Abstract
Ceftaroline is the first member of a novel class of cephalosporins approved for use in the United States. Although prior studies have identified eight ceftaroline-resistant methicillin-resistant Staphylococcus aureus (MRSA) isolates in Europe and Asia with MICs ranging from 4 to 8 mg/liter, high-level resistance to ceftaroline (>32 mg/liter) has not been described in MRSA strains isolated in the United States. We isolated a ceftaroline-resistant (MIC > 32 mg/liter) MRSA strain from the blood of a cystic fibrosis patient and five MRSA strains from the respiratory tract of this patient. Whole-genome sequencing identified two amino acid-altering mutations uniquely present in the ceftaroline-binding pocket of the transpeptidase region of penicillin-binding protein 2a (PBP2a) in ceftaroline-resistant isolates. Biochemical analyses and the study of isogenic mutant strains confirmed that these changes caused ceftaroline resistance. Thus, we identified the molecular mechanism of ceftaroline resistance in the first MRSA strain with high-level ceftaroline resistance isolated in the United States.
INTRODUCTION
Ceftaroline, a new cephalosporin with activity against methicillin-resistant Staphylococcus aureus (MRSA) strains, was approved for use by the United States Food and Drug Administration (FDA) in 2010 (http://www.fda.gov/newsevents/newsroom/pressannouncements/ucm231594.htm). Its novel structure resists degradation by some beta-lactamases and allows a high affinity of binding to penicillin-binding protein 2a (PBP2a), present in MRSA strains (1–4). The FDA has established a susceptibility breakpoint of ≤1 mg/liter for S. aureus, and the Clinical and Laboratory Standards Institute (CLSI) has established breakpoints of ≤1 mg/liter for susceptible, 2 mg/liter for intermediate, and ≥4 mg/liter for resistant (5). Here the terms susceptible, intermediate, and resistant will be used as defined by the CLSI guidelines.
International surveillance studies of S. aureus initially identified only one instance of four isolates from a single hospital in Greece with ceftaroline MICs of 4 mg/liter (6–9). The Greek isolates belonged to sequence type (ST) 239 (ST239), a MRSA clonal type that is distributed worldwide but is predominantly found in Asia, Europe, and South America (10, 11). Recent work by Alm et al. identified 4 isolates with ceftaroline MICs of 8 mg/liter isolated from patients in Thailand and Spain (12). These isolates belonged to ST228, another MRSA clonal type commonly found in Europe (13). No S. aureus clinical isolate with a ceftaroline MIC greater than 8 mg/liter has been reported, and all ceftaroline-resistant clinical isolates recovered to date have been isolated outside the Western Hemisphere.
CASE REPORT
A 20-year-old male with cystic fibrosis was transferred to Houston Methodist Hospital in January 2013 to be evaluated for lung and kidney transplant. Patients with cystic fibrosis are known to develop chronic lung infections that adapt over time to this unique anatomic niche (14, 15). His complicated medical history included pancreatic insufficiency, liver transplantation in 2004, steroid-induced diabetes, end-stage renal disease, and testicular cancer. He had a long history of respiratory infections with several multidrug-resistant bacteria, including MRSA. He was treated with ceftaroline at an outside hospital immediately prior to transfer to Houston Methodist Hospital. The patient was periodically hospitalized from January to July 2013 and was treated for recurrent respiratory and catheter-related infections caused by MRSA and multidrug-resistant Pseudomonas aeruginosa. His antibiotic exposure included long treatment courses with various agents, including meropenem, ceftazidime, doxycycline, vancomycin, linezolid, cefepime, ciprofloxacin, and inhaled and systemic colistin and tobramycin. Shortly after being readmitted to our hospital in June 2013, MRSA was grown from cultures of blood and respiratory specimens. These two isolates were resistant to clindamycin, linezolid, oxacillin, and trimethoprim-sulfamethoxazole and susceptible to minocycline, rifampin, and vancomycin. His blood isolate grew confluently around the ceftaroline Etest strip, yielding an MIC of >32 mg/liter. Five additional S. aureus respiratory tract isolates were available for further study. All MRSA isolates from this patient had a small-colony-variant (SCV) phenotype.
MATERIALS AND METHODS
Bacterial isolates.
Patient isolates were grown on tryptic soy agar supplemented with 5% sheep blood. Five of the isolates grew from expectorated sputum, and the sixth isolate was obtained from an aerobic blood culture bottle. The isolates were cultured by the Diagnostic Microbiology Laboratory at Houston Methodist Hospital. The intermediate (strain TMHS-4519) and highly resistant (strains TMHS-3125, TMHS-5006, and TMHS-5007) isolates have been deposited with the American Type Culture Collection. The study protocol was approved by the Houston Methodist Research Institute Institutional Review Board (protocol IRB1010-0199).
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing was performed with the BD Phoenix (Becton Dickson, Franklin Lakes, NJ), Etest (bioMérieux, Crappone, France), or Kirby-Bauer disc diffusion methods, as indicated below.
Genome sequencing and bioinformatic analyses.
