Abstract
Intermittent preventive treatment of infants (IPTi) reduces early childhood malaria-related morbidity. While genotypic drug resistance markers have proven useful in predicting the efficacy of antimalarial drugs in case management, there are few equivalent data relating to their protective efficacy when used as IPTi. The present data from an IPTi trial in Papua New Guinea demonstrate how these markers can predict protective efficacy of IPTi for both Plasmodium falciparum and Plasmodium vivax.
TEXT
Intermittent preventive treatment of infants (IPTi) has reduced early childhood malaria-related morbidity in many settings (1, 2). Although its precise mechanism of action has been debated, it is now generally agreed that the intermittent short courses of slowly eliminated antimalarial drugs provide protection due to prolonged prophylactic activity (2). Therefore, the pharmacokinetic and pharmacodynamic properties that determine a drug's suitability for IPTi may differ from those important for therapeutic efficacy in case management.
Genotypic drug resistance markers correlate well with in vivo therapeutic response in the case management of acute infections. Mutations of Plasmodium falciparum and Plasmodium vivax dihydrofolate reductase (pfdhfr and pvdhfr) and dihydropteroate synthase (pfdhps and pvdhps) genes are associated with resistance to pyrimethamine and sulfadoxine, respectively (3–6). Parasite resistance to 4-aminoquinolines is associated with mutations in the chloroquine resistance transporter gene (pfcrt) (7) and multidrug resistance gene 1 (pfmdr1) in P. falciparum infections and with P. vivax multidrug resistance gene 1 (pvmdr1) mutations in some studies of vivax malaria (8, 9) but not others (10).
IPTi effectiveness is likely to be highly dependent on drug resistance patterns in local parasite populations. Current IPTi regimens utilize either 4-aminoquinoline or antifolate drugs (2), and so existing genotypic resistance markers should predict protective efficacy in a given setting without the need for large expensive field-based efficacy studies. However, data directly relating drug resistance genotypes to IPTi efficacy is limited to African IPTi trials of sulfadoxine-pyrimethamine (SP) dual therapy, which have demonstrated that the pfdhfr triple mutant genotype predicts reduced efficacy (11, 12). No studies have examined the relationship between IPTi efficacy and genotypic markers in P. vivax infections nor between 4-aminoquinoline resistance markers and IPTi efficacy in falciparum malaria. In addition, there are concerns that the high burden of antimalarial drug use through IPTi will drive the selection and spread of parasite drug resistance, with a consequent reduction in public health benefit (13). Serial monitoring of validated genotypic markers could help identify this phenomenon at an early stage (12).
In Papua New Guinea (PNG), an IPTi study of 1,121 infants conducted from 2006 to 2010 showed that amodiaquine plus SP (AQ-SP) reduced P. falciparum and P. vivax incidence by 35% and 23%, respectively, compared with placebo (14). Artesunate-SP (ART-SP) had a similar efficacy (31%) against P. falciparum but none against P. vivax infections. Protective efficacy of both SP-containing regimens therefore fell in the midrange of comparative African trials of IPTi using SP (median protective efficacy, 30.3%; range, 20 to 59%) (1), suggesting a likelihood of highly cost-effective public health benefit. However, protective efficacy against P. vivax was dependent on the partner drug used alongside SP. We therefore hypothesized that therapy-specific efficacy in P. falciparum and P. vivax infections reflects both pharmacokinetic differences between ART and AQ and intrinsic differences in the sensitivities of P. falciparum and P. vivax parasites to SP. In the present study, we investigated whether the frequency of mutations associated with 4-aminoquinoline and SP resistance could explain differences in IPTi efficacy of AQ-SP and ART-SP and explored the impact of different IPTi regimens on resistance marker prevalence.
The IPTi trial study sample and clinical methods have been described in detail elsewhere (14). All participants were enrolled at age 3 months and had blood samples collected at 3, 6, 9, and 12 months after enrollment (i.e., passive detection in children aged 6 to 15 months) in each study arm (AQ-SP, ART-SP, and control). The study was approved by the PNG Medical Research Advisory Committee (approval no. 05.20) and the PNG Institute of Medical Research Institutional Review Board (approval no. 06.01).
