Abstract
Glycosylation is one of the common posttranslational modifications in eukaryotes. Recently, glycosylated proteins have also been identified in prokaryotes. A few glycosylated proteins, including gingipains, have been identified in Porphyromonas gingivalis, a major pathogen associated with chronic periodontitis. However, no other glycosylated proteins have been found. The present study identified glycoproteins in P. gingivalis cell lysates by lectin blotting. Whole-cell lysates reacted with concanavalin A (ConA), Lens culinaris agglutinin (LCA), Phaseolus vulgaris erythroagglutinin (PHA-E4), and wheat germ agglutinin (WGA), suggesting the presence of mannose-, N-acetylgalactosamine-, or N-acetylglucosamine (GlcNAc)-modified proteins. Next, glycoproteins were isolated by ConA-, LCA-, PHA-E4-, or WGA-conjugated lectin affinity chromatography although specific proteins were enriched only by the WGA column. Mass spectrometry analysis showed that an OmpA-like, heterotrimeric complex formed by Pgm6 and Pgm7 (Pgm6/7) was the major glycoprotein isolated from P. gingivalis. Deglycosylation experiments and Western blotting with a specific antibody indicated that Pgm6/7 was modified with O-GlcNAc. When whole-cell lysates from P. gingivalis mutant strains with deletions of Pgm6 and Pgm7 were applied to a WGA column, homotrimeric Pgm7, but not Pgm6, was isolated. Heterotrimeric Pgm6/7 had the strongest affinity for fibronectin of all the extracellular proteins tested, whereas homotrimeric Pgm7 showed reduced binding activity. These findings suggest that the heterotrimeric structure is important for the biological activity of glycosylated WGA-binding OmpA-like proteins in P. gingivalis.
INTRODUCTION
Porphyromonas gingivalis, a Gram-negative, black-pigmented, obligate anaerobe, is the major etiological agent in chronic periodontitis (1, 2). It is also thought to be associated with systemic diseases, including cardiovascular disease (3), rheumatoid arthritis (4), and nonalcoholic fatty liver disease (5). The virulence of P. gingivalis has been attributed to a variety of factors associated with its cell surface (6); indeed, we previously identified several surface components, including the outer membrane proteins (7). The OmpA-like outer membrane proteins of P. gingivalis, designated Pgm6 and Pgm7, form a heterotrimeric structure (Pgm6/7) that is responsible for the maintenance and stability of the outer membrane (8, 9).
Glycosylation is one of the most common posttranslational modifications and was once thought to be restricted to eukaryotes (10). However, glycosylated proteins have also been identified in prokaryotes (11–13). The most studied system is N-glycosylation in Campylobacter (14, 15). Pilin, isolated from Neisseria, was one of the first examples of an O-glycosylated glycoprotein in a bacterial pathogen (16, 17). More recently, general O-glycosylation systems have been identified in pathogenic Neisseria strains and in the major human intestinal symbiont, Bacteroides fragilis (18–20). In P. gingivalis, gingipains (21), HBP35 (22), OMP85 (23), and Mfa1 (24) are glycosylated proteins; however, it is unclear whether other proteins are glycosylated.
Here, we attempted to detect glycoproteins in lysates of P. gingivalis cells by lectin blotting. We then isolated the glycoproteins by lectin affinity chromatography and identified them using mass spectrometry. We found that the OmpA-like proteins, Pgm6 and Pgm7, were major glycoproteins within P. gingivalis and characterized the type of glycosylation and possible saccharide modifications. Moreover, we showed the biological importance of the heterotrimeric structure of glycosylated Pgm6/7 by examining binding of the complex to extracellular matrix (ECM) proteins.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are shown in Table 1 (9, 21, 25–27). The P. gingivalis KDP390 strain was a kind gift from K. Nakayama (Nagasaki University Graduate School of Biomedical Sciences, Nagasaki, Japan). The P. gingivalis W50 PorR and W50 WbpB strains were kind gifts from M. A. Curtis (Barts and the London, Queen Mary's School of Medicine and Dentistry, London, United Kingdom). All P. gingivalis strains were grown at 37°C under anaerobic conditions (10% [vol/vol] CO2, 10% [vol/vol] H2, and 80% [vol/vol] N2) in Trypticase soy broth (BD, Franklin Lakes, NJ, USA) supplemented with 2.5 mg ml−1 yeast extract, 2.5 μg ml−1 hemin, 5 μg ml−1 menadione, and 0.1 mg ml−1 dithiothreitol. Bacterial growth was monitored by measuring optical density at 660 nm (OD660).
TABLE 1.
Bacterial strains used in the study
| Porphyromonas gingivalis strain | Relevant characteristic(s)a | Source or referenceb |
|---|---|---|
| W50 | Wild type | Laboratory stock |
| ATCC 33277 | Wild type, type strain | ATCC |
| Δ694 | PG0694 (PGN0728) deletion mutant of ATCC 33277, Cmr | 9 |
| Δ695 | PG0695 (PGN0729) deletion mutant of ATCC 33277, Cmr | 9 |
| Δ695-Δ694 | PG0695 (PGN0729) and PG0694 (PGN0728) deletion mutant of ATCC 33277, Cmr | 9 |
| W50 PorR | Mutant of W50, porR (PG1138)::erm, Emr | 21 |
| W50 WbpB | Mutant of W50, wbpB (PG2199)::erm, Emr | 21 |
| Prfa1 | Mutant of ATCC 33277, rfa (PG1155 [PGN2155])::erm, Emr | 26 |
| Pugd1 | Mutant of ATCC 33277, ugdA (PG1277 [PGN0613])::erm, Emr | 26 |
| RE1 | Mutant of 381, gtfA (PG0750 [PGN0777])::erm, Emr | 25 |
| KDP390 | Mutant of ATCC 33277, gtfB (PG1149 [PGN1251])::Tn4400′, Tcr | 27 |
Locus tags from ATCC 33277 are identified by a “PGN” prefix; protein coding sequences from strain W83 are identified by a “PG” prefix. The ATCC 33277 and 381 strains have similar genetic backgrounds. Cmr, chloramphenicol resistant; Emr, erythromycin resistant; Tcr, tetracycline resistant.
ATCC, American Type Culture Collection.
Cell fractionation, SDS-PAGE, and Western blotting.
Preparation of bacterial whole-cell lysates, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Western blot analyses were performed essentially as described previously (7, 9). The gels were stained with Coomassie brilliant blue R-250 (CBB) or SyproRuby (Molecular Probes, Eugene, OR, USA) to detect proteins or with the Pro-Q emerald 300 fluorescent stain (Molecular Probes), which reacts with periodate-oxidized carbohydrate groups (28), according to the manufacturer's protocol. Antiserum specific for the Pgm6/7 protein derived from the ATCC 33277 strain was used as the primary antibody for Western blotting (29).
Lectin blotting.
Proteins were separated by SDS-PAGE and electrophoretically transferred to a nitrocellulose membrane. Staining with horseradish peroxidase-conjugated lectins (concanavalin A [ConA], Dolichos biflorus agglutinin [DBA], Lens culinaris agglutinin [LCA], Phaseolus vulgaris erythroagglutinin [PHA-E4], peanut agglutinin [PNA], Ricinis communis agglutinin [RCA120], Ulex europaeus agglutinin [UEA-I], and wheat germ agglutinin [WGA]) (Table 2) was performed according to the manufacturer's recommendations (J-Oil Mills, Tokyo, Japan). In brief, the membrane was blocked in 10 mM Tris-HCl (pH 7.4) containing 0.15 M NaCl and 0.05% (vol/vol) Tween 20 (TBS-T) at 4°C for 1 h. The membrane was then incubated with peroxidase-conjugated lectin (diluted 1:100 in TBS-T) at 20°C for 3 h. After excess lectin was removed by rinsing with TBS, the membrane was developed using 3,3′-diaminobenzidine (DAB).
TABLE 2.
