Abstract
Antibiotic resistance, especially due to β-lactamases, has become one of the main obstacles in the correct treatment of Salmonella infections; furthermore, antibiotic resistance determines a gain of function that may encompass a biological cost, or fitness reduction, to the resistant bacteria. The aim of this work was to determine in vitro if the production of the class B β-lactamase VIM-2 determined a fitness cost for Salmonella enterica serovar Typhimurium. To that end the gene blaVIM-2 was cloned into the virulent strain S. Typhimurium SL1344, using both the tightly regulated pBAD22 vector and the natural plasmid pST12, for inducible and constitutive expression, respectively. Fitness studies were performed by means of motility, growth rate, invasiveness in epithelial cells, and plasmid stability. The expression of blaVIM-2 was accompanied by alterations in micro- and macroscopic morphology and reduced growth rate and motility, as well as diminished invasiveness in epithelial cells. These results suggest that VIM-2 production entails a substantial fitness cost for S. Typhimurium, which in turn may account for the extremely low number of reports of metallo-β-lactamase-producing Salmonella spp.
INTRODUCTION
The genus Salmonella includes pathogenic serovars that produce a variety of diseases in humans and animals (1). Antibiotic treatment of nontyphoidal salmonellosis is usually indicated for severe infections in children, elderly people, and immunocompromised patients; therapeutic options include folate inhibitors, fluoroquinolones, and oxyiminocephalosporins (2). The development of strains with increasing levels of resistance due to widespread use of antibiotics has been one of the main setbacks for the effective treatment of Salmonella infections (3). Most of the clinical isolates of β-lactamase-producing Salmonella synthesize class A β-lactamases (4), and the occurrence of isolates carrying plasmidic class C β-lactamases has also been reported worldwide (5, 6). Regarding class D β-lactamases, only a few variants have been reported in Salmonella serotypes (e.g., OXA-1, -2, -53, -129, and -48) (7, 8, 9). In addition, the occurrence of metallo-β-lactamase (MBL)-producing salmonellae is an infrequent event. In recent years, a few clinical cases have been reported, all of them corresponding to NDM-producing Salmonella (10, 11, 12). Interestingly, VIM alleles were not reported in Salmonella isolates until 2013 (9, 13).
Other authors have studied the biological cost for Salmonella enterica serovar Typhimurium associated with the production of class A, C, and D β-lactamases (14); nevertheless, there is no information regarding the effects of the synthesis of MBL in such microorganisms.
In this work, we hypothesized that the production of VIM-2 entailed a biological cost for Salmonella. To address this question, we analyzed the effects caused by VIM-2 production in S. Typhimurium on parameters such as motility, growth rate, and eukaryotic cell invasiveness. The class D carbapenemase OXA-66 was used as a control since it showed no deleterious effects on the fitness of S. Typhimurium.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
S. Typhimurium SL5338 (15), a strain deficient in all three restriction systems, was the mediator to transform plasmid DNA into strain S. Typhimurium SL1344 (16), used for all the in vitro experiments. The Escherichia coli strains used were DH5α (17) and MC1061 (18) and clinical isolate NF7 (19). Genes blaOXA-66 and blaVIM-2 were obtained from Acinetobacter baumannii strain 39AT (20) and Pseudomonas aeruginosa strain I42, respectively (21); the latter strain was kindly provided by Juan Ayala (Centro de Biología Molecular Severo Ochoa, Universidad Autónoma de Madrid, Spain). A detailed description of the bacterial strains used in this study is given in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| S. Typhimurium strains | ||
| SL1344 | xyl hisG rpsL Smr | 16 |
| SL5338 | galE r− m+ | 15 |
| E. coli strains | ||
| DH5α | F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG φ80dlacZΔM15 Δ(lacZYA-argF)U169 hsdR17(rK− mK+) λ− | 17 |
| MC1061 | F− Δ(ara-leu)7697 [araD139]B/r Δ(codB-lacI)3 galK16 galE15 λ− e14− mcrA0 relA1 rpsL150 (strR) spoT1 mcrB1 hsdR2(r− m+) | 18 |
| NF7 | Uropathogenic E. coli clinical isolate, receptor strain | 19 |
| P. aeruginosa strain I42 | Template strain, blaVIM-2 | 21 |
| Acinetobacter calcoaceticus-A. baumannii complex strain 39AT | Template strain, blaOXA-66 | 20 |
| Plasmids | ||
| pBAD22 | Expression vector with PBAD and AraC control (Apr) | 23 |
| pLV1 | blaVIM-2 inserted between sites EcoRI and HindIII of pBAD22 | This work |
| pLO5 | blaOXA-66 inserted between sites EcoRI and XbaI of pBAD22 | This work |
| pST12 | Environmental plasmid with blaTEM-144 under the control of P3 promoter | 25 |
| pSTVIM | blaVIM-2 inserted between sites PstI of plasmid pST12 | This work |
| pKD4 | Template plasmid, pANTSγ derivative (Apr Knr) | 24 |
| pBAD22 blaTEM-1::kan | aph(3′) inserted in site PvuI of pBAD22 (Knr) | This work |
| pLO5 blaTEM-1::kan | aph(3′) inserted in site PvuI of pLO5 | This work |
| pLV1 blaTEM-1::kan | aph(3′) inserted in site PvuI of pLV1 | This work |
Smr, streptomycin resistant; Apr, ampicillin resistant; Knr, kanamycin resistant.
All strains were grown aerobically at 37°C in Luria-Bertani (LB) broth or agar (Amresco, Solon, OH) supplemented with 100 μg/ml ampicillin (AMP) or 25 μg/ml kanamycin (KAN) (Sigma-Aldrich, St. Louis, MO) when required.
DNA manipulation.
Genetic manipulation was performed using standard laboratory techniques. Plasmids and chromosomal DNA were purified following the manufacturer's recommendations (QIAprep Spin miniprep kit and DNeasy blood and tissue kit; Qiagen Sample Assay Technologies). DNA restriction and ligation assays were carried out under standard conditions per the manufacturer's instruction (Fermentas, Invitrogen). Preparation of competent E. coli and electrocompetent S. Typhimurium cells and DNA transformation were performed as previously described (22).