Drug resistance genotyping was performed using a ligase detection reaction-fluorescent microsphere assay (LDR-FMA). Briefly, fragments of pfcrt, pfmdr1, pfdhfr, and pfdhps and fragments of pvmdr1, pvdhfr, and pvdhps were amplified from P. falciparum- and P. vivax-positive samples, respectively. LDR was performed using allele-specific primers as previously described (15–17). Products were run through a Bio-Plex 200 reader, and fluorescence signals were analyzed to determine which alleles were present in each sample. A positivity cutoff was determined for each locus by the use of methodology developed previously to account for an allele-specific background (15, 18). Positive controls (genomic DNA [gDNA] from P. falciparum reference strains or P. vivax mdr1, dhfr, and dhps plasmids) displaying wild -type and mutant alleles were run in each assay. Statistical analyses were performed using R (BiostaTGV) to compare genotypic frequencies in each treatment group, using chi-square and Fisher's exact tests.
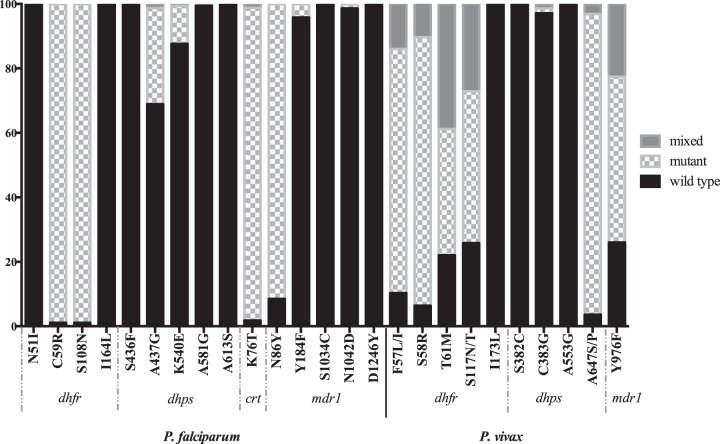
The proportions of individuals with infections with wild-type and mutant genotypes of the different parasite genes are shown in Fig. 1. The pfcrt 76T and pfmdr 86Y mutations appeared close to fixation in P. falciparum, and the proportion of P. vivax infections with the pvmdr1 976F mutation was 73.8%, suggesting compromised AQ efficacy against P. falciparum (17, 19, 20) and possibly (given the currently uncertain functional significance of pvmdr1 [8–10]) against P. vivax.
FIG 1.
Proportions of infections with wild-type and mutant genotypes in all P. falciparum and P. vivax infections.
While 98.9% of P. falciparum-infected individuals carried at least one pfdhfr mutation, only mutations at codons 59 (C→R) and 108 (S→N) were identified. The 51I mutant was not detected, so none had the pfdhfr triple mutation (51I-59R-108N) nor the pfdhfr/pfdhps quintuple mutant haplotype associated with high-level SP resistance in Africa (20). Similarly, the pfdhfr I164L mutation which is associated with high-level SP resistance in Africa, even when present as a single mutation (21), was not detected. Mutations in pfdhps (codons 437, 540, and 581) were present in 29.8% of the infected individuals, with 17.5% and 11.7% of monoclonal infections having pfdhfr or pfdhps triple and quadruple mutant haplotypes, respectively (see Table S1 in the supplemental material).
For P. vivax, 57.3% of the individuals with monoclonal infections showed the quadruple mutant genotype (57L-58R-61M-117T) associated with very high rates of in vivo SP treatment failure (22, 23) (see Table S1 in the supplemental material). Only 3.0% of all P. vivax infections showed a mutation at pvdhps codons 383 and/or 553.