Sugar specificity of the lectins used in the study
| Lectin | Full name | Sugar specificitya |
|---|---|---|
| AAL | Aleuria aurantia lectin | α-l-Fucose |
| ConA | Concanavalin A | α-d-Mannose, α-d-glucose |
| DBA | Dolichos biflorus agglutinin | α-d-GalNAc |
| DSA | Datura stramonium agglutinin | β-d-GlcNAc |
| ECA | Erythrina cristagalli agglutinin | d-GalNAc |
| LCA | Lens culinaris agglutinin | α-d-Mannose, α-d-glucose |
| Lotus | Lotus tetragonolobus | α-l-Fucose |
| MAM | Maackia amurensis agglutinin | Sia α2-3-galactose |
| PHA-E4 | Phaseolus vulgaris erythroagglutinin | d-GalNAc |
| PHA-L4 | Phaseolus vulgaris leucoagglutinin | d-GalNAc |
| PNA | Peanut agglutinin | β-d-Galactose |
| PSA | Pisum sativum agglutinin | α-d-Mannose, α-d-glucose |
| PWM | Poke weed mitogen | (β-d-GlcNAc)n |
| RCA 120 | Ricinis communis agglutinin | β-d-Galactose |
| SBA | Soybean agglutinin | α-d-GalNAc |
| SSA | Sambucus sieboldiana lectin | Sia α2-6-galactose/GalNAc |
| UEA-I | Ulex europaeus agglutinin | α-l-Fucose |
| WGA | Wheat germ agglutinin | β-d-GlcNAc |
GalNAc, N-acetylgalactosamine; GlcNAc, N-acetylglucosamine; Sia, sialic acid (N-acetylneuraminic acid).
Lectin affinity chromatography.
All steps were performed at 4°C. Whole-cell lysates derived from P. gingivalis were solubilized with 1% dodecyl maltoside (DM). Solubilized proteins (5 mg) were applied to a lectin-conjugated agarose minicolumn (17 lectins; J-Oil Mills) (Table 2) and then washed with 10 bed volumes of 10 mM Tris-HCl (pH 7.4) containing 0.15 M NaCl and 0.03% DM. The bound proteins were eluted with corresponding inhibitory sugars in the same buffer according to the manufacturer's instructions. The eluted proteins were extensively dialyzed against 10 mM Tris-HCl (pH 7.4) and concentrated using Ficoll PM400 (GE Healthcare, Uppsala, Sweden). The concentrated proteins were then subjected to SDS-PAGE.
Protein analysis by MS.
Isolated proteins were identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) as described previously (30, 31). CBB- or SyproRuby-stained protein bands were excised from the SDS-PAGE gels and digested with trypsin. The resulting peptides were extracted, concentrated, and analyzed in a 4800 MALDI-tandem TOF (TOF/TOF) analyzer (Applied Biosystems, Foster City, CA, USA). The proteins were deduced from the MS peaks using the peptide mass fingerprinting methods within MASCOT (Matrix Science, Boston, MA) and identified according to the significance criteria set by the search program (P < 0.05).
Protein modifications were identified at the Proteomics/Mass Spectrometry Laboratory at the University of California, Berkeley. Tryptic peptides were analyzed by one-dimension liquid chromatography coupled with tandem MS (LC-MS/MS) using an Agilent 1100 series high performance LC (Palo Alto, CA, USA) coupled to a Thermo LTQ XL linear ion trap mass spectrometer with an electrospray ionization source (Waltham, MA, USA). The programs SEQUEST and DTASelect were used to identify peptides and proteins from the database (32, 33).
Deglycosylation assay.
Chemical deglycosylation of OmpA-like proteins was performed using anhydrous trifluoromethanesulfonic acid (TFMS) containing 10% anisole (23). Enzymatic deglycosylation of OmpA-like proteins was performed using a commercial kit (Prozyme, San Leandoro, CA, USA). N-Glycanase (peptide-N-glycosidase F), sialidase A, and O-glycanase (endo-α-N-acetylgalactosaminidase) were used according to the manufacturer's protocols. Bovine fetuin was included as a positive control. Deglycosylation efficiency was monitored by SDS-PAGE, followed by CBB and Pro-Q emerald staining.
Detection of possible O-GlcNAc modifications.
A monoclonal antibody highly specific for O-linked N-acetylglucosamine (O-GlcNAc) (clone CTD110.6; Abcam, Cambridge, United Kingdom) was used to probe the transferred proteins (34). Blots were incubated with horseradish peroxidase-linked goat anti-mouse IgM (Dako, Glostrup, Denmark) and developed with an ECL Plus Western blotting detection system (GE Healthcare). α-Crystallin (Sigma-Aldrich, St. Louis, MO, USA), which contains O-GlcNAc, was used as a control protein (35).
Binding assay.
Binding of isolated OmpA-like proteins to ECM proteins was examined in an enzyme-linked immunosorbent assay (ELISA) performed in polystyrene microtiter plates (96-well MaxiSorp; Nunc, Roskilde, Denmark). The plates were coated with fibronectin, laminin, collagen type I, or collagen type IV (Sigma-Aldrich) (20 μg ml−1 protein dissolved in 10 mM Tris-HCl, pH 8.0; 100 μl/well) at 4°C overnight (36). After three washes with Dulbecco's phosphate-buffered saline (PBS; pH 7.4) containing 0.05% (vol/vol) Tween 20, the wells were blocked with 300 μl of 1% (wt/vol) bovine serum albumin (BSA) in PBS at room temperature for 2 h, washed again, and then incubated with 100 μl of isolated OmpA-like proteins at room temperature for 2 h, followed by a further incubation at 4°C overnight. After another washing step, the wells were treated with 100 μl of anti-Pgm6/7 antibody (1:10,000 dilution) followed by peroxidase-conjugated goat anti-rabbit IgG (1:5,000 dilution). Finally, a TMB (3,3′,5,5′-tetramethylbenzidine) peroxidase enzyme immunoassay (EIA) substrate (100 μl; Bio-Rad, Hercules, CA, USA) was added to each well. The reaction was stopped by the addition of 100 μl of 0.5 M H2SO4. The binding activities were assessed by measuring the OD450 values in a microplate reader (model 680; Bio-Rad). All assays were carried out in triplicate, and the standard deviations were determined. PBS (100 μl) alone was used as a negative control. The mean OD450 value of the negative control was subtracted from that of triplicate wells containing OmpA-like proteins, and the resulting value was defined as net binding. Preliminary experiments were performed to check that the anti-Pgm6/7 antibody showed similar reactivity against heterotrimeric Pgm6/7 and homotrimeric Pgm6 and Pgm7.
Bacterial adhesion assay.
The bacterial adhesion assay has been described previously (25). Briefly, P. gingivalis cells (OD660 adjusted to 1.0 with PBS; 100 μl) were added to the wells of a 96-well polystyrene plastic plate coated with ECM molecules as described above. After an anaerobic incubation lasting for 3 h, the plate was washed with PBS. Adherent cells were stained with 1% crystal violet and then washed with water. Adhesion was evaluated by measuring the OD570 after ethanol elution of the crystal violet. All assays were carried out in triplicate, and the standard deviations were determined. PBS alone was used as a negative control. The mean OD570 value of the negative control was subtracted from that of triplicate wells containing P. gingivalis cells, and the resulting value was defined as net binding.
Statistical analysis.
Student's t test was used for data analysis. Differences were considered significant at a P value of <0.05.
RESULTS
Lectin blotting.
To identify and characterize putative P. gingivalis glycoproteins, whole-cell lysates of P. gingivalis W50 were separated by SDS-PAGE and blotted onto nitrocellulose membranes. Blots were probed individually with an array of peroxidase-conjugated lectins with different specificities (Table 2). Only ConA, LCA, PHA-E4, and WGA reacted with a number of P. gingivalis proteins (Fig. 1). The sugar specificity of the lectins indicated that mannose, N-acetylgalactosamine, and GlcNAc residues were present on putative P. gingivalis glycoproteins. The binding of peroxidase-conjugated lectins to P. gingivalis ATCC 33277 was similar to that of W50 (data not shown).
FIG 1.