In order to assess the effects of VIM-2 production on bacterial fitness, we cloned the blaVIM-2 gene into pBAD22 under the control of the arabinose-regulated PBAD promoter (23); additionally, blaOXA-66 was cloned into pBAD22 as a control, thus obtaining plasmids pLV1 and pLO5 (Table 1). Construction of pLO5 and pLV1 was performed as follows. Genes blaOXA-66 and blaVIM-2 were amplified using the primers oxaF and oxaR and vimF and vimR (see Table S1 in the supplemental material), using whole genomic DNA from A. baumannii strain 39AT and P. aeruginosa I42, respectively, as the template. PCR products of blaOXA-66 and blaVIM-2 were then doubly digested with EcoRI/XbaI and EcoRI/HindIII (Fermentas Life Sciences, Vilnius, Lithuania), respectively, and cloned into plasmid pBAD22. Insert identity was assessed by sequencing with primers pBAD5 and pBADrev (see Table S1 in the supplemental material).
Later, the blaTEM gene endogenous to pBAD22 was inactivated to avoid possible interferences between the β-lactamases in the resulting constructs, thus yielding pBAD22 blaTEM-1::kan, pLO5 blaTEM-1::kan, and pLV1 blaTEM-1::kan. These plasmids were constructed by inserting the kanamycin resistance gene aph(3′) into a PvuI recognition site in the blaTEM-1 gene present in pBAD22, pLO5, and pLV1, respectively (Table 1). Plasmid pKD4 (24) served as a template for PCR amplification of aph(3′) using primers P1pvu and P2pvu (see Table S1 in the supplemental material).
Additionally, constitutive blaVIM-2 expression was achieved by cloning said gene into a natural plasmid. Plasmid pST12 (accession number HG428760) is a ColE1-like plasmid obtained from an environmental Salmonella enterica serovar Derby isolate, featuring blaTEM-144 under the regulation of promoter P3 and three recognition sites for the restriction enzyme PstI (25). Plasmid pSTVIM is a derivative of pST12 carrying blaVIM-2 in place of blaTEM-144. Briefly, plasmid pST12 was digested with PstI, generating three fragments of 4.76 kb (containing the backbone of pST12), 2.85 kb (containing the extended-spectrum β-lactamase [ESBL] gene tnpR and part of tnpA), and 0.66 kb (containing another fragment of tnpA); the 4.76-kb fragment was recovered by gel excision and treated with alkaline phosphatase to prevent religation. Then a P3 promoter-blaVIM-2 fusion was created; P3 and blaVIM-2 were amplified with primers prmF and pvmRV and vmpFW and vmpRV, respectively (see Table S1 in the supplemental material). Primer pvmRV has a 3′ 20-mer region exhibiting perfect homology with the 5′ region of primer vmpFW. Next, an equimolar mixture of both PCR products was used as the template for PCR fusion of P3-blaVIM-2; after 2 cycles of amplification aimed at the hybridization of the promoter with the MBL-coding gene, primers prmF and vmpRV were added to the reaction mix, allowing the PCR to proceed for another 30 cycles. The P3-blaVIM-2 hybrid was treated with PstI and then ligated to the 4.7-kb fragment of pST12. Sealing of 5′ nicks was achieved by transforming pSTVIM into E. coli MC1061 prior to its final electroporation into S. Typhimurium SL1344. Insert identity was confirmed by sequencing using primers pSTf and pSTr (see Table S1 in the supplemental material).
Finally, for comparison purposes, plasmids pBAD22 blaTEM-1::kan, pLO5 blaTEM-1::kan, pLV1 blaTEM-1::kan, pST12, and pSTVIM, were also electroporated into E. coli strain NF7.
Analysis of β-lactamase production and determination of β-lactamase activity.
Crude extracts were obtained from strains SL1344(pBAD22 blaTEM-1::kan), SL1344(pLO5 blaTEM-1::kan), and SL1344(pLV1 blaTEM-1::kan), both in the presence and in the absence of 0.2% (wt/vol) l-arabinose, to analyze enzyme production and to assess β-lactamase activity of the different constructs. Briefly, overnight (ON) cultures in LB broth with kanamycin were diluted 1/100-fold in fresh preheated LB with kanamycin and incubated for another 90 min; then l-arabinose was added and induced cultures were further incubated for 2 h at 37°C with shaking (200 rpm). Strain SL1344(pBAD22) was cultivated in parallel in the presence of ampicillin. Cells were then harvested at 4,000 rpm, washed with sterile phosphate-buffered saline (PBS), and resuspended in buffer (Tris 10 mM and NaCl 50 mM [pH 7.5]), and sonicated at 0°C. Finally, the extracts were centrifuged for 30 min at 10,000 rpm at 4°C, and the supernatant was recovered.
Protein concentration was estimated by Bradford's method (26), following the manufacturer's instructions (Sigma, Saint Louis, MO), and ∼50 μg of proteins of each crude extract was analyzed by 15% SDS-PAGE with Coomassie stain. Accordingly, the crude extract of induced strain SL1344(pLV1 blaTEM-1::kan) showed an increase in intensity of an ∼28-kDa band, compatible with VIM-2 (predicted molecular weight, 28.3 kDa) and suggestive of the induced synthesis of such enzymes.
β-Lactamase activity was then assessed by the iodometric overlay method (27) using 500 μg/ml ampicillin as the substrate; while the crude extract of the induced strain SL1344(pLV1 blaTEM-1::kan) showed immediate β-lactamase activity (∼30 s), no activity was detected with the extract of the uninduced strain SL1344(pLV1 blaTEM-1::kan), indicating arabinose-regulated VIM-2 production. On the other hand, the crude extracts of control strains SL1344(pBAD22) and induced SL1344(pLO5 blaTEM-1::kan) also displayed β-lactamase activity, albeit not as readily as SL1344(pLV1 blaTEM-1::kan). The crude extract of strain SL1344(pBAD22 blaTEM-1::kan) showed no activity, with or without induction.
Antibiotic susceptibility testing.
Antibiotic susceptibility profiles of strains harboring the various constructs were determined by disk diffusion assay following the Clinical and Laboratory Standards Institute (CLSI) guidelines (28). MIC values for ceftazidime and imipenem were determined by Etest (bioMérieux, Marcy l'Etoile, France), following the manufacturer's instructions. Results were interpreted according to CLSI breakpoints (28). When necessary, Mueller-Hinton plates for antibiograms and MIC assays were supplemented with 0.05% (wt/vol) l-arabinose. Additionally, EDTA disks (2.5 μM) were also used to assess the inhibitory effects on the MBL VIM-2. S. Typhimurium strains SL1344 wild type (WT), SL1344(pBAD22), SL1344(pBAD22 blaTEM-1::kan), and SL1344(pST12) were included as controls. As expected, the VIM-2-producing strains SL1344(pLV1 blaTEM-1::kan) (inducible) and SL1344(pSTVIM) (constitutive) showed activity against oxyiminocephalosporins and carbapenems; additionally, a characteristic synergy effect between the EDTA disk and β-lactam-containing disks was observed. On the other hand, strain SL1344(pBAD blaTEM-1::kan) showed resistance only to kanamycin, whereas strain SL1344(pLO5 blaTEM-1::kan) showed resistance only to ampicillin and cephalothin (data not shown). MIC results are displayed in Table 2.
TABLE 2.
MICs to ceftazidime and imipenem for inducible and constitutive VIM-2 producing strains
| Strain | MIC (mg/liter) |
|
|---|---|---|
| Uninduced | Induced (0.05% l-arabinose) | |
| SL1344(pBAD22 blaTEM-1::kan) | ||
| Ceftazidime | 0.25 | 0.25 |
| Imipenem | 0.023 | 0.023 |
| SL1344(pLV1 blaTEM-1::kan) | ||
| Ceftazidime | 0.5 | 1 |
| Imipenem | 0.032 | 0.75 |
| SL1344(pLV1 blaTEM-1::kan) [mRNA (2ΔCT)] | 0.15 | 78.61 |
| SL1344(pST12) | 64a | 0.035b |
| SL1344(pSTVIM) | 12a | 0.125b |
MIC of ceftazidime.
MIC of imipenem.
Detection of alterations in colony morphology.
Ten microliters of bacterial suspensions of strains SL1344(pBAD22), SL1344(pLO5), and SL1344(pLV1) containing 1.5 × 104 CFU/ml (∼150 CFU) was streaked onto 5% sheep blood agar containing increasing concentrations of l-arabinose (0%, 0.05%, and 0.1%) and supplemented with 100 μg/ml ampicillin. Plates were incubated aerobically ON at 37°C. Similarly, strains SL1344(pST12) and SL1344(pSTVIM) were streaked onto 5% sheep blood agar plates without l-arabinose.
Motility tests.
Swimming motility assays were performed for strains SL1344(pBAD22 blaTEM-1::kan), SL1344(pLO5 blaTEM-1::kan), and SL1344(pLV1 blaTEM-1::kan), according to Cordeiro et al. (6), in LB plates containing 0.3% agar and increasing concentrations of l-arabinose (0%, 0.05% and 0.1%) and were incubated for 6 h at 37°C. Motility assays for strains SL1344(pST12) and SL1344(pSTVIM) were performed similarly, albeit with plating onto 0.3% LB agar without l-arabinose. The diameter of the motility halo was measured after 6 h of incubation. Assays were performed in triplicate.
Growth curves.
Growth curves for β-lactamase-producing strains were performed as previously described (6) for strains SL1344(pBAD22 blaTEM-1::kan), SL1344(pLO5 blaTEM-1::kan), and SL1344(pLV1 blaTEM-1::kan) in LB supplemented with kanamycin and with and without 0.05% (wt/vol) l-arabinose. Growth curves for strains SL1344(pST12) and SL1344(pSTVIM) were performed with LB broth supplemented with ampicillin, and no l-arabinose was added.
Additionally, growth curves for strains NF7(pBAD22 blaTEM-1::kan) and NF7(pLV1 blaTEM-1::kan) were performed in kanamycin-supplemented LB with and without arabinose, whereas those for strains NF7(pST12) and NF7(pSTVIM) were performed in ampicillin-supplemented LB broth.
In all cases, measurements of the optical density at 600 nm (OD600) were taken at regular intervals (30 min) until the stationary growth phase. Cultures were diluted every 60 min with fresh preheated LB (supplemented with kanamycin or ampicillin and arabinose) to maintain the cultures in the exponential phase for approximately 3 h. All assays were performed in triplicate.
Cell invasion assays.
Bacterial viability in cell tissue culture medium, as well as susceptibility to gentamicin, was determined for every strain prior to the invasion assays in the Caco-2 cell line. Caco-2 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and were maintained in minimal essential medium with Earle's salts (high glucose, 4.5 g/liter) (PAA, Pasching, Austria) supplemented with 2 mM l-glutamine (Sigma-Aldrich) and 15% fetal bovine serum (PAA, Pasching, Austria). Cells were grown at 37°C in 5% CO2, at up to 80% confluence. Caco-2 invasion assays were performed as previously described (6). Strains SL1344(pBAD22), SL1344(pLO5), SL1344(pLV1), SL344(pST12), and SL1344(pSTVIM) were grown accordingly until the log phase (OD600, ∼0.4), as described in the growth curves section. Each strain was tested in triplicate in three independent experiments.
Quantitative real-time PCR of blaOXA-66 and blaVIM-2.
Fresh cultures of strains SL1344(pBAD22), SL1344(pLO5), and SL1344(pLV1) in LB containing ampicillin were induced with 0.05% (wt/vol) l-arabinose during 60 min (up to an OD600 of ∼0.4 to 0.8), and total RNA extraction was performed using an RNeasy Protect bacterial kit (Qiagen) according to manufacturer recommendations. Cultures grown in the same conditions but without arabinose were grown in parallel. Strains SL1344(pSTVIM) and S. Typhimurium SL1344 WT were incubated similarly but without l-arabinose (the latter in the absence of ampicillin).
