Abstract
β-Lactam antibiotics were the earliest discovered and are the most widely used group of antibiotics that work by inactivating penicillin-binding proteins to inhibit peptidoglycan biosynthesis. As one of the most efficient defense strategies, many bacteria produce β-lactam-degrading enzymes, β-lactamases, whose biochemical functions and regulation have been extensively studied. A signal transduction pathway for β-lactamase induction by β-lactam antibiotics, consisting of the major peptidoglycan recycling enzymes and the LysR-type transcriptional regulator, AmpR, has been recently unveiled in some bacteria. Because inactivation of some of these proteins, especially the permease AmpG and the β-hexosaminidase NagZ, results in substantially elevated susceptibility to the antibiotics, these have been recognized as potential therapeutic targets. Here, we show a contrasting scenario in Shewanella oneidensis, in which the homologue of AmpR is absent. Loss of AmpG or NagZ enhances β-lactam resistance drastically, whereas other identified major peptidoglycan recycling enzymes are dispensable. Moreover, our data indicate that there exists a parallel signal transduction pathway for β-lactamase induction, which is independent of either AmpG or NagZ.
INTRODUCTION
The production of β-lactamase is one of the most important mechanisms for Gram-negative bacteria to cope with the threat derived from β-lactams. The β-lactam antibiotics, which mimic the d-Ala-d-Ala dipeptide in an elongated conformation, covalently bind to the serine active center of penicillin-binding proteins (PBPs), eventually blocking the formation of cross-links during peptidoglycan synthesis (1). β-Lactamases share an ancestor with PBPs but function to degrade β-lactams (2). Due to the prolonged use of β-lactams, β-lactamase production has been acquired by many bacteria, especially pathogens, leading to therapeutic failures.
Based on the amino acid sequence, β-lactamases can be classified as types A to D. Classes A, C, and D are serine β-lactamases, while class B is a metallo-β-lactamase (3). To date, studies on the regulation of β-lactamase expression have been restricted to AmpC, an inducible chromosomal-mediated class C cephalosporinase, as in Enterobacter cloacae, Citrobacter freundii, and Pseudomonas aeruginosa (4, 5). AmpR, a LysR-family transcriptional regulator, plays a central role in regulating expression of AmpC. Generally, genes encoding AmpR and AmpC are found in a divergent operon, and the DNA-binding domain of AmpR interacts with the intergenic region and influences the transcription of both genes. The effector-binding domain of AmpR has the ability to work with two regulatory ligands: an activator ligand (AL) and a repressor ligand (RL). Both ligands are intermediate products derived from peptidoglycan recycling. Therefore, the induction of AmpC is intimately linked to peptidoglycan recycling (5–7).
Under normal conditions, GlcNAc-1,6-anhydromuropeptides, products of peptidoglycan-hydrolyzing enzymes, enter the cytoplasm through the inner membrane permease, AmpG, and are then processed by NagZ and AmpD to generate GlcNAc, anhydro-N-acetylmuramic acid (anhMurNAc), and the tripeptide l-Ala-d-Glu-meso-diaminopimelate (meso-A2pm). Both the tripeptide and dipeptide d-Ala-d-Ala can be further added onto UDP-MurNAc successively by Mpl and MurF, leading to formation of UDP-MurNAc pentapeptide for peptidoglycan precursor synthesis (8–10). UDP-MurNAc pentapeptide serves as an RL to mediate the repression of AmpC. In the presence of an inducer (β-lactams), perturbation of peptidoglycan synthesis sharply increases the amount of GlcNAc-anhMurNAc peptides, and, consequently, anhMurNAc peptides accumulate in the cytoplasm. The anhMurNAc peptides act as an AL for AmpC induction, because they could displace an RL from the effector-binding domain of AmpR, resulting in the activation of AmpR (5–7, 11). Induction of AmpC also requires a cytoplasmic membrane protein AmpE, which is encoded by the ampE gene from the ampDE operon (12, 13). Furthermore, recent studies demonstrate that periplasmic peptidoglycan-remodeling enzymes also modulate the expression of AmpC, such as low-molecular-weight PBP4 and lytic transglycosylases (LTs) in P. aeruginosa (14, 15).
The paradigm of AmpC induction is applicable to other classes of β-lactamases, for example, class A β-lactamase L2 and class B β-lactamase L1 in Stenotrophomonas maltophilia (16). In addition, peptidoglycan fragments are found to be involved in the regulation of β-lactamase expression in Gram-positive bacteria, although the regulatory mechanism is quite different from that of AmpC (17). These observations suggest that the involvement of peptidoglycan recycling is a generic mechanism in β-lactamase expression.
Shewanella species are Gram-negative facultative anaerobes that have a wide distribution in marine and freshwater environments. Because of the metabolic versatility, bacteria in the genus have been intensively studied, especially the model strain Shewanella oneidensis MR-1 (18). Meanwhile, Shewanella strains are gradually emerging as human and animal pathogens as reports of Shewanella infections have been increasing in recent years (19, 20). In addition, Shewanella species are regarded as a reservoir of antibiotic resistance determinants because a number of naturally occurring β-lactamases and Qnr-type quinolone resistance cassettes have been isolated (21–25). Noticeably, S. oneidensis possesses seven genes predicted to encode β-lactamases, including AmpC and class D β-lactamase BlaA (also referred to as OXA-54) (22, 26). BlaA, along with its analogue from Shewanella xiamenensis (OXA-181), is proposed to be the progenitor of carbapenem-hydrolyzing oxacillinases, which compromise the therapeutic efficiency of carbapenems in clinically relevant Gram-negative pathogens (22, 25). While the physiological relevance of AmpC is yet to be determined, BlaA is the key enzyme conferring S. oneidensis resistance to some β-lactams (26). The S. oneidensis blaA gene is inducible by ampicillin at high levels, like ampC genes in other enterobacteria, but the former differs from the latter in that it does not form a divergon with the gene coding for the transcriptional regulator. Besides, the gene encoding an AmpR homologue is absent in the S. oneidensis genome, implicating a possibility of an uncommon regulatory mechanism (26, 27).