Overall parasite genotype patterns suggested some retention of SP efficacy against P. falciparum, given the absence of the quintuple mutant haplotype and pfdhfr I164L mutant. The high prevalence of the pvdhfr quadruple mutant genotype suggested poor or absent SP efficacy against P. vivax. Any AQ-SP efficacy against P. falciparum would therefore be due largely to the SP component, and conversely, any efficacy against P. vivax would result from the use of AQ. For ART-SP, given the negligible posttreatment prophylactic duration of ART, genotypic prevalence predicts retention of some SP efficacy against P. falciparum but virtually none against P. vivax, consistent with the differential protective efficacies against the two species demonstrated in the IPTi trial (14). The present genotypic data are therefore concordant with the observed clinical outcomes (14), explaining both the similar protective efficacies of AQ-SP and ART-SP against P. falciparum and the discordant efficacies of AQ-SP and ART-SP against P. vivax.
There were no differences between P. falciparum mutant genotype frequencies in the control AQ-SP and ART-SP study arms that might have suggested selection of more resistant strains (Table 1). However, the low population diversity shown by the dominance of one or two pfdhfr, pfdhps, pfcrt, and pfmdr1 alleles means that few of the genes evaluated would be under selection pressure. Therefore, we cannot exclude selection of resistance in other contexts, with different balances of parasite genotypes. Indeed, selection of P. falciparum drug-resistant mutants has previously been shown following single- or three-dose SP IPTi in Africa (13, 24). No between-group differences were seen for pvdhfr or pvdhps mutations (Table 1).
TABLE 1.
Proportions of individuals infected with wild-type and mutant P. falciparum or P. vivax genotypesa
| Sample type and gene | Codon(s) | Amino acid(s)b | % of individuals infected with mutant genotype from the indicated study arm |
P valued | ||
|---|---|---|---|---|---|---|
| Placebo | AQ-SP | ART-SP | ||||
| P. falciparum | ||||||
| n = 70 | n = 59 | n = 49 | ||||
| pfdhfr | 51 | Nc | 100 | 100 | 100 | NS |
| 59 | C/R | 1.4/98.6 | 1.7/98.3 | 0/100 | NS | |
| 108 | S/N | 1.4/98.6 | 1.7/98.3 | 0/100 | NS | |
| 164 | Ic | 100 | 100 | 100 | NS | |
| n = 63 | n = 51 | n = 47 | ||||
| pfdhps | 436–437 | SA/SG | 69.8/30.2 | 64.7/37.3 | 76.6/25.5 | NS |
| 540 | K/E | 87.3/12.7 | 86.3/13.7 | 91.5/10.6 | NS | |
| 581 | A/G | 100/0 | 100/2.0 | 100/0 | NS/NS | |
| 613 | A | 100 | 100 | 100 | NS | |
| n = 66 | n = 55 | n = 44 | ||||
| pfcrt | 72–76 | CVMNK/SVMNT/CVIET | 1.5/98.5/0 | 3.6/96.4/1.8 | 4.5/100/0 | NS |
| n = 59 | n = 44 | n = 39 | ||||
| pfmdr1 | 86 | N/Y | 10.2/89.8 | 9.1/90.9 | 5.1/94.9 | NS |
| 184 | Y/F | 93.2/6.8 | 100/0 | 100/0 | NS | |
| 1034 | Sc | 100 | 100 | 100 | NS | |
| 1042 | N/D | 96.6/3.4 | 100/0 | 100/0 | NS | |
| 1246 | Dc | 100 | 100 | 100 | NS | |
| P. vivax | ||||||
| n = 177 | n = 167 | n = 205 | ||||
| pvdhfr | 57/58/61 | FST/FRT/LRT/LRM | 15.3/11.3/41.8/66.1 | 18.0/7.2/47.9/60.5 | 16.6/7.8/46.3/56.1 | NS |
| 117 | S/N/T | 51.4/9.6/74.6 | 55.7/6.0/68.9 | 52.2/6.8/66.3 | NS | |
| 173 | Ic | 100 | 100 | 100 | NS | |
| n = 170 | n = 160 | n = 195 | ||||
| pvdhps | 382–383 | SC/SG/CC | 98.8/3.5/0 | 98.1/1.9/0.6 | 98.5/3.1/0 | NS |
| 553 | A/G | 100/0 | 100/0.6 | 100/0 | NS/NS | |
| 647 | A/S/P | 8.8/1.2/93.5 | 5.0/0/96.9 | 6.2/0.5/97.9 | NS | |
| n = 173 | n = 170 | n = 211 | ||||
| pvmdr1 | 976 | Y/F | 46.2/74.0 | 54.1/72.9 | 46.0/74.4 | NS |
n values represent the numbers of individuals in each group.
Boldface indicates mutant amino acids.
pfdhfr 51N and 164L, pfdhps 436–437FG and 613S, pfmdr1 1034C and 1246Y, and pvdhfr 173L genotypes were not observed.
NS, not significant.
We have demonstrated concordance of P. falciparum and P. vivax drug resistance genotypic prevalence with the protective efficacy of two IPTi drug combinations. Drug resistance genotyping is a valuable public health tool for monitoring antimalarial drug resistance and the early detection of compromised therapeutic efficacy in case management. Analysis of population prevalence of genotypic markers could similarly help predict the protective efficacy of IPTi drug regimens in a wide variety of settings where malaria is endemic. It is clear from the present and previous studies that the efficacy of IPTi with SP is highly dependent on prevalence of SP resistance at a population level (2, 11, 12). However, the complex genetics of antifolate drug resistance makes it challenging to establish clear, simple guidelines to help policy makers decide on the appropriateness or other aspects of using SP for IPTi in their national programs.
To be useful in practice, such guidelines require a clear cutoff to recommend its use (2). Current WHO guidelines use a threshold of pfdhps codon 540 mutant prevalence of ≤50% to endorse SP use in IPTi in areas of moderate to high P. falciparum transmission (25). These recommendations would therefore have endorsed the use of SP in IPTi in our PNG setting, and this is supported by the results of the clinical component of this study. However, more sophisticated guidelines will be needed for situations in which 4-aminoquinolines are considered for IPTi in settings in which the pfdhfr I164L mutation is prevalent and for settings outside Africa where P. vivax is endemic. In order to enhance guideline development, it is important, therefore, that drug resistance markers be considered a routine assessment in future IPTi efficacy evaluations.
Supplementary Material
ACKNOWLEDGMENTS
We thank all the field team members who helped collect the samples used in this analysis. We also thank all the study participants.
This study was supported by grants from the Bill & Melinda Gates Foundation (grant no. 34678) and the Australian National Health and Medical Research Council (grant no. 1010203). T.M.D. was supported by an NHMRC Practitioner Fellowship (no. 1058260), I.M. by an NHMRC Senior Research Fellowship (no. 1043345), and H.K. by an NHMRC Career Development Fellowship (no. 1064772).
Footnotes
Published ahead of print 25 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03323-14.
REFERENCES
- 1.Aponte JJ, Schellenberg D, Egan A, Breckenridge A, Carneiro I, Critchley J, Danquah I, Dodoo A, Kobbe R, Lell B, May J, Premji Z, Sanz S, Sevene E, Soulaymani-Becheikh R, Winstanley P, Adjei S, Anemana S, Chandramohan D, Issifou S, Mockenhaupt F, Owusu-Agyei S, Greenwood B, Grobusch MP, Kremsner PG, Macete E, Mshinda H, Newman RD, Slutsker L, Tanner M, Alonso P, Menendez C. 2009. Efficacy and safety of intermittent preventive treatment with sulfadoxine-pyrimethamine for malaria in African infants: a pooled analysis of six randomised, placebo-controlled trials. Lancet 374:1533–1542. 10.1016/S0140-6736(09)61258-7. [DOI] [PubMed] [Google Scholar]
- 2.Gosling RD, Cairns ME, Chico RM, Chandramohan D. 2010. Intermittent preventive treatment against malaria: an update. Expert Rev. Anti Infect. Ther. 8:589–606. 10.1586/eri.10.36. [DOI] [PubMed] [Google Scholar]
- 3.de Pecoulas PE, Basco LK, Tahar R, Ouatas T, Mazabraud A. 1998. Analysis of the Plasmodium vivax dihydrofolate reductase-thymidylate synthase gene sequence. Gene 211:177–185. 10.1016/S0378-1119(98)00118-8. [DOI] [PubMed] [Google Scholar]