Next, 100 ng of the resulting RNA was treated with DNase (Invitrogen, Carlsbad, CA) following the manufacturer's instructions and further reverse transcribed using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Invitrogen) and random hexamer primers in a 20-μl reaction mixture. Then 2 μl of a 1/16 dilution of this mixture was used for quantitative real-time PCR (qRT-PCR) using SYBR green (QuantiTect; Qiagen) in a Corbett Rotor-Gene 6000 (Qiagen). Primer sequences are detailed in Table S1 in the supplemental material. The cycling program was 15 min at 95°C and 45 cycles of 15 s at 94°C, 30 s at 57°C, and 30 s at 72°C. The comparative threshold cycle (CT) method was used for relative mRNA quantification (29) using the blaTEM-1 gene present on pBAD22 and derivatives as an endogenous reference to avoid errors derived from plasmid copy number variations. Briefly, for strains SL1344(pLV1) and SL1344(pLO5), the increase in mRNA for blaVIM-2 and blaOXA-66, respectively, was calculated as the 2ΔΔCT or fold change [2ΔΔCT = 2ΔCT (induced)/2ΔCT (uninduced)]. For strain SL1344(pSTVIM) (constitutive expression), the ΔCT was calculated as ΔCT = CT16S − CTVIM, where CT16S is the threshold cycle obtained for the endogenous control (16S rRNA gene) and CTVIM is the threshold cycle obtained for blaVIM-2.
In this regard, we detected a 520-fold increase of blaVIM-2 mRNA for strain SL1344(pLV1) and a 310-fold increase of blaOXA-66 mRNA for strain SL1344(pLO5) when strains were grown in the presence of arabinose relative to the absence of induction. Constitutive production of VIM-2 [strain SL1344(pSTVIM)] was also measured by qRT-PCR. In the absence of an induced and uninduced condition (i.e., constitutive expression), we calculated the 2−ΔCT value instead of the more commonly used 2−ΔΔCT (fold change). cDNA from strains S. Typhimurium SL1344 WT and SL1344(pST12) were used as negative controls. In this regard, the 2−ΔCT value obtained for strain SL1344(pSTVIM) was 3.25E-4 ± 1.33E-4; this value was two orders of magnitude lower than that obtained for strain SL1344(pLV1) (data not shown). Such differences occurred because blaVIM-2 in the plasmid pSTVIM is under the control of a weak promoter (30).
Plasmid stability assay.
Plasmid stability assays were performed following a modified version of the procedure described by Medina et al. (31). Briefly, ON cultures in LB broth supplemented with kanamycin were diluted 1/200-fold in preheated LB broth; dilutions were incubated for 60 min at 37°C with shaking (200 rpm). Each culture was then fractioned in two; one half was induced with 0.05% (wt/vol) l-arabinose, whereas the other was repressed with 0.05% (wt/vol) glucose. Both sets of cultures were further incubated for another 11 h. Cultures were then diluted 1/200-fold every 12 h two times in fresh preheated LB broth with the corresponding amounts of l-arabinose or glucose. Strains SL1344(pST12) and SL1344(pSTVIM) were grown similarly, albeit without arabinose or glucose. Finally, samples were collected at an OD600 of ∼0.8 (23 generations estimated) and diluted in sterile PBS, and equal volumes (10 μl) were plated in LB agar with and without kanamycin. The proportion of plasmid-carrying bacteria was calculated as the number of CFU obtained in LB agar supplemented with kanamycin versus the CFU obtained in LB agar without antibiotic. All assays were performed in triplicate.
Statistical analysis.
Numerical results obtained during this study were analyzed with the aid of SPSS 17.0 software (SPSS, Inc., Chicago, IL). Differences in motility, growth curves, and invasiveness in Caco-2 cells were compared by one-way analysis of variance (ANOVA). The Bonferroni adjustment was used as a post hoc test. Homoscedasticity was assessed by Levene's test of homogeneity of variances, and the Welch test was used whenever data showed heteroscedasticity. For all tests, differences were considered statistically significant for P values of <0.05 (two-tailed).
RESULTS
Effects of VIM-2 production on macroscopic and cell morphology.
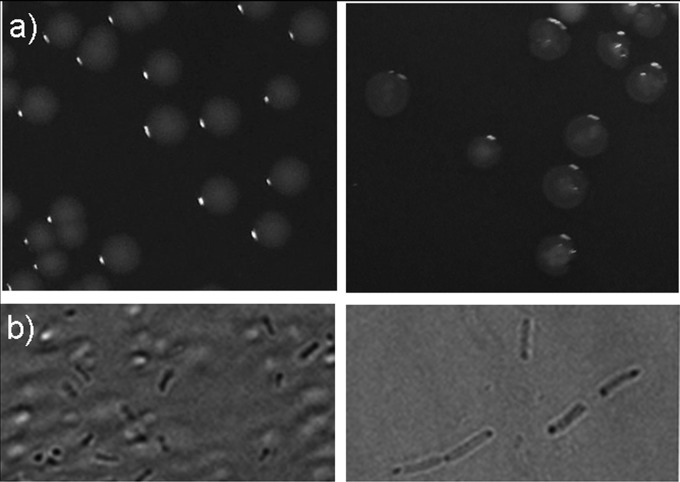
In the presence of 0.05% l-arabinose, strain SL1344(pLV1) produced transparent and flattened colonies with irregular edges (Fig. 1a, right). Likewise, wet-mount microscopy of log-phase cultures showed that cells producing VIM-2 appeared lysed and larger than the control strain and the uninduced strain, suggesting a defect in septation and/or segregation as well as an increased susceptibility to osmotic lysis (Fig. 1b, right). Constitutive production of VIM-2 [strain SL1344(pSTVIM)] was accompanied by alterations of cellular and colony morphology similar to those described for the induced strain SL1344(pLV1) (data not shown). Conversely, strains SL1344(pBAD22) and SL1344(pLO5) did not show any micro- or macroscopic alterations when grown in the presence or absence of l-arabinose (data not shown).
FIG 1.
(a) Macroscopic morphology of S. Typhimurium SL1344 WT (left) and VIM-2-producing strain SL1344(pLV1) in the presence of arabinose (right). (b) Wet-mount microscopy of log-phase cultures of S. Typhimurium SL1344 WT (left) and induced strain SL1344(pLV1) (right).
Effect of VIM-2 production on Salmonella motility.