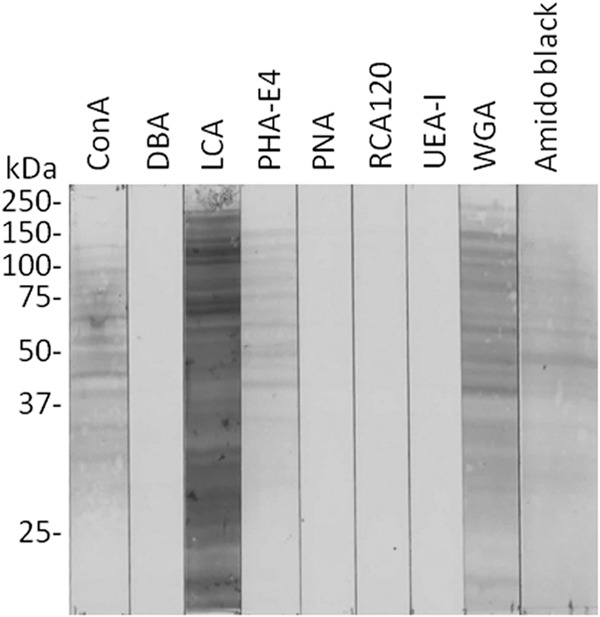
Detection of glycoproteins in P. gingivalis whole-cell lysates by lectin blotting. P. gingivalis W50 whole-cell lysates were separated by SDS-PAGE under reducing conditions and electrophoretically transferred to a nitrocellulose membrane. The membrane was blocked in TBS containing 0.05% Tween 20 for 1 h at 4°C. The membrane was then incubated with peroxidase-conjugated lectin (1:100 in TBS containing 0.05% Tween 20) for 3 h at 20°C. After excess lectin was removed by rinsing the membrane with TBS, it was developed using 3,3′-dimethylbenzidine. Transferred proteins were also stained with amido black as a loading control.
Lectin affinity chromatography.
P. gingivalis glycoproteins were isolated by lectin affinity chromatography. P. gingivalis W50 whole-cell lysates, solubilized in DM, were loaded onto lectin-conjugated agarose columns comprising ConA, LCA, PHA-E4, or WGA. Following extensive washing, the lectin-binding components were eluted with inhibitory sugars. Concentrated eluates were subjected to SDS-PAGE and stained with SyproRuby for sensitive detection (Fig. 2). Eluates from the WGA column yielded a specific broad 40-kDa band (Fig. 2, band 7) and several weaker bands. Eluates from the PHA-E4, Datura stramonium agglutinin (DSA), and Maackia amurensis agglutinin (MAM) columns yielded a few weak bands. However, the ConA and LCA columns showed very weak glycoprotein adsorption. The binding of some lectins was not consistent between the blotting and chromatography experiments. This may be because the molecular structure was stricter in binding to lectin beads. Since WGA and DSA have overlapping specificities for GlcNAc residues, it was likely that the 40-kDa glycoprotein recognized by these lectins contained GlcNAc residues. The stained bands were cut out from the gel, trypsin digested, and then subjected to MALDI-TOF MS. Proteins were identified by peptide mass fingerprinting and database searching using the MASCOT search engine. The major 40-kDa protein eluted from the WGA and DSA columns (Fig. 2, bands 7 and 11, respectively) was identified as OmpA-like protein Pgm6/7. A list of identified glycoproteins is presented in Table 3.
FIG 2.
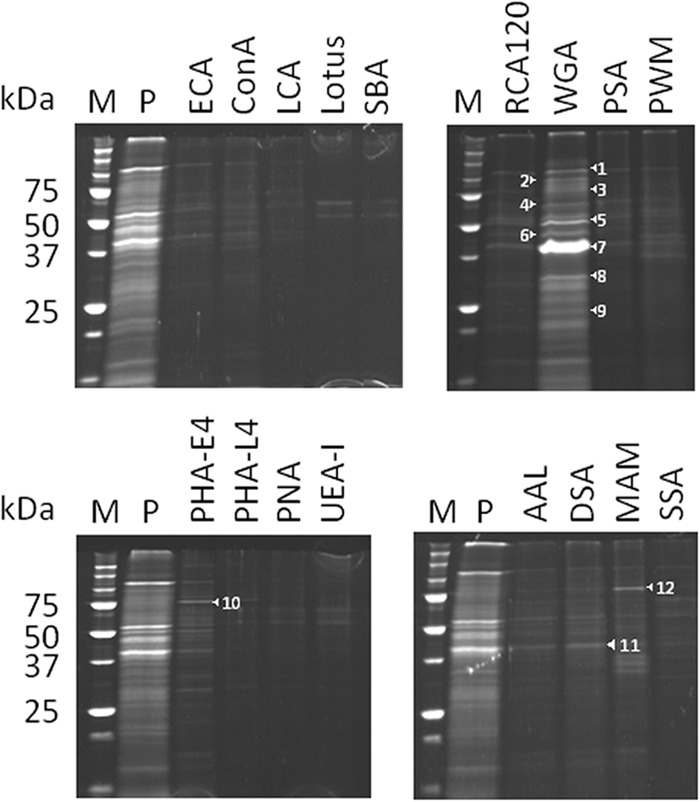
Isolation of glycoproteins from P. gingivalis whole-cell lysates by lectin affinity chromatography. Whole-cell lysates from P. gingivalis W50 were solubilized with 1% DM. A total of 5 mg of the solubilized proteins was applied to a lectin-conjugated agarose minicolumn overnight at 4°C and then washed with 10 bed volumes of 10 mM Tris-HCl (pH 7.4) containing 0.15 M NaCl and 0.03% DM. After the bound proteins were eluted with corresponding inhibitory (hapten) sugars in the same buffer, the eluted proteins were extensively dialyzed against 10 mM Tris-HCl (pH 7.4) and concentrated by Ficoll dialysis. The concentrated proteins were subjected to SDS-PAGE under reducing conditions. The gels were stained with SyproRuby. Numbered protein bands are identified in Table 3. Lane M, molecular marker; lane P, whole-cell lysate prior to application to the column.
TABLE 3.
Glycoproteins isolated by lectin affinity chromatography
| Lectin | Protein banda | Identified protein | CDS no.b |
|---|---|---|---|
| WGA | 1 | RagA | PG0185 |
| 2 | Peptidylarginine deiminase | PG1424 | |
| 3 | TPR domain proteinc | PG1028 | |
| 4 | RagB | PG0186 | |
| 5 | Lys-gingipain | PG1844 | |
| 6 | Arg-gingipain | PG2024 | |
| 7 | Pgm6/7 | PG0695/PG0694 | |
| 8 | Pgm7 | PG0694 | |
| 9 | Pgm6 | PG0695 | |
| PHA-E4 | 10 | Hypothetical protein | PG0491 |
| DSA | 11 | Pgm6/7 | PG0695/PG0694 |
| MAM | 12 | Hypothetical protein | PG0491 |
The protein bands are identified by number in Fig. 2.
Protein-coding sequence (CDS) number of P. gingivalis W83 in the genome database.
TPR, tetratricopeptide repeat.
Deglycosylation assay.
To verify that OmpA-like proteins were carbohydrate modified, OmpA-like protein Pgm6/7 was isolated from P. gingivalis ATCC 33277 using a WGA column, and both chemical and enzymatic deglycosylation assays were performed. Fetuin, a well-known glycoprotein, was used as a positive control. Chemical treatment with TFMS effectively removed the glycans from fetuin as its apparent molecular mass was reduced, and it lost its reactivity with Pro-Q emerald (Fig. 3A). Intact Pgm6/7 appeared as several diffuse bands around 40 kDa upon Pro-Q emerald staining. Similar to fetuin, Pgm6/7 lost its reactivity with Pro-Q emerald after TFMS treatment although there was no change in the apparent molecular mass. Although the same amounts of proteins were loaded onto the SDS-PAGE gels both before and after treatment with TFMS, for some unknown reason the protein recovered after treatment was not stained well by CBB. These results suggest that Pgm6/7 may be modified by glycosylations. Further enzymatic treatment showed that fetuin was N-glycosylated and sialylated (Fig. 3B). Presumably, the band observed at about 35 kDa upon CBB staining represented excess N-glycanase. In contrast, neither N-glycanase nor O-glycanase affected the Pgm6/7 protein. According to the manufacturer, N-glycanase treatment is the most effective method of removing virtually all N-linked oligosaccharides from a glycoprotein. However, the O-glycanase used in this assay removes only O-linked core Galβ(1-3)GalNAc when the unmodified core structure is attached to Ser or Thr residues. Therefore, we could almost exclude the presence of N-glycosylations but could not rule out the possibility that Pgm6/7 carried O-glycosylation.