- 4.de Pecoulas PE, Tahar R, Ouatas T, Mazabraud A, Basco LK. 1998. Sequence variations in the Plasmodium vivax dihydrofolate reductase-thymidylate synthase gene and their relationship with pyrimethamine resistance. Mol. Biochem. Parasitol. 92:265–273. 10.1016/S0166-6851(97)00247-8. [DOI] [PubMed] [Google Scholar]
- 5.Korsinczky M, Fischer K, Chen N, Baker J, Rieckmann K, Cheng Q. 2004. Sulfadoxine resistance in Plasmodium vivax is associated with a specific amino acid in dihydropteroate synthase at the putative sulfadoxine-binding site. Antimicrob. Agents Chemother. 48:2214–2222. 10.1128/AAC.48.6.2214-2222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sibley CH, Hyde JE, Sims PF, Plowe CV, Kublin JG, Mberu EK, Cowman AF, Winstanley PA, Watkins WM, Nzila AM. 2001. Pyrimethamine-sulfadoxine resistance in Plasmodium falciparum: what next? Trends Parasitol. 17:582–588. 10.1016/S1471-4922(01)02085-2. [DOI] [PubMed] [Google Scholar]
- 7.Sidhu AB, Verdier-Pinard D, Fidock DA. 2002. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science 298:210–213. 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brega S, Meslin B, de Monbrison F, Severini C, Gradoni L, Udomsangpetch R, Sutanto I, Peyron F, Picot S. 2005. Identification of the Plasmodium vivax mdr-like gene (pvmdr1) and analysis of single-nucleotide polymorphisms among isolates from different areas of endemicity. J. Infect. Dis. 191:272–277. 10.1086/426830. [DOI] [PubMed] [Google Scholar]
- 9.Sá JM, Nomura T, Neves J, Baird JK, Wellems TE, del Portillo HA. 2005. Plasmodium vivax: allele variants of the mdr1 gene do not associate with chloroquine resistance among isolates from Brazil, Papua, and monkey-adapted strains. Exp. Parasitol. 109:256–259. 10.1016/j.exppara.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Barnadas C, Ratsimbasoa A, Tichit M, Bouchier C, Jahevitra M, Picot S, Menard D. 2008. Plasmodium vivax resistance to chloroquine in Madagascar: clinical efficacy and polymorphisms in pvmdr1 and pvcrt-o genes. Antimicrob. Agents Chemother. 52:4233–4240. 10.1128/AAC.00578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin JT, Cairns M, Ghani AC, Roper C, Schellenberg D, Carneiro I, Newman RD, Grobusch MP, Greenwood B, Chandramohan D, Gosling RD. 2010. Protective efficacy of intermittent preventive treatment of malaria in infants (IPTi) using sulfadoxine-pyrimethamine and parasite resistance. PLoS One 5:e12618. 10.1371/journal.pone.0012618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venkatesan M, Alifrangis M, Roper C, Plowe CV. 2013. Monitoring antifolate resistance in intermittent preventive therapy for malaria. Trends Parasitol. 29:497–504. 10.1016/j.pt.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marks F, von Kalckreuth V, Kobbe R, Adjei S, Adjei O, Horstmann RD, Meyer CG, May J. 2005. Parasitological rebound effect and emergence of pyrimethamine resistance in Plasmodium falciparum after single-dose sulfadoxine-pyrimethamine. J. Infect. Dis. 192:1962–1965. 10.1086/497698. [DOI] [PubMed] [Google Scholar]