Strain SL1344(pLV1 blaTEM-1::kan) displayed a statistically significant reduction in motility when grown in inducing versus noninducing conditions (0% l-arabinose, 44.5 ± 4.5 mm; 0.05% l-arabinose, 26.3 ± 4.1 mm), suggesting that the production of VIM-2 has a negative effect on the motility of S. Typhimurium. In opposition, strains SL1344(pBAD22 blaTEM-1::kan) and SL1344(pLO5 blaTEM-1::kan) showed similar motility values, either in the absence or the presence of arabinose, in relation to strain S. Typhimurium SL1344 WT (data not shown).
Constitutive expression of blaVIM-2 in strain SL1344(pSTVIM) was also shown to negatively affect the motility of S. Typhimurium, whereas strain SL1344(pST12) displayed behavior similar to that of strain Typhimurium SL1344 WT [SL1344(pSTVIM), 16 ± 4.1 mm; SL1344(pST12), 39 ± 7.1 mm; SL1344 WT, 39.3 ± 5.7 mm]. This suggests that the reduction in motility could be associated with the expression of blaVIM-2 and not with the plasmid per se.
Effect of VIM-2 production on Salmonella growth rate.
In vitro growth rates, measured by changes in OD600, were nearly identical for variants producing the OXA enzyme (induced or uninduced) and strain S. Typhimurium SL1344 WT as well [doubling times, SL1344 WT, 25.7 ± 0.6 min; SL1344(pBAD22 blaTEM-1::kan), 26.2 ± 1.7 min; induced SL1344(pBAD22 blaTEM-1::kan), 25.8 ± 1.2 min; SL1344(pLO5 blaTEM-1::kan), 26.4 ± 1.3 min; induced SL1344(pLO5 blaTEM-1::kan), 25.9 ± 1 min]. On the other hand, strain SL1344(pLV1 blaTEM-1::kan) displayed a lower growth rate, which decreased slightly in the presence of l-arabinose [SL1344(pLV1 blaTEM-1::kan), 28.5 ± 0.6 min; induced SL1344(pLV1 blaTEM-1::kan), 29.5 ± 0.2 min]. Interestingly, although such differences were not statistically significant, when induced, strain SL1344(pLV1 blaTEM-1::kan) reached a stationary growth phase at significantly lower OD600 values than the other strains (Fig. 2a). In the absence of induction, strain SL1344(pLV1 blaTEM-1::kan) showed a similar behavior to the rest of the strains.
FIG 2.
(a) Growth curves for S. Typhimurium transformed with the different constructs, in presence and in absence of induction. (b) Growth curves for the constitutive VIM-2-producing strain. In all cases OD600 values are adjusted according to the dilutions performed during the assays, and the mean of three independent assays is plotted.
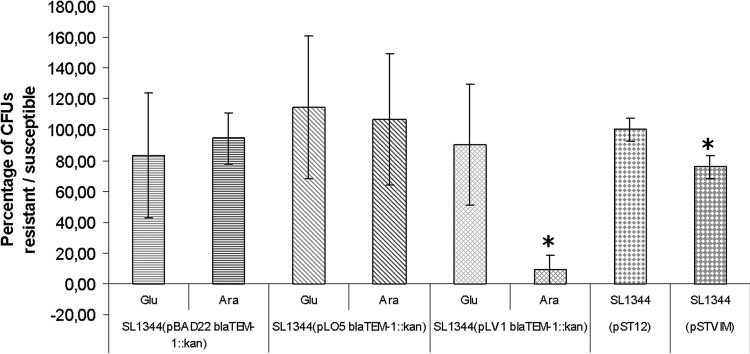
Similar to the results above, constitutive expression of blaVIM-2 encoded in plasmid pSTVIM had a biological cost for Salmonella in terms of growth rate; OD600 values were significantly lower for strain SL1344(pSTVIM) than those obtained for strains SL1344(pST12) and S. Typhimurium SL1344 WT (Fig. 2b). Interestingly, after 5 h of incubation, growth curves of strain LVR48 reached a turning point, and after 8 h of incubation the OD600 value for this strain was similar to those for SL1344(pST12) and SL1344 WT; this behavior might be partially explained by the loss of the resistance plasmid by strain SL1344(pSTVIM) (Fig. 3).
FIG 3.
Plasmid stability in the absence of antibiotic selective pressure. The percentage of plasmid-bearing bacteria (kanamycin resistance), in either the absence or presence of inductor, is plotted for each strain. Values over 100% are probably due to technique variations. Asterisks indicate a statistically significant difference in plasmid stability between the repressed and induced states and between the clone carrying pST12 and the clone carrying pSTVIM.
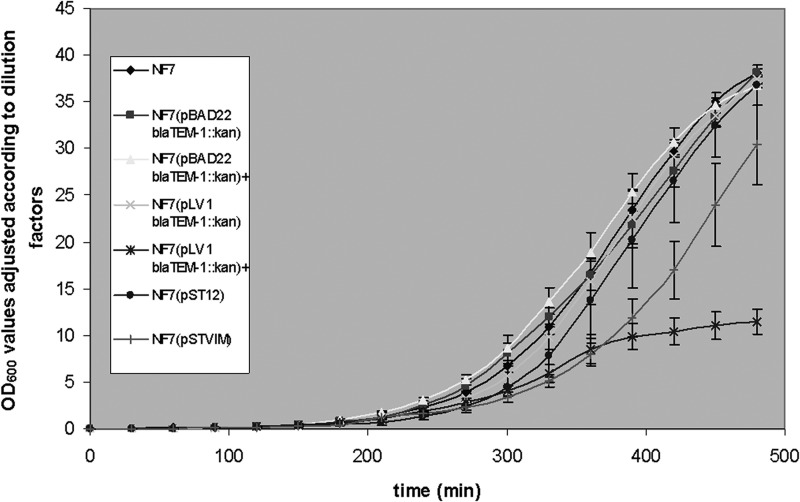
On the other hand, induced and constitutive expression by E. coli NF7 of blaVIM-2 showed results similar to those obtained with S. Typhimurium (Fig. 4).
FIG 4.
Growth curves in linear scale for E. coli strain NF7 transformed with the different constructs for inducible and constitutive VIM-2 production. Plus (+) symbols indicate growth in the presence of inductor. Please note that OD600 values are adjusted according to the dilutions performed during the assays. The means of three independent assays are plotted; vertical bars indicate standard deviations.