FIG 3.
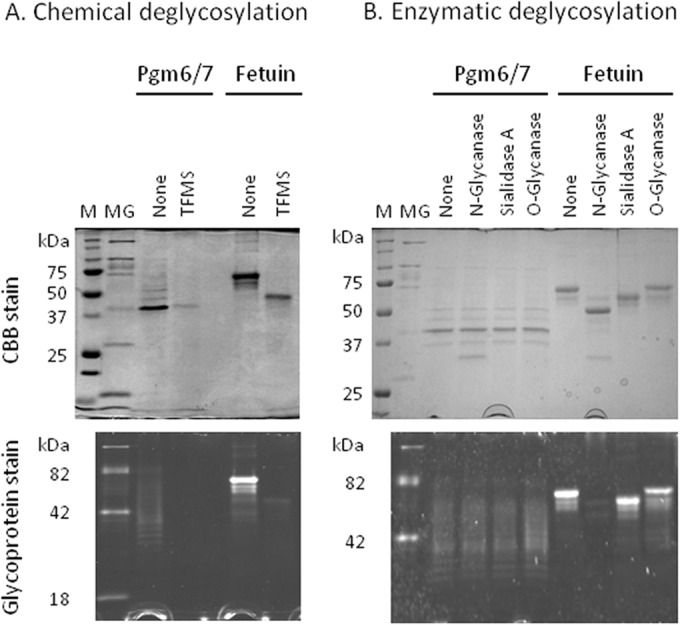
Deglycosylation of OmpA-like proteins derived from P. gingivalis. (A) Chemical deglycosylation of OmpA-like proteins from P. gingivalis ATCC 33277 was performed using anhydrous trifluoromethanesulfonic acid (TFMS) containing 10% anisole. (B) Enzymatic deglycosylation of OmpA-like proteins was performed using a commercial kit. Fetuin was included as a positive control. The deglycosylation efficiency was monitored by SDS-PAGE under reducing conditions, followed by staining with CBB or Pro-Q emerald. Lane M, molecular marker for proteins; lane MG, molecular marker for glycoproteins.
Detection of possible O-GlcNAc modifications using a specific antibody.
Since Pgm6/7 has affinity for the WGA lectin and carries O-glycosylations, the presence of O-GlcNAc modifications was assessed by immunoblotting with a monoclonal antibody specific for O-GlcNAc. α-Crystallin was used as a positive control. The O-GlcNAc-modified protein yielded an immunoreactive band of approximately 20 kDa (Fig. 4, arrowhead). Pgm6/7 from P. gingivalis ATCC 33277 also showed a positive band at 40 kDa (Fig. 4, arrow). Whole-cell lysates yielded several strongly positive bands, including Pgm6/7, suggesting that O-GlcNAc modification of P. gingivalis proteins is common.
FIG 4.
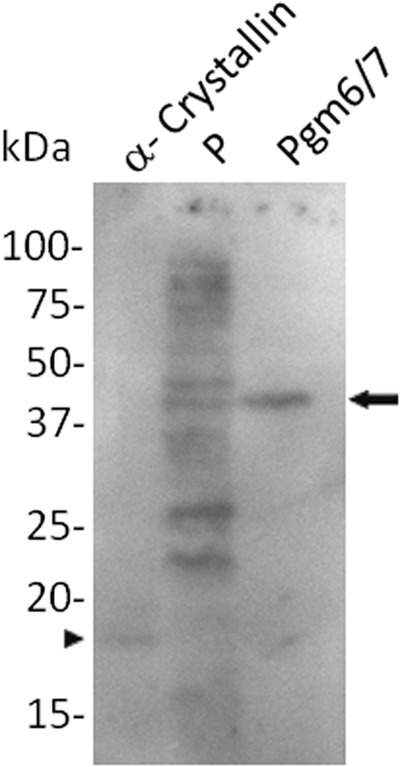
Detection of possible O-GlcNAc glycosylations on OmpA-like proteins derived from P. gingivalis. Samples were subjected to SDS-PAGE under reducing conditions. A monoclonal anti-O-GlcNAc antibody was used to probe the transferred proteins. Blots were incubated with peroxidase-linked goat anti-mouse IgM and developed with an ECL Plus system. α-Crystallin was used as a control protein. The arrow and arrowhead indicate positive bands of Pgm6/7 and α-crystallin, respectively. Lane P, whole-cell lysate prior to application to the column.
Isolation of OmpA-like proteins from wild-type and mutant P. gingivalis strains.
Because OmpA-like protein Pgm6/7 derived from wild-type ATCC 33277 was purified to near-homogeneity by one-step WGA affinity chromatography, we examined whether it was possible to isolate OmpA-like proteins (Pgm6 and Pgm7) from mutant strains. Lysates were prepared from wild-type and mutant cells and loaded onto a WGA column. The eluate from the column was concentrated and subjected to SDS-PAGE (Fig. 5A). Staining the gels with CBB revealed a band of approximately 120 kDa in both the wild-type and ATCC 33277 with a deletion of the PG0695 gene (Δ695 mutant) under nonreducing conditions, suggesting that heterotrimeric and homotrimeric structures are formed, respectively (9). These bands reacted with anti-Pgm6/7 antibody (Fig. 5B). However, no obvious band was obtained from ATCC 33277 strain with a deletion of the PG0694 gene (Δ694 mutant). MALDI-TOF MS confirmed that the band isolated from the Δ695 mutant was Pgm7 (data not shown). No bands corresponding to Pgm6 and Pgm7 were obtained from the Δ694-Δ695 negative-control double mutant (data not shown).
FIG 5.
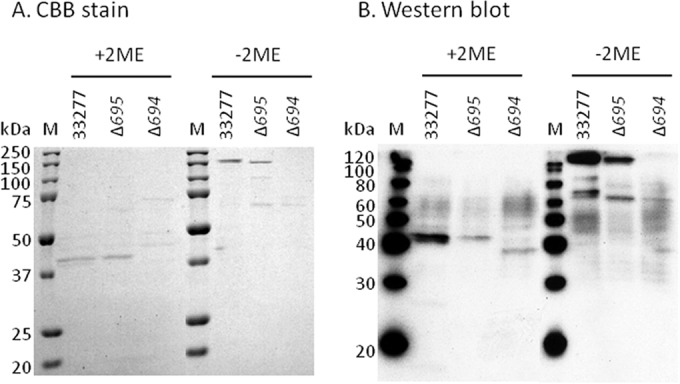
Enrichment of OmpA-like glycoproteins from P. gingivalis wild-type and mutant strains by WGA column chromatography. Whole-cell lysates from P. gingivalis wild-type and mutant strains were solubilized with 1% DM and applied to a WGA-agarose minicolumn overnight at 4°C and then washed with 10 bed volumes of 10 mM Tris-HCl (pH 7.4) containing 0.15 M NaCl and 0.03% DM. The bound proteins were eluted with GlcNAc in the same buffer and then extensively dialyzed against 10 mM Tris-HCl (pH 7.4) and concentrated. The concentrated proteins were subjected to SDS-PAGE under nonreducing and reducing conditions and stained with CBB (A) and then further analyzed by Western blotting with an anti-Pgm6/7 antibody (B). Lane M, molecular marker. 2ME, 2-mercaptoethanol.
Identification of glycosylation sites.
Heterotrimeric OmpA-like proteins isolated from wild-type P. gingivalis were subjected to LC-MS/MS analysis to identify potential glycosylation sites. Tryptic peptides from corresponding SDS-PAGE gel bands were analyzed. Ion signals from 269PVSCPECPEVTPVTK283 within Pgm6 (PG0695) were observed at m/z 1,862.591 and 1,700.12 (Fig. 6). The former was 162.1 Da larger because of a hexose modification at Ser271. No N-acetyl hexosamine (203 Da) or sialic acid (291.3 Da) modifications were found. Surprisingly, no modifications were detected within Pgm7 (PG0694). The same result was obtained when we analyzed homotrimeric Pgm7 isolated from the Δ695 mutant (data not shown). Further investigations are required to determine whether any GlcNAc residues are present.
