Abstract
Streptococcus pneumoniae is a leading cause of infectious disease globally. Nasopharyngeal colonization occurs in biofilms and precedes infection. Prior studies have indicated that biofilm-derived pneumococci are avirulent. However, influenza A virus (IAV) infection releases virulent pneumococci from biofilms in vitro and in vivo. Triggers of dispersal include IAV-induced changes in the nasopharynx, such as increased temperature (fever) and extracellular ATP (tissue damage). We used whole-transcriptome shotgun sequencing (RNA-seq) to compare the S. pneumoniae transcriptome in biofilms, bacteria dispersed from biofilms after exposure to IAV, febrile-range temperature, or ATP, and planktonic cells grown at 37°C. Compared with biofilm bacteria, actively dispersed S. pneumoniae, which were more virulent in invasive disease, upregulated genes involved in carbohydrate metabolism. Enzymatic assays for ATP and lactate production confirmed that dispersed pneumococci exhibited increased metabolism compared to those in biofilms. Dispersed pneumococci also upregulated genes associated with production of bacteriocins and downregulated colonization-associated genes related to competence, fratricide, and the transparent colony phenotype. IAV had the largest impact on the pneumococcal transcriptome. Similar transcriptional differences were also observed when actively dispersed bacteria were compared with avirulent planktonic bacteria. Our data demonstrate complex changes in the pneumococcal transcriptome in response to IAV-induced changes in the environment. Our data suggest that disease is caused by pneumococci that are primed to move to tissue sites with altered nutrient availability and to protect themselves from the nasopharyngeal microflora and host immune response. These data help explain pneumococcal virulence after IAV infection and have important implications for studies of S. pneumoniae pathogenesis.
INTRODUCTION
Streptococcus pneumoniae (the pneumococcus) is a Gram-positive organism that asymptomatically colonizes the upper respiratory tract. Approximately 10 to 50% of American and European children less than 3 years of age are colonized with S. pneumoniae (1–4). The prevalence of S. pneumoniae colonization is even higher in resource-poor settings, where 66% to 97% of children and over half of adults are colonized (5–8). While asymptomatic colonization is the most common outcome after acquisition of S. pneumoniae, this organism is a leading cause of acute otitis media, pneumonia, and meningitis globally (9, 10). Estimates indicate that in 2011, S. pneumoniae caused 585,000 severe episodes of pneumonia and 411,000 deaths worldwide in children less than 5 years of age (11). Researchers have thus far been unable to fully explain why some individuals colonized by S. pneumoniae progress to disease and others do not.
Epidemiologic and laboratory data indicate that infection with influenza A virus (IAV) is an important risk factor for disease due to S. pneumoniae (12–14). Infection with IAV is associated with an increase in susceptibility to pneumococcal pneumonia of approximately 100-fold for a brief period of time (12). Children under 4 years of age experienced an approximately 15-fold higher risk of pneumococcal bloodstream infections during the 2009-2010 H1N1 IAV pandemic, compared to rates during the summer months in 2006 to 2008 (15). Mechanisms of pathogenesis include IAV destruction of respiratory epithelial cells with subsequent impairment of mucociliary bacterial clearance, upregulation or exposure of receptors for pneumococcal adhesion, and IAV-induced suppression of innate and adaptive immune responses to S. pneumoniae (13, 16–18).
S. pneumoniae forms multicellular biofilms within the upper respiratory tract in humans and in animal models (19–22). Several studies have suggested that pneumococci in biofilms exhibit attenuated virulence in comparison to planktonic cells (23, 24). Weimer et al. showed that biofilm formation in the middle ear inhibits progression to invasive disease (25). In contrast, other research has shown that some genes that are important for biofilm formation are also critical for progression to disease (26). Prior research by members of our group demonstrated that IAV infection and specific environmental signals elicit active release of invasive pneumococci from colonizing biofilms (27).
S. pneumoniae transcriptional profiles change in response to environmental stimuli and differ between the upper respiratory tract and other tissue sites (28, 29). Understanding how transcriptional profiles are altered under various conditions will provide a critical step to understanding mechanisms that underlie progression from biofilm colonization to disease. Whole-transcriptome shotgun sequencing (RNA-seq) allows for deep sequencing of cDNA, identification of rare mRNA transcripts, accurate mapping of transcripts to reference genomes, and quantitative comparison of transcript levels in different samples (30, 31).
In the current study, we used RNA-seq to compare differences in the S. pneumoniae transcriptome under five conditions: (i) bacterial biofilms; (ii) bacteria actively dispersed from biofilms after exposure to IAV, to model a known risk factor for pneumococcal disease; (iii) heat to simulate fever during IAV infection (32); (iv) extracellular ATP, which is released from damaged host cells following IAV infection (33, 34); as well as (v) planktonic cells grown in laboratory medium at 37°C. Each of these five conditions represents bacterial populations that differ markedly in their ability to cause disease. Our data indicate that a complex pattern of interrelated transcriptional changes is involved in the initial stages of pneumococcal virulence.
MATERIALS AND METHODS
Ethics statement.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the University at Buffalo, Buffalo, NY. All bacterial inoculations and treatments were performed under conditions to minimize any potential suffering of the animals.
Reagents.
Cell culture reagents were from Invitrogen, Carlsbad, CA. Bacterial and cell culture media and reagents were from VWR Inc., Radnor, PA. Chemically defined bacterial growth medium (CDM) was obtained from JRH Biosciences, Lexera, KS. Sheep blood was purchased from BioLink, Inc., Liverpool, NY. All remaining reagents were purchased from Sigma-Aldrich, St. Louis, MO.
Cells, bacteria, and virus strains.
We used NCI-H292 bronchial carcinoma cells (ATCC CCL-1848) as our human respiratory epithelial cell (HREC) line for all biofilm experiments. HRECs were grown as previously described (35). The S. pneumoniae strain EF3030 was used for all experiments (36). EF3030 is an otitis media isolate that expresses 19F capsule and is sequence type (ST) 43. The H1N1 IAV strain A/PR8/34 was used for respiratory virus infection experiments, and titers were determined by plaque assays (37). Virus stocks were kindly provided by Paul Knight, University at Buffalo, Buffalo, NY.
Infection experiments.
To induce septicemia, BALB/cByJ mice (Jackson Laboratories, Bar Harbor, ME) were injected intraperitoneally with 5 × 105 to 1 × 106 CFU of pneumococci. Mice were inspected six times daily over a period of 24 h for illness and morbidity by monitoring the presence of huddling, ruffled fur, lethargy, and abdominal surface temperature. Mice found to be moribund before the end of the 24-h infection period were euthanized. Blood samples were collected and bacterial burdens were determined by plating serial dilutions on tryptic soy agar–5% blood agar plates as described previously (27). For infections of mice with IAV-dispersed pneumococci, the bacterial population was washed three times in phosphate-buffered saline (PBS) to remove free virus before injection. This group of mice was also tested for IAV viremia via a plaque assay on Madin-Darby canine kidney cells, as described previously (37).
Biofilm model, active dispersal, and planktonic growth conditions.
To model IAV-induced dispersal of pneumococci, the experiments were conducted on live epithelial cells, because live cells are required for productive IAV infection. CDM-grown pneumococci were seeded onto confluent HRECs prefixed in 4% buffered paraformaldehyde at 34°C for 48 hours, with change of medium occurring every 12 h (20). The biofilms that formed on the fixed epithelial cells were gently pipetted off and resuspended in fresh antibiotic-free RPMI 1640 medium (Invitrogen, Carlsbad, CA) with 2% fetal bovine serum, diluted 1:10, and “transplanted” onto live HREC cultures grown to confluence in 24-well plates. This procedure was used because broth-grown bacteria are toxic to epithelial cells. Once the biofilms had reorganized and matured on the live epithelial cells (biofilm maturation occurred approximately 24 h after transplantation), the epithelial biofilm cultures were inoculated with IAV at a multiplicity of infection of 1 virus/cell for 1 h. Virus was removed, and fresh medium was added. Biofilms were incubated in the presence of IAV-infected epithelial cells for 24 h, and medium was changed every 4 h. S. pneumoniae actively dispersed from the biofilm were collected during the last 4 h of the 24-h incubation, prior to noticeable cytopathic changes in the epithelial substratum (27).
Dispersal of cells with ATP and heat was performed on fixed cells to ensure that pneumococci were responding to the specific environmental conditions and not products released from eukaryotic cells. CDM-grown pneumococci were seeded onto confluent HRECs prefixed in 4% buffered paraformaldehyde, at 34°C for 48 h, with changes of medium every 12 h (20). To disperse biofilms with heat and ATP, the biofilms were washed in fresh CDM and pneumococci were dispersed by incubation in the presence of 10 μM ATP or exposure to 38.5°C for 4 h.
Planktonic pneumococci were grown in culture CDM (38) or in RPMI 1640 with 2% fetal bovine serum at 37°C until they reached the mid- to late-logarithmic phase (optical density at 600 nm [OD600] of approximately 0.5). We examined pneumococci that were planktonically grown in broth so that we could study S. pneumoniae grown under the traditional laboratory conditions. We also used S. pneumoniae grown planktonically in RPMI cell culture medium in our murine infection experiments to ensure that the medium did not produce phenotypic changes in virulence.
RNA isolation, high-throughput strand-specific RNA library preparation and quantitative reverse transcription-PCR (qRT-PCR).
With the exception of S. pneumoniae grown planktonically in RPMI, bacterial pellets from each population described above were resuspended in 0.5 ml of 0.9% NaCl, 1 ml of RNAprotect (Qiagen, Valencia, CA), and the mixture was incubated at room temperature for 5 min. Cells were then pelleted at 9,000 × g for 2 min at room temperature, and RNA was purified by using Qiashredder columns and the RNeasy minikit (38). Fifty biofilms were used to extract RNA from mature biofilms; 200 to 800 biofilms were needed to obtain enough RNA from the dispersed populations for all measurements, and three independent biological samples were collected and pooled for the experimental broth-grown population.
To analyze the relative gene expression, qRT-PCR was performed using the primers in Table S8 in the supplemental material, and the resulting data were analyzed by using the 2–ΔΔCT method (39). Gyrase A (gyrA) was chosen as the reference gene for data normalization, as it did not exhibit any significant change in expression in the RNA-seq analysis.
