Abstract
This study examined molecular and epidemiologic factors associated with Escherichia coli sequence type 131 (ST131) among hospitalized patients colonized intestinally with fluoroquinolone (FQ)-resistant E. coli between 2002 and 2004. Among 86 patients, 21 (24%) were colonized with ST131. The proportion of ST131 isolates among colonizing isolates increased significantly over time, from 8% in 2002 to 50% in 2004 (P = 0.003). Furthermore, all 19 clonally related isolates were ST131. Future studies should identify potential transmissibility differences between ST131 and non-ST131 strains.
TEXT
The increase in fluoroquinolone (FQ)-resistant Escherichia coli (FQREC) over the past decade has been attributed mainly to the widespread emergence of a single disseminated E. coli clonal group, sequence type 131 (ST131) (1–4), which frequently exhibits multidrug resistance, most notably to FQs and extended-spectrum cephalosporins (ESCs) (5). E. coli isolates within phylogenetic group B2, including specifically E. coli ST131, may have a greater capacity for successful and persistent colonization of the gastrointestinal tract (6, 7). However, FQ resistance mechanisms associated with ST131 status have only been characterized in clinical isolates (8). In addition, few data exist on epidemiologic risk factors for colonization or infection with E. coli ST131 (8–10). Therefore, we sought to evaluate the association between E. coli ST131 and molecular and epidemiologic characteristics among intestine-colonizing FQREC isolates from a previously described population of hospitalized patients (11, 12) that were assessed during the initial rapid rise in prevalence of E. coli ST131.
As previously described (11, 12), three annual fecal surveillance surveys were performed hospital wide at two university-affiliated hospitals during 2002, 2003, and 2004. For the present study, each subject could be included only once, with inclusion of only the first sample for each subject. The University of Pennsylvania Institutional Review Board approved the study.
FQREC was defined as isolates exhibiting a levofloxacin MIC of ≥8 μg/ml. Detection of FQREC from fecal samples and evaluation for specific mechanisms of FQ resistance were performed as previously described (11–13). Overexpression of the multidrug efflux pump AcrAB was measured using the organic solvent tolerance (OST) assay (14, 15). Genetic relatedness of E. coli isolates was determined by pulsed-field gel electrophoresis (PFGE) analysis (11), with profiles analyzed using Fingerprinting II Informatix software v3.0 (Bio-Rad Laboratories, Inc., Hercules, CA) and interpreted according to established criteria (16). Clonal relatedness was defined as >80% similarity.
The major E. coli phylogenetic group was determined by triplex PCR (17). Group B2 isolates were evaluated for ST131 status by detection of ST131-specific single-nucleotide polymorphisms (SNPs) in mdh and gyrB (18) and for the O25b rfb genotype (19). The H30 ST131 subclone was identified by established PCR-based detection of subclone-specific SNPs in fimH (5, 8). For the (CTX-M-15-associated) H30-Rx subclone within the H30 ST131 subclone (20), primers were used that detect an H30-Rx-specific SNP (G723A) within the allantoin protein-encoding gene, ybbW (21). Isolates were screened for blaCTX-M-15 by PCR (21), tested for susceptibility to antibiotic agents by use of the semiautomated Vitek 2 identification and susceptibility system (bioMérieux, Durham, NC), and interpreted according to Clinical and Laboratory Standards Institute criteria. Clinical data were abstracted as previously described from the Pennsylvania Integrated Clinical and Administrative Research Database (13).
Associations between E. coli ST131 and molecular and clinical characteristics were determined. Categorical variables were compared using the Fisher exact test, and continuous variables were compared using the Wilcoxon rank sum test. Multivariable analyses were performed using multiple logistic regression (22), with calculation of adjusted odds ratios (ORs) with 95% confidence intervals (CIs). A stepwise selection procedure was used, with variables with P values of <0.20 on bivariable analyses considered candidate variables and maintained in the final model if their inclusion was statistically significant on likelihood ratio testing (23). For all calculations, a two-tailed P value of <0.05 was considered significant. All calculations were performed using STATA v13.0 (StataCorp LP, College Station, TX).
Over the 3-year study period, a total of 89 (11.5%) of 774 unique subjects were colonized with FQREC (12, 13). For the present study, 86 FQREC isolates (each representing a unique study patient) constituted the study population and were characterized further. Of these, 21 (24%) were identified as ST131 by dual-SNP PCR (Table 1). All 21 ST131 isolates represented the H30 fimH-based subclone within ST131, and 7 (33%) of these belonged specifically to the (CTX-M-15-associated) H30-Rx subset within H30.
TABLE 1.