Effect of VIM-2 production on Salmonella invasion of epithelial cells.
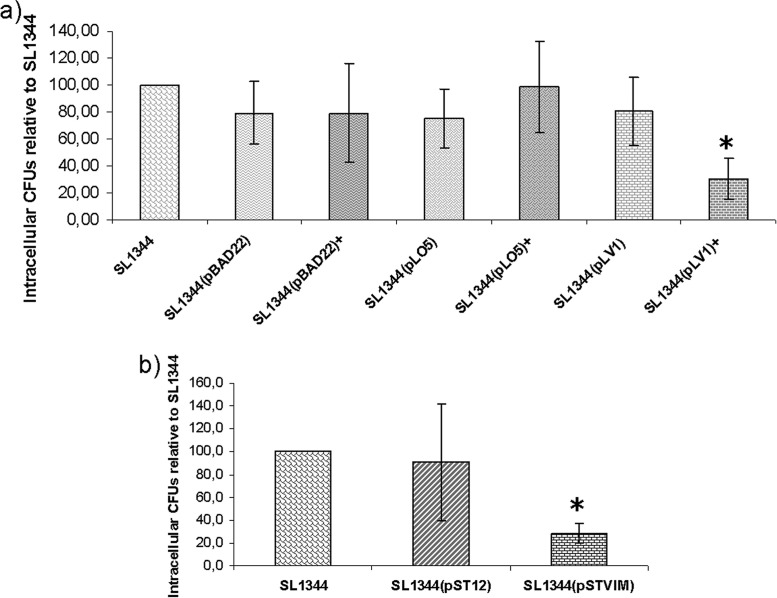
To gain some insight into the influence of blaVIM-2 expression on the natural life cycle of Salmonella, invasion assays were performed in Caco-2 cells. As shown in Fig. 5a, induced synthesis of VIM-2 produced a significant reduction in the invasiveness of strain SL1344(pLV1) compared to the invasiveness of strains carrying blaOXA-66 or the empty pBAD22 vector or S. Typhimurium SL1344 WT. On the other hand, in the absence of l-arabinose, invasion levels for strain SL1344(pLV1) were similar to those of the control strains.
FIG 5.
Invasion assays in Caco-2 cells for the inducible VIM-2-producing strain (a) and for the constitutive strain (b). Data are expressed as the means of intracellular CFU of three independent assays, relative to control strain S. Typhimurium SL1344. Vertical bars indicate standard deviations. The asterisk indicates a statistically significant difference in invasiveness between the induced or constitutive VIM-2-producing strain and the other strains.
We also determined whether constitutive expression of blaVIM-2 affected the ability of S. Typhimurium to invade epithelial cells. In this sense, strain SL1344(pSTVIM) showed a significant reduction in invasiveness, being approximately 70% less invasive than strain S. Typhimurium SL1344 WT (Fig. 5b).
Effect of VIM-2 production on plasmid stability.
To assess whether the expression of blaVIM-2 affected the stability of the plasmid encoding this β-lactamase, strains SL1344(pBAD22 blaTEM-1::kan), SL1344(pLO5 blaTEM-1::kan), SL1344(pLV1 blaTEM-1::kan), SL1344(pST12), and SL1344(pSTVIM) were grown during ca. 23 generations in the absence of antibiotic selective pressure and in the presence or absence of inductor and then plated on LB agar with and without antibiotic. Under these conditions, plasmids pBAD22 blaTEM-1::kan and pLO5 blaTEM-1::kan were stably maintained in their respective hosts in either the presence or the absence of inductor.
However, in the absence of antibiotic selective pressure, induced synthesis of VIM-2 determined the loss of plasmid pLV1 blaTEM-1::kan in ∼90% of the bacterial population, whereas constitutive production of said enzyme (albeit at low levels) resulted in the loss of plasmid pSTVIM in ∼25% of the bacterial population, clearly indicating a deleterious effect associated with the production of that MBL (Fig. 3).
DISCUSSION
Although the biological cost of expressing β-lactamases for Salmonella has already been studied (14), this work constitutes the first attempt to evaluate the possible alterations associated with MBL production in this important pathogen.
Previously, Morosini et al. showed that the introduction of a plasmidic ampC allele into S. Typhimurium resulted in a lower growth rate and less invasiveness in MDCK cells (14).
In our work, two separate strategies were followed, controlled synthesis of VIM-2 by means of an inducible expression vector and weak constitutive production of said enzyme by means of a naturally occurring plasmid obtained from an environmental S. Derby isolate.
The different assays in LB media and Caco-2 cells showed a significant fitness reduction for S. Typhimurium associated with the production of VIM-2, reflected by a lower growth rate and less motility and invasiveness than the control strains [S. Typhimurium SL1344 WT, SL1344(pBAD22), SL1344(pLO5), SL1344(pBAD22 blaTEM-1::kan), SL1344(pLO5 blaTEM-1::kan), and SL1344(pST12)], as well as alterations in cell and colony morphology. Although these deleterious effects might be related to plasmid maintenance functions, the strain carrying the empty vector (i.e., no insert) as well as that carrying plasmid pLO5 showed similar behavior to S. Typhimurium SL1344 WT. Similarly, many of the phenotypical studies in this work were carried out in the absence of β-lactams, ruling out the possibility of such antibiotics being responsible for the fitness reduction (on account of their effect on the bacterial cell wall).
Our results differ widely from those obtained by Fernández et al. (32). These authors reported that the synthesis of OXA-10-like, OXA-24, and SFO β-lactamases in E. coli is accompanied by changes in peptidoglycan composition and a fitness reduction (i.e., lower growth rate in competition assays); on the other hand, these authors found no alterations when E. coli expressed VIM-1. Besides the evaluation of different parameters, two possible explanations might account for the discrepancies between this study and the one previously mentioned: (i) differences intrinsic to the microorganisms used as the final receptor (S. Typhimurium versus E. coli) and to the expression vectors (pBAD22 versus pBGS18-pCT) and (ii) functional properties specific to the enzymes used in each work (VIM-2 versus VIM-1 and OXA-66 versus OXA-24/OXA-10-like). In this regard, the growth curves obtained for VIM-2-producing E. coli and S. Typhimurium showed similar behaviors, thus suggesting that hypothesis 1 is quite unlikely.