Genomic DNA for whole-genome sequencing was isolated from multiple colonies grown overnight in tryptic soy broth. The cells were lysed using matrix B in a FastPrep-96 instrument (MPBio, Santa Ana, CA), and genomic DNA was extracted using a Qiagen DNeasy blood and tissue kit (Qiagen, Venlo, Netherlands). Sequencing libraries were prepared using an Illumina NexteraXT library preparation kit (Illumina, San Diego, CA). Genomes were sequenced with an Illumina MiSeq instrument (Illumina, San Diego, CA), using the Generate FASTQ work flow. All sequence reads have been deposited with NCBI under BioProject accession number PRJNA244946. Bioinformatic analysis was performed using the Velvet algorithm (v. 1.2.10) for assembly and the VAAL software package for identification of single nucleotide polymorphisms (SNPs) (16, 17). The N315 reference genome was utilized for all comparisons (18). Velvet was run using the short read parameter and a k-mer size of 31. VAAL was run using default settings without the vecseq vector. Phylogenetic trees were constructed by first processing the VAAL K28 output files with the in-house-developed scripts Prephix (v3.1.1) and Phrecon (v4.1) to generate a multi-FASTA file for phylogenetic inference. A neighbor-joining phylogenetic tree was constructed using the SplitsTree4 (v4.13.1) program (19). SNP comparisons were performed with the snp_compare script, which is a part of the Prephix package. Analysis of the inferred effect of SNPs was achieved with the SNPeffect database. The Prephix, Phrecon, snp_compare, and SNPeffect packages are available online at https://github.com/codinghedgehog/. MLST typing was performed using the Short Read Sequence Typing for Bacterial Pathogens (SRST2) program https://github.com/katholt/srst2) and the ST database from http://saureus.mlst.net/.
Construction of PBP2a mutants.
Wild-type PBP2a lacking the N-terminal membrane-spanning region was previously cloned into pET-29a(+) from the mecA sequence (residues 23 to 668) with an N-terminal 6× His tag. The mecA E239K, Y446N, and E447K single mutants, the Y446N-E447K double mutant, and the E239K-Y446N-E447K triple mutant were created through QuikChange PCR using mutagenesis primers (see Table S1 in the supplemental material). The introduction of the mutations was verified by DNA sequencing.
In vitro PBP2a-ceftaroline binding studies.
In vitro ceftaroline binding assays were done with soluble, purified recombinant PBP2a proteins (residues 23 to 668). Bocillin, a fluorescent penicillin, was used in an in vitro competition assay (20). In short, wild-type PBP2a, the E239K, Y446N, and E447K single mutants, the Y446N-E447K double mutant, and the E239K-Y446N-E447K triple mutant, each at a final concentration of 1 μM, were incubated with various concentrations of ceftaroline for 30 min at room temperature. Bocillin was added to each reaction mixture, and samples were incubated for 30 min. To obtain a comparable fluorescence signal from each analyte, wild-type PBP2a and the E447K and Y446N-E447K proteins were incubated with 25 μM Bocillin, the E239K protein was incubated with 50 μM Bocillin, the E239K-Y446N-E447K triple mutant was incubated with 100 μM Bocillin, and the Y446N mutant was incubated with 300 μM Bocillin. Subsequently, the reaction mixtures were denatured and subjected to SDS-PAGE. The proteins were detected with a ChemiDoc XRS imager using a fluorescein emission filter (520 nm). The fluorescence intensity of each band was quantified using ImageJ software, and the 50% inhibitory concentration (IC50) was calculated using GraphPad Prism software (v5.01).
Construction of isogenic mutant strains.
Insertional inactivation of mecA was performed as previously described (21) by phage 80-mediated transduction from strain S. aureus COLΔmecA into clinical strain TMHS-5007. In these strains, the mecA gene is interrupted after 910 bp by tetM (5′ to the penicillin-binding domain). Integration of tetM into mecA in TMHS-5007 was confirmed by PCR with primers F1 (CGTGAGCAATGAACTGATTATAC) and F2 (CATAGTAACGTAGACCGTAC), located outside on the chromosomal regions upstream and downstream of the cloned fragment.
Transcomplementation of mecA was conducted with a construct encompassing the complete mecA gene from strain TMHS-5007 containing the mecA gene encoding mutant amino acids (E239K, Y446N, E447K) and the upstream region (236 bp) that includes the putative ribosomal binding site and promoter. The mecA primers used were HindIII-mecAG (GTGGTAAGCTTATGACTAACCGAAGAAGTCGTGT) and mecA-R2 EcoRI-MecA-RL (GGTGGTGAATTCGACTCGTTACAGTGTCACTTTCAAC), where the underlined sequences indicate the HindIII and EcoRI restriction endonuclease sites. The resulting 2.5-kb PCR product was digested with HindIII and EcoRI, purified, and ligated into Escherichia coli/S. aureus shuttle plasmid pCL15 (pSpac promoter IPTG [isopropyl-β-d-thiogalactopyranoside]-inducible S. aureus replicon) (22). This construct was designated pCL15-MUT mecA. The mecA gene in plasmid pCL15-MUT was confirmed to have the correct mutations by DNA sequencing. The plasmid was electroporated into strain RN4220 and subsequently transduced into mecA-null strain TMHS-5007 ΔmecA by phage 80-mediated transduction. An analogous strategy was used to clone the wild-type mecA gene from S. aureus strain N315 into pCL15 mecA. All genetic constructs were verified by restriction endonuclease analysis and DNA sequencing. Determination of the MICs of ceftaroline in all constructs was performed by Etest and broth dilution assay.