FIG 6.
Identification of glycosylation sites. OmpA-like proteins isolated from the P. gingivalis wild-type strain by WGA column chromatography were subjected to LC-MS/MS analysis to identify the glycosylation sites. Tryptic peptides from corresponding SDS-PAGE gel bands were analyzed. Peptide sequences and posttranslational modifications were determined using the SEQUEST and DTASelect programs. (A) Amino acid sequences of Pgm6 and Pgm7. Boldface letters indicate peptides identified by MS, and the underlined peptide is glycosylated. Peptides not identified by MS are indicated in lightface. The sequence coverage of Pgm6 and Pgm7 was 58.3% and 55.5%, respectively. (B) Detection of glycosylation sites in Pgm6. Modified amino acids are underlined.
Binding assay.
We next examined the binding of OmpA-like proteins eluted from the WGA column to several ECM proteins (Fig. 7). Pgm6/7 isolated from the wild-type ATCC 33277 strain showed the strongest binding to fibronectin, followed by laminin and collagen type I. It also bound weakly to collagen type IV. Pgm7 isolated from the Δ695 mutant showed weaker binding to all of the ECM molecules tested than the wild-type Pgm6/7. Thus, the heterotrimeric structure of Pgm6/7 may be important for binding.
FIG 7.
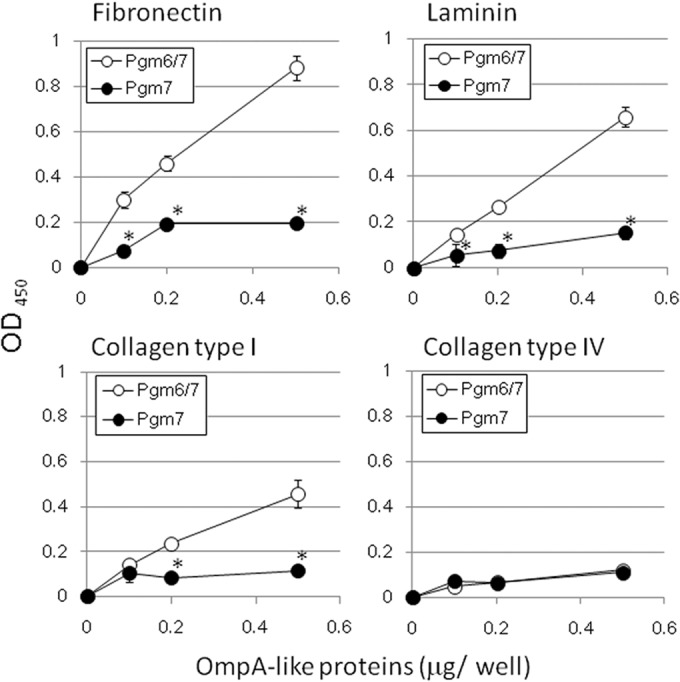
Binding of OmpA-like glycoproteins from P. gingivalis to extracellular matrix (ECM) molecules. Purified P. gingivalis OmpA-like proteins were applied to a polystyrene plastic plate coated with ECM molecules and incubated at room temperature for 2 h and then at 4°C overnight. After being washed, the wells were treated with an anti-Pgm6/7 antibody (1:10,000), followed by peroxidase-conjugated goat anti-rabbit IgG (1:5,000). EIA peroxidase substrate, 3,3′,5,5′-tetramethylbenzidine, was then added to each well. The reactions were stopped by the addition of 100 μl of 0.5 M H2SO4. The binding activity was assessed by measuring the OD450 values in a microplate reader. All assays were performed in triplicate, and the standard deviations were determined. *, significantly different from Pgm6/7 (P < 0.05).
Bacterial adhesion assay.
Next, we examined the adhesion of whole bacteria to plates coated with ECM molecules. Wild-type ATCC 33277 cells adhered strongly to all ECM molecules tested. Although fewer Δ695-Δ694 cells tended to adhere to the plates (Fig. 8), a good level of adhesion was still observed, presumably due to the expression of adhesion molecules other than Pgm6/7. Significantly more wild-type cells than mutant cells adhered to plates coated with fibronectin, collagen type I, and collagen type IV.
FIG 8.
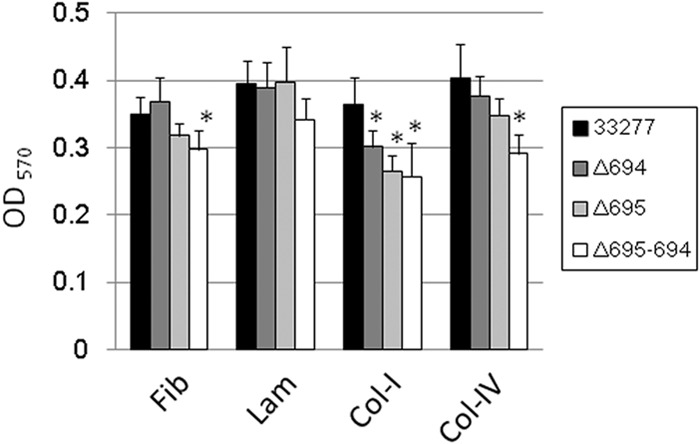
Adhesion of P. gingivalis wild-type and mutant bacteria to extracellular matrix (ECM) molecules. P. gingivalis (OD660 adjusted to 1.0) cells were applied onto a polystyrene plastic plate coated with ECM molecules. After incubation under anaerobic conditions for 3 h, the plate was washed with PBS. Adherent cells were stained with 1% crystal violet and washed with water. Adherence was evaluated by measuring the OD570 after elution of the crystal violet with ethanol. All assays were performed in triplicate, and the standard deviations were determined. Typical results are shown. *, significantly different from ATCC 33277 (P < 0.05). Fib, fibronectin; Lam, laminin; Col-I, collagen type I; Col-IV, collagen type IV.
Effect of mutating glycosylation-related genes on the glycosylation of OmpA-like proteins that react with WGA.
To examine whether glycosylation-related genes are responsible for the WGA-reactive glycosylations expressed on OmpA-like proteins, we tested P. gingivalis mutants of Rfa, UgdA, GtfA, GtfB, PorR, and WbpB (Table 1). Cell lysates were prepared and subjected to affinity chromatography on a WGA column, followed by SDS-PAGE. Eluates derived from all of the mutants yielded a 40-kDa band on SDS-PAGE gels (Fig. 9). MALDI-TOF MS identified this band as Pgm6/7 (data not shown). Pro-Q emerald staining confirmed that the bands derived from Rfa, UgdA, GtfA, and GtfB mutants were glycosylated. These data indicate that glycosylation of Pgm6/7 was unaffected in these mutants. Thus, modification of OmpA-like proteins in P. gingivalis may be mediated by other as yet unidentified glycosyltransferases.
FIG 9.
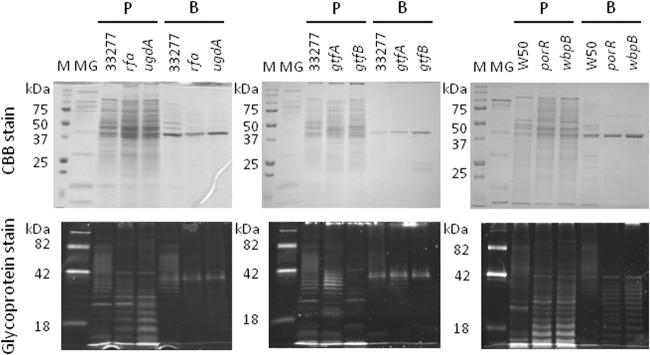
Effect of mutating glycosylation-related genes on the glycosylation of WGA-reactive OmpA-like proteins derived from P. gingivalis. Whole-cell lysates and the WGA lectin-bound fraction derived from P. gingivalis wild-type and mutant strains were subjected to SDS-PAGE under reducing conditions and stained with CBB or Pro-Q emerald. The following strains were used: ATCC 33277 (wild type), Prfa (rfa), PugdA (ugdA), RE1 (gtfA), KDP390 (gtfB), W50 (wild type), W50 PorR (porR), and W50 WbpB (wbpB). Lane M, molecular marker; lane MG, molecular marker for glycoproteins; lanes P, whole-cell lysates prior to application to the column; lanes B, proteins bound to the WGA column.
DISCUSSION
Here, we showed that affinity chromatography using a WGA column efficiently enriched glycosylated OmpA-like proteins Pgm6 and Pgm7 from P. gingivalis. A previous study showed that molecules expressed on the P. gingivalis cell surface bound to WGA (37). Thus, we speculated that proteins expressed by P. gingivalis may harbor GlcNAc modifications. In the present study, we used an O-GlcNAc-specific antibody to show that OmpA-like proteins carry O-GlcNAc moieties. To date, glycosylation of OmpA-like proteins has been reported only in Flavobacterium psychrophilum (38, 39); however, the type of modified saccharides expressed remains unknown.
We then used a series of mutants of known glycosyltransferases in P. gingivalis (GtfA, GtfB, Rfa, UgdA, PorR, and WbpB); however, all retained affinity for WGA and reactivity with Pro-Q emerald. Although the role of GtfA in glycan biosynthesis was not examined, several studies show that GtfB, Rfa, and UgdA are involved in lipopolysaccharide and anionic polysaccharide biosynthesis (25–27). PorR and WbpB are involved in the synthesis of anionic polysaccharides (21, 40). These results suggest that O-GlcNAc modification is independent from the synthesis of extracellular polysaccharides; therefore, other glycosyltransferases would be involved in the glycosylation of Pgm6/7. Moreover, Western blotting with an O-GlcNAc specific antibody suggested that proteins other than Pgm6/7 might carry O-GlcNAc moieties. These findings suggest that P. gingivalis possesses a general O-glycosylation system.
Previous studies identified a general O-glycosylation system in Bacteroides fragilis (19, 41) although no glycosylated OmpA-like proteins from B. fragilis were found; however the putative glycosylation motif D(S/T)(A/I/L/M/T/V) was described. Recently, the same O-glycosylation motif was proposed for S-layer glycoproteins derived from Tannerella forsythia (42). Since P. gingivalis belongs to the phylum Bacteroidetes, we examined whether this motif was present in the OmpA-like proteins. We identified the motif DST at amino acid residues 372 to 374 in Pgm6 but not in Pgm7. This may be because glycoproteins from B. fragilis and S-layer glycoproteins from T. forsythia are modified by glycans that contain fucose (19, 41, 42); therefore, the three-residue motif D(S/T)(A/I/L/M/T/V) within P. gingivalis Pgm6/7 may not be suitable for modification.
In our preliminary experiment, we found that WGA affinity chromatography also enriched glycosylated OmpA-like proteins from Tannerella and Bacteroides species, which also belong to the phylum Bacteroidetes (data not shown). These findings suggest that glycosylation of OmpA-like proteins may be a common phenomenon; however, further examinations are required.
The O-GlcNAc modification is an important dynamic regulatory modification in eukaryotic cells (43). However, few studies report this type of modification in prokaryotes. For example, the flagellum protein of Listeria monocytogenes is modified with O-GlcNAc (44, 45). A very recent study demonstrated that the major autolysin Acm2 from Lactobacillus plantarum is also modified with O-GlcNAc (46). Presumably, the bacterial O-GlcNAc moiety may be involved in virulence, as well as having a regulatory function.
We used a newly developed prediction program called OGlcNAcScan to identify putative O-GlcNAc attachment sites. The program was trained using protein sequences bearing known O-GlcNAcylations (47). This tool (http://cbsb.lombardi.georgetown.edu/hulab/OGAP.html) predicted potential O-GlcNAc modification sites in Pgm6/7 at the default threshold. Pgm6 contains two putative sites at Thr282 and Ser343. Pgm7 also contains two putative sites, at Ser181 and Ser349.
We then attempted to analyze trypsin-digested OmpA-like proteins derived from P. gingivalis by MS to confirm the presence of O-GlcNAc modifications; however, we obtained sequence coverage of around only 50% and so were not able to clearly identify any such sites. This approach is complicated by several obstacles. First, many glycoproteins are resistant to trypsin, and the resulting glycopeptides are often too large for MS analysis (48). Second, nonglycosylated peptides within tryptic digests tend to suppress the signal generated by glycopeptides, which have much poorer ionization efficiency (49). Third, O-GlcNAc glycosylation shows very low stoichiometry, and the sugar-protein linkage is highly labile (50).
We must also consider the affinity of the lectin used in this study. Although WGA is thought to be specific for O-GlcNAc, it also binds sialic acid and (albeit weakly) glucose (51, 52). It may be that metabolic labeling methods developed in mammalian cells might be useful tools for identifying of O-GlcNAc modifications on OmpA-like proteins (53).
We also used WGA lectin to isolate OmpA-like proteins from mutant strains of P. gingivalis. We succeeded in obtaining homotrimeric Pgm7 but not Pgm6. Previously, we showed that homotrimeric Pgm6 was prone to degradation (9). During the isolation step, we found that the unstable Pgm6 derived from the Δ694 mutant strain did not retain its native glycoprotein structure and lost affinity for the lectin. Indeed, Western blotting with an anti-Pgm6/7 antibody identified a band of less than 40 kDa under reducing conditions and a faint 70-kDa band (presumable a homodimer) under nonreducing conditions (Fig. 5B). A recent study showed that the Pro-Q emerald glycoprotein stain detected glycosylations carried by Pgm6 (PGN0729) after two-dimensional PAGE of crude whole-cell proteins derived from wild-type bacteria (54).
Isolated heterotrimeric Pgm6/7 showed higher binding to ECM proteins including fibronectin than homotrimeric Pgm7. Thus, the heterotrimer appears to be the functional unit. To the best of our knowledge, this is the first report to use isolated bacterial proteins to show the biological importance of heterotrimeric structure of Pgm6/7. Several studies suggest that OmpA-like proteins from a variety of Gram-negative bacteria bind fibronectin (55–59). In previous studies, we examined deletion mutants of OmpA-like proteins and found that heterotrimeric Pgm6/7 was also important for maintaining the integrity of the outer membrane (8, 9).
To summarize, we demonstrated that the glycosylation of OmpA-like proteins with WGA-reactive moiety is found in P. gingivalis. Further studies of the potential role of glycosylated proteins and identification of the enzymes responsible for glycosylation in P. gingivalis are under way. Identifying the relationship between glycoprotein expression and pathogenicity may aid the diagnosis and treatment of periodontal pathogen-related diseases.
ACKNOWLEDGMENTS
We thank H. Nikaido (Department of Molecular and Cell Biology, University of California) for valuable suggestions. We also thank L. Kohlstaedt (Proteomics/Mass Spectrometry Laboratory, University of California, Berkeley, CA) for performing LC-MS/MS analysis.