RNA samples were submitted for 75-bp single-end sequencing on the Illumina HiSeq apparatus at the Yale Center for Genome Analysis. The quality and concentration of RNA were determined by running total RNA on an Agilent RNA Nano bioanalyzer chip. rRNA transcripts were depleted from total RNA by using a prokaryote-specific Ribo-Zero magnetic kit (Epicentre). A strand-specific library was made using a modified version of the protocol described by Zhong et al. (40). Following selection, the RNA was fragmented in first-strand buffer to an average size of approximately 140 bp. SuperScript III reverse transcriptase was used for first-strand synthesis using random hexamers (Invitrogen, Carlsbad, CA). During second-strand synthesis with DNA polymerase I, dTTP was replaced by dUTP in the nucleotide pool. Magnetic AMPure XP beads (Beckman Coulter) were used to purify the cDNA. Following each library process step, DNA was selectively precipitated by weight and rebound to the beads through the addition of a 20% polyethylene glycol, 2.5 M NaCl solution. T4 DNA polymerase and T4 polynucleotide kinase were used to blunt end and phosphorylate the fragments. The large Klenow fragment was used to add a single adenine residue to the 3′ end of each fragment, and custom adapters (IDT) were ligated using T4 DNA ligase. Adapter-ligated DNA fragments were then treated with uracil-DNA glycosylase to remove uracil added during second-strand synthesis. The first strand was then PCR amplified using custom-made primers (IDT). During PCR, a unique 6-base identifier was inserted at the end of each DNA fragment to allow for multiplexing of samples during sequencing.
RNA-seq analyses.
The pipeline for RNA-seq analysis consisted of preprocessing raw reads using Btrim, mapping the EF3030 reads to reference genomes, and storage of mapping information in SAMtools bam format (41, 42). Full-genome sequence data are not currently available for EF3030. Reads were mapped to SPnINV200 (EMBL/GenBank accession number FQ312029), a serotype 14 ST9 strain that has three multilocus sequence type alleles in common with EF3030. We also mapped EF3030 RNA transcripts to ST556, which is a 19F middle ear isolate whose genome has recently been sequenced (accession number CP003357) (43). Alignments were done using Bowtie (44). Two mismatches were allowed. Summary statistics were calculated using an in-house program to describe the total number of reads, number of reads mapped, and reads mapped to coding sequences. Due to the slightly better coverage, data are presented for mapping to SPnINV200 as the reference genome. DESeq was used to identify differentially expressed genes (45). DESeq allows for the calculation of P values adjusted for multiple testing. Genes with an adjusted P value of <0.05 were considered significantly upregulated or downregulated. Alignments were verified and visualized using the integrative genomics viewer (46, 47). Heat maps were generated from the log2 fold changes of gene expression levels by using the heatmap.2 function in the R package gplots (48).
Enzyme assays for ATP and lactate production.
The biofilm, dispersed, and planktonic bacteria grown in CDM were washed twice in PBS by centrifugation at 8,000 × g for 2 min and resuspended in PBS to the original volume, and metabolism was induced by addition of 25 mM glucose. At various times after glucose addition, intracellular ATP concentration and lactate secretion were measured. ATP production was determined using the ATP determination kit (Invitrogen) according to the manufacturer's instructions with the exception that 0.5% Triton X-100 was added to lyse S. pneumoniae (49). Lactate produced by the bacteria was determined essentially as described previously (50).
RESULTS
Virulence of pneumococcal populations in a murine septicemia model.
In a previous study we established that specific environmental signals elicit active dispersal of invasive pneumococci from colonizing biofilms (27). To better understand the phenotypic changes associated with active dispersal, and how it relates to virulence, we tested various bacterial populations for their ability to induce septicemia in mice (Fig. 1). Broth-grown EF3030 pneumococci were used, as they are known to cause nondisseminating lobar pneumonia but are generally unable to cause septicemia in mice (51, 52). Mice were injected intraperitoneally with 5 ×105 CFU of in vitro-grown biofilm bacteria or bacteria actively released from biofilms in vitro by IAV, heat, or ATP. As IAV-released bacteria were grown in RPMI cell culture medium, we also injected mice intraperitoneally with EF3030 bacteria planktonically grown in RPMI to control for the medium conditions. Biofilm, broth-grown, and cell culture medium-grown bacteria were cleared from the blood within 24 h (Fig. 1). However, bacteria actively released from biofilms in vitro by IAV, heat, and ATP all caused bacteremia. The most virulent phenotype was observed for IAV-dispersed bacteria; all 8 mice in this experimental group presented with septicemia, indicated by ruffled fur, lethargy, and fever. Five out of the 8 mice had to be euthanized before the end of the 24-hour experimental period. None of the mice had detectable levels of IAV in the blood at the time of blood culture.
FIG 1.
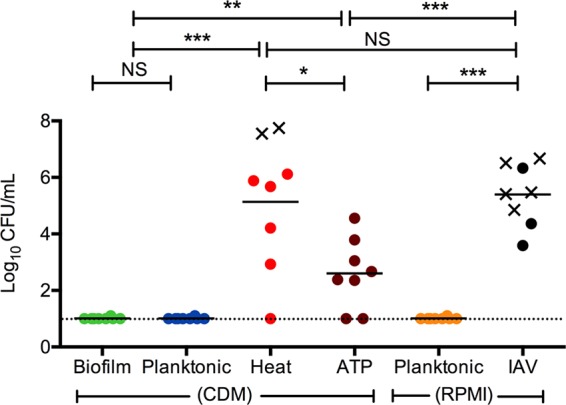
Virulence of bacterial populations after intraperitoneal inoculation. Individual 6-week-old female BALB/cByJ mice were inoculated intraperitoneally with biofilm bacteria (biofilm), broth-grown planktonic bacteria (planktonic, CDM), populations actively dispersed from biofilms grown on fixed epithelial cells in CDM after exposure to increased temperature (heat) or ATP (ATP), or with planktonic bacteria grown in cell culture medium (planktonic RPMI) or bacteria dispersed from biofilms grown on live epithelial cells after infection with influenza A virus (IAV), to determine their respective virulence once reaching the vascular compartment. Blood was collected at 24 h postinfection, or when the animals were euthanized based on being moribund. Each dot in the graphs represents an individual mouse. An X represents a mouse that became moribund and required euthanasia before the end of the experiment. The dotted line represents the limit of detection of bacterial burden. Each experiment included at least 8 mice. Statistical analysis was performed using the Student t test. NS, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Heat-dispersed pneumococci behaved similarly to IAV-dispersed bacteria and caused septicemia in all except one mouse in the experimental group with a bacterial burden that was not significantly different from the IAV-dispersed group. Three out of the 8 mice had to be euthanized before the end of the experimental period. Pneumococci dispersed with ATP also caused bacteremia; however, these mice all survived the experimental period and had significantly lower bacterial loads in the bloodstream than did those in the IAV and heat dispersal groups (P < 0.001 for both comparisons) (Fig. 1). These data demonstrated that the populations of EF3030 actively dispersed from biofilms were virulent in a septicemia model and that virulence differed depending on the dispersal agent, with IAV causing the most severe disease. The results also highlighted a dramatic difference in virulence between broth-grown bacteria, which are commonly used in experimental infection, and bacteria actively dispersed from biofilms by environmental factors associated with IAV infection.
RNA-seq read mapping and qRT-PCR verification.
Based on the observed differences in virulence, RNA-seq analysis was performed to determine the transcriptional differences between the pneumococcal populations. We obtained between 35,797,340 and 70,482,854 high-quality reads for each experimental condition (see Table S1 in the supplemental material). With the exception of a replicate sequence run of heat-dispersed bacteria, over 90% of the reads were successfully mapped to the reference genome SPnINV200. rRNA removal was less effective for a replicate heat-dispersed library, where rRNA comprised 8.5% of reads, compared to a range of 0.2% to 3.2% of reads in all other experimental samples. As a result, 58.9% of reads were mapped to SPnINV200. The data from both heat-dispersed replicates were included due to the high correlation (r = 0.895 for replicate heat experiments, compared with r = 0.993 for biofilm replicates). Moreover, inclusion of the second run would not introduce many false positives but would potentially introduce some false negatives, due to the large within-condition variance. To verify the RNA-seq results, qRT-PCR was conducted on a subset of 10 genes with increased or decreased expression for all comparisons (see Fig. S1 in the supplemental material). The qRT-PCR analysis showed a good correlation with the RNA-seq data and also clearly highlighted the differences in expression profiles between the populations.
Transcriptional differences in biofilm versus IAV-dispersed pneumococci.
IAV infection is associated with increased risk of secondary pneumococcal infection (12, 53, 54). Thus, we first compared the expression of genes in biofilm bacteria associated with colonization (20) to expression of disease-causing IAV-dispersed bacteria. Of 1,988 expressed genes, 1,159 (58%) were significantly and differentially expressed, with 604 upregulated and 554 downregulated in IAV-dispersed bacteria.
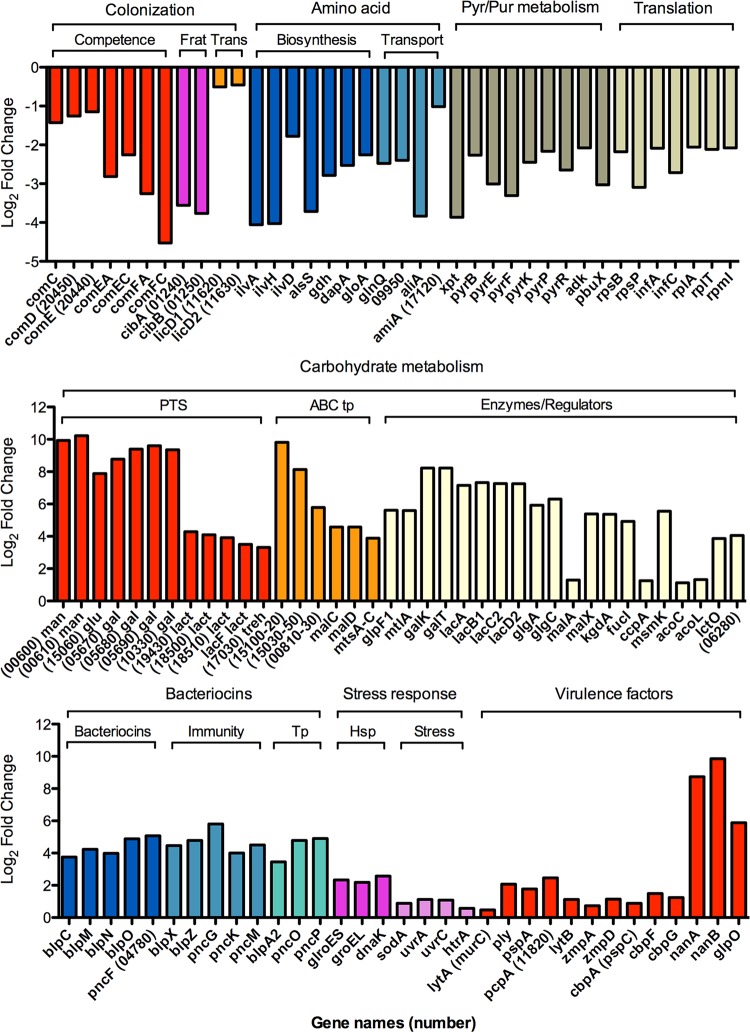
Genes highly upregulated in the IAV-dispersed bacteria were predominantly associated with carbohydrate metabolism and bacteriocin production, although several well-established virulence factors were also upregulated (Fig. 2; see also Table S2 in the supplemental material). A high level of upregulation of PTS transporters for rapid uptake of carbohydrates, such as mannose/fructose, glucose, and galactitol (range, 237- to 1,199-fold), with a slightly lower upregulation of PTS systems for lactose and trehalose (range, 10- to 20-fold) was observed. Similarly, several ABC transport systems with potential specificity for carbohydrates and metal ions, as well as transporters for uptake of glycerol (glpF1) and mannitol (mtlA), were highly upregulated. Additionally, genes involved in galactose, sucrose, maltose, pentose, and fucose metabolism, including alcohol dehydrogenases (adh), were upregulated between 30- and 300-fold (Fig. 2). Some of the changes in expression of metabolic genes may be associated with an upregulation of the catabolite control protein A (ccpA) gene, which has been shown to induce glycolysis as well as expression of the ATP-binding protein MsmK (55), which regulates the 2-oxoacid dehydrogenase complex, lactate oxidase, and malA. The IAV-dispersed cells also upregulated glpO (α-glycerophosphate oxidase), which was recently shown to act as a virulence factor during meningitis (56). Genes encoding neuraminidases A and B (nanA and nanB), enzymes involved in production of UDP-sugars and glycerolipid metabolism, were among the most highly upregulated genes observed (428- and 924-fold, respectively).
FIG 2.
Relative expression of representative genes in IAV-dispersed cells compared to biofilm bacteria. The upper graph shows log2 fold changes in expression of representative genes involved in colonization, including competence, fratricide (Frat), and transparent phenotype (Trans), amino acid biosynthesis and transport, pyrimidine and purine metabolism, and translation, including both ribosomal proteins and other translational proteins. The middle graph shows log2 fold changes in expression of genes involved in carbohydrate metabolism, including PTSs, ABC systems (ABC tp), and enzymes and regulators involved in metabolism of various carbon sources. The lower graph shows log2 fold changes in expression of genes involved in bacteriocin function, including biosynthesis, immunity, and transport (Tp), stress response proteins, including heat shock proteins (Hsp) and other stress response genes, and common virulence factors.
Genes involved in bacteriocin production were upregulated 11- to 33-fold in IAV-dispersed cells compared to biofilm bacteria. These genes included bacteriocins, their immunity proteins, and export mechanisms (Fig. 2). There was a strong stress response in the IAV-dispersed bacteria, as evidenced by an upregulation of genes encoding heat shock proteins and other proteins involved in DNA repair and response to reactive oxygen species. With regard to established virulence factors, the major autolysin (lytA) was only slightly upregulated (1.4-fold) and pneumolysin (ply) showed a higher increased expression, as did pneumococcal surface protein A (pspA), pneumococcal choline-binding protein A (pcpA), lytB, and the IgA proteases zmpA and zmpD. Interestingly, three choline-binding proteins (cbpA or pspC, cbpF, and cbpG) that have been associated with colonization (57, 58) and would be expected to be upregulated in biofilm bacteria had slightly higher expression in IAV-dispersed cells than in biofilm bacteria.
Genes that were highly downregulated in IAV-dispersed cells included those associated with competence induction, fratricide, and the transparent phenotype, which have all been associated with colonization in prior studies (23, 59–61). Furthermore, amino acid biosynthesis, amino acid and oligopeptide transport, pyrimidine and purine biosynthesis, and proteins involved in translation were significantly downregulated (4.5- to 15-fold). Representative genes for these pathways are shown in Fig. 2, and a full representation of differentially regulated genes can be found in Table S2 in the supplemental material.
Transcriptional differences in biofilm bacteria versus IAV-, heat-, and ATP-dispersed pneumococci.
We next examined transcriptional profiles of heat- and ATP-dispersed pneumococci grown on fixed epithelial cells in an attempt to identify genes whose differential expression was a consequence of environmental exposures associated with IAV infection. We used biofilm bacteria as the reference group for each of these comparisons. The most highly downregulated genes in heat- and ATP-dispersed cells exhibited 16-fold and 47-fold changes in expression, respectively. This was similar in scale to the 23-fold decrease observed for the most highly downregulated gene (comFC) in IAV-dispersed cells. However, neither heat-dispersed (7.7-fold) nor ATP-dispersed (15-fold) pneumococci showed upregulation of genes anywhere near the 1,199-fold change in expression observed for SPNINV_00610, the most highly upregulated gene in IAV-dispersed cells.
Although heat-dispersed pneumococci were almost as virulent as IAV-dispersed pneumococci in vivo (Fig. 1), fewer genes were differentially expressed in heat-dispersed cells. Only 134 out of 1,991 (7%) expressed genes were significantly differentially regulated (see Table S3 in the supplemental material). Of these, 40 genes were upregulated and 90 were downregulated. As for IAV-dispersed bacteria, heat-dispersed pneumococci showed higher expression of genes related to carbohydrate transport and utilization (although fewer changes in expression of genes encoding PTSs were observed) and bacteriocin production. Genes encoding pspA and adh were also upregulated. Similar to IAV-dispersed pneumococci, competence and fratricide genes, as well as genes involved in amino acid, purine, and pyrimidine biosynthesis and import, were downregulated in comparison to biofilm bacteria, whereas no downregulation was observed for translational proteins. In contrast to IAV-dispersed cells, heat-dispersed pneumococci did not exhibit any major changes in the expression of the virulence-associated genes pcpA, lytA, and pneumolysin. To our surprise, stress response genes were not upregulated in heat-dispersed cells. While exposure of S. pneumoniae to 38.5°C is sufficient to produce egress of S. pneumoniae from biofilms, this temperature shift may not be sufficient to induce the full range of heat shock proteins induced by temperatures in the range of 40 to 42°C that are regularly used to investigate heat shock responses (62, 63).
ATP-dispersed pneumococci were less virulent than IAV-dispersed and heat-dispersed bacteria in the septicemia model (Fig. 1). Of the 567 out of 1,990 (29%) expressed genes that were significantly differentially regulated, 291 were upregulated and 276 were downregulated (see Table S4 in the supplemental material). ATP-dispersed cells also showed a similar upregulation of PTSs and carbohydrate utilization systems, neuraminidases, and alcohol dehydrogenases as IAV-dispersed cells, but similar to heat-dispersed bacteria, no upregulation of lytA and pneumolysin was observed. Similar to the expression profile in IAV-dispersed cells, competence genes and genes involved in amino acid, purine, and pyrimidine biosynthesis and import were downregulated in ATP-dispersed cells (see Table S4). Interestingly, and in contrast to heat-dispersed pneumococci, genes involved in translation were downregulated to the same degree as in IAV-dispersed cells.
In an attempt to elucidate the pneumococcal core genes involved in the increased virulence of our dispersed cells, we identified genes that were differentially expressed under each of the three conditions (i.e., IAV, heat, and ATP) compared to biofilm bacteria. In total, 90 genes were differentially regulated (Table 1). These genes are visually represented in a heat map in Fig. S2 in the supplemental material. Seven genes were upregulated and 63 genes were downregulated in actively dispersed cells compared to biofilm bacteria. The consistently upregulated genes in each of the three dispersed populations were predominantly associated with carbohydrate metabolism. Competence genes and genes involved in amino acid, purine, and pyrimidine metabolism, as well as a few regulatory genes, sensory kinases, and hypothetical genes, were downregulated.
TABLE 1.