RESULTS
Characterization of strains.
The Etest indicating the blood isolate's MIC of >32 mg/liter was repeated two more times in our laboratory, with identical results being obtained. The ceftaroline MIC was confirmed by an external laboratory using a different lot of ceftaroline Etest strips. The five sputum MRSA isolates were tested retrospectively for ceftaroline resistance by the Diagnostic Microbiology Laboratory of Houston Methodist Hospital. The patient's initial sputum isolate collected shortly after transfer to Houston Methodist Hospital in January had high-level ceftaroline resistance (MIC > 32 mg/liter). In contrast, the three sputum isolates collected in March and May each had ceftaroline MIC values ranging from 1 mg/liter to 1.5 mg/liter. The isolates with MICs of 1 mg/liter were considered susceptible according to the FDA and CLSI breakpoints. The isolate with an MIC of 1.5 mg/liter was considered nonsusceptible by the FDA breakpoints but intermediate by the current CLSI breakpoints (5). The sputum isolate collected 1 day after the ceftaroline-resistant isolate was cultured from blood had an MIC of >32 mg/liter, which was confirmed by broth dilution assay. As noted previously, all six isolates had an SCV phenotype. The MICs of the other antibiotics are listed in Table 1.
TABLE 1.
Antibiotic susceptibility resultsa
| Strain | Collection date (day/mo/yr) | Source | Resistance (MIC [mg/liter]) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ceftaroline | Clindamycin | Daptomycin | Erythromycin | Levofloxacin | Linezolid | Methicillin | Minocycline | Penicillin G | Rifampin | Tetracycline | Telavancin | TMP-SMX | Vancomycin | |||
| TMHS-3125 | 1/21/2013 | Sputum | R (>32) | R (>2) | S (1) | R (>4) | NP | R (>4) | R (>2) | S (2) | R (>1) | S (≤0.5) | S (2) | NP | S (≤0.5/9.5) | S (1) |
| TMHS-3957 | 3/21/2013 | Sputum | S (1) | R (>2) | S (1) | R (>4) | NP | R (>4) | R (>2) | S (2) | R (>1) | S (≤0.5) | S (0.5) | NP | S (≤0.5/9.5) | S (1.5) |
| TMHS-4147 | 3/28/2013 | Sputum | S (1) | R (>2) | S (1) | R (>4) | NP | R (>4) | R (>2) | S (≤1) | R (>1) | S (≤0.5) | S (0.5) | NP | S (≤0.5/9.5) | S (1.5) |
| TMHS-4519 | 5/9/2013 | Sputum | I (1.5) | R (>2) | S (1) | R (>4) | NP | R (>4) | R (>2) | S (≤1) | R (>1) | S (1) | S (2) | NP | S (≤0.5/9.5) | S (1) |
| TMHS-5006 | 6/19/2013 | Blood | R (>32) | R | NS (1.5) | R | R | R | R | S | R | S | S (1.5) | 0.19 | R | S (1) |
| TMHS-5007 | 6/20/2013 | Sputum | R (>32) | R (>2) | S (1) | R (>4) | NP | R (>4) | R (>2) | S (≤1) | R (>1) | S (≤0.5) | S (1.5) | NP | S (≤0.5/9.5) | S (1) |
Results for ceftaroline, telavancin, tetracycline, vancomycin, and daptomycin were determined by Etest. All other antibiotics were tested with the BD Phoenix system, except for tests with TMHS-5006 for which MICs are not provided, for which the Kirby-Bauer disc diffusion method was used. S, susceptible; I, intermediate; R, resistant; NS, nonsusceptible; NP, not performed. CLSI breakpoints for ceftaroline were used. No susceptible, intermediate, or resistant result is listed for telavancin, as CLSI breakpoints for telavancin have not been established. TMP-SMX, trimethoprim-sulfamethoxazole.
Genome sequencing and analysis.
