Abstract
The cfr gene was identified in three linezolid-resistant USA300 methicillin-resistant Staphylococcus aureus (MRSA) isolates collected over a 3-day period at a New York City medical center in 2011 as part of a routine surveillance program. Each isolate possessed a plasmid containing a pSCFS3-like cfr gene environment. Transformation of the cfr-bearing plasmids into the S. aureus ATCC 29213 background recapitulated the expected Cfr antibiogram, including resistance to linezolid, tiamulin, clindamycin, and florfenicol and susceptibility to tedizolid.
TEXT
The cfr multidrug resistance gene represents the first horizontally transferable resistance determinant for linezolid (LZD) (1). Methylation of the 23S rRNA base A2503 by the Cfr methyltransferase confers resistance to 6 classes of drugs that target the peptidyl transferase center in the 50S ribosomal subunit, i.e., phenicols, lincosamides, oxazolidinones, pleuromutilins, streptogramin A, and 16-member-ring macrolides (2, 3). Therefore, use of any one of these classes can select for retention of the cfr gene. The cfr gene has been found globally (4); in the United States, it has been identified in isolates from California (5), Ohio (6), Arizona (6), Utah (7), Michigan (8), Indiana (9), Missouri (7), Kentucky (8), Maryland (10), and Illinois (11). U.S. surveillance studies have shown that the frequency of LZD resistance among Gram-positive pathogens from 2004 to 2012 was <0.5%, and of these isolates, ∼5 to 16% possessed the cfr gene (8, 12, 13).
Other than cfr, linezolid resistance has primarily been associated with rare, chromosomal mutations in genes encoding 23S rRNA or ribosomal proteins L3 and L4 (14, 15). While the novel oxazolidinone tedizolid (TZD) (16) is also impacted by these chromosomal mutations, it retains antimicrobial activity against LZDr cfr-positive strains without chromosomal mutations due to structural features that increase its binding site affinity and reduce steric clash with the Cfr-modified A2503 residue compared to linezolid (17, 18).
(Portions of this work were presented at the 53rd Interscience Conference on Antimicrobial Agents and Chemotherapy [19].)
A 2011 TZD surveillance study of 3,817 Gram-positive isolates (20) included Staphylococcus aureus strains 2823634, 2823586, and 2823605, which were collected over a 3-day period from 3 different patients at the New York Presbyterian Hospital/Weill Cornell Medical Center (New York, NY) (Table 1). These clinical isolates, S. aureus strain ATCC 29213, and transformants of the latter were cultured aerobically at 37°C on cation-adjusted Mueller-Hinton II agar (MHA; Becton Dickinson, Franklin Lakes, NJ) or in MH broth (MHB). MIC values were assessed by broth microdilution as previously described (21, 22); daptomycin MIC values were assessed in MHB supplemented with 50 mg/liter Ca2+. Each of these three clinical isolates was resistant to LZD (MIC = 16 μg/ml) but susceptible to TZD, with an MIC value of 0.5 μg/ml (Table 2), equivalent to the S. aureus wild-type MIC90 for TZD (23), prompting further genotypic analyses of their relatedness and underlying resistance mechanisms.
TABLE 1.
MRSA surveillance isolates analyzed in this study
| Strain | Source | Isolation date (mo/day/yr) | Presence of cfr | Notes |
|---|---|---|---|---|
| 2823634 | Blood | 3/11/2011 | + | Recurrent bacteremia with probable hVISA,a not treated with linezolid |
| 2823586 | Blood | 3/12/2011 | + | Line-associated bacteremia, treated with linezolid for earlier MRSA bacteremia |
| 2823605 | Wound | 3/13/2011 | + | Line-associated bacteremia, treated with linezolid for earlier MRSA bacteremia |
| 2823611 | Urine | 4/12/2011 | − | Not treated with linezolid |
hVISA, heterogeneous vancomycin-intermediate Staphylococcus aureus.
TABLE 2.