Microbiologic characteristics of 86 ST131 and non-ST131 E. coli fecal isolates from patients colonized with fluoroquinolone-resistant E. colia
| Variable | Result for: |
P value | |
|---|---|---|---|
| ST131 (n = 21) | Non-ST131 (n = 65) | ||
| No. (%) with H30 | 21 (100) | NA | |
| No. (%) with H30-Rx | 7 (33) | NA | |
| Median levofloxacin MIC (IQR) | 32 (32–32) | 32 (32–32) | 0.64 |
| Median no. of gyrA mutations (IQR) | 2 (2–2) | 2 (2–2) | 0.42 |
| Median no. of parC mutations (IQR) | 2 (2–2) | 1 (1–1) | <0.001 |
| No. (%) with OST-positive status | 3 (14) | 38 (59) | <0.001 |
| No. (%) with blaCTX-M-15 | 0 (0) | 1 (2) | >0.99 |
Data are presented as number (percentage) of isolates unless noted otherwise. NA, not applicable; IQR, interquartile range; OST, organic solvent tolerance.
ST131 and non-ST131 isolates did not differ significantly for the levofloxacin MIC or for the number of mutations in gyrA (Table 1). However, these groups differed for specific gyrA mutations; all 21 ST131 isolates, but only 51 (79%) of non-ST131 isolates, exhibited Ser83→Leu and Asp87→Asn mutations (P = 0.02). ST131 isolates had a greater number of replacement parC mutations compared to non-ST131 isolates. The most common combination of replacement parC mutations among ST131 isolates (88%) was Ser80→Ile and Glu84→Val. Results of antibiotic susceptibility testing are shown in Table 2. ST131 isolates demonstrated a low prevalence of ESC resistance, with the single ESC-resistant isolate belonging to the H30-Rx subgroup within H30.
TABLE 2.
Antibiotic resistance in relation to ST131 status among 86 fluoroquinolone-resistant fecal Escherichia coli isolates from colonized hospital inpatients
| Antibiotic | Prevalence of resistance, no. of isolates (column %) |
P value | |
|---|---|---|---|
| ST131 (n = 21) | Non-ST131 (n = 65) | ||
| Ampicillin-sulbactam | 19 (92) | 46 (72) | 0.08 |
| Cefazolin | 6 (29) | 20 (32) | >0.99 |
| Ceftazidime | 1 (5) | 12 (19) | 0.17 |
| Ceftriaxone | 1 (5) | 11 (17) | 0.28 |
| Gentamicina | 7 (33) | 22 (34) | >0.99 |
| Imipenem | 1 (5) | 0 (0) | 0.24 |
| Piperacillin-tazobactam | 3 (14) | 4 (6) | 0.35 |
| Trimethoprim-sulfamethoxazole | 12 (57) | 39 (60) | >0.99 |
Isolate resistance profiles were the same for tobramycin as for gentamicin.
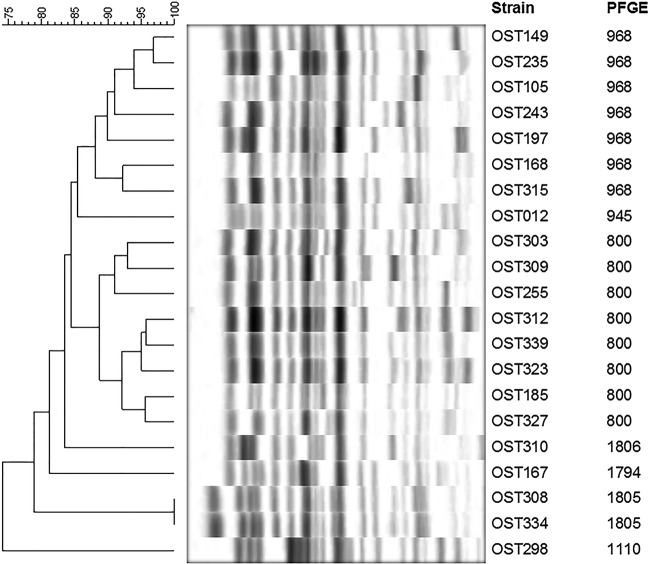
Among the 71 FQREC isolates that could be successfully characterized by PFGE (13), 19 demonstrated clonal relatedness; all of these were ST131. Thus, 19 (90%) of 21 ST131 isolates were clonally related, compared to 0 (0%) of 50 non-ST131 isolates (P < 0.001). The 21 ST131 study isolates' PFGE profiles were subsequently compared to an existing reference library comprising PFGE profiles from 1,292 ST131 isolates (mostly human clinical isolates) (24). The predominant pulsotypes (defined as having ≥94% similarity to an index isolate) represented among the present ST131 colonizing isolates were 968 and 800 (33% and 38% of isolates, respectively) (Fig. 1). Pulsotypes 968 and 800 were also the two most common pulsotypes in the reference library (29% and 12% of isolates, respectively).