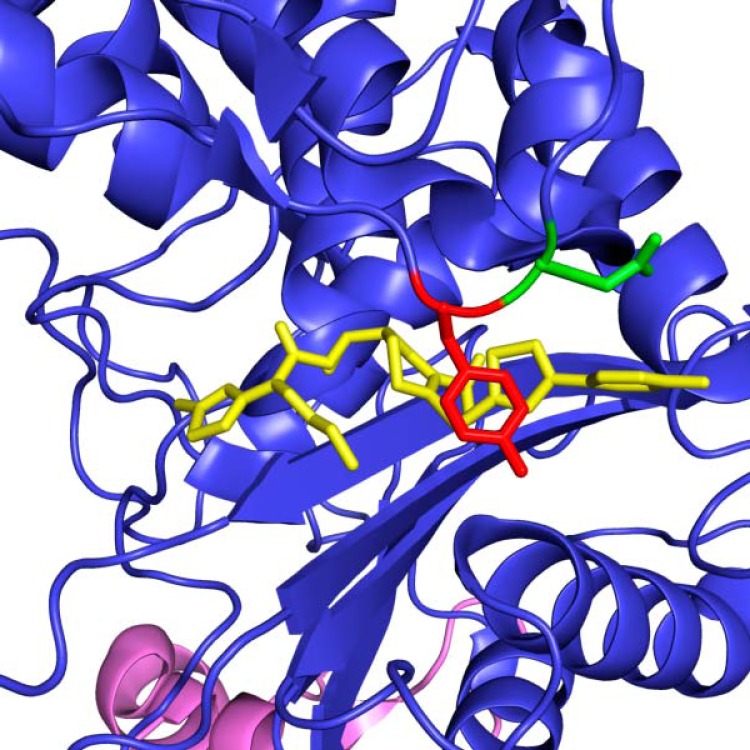
To begin to unravel the molecular mechanism of high-level ceftaroline resistance in these strains, whole-genome sequencing of the patient's six MRSA isolates was performed. A total of 4.3 million paired-end 250-bp reads were generated, which translates to an average coverage of 128-fold per strain. The strains were identified as belonging to ST5 using SRST2. SNPs were identified using VAAL, with the genome of strain N315 (NCBI accession number NC_002745) used as a reference. Compared to the reference genome, the resistant strains had a total of 508 SNPs in common. Comparison of these 508 SNPs with the SNPs in the genome of the most distantly related ceftaroline-susceptible strain (TMHS-3957) cultured from this patient identified 51 SNPs uniquely present in the resistant isolates (see Fig. S1 and Table S2 in the supplemental material). SNPs were verified by Sanger sequencing. Of the 51 SNPs uniquely present in all three ceftaroline-resistant isolates compared to the sequence of reference strain N315, 14 were synonymous SNPs (that is, they caused no amino acid change) and were excluded from further analysis. Of the remaining 37 SNPs, 9 were in intergenic regions and 28 were nonsynonymous SNPs (that is, they resulted in an amino acid replacement). Inspection of these 28 nonsynonymous SNPs identified 2 SNPs located in contiguous codons in the gene encoding PBP2a (see Table S3 in the supplemental material). These two SNPs resulted in replacement of amino acids (Y446N and E447K) located directly in the penicillin-binding pocket of the transpeptidase region of PBP2a (23). SNPs were verified by Sanger sequencing. The Y446N replacement is predicted to be a radical change (24). Importantly, very recent structural studies reported by Otero et al. (25) described the Y446 residue to be the gatekeeper to the transpeptidase active site. The E447K replacement results in a change from a negatively charged glutamic acid side chain to a positively charged lysine. On the basis of the crystal structure of PBP2a complexed with ceftaroline, these two amino acid replacements occur at the corner of the active site where the positively charged pyridine ring of ceftaroline binds to PBP2a (Fig. 1). It is important to note that amino acid changes at the Y446 and E447 residues have been implicated in resistance to ceftobiprole (a related anti-MRSA cephalosporin) derived after serial passaging in vitro (26). A third amino acid replacement (E239K) was also identified in isolate TMHS-5007 (Fig. 2). This change maps very close to the allosteric site (25).
FIG 1.
Active site of acyl-PBP2a from MRSA with bound ceftaroline and the two contiguous amino acids that are altered (Y446N and E447K) in the resistant organisms. This view of the PBP2a transpeptidase active site shows the Y446 (red) and E447 (green) amino acids highlighted in front of a bound molecule of ceftaroline (yellow). The transpeptidase portion of the molecule is colored blue, and the allosteric portion of PBP2a is colored violet. The structure has PDB accession number 3ZG0 and was published by Otero et al. (25).
FIG 2.
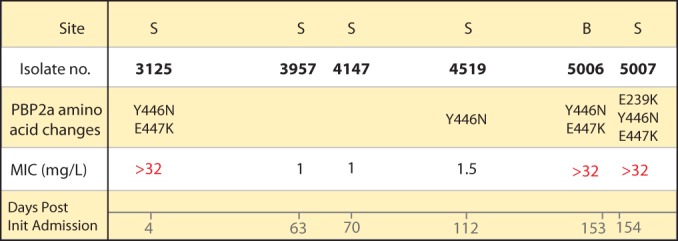
Natural history of six MRSA clinical isolates collected over 154 days with their PBP2a active-site amino acid mutations and ceftaroline MICs. S, sputum samples; B, blood isolate. The highly resistant isolates have MICs of >32 mg/liter, which are marked in red. Init, initial.
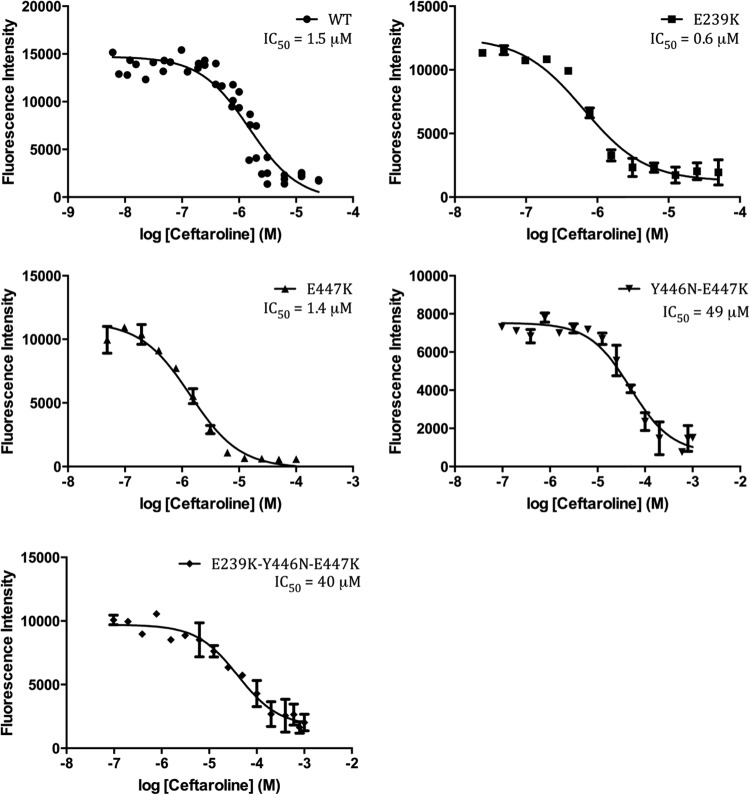
Inspection of the raw sequencing reads from all six genomes revealed that the susceptible isolates (TMHS-3957 and TMHS-4147) represented mixed populations. Approximately half the sequencing reads of each isolate had the E447K PBP2a active-site mutation without the corresponding Y446N change. Isolate TMHS-4519 is ceftaroline intermediate (MIC = 1.5 mg/liter). This isolate had a PBP2a containing the Y446N polymorphism without the corresponding E447K change. These findings suggest that the Y446N mutation in isolation is sufficient to cause an increased MIC of ceftaroline (1.5 mg/liter) but that the presence of both PBP2a amino acid changes (Y446N and E447K) is required to produce the observed high-level resistance (MIC > 32 mg/liter) (Fig. 2). This hypothesis was supported by results from in vitro binding studies conducted with ceftaroline and recombinant purified wild-type and mutant PBP2a enzyme (residues 23 to 668; see Materials and Methods). Ceftaroline had an IC50 of 1.5 μM for wild-type PBP2a (Fig. 3; see also Table S4 in the supplemental material), in agreement with the previously reported values for ceftaroline with both recombinant purified PBP2a and PBP2a membrane fractions (27). The E239K mutation, which is very close to the recently reported allosteric site (25), resulted in a 2-fold reduction in the IC50 of ceftaroline. Even at 1 mM ceftaroline, the Y446N mutant did not show a reduced fluorescence intensity. Thus, we estimate it to have an IC50 of greater than 500 μM. The E447K amino acid replacement alone did not impact inhibition by ceftaroline, as shown by its IC50 of 1.4 μM, which is very similar to that for the wild-type enzyme. The Y446N-E447K double mutant had an IC50 approximately 30-fold higher than that for the wild type at 49 μM, whereas the E239K-Y446N-E447K triple mutant had an IC50 of 40 μM, which is slightly lower than that for the double mutant but approximately 25-fold higher than that for the wild type.
FIG 3.
Bocillin competition assay to determine the IC50 of ceftaroline for the PBP2a wild type and mutants. The y axis represents the residual fluorescence intensity of Bocillin, while the x axis represents the logarithmic ceftaroline concentration (in molar). The Y446N mutant did not show any decrease in fluorescence intensity even with the highest concentration of inhibitor tested, and thus, the results for that mutant are not represented here.
Analysis of isogenic mutant strains.
To definitively demonstrate that the identified amino acid changes caused high-level resistance to ceftaroline, we constructed isogenic mutant strains of our clinical strain S. aureus TMHS-5007 (see the Methods in the supplemental material). The mecA gene was first genetically inactivated in this strain, resulting in enhanced susceptibility to ceftaroline (MIC < 0.5 mg/liter), as anticipated. Next, this strain was altered by genetic complementation to contain the mutant mecA gene containing only the three amino acid replacements (E239K, Y446N, E447K) identified in the high-level ceftaroline-resistant strain. This was accomplished by complementing the TMHS-5007 ΔmecA mutant strain with a vector containing either full-length wild-type mecA or E239K-Y446N-E447K-mutated mecA. Importantly, the resulting complemented strain containing the mutant mecA gene had high-level ceftaroline resistance (MIC > 32 mg/liter), whereas the mutant strain complemented with the wild-type mecA gene remained susceptible to ceftaroline (MIC = 0.5 mg/liter).
Finally, we inspected the genome sequence data to identify mutations that may contribute to the SCV phenotype. All six strains had an SNP that resulted in an amino acid replacement (S33P) in ThyA, a protein involved in thymidine metabolism. Polymorphisms in thyA can cause the SCV phenotype in S. aureus, particularly in strains cultured from cystic fibrosis patients (28, 29). We hypothesize that this thyA amino acid change is responsible for the SCV phenotype observed in these strains. No SNPs were present in other genes implicated in the SCV phenotype (30–32).
DISCUSSION
Here we report and experimentally confirm a previously undescribed high-level antibiotic resistance mechanism in clinical isolates of MRSA in the United States. Ceftaroline was only recently approved by the FDA in October 2010, and no high-level resistance has been reported, despite the study of thousands of clinical isolates (6–9, 12, 33). In the aggregate, our data show that a combination of two contiguous amino acid changes (Y446N and E447K) in PBP2a renders S. aureus highly resistant to ceftaroline (MIC > 32 mg/liter). While previous studies have found ceftaroline-resistant isolates (MICs = 4 to 8 mg/liter) in clonal types ST239 and ST228, which are common to Europe, Asia, and South America, our isolates belong to ST5, a common clonal type in the United States (8, 12, 34).
PBP2a is subject to allosteric regulation, where ceftaroline or peptidoglycan binds at an allosteric region some 60 Å away from the active site, causing the molecule to open and facilitating ceftaroline binding at the active site (25). Only one strain (TMHS-5007) had a variant amino acid (E239K) at the allosteric regulatory site. This E239K mutation has been implicated in the ceftobiprole resistance that arose during in vitro passage and selection (26). Our in vitro biochemical inhibition results strongly suggest that the Y446N amino acid replacement is the main contributor of resistance to ceftaroline in the Y446N-E447K double mutant. This result is in agreement with the recent structural studies that implicated Y446N as the gatekeeper to the active site (25). The E447K mutant had an IC50 very similar to that of the wild type, and thus, it may play a role in either stabilizing the double mutant or facilitating the transpeptidation reaction, despite the change in the active site. The Y446N mutant has a ceftaroline IC50 estimated to be >500 mg/liter in our in vitro assay, which contrasts with the observed MIC of 1.5 mg/liter in vivo for TMHS-4519. We speculate that the most likely cause of this discrepancy is a lack of transpeptidase function in the Y446N mutant PBP2a. As a result, while ceftaroline would not bind the nonfunctional Y446N mutant PBP2a, it would continue to bind to other PBP molecules, as has been described, resulting in a ceftaroline-susceptible phenotype (8). The E239K-Y446N-E447K triple mutant had a reduced IC50 compared to that of the double mutant; it was, however, still resistant, unlike the wild type. This reduction in IC50 may be due to the presence of the E239K mutation, which potentiates ceftaroline binding. The combination of these antagonistic mutations (to form the E239K-Y446N-E447K triple mutant) results in a decrease in the IC50 for ceftaroline to 40 μM, whereas the IC50 for ceftaroline is 49 μM for the double mutant. Although this decrease in the IC50 for the triple mutant in our in vitro assay is not drastic, it might be interesting to test if the combination of ceftaroline with ceftobiprole shows some synergy in vivo. After completion of our work, Alm et al. reported the combination of E239K and E447K amino acid changes in four clinical isolates with ceftaroline MICs of 8 mg/liter (12). Their work suggests that a high-level-resistance phenotype may exist for strains with the E239K and E447K mutations in combination; however, isogenic mutants were not tested to confirm this correlation.
We hypothesize the following scenario to explain the origin and emergence of this drug-resistant bloodstream isolate. It is reasonable to think that this resistant isolate initially emerged under the selective pressure of ceftaroline therapy given at an outside hospital. When the patient was transferred to our hospital and ceftaroline therapy was stopped, we believe that this isolate receded into the S. aureus population present in the patient's lung, while other ceftaroline-susceptible isolates rose to prominence. The precise events leading to the reemergence of the ceftaroline-resistant strains in June are unclear.
Strains from this patient also have a nonsynonymous SNP in thyA that likely causes their SCV phenotype. Mutations in thyA have previously been described to occur in SCVs from cystic fibrosis patients (28, 29). Of note, SCV strains of S. aureus have been reported to have a hypermutable phenotype caused by mutations in mutS and mutL. All six isolates that we studied have a 12-bp in-frame deletion in mutS (deletion of residues 1001 to 1012 [AACGTCTTGTTG]) and a single base frameshift deletion in mutL (deletion of an A residue at position 1028). It is possible that an increased mutation rate may have aided the development of ceftaroline resistance in this patient, but this is speculative.
The case described here illustrates the rapidly emerging clinical utility of whole-genome sequencing in the diagnostic microbiology laboratory (35). Although these isolates were sequenced retrospectively to identify the polymorphism(s) responsible for their observed phenotype, one can readily envision a day in the near future when low-cost, high-throughput genome sequencing facilitates the routine identification of organisms and antibiotic resistance in a prospective fashion.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funds from the Houston Methodist Hospital and the Fondren Foundation. Work in T. Palzkill's laboratory was supported by NIH grant AI32956, and that in A. E. Rosato's laboratory was supported by NIH grant 5R01AI080688-05.
We thank Kathryn Stockbauer of the Office of Academic Development in the Department of Pathology and Genomic Medicine for editorial assistance; Andrew Pann for his role in developing the Prephix, Phrecon, and snp_compare computer scripts; Sabrina Badgett of the Molecular Diagnostics Laboratory at Houston Methodist Hospital for preparing genome libraries and sequencing the isolates; Conception Cantu and the Diagnostic Microbiology Laboratory at Houston Methodist Hospital for collecting and banking S. aureus isolates; and Forest Laboratories for providing ceftaroline analytical powder.
Footnotes
Published ahead of print 25 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03622-14.
REFERENCES
- 1.Bush K, Heep M, Macielag MJ, Noel GJ. 2007. Anti-MRSA beta-lactams in development, with a focus on ceftobiprole: the first anti-MRSA beta-lactam to demonstrate clinical efficacy. Expert Opin. Investig. Drugs 16:419–429. 10.1517/13543784.16.4.419. [DOI] [PubMed] [Google Scholar]
- 2.Jacqueline C, Caillon J, Le Mabecque V, Miegeville AF, Hamel A, Bugnon D, Ge JY, Potel G. 2007. In vivo efficacy of ceftaroline (PPI-0903), a new broad-spectrum cephalosporin, compared with linezolid and vancomycin against methicillin-resistant and vancomycin-intermediate Staphylococcus aureus in a rabbit endocarditis model. Antimicrob. Agents Chemother. 51:3397–3400. 10.1128/AAC.01242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parish D, Scheinfeld N. 2008. Ceftaroline fosamil, a cephalosporin derivative for the potential treatment of MRSA infection. Curr. Opin. Investig. Drugs 9:201–209. [PubMed] [Google Scholar]
- 4.Sader HS, Fritsche TR, Jones RN. 2008. Antimicrobial activities of ceftaroline and ME1036 tested against clinical strains of community-acquired methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 52:1153–1155. 10.1128/AAC.01351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. CLSI document M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 6.Jones RN, Mendes RE, Sader HS. 2010. Ceftaroline activity against pathogens associated with complicated skin and skin structure infections: results from an international surveillance study. J. Antimicrob. Chemother. 65(Suppl 4):S17–S31. 10.1093/jac/dkq252. [DOI] [PubMed] [Google Scholar]
- 7.Jones RN, Farrell DJ, Mendes RE, Sader HS. 2011. Comparative ceftaroline activity tested against pathogens associated with community-acquired pneumonia: results from an international surveillance study. J. Antimicrob. Chemother. 66(Suppl 3):S69–S80. 10.1093/jac/dkr101. [DOI] [PubMed] [Google Scholar]
- 8.Mendes RE, Tsakris A, Sader HS, Jones RN, Biek D, McGhee P, Appelbaum PC, Kosowska-Shick K. 2012. Characterization of methicillin-resistant Staphylococcus aureus displaying increased MICs of ceftaroline. J. Antimicrob. Chemother. 67:1321–1324. 10.1093/jac/dks069. [DOI] [PubMed] [Google Scholar]
- 9.Flamm RK, Sader HS, Farrell DJ, Jones RN. 2012. Summary of ceftaroline activity against pathogens in the United States, 2010: report from the Assessing Worldwide Antimicrobial Resistance Evaluation (AWARE) surveillance program. Antimicrob. Agents Chemother. 56:2933–2940. 10.1128/AAC.00330-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smyth DS, McDougal LK, Gran FW, Manoharan A, Enright MC, Song JH, de Lencastre H, Robinson DA. 2010. Population structure of a hybrid clonal group of methicillin-resistant Staphylococcus aureus, ST239-MRSA-III. PLoS One 5:e8582. 10.1371/journal.pone.0008582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474. 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alm RA, McLaughlin RE, Kos VN, Sader HS, Iaconis JP, Lahiri SD. 2014. Analysis of Staphylococcus aureus clinical isolates with reduced susceptibility to ceftaroline: an epidemiological and structural perspective. J. Antimicrob. Chemother. 69:2065–2075. 10.1093/jac/dku114. [DOI] [PubMed] [Google Scholar]