Several factors might be responsible for the fitness reduction for Salmonella expressing blaVIM-2. Intuitively, we could adjudicate the loss of motility and invasiveness and the diminished growth rate to saturation of the synthetic pathway due to VIM-2 overproduction. Nevertheless, we observed the same alterations when blaVIM-2 was under the control of the weak promoter P3 (pSTVIM). In addition, blaOXA-66 mRNA levels in induced strain SL1344(pLO5) were higher than blaVIM-2 levels in strain SL1344(pSTVIM); however, no alterations associated with the former were observed.
Alternatively, Marciano et al. suggested that the signal peptide of the class A carbapenemase SME-1 might be responsible for cell lysis and plasmid rearrangements in an E. coli strain, either by forming pores on the bacterial cell wall or by hijacking the protein synthesis machinery (33). Whether the signal peptide of VIM-2 is accountable for the deleterious effects observed in S. Typhimurium remains to be investigated.
We report in this work that VIM-2 production, either induced or constitutive (strong and weak promoter, respectively), reduces the ability of S. Typhimurium to invade eukaryotic cells. This may be partially explained by the occurrence of filamentous bacteria. The internalization of Salmonella in nonphagocytic cells is a highly coordinated event, mediated by flagella, fimbriae, and the SPI-1-encoded type III secretion system (34). Nonmotile mutant salmonellae have been reported to display reduced invasiveness in epithelial cells (35). In this sense, the reduction in the motility of VIM-2-producing S. Typhimurium might also account for the aforementioned loss of invasiveness.
Currently, VIM-2 is the most frequently detected transferable metallo-β-lactamase worldwide (36); nevertheless, there have been only a few reports of the occurrence of VIM enzymes in Salmonella isolates.
Furthermore, only a few reports concerning the occurrence of MBL in clinical isolates of this genus have been published; although epidemiologically unrelated, such isolates carried all blaNDM-1 (9, 10, 11), and recently, VIM-2. Consequently, the extremely low frequency of class B β-lactamase-producing salmonellae, together with the data presented in this work, support the hypothesis that the expression of such enzymes in this particular microorganism might be accompanied by an important fitness reduction. This biological cost may, in turn, negatively affect the ability of Salmonella to colonize the human gut, thus limiting the dissemination of resistant clones. In addition, plasmid stability assays suggest a strong bias toward the loss of the resistant phenotype instead of the development of compensatory mutations alleviating such biological cost; this may also account for the relative worldwide absence of MBL-producing salmonellae.
Our results may partially explain the low frequency of VIM-2-producing salmonellae; however, recent reports of Salmonella carrying blaNDM (9, 10, 11) suggest that the latter might impose a lower biological cost on its host. Further studies are required in order to confirm or rule out such a hypothesis.
Supplementary Material
ACKNOWLEDGMENT
This work was partially supported by grants from CSIC (Comisión Sectorial de Investigación Científica, Uruguay) to R.V.
Footnotes
Published ahead of print 18 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02847-14.
REFERENCES
- 1.Pegues DA, Ohl ME, Millar SI. 2005. Salmonella species, including Salmonella typhi. In Mandell GL, Bennett JE, Dolin R. (ed), Principles and practice of infectious diseases, 7th ed, vol 2 Elsevier, Philadelphia, Pa. [Google Scholar]
- 2.Guerrant RL, Van Gilder T, Steiner TS, Thielman NM, Slutsker L, Tauxe RV, Hennessy T, Griffin PM, DuPont H, Sack RB, Tarr P, Neill M, Nachamkin I, Reller LB, Osterholm MT, Bennish ML, Pickering LK. 2001. Practice guidelines for the management of infectious diarrhea. Clin. Infect. Dis. 32:331–350. 10.1086/318514. [DOI] [PubMed] [Google Scholar]
- 3.Boyle EC, Bishop JL, Grassl GA, Finlay BB. 2007. Salmonella: from pathogenesis to therapeutics. J. Bacteriol. 189:489–1495. 10.1128/JB.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arlet G, Barrett TJ, Butaye P, Cloeckaert A, Mulvey MR, White DG. 2006. Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes Infect. 8:1945–1954. 10.1016/j.micinf.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 5.Jacoby GA. 2009. AmpC β-lactamases. Clin. Microbiol. Rev. 22:161–182. 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cordeiro NF, Yim L, Betancor L, Cejas D, García-Fulgueiras V, Mota MI, Varela G, Anzalone L, Algorta G, Gutkind G, Ayala JA, Chabalgoity JA, Vignoli R. 2013. Identification of the first blacmy-2 gene in Salmonella enterica serovar Typhimurium isolates obtained from cases of paediatric diarrhoea illness detected in South America. J. Global Antimicrobial Resistance. 1:143–148. 10.1016/j.jgar.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Mulvey MR, Boyd DA, Baker L, Mykytczuk O, Reis EM, Asensi MD, Rodrigues DP, Ng LK. 2004. Characterization of a Salmonella enterica serovar Agona strain harbouring a class 1 integron containing a novel OXA-type β-lactamase (blaOXA-53) and 6′-N-aminoglycoside acetyltransferase genes [aac(6′)-I30]. J. Antimicrob. Chemother. 54:354–359. 10.1093/jac/dkh347. [DOI] [PubMed] [Google Scholar]
- 8.Michael GB, Cardoso M, Schwarz S. 2008. Molecular analysis of multiresistant porcine Salmonella enterica subsp. enterica serovar Bredeney isolates from Southern Brazil: identification of resistance genes, integrons and a group II intron. Int. J. Antimicrob. Agents 32:120–129. 10.1016/j.ijantimicag.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 9.Le Hello S, Harrois D, Bouchrif B, Sontag L, Elhani D, Guibert V, Zerouali K, Weill FX. 2013. Highly drug-resistant Salmonella enterica serotype Kentucky ST198-X1: a microbiological study. Lancet Infect. Dis. 13:672–679. 10.1016/S1473-3099(13)70124-5. [DOI] [PubMed] [Google Scholar]
- 10.Savard P, Gopinath R, Wenming Z, Kitchel B, Rasheed JK, Tekle T, Roberts A, Ross T, Razeq J, Landrum BM, Wilson LE, Limbago B, Perl TM, Carroll KC. 2011. First NDM-positive Salmonella sp. strain identified in the United States. Antimicrob. Agents Chemother. 55:5957–5958. 10.1128/AAC.05719-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabanes F, Lemant J, Picot S, Simac C, Cousty J, Jalin L, Naze F, Boisson V, Cresta MP, André H, Thibault L, Tixier F, Winer A, Antok E, Michault A. 2012. Emergence of Klebsiella pneumoniae and Salmonella metallo-β-lactamase (NDM-1) producers on Reunion Island. J. Clin. Microbiol. 50:3812–3813. 10.1128/JCM.01029-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J, Wang M, Ding H, Ye M, Hu F, Guo Q, Xu X, Wang M. 2013. New Delhi metallo-β-lactamase-1 in carbapenem-resistant Salmonella strain, China. Emerg. Infect. Dis. 19:2049–2051. 10.3201/eid1912.130051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer J, Rodríguez I, Schmoger S, Friese A, Roesler U, Helmuth R, Guerra B. 2013. Salmonella enterica subsp. enterica producing VIM-1 carbapenemase isolated from livestock farms. J. Antimicrob. Chemother. 68:478–480. 10.1093/jac/dks393. [DOI] [PubMed] [Google Scholar]
- 14.Morosini MI, Ayala JA, Baquero F, Martinez JL, Blázquez J. 2000. Biological cost of AmpC production for Salmonella enterica serotype Typhimurium. Antimicrob. Agents Chemother. 44:3137–3143. 10.1128/AAC.44.11.3137-3143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bullas LR, Ryu J. 1983. Salmonella Typhimurium LT2 strains which are r{minus] m+ for all three chromosomally located systems of DNA restriction and modification. J. Bacteriol. 156:471–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoiseth SK, Stocker BA. 1981. Aromatic-dependent Salmonella Typhimurium are non-virulent and effective as live vaccines. Nature 219:238–239. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan D. 1985. Techniques for transformation of E. coli, p. 109–135 In DM Glover. (ed.), DNA cloning: a practical approach, vol. 1 IRL Press, McLean, VA. [Google Scholar]
- 18.Casadaban MJ, Cohen SH. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179–207. 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 19.Robino L, Scavone P, Araujo L, Algorta G, Zunino P, Vignoli R. 2013. Detection of intracellular bacterial communities in a child with Escherichia coli recurrent urinary tract infections. Pathog. Dis. 68:78–81. 10.1111/2049-632X.12047. [DOI] [PubMed] [Google Scholar]
- 20.Medina JC, Seija V, Vignoli R, Pontet J, Robino L, Cordeiro NF, Bado I, García-Fulgueiras V, Berro M, Bazet C, Savio E, Rieppi G. 2013. Polyclonal endemicity of Acinetobacter baumannii in ventilated patients in an intensive care unit in Uruguay. Int. J. Infect. Dis. 17:422–427. 10.1016/j.ijid.2012.12.025. [DOI] [PubMed] [Google Scholar]
- 21.Toleman MA, Vinodh H, Sekar U, Kamat V, Walsh TR. 2007. blaVIM-2-harboring integrons isolated in India, Russia, and the United States arise from an ancestral class 1 integron predating the formation of the 3′ conserved sequence. Antimicrob. Agents Chemother. 51:2636–2638. 10.1128/AAC.01043-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 23.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vignoli R, Cordeiro NF, García V, Mota MI, Betancor L, Power P, Chabalgoity JA, Schelotto F, Gutkind G, Ayala JA. 2006. New TEM-derived extended-spectrum β-lactamase and its genomic context in plasmids from Salmonella enterica serovar Derby isolates from Uruguay. Antimicrob. Agents Chemother. 50:781–784. 10.1128/AAC.50.2.781-784.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 27.Oberhofer TR, Towle DW. 1982. Evaluation of the rapid penicillinase paper strip for detection of β-lactamase. J. Clin. Microbiol. 15:196–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing: 24th informational supplement M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 29.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Goussard S, Courvalin P. 1999. Updated sequence information for TEM β-lactamase genes. Antimicrob. Agents Chemother. 43:367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Medina C, Camacho EM, Flores A, Mesa-Pereira B, Santero E. 2011. Improved expression systems for regulated expression in Salmonella infecting eukaryotic cells. PLoS One 6:e23055. 10.1371/journal.pone.0023055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernández A, Pérez A, Ayala JA, Mallo S, Rumbo-Feal S, Tomás M, Poza M, Bou G. 2012. Expression of OXA-type and SFO-1 β-lactamases induces changes in peptidoglycan composition and affects bacterial fitness. Antimicrob. Agents Chemother. 56:1877–1884. 10.1128/AAC.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marciano DC, Karkouti OY, Palzkill T. 2007. A fitness cost associated with the antibiotic resistance enzyme SME-1 β-lactamase. Genetics 176:2381–2392. 10.1534/genetics.106.069443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Misselwitz B, Barrett N, Kreibich S, Vonaesch P, Andritschke D, Rout S, Weidner K, Sormaz M, Songhet P, Horvath P, Chabria M, Vogel V, Spori DM, Jenny P, Hardt WF. 2012. Near surface swimming of Salmonella Typhimurium explains target-site selection and cooperative invasion. PLoS Pathog. 8:e1002810. 10.1371/journal.ppat.1002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yim L, Betancor L, Martínez A, Bryant C, Maskell D, Chabalgoity JA. 2011. Naturally occurring motility-defective mutants of Salmonella enterica serovar Enteritidis isolated preferentially from nonhuman rather than human sources. Appl. Environ. Microbiol. 77:7740–7748. 10.1128/AEM.05318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borgianni L, Vandenameele J, Matagne A, Bini L, Bonomo RA, Frère JM, Rossolini GM, Docquier JD. 2010. Mutational analysis of Vim-2 reveals an essential determinant for metallo-β-lactamase stability and folding. Antimicrob. Agents Chemother. 54:3197–3204. 10.1128/AAC.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.