In this study, we made attempts to investigate the roles of peptidoglycan recycling proteins in the susceptibility of S. oneidensis to β-lactams. We found that inactivation of NagZ and AmpG elevated the resistance dramatically, contrasting with the reduced resistance associated with their counterparts characterized thus far. The unexpected phenotype was then linked to BlaA, indicating that the induction of the S. oneidensis β-lactamase is also modulated by peptidoglycan recycling enzymes. Unlike the AmpG of P. aeruginosa, the S. oneidensis counterpart does not appear to be a primary target of carbonyl cyanide m-chlorophenylhydrazone (CCCP), an inhibitor of the proton motive force. Furthermore, cells lacking either NagZ or AmpG retained the ability to respond to ampicillin, suggesting the presence of a parallel pathway that is independent of these two proteins for BlaA induction in S. oneidensis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Information for all of the primers used in this study is available upon request. All chemicals were acquired from Sigma (Shanghai, China) unless otherwise noted. For genetic manipulation, S. oneidensis and Escherichia coli were cultivated aerobically in Luria-Bertani (LB) medium (Difco, Detroit, MI) at 30°C and 37°C, respectively. When needed, the growth medium was supplemented with chemicals at the following concentrations: 2,6-diaminopimelic acid, 0.3 mM; ampicillin, 100 mg/ml; kanamycin, 50 mg/ml; and gentamicin, 15 mg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Organism and strain | ||
| E. coli | ||
| DH5α | Host strain for plasmids | Lab stock |
| WM3064 | Donor strain for conjugation; ΔdapA | W. Metcalf, UIUCa |
| P. aeruginosa | ||
| PAO1 | Reference strain | 36 |
| PAO1ΔDDh2Dh3 | PAO1 with the three ampD homologues deleted | 36 |
| PAO1ΔG | PAO1 with ampG (PA4393) deleted | 37 |
| S. oneidensis | ||
| MR-1 | Wild type | Lab stock |
| HG0421 | ΔampD derived from MR-1 | This study |
| HG0422 | ΔampE derived from MR-1 | This study |
| HG0837 | ΔblaA derived from MR-1 | 26 |
| HG2250 | ΔnagZ derived from MR-1 | This study |
| HG3814 | ΔampG derived from MR-1 | This study |
| HG2250–0837 | ΔnagZΔblaA derived from MR-1 | This study |
| HG3814–0837 | ΔampGΔblaA derived from MR-1 | This study |
| HG0421–2250 | ΔampDΔnagZ derived from MR-1 | This study |
| Plasmids | ||
| pHGM01 | Apr Gmr Cmr suicide vector | 29 |
| pHG101 | Promoterless vector for complementation | 30 |
| pHGEI01 | Integrative lacZ reporter vector | 31 |
| pTP327-PblaA | pTP327 containing the blaA promoter | 26 |
| pHGEI01-PblaA | pHGI01 containing the blaA promoter | This study |
UIUC, University of Illinois at Urbana-Champaign.
Antibiotic susceptibility assay.
The MICs of the antibiotics were determined by the broth microdilution method as recommended by the Clinical and Laboratory Standards Institute (CLSI) (28). The antibiotics used in the susceptibility assay were ampicillin (AMP), cefotaxime (CTX), and imipenem (IPM). All MICs were determined at least in triplicate.
In-frame deletion mutagenesis and complementation.
The in-frame deletion strains of S. oneidensis were constructed by the att-based fusion PCR method (29). In brief, two fragments flanking the targeted gene were amplified by PCR with primers containing attB and a gene-specific sequence and then were joined by a second round of PCR (fusion PCR). The fusion fragments were introduced into plasmid pHGM01 by site-specific recombination using BP Clonase (Invitrogen) and then transformed into E. coli WM3064. The resulting mutagenesis vectors were transferred into proper S. oneidensis strains via conjugation. Integration of the mutagenized constructs into the chromosome was selected by resistance to gentamicin and verified by PCR. The correct transconjugants were grown in LB broth in the absence of NaCl and plated onto LB medium supplemented with 10% sucrose. Gentamicin-sensitive and sucrose-resistant colonies were screened by PCR for deletion of the target gene. All mutants were verified by sequencing of the mutated regions.
The plasmid pHG101 was used for the genetic complementation of mutants (30). A fragment containing the gene of interest and its native promoter was amplified by PCR and cloned into pHG101. The resulting complementation vector was transferred into its corresponding mutant strain via conjugation, and its presence was confirmed by plasmid purification and restriction enzyme digestion.
Promoter activity assay.
The activity of the PblaA promoter within a multicopy plasmid was assayed previously (26). In this study, we moved the transcriptional lacZ fusion into a markerless integrative reporter system, allowing us to examine the activity of the promoter without interferences from the copy number difference and the antibiotic resistance gene (31). The sequence of the target promoter (∼400 bp) was amplified and cloned into the transcriptional fusion vector pHGEI01 (31). The resultant plasmid was then transferred into S. oneidensis strains via conjugation. Proper integration of the promoter fusion constructs was confirmed by PCR. Overnight cultures were diluted 1:100 in 3 ml fresh LB medium in the absence and presence of ampicillin at 2.5 μg/ml or 50 μg/ml. The cultures were allowed to incubate for an additional 3 h at 30°C and 200 rpm. Following incubation, cells were harvested by centrifugation at 4°C, washed with phosphate-buffered saline (PBS), and treated with lysis buffer (0.25 M Tris-HCl [pH 7.5], 0.5% Triton X-100). The protein concentration of the cell lysates was determined using a Bradford assay with bovine serum albumin (BSA) as a standard (Bio-Rad). β-Galactosidase activity was determined by monitoring color development at 420 nm using a Sunrise microplate reader (Tecan) as described previously (30).
BlaA β-lactamase activity assay.
The BlaA β-lactamase activity was measured by nitrocefin hydrolysis as described previously (15). Overnight bacterial cultures were diluted 1:100 in 3 ml fresh LB medium and grown at 30°C and 200 rpm. Two concentrations of ampicillin (50 and 200 μg/ml, final concentration) were added to the early-exponential-phase cultures (optical density at 600 nm [OD600] of ≈ 0.2) for blaA induction. The cultures were incubated for an additional 2 h at 30°C and 200 rpm. After incubation, 1 ml culture was centrifuged at 2,500 × g for 5 min, washed once with 1 ml 50 mM phosphate buffer (pH 7.0), and resuspended in 1 ml of the same buffer. Crude cell extracts were prepared by sonication (pulse on, 3 s; pulse off, 4 s; 10 times). The protein content of the crude extracts was determined using a Bradford assay with BSA as a standard (Bio-Rad). The nitrocefin hydrolysis assays were performed in 200-μl reaction mixtures containing 8 μg total protein and 4 μg nitrocefin (Calbiochem, San Diego, CA). Nitrocefin hydrolysis was measured every minute for 10 min at 25°C by absorbance at 486 nm. The specific β-lactamase activity was expressed as nanomoles of nitrocefin hydrolyzed per minute per milligram of protein. All induction experiments were performed in triplicate, and the results presented are averages from the three experiments.
Other analyses.
The topology of the protein of interest was predicted by using TMHMM (32). The experimental values were subjected to statistical analyses, and the results are presented as means ± standard deviations. The Student's t test was performed for pairwise comparisons of groups.
RESULTS
Peptidoglycan recycling enzymes in S. oneidensis.
Most of the functionally characterized peptidoglycan recycling enzymes are conserved in Gram-negative bacteria. Not surprisingly, a set of enzymes involved in the peptidoglycan recycling are present in all of the Shewanella strains sequenced so far. These include AmpG, AmpD, Mpl, NagZ, NagK, AnmK, and SltY, but not AmiD, LdcA, MurQ, MpaA, or YcjG (Table 2) (9). This suggests that the pathway for peptidoglycan recycling in Shewanella species consists of the core components documented in the paradigm, but at the same time deviation appears significant. Interestingly, S. oneidensis lacks a homologue of AmpR (26, 27).
TABLE 2.
Peptidoglycan recycling enzymes in S. oneidensis
| Locus | Gene | Location | Product | Identity (%)a |
|---|---|---|---|---|
| SO0421 | ampD | Cytoplasm | N-Acetyl-anhydromuramyl-l-Ala amidase | 63 |
| SO1313 | anmK | Cytoplasm | anhMurNAc kinase | 49 |
| SO1656 | nagK | Cytoplasm | GlcNAc kinase | 41 |
| SO2040 | sltY | Periplasm | Lytic transglycosylase | 29 |
| SO2250 | nagZ | Cytoplasm | exo-N-Acetyl-β-glucosaminidase | 50 |
| SO3805 | mpl | Cytoplasm | Mpl proteinb | 64 |
| SO3814 | ampG | CMc | GlcNAc-anhMurNAc permease | 34 |
Compared to E. coli counterparts.
UDP-N-acetylmuramate: l-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase.
CM, cytoplasmic membrane.
In S. oneidensis, the SO3814 gene encodes a protein of 461 amino acids (aa), sharing 34% sequence identity and 53% similarity with E. coli AmpG. Like its counterparts in other bacteria, SO3814 is predicted to be an integral membrane protein with 12 transmembrane segments. In parallel, there is 63% homology between SO0421 and E. coli AmpD, suggesting that SO0421 may be the cytoplasmic anhydro-N-acetyl-muramyl-l-Ala amidase. Generally, genes encoding AmpD and AmpE form an operon in enterobacteria (13). However, whether this is the case in S. oneidensis is uncertain, given that the intergenic region between SO0421 and SO0422 (predicted to encode the homologue of AmpE) is rather lengthy (270 bp). The SO3805 gene product shares 64% homology with E. coli Mpl, a specific enzyme for the ligation of tripeptide l-Ala-d-Glu-meso-A2pm with UDP-MurNAc during peptidoglycan recycling (8). In addition, some enzymes for recycling amino sugars are also found in S. oneidensis. For example, SO1313, SO1656, SO2040, and SO2250 share 49%, 41%, 29%, and 50% sequence identity with AnmK, NagK, SltY, and NagZ of E. coli, respectively. Based on the high levels of sequence identity, we designated SO3814, SO0421, SO0422, SO3805, SO1313, SO1656, SO2040, and SO2250 of S. oneidensis as ampG, ampD, ampE, mpl, anmK, nagK, sltY, and nagZ, respectively. Given that the AmpG-AmpD-NagZ network is intimately involved in the regulation of AmpC expression, we focus on these proteins in this study.
Deletion of nagZ or ampG increases β-lactam resistance.
S. oneidensis is well known to be naturally resistant to ampicillin, which is mainly attributed to the production of BlaA (26). To study whether the expression of BlaA is related to peptidoglycan recycling, strains lacking the ampD, ampE, nagZ, and ampG genes were constructed and subjected to MIC determination for ampicillin. Deletion of the ampD and ampE genes resulted in susceptibility to ampicillin at levels comparable to that of the wild type. In contrast, both the ΔnagZ and ΔampG strains exhibited significantly improved resistance to ampicillin (especially the latter, which had the strongest resistance among all tested strains). Compared to the wild type, the ΔampG and ΔnagZ strains had 32- and 8-fold increases in ampicillin MICs, respectively (Table 3).
TABLE 3.
MICs of β-lactams and activities of BlaA in S. oneidensis and P. aeruginosa
| Strain or mutationa | MIC (μg/ml) forb: |
β-Lactamase activity (mean ± SD) withc: |
||||
|---|---|---|---|---|---|---|
| AMP | CTX | IMP | No inducer | Inducer (50 μg/ml) | Inducer (200 μg/ml) | |
| WT | 4 | 0.02 | 0.5 | 7 ± 1.1 | 25 ± 1.8 | 71 ± 7.6 |
| ΔampD | 4 | 0.02 | 0.5 | 7 ± 1.7 | 29 ± 3.3 | 68 ± 10 |
| ΔampE | 4 | 0.02 | 0.5 | 6 ± 0.6 | 22 ± 3 | 76 ± 10 |
| ΔnagZ | 32 | 0.04 | 2 | 11 ± 1.2 | 54 ± 2.7 | 147 ± 14 |
| ΔampG | 128 | 0.08 | 2 | 13 ± 2.2 | 131 ± 10 | 168 ± 8 |
| ΔblaA | <0.5 | <0.005 | <0.06 | 2 ± 0.3 | 0 | 0 |
| ΔampDΔnagZ | 32 | 0.04 | 2 | 7 ± 0.2 | 75 ± 4.9 | 150 ± 10 |
| ΔnagZ/pHG101 | 32 | 0.04 | 2 | —d | — | — |
| ΔampG/pHG101 | 128 | 0.08 | 2 | — | — | — |
| ΔnagZ/nagZ | 4 | 0.02 | 0.5 | 8 ± 0.7 | 19 ± 1.4 | 59 ± 0.7 |
| ΔampG/ampG | 4 | 0.02 | 0.5 | 6 ± 0.2 | 8 ± 0.9 | 45 ± 3.3 |
| ΔnagZΔblaA | <0.5 | <0.005 | <0.06 | 2 ± 0.4 | 0 | 0 |
| ΔampGΔblaA | <0.5 | <0.005 | <0.06 | 1 ± 0.5 | 0 | 0 |
| ΔnagZ/nagZEC | 4 | 0.02 | 0.5 | — | — | — |
| ΔampG/ampGEC | 4 | 0.04 | 1 | — | — | — |
| PAO1 | 256 | — | — | — | — | — |
| PAO1ΔDDh2Dh3 | 1,024 | — | — | — | — | — |
| PAO1ΔDDh2Dh3/ampDSO | 256 | — | — | — | — | — |
| PAO1ΔG | 32 | — | — | — | — | — |
| PAO1ΔG/ampGSO | 256 | — | — | — | — | — |
ΔnagZ/nagZ and ΔampG/ampG represent ΔnagZ and ΔampG strains, respectively, that were complemented with S. oneidensis genes in trans. ΔnagZ/nagZEC and ΔampG/ampGEC represent ΔnagZ and ΔampG strains, respectively, that were complemented with E. coli genes in trans. PAO1ΔDDh2Dh3/ampDSO and PAO1ΔG/ampGSO represent PAO1ΔDDh2Dh3 and PAO1ΔG, respectively, that were complemented with S. oneidensis genes in trans.
AMP, ampicillin; CTX, cefotaxime; IMP, imipenem.
Nanomoles of nitrocefin hydrolyzed per minute per milligram of protein. Induction was carried out with ampicillin for 2 h.
—, not tested.
To evaluate the effects of these peptidoglycan recycling enzymes on resistance to other β-lactams, the MICs for cefotaxime and imipenem were determined (Table 3). Consistently, deletion of the ampG and nagZ genes improved the resistance to both of these β-lactams, while the resistance of strains lacking AmpD or AmpE was not significantly altered. Compared to the wild type, the ΔampG strain had a 4-fold increase in resistance to both β-lactams, while the ΔnagZ strain had 4- and 2-fold increases in resistance to cefotaxime and imipenem, respectively.
More importantly, expression of the nagZ and ampG genes under the control of their native promoters in pHG101 reversed all β-lactam susceptibility to the level of the wild type, indicating that the phenotypes observed in the ΔnagZ and ΔampG strains were due to the mutation per se (Table 3). These data, collectively, indicate that NagZ and AmpG are important players mediating the β-lactam resistance in S. oneidensis, whereas AmpD and AmpE hardly influence the process.
Increased β-lactam resistance is dependent on BlaA.
Given that BlaA is the main cause of the resistance of S. oneidensis to ampicillin, we reasoned that the improved β-lactam resistance of the ΔnagZ and ΔampG strains likely results from enhanced expression of the blaA gene. To validate this, the blaA gene was deleted from the ΔnagZ and ΔampG strains. As expected, the loss of BlaA completely abolished the β-lactam resistance of the ΔnagZ and ΔampG strains (Table 3).
Expression of the blaA gene is induced by ampicillin only at high levels (26). To monitor the impact of the loss of NagZ or AmpG on expression of the blaA gene, a markerless integrative lacZ-reporter system was employed to assess the activity of the blaA promoter (PblaA) in the absence and presence of ampicillin (31). As shown in Fig. 1, the expression of β-galactosidase driven by the blaA promoter in the ΔampD and ΔampE strains was similar to that in the wild type under all conditions. However, deletion of the nagZ and ampG genes conclusively resulted in increased activities of PblaA. Nonetheless, the induction effect of ampicillin was still apparent. In the presence of 2.5 μg/ml ampicillin, PblaA activity in the ΔnagZ and ΔampG strains equated approximately to that in the wild type induced by ampicillin at 50 μg/ml. Notably, the ΔampG strain had the highest activity when induced by 50 μg/ml ampicillin.
FIG 1.
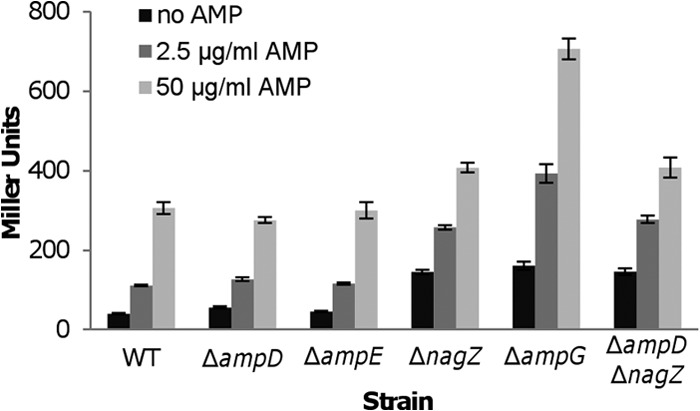
Improved ampicillin resistance in nagZ and ampG mutants is dependent on the expression of BlaA. PblaA promoter activities were determined by measuring β-galactosidase (in Miller units) using the PblaA-lacZ reporter system in ΔampD, ΔampE, ΔnagZ, ΔampG, and ΔampDΔnagZ strains in the absence and presence of ampicillin at 50 μg/ml and 2.5 μg/ml. Results are averages from at least three replicates, and the error bars represent standard deviations (SD). WT, wild type.
To confirm, BlaA activity was measured directly by nitrocefin hydrolysis. This method is specific for BlaA as the activity was hardly detected in strains without BlaA (Table 3). Consistent with the data from the lacZ reporter shown above, the basal levels of BlaA activity in the ΔnagZ and ΔampG strains were significantly higher (to ∼2-fold) than those in the wild-type and other tested mutant strains. By induction, the activities of BlaA increased proportionally with ampicillin concentrations in all strains. In the wild-type, ΔampD, and ΔampE strains, induction by ampicillin at 50 μg/ml and 200 μg/ml led to ∼3.6-fold and ∼10-fold increases, respectively, compared to the corresponding basal levels. On the contrary, the BlaA activities in the ΔnagZ strain were ∼2-fold higher than those in the wild-type strain when induced by ampicillin at the same concentrations. The effect of the ampicillin induction on the BlaA activities in the ΔampG strain was the most drastic, especially with the antibiotic dose of 50 μg/ml (∼5.2-fold compared to that in the wild type). In addition, expression of the nagZ and ampG genes in trans conferred on the corresponding mutants the lowest levels of BlaA activity under all tested conditions. This can be readily explained by overexpression, an intrinsic feature of the multiple-copy vector pHG101 (31, 33, 34). Based on these results, we conclude that NagZ and AmpG exert a repressive effect on the blaA gene, leading to the elevated resistance to ampicillin at all levels. More importantly, it is evident that the blaA gene in the ΔnagZ and ΔampG strains is still responsive to ampicillin, indicating the presence of an alternative pathway for the induction of the blaA gene in S. oneidensis.
Peptidoglycan recycling enzymes of Gram-negative bacteria are exchangeable in functionality.
The effects of NagZ, AmpG, and AmpD on β-lactamase expression in S. oneidensis are in contrast to those in other ampR-ampC-containing bacteria, prompting us to ascertain whether these peptidoglycan recycling enzymes are actually the S. oneidensis counterparts. At the beginning, the nagZ and ampG genes from E. coli (nagZEC and ampGEC, respectively) were cloned into pHG101 and expressed in the corresponding mutants of S. oneidensis. Results showed that both ΔnagZ and ΔampG strains harboring their E. coli homologues decreased the resistance to β-lactams (Table 3), indicating that NagZEC and AmpGEC are functional in S. oneidensis.
However, E. coli is not applicable for testing the function of enzymes from S. oneidensis, since AmpR, the regulator of ampC induction, is absent from the bacterium (35). Instead, the whole ampR-ampC operon is found in P. aeruginosa, which shares conserved peptidoglycan recycling enzymes with E. coli (9, 36, 37). Therefore, the ampD and ampG genes from S. oneidensis (ampDSO and ampGSO, respectively) were expressed in the P. aeruginosa PAO1ΔDDh2Dh3 and PAO1ΔG strains, respectively (36, 37). As expected, the expression of AmpDSO and AmpGSO restored the ampicillin resistance comparable to that of the parent strain (PAO1) (Table 3), indicating that AmpDSO and AmpGSO are functional replacements for P. aeruginosa AmpD and AmpG. Taken together, these results confirm that NagZ, AmpG, and AmpD in S. oneidensis, like their counterparts in other Gram-negative bacteria, are major peptidoglycan recycling enzymes.
Effect of CCCP on ampicillin resistance.
In some enterobacteria, AmpG is a permease responsible for the transportation of signal molecules for ampC induction; thus, inactivation of AmpG drastically reduces the β-lactam resistance (37). In P. aeruginosa, the proton motive force inhibitor CCCP can effectively annul the function of AmpG, leading to an increased sensitivity to β-lactams (37, 38). As our data demonstrate improved β-lactam resistance in the absence of AmpG in S. oneidensis, whether CCCP inhibits S. oneidensis AmpG deserves verification. Interestingly, CCCP exhibited excellent antimicrobial properties against S. oneidensis, with an MIC of <5 μM compared to 250 μM for P. aeruginosa PAO1 (data not shown) (37). The addition of CCCP at subinhibitory concentrations did not elicit a noticeable phenotypic change with respect to ampicillin susceptibility in the wild-type, ΔampG, and ΔampG strains carrying a copy of the ampG gene for complementation (Table 4). Consistently, the BlaA activity was not altered in these strains in the presence of CCCP (Table 4). Although it is possible that the concentration of CCCP used in the test is too low to inhibit AmpG, the results imply that the AmpG of S. oneidensis, unlike its counterpart in P. aeruginosa, is not a primary target of CCCP.
TABLE 4.
Effects of CCCP (1 μM) on ampicillin resistance and BlaA expression in S. oneidensis wild type and its derivate strains
| Strain or mutationa | MIC (μg/ml) | β-Lactamase activity (mean ± SD) withb: |
|
|---|---|---|---|
| No inducer | Inducer (200 μg/ml) | ||
| WT | 16 | 7 ± 1.3 | 69 ± 6.4 |
| ΔampG | 128 | 12 ± 2.7 | 178 ± 23 |
| ΔampG/ampG | 16 | 8 ± 1.6 | 58 ± 7 |
ΔampG/ampG represents the ΔampG strain that was complemented with S. oneidensis genes in trans.
Nanomoles of nitrocefin hydrolyzed per minute per milligram of protein. Induction was carried out with ampicillin for 2 h.
NagZ functionally overwhelms AmpD in induction of BlaA.
Both AmpD and NagZ have the ability to catalyze the hydrolysis of anhydromuropeptides in the cytoplasm (6, 39). Deletion of the ampD gene produces a negligible impact on the S. oneidensis resistance to β-lactams; however, this may be due to the presence of the more robust enzyme NagZ. To test this notion, the ampD gene was removed from the ΔnagZ strain, and the resulting double mutant was subjected to an antibiotic susceptibility test. As shown in Table 3, the MICs of β-lactams for the ΔampDΔnagZ strain were also the same as those for the ΔnagZ mutant, which had 8-, 4-, and 2-fold increases in ampicillin, imipenem, and cefotaxime resistance, respectively, compared to those for the wild type (Table 3), suggesting that AmpD does not play an indispensable role in the absence of NagZ. Furthermore, the promoter activity of the blaA gene and the BlaA activity in the ΔampDΔnagZ strain were not significantly different from those in the ΔnagZ strain (Fig. 1 and Table 3). Based on these data, we conclude that NagZ plays a dominant role in BlaA induction, whereas AmpD is largely redundant, at least in the context of β-lactam resistance and BlaA induction.
DISCUSSION
Peptidoglycan turnover and recycling are processes that occur throughout the whole bacterial life cycle, including elongation and septation. Nearly half of the E. coli peptidoglycan is recycled per generation, and as a result, a large number of peptidoglycan fragments are produced (9). It has been suggested that peptidoglycan recycling is not essential for bacterial growth, at least under laboratory conditions. Instead, the primary function for the recycling pathway is involvement in the regulation of resistance and virulence response (40, 41). In ampR-ampC divergon-containing bacteria such as several Enterobacteriaceae and P. aeruginosa, the induction of AmpC, a class C β-lactamase, triggered by peptidoglycan recycling intermediates, plays a major role in β-lactam resistance (5–7). Here we present evidence to demonstrate that the induction of class D β-lactamase BlaA of S. oneidensis, a bacterium lacking an AmpR homologue, is also intimately linked to peptidoglycan recycling. However, the effects of the major peptidoglycan recycling enzymes on BlaA expression are distinct from the paradigm of AmpC induction. Loss of AmpG or NagZ but not of AmpD or AmpE significantly increased BlaA expression.
One of the remarkable differences is that inactivation of AmpG results in increased expression of BlaA in S. oneidensis, conferring on the bacterium high-level resistance to β-lactams. AmpG proteins, imbedded in the inner membrane with at least 10 transmembrane-spanning segments (as in E. coli and P. aeruginosa), are responsible for the transportation of the major peptidoglycan recycling products, GlcNAc-anhMurNAc peptides, into the cytoplasm (42, 43). GlcNAc-anhMurNAc peptides are precursors of the signal molecule for AmpC induction (38). The loss of AmpG blocks the induction pathway and causes increased β-lactam sensitivity (37, 38, 44). Strikingly, our results are in contrast to the AmpC paradigm although S. oneidensis AmpG shares a similar topological structure and can functionally replace its E. coli and P. aeruginosa counterparts. Some bacteria harbor more than one AmpG-like protein, implicating multiple pathways for GlcNAc-anhMurNAc peptides. For instance, an additional AmpG homologue is required for maximum induction of chromosomal β-lactamase in P. aeruginosa (37, 43). Given that S. oneidensis, like most other bacteria hosting AmpG, encodes only one such protein, the BlaA hyperexpression in the ΔampG strain likely results from the blockage of GlcNAc-anhMurNAc peptides.
Another distinction between induction of S. oneidensis BlaA and the paradigmatic AmpC relates to the effect of AmpD. AmpD is a cytoplasmic amidase highly specific for cleavage of the anhMurNAc tripeptide, which functions as an AL for AmpC induction (6). Consequently, inactivation of AmpD derepresses AmpC expression (45). However, the loss of AmpD has no significant effect on BlaA expression in S. oneidensis. In agreement with the studies for AmpC induction (12, 13), deletion of the ampE gene alone did not change β-lactamase expression and β-lactam resistance.
The role of NagZ in BlaA expression is also different from that in the AmpC system. In P. aeruginosa, the loss of NagZ attenuates β-lactam resistance even in AmpC hyperexpression strains (ΔampD and/or ΔdacB) (39, 46). In contrast, deletion of the S. oneidensis nagZ gene increases β-lactam resistance via enhancement of both basal level and induced levels of BlaA expression. Compared with the ΔampG strain, the ΔnagZ strain has slightly lower levels of BlaA expression and β-lactam resistance, suggesting the possibility that these two proteins function in the same signal transduction pathway. This proposal is supported by the finding that both the ΔnagZ and ΔampG strains remain inducible by ampicillin. Therefore, although the physiological effects of S. oneidensis AmpG and NagZ are opposite to those of the paradigm counterparts, they may still work as vital components of the same pathway.
Since the loss of AmpG increases the basal level of BlaA expression in S. oneidensis, the accumulation of GlcNAc-anhMurNAc peptides in the cytoplasm may trigger the repression of BlaA expression. In strains lacking NagZ rather than AmpD, BlaA induction is affected, implying that the products of NagZ, GlcNAc, or anhMurNAc peptides are likely to be the actual signals for BlaA repression. The production of these repressed signals has been demonstrated to be derived not only from the catalysis of NagZ but also from the periplasm via AmpG permease (9, 38). In line with this, we found that the ΔampG strain has the strongest β-lactamase expression among all tested strains, indicating that the cytoplasmic levels of the repressed signals in the ΔampG strain are lower than those in the ΔnagZ strain in the presence of an inducer (ampicillin).
S. oneidensis apparently possesses a parallel pathway for BlaA induction, which is independent of NagZ and AmpG. A similar scenario has been recently reported in Shewanella maltophilia, which hosts NagZ-dependent and NagZ-independent pathways (47). However, S. maltophilia differs from S. oneidensis in that its two pathways still rely on AmpG, suggesting that both pathways share the same precursors for signal molecules (47, 48). In S. oneidensis, on the contrary, the presence of the AmpG-independent BlaA induction pathway implies that the signal molecules for this pathway might reside in the periplasm. This would require a two-component system (TCS), which can sense signals in the periplasm and pass them to a DNA-binding response regulator in the cytoplasm for regulation of target genes. One such example is CreBC (or BlrAB) in P. aeruginosa, which is involved in the AmpC expression (14). Given that S. oneidensis encodes a large number of TCS and many remain uncharacterized, the possibility of TCS-mediated signal transduction cannot be excluded. Meanwhile, the possibility that the actual signal molecules are derived from short peptides and free sugars involved in peptidoglycan recycling, whose transportation is generally independent of AmpG permease, cannot be ruled out (9).
Apart from these scenarios, the ampR-blaA divergon of the AmpC paradigm is absent in all members of Shewanella sequenced so far, manifesting another significant difference. We do not yet know whether a homologue of AmpR is encoded in the S. oneidensis genome. However, given that AmpR is a LysR-type transcriptional regulator and large numbers of this same type of regulator are present in S. oneidensis, the possibility of the presence of an AmpR homologue cannot be superficially excluded. We are working to identify the regulators involved in the process.
Based on the AmpC model in Enterobacteriaceae and P. aeruginosa, AmpG and NagZ have been proposed as candidate targets for prevention of β-lactam resistance. In agreement with the hypersusceptibility of the nagZ mutant to β-lactams, the addition of NagZ inhibitors attenuates β-lactam resistance in the AmpC-hyperproducing strains (39, 46, 49, 50). In parallel, CCCP reduces resistance to β-lactams in clinical isolates of P. aeruginosa by inhibiting AmpG (37, 38). However, these means are not practicable for S. oneidensis and probably many more organisms that share features similar to those revealed in this study. Moreover, the class D β-lactamase BlaA represents a group of carbapenem-hydrolyzing oxacillinases that have been acquired by clinically relevant Gram-negative species such as Acinetobacter baumannii and Klebsiella pneumonia (22, 27). It is therefore of particular importance to understand the regulation of β-lactamases in bacteria that are phylogenetically more diverse.
ACKNOWLEDGMENTS
This research was supported by the National Natural Science Foundation of China (31270097), the Major State Basic Research Development Program (973 Program 2010CB833803), and the Doctoral Fund of the Ministry of Education of China (20130101110142).
Footnotes
Published ahead of print 18 August 2014
REFERENCES
- 1.Zapun A, Contreras-Martel C, Vernet T. 2008. Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol. Rev. 32:361–385. 10.1111/j.1574-6976.2007.00095.x. [DOI] [PubMed] [Google Scholar]
- 2.Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. 2006. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 30:673–691. 10.1111/j.1574-6976.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 3.Bush K, Jacoby GA. 2010. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 54:969–976. 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindberg F, Westman L, Normark S. 1985. Regulatory components in Citrobacter freundii ampC beta-lactamase induction. Proc. Natl. Acad. Sci. U. S. A. 82:4620–4624. 10.1073/pnas.82.14.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobs C, Huang L, Bartowsky E, Normark S, Park J. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. EMBO J. 13:4684–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobs C, Joris B, Jamin M, Klarsov K, Beeumen J, Mengin-Lecreulx D, Heijenoort J, Park JT, Normark S, Frère JM. 1995. AmpD, essential for both beta-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-l-alanine amidase. Mol. Microbiol. 15:553–559. 10.1111/j.1365-2958.1995.tb02268.x. [DOI] [PubMed] [Google Scholar]
- 7.Jacobs C, Frère J-M, Normark S. 1997. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible beta-lactam resistance in gram-negative bacteria. Cell 88:823–832. 10.1016/S0092-8674(00)81928-5. [DOI] [PubMed] [Google Scholar]
- 8.Mengin-Lecreulx D, van Heijenoort J, Park JT. 1996. Identification of the mpl gene encoding UDP-N-acetylmuramate: l-alanyl-gamma-d-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J. Bacteriol. 178:5347–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park JT, Uehara T. 2008. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 72:211–227. 10.1128/MMBR.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsutsumi Y, Tomita H, Tanimoto K. 2013. Identification of novel genes responsible for overexpression of ampC in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 57:5987–5993. 10.1128/AAC.01291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dietz H, Pfeifle D, Wiedemann B. 1997. The signal molecule for beta-lactamase induction in Enterobacter cloacae is the anhydromuramyl-pentapeptide. Antimicrob. Agents Chemother. 41:2113–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honoré N, Nicolas MH, Cole S. 1989. Regulation of enterobacterial cephalosporinase production: the role of a membrane-bound sensory transducer. Mol. Microbiol. 3:1121–1130. 10.1111/j.1365-2958.1989.tb00262.x. [DOI] [PubMed] [Google Scholar]
- 13.Juan C, Maciá MD, Gutiérrez O, Vidal C, Pérez JL, Oliver A. 2005. Molecular mechanisms of beta-lactam resistance mediated by AmpC hyperproduction in Pseudomonas aeruginosa clinical strains. Antimicrob. Agents Chemother. 49:4733–4738. 10.1128/AAC.49.11.4733-4738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moya B, Dötsch A, Juan C, Blázquez J, Zamorano L, Haussler S, Oliver A. 2009. Beta-lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog. 5:e1000353. 10.1371/journal.ppat.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavallari JF, Lamers RP, Scheurwater EM, Matos AL, Burrows LL. 2013. Changes to its peptidoglycan-remodeling enzyme repertoire modulate beta-lactam resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 57:3078–3084. 10.1128/AAC.00268-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okazaki A, Avison MB. 2008. Induction of L1 and L2 beta-lactamase production in Stenotrophomonas maltophilia is dependent on an AmpR-type regulator. Antimicrob. Agents Chemother. 52:1525–1528. 10.1128/AAC.01485-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amoroso A, Boudet J, Berzigotti S, Duval V, Teller N, Mengin-Lecreulx D, Luxen A, Simorre J-P, Joris B. 2012. A peptidoglycan fragment triggers beta-lactam resistance in Bacillus licheniformis. PLoS Pathog. 8:e1002571. 10.1371/journal.ppat.1002571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, Nealson KH, Osterman AL, Pinchuk G, Reed JL. 2008. Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 6:592–603. 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 19.Héritier C, Poirel L, Nordmann P. 2004. Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing Ambler class D beta-lactamase from Shewanella algae. Antimicrob. Agents Chemother. 48:1670–1675. 10.1128/AAC.48.5.1670-1675.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Qiao G, Li Q., Zhou W, Won KM, Xu DH, Park SI. 2010. Biological characteristics and pathogenicity of a highly pathogenic Shewanella marisflavi infecting sea cucumber, Apostichopus japonicus. J. Fish Dis. 33:865–877. 10.1111/j.1365-2761.2010.01189.x. [DOI] [PubMed] [Google Scholar]
- 21.Wang D, Wang Y, Huang H, Lin J, Xiao D, Kan B. 2013. Identification of tetrodotoxin-producing Shewanella spp. from feces of food poisoning patients and food samples. Gut Pathog. 5:15. 10.1186/1757-4749-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel L, Héritier C, Nordmann P. 2004. Chromosome-encoded Ambler class D beta-lactamase of Shewanella oneidensis as a progenitor of carbapenem-hydrolyzing oxacillinase. Antimicrob. Agents Chemother. 48:348–351. 10.1128/AAC.48.1.348-351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poirel L, Héritier C, Nordmann P. 2005. Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella livingstonensis (SLB-1) and Shewanella frigidimarina (SFB-1). J. Antimicrob. Chemother. 55:680–685. 10.1093/jac/dki065. [DOI] [PubMed] [Google Scholar]
- 24.Poirel L, Rodriguez-Martinez J-M, Mammeri H, Liard A, Nordmann P. 2005. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob. Agents Chemother. 49:3523–3525. 10.1128/AAC.49.8.3523-3525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potron A, Poirel L, Nordmann P. 2011. Origin of OXA-181, an emerging carbapenem-hydrolyzing oxacillinase, as a chromosomal gene in Shewanella xiamenensis. Antimicrob. Agents Chemother. 55:4405–4407. 10.1128/AAC.00681-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin J, Sun L, Dong Y, Chi X, Zhu W, Qi S-h, Gao H. 2013. Expression of blaA underlies unexpected ampicillin-induced cell lysis of Shewanella oneidensis. PLoS One 8:e60460. 10.1371/journal.pone.0060460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walther-Rasmussen J, Hoiby N. 2006. OXA-type carbapenemases. J. Antimicrob. Chemother. 57:373–383. 10.1093/jac/dki482. [DOI] [PubMed] [Google Scholar]
- 28.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. CLSI document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 29.Jin M, Jiang Y, Sun L, Yin J, Fu H, Wu G, Gao H. 2013. Unique organizational and functional features of the cytochrome c maturation system in Shewanella oneidensis. PLoS One 8:e75610. 10.1371/journal.pone.0075610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu L, Wang J, Tang P, Chen H, Gao H. 2011. Genetic and molecular characterization of flagellar assembly in Shewanella oneidensis. PLoS One 6:e21479. 10.1371/journal.pone.0021479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu H, Jin M, Ju L, Mao Y, Gao H. 2014. Evidence for function overlapping of CymA and the cytochrome bc1 complex in the Shewanella oneidensis nitrate and nitrite respiration. Environ. Microbiol., in press. 10.1111/1462-2920.12457. [DOI] [PubMed] [Google Scholar]
- 32.Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580. 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 33.Dong Y, Wang J, Fu H, Zhou G, Shi M, Gao H. 2012. A Crp-dependent two-component system regulates nitrate and nitrite respiration in Shewanella oneidensis. PLoS One 7:e51643. 10.1371/journal.pone.0051643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun L, Dong Y, Shi M, Zhou Q, Luo Z-Q, Gao H. 2014. Two residues predominantly dictate functional difference in motility between Shewanella oneidensis flagellins FlaA and FlaB. J. Biol. Chem. 289:14547–14559. 10.1074/jbc.M114.552000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honoré N, Nicolas MH, Cole S. 1986. Inducible cephalosporinase production in clinical isolates of Enterobacter cloacae is controlled by a regulatory gene that has been deleted from Escherichia coli. EMBO J. 5:3709–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Juan C, Moyá B, Pérez JL, Oliver A. 2006. Stepwise upregulation of the Pseudomonas aeruginosa chromosomal cephalosporinase conferring high-level beta-lactam resistance involves three AmpD homologues. Antimicrob. Agents Chemother. 50:1780–1787. 10.1128/AAC.50.5.1780-1787.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Bao Q, Gagnon LA, Huletsky A, Oliver A, Jin S, Langaee T. 2010. ampG gene of Pseudomonas aeruginosa and its role in beta-lactamase expression. Antimicrob. Agents Chemother. 54:4772–4779. 10.1128/AAC.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng Q, Park JT. 2002. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J. Bacteriol. 184:6434–6436. 10.1128/JB.184.23.6434-6436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asgarali A, Stubbs KA, Oliver A, Vocadlo DJ, Mark BL. 2009. Inactivation of the glycoside hydrolase NagZ attenuates antipseudomonal beta-lactam resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:2274–2282. 10.1128/AAC.01617-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Folkesson A, Eriksson S, Andersson M, Park JT, Normark S. 2005. Components of the peptidoglycan-recycling pathway modulate invasion and intracellular survival of Salmonella enterica serovar Typhimurium. Cell. Microbiol. 7:147–155. 10.1111/j.1462-5822.2004.00443.x. [DOI] [PubMed] [Google Scholar]
- 41.Boudreau MA, Fisher JF, Mobashery S. 2012. Messenger functions of the bacterial cell wall-derived muropeptides. Biochemistry 51:2974–2990. 10.1021/bi300174x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chahboune A, Decaffmeyer M, Brasseur R, Joris B. 2005. Membrane topology of the Escherichia coli AmpG permease required for recycling of cell wall anhydromuropeptides and AmpC beta-lactamase induction. Antimicrob. Agents Chemother. 49:1145–1149. 10.1128/AAC.49.3.1145-1149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong K-F, Aguila A, Schneper L, Mathee K. 2010. Pseudomonas aeruginosa beta-lactamase induction requires two permeases, AmpG and AmpP. BMC Microbiol. 10:328. 10.1186/1471-2180-10-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korfmann G, Sanders C. 1989. ampG is essential for high-level expression of AmpC beta-lactamase in Enterobacter cloacae. Antimicrob. Agents Chemother. 33:1946–1951. 10.1128/AAC.33.11.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindberg F, Lindquist S, Normark S. 1987. Inactivation of the ampD gene causes semiconstitutive overproduction of the inducible Citrobacter freundii beta-lactamase. J. Bacteriol. 169:1923–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zamorano L, Reeve TM, Deng L, Juan C, Moyá B, Cabot G, Vocadlo DJ, Mark BL, Oliver A. 2010. NagZ inactivation prevents and reverts beta-lactam resistance, driven by AmpD and PBP 4 mutations, in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 54:3557–3563. 10.1128/AAC.00385-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y-W, Hu R-M, Lin C-W, Chung T-C, Yang T-C. 2012. NagZ-dependent and NagZ-independent mechanisms for beta-lactamase expression in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 56:1936–1941. 10.1128/AAC.05645-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin C-W, Lin H-C, Huang Y-W, Chung T-C, Yang T-C. 2011. Inactivation of mrcA gene derepresses the basal-level expression of L1 and L2 beta-lactamases in Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 66:2033–2037. 10.1093/jac/dkr276. [DOI] [PubMed] [Google Scholar]
- 49.Stubbs KA, Balcewich M, Mark BL, Vocadlo DJ. 2007. Small molecule inhibitors of a glycoside hydrolase attenuate inducible AmpC-mediated beta-lactam resistance. J. Biol. Chem. 282:21382–21391. 10.1074/jbc.M700084200. [DOI] [PubMed] [Google Scholar]
- 50.Mark BL, Vocadlo DJ, Oliver A. 2011. Providing beta-lactams a helping hand: targeting the AmpC beta-lactamase induction pathway. Future Microbiol. 6:1415–1427. 10.2217/fmb.11.128. [DOI] [PubMed] [Google Scholar]