- 13.Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, Chow H, Ip M, Jatzwauk L, Jonas D, Kadlec K, Kearns A, Laurent F, O'Brien FG, Pearson J, Ruppelt A, Schwarz S, Scicluna E, Slickers P, Tan H-L, Weber S, Ehricht R. 2011. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6:e17936. 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 103:8487–8492. 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McAdam PR, Holmes A, Templeton KE, Fitzgerald JR. 2011. Adaptive evolution of Staphylococcus aureus during chronic endobronchial infection of a cystic fibrosis patient. PLoS One 6:e24301. 10.1371/journal.pone.0024301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nusbaum C, Ohsumi TK, Gomez J, Aquadro J, Victor TC, Warren RM, Hung DT, Birren BW, Lander ES, Jaffe DB. 2009. Sensitive, specific polymorphism discovery in bacteria using massively parallel sequencing. Nat. Methods 6:67–69. 10.1038/nmeth.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225–1240. 10.1016/S0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 19.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267. 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 20.Zhao G, Meier TI, Kahl SD, Gee KR, Blaszczak LC. 1999. BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob. Agents Chemother. 43:1124–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finan JE, Rosato AE, Dickinson TM, Ko D, Archer GL. 2002. Conversion of oxacillin-resistant staphylococci from heterotypic to homotypic resistance expression. Antimicrob. Agents Chemother. 46:24–30. 10.1128/AAC.46.1.24-30.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luong TT, Lee CY. 2006. The arl locus positively regulates Staphylococcus aureus type 5 capsule via an mgrA-dependent pathway. Microbiology 152:3123–3131. 10.1099/mic.0.29177-0. [DOI] [PubMed] [Google Scholar]
- 23.Lim D, Strynadka NCJ. 2002. Structural basis for the β lactam resistance of PBP 2a from methicillin-resistant Staphylococcus aureus. Nat. Struct. Mol. Biol. 9:870–876. 10.1038/nsb858. [DOI] [PubMed] [Google Scholar]
- 24.Grantham R. 1974. Amino acid difference formula to help explain protein evolution. Science 185:862–864. 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 25.Otero LH, Rojas-Altuve A, Llarrull LI, Carrasco-López C, Kumarasiri M, Lastochkin E, Fishovitz J, Dawley M, Hesek D, Lee M, Johnson JW, Fisher JF, Chang M, Mobashery S, Hermoso JA. 2013. How allosteric control of Staphylococcus aureus penicillin binding protein 2a enables methicillin resistance and physiological function. Proc. Natl. Acad. Sci. U. S. A. 110:16808–16813. 10.1073/pnas.1300118110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerjee R, Gretes M, Basuino L, Strynadka N, Chambers HF. 2008. In vitro selection and characterization of ceftobiprole-resistant methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 52:2089–2096. 10.1128/AAC.01403-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moisan H, Pruneau M, Malouin F. 2010. Binding of ceftaroline to penicillin-binding proteins of Staphylococcus aureus and Streptococcus pneumoniae. J. Antimicrob. Chemother. 65:713–716. 10.1093/jac/dkp503. [DOI] [PubMed] [Google Scholar]
- 28.Besier S, Zander J, Kahl BC, Kraiczy P, Brade V, Wichelhaus TA. 2008. The thymidine-dependent small-colony-variant phenotype is associated with hypermutability and antibiotic resistance in clinical Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 52:2183–2189. 10.1128/AAC.01395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chatterjee I, Kriegeskorte A, Fischer A, Deiwick S, Theimann N, Proctor RA, Peters G, Herrmann M, Kahl BC. 2008. In vivo mutations of thymidylate synthase (encoded by thyA) are responsible for thymidine dependency in clinical small-colony variants of Staphylococcus aureus. J. Bacteriol. 190:834–842. 10.1128/JB.00912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lannergard J, von Eiff C, Sander G, Cordes T, Seggewiss J, Peters G, Proctor RA, Becker K, Hughes D. 2008. Identification of the genetic basis for clinical menadione-auxotrophic small-colony variant isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 52:4017–4022. 10.1128/AAC.00668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sendi P, Proctor RA. 2009. Staphylococcus aureus as an intracellular pathogen: the role of small colony variants. Trends Microbiol. 17:54–58. 10.1016/j.tim.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4:295–305. 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Xiao M, Yang Q-W, Wang Y, Wang H, Zhao Y, Brown M, Zhao H-R, Kong F, Xu Y-C. 2013. High ceftaroline non-susceptibility in Staphylococcus aureus isolated from acute skin infections in 15 tertiary hospitals in China. J. Med. Microbiol. 62:496–497. 10.1099/jmm.0.052522-0. [DOI] [PubMed] [Google Scholar]
- 34.Hudson LO, Reynolds C, Spratt BG, Enright MC, Quan V, Kim D, Hannah P, Mikhail L, Alexander R, Moore DF, Godoy D, Bishop CJ, Huang SS. 2013. Diversity of methicillin-resistant Staphylococcus aureus strains isolated from residents of 26 nursing homes in Orange County, California. J. Clin. Microbiol. 51:3788–3795. 10.1128/JCM.01708-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olsen RJ, Long SW, Musser JM. 2012. Bacterial genomics in infectious disease and the clinical pathology laboratory. Arch. Pathol. Lab. Med. 136:1414–1422. 10.5858/arpa.2012-0025-RA. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.