Genes differentially regulated under biofilm versus active dispersed conditions
| Function group and gene | Description | IAV dispersed |
Heat dispersed |
ATP dispersed |
|||
|---|---|---|---|---|---|---|---|
| Log2 fold change | P value | Log2 fold change | P value | Log2 fold change | P value | ||
| Competence/DNA transformation | |||||||
| comFC | Competence protein | −4.5 | <0.001 | −3.2 | <0.001 | −3.9 | <0.001 |
| comEC | Putative competence protein | −2.3 | 0.009 | −3.0 | <0.001 | −3.0 | 0.008 |
| SPNINV200_16350 | Type IV prepilin peptidase family protein | −2.2 | <0.001 | −1.4 | 0.006 | −2.0 | <0.001 |
| cinA | CinA-like protein | −1.2 | 0.008 | −1.3 | 0.020 | −1.6 | 0.008 |
| SPNINV200_18610 | Competence protein ComGF | −3.0 | <0.001 | −2.6 | <0.001 | −3.8 | <0.001 |
| SPNINV200_18630 | Putative competence protein | −3.0 | <0.001 | −2.6 | <0.001 | −3.9 | <0.001 |
| Comic | Putative competence protein | −2.9 | <0.001 | −2.9 | <0.001 | −4.0 | <0.001 |
| SPNINV200_18650 | Putative competence protein | −3.1 | <0.001 | −2.9 | <0.001 | −3.8 | <0.001 |
| SPNINV200_18660 | Putative competence protein | −3.3 | 0.023 | −3.0 | 0.011 | −3.7 | 0.039 |
| comFA | Late competence protein | −3.3 | <0.001 | −2.8 | <0.001 | −3.4 | <0.001 |
| comC2 | Competence-stimulating peptide (Csp-2) | −1.4 | <0.001 | −2.2 | <0.001 | −5.1 | <0.001 |
| Bacteriocins/immunity | |||||||
| SPNINV200_01250 | Class IIb bacteriocin | −3.8 | <0.001 | −3.7 | <0.001 | −5.6 | <0.001 |
| blpC | Peptide pheromone BlpC | 3.8 | <0.001 | 1.5 | 0.002 | −0.88 | 0.004 |
| blpB | ABC transporter BlpB | 3.7 | <0.001 | 1.4 | 0.003 | −0.91 | <0.001 |
| blpA2 | Bacteriocin transporter BlpA2 | 3.4 | <0.001 | 1.2 | 0.034 | −0.89 | <0.001 |
| pncG | Putative immunity protein | 5.8 | <0.001 | 2.2 | <0.001 | −0.72 | 0.045 |
| pncM | Putative immunity protein | 4.5 | <0.001 | 1.7 | <0.001 | −1.3 | 0.042 |
| blpX | Putative immunity protein BlpX | 4.5 | <0.001 | 1.7 | <0.001 | −1.4 | 0.020 |
| pncO | Bacteriocin ABC transporter transmembrane domain BlpY | 4.8 | <0.001 | 1.7 | <0.001 | −1.1 | <0.001 |
| blpZ | Putative immunity protein BlpZ | 4.8 | <0.001 | 1.6 | <0.001 | −0.94 | <0.001 |
| pncP | ABC transporter ATP-binding domain PncP | 4.9 | <0.001 | 1.5 | 0.002 | −0.99 | <0.001 |
| Amino acid synthesis and acquisition | |||||||
| aliA | Extracellular oligopeptide-binding protein | −3.8 | <0.001 | −1.3 | 0.002 | −1.1 | <0.001 |
| alsS | Acetolactate synthase large subunit | −3.7 | <0.001 | −1.5 | <0.001 | −1.6 | 0.003 |
| ilvH | Acetolactate synthase small subunit | −4.0 | <0.001 | −1.6 | <0.001 | −1.7 | 0.002 |
| ilvC | Ketol-acid reductoisomerase | −3.9 | <0.001 | −1.7 | <0.001 | −1.4 | <0.001 |
| ilvA | Threonine dehydratase | −4.1 | <0.001 | −1.7 | <0.001 | −1.4 | <0.001 |
| ilvD | Dihydroxy-acid dehydratase | −1.8 | <0.001 | −1.2 | 0.007 | −0.69 | 0.004 |
| SPNINV200_06610 | Branched-chain amino acid ABC transporter | −2.0 | <0.001 | −1.1 | 0.025 | −0.77 | 0.001 |
| SPNINV200_06620 | Branched-chain amino acid transport system permease | −2.0 | <0.001 | −1.0 | 0.039 | −0.79 | 0.037 |
| SPNINV200_07520 | Putative branched-chain-amino acid aminotransferase | −2.4 | <0.001 | −1.0 | 0.019 | −0.94 | <0.001 |
| Nucleotide metabolic processes | |||||||
| SPNINV200_02690 | Putative guanine/hypoxanthine permease | −1.7 | <0.001 | −1.8 | 0.012 | −3.6 | <0.001 |
| pyrF | Orotidine 5′-phosphate decarboxylase | −3.3 | <0.001 | −3.2 | <0.001 | −2.7 | <0.001 |
| pyrE | Orotate phosphoribosyltransferase | −3.0 | <0.001 | −2.7 | <0.001 | −2.4 | <0.001 |
| pyrK | Dihydroorotate dehydrogenase electron transfer subunit | −2.4 | <0.001 | −1.6 | 0.002 | −2.2 | <0.001 |
| pyrD2 | Dihydroorotate dehydrogenase, catalytic subunit | −2.14 | <0.001 | −1.6 | 0.003 | −1.7 | <0.001 |
| SPNINV200_09870 | GMP reductase, purine nucleotide metabolic process | −2.6 | <0.001 | −2.2 | <0.001 | −2.8 | <0.001 |
| carA | Carbamoyl-phosphate synthase small chain | −1.9 | <0.001 | −1.8 | <0.001 | −1.5 | <0.001 |
| pyrB | Aspartate carbamoyltransferase | −2.3 | <0.001 | −2.3 | <0.001 | −1.8 | <0.001 |
| pyrR | Bifunctional protein: pyrimidine operon regulatory protein, uracil phosphoribosyltransferase | −2.6 | <0.001 | −2.7 | <0.001 | −1.9 | <0.001 |
| pyrP | Uracil permease | −2.2 | <0.001 | −1.5 | 0.003 | −1.4 | <0.001 |
| xpt | Putative xanthine phosphoribosyltransferase | −3.9 | <0.001 | −2.3 | <0.001 | −3.4 | <0.001 |
| pbuX | Putative xanthine permease | −3.0 | <0.001 | −1.9 | <0.001 | −3.4 | <0.001 |
| Translation | <0.001 | <0.001 | |||||
| rpsP | 30S ribosomal protein S16 | −3.1 | <0.001 | −1.2 | 0.013 | −2.1 | <0.001 |
| SPNINV200_07580 | 30S ribosomal protein S1 | −2.2 | <0.001 | −1.0 | 0.034 | −1.8 | <0.001 |
| Carbohydrate/energy metabolism | |||||||
| SPNINV200_02290 | DeoR family, glucitol operon repressor | −1.8 | <0.001 | −1.6 | <0.001 | −1.0 | <0.001 |
| SPNINV200_02300 | Putative sugar-binding regulatory protein, Sor operon | −1.4 | <0.001 | −1.4 | 0.003 | −1.0 | <0.001 |
| fhs1 | Formate-tetrahydrofolate ligase | −1.1 | <0.001 | −1.6 | 0.003 | −2.2 | <0.001 |
| eno | Enolase | 0.60 | 0.003 | 1.4 | 0.003 | 0.8 | <0.001 |
| dexB | Glucan 1,6-α-glucosidase | 1.5 | <0.001 | 1.2 | 0.011 | 1.8 | <0.001 |
| pgk | Phosphoglycerate kinase | 1.1 | <0.001 | 1.4 | 0.003 | 1.4 | <0.001 |
| glpF1 | Glycerol uptake facilitator protein 1 | 5.6 | <0.001 | −1.2 | 0.023 | 1.4 | <0.001 |
| glpO | α-Glycerophosphate oxidase | 5.9 | <0.001 | −1.2 | 0.023 | 1.2 | <0.001 |
| glpK | Glycerol kinase | 5.4 | <0.001 | −1.4 | 0.003 | 0.90 | <0.001 |
| Additional genes | |||||||
| gdh | NADP-specific glutamate dehydrogenase | −2.8 | <0.001 | −2.0 | <0.001 | −0.89 | <0.001 |
| SPNINV200_12650 | Nicotinate phosphoribosyltransferase (NAPRTase) family protein | 0.56 | 0.010 | 1.1 | 0.015 | 1.6 | <0.001 |
| SPNINV200_13080 | ABC transporter | 3.3 | <0.001 | 1.5 | 0.002 | 1.0 | <0.001 |
| SPNINV200_14190 | DEAD box helicase family protein | −2.7 | <0.001 | −1.3 | 0.002 | −1.7 | <0.001 |
| SPNINV200_14200 | Major facilitator superfamily protein | −2.9 | <0.001 | −2.1 | <0.001 | −3.6 | <0.001 |
| mtsB | Metal cation ABC transporter ATP-binding protein | 3.7 | <0.001 | −1.3 | 0.031 | 0.70 | 0.002 |
| SPNINV200_15390 | ABC transporter, ATP-binding protein | 0.77 | <0.001 | −1.1 | 0.015 | −2.8 | <0.001 |
| SPNINV200_15760 | Putative sodium:dicarboxylate symporter family protein | −2.1 | <0.001 | −1.5 | <0.001 | −1.1 | <0.001 |
| SPNINV200_16750 | Zinc-binding dehydrogenase | 4.6 | <0.001 | 1.3 | 0.016 | 2.6 | <0.001 |
| SPNINV200_17300 | Putative single-stranded DNA-binding protein | −3.3 | <0.001 | −2.8 | <0.001 | −3.7 | <0.001 |
| SPNINV200_18310 | Putative nucleotide-binding protein | −1.5 | <0.001 | −1.1 | 0.039 | −2.0 | <0.001 |
| SPNINV200_18590 | Putative methyltransferase (pseudogene) | −3.0 | <0.001 | −2.6 | <0.001 | −3.8 | <0.001 |
| SPNINV200_19360 | Putative DNA-binding protein | −2.9 | <0.001 | −1.5 | 0.002 | −1.0 | 0.005 |
| SPNINV200_19680 | Probable alcohol dehydrogenase | 4.9 | <0.001 | 1.6 | 0.002 | 2.5 | <0.001 |
| SPNINV200_20430 | TetR family regulatory protein | −2.1 | <0.001 | −1.3 | 0.014 | −1.6 | <0.001 |
| SPNINV200_20440 | Putative response regulator protein | −1.2 | <0.001 | −2.1 | <0.001 | −5.2 | <0.001 |
| SPNINV200_20450 | Putative sensor histidine kinase | −1.3 | <0.001 | −2.4 | <0.001 | −5.1 | <0.001 |
| Unknown function and hypothetical proteins | |||||||
| SPNINV200_00180 | Hypothetical protein | −0.69 | 0.006 | −1.8 | <0.001 | −3.2 | <0.001 |
| SPNINV200_01240 | Putative membrane protein | −3.6 | <0.001 | −4.0 | <0.001 | −5.4 | <0.001 |
| SPNINV200_01590 | Putative membrane protein | −0.85 | <0.001 | −1.9 | <0.001 | −0.6 | 0.012 |
| SPNINV200_01610 | Putative integral membrane protein | −4.3 | <0.001 | −1.9 | <0.001 | −0.74 | 0.006 |
| SPNINV200_02700 | Putative CAAX amino terminal protease family membrane protein | −2.8 | <0.001 | −1.8 | 0.014 | −3.7 | <0.001 |
| SPNINV200_03320 | Conserved hypothetical protein | −0.85 | 0.011 | −1.2 | 0.045 | −1.1 | 0.021 |
| SPNINV200_03890 | Hypothetical protein | 0.58 | 0.005 | −1.8 | <0.001 | −4.4 | <0.001 |
| SPNINV200_03900 | Putative membrane protein | 0.98 | <0.001 | −1.9 | <0.001 | −4.1 | <0.001 |
| SPNINV200_03910 | Hypothetical protein | 0.96 | <0.001 | −1.8 | <0.001 | −4.1 | <0.001 |
| SPNINV200_04070 | Hypothetical protein | −3.7 | <0.001 | −1.7 | <0.001 | −1.2 | 0.005 |
| SPNINV200_04080 | Hypothetical protein | −3.1 | <0.001 | −1.7 | <0.001 | −1.1 | 0.008 |
| SPNINV200_04640 | Hypothetical protein | 6.1 | <0.001 | 1.7 | 0.042 | −0.94 | 0.013 |
| SPNINV200_06850 | Hypothetical protein | −2.7 | <0.001 | −1.4 | <0.001 | −1.6 | <0.001 |
| SPNINV200_06960 | Putative permease | 0.49 | <0.001 | −1.3 | 0.015 | −0.86 | <0.001 |
| SPNINV200_15770 | Putative membrane protein | −1.5 | <0.001 | −1.1 | 0.023 | −0.60 | 0.026 |
| SPNINV200_18600 | Putative exported protein | −2.9 | <0.001 | −2.6 | <0.001 | −3.8 | <0.001 |
| SPNINV200_18620 | Putative membrane protein | −2.9 | <0.001 | −2.6 | <0.001 | −3.6 | <0.001 |
| SPNINV200_19370 | Conserved hypothetical protein | −3.1 | <0.001 | −1.3 | 0.002 | −0.74 | 0.003 |
| SPNINV200_19380 | Hypothetical protein | −1.8 | <0.001 | −1.2 | 0.011 | −0.85 | <0.001 |
| SPNINV200_19920 | Hypothetical protein | 5.7 | <0.001 | −1.5 | 0.014 | 1.5 | <0.001 |
Twenty additional genes were significantly differentially regulated in biofilm versus IAV-, heat-, and ATP-dispersed bacteria, but the direction was not the same under each dispersal condition. Notably, 8 of these 20 genes were involved in bacteriocin production and secretion. These genes were downregulated in ATP-dispersed pneumococci and upregulated in the IAV- and heat-dispersed populations, the two populations that were most virulent in the infection model.