MICs of S. aureus clinical isolates and ATCC 29213 cfr-bearing plasmid transformants thereof
| Origin | Strain | Presence of cfr | MIC (μg/ml)a |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TZD | LZD | TIA | FFC | CLI | ERY | GEN | OXA | CIP | TMP | TET | VAN | DAP | |||
| Clinical | 2823611 | − | 0.5 | 2 | 0.5 | 8 | 0.13 | 16 | >128 | 64 | 1 | 2 | 1 | 1 | 0.25 |
| 2823634 | + | 0.5 | 16 | >128 | >128 | >128 | 16 | >128 | 64 | 1 | 1 | 1 | 1 | 0.25 | |
| 2823586 | + | 0.5 | 16 | >128 | >128 | >128 | 16 | >128 | 64 | 1 | 1 | 1 | 1 | 0.25 | |
| 2823605 | + | 0.5 | 16 | >128 | >128 | >128 | 16 | >128 | 64 | 1 | 1 | 1 | 1 | 0.25 | |
| Laboratory | 29213 | − | 0.5 | 2 | 0.5 | 8 | 0.13 | 0.5 | 1 | 0.5 | 0.5 | 2 | 1 | 1 | 0.25 |
| 29213(p2823634) | + | 0.5 | 16 | >128 | >128 | >128 | 0.25 | 1 | 0.25 | 0.5 | 4 | 1 | 1 | 0.25 | |
| 29213(p2823586) | + | 0.5 | 16 | >128 | >128 | >128 | 0.25 | 1 | 0.25 | 0.5 | 4 | 1 | 1 | 0.25 | |
| 29213(p2823605) | + | 0.5 | 16 | >128 | >128 | >128 | 0.25 | 1 | 0.25 | 0.5 | 4 | 1 | 1 | 0.25 | |
TZD, tedizolid; LZD, linezolid; TIA, tiamulin; FFC, florfenicol; CLI, clindamycin; ERY, erythromycin; GEN, gentamicin; OXA, oxacillin; CIP, ciprofloxacin; TMP, trimethoprim; TET, tetracycline; VAN, vancomycin; DAP, daptomycin.
The typing and relatedness of isolates were assessed by pulsed-field gel electrophoresis (PFGE). Chromosomal DNA was prepared in agarose plugs, digested with SmaI restriction endonuclease, and analyzed as previously described (24). A USA300 control strain was included as a reference. All 3 strains had USA300-like profiles, suggesting a common genetic background (Fig. 1). An additional group of 7 methicillin-resistant S. aureus (MRSA) isolates collected at this site (up to 2 months prior and 1 month after) were also analyzed by PFGE, revealing an LZDs isolate (2823611) with a nearly identical profile (Fig. 1) (Table 2). The other 6 MRSA isolates had PFGE profiles unrelated to this cluster of 4 isolates (data not shown).
FIG 1.
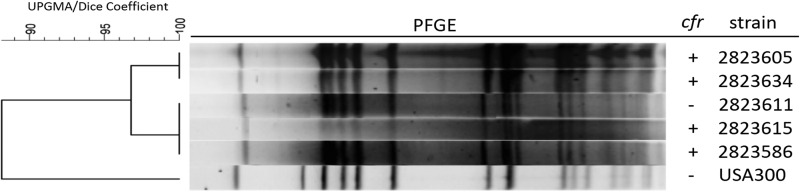
PFGE profiles of cfr-positive (+) and cfr-negative (−) isolates collected from the same hospital and a USA300 reference strain.
The genetic basis of LZD resistance was determined by amplification and sequence analysis of genes encoding 23S rRNA, ribosomal proteins L3 (rplC) and L4 (rplD), and Cfr as previously described (25). Sequencing data were analyzed using Vector NTI Advance 11 software (Invitrogen, Carlsbad, CA). All three of the LZDr isolates possessed the cfr gene and carried wild-type alleles for 23S rRNA, rplC, and rplD. These data and the 32-fold MIC value differential between TZD and LZD are consistent with previous reports for S. aureus possessing the cfr gene and lacking chromosomal resistance mutations (17, 25). The observed resistance to tiamulin, florfenicol, and clindamycin is also consistent with the presence of cfr (Table 2). As expected, the LZDs 2823611 isolate was PCR negative for cfr and possessed wild-type alleles for all chromosomal genes sequenced.