- 14.Senn N, Rarau P, Stanisic DI, Robinson L, Barnadas C, Manong D, Salib M, Iga J, Tarongka N, Ley S, Rosanas-Urgell A, Aponte JJ, Zimmerman PA, Beeson JG, Schofield L, Siba P, Rogerson SJ, Reeder JC, Mueller I. 2012. Intermittent preventive treatment for malaria in Papua New Guinean infants exposed to Plasmodium falciparum and P. vivax: a randomized controlled trial. PLoS Med. 9:e1001195. 10.1371/journal.pmed.1001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barnadas C, Kent D, Timinao L, Iga J, Gray LR, Siba P, Mueller I, Thomas PJ, Zimmerman PA. 2011. A new high-throughput method for simultaneous detection of drug resistance associated mutations in Plasmodium vivax dhfr, dhps and mdr1 genes. Malar. J. 10:282. 10.1186/1475-2875-10-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carnevale EP, Kouri D, DaRe JT, McNamara DT, Mueller I, Zimmerman PA. 2007. A multiplex ligase detection reaction-fluorescent microsphere assay for simultaneous detection of single nucleotide polymorphisms associated with Plasmodium falciparum drug resistance. J. Clin. Microbiol. 45:752–761. 10.1128/JCM.01683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong RP, Karunajeewa H, Mueller I, Siba P, Zimmerman PA, Davis TM. 2011. Molecular assessment of Plasmodium falciparum resistance to antimalarial drugs in Papua New Guinea using an extended ligase detection reaction fluorescent microsphere assay. Antimicrob. Agents Chemother. 55:798–805. 10.1128/AAC.00939-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DaRe JT, Kouri DP, Zimmerman PA, Thomas PJ. 2010. Differentiating Plasmodium falciparum alleles by transforming Cartesian X,Y data to polar coordinates. BMC Genet. 11:57. 10.1186/1471-2350-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marfurt J, Muller I, Sie A, Oa O, Reeder JC, Smith TA, Beck HP, Genton B. 2008. The usefulness of twenty-four molecular markers in predicting treatment outcome with combination therapy of amodiaquine plus sulphadoxine-pyrimethamine against falciparum malaria in Papua New Guinea. Malar. J. 7:61. 10.1186/1475-2875-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Picot S, Olliaro P, de Monbrison F, Bienvenu AL, Price RN, Ringwald P. 2009. A systematic review and meta-analysis of evidence for correlation between molecular markers of parasite resistance and treatment outcome in falciparum malaria. Malar. J. 8:89. 10.1186/1475-2875-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynch C, Pearce R, Pota H, Cox J, Abeku TA, Rwakimari J, Naidoo I, Tibenderana J, Roper C. 2008. Emergence of a dhfr mutation conferring high-level drug resistance in Plasmodium falciparum populations from southwest Uganda. J. Infect. Dis. 197:1598–1604. 10.1086/587845. [DOI] [PubMed] [Google Scholar]
- 22.Hastings MD, Porter KM, Maguire JD, Susanti I, Kania W, Bangs MJ, Sibley CH, Baird JK. 2004. Dihydrofolate reductase mutations in Plasmodium vivax from Indonesia and therapeutic response to sulfadoxine plus pyrimethamine. J. Infect. Dis. 189:744–750. 10.1086/381397. [DOI] [PubMed] [Google Scholar]
- 23.Tjitra E, Baker J, Suprianto S, Cheng Q, Anstey NM. 2002. Therapeutic efficacies of artesunate-sulfadoxine-pyrimethamine and chloroquine-sulfadoxine-pyrimethamine in vivax malaria pilot studies: relationship to Plasmodium vivax dhfr mutations. Antimicrob. Agents Chemother. 46:3947–3953. 10.1128/AAC.46.12.3947-3953.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayor A, Serra-Casas E, Sanz S, Aponte JJ, Macete E, Mandomando I, Puyol L, Berzosa P, Dobano C, Aide P, Sacarlal J, Benito A, Alonso P, Menendez C. 2008. Molecular markers of resistance to sulfadoxine-pyrimethamine during intermittent preventive treatment for malaria in Mozambican infants. J. Infect. Dis. 197:1737–1742. 10.1086/588144. [DOI] [PubMed] [Google Scholar]
- 25.World Health Organization. 2010. Policy recommendation on intermittent preventive treatment during infancy with sulphadoxine-pyrimethamine (SP-IPTi) for Plasmodium falciparum malaria control in Africa. World Health Organization, Geneva, Switzerland. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.