Transcriptional differences in broth-grown versus actively dispersed pneumococci.
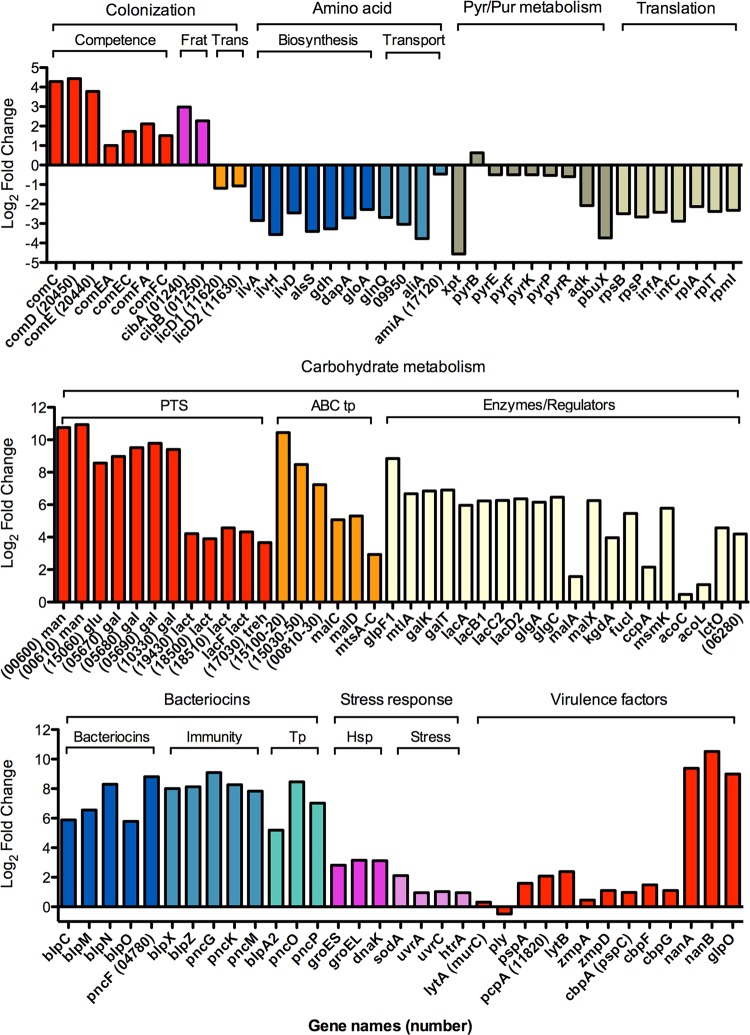
As broth-grown bacteria, like biofilm bacteria, were unable to persist in the bloodstream, we were interested in comparing the gene expression profiles between actively dispersed populations of pneumococci and broth-grown bacteria to further understand what genes may contribute to virulence. Comparison of broth-grown bacteria with IAV-dispersed pneumococci resulted in 1,186 of 1,988 (60%) expressed genes being significantly differentially regulated, with 543 genes being downregulated and 643 genes upregulated in IAV-dispersed pneumococci (see Table S5 in the supplemental material). To compare these data with the comparisons above using biofilm bacteria as a reference group (Fig. 2), the data for the same representative genes are presented in Fig. 3. Similar to the comparison with biofilms, genes associated with carbohydrate metabolism, bacteriocin production and transport, stress response genes, and well-known virulence factors were upregulated in IAV-dispersed cells compared with broth-grown bacteria. Genes encoding proteins involved in amino acid metabolism, translation, and purine and pyrimidine metabolism were downregulated in IAV-dispersed bacteria compared to broth-grown bacteria. In contrast, downregulation of components of the F(0)F(1)-H+-ATPase and ply was observed, and competence genes and fratricide genes were upregulated in dispersed cells, due to their high level of downregulation in broth-grown bacteria.
FIG 3.
Relative expression of representative genes in IAV-dispersed cells compared to broth-grown bacteria. The upper graph shows log2 fold changes in expression of representative genes involved in colonization, including competence, fratricide (Frat), and transparent phenotype (Trans), amino acid biosynthesis and transport, pyrimidine and purine metabolism, and translation, including both ribosomal proteins and other translational proteins. The middle graph shows log2 fold changes in expression of genes involved in carbohydrate metabolism, including PTSs, ABC systems (ABC tp), and enzymes and regulators involved in metabolism of various carbon sources. The lower graph shows log2 fold changes in expression of genes involved in bacteriocin function, including biosynthesis, immunity, and transport (Tp), stress response proteins, including heat shock proteins (Hsp) and other stress response genes, and common virulence factors.
Comparison of broth-grown to heat-dispersed pneumococci showed that 224 out of 1,991 (11%) expressed genes were differentially regulated, with 82 upregulated and 142 downregulated in heat-dispersed cells (see Table S6 in the supplemental material). When comparing broth-grown bacteria to ATP-dispersed pneumococci, 725 of 1,992 (38%) expressed genes were differentially regulated, with 356 downregulated and 369 upregulated in ATP-dispersed cells (see Table S7 in the supplemental material). Collectively, these comparisons indicated that all three actively dispersed populations showed a major change in metabolism, with decreased amino acid and purine and pyrimidine metabolism in favor of increased carbohydrate metabolism. Additionally, all three dispersed populations showed an increase in the virulence factor-encoding genes nanA, glpO, and lytB, with IAV-dispersed bacteria showing increased expression of additional genes, such as nanB, pcpA, pspA, prtA, htrA, and the IgA proteases zmpB and zmpD. Although all of the populations showed increased bacteriocin production, heat-dispersed (7- to 80-fold) and IAV-dispersed (16- to 544-fold) bacteria showed a much higher increase than did ATP-dispersed pneumococci (2- to 7-fold). These data are similar to the biofilm versus actively dispersed experimental comparisons.
Enzymatic assays for ATP and lactate production.
Collectively, our data indicate that all actively dispersed populations showed a major change in metabolism compared to biofilm and planktonic bacteria. To functionally confirm the RNA-seq data, we measured the ability of each bacterial population to metabolize glucose, the main nutrient source in the bloodstream. Glucose was added as the sole carbon source, and pneumococcal production of intracellular ATP and lactate secretion was measured over time (Fig. 4). Biofilm bacteria were unable to effectively produce ATP in the presence of glucose over 30 min (Fig. 4). The baseline level of ATP in the cells was approximately 10-fold lower than in all other bacterial populations (2 nM compared to 10 to 35 nM per 106 cells in the remaining populations), suggesting that biofilm bacteria have low metabolic activity. In contrast, IAV-dispersed cells rapidly produced ATP, with a peak production after 30 min of 334 nM per 106 cells (161 times that of the biofilm peak) and an initial rate of intracellular ATP production of 67 nM per minute, which was 252 times faster than for the biofilm bacteria (Fig. 4A). Similar trends were observed when measuring lactate production, which is the main end product of pneumococcal fermentation from glucose. Extracellular lactate secretion was initially 22 times higher in IAV-dispersed cells than in biofilm cells and was 21 times higher over 30 min (Fig. 4B).
FIG 4.
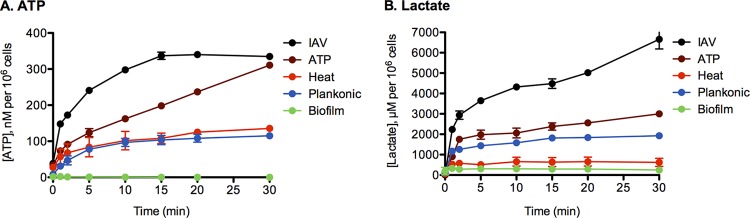
Glucose metabolism of bacterial populations. Biofilm bacteria, broth-grown planktonic bacteria (planktonic), or populations actively dispersed from biofilms after infection with IAV or exposed to increased temperature or ATP were washed in PBS to eliminate any carbon source, and 25 mM glucose was added at time zero. Intracellular ATP (A) and secretion of lactate (B) were determined 1, 2, 5, 10, 15, 20, 15, and 30 min after addition of glucose. Two separate experiments were performed in duplicate, and the mean values with standard deviations are depicted in the graphs.
Although heat-dispersed bacteria had a 65-fold increased ATP production compared with biofilm bacteria, these bacteria secreted only 2-fold higher levels of lactate. This suggests that metabolism of glucose in these cells that leads to ATP production does not occur through a major induction of glycolysis. This is consistent with the RNA-seq data, which showed no upregulation of PTSs responsible for rapid uptake and shuttling of glucose to the glycolytic pathway (e.g., SPNINV_06700) and no upregulation of ccpA in heat-treated cells (64).
ATP-dispersed cells showed a similar pattern of upregulation of PTSs and carbohydrate metabolism genes as the IAV-dispersed cells and showed a similar, although not as pronounced, increased production of ATP and lactate. Compared to biofilm bacteria, ATP-dispersed pneumococci had an initial rate of intracellular ATP production that was 72-fold higher (19.3 mM per minute) and a 149-fold-higher peak after 30 min (311 nM per 106 cells). Similarly, the initial rate of lactate production was 18-fold higher, and levels were 9-fold higher after 30 min.
Avirulent broth-grown bacteria showed ATP and lactate production levels that were higher than in biofilm bacteria. Interestingly, the ATP production of these bacteria was similar to that in heat-dispersed pneumococci but lower than in ATP- or IAV-dispersed pneumococci, and the lactate production was higher than in heat-dispersed bacteria and lower than in ATP- and IAV-dispersed bacteria. Although this supports the RNA-seq results for glucose metabolism, which was the purpose of the experiment, it also means that in vitro glucose metabolism is not the sole predictor of virulence. This is not surprising, as metabolism is intricately regulated and additional factors are likely to contribute to the observed virulence differences in Fig. 1.
DISCUSSION
We used RNA-seq and a novel biofilm model of coinfection to identify transcriptional changes during active dispersal of S. pneumoniae as it transitions from asymptomatic colonization in biofilms into an invasive pathogen. We identified changes in several gene categories, including those associated with metabolism, colonization, and disease. IAV had the largest impact on the pneumococcal transcriptome, both in terms of the number of genes differentially regulated and in the range of fold changes. These data help explain the strong epidemiological link between respiratory virus infection and pneumococcal disease and have important implications for laboratory studies of pneumococcal pathogenesis.
Moreover, these data challenge the thinking that regards pathogenic organisms as organisms expressing a specific set of virulence factors that are turned on or off when needed, and instead our data emphasize multifactorial, adaptive changes to the host environment as a way for these bacteria to survive and thrive during the course of infection. Mutation of individual genes or pathways will therefore be unlikely to produce complete answers about the nature of virulence, especially since the organisms will most certainly further change and adapt as disease progresses. However, our results provide an initial global look at the complexity of transcriptional changes associated with virulence and a starting point to further elucidate the key mechanisms involved.
Virulent S. pneumoniae upregulated genes involved in uptake and utilization of carbohydrates through glycolysis and other pathways.
S. pneumoniae relies solely on carbohydrates as a carbon source (65). S. pneumoniae contains genes encoding at least 20 carbohydrate transporters in the core genome (66) and can ferment >30 different carbohydrates (67). Genes regulating carbohydrate metabolism, PTS, and ABC transporters have been identified as important during tissue-specific disease (29, 68, 69). Carbohydrate metabolism may provide selective advantages in different host niches, as the carbon source availability in the nasopharynx will be drastically different from other body sites. As an example, glpO was upregulated 60-fold and 509-fold in IAV-dispersed cells compared to biofilm and 37°C planktonic cells, respectively. glpO encodes a surface-expressed α-glycerophosphate oxidase involved in glycerol metabolism and was recently identified as a promising vaccine antigen in a screen for S. pneumoniae genes expressed during meningitis (56). Glycerol is more abundant than glucose in the brain, whereas glucose is more abundant in the blood (70, 71). We were able to verify the RNA-seq expression data by measuring glucose metabolism of the various pneumococcal populations in vitro. Biofilm bacteria were unable to effectively metabolize glucose, whereas the dispersed populations all metabolized glucose effectively and were closely associated with expression of genes involved in glucose metabolism. While general metabolic differences were observed following the addition of glucose, there was not a direct correlation between glucose metabolism and virulence. This was not unexpected, given that the transcriptional shift in response to environmental changes is complex and virulence is multifaceted.
NanA and NanB are important for colonization, pneumonia, and sepsis (72). S. pneumoniae is viable with sialic acid as its only carbon source (73). Provision of sialic acid in a murine model of carriage led to a 10- to 1,000-fold increase in colonizing S. pneumoniae and secondary spread to the lungs (74). The 428- and 923-fold increased expression levels of nanA and nanB in S. pneumoniae cells dispersed by IAV infection was particularly interesting, given prior work by McCullers et al. on increased virulence of S. pneumoniae following IAV infection (18, 75). Their model of IAV-S. pneumoniae synergism suggests that IAV-encoded neuraminidases remove sialic acid from eukaryotic cellular surfaces, thereby exposing receptors used for S. pneumoniae adherence. Our data implicate an additional role for sialic acid in the synergism between IAV and S. pneumoniae; IAV- and S. pneumoniae-released sialic acid could also be used as a carbon source for pneumococci and a signal for increased invasiveness.
IAV- and heat-dispersed S. pneumoniae were more virulent than ATP-dispersed cells in our septicemia model. Genes related to bacteriocin production were upregulated in IAV- and heat-dispersed cells and downregulated in ATP-dispersed cells. Bacteriocins are small antimicrobial peptides that are active against closely related bacteria (76). S. pneumoniae isolates that produce bacteriocins also express immunity proteins to protect them from the effects of their own bacteriocins. The blp locus, which was upregulated in our IAV- and heat-dispersed cells, has been shown to be important for intraspecies competition during S. pneumoniae cocolonization in a murine model (77). While typically thought to be important for colonization, our data suggest that bacteriocin production may give newly dispersed cells a competitive advantage immediately after leaving the biofilm and in the initial stages of dissemination and infection. In support of our hypothesis, BlpR, the response regulator that controls bacteriocin production (77), is highly expressed in vivo in murine brain, lung, and blood (78). While a clear role for S. pneumoniae bacteriocins in invasive disease has yet to be identified, bacteriocin receptors in Gram-negative bacteria (e.g., Escherichia coli) have been shown to play a multifunctional role in the uptake of nutrients (79). Future research should examine whether S. pneumoniae bacteriocin receptors are multifunctional and elucidate the role of bacteriocins during different stages of S. pneumoniae infection.
Of the triggers studied here to disperse pneumococcal biofilms, fever is a general response to acute infections (32). ATP is generally absent in the extracellular environment in healthy tissues, and its release in the extracellular environment is likely a general response to infection with several respiratory viruses, including adenovirus and respiratory syncytial virus (80–82), that serves as a warning signal that cells are stressed. Therefore, our observation of changes in pneumococcal virulence in response to heat and ATP may have implications for respiratory tract infections due to viruses other than IAV. Our data also have important implications for experimental studies of S. pneumoniae pathogenesis. The majority of in vitro biofilm research has examined the growth of S. pneumoniae biofilms on abiotic surfaces (78, 83). Biofilms formed on abiotic surfaces differ from those formed in vivo (24). Biofilms formed on epithelial cells at 34°C have been shown to mimic the in vivo environment more accurately than those formed on abiotic surfaces (20). EF3030 is a generally noninvasive strain in mice, and exposure to stimuli such as IAV and ATP can make EF3030 invasive. Adoption of methods described in this paper could potentially expand the number of S. pneumoniae strains that can be studied in animal models and produce infection more similar to human disease. Our models could therefore be used to study a wider range of pneumococcal strains, including clinical isolates that are commonly not virulent in mice, as well as other bacterial species colonizing the nasopharyngeal tract, and mechanisms of coinfection with other respiratory viruses.
It is important to note that the transcriptional changes we observed involve initial responses of pneumococci to changes in their environment. As S. pneumoniae invades the middle ear, the lungs, or the bloodstream, its expression profiles are bound to change further. In addition, the populations of pneumococci were grown using different models (e.g., IAV infection was performed on live epithelial cells, while infections with heat- and ATP-dispersed bacteria were performed on fixed cells). While our findings should be interpreted with this caveat in mind, the various experimental conditions should not present a major limitation, because each of these experimental conditions produced populations of S. pneumoniae that differed in virulence in our murine model. Our data are meant to identify transcriptional changes that explain the observed differences in virulence. These data also highlight important areas for future research, including the role of carbohydrate metabolism in tissue-specific virulence and the role of bacteriocins in pneumococcal disease. Greater understanding of pneumococcal bacteriocins may provide additional avenues for the development of therapeutics (76). Genes that encode surface-exposed proteins and are significantly upregulated in invasive disease may represent promising vaccine targets (e.g., glpO).
Supplementary Material
ACKNOWLEDGMENTS
Funding for this research was provided by the National Institute on Deafness and Communication Disorders (R21DC011667 to M.M.P. and R01DC013554 to A.P.H.), the National Institute of General Medical Sciences (T32GM099607 to H.H.), and the Department of Microbiology and Immunology, School of Medicine and Biomedical Sciences, University at Buffalo, NY (A.P.H.).
We thank C. Tschudi for advice on RNA-seq analyses and R. Reddinger for growth of biofilms for the glucose experiments. We also acknowledge J. Overton and the Yale Center for Genome Analysis for cDNA library preparation and the RNA-sequencing.
Footnotes
Published ahead of print 18 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02225-14.
REFERENCES
- 1.Aniansson G, Alm B, Andersson B, Larsson P, Nylen O, Peterson H, Rigner P, Svanborg M, Svanborg C. 1992. Nasopharyngeal colonization during the first year of life. J. Infect. Dis. 165:S38–S42. 10.1093/infdis/165-Supplement_1-S38. [DOI] [PubMed] [Google Scholar]
- 2.Huang SS, Hinrichsen VL, Stevenson AE, Rifas-Shiman SL, Kleinman K, Pelton SI, Lipsitch M, Hanage WP, Lee GM, Finkelstein JA. 2009. Continued impact of pneumococcal conjugate vaccine on carriage in young children. Pediatrics 124:e1–e11. 10.1542/peds.2008-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Revai K, Mamidi D, Chonmaitree T. 2008. Association of nasopharyngeal bacterial colonization during upper respiratory tract infection and the development of acute otitis media. Clin. Infect. Dis. 46:e34–e37. 10.1086/525856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gray BM, Converse GM, III, Dillon HC., Jr 1980. Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J. Infect. Dis. 142:923–933. 10.1093/infdis/142.6.923. [DOI] [PubMed] [Google Scholar]
- 5.Hill PC, Akisanya A, Sankareh K, Cheung YB, Saaka M, Lahai G, Greenwood BM, Adegbola RA. 2006. Nasopharyngeal carriage of Streptococcus pneumoniae in Gambian villagers. Clin. Infect. Dis. 43:673–679. 10.1086/506941. [DOI] [PubMed] [Google Scholar]
- 6.Kwambana BA, Barer MR, Bottomley C, Adegbola RA, Antonio M. 2011. Early acquisition and high nasopharyngeal co-colonisation by Streptococcus pneumoniae and three respiratory pathogens amongst Gambian new-borns and infants. BMC Infect. Dis. 11:175. 10.1186/1471-2334-11-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coles CL, Labrique A, Saha SK, Ali H, Al-Emran H, Rashid M, Christian P, West KP, Jr, Klemm R. 2011. Newborn vitamin A supplementation does not affect nasopharyngeal carriage of Streptococcus pneumoniae in Bangladeshi infants at age 3 months. J. Nutr. 141:1907–1911. 10.3945/jn.111.141622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdullahi O, Karani A, Tigoi CC, Mugo D, Kungu S, Wanjiru E, Jomo J, Musyimi R, Lipsitch M, Scott JA. 2012. The prevalence and risk factors for pneumococcal colonization of the nasopharynx among children in Kilifi District, Kenya. PLoS One 7:e30787. 10.1371/journal.pone.0030787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T, Hib Pneumococcal Global Burden of Disease Study Team 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902. 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 10.Casey JR, Adlowitz DG, Pichichero ME. 2010. New patterns in the otopathogens causing acute otitis media six to eight years after introduction of pneumococcal conjugate vaccine. Pediatr. Infect. Dis. 29:304–309. 10.1097/INF.0b013e3181c1bc48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker CL, Rudan I, Liu L, Nair H, Theodoratou E, Bhutta ZA, O'Brien KL, Campbell H, Black RE. 2013. Global burden of childhood pneumonia and diarrhoea. Lancet 381:1405–1416. 10.1016/S0140-6736(13)60222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shrestha S, Foxman B, Weinberger DM, Steiner C, Viboud C, Rohani P. 2013. Identifying the interaction between influenza and pneumococcal pneumonia using incidence data. Sci. Transl. Med. 5:191ra184. 10.1126/scitranslmed.3005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCullers JA. 2014. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat. Rev. Microbiol. 12:252–262. 10.1038/nrmicro3231. [DOI] [PubMed] [Google Scholar]
- 14.Morens DM, Taubenberger JK, Fauci AS. 2008. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J. Infect. Dis. 198:962–970. 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tasher D, Stein M, Simoes EA, Shohat T, Bromberg M, Somekh E. 2011. Invasive bacterial infections in relation to influenza outbreaks, 2006–2010. Clin. Infect. Dis. 53:1199–1207. 10.1093/cid/cir726. [DOI] [PubMed] [Google Scholar]
- 16.Koppe U, Suttorp N, Opitz B. 2012. Recognition of Streptococcus pneumoniae by the innate immune system. Cell. Microbiol. 14:460–466. 10.1111/j.1462-5822.2011.01746.x. [DOI] [PubMed] [Google Scholar]
- 17.Sun K, Metzger DW. 2008. Inhibition of pulmonary antibacterial defense by interferon-gamma during recovery from influenza infection. Nat. Med. 14:558–564. 10.1038/nm1765. [DOI] [PubMed] [Google Scholar]
- 18.McCullers JA, Bartmess KC. 2003. Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J. Infect. Dis. 187:1000–1009. 10.1086/368163. [DOI] [PubMed] [Google Scholar]
- 19.Munoz-Elias EJ, Marcano J, Camilli A. 2008. Isolation of Streptococcus pneumoniae biofilm mutants and their characterization during nasopharyngeal colonization. Infect. Immun. 76:5049–5061. 10.1128/IAI.00425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marks LR, Parameswaran GI, Hakansson AP. 2012. Pneumococcal interactions with epithelial cells are crucial for optimal biofilm formation and colonization in vitro and in vivo. Infect. Immun. 80:2744–2760. 10.1128/IAI.00488-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoa M, Tomovic S, Nistico L, Hall-Stoodley L, Stoodley P, Sachdeva L, Berk R, Coticchia JM. 2009. Identification of adenoid biofilms with middle ear pathogens in otitis-prone children utilizing SEM and FISH. Int. J. Pediatr. Otorhinolaryngol. 73:1242–1248. 10.1016/j.ijporl.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 22.Hall-Stoodley L, Hu FZ, Gieseke A, Nistico L, Nguyen D, Hayes J, Forbes M, Greenberg DP, Dice B, Burrows A, Wackym PA, Stoodley P, Post JC, Ehrlich GD, Kerschner JE. 2006. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296:202–211. 10.1001/jama.296.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez CJ, Kumar N, Lizcano A, Shivshankar P, Dunning Hotopp JC, Jorgensen JH, Tettelin H, Orihuela CJ. 2011. Streptococcus pneumoniae in biofilms are unable to cause invasive disease due to altered virulence determinant production. PLoS One 6:e28738. 10.1371/journal.pone.0028738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanchette-Cain K, Hinojosa CA, Akula Suresh Babu R, Lizcano A, Gonzalez-Juarbe N, Munoz-Almagro C, Sanchez CJ, Bergman MA, Orihuela CJ. 2013. Streptococcus pneumoniae biofilm formation is strain dependent, multifactorial, and associated with reduced invasiveness and immunoreactivity during colonization. mBio 4(5):e00745–13. 10.1128/mBio.00745-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weimer KE, Armbruster CE, Juneau RA, Hong W, Pang B, Swords WE. 2010. Coinfection with Haemophilus influenzae promotes pneumococcal biofilm formation during experimental otitis media and impedes the progression of pneumococcal disease. J. Infect. Dis. 202:1068–1075. 10.1086/656046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trappetti C, Ogunniyi AD, Oggioni MR, Paton JC. 2011. Extracellular matrix formation enhances the ability of Streptococcus pneumoniae to cause invasive disease. PLoS One 6:e19844. 10.1371/journal.pone.0019844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marks LR, Davidson BA, Knight PR, Hakansson AP. 2013. Interkingdom signaling induces Streptococcus pneumoniae biofilm dispersion and transition from asymptomatic colonization to disease. mBio 4(4):e00438–13. 10.1128/mBio.00438-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orihuela CJ, Radin JN, Sublett JE, Gao G, Kaushal D, Tuomanen EI. 2004. Microarray analysis of pneumococcal gene expression during invasive disease. Infect. Immun. 72:5582–5596. 10.1128/IAI.72.10.5582-5596.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogunniyi AD, Mahdi LK, Trappetti C, Verhoeven N, Mermans D, Van der Hoek MB, Plumptre CD, Paton JC. 2012. Identification of genes that contribute to the pathogenesis of invasive pneumococcal disease by in vivo transcriptomic analysis. Infect. Immun. 80:3268–3278. 10.1128/IAI.00295-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Croucher NJ, Fookes MC, Perkins TT, Turner DJ, Marguerat SB, Keane T, Quail MA, He M, Assefa S, Bahler J, Kingsley RA, Parkhill J, Bentley SD, Dougan G, Thomson NR. 2009. A simple method for directional transcriptome sequencing using Illumina technology. Nucleic Acids Res. 37:e148. 10.1093/nar/gkp811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliver HF, Orsi RH, Ponnala L, Keich U, Wang W, Sun Q, Cartinhour SW, Filiatrault MJ, Wiedmann M, Boor KJ. 2009. Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics 10:641. 10.1186/1471-2164-10-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hasday JD, Fairchild KD, Shanholtz C. 2000. The role of fever in the infected host. Microbes Infect. 2:1891–1904. 10.1016/S1286-4579(00)01337-X. [DOI] [PubMed] [Google Scholar]
- 33.Grebe KM, Takeda K, Hickman HD, Bailey AL, Embry AC, Bennink JR, Yewdell JW. 2010. Cutting edge: sympathetic nervous system increases proinflammatory cytokines and exacerbates influenza A virus pathogenesis. J. Immunol. 184:540–544. 10.4049/jimmunol.0903395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lietzen N, Ohman T, Rintahaka J, Julkunen I, Aittokallio T, Matikainen S, Nyman TA. 2011. Quantitative subcellular proteome and secretome profiling of influenza A virus-infected human primary macrophages. PLoS Pathog. 7:e1001340. 10.1371/journal.ppat.1001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Schilfgaarde M, van Alphen L, Eijk P, Everts V, Dankert J. 1995. Paracytosis of Haemophilus influenzae through cell layers of NCI-H292 lung epithelial cells. Infect. Immun. 63:4729–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersson B, Dahmen J, Frejd T, Leffler H, Magnusson G, Noori G, Eden CS. 1983. Identification of an active disaccharide unit of a glycoconjugate receptor for pneumococci attaching to human pharyngeal epithelial cells. J. Exp. Med. 158:559–570. 10.1084/jem.158.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baumgarth N, Herman OC, Jager GC, Brown LE, Herzenberg LA, Chen J. 2000. B-1 and B-2 cell-derived immunoglobulin M antibodies are nonredundant components of the protective response to influenza virus infection. J. Exp. Med. 192:271–280. 10.1084/jem.192.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyx RE, Roche-Hakansson H, Hakansson AP. 2011. Role of dihydrolipoamide dehydrogenase in regulation of raffinose transport in Streptococcus pneumoniae. J. Bacteriol. 193:3512–3524. 10.1128/JB.01410-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3:1101–1108. 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 40.Zhong S, Joung J-G, Zheng Y, Chen Y-r, Liu B, Shao Y, Xiang JZ, Fei Z, Giovannoni JJ. 2011. High-throughput Illumina strand-specific RNA sequencing library preparation. Cold Spring Harbor Protoc. 2011:940–949. 10.1101/pdb.prot5652. [DOI] [PubMed] [Google Scholar]
- 41.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong Y. 2011. Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 98:152–153. 10.1016/j.ygeno.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 43.Li G, Hu FZ, Yang X, Cui Y, Yang J, Qu F, Gao GF, Zhang JR. 2012. Complete genome sequence of Streptococcus pneumoniae strain ST556, a multidrug-resistant isolate from an otitis media patient. J. Bacteriol. 194:3294–3295. 10.1128/JB.00363-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106. 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14:178–192. 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warnes GR. gplots: various R programming tools for plotting data, version 2.6.0. inside-R, Revolution Analytics, Mountain View, CA: http://www.inside-r.org/packages/plots. [Google Scholar]
- 49.Simpson WJ, Hammond JR. 1991. The effect of detergents on firefly luciferase reactions. J. Biolumin. Chemilumin. 6:97–106. 10.1002/bio.1170060207. [DOI] [PubMed] [Google Scholar]
- 50.Lundholm L, Mohme-Lundholm E, Vamos N. 1963. Lactic acid assay with L(+)lactic acid dehydrogenase from rabbit muscle. Acta Physiol. Scand. 58:243–249. 10.1111/j.1748-1716.1963.tb02645.x. [DOI] [PubMed] [Google Scholar]
- 51.Henken S, Bohling J, Ogunniyi AD, Paton JC, Salisbury VC, Welte T, Maus UA. 2010. Evaluation of biophotonic imaging to estimate bacterial burden in mice infected with highly virulent compared to less virulent Streptococcus pneumoniae serotypes. Antimicrob. Agents Chemother. 54:3155–3160. 10.1128/AAC.00310-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Briles DE, Hollingshead SK, Paton JC, Ades EW, Novak L, van Ginkel FW, Benjamin WH., Jr 2003. Immunizations with pneumococcal surface protein A and pneumolysin are protective against pneumonia in a murine model of pulmonary infection with Streptococcus pneumoniae. J. Infect. Dis. 188:339–348. 10.1086/376571. [DOI] [PubMed] [Google Scholar]
- 53.McCullers JA. 2006. Insights into the interaction between influenza virus and pneumococcus. Clin. Microbiol. Rev. 19:571–582. 10.1128/CMR.00058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weinberger DM, Simonsen L, Jordan R, Steiner C, Miller M, Viboud C. 2012. Impact of the 2009 influenza pandemic on pneumococcal pneumonia hospitalizations in the United States. J. Infect. Dis. 205:458–465. 10.1093/infdis/jir749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iyer R, Baliga NS, Camilli A. 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 187:8340–8349. 10.1128/JB.187.24.8340-8349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahdi LK, Wang H, Van der Hoek MB, Paton JC, Ogunniyi AD. 2012. Identification of a novel pneumococcal vaccine antigen preferentially expressed during meningitis in mice. J. Clin. Invest. 122:2208–2220. 10.1172/JCI45850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gosink KK, Mann ER, Guglielmo C, Tuomanen EI, Masure HR. 2000. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect. Immun. 68:5690–5695. 10.1128/IAI.68.10.5690-5695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balachandran P, Brooks-Walter A, Virolainen-Julkunen A, Hollingshead SK, Briles DE. 2002. Role of pneumococcal surface protein C in nasopharyngeal carriage and pneumonia and its ability to elicit protection against carriage of Streptococcus pneumoniae. Infect. Immun. 70:2526–2534. 10.1128/IAI.70.5.2526-2534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trappetti C, Potter AJ, Paton AW, Oggioni MR, Paton JC. 2011. LuxS mediates iron-dependent biofilm formation, competence, and fratricide in Streptococcus pneumoniae. Infect. Immun. 79:4550–4558. 10.1128/IAI.05644-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vidal JE, Ludewick HP, Kunkel RM, Zahner D, Klugman KP. 2011. The LuxS-dependent quorum-sensing system regulates early biofilm formation by Streptococcus pneumoniae strain D39. Infect. Immun. 79:4050–4060. 10.1128/IAI.05186-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim JO, Weiser JN. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J. Infect. Dis. 177:368–377. 10.1086/514205. [DOI] [PubMed] [Google Scholar]
- 62.Robertson GT, Ng WL, Foley J, Gilmour R, Winkler ME. 2002. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J. Bacteriol. 184:3508–3520. 10.1128/JB.184.13.3508-3520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee MR, Bae SM, Kim TS, Lee KJ. 2006. Proteomic analysis of protein expression in Streptococcus pneumoniae in response to temperature shift. J. Microbiol. 44:375–382. [PubMed] [Google Scholar]
- 64.Deutscher J, Aké FMD, Derkaoui M, Zébré AC, Cao TN, Bouraoui H, Kentache T, Mokhtari A, Milohanic E, Joyet P. 2014. The bacterial phosphoenolpyruvate: carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol. Mol. Biol. Rev. 78:231–256. 10.1128/MMBR.00001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buckwalter CM, King SJ. 2012. Pneumococcal carbohydrate transport: food for thought. Trends Microbiol. 20:517–522. 10.1016/j.tim.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Obert C, Sublett J, Kaushal D, Hinojosa E, Barton T, Tuomanen EI, Orihuela CJ. 2006. Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect. Immun. 74:4766–4777. 10.1128/IAI.00316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bidossi A, Mulas L, Decorosi F, Colomba L, Ricci S, Pozzi G, Deutscher J, Viti C, Oggioni MR. 2012. A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS One 7:e33320. 10.1371/journal.pone.0033320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Orihuela CJ, Gao G, Francis KP, Tuomanen EI. 2004. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J. Infect. Dis. 190:1661–1669. 10.1086/424596. [DOI] [PubMed] [Google Scholar]
- 69.Iyer R, Camilli A. 2007. Sucrose metabolism contributes to in vivo fitness of Streptococcus pneumoniae. Mol. Microbiol. 66:1–13. 10.1111/j.1365-2958.2007.05878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kiyoshima A, Kudo K, Nishida N, Ikeda N. 2002. HPLC simultaneous determination of glycerol and mannitol in human tissues for forensic analysis. Forensic Sci. Int. 125:127–133. 10.1016/S0379-0738(01)00624-7. [DOI] [PubMed] [Google Scholar]
- 71.Reinstrup P, Stahl N, Mellergard P, Uski T, Ungerstedt U, Nordstrom CH. 2000. Intracerebral microdialysis in clinical practice: baseline values for chemical markers during wakefulness, anesthesia, and neurosurgery. Neurosurgery 47:701–709. 10.1097/00006123-200009000-00035. [DOI] [PubMed] [Google Scholar]
- 72.Manco S, Hernon F, Yesilkaya H, Paton JC, Andrew PW, Kadioglu A. 2006. Pneumococcal neuraminidases A and B both have essential roles during infection of the respiratory tract and sepsis. Infect. Immun. 74:4014–4020. 10.1128/IAI.01237-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marion C, Burnaugh AM, Woodiga SA, King SJ. 2011. Sialic acid transport contributes to pneumococcal colonization. Infect. Immun. 79:1262–1269. 10.1128/IAI.00832-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trappetti C, Kadioglu A, Carter M, Hayre J, Iannelli F, Pozzi G, Andrew PW, Oggioni MR. 2009. Sialic acid: a preventable signal for pneumococcal biofilm formation, colonization, and invasion of the host. J. Infect. Dis. 199:1497–1505. 10.1086/598483. [DOI] [PubMed] [Google Scholar]
- 75.Peltola VT, McCullers JA. 2004. Respiratory viruses predisposing to bacterial infections: role of neuraminidase. Pediatr. Infect. Dis. 23:S87–97. 10.1097/01.inf.0000108197.81270.35. [DOI] [PubMed] [Google Scholar]
- 76.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins: a viable alternative to antibiotics? Nat. Rev. Microbiol. 11:95–105. 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 77.Dawid S, Roche AM, Weiser JN. 2007. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect. Immun. 75:443–451. 10.1128/IAI.01775-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oggioni MR, Trappetti C, Kadioglu A, Cassone M, Iannelli F, Ricci S, Andrew PW, Pozzi G. 2006. Switch from planktonic to sessile life: a major event in pneumococcal pathogenesis. Mol. Microbiol. 61:1196–1210. 10.1111/j.1365-2958.2006.05310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Govan JR. 1986. In vivo significance of bacteriocins and bacteriocin receptors. Scand. J. Infect. Dis. Suppl. 49:31–37. [PubMed] [Google Scholar]
- 80.Di Virgilio F. 2007. Liaisons dangereuses: P2X(7) and the inflammasome. Trends Pharmacol. Sci. 28:465–472. 10.1016/j.tips.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 81.Lee BH, Hwang DM, Palaniyar N, Grinstein S, Philpott DJ, Hu J. 2012. Activation of P2X(7) receptor by ATP plays an important role in regulating inflammatory responses during acute viral infection. PLoS One 7:e35812. 10.1371/journal.pone.0035812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Okada SF, Zhang L, Kreda SM, Abdullah LH, Davis CW, Pickles RJ, Lazarowski ER, Boucher RC. 2011. Coupled nucleotide and mucin hypersecretion from goblet-cell metaplastic human airway epithelium. Am. J. Respir. Cell Mol. Biol. 45:253–260. 10.1165/rcmb.2010-0253OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Allegrucci M, Hu FZ, Shen K, Hayes J, Ehrlich GD, Post JC, Sauer K. 2006. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J. Bacteriol. 188:2325–2335. 10.1128/JB.188.7.2325-2335.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.