To assess whether the cfr gene was plasmid borne, total plasmid DNA was isolated from each strain and transformed into S. aureus ATCC 29213 as previously described (26, 27). Putative cfr-positive transformant colonies that grew on MHA medium containing 5 μg/ml of tiamulin were confirmed through PCR amplification of the cfr gene. The antibiogram of these isogenic 29213 transformant strains matched the profile of the parent strains, providing further evidence that the cfr gene wholly accounted for the LZDr phenotype observed (Table 2). Within the range of drugs tested, no additional drug resistance was conferred by the cfr-bearing plasmids (p2823634, p2823586, and p2823605). The consistency in MIC values for the non-Cfr-impacted drugs tested between the cfr-negative 2823611 isolate and the three cfr-positive isolates supports the possibility that this endemic hospital strain acquired the cfr-bearing plasmid.
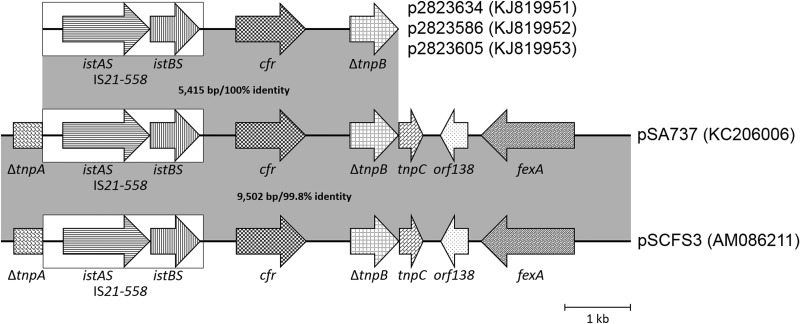
The relatedness of the p2823634, p2823586, and p2823605 plasmids was investigated through single-primer PCR amplification (28) and sequence analysis of the immediate cfr gene flanking region using plasmid DNA reisolated from each of the 29213 cfr-positive transformants. Each plasmid possessed an identical 5.4-kb region containing the IS21-558 element upstream of cfr and a truncated tnpB gene downstream (Fig. 2). This proximal cfr environment for the plasmids contained in these three isolates is 99.7% identical to that found in the pSCFS3 plasmid identified in German S. aureus and Staphylococcus lentus veterinary isolates from the early 2000s (AM086211) (29) and 100% identical to the pSA737 plasmid from the 2007 Ohio S. aureus isolate 004-737X (KC206006) (6, 30).
FIG 2.
Schematic comparison of cfr gene environments found in p2823634, p2823586, and p2823605 with those previously described for S. aureus plasmids pSA737 (30) and pSCFS3 (29).
Although there is limited information on patient medical history, the use of LZD was a commonality for two patients with cfr-positive isolates. Because cfr has a low fitness cost (31) and can be selected for by any drug within the Cfr resistance spectrum, it is not unexpected that a cfr-positive isolate was recovered from a patient not receiving LZD therapy (Table 1).
This report is the first documentation of the cfr gene in clinical isolates from New York State. The presence of cfr within the epidemic USA300 genetic background suggests the possibility of further dissemination. Continued LZD resistance surveillance efforts that incorporate identification of cfr by PCR can readily monitor the frequency and location of strains carrying this gene. Armed with this information, appropriate drug selection strategies can be utilized to combat the potential spread of cfr.
Nucleotide sequence accession numbers.
Sequences of the 5,415-bp cfr gene environments analyzed for p2823634, p2823586, and p2823605 were deposited into the NCBI database under GenBank accession numbers KJ819951, KJ819952, and KJ819953, respectively.
ACKNOWLEDGMENTS
In part, this research was funded and conducted by Trius Therapeutics, Inc.
Jeffrey B. Locke is an employee of Cubist. Douglas E. Zuill and Karen J. Shaw were employees of Trius Therapeutics, Inc., at the time this research was conducted and the paper was drafted. All authors had full access to the data and had final responsibility for the decision to submit for publication.
Footnotes
Published ahead of print 18 August 2014
REFERENCES
- 1.Toh SM, Xiong L, Arias CA, Villegas MV, Lolans K, Quinn J, Mankin AS. 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64:1506–1514. 10.1111/j.1365-2958.2007.05744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50:2500–2505. 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith LK, Mankin AS. 2008. Transcriptional and translational control of the mlr operon, which confers resistance to seven classes of protein synthesis inhibitors. Antimicrob. Agents Chemother. 52:1703–1712. 10.1128/AAC.01583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen J, Wang Y, Schwarz S. 2013. Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J. Antimicrob. Chