. Invest. 115:2499–2507. 10.1172/JCI23829. [DOI] [PMC free article] [PubMed] [Google Scholar]