This work was supported by Grants-in-Aid for Scientific Research (20592165 and 23592720 to Y.M. and 20890248 and 22791783 to Y.H.) from the Japan Society for the Promotion of Science, and the Strategic Research AGU-Platform Formation and Assistance for Development of Graduate School Infrastructure and Focus from The Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
Published ahead of print 18 August 2014
REFERENCES
- 1.Holt SC, Ebersole JL. 2005. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex,” a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2000 38:72–122. 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- 2.Pathirana RD, O'Brien-Simpson NM, Reynolds EC. 2010. Host immune responses to Porphyromonas gingivalis antigens. Periodontol. 2000 52:218–237. 10.1111/j.1600-0757.2009.00330.x. [DOI] [PubMed] [Google Scholar]
- 3.Demmer RT, Desvarieux M. 2006. Periodontal infections and cardiovascular disease: the heart of the matter. J. Am. Dent. Assoc. 137(Suppl):14S–20S. 10.14219/jada.archive.2006.0402. [DOI] [PubMed] [Google Scholar]
- 4.Detert J, Pischon N, Burmester GR, Buttgereit F. 2010. The association between rheumatoid arthritis and periodontal disease. Arthritis Res. Ther. 12:218. 10.1186/ar3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoneda M, Naka S, Nakano K, Wada K, Endo H, Mawatari H, Imajo K, Nomura R, Hokamura K, Ono M, Murata S, Tohnai I, Sumida Y, Shima T, Kuboniwa M, Uemura K, Kamisaki Y, Amano A, Okanoue T, Ooshima T, Nakajima A. 2012. Involvement of a periodontal pathogen, Porphyromonas gingivalis on the pathogenesis of non-alcoholic fatty liver disease. BMC Gastroenterol. 12:16. 10.1186/1471-230X-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshimura F, Murakami Y, Nishikawa K, Hasegawa Y, Kawaminami S. 2009. Surface components of Porphyromonas gingivalis. J. Periodont. Res. 44:1–12. 10.1111/j.1600-0765.2008.01135.x. [DOI] [PubMed] [Google Scholar]
- 7.Murakami Y, Imai M, Nakamura H, Yoshimura F. 2002. Separation of the outer membrane and identification of major outer membrane proteins from Porphyromonas gingivalis. Eur. J. Oral Sci. 110:157–162. 10.1034/j.1600-0722.2002.11171.x. [DOI] [PubMed] [Google Scholar]
- 8.Iwami J, Murakami Y, Nagano K, Nakamura H, Yoshimura F. 2007. Further evidence that major outer membrane proteins homologous to OmpA in Porphyromonas gingivalis stabilize bacterial cells. Oral Microbiol. Immunol. 22:356–360. 10.1111/j.1399-302X.2007.00363.x. [DOI] [PubMed] [Google Scholar]
- 9.Nagano K, Read EK, Murakami Y, Masuda T, Noguchi T, Yoshimura F. 2005. Trimeric structure of major outer membrane proteins homologous to OmpA in Porphyromonas gingivalis. J. Bacteriol. 187:902–911. 10.1128/JB.187.3.902-911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hitchen PG, Dell A. 2006. Bacterial glycoproteomics. Microbiology 152:1575–1580. 10.1099/mic.0.28859-0. [DOI] [PubMed] [Google Scholar]
- 11.Abu-Qarn M, Eichler J, Sharon N. 2008. Not just for Eukarya anymore: protein glycosylation in Bacteria and Archaea. Curr. Opin. Struct. Biol. 18:544–550. 10.1016/j.sbi.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Messner P. 2009. Prokaryotic protein glycosylation is rapidly expanding from “curiosity” to “ubiquity.” Chembiochem 10:2151–2154. 10.1002/cbic.200900388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szymanski CM, Wren BW. 2005. Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. Microbiol. 3:225–237. 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- 14.Linton D, Allan E, Karlyshev AV, Cronshaw AD, Wren BW. 2002. Identification of N-acetylgalactosamine-containing glycoproteins PEB3 and CgpA in Campylobacter jejuni. Mol. Microbiol. 43:497–508. 10.1046/j.1365-2958.2002.02762.x. [DOI] [PubMed] [Google Scholar]
- 15.Szymanski CM, Yao R, Ewing CP, Trust TJ, Guerry P. 1999. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32:1022–1030. 10.1046/j.1365-2958.1999.01415.x. [DOI] [PubMed] [Google Scholar]
- 16.Power PM, Roddam LF, Dieckelmann M, Srikhanta YN, Tan YC, Berrington AW, Jennings MP. 2000. Genetic characterization of pilin glycosylation in Neisseria meningitidis. Microbiology 146:967–979. [DOI] [PubMed] [Google Scholar]
- 17.Stimson E, Virji M, Makepeace K, Dell A, Morris HR, Patne G, Saunders JR, Jennings MP, Barker S, Panico M, Blench I, Moxon ER. 1995. Meningococcal pilin: a glycoprotein substituted with digalactosyl 2,4-diacetamido-2,4,6-trideoxyhexose. Mol. Microbiol. 17:1201–1214. 10.1111/j.1365-2958.1995.mmi_17061201.x. [DOI] [PubMed] [Google Scholar]
- 18.Børud B, Aas FE, Vik Å, Winther-Larsen HC, Egge-Jacobsen W, Koomey M. 2010. Genetic, structural, and antigenic analyses of glycan diversity in the O-linked protein glycosylation systems of human Neisseria species. J. Bacteriol. 192:2816–2829. 10.1128/JB.00101-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fletcher CM, Coyne MJ, Villa OF, Chatzidaki-Livanis M, Comstock LE. 2009. A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell 137:321–331. 10.1016/j.cell.2009.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vik Å, Aas FE, Anonsen JH, Bilsborough S, Schneider A, Egge-Jacobsen W, Koomey M. 2009. Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U. S. A. 106:4447–4452. 10.1073/pnas.0809504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallagher A, Aduse-Opoke J, Rangarajan M, Slaney JM, Curtis MA. 2003. Glycosylation of the Arg-gingipains of Porphyromonas gingivalis and comparison with glycoconjugate structure and synthesis in other bacteria. Curr. Protein Pept. Sci. 4:427–441. 10.2174/1389203033486974. [DOI] [PubMed] [Google Scholar]
- 22.Shoji M, Sato K, Yukitake H, Kondo Y, Narita Y, Kadowaki T, Naito M, Nakayama K. 2011. Por secretion system-dependent secretion and glycosylation of Porphyromonas gingivalis hemin-binding protein 35. PLoS One 6:e21372. 10.1371/journal.pone.0021372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakao R, Tashiro Y, Nomura N, Kosono S, Ochiai K, Yonezawa H, Watanabe H, Senpuku H. 2008. Glycosylation of the OMP85 homolog of Porphyromonas gingivalis and its involvement in biofilm formation. Biochem. Biophys. Res. Commun. 365:784–789. 10.1016/j.bbrc.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 24.Zeituni AE, McCaig W, Scisci E, Thanassi DG, Cutler CW. 2010. The native 67-kilodalton minor fimbria of Porphyromonas gingivalis is a novel glycoprotein with DC-SIGN-targeting motifs. J. Bacteriol. 192:4103–4110. 10.1128/JB.00275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narimatsu M, Noiri Y, Itoh S, Noguchi N, Kawahara T, Ebisu S. 2004. Essential role for the gtfA gene encoding a putative glycosyltransferase in the adherence of Porphyromonas gingivalis. Infect. Immun. 72:2698–2702. 10.1128/IAI.72.5.2698-2702.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato K, Kido N, Murakami Y, Hoover CI, Nakayama K, Yoshimura F. 2009. Lipopolysaccharide biosynthesis-related genes are required for colony pigmentation of Porphyromonas gingivalis. Microbiology 155:1282–1293. 10.1099/mic.0.025163-0. [DOI] [PubMed] [Google Scholar]
- 27.Yamaguchi M, Sato K, Yukitake H, Noiri Y, Ebisu S, Nakayama K. 2010. A Porphyromonas gingivalis mutant defective in a putative glycosyltransferase exhibits defective biosynthesis of the polysaccharide portions of lipopolysaccharide, decreased gingipain activities, strong autoaggregation, and increased biofilm formation. Infect. Immun. 78:3801–3812. 10.1128/IAI.00071-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinberg TH, Pretty On Top K, Berggren KN, Kemper C, Jones L, Diwu Z, Haugland RP, Patton WF. 2001. Rapid and simple single nanogram detection of glycoproteins in polyacrylamide gels and on electroblots. Proteomics 1:841–855. . [DOI] [PubMed] [Google Scholar]
- 29.Murakami Y, Masuda T, Imai M, Iwami J, Nakamura H, Noguchi T, Yoshimura F. 2004. Analysis of major virulence factors in Porphyromonas gingivalis under various culture temperatures using specific antibodies. Microbiol. Immunol. 48:561–569. 10.1111/j.1348-0421.2004.tb03552.x. [DOI] [PubMed] [Google Scholar]
- 30.Hasegawa Y, Iwami J, Sato K, Park Y, Nishikawa K, Atsumi T, Moriguchi K, Murakami Y, Lamont RJ, Nakamura H, Ohno N, Yoshimura F. 2009. Anchoring and length regulation of Porphyromonas gingivalis Mfa1 fimbriae by the downstream gene product Mfa2. Microbiology 155:3333–3347. 10.1099/mic.0.028928-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuda T, Murakami Y, Noguchi T, Yoshimura F. 2006. Effects of various growth conditions in a chemostat on expression of virulence factors in Porphyromonas gingivalis. Appl. Environ. Microbiol. 72:3458–3467. 10.1128/AEM.72.5.3458-3467.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eng JK, MacCormak AL, Yates JR., III 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976–989. 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 33.Tabb DL, McDonald WH, Yates JR., III 2002. DTASelect and Contrast: tools for assembling and comparing protein identification from shotgun proteomics. J. Proteome Res. 1:21–26. 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Comer FI, Vosseller K, Wells L, Accavitti MA, Hart GW. 2001. Characterization of a mouse monoclonal antibody specific for O-linked N-acetylglucosamine. Anal. Biochem. 293:169–177. 10.1006/abio.2001.5132. [DOI] [PubMed] [Google Scholar]
- 35.Roquemore EP, Dell A, Morris HR, Panico M, Reason AJ, Savoy LA, Wistow GJ, Zigler JS, Earles BJ, Hart GW. 1992. Vertebrate lens α-crystallins are modified by O-linked N-acetylglucosamine. J. Biol. Chem. 267:555–563. [PubMed] [Google Scholar]
- 36.Farfan M, Inman KG, Nataro JP. 2008. The major pilin subunit of the AAF/II fimbriae from enteroaggregative Escherichia coli mediates binding to extracellular matrix proteins. Infect. Immun. 76:4378–4384. 10.1128/IAI.00439-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grenier D, Labbé S, Mouton C, Mayrand D. 1994. Hydrolytic enzymes and lectin-binding activity of black-pigmented anaerobic rods. Microbiology 140:873–878. 10.1099/00221287-140-4-873. [DOI] [PubMed] [Google Scholar]
- 38.Dumetz F, LaPatra SE, Duchaud E, Claverol S, Hénaff ML. 2007. The Flavobacterium psychrophilum OmpA, an outer membrane glycoprotein, induces a humoral response in rainbow trout. J. Appl. Microbiol. 103:1461–1470. 10.1111/j.1365-2672.2007.03359.x. [DOI] [PubMed] [Google Scholar]
- 39.Merle C, Faure D, Urdaci MC, Hénaff ML. 2003. Purification and characterization of a membrane glycoprotein from the fish pathogen Flavobacterium psychrophilum. J. Appl. Microbiol. 94:1120–1127. 10.1046/j.1365-2672.2003.01946.x. [DOI] [PubMed] [Google Scholar]
- 40.Slaney JM, Gallagher A, Aduse-Opoke J, Pell K, Curtis MA. 2006. Mechanisms of resistance of Porphyromonas gingivalis to killing by serum complement. Infect. Immun. 74:5352–5361. 10.1128/IAI.00304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fletcher CM, Coyne MJ, Comstock LE. 2011. Theoretical and experimental characterization of the scope of protein O-glycosylation in Bacteroides fragilis. J. Biol. Chem. 286:3219–3226. 10.1074/jbc.M110.194506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Posch G, Pabst M, Brecker L, Altmann F, Messner P, Schäffer C. 2011. Characterization and scope of S-layer protein O-glycosylation in Tannerella forsythia. J. Biol. Chem. 286:38714–38724. 10.1074/jbc.M111.284893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zachara NE, Hart GW. 2006. Cell signaling, the essential role of O-GlcNAc! Biochim. Biophys. Acta 1761:599–617. 10.1016/j.bbalip.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 44.Schirm M, Kalmokoff M, Aubry A, Thibault P, Sandoz M, Logan SM. 2004. Flagellin from Listeria monocytogenes is glycosylated with β-O-linked N-acetylglucosamine. J. Bacteriol. 186:6721–6727. 10.1128/JB.186.20.6721-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen A, Kamp HD, Gründling A, Higgins DE. 2006. A bifunctional O-GlcNAc transferase governs flagellar motility through anti-repression. Genes Dev. 20:3283–3295. 10.1101/gad.1492606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fredriksen L, Mathiesen G, Moen A, Bron PA, Kleerebezem M, Eijsink VGH, Egge-Jacobsen W. 2012. The major autolysin Acm2 from Lactobacillus plantarum undergoes cytoplasmic O-glycosylation. J. Bacteriol. 194:325–333. 10.1128/JB.06314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Torii M, Liu H, Hart GW, Hu Z-Z. 2011. dbOGAP—an integrated bioinformatics resource for protein O-GlcNAcylation. BMC Bioinformatics 12:91. 10.1186/1471-2105-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.An HJ, Froehlich JW, Lebrilla CB. 2009. Determination of glycosylation sites and site-specific heterogeneity in glycoproteins. Curr. Opin. Chem. Biol. 13:421–426. 10.1016/j.cbpa.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larsen MR, Højrup P, Roepstorff P. 2005. Characterization of gel-separated glycoproteins using two-step proteolytic digestion combined with sequential microcolumns and mass spectrometry. Mol. Cell. Proteomics 4:107–119. 10.1074/mcp.M400068-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.North SJ, Hitchen PG, Haslam SM, Dell A. 2009. Mass spectrometry in the analysis of N-linked and O-linked glycans. Curr. Opin. Struct. Biol. 19:498–506. 10.1016/j.sbi.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagata Y, Burger MM. 1974. Wheat germ agglutinin. Molecular characteristics and specificity for sugar binding. J. Biol. Chem. 249:3116–3122. [PubMed] [Google Scholar]
- 52.Peters BP, Ebisu S, Goldstein IJ, Flashner M. 1979. Interaction of wheat germ agglutinin with sialic acid. Biochemistry 18:5505–5511. 10.1021/bi00591a038. [DOI] [PubMed] [Google Scholar]
- 53.Vocadlo DJ, Hang HC, Kim E-J, Hanover JA, Bertozzi CR. 2003. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc. Natl. Acad. Sci. U. S. A. 100:9116–9121. 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kishi M, Hasagawa Y, Nagano K, Nakamura H, Murakami Y, Yoshimura F. 2012. Identification and characterization of novel glycoproteins involved in growth and biofilm formation by Porphyromonas gingivalis. Mol. Oral Microbiol. 27:458–470. 10.1111/j.2041-1014.2012.00659.x. [DOI] [PubMed] [Google Scholar]
- 55.Abe T, Murakami Y, Nagano K, Hasegawa Y, Moriguchi K, Ohno N, Shimozato K, Yoshimura F. 2011. OmpA-like protein influences cell shape and adhesive activity of Tannerella forsythia. Mol. Oral Microbiol. 26:374–387. 10.1111/j.2041-1014.2011.00625.x. [DOI] [PubMed] [Google Scholar]
- 56.Dabo SM, Confer AW, Quijano-Blas RA. 2003. Molecular and immunological characterization of Pasteurella multocida serotype A:3 OmpA: evidence of its role in P. multocida interaction with extracellular matrix. Microb. Pathog. 35:147–157. 10.1016/S0882-4010(03)00098-6. [DOI] [PubMed] [Google Scholar]
- 57.Lo RY, Sorensen LS. 2007. The outer membrane protein OmpA of Mannheimia haemolytica A1 is involved in the binding of fibronectin. FEMS Microbiol. Lett. 274:226–231. 10.1111/j.1574-6968.2007.00830.x. [DOI] [PubMed] [Google Scholar]
- 58.Oliveira R, de Morais ZM, Gonçales AP, Romero EC, Vasconcellos SA, Nasciment AL. 2011. Characterization of novel OmpA-like protein of Leptospira interrogans that binds extracellular matrix molecules and plasminogen. PLoS One 6:e21962. 10.1371/journal.pone.0021962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scott NE, Marzook NB, Deutscher A, Falconer L, Crossett B, Djordjevic SP, Cordwell SJ. 2010. Mass spectrometric characterization of the Campylobacter jejuni adherence factor CadF reveals posttranslational processing that removes immunogenicity while retaining fibronectin binding. Proteomics 10:277–288. 10.1002/pmic.200900440. [DOI] [PubMed] [Google Scholar]