FIG 1.
Pulsed-field gel electrophoresis (PFGE) profiles of 21 ST131 fluoroquinolone-resistant fecal Escherichia coli isolates. Data columns to the right of the PFGE profiles show the isolate number (left) and pulsotype (right). Pulsotypes were assigned by comparison to an existing large private PFGE profile reference library (24). Pulsotypes 968, 945, 800, and 1110 (n = 17 isolates) were already established within the reference library; pulsotypes 1806, 1794, and 1805 (n = 4 isolates) were newly assigned here. Note that since profiles were classified as to pulsotype based on ≥94% profile similarity to an index isolate's profile, within the same pulsotype, certain profiles could exhibit as little as 88% similarity.
The by-year prevalence of ST131 rose steadily across the study period (8% in 2002, 19% in 2003, and 50% in 2004; P = 0.003, chi-square test for trend) (Table 3). In multivariable analyses (Table 4), only study year was an independent risk factor for having an ST131 isolate (OR, 3.89; 95% CI, 1.71 to 8.81; P = 0.001). The dramatically increasing proportion of ST131 colonizing isolates over time, which resulted in ST131 comprising half of FQREC isolates during the final year, is in concordance with the increasing prevalence of E. coli ST131 among clinical isolates over the past decade (1–3). This suggests that ST131 has emerged as both a highly prevalent pathogen causing clinical infections and a successful colonizer in the hospital setting.
TABLE 3.
Bivariable analyses of risk factors for ST131 among hospital inpatients colonized intestinally with fluoroquinolone-resistant E. colia
| Variable | Result for: |
OR (95% CI) | P value | |
|---|---|---|---|---|
| ST131 (n = 21) | Non-ST131 (n = 65) | |||
| Yr of culture: | ||||
| 2002 | 2 (8) | 23 (92) | ||
| 2003 | 7 (19) | 30 (81) | 0.003 | |
| 2004 | 12 (50) | 12 (50) | ||
| Mean age (SD), yr | 61.0 (15) | 63.8 (16) | 0.53 | |
| No. (%) female | 6 (29) | 28 (43) | 0.53 (0.15–1.69) | 0.31 |
| No. (%) nonwhite | 10 (48) | 39 (60) | 0.61 (0.20–1.84) | 0.45 |
| No. (%) in surgical service | 9 (43) | 17 (26) | 2.12 (0.66–6.60) | 0.18 |
| No. (%) with nosocomial onset | 14 (74) | 41 (71) | 1.16 (0.33–4.77) | >0.99 |
| No. (%) with admission to hospital 2 | 5 (24) | 20 (31) | 0.70 (0.18–2.40) | 0.59 |
| Mean (SD) hospital day of sampling | 14.4 (14) | 13.4 (24) | 0.15 | |
| No. (%) in ICU on culture date | 3 (14) | 11 (17) | 0.82 (0.13–3.60) | >0.99 |
| No. (%) with diabetes mellitus | 5 (24) | 26 (40) | 0.47 (0.12–1.57) | 0.20 |
| No. (%) with malignancy | 5 (24) | 21 (32) | 0.65 (0.17–2.22) | 0.59 |
| No. (%) with renal insufficiency | 2 (10) | 13 (20) | 0.42 (0.04–2.16) | 0.34 |
| Mean (SD) Charlson comorbidity score | 2.2 (2) | 3.9 (4) | 0.09 | |
| No. (%) with chemotherapy ≤30 days prior to sampling | 3 (14) | 5 (8) | 2.00 (0.28–11.4) | 0.40 |
| No. (%) with immunosuppression ≤30 days prior to sampling | 3 (14) | 10 (15) | 0.92 (0.15–4.12) | >0.99 |
| No. (%) taking antibiotics ≤30 days prior to samplingb | ||||
| Any antibiotic | 13 (62) | 35 (54) | 1.39 (0.46–4.42) | 0.62 |
| 1st-generation cephalosporinc | 7 (33) | 12 (19) | 2.21 (0.61–7.47) | 0.22 |
| Levofloxacin | 10 (48) | 13 (20) | 3.64 (1.11–11.7) | 0.02 |
Data are presented as numbers (percentages) except where noted. OR, odds ratio; CI, confidence interval; SD, standard deviation; ICU, intensive care unit.
Only results for antibiotics with a P value of <0.30 are shown.
Cefazolin or cephalexin.
TABLE 4.
Multivariable model of risk factors for recovery of ST131 isolates among hospital inpatients colonized with fluoroquinolone-resistant E. coli
| Variable | OR (95% CI)a | P value |
|---|---|---|
| Receipt of levofloxacin ≤30 days prior to sampling | 2.28 (0.68–7.68) | 0.18 |
| Yr of culture | 3.89 (1.71–8.81) | 0.001 |
| Charlson comorbidity score | 0.80 (0.64–1.01) | 0.055 |
OR, odds ratio; CI, confidence interval.
A majority of ST131 isolates exhibited a specific combination of gyrA and parC replacement mutations, which confirms a close association of the H30 ST131 subclone with a distinctive gyrA and parC allele combination among FQREC clinical isolates, consistent with the largely single-strain origin of FQ resistance within ST131 E. coli isolates (8). Furthermore, all 19 clonally related study isolates were ST131, suggesting that ST131 isolates, or those of the particular pulsotypes observed here, may be characterized by increased transmissibility compared with other E. coli strains. However, future studies are needed to evaluate the potential increased risk of dissemination in the clinical setting. In addition, the predominance of pulsotype 968 (which is associated specifically with FQ resistance) among both colonizing and clinical isolates (24) suggests that this particular pulsotype may exhibit greater intestinal fitness than others. This possible intestinal fitness advantage of pulsotype 968, which has become predominant in the later years in the emergence of ST131 may have contributed to the recent expansion of ST131 in the clinical setting (24). However, further research is needed to assess this hypothesized association.
In conclusion, our findings demonstrate a dramatically increasing prevalence of ST131 over time (2002 to 2004) among hospitalized patients with FQREC gastrointestinal tract colonization. Our results highlight the importance of evaluation for potentially increased transmissibility of ST131 isolates, including specifically in the hospital setting, and of the development and implementation of infection control interventions to reduce such spread if it is documented.
ACKNOWLEDGMENTS
Technical assistance by Charles Garrigan is greatly appreciated.
This article is based upon work supported by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs, grant 1 I01 CX000192 01 (to J.R.J.). This work was also supported by a Public Health Service grant from the Centers for Disease Control and Prevention (RS1/CCR320627-01 to E.L.), the National Institutes of Health (K01-AI103028 to J.H.H. and K24-AI080942 to E.L.), a Commonwealth Universal Research Enhancement Program grant from the Pennsylvania State Department of Health (to E.L.), and the CDC Prevention Epicenters Program (U54-CK000163 to E.L.).
The funding agencies had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data or preparation, review, or approval of the manuscript.
J.R.J. has had research grants or contracts with ICET, Merck, Rochester Medical, Crucell, and Syntiron, and has patent applications for diagnostic tests to detect ST131 and other E. coli clonal groups.
Footnotes
Published ahead of print 8 September 2014
REFERENCES
- 1.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin. Infect. Dis. 51:286–294. 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- 2.Peirano G, Pitout JD. 2010. Molecular epidemiology of Escherichia coli producing CTX-M beta-lactamases: the worldwide emergence of clone ST131 O25:H4. Int. J. Antimicrob. Agents 35:316–321. 10.1016/j.ijantimicag.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66:1–14. 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 4.Lautenbach E. 2013. Flying under the radar: the stealth pandemic of Escherichia coli sequence type 131. Clin. Infect. Dis. 57:1266–1269. 10.1093/cid/cit505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colpan A, Johnston B, Porter S, Clabots C, Anway R, Thao L, Kuskowski MA, Tchesnokova V, Sokurenko EV, Johnson JR. 2013. Escherichia coli sequence type 131 (ST131) subclone H30 as an emergent multidrug-resistant pathogen among U.S. veterans. Clin. Infect. Dis. 57:1256–1265. 10.1093/cid/cit503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nowrouzian FL, Adlerberth I, Wold AE. 2006. Enhanced persistence in the colonic microbiota of Escherichia coli strains belonging to phylogenetic group B2: role of virulence factors and adherence to colonic cells. Microbes Infect. 8:834–840. 10.1016/j.micinf.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Vimont S, Boyd A, Bleibtreu A, Bens M, Goujon JM, Garry L, Clermont O, Denamur E, Arlet G, Vandewalle A. 2012. The CTX-M-15-producing Escherichia coli clone O25b:H4-ST131 has high intestine colonization and urinary tract infection abilities. PLoS One 7:e46547. 10.1371/journal.pone.0046547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson JR, Tchesnokova V, Johnston B, Clabots C, Roberts PL, Billig M, Riddell K, Rogers P, Qin X, Butler-Wu S, Price LB, Aziz M, Nicolas-Chanoine MH, DebRoy C, Robicsek A, Hansen G, Urban C, Platell J, Trott DJ, Zhanel G, Weissman SJ, Cookson BT, Fang FC, Limaye AP, Scholes D, Chattopadhyay S, Hooper DC, Sokurenko EV. 2013. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J. Infect. Dis. 207:919–928. 10.1093/infdis/jis933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banerjee R, Johnston B, Lohse C, Porter SB, Clabots C, Johnson JR. 2013. Escherichia coli sequence type 131 is a dominant, antimicrobial-resistant clonal group associated with healthcare and elderly hosts. Infect. Control Hosp. Epidemiol. 34:361–369. 10.1086/669865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung HC, Lai CH, Lin JN, Huang CK, Liang SH, Chen WF, Shih YC, Lin HH, Wang JL. 2012. Bacteremia caused by extended-spectrum-beta-lactamase-producing Escherichia coli sequence type ST131 and non-ST131 clones: comparison of demographic data, clinical features, and mortality. Antimicrob. Agents Chemother. 56:618–622. 10.1128/AAC.05753-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lautenbach E, Fishman NO, Metlay JP, Mao X, Bilker WB, Tolomeo P, Nachamkin I. 2006. Phenotypic and genotypic characterization of fecal Escherichia coli isolates with decreased susceptibility to fluoroquinolones: results from a large hospital-based surveillance initiative. J. Infect. Dis. 194:79–85. 10.1086/503046. [DOI] [PubMed] [Google Scholar]
- 12.Lautenbach E, Metlay JP, Weiner MG, Bilker WB, Tolomeo P, Mao X, Nachamkin I, Fishman NO. 2009. Gastrointestinal tract colonization with fluoroquinolone-resistant Escherichia coli in hospitalized patients: changes over time in risk factors for resistance. Infect. Control Hosp. Epidemiol. 30:18–24. 10.1086/592703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han JH, Nachamkin I, Tolomeo P, Mao X, Bilker WB, Lautenbach E. 2012. Risk factors for efflux pump overexpression in fluoroquinolone-resistant Escherichia coli. J. Infect. Dis. 206:1597–1603. 10.1093/infdis/jis567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White DG, Goldman JD, Demple B, Levy SB. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179:6122–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Dzink-Fox JL, Chen M, Levy SB. 2001. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob. Agents Chemother. 45:1515–1521. 10.1128/AAC.45.5.1515-1521.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goering RV, Tenover FC. 1997. Epidemiological interpretation of chromosomal macro-restriction fragment patterns analyzed by pulsed-field gel electrophoresis. J. Clin. Microbiol. 35:2432–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558. 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson JR, Menard M, Johnston B, Kuskowski MA, Nichol K, Zhanel GG. 2009. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002 to 2004. Antimicrob. Agents Chemother. 53:2733–2739. 10.1128/AAC.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clermont O, Lavollay M, Vimont S, Deschamps C, Forestier C, Branger C, Denamur E, Arlet G. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J. Antimicrob. Chemother. 61:1024–1028. 10.1093/jac/dkn084. [DOI] [PubMed] [Google Scholar]
- 20.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 4(6):e00377–13. 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banerjee R, Robicsek A, Kuskowski MA, Porter S, Johnston BD, Sokurenko E, Tchesnokova V, Price LB, Johnson JR. 2013. Molecular epidemiology of Escherichia coli sequence type 131 and its H30 and H30-Rx subclones among extended-spectrum-beta-lactamase-positive and -negative E. coli clinical isolates from the Chicago region, 2007 to 2010. Antimicrob. Agents Chemother. 57:6385–6388. 10.1128/AAC.01604-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosmer DW, Lemeshow S. 1989. Applied logistic regression. Wiley and Sons, New York, NY. [Google Scholar]
- 23.Mickey RM, Greenland S. 1989. The impact of confounder selection criteria on effect estimation. Am. J. Epidemiol. 129:125–137. [DOI] [PubMed] [Google Scholar]
- 24.Johnson JR, Nicolas-Chanoine MH, DebRoy C, Castanheira M, Robicsek A, Hansen G, Weissman S, Urban C, Platell J, Trott D, Zhanel G, Clabots C, Johnston BD, Kuskowski MA. 2012. Comparison of Escherichia coli ST131 pulsotypes, by epidemiologic traits, 1967–2009. Emerg. Infect. Dis. 18:598–607. 10.3201/eid1804.111627. [DOI] [PMC free article] [PubMed] [Google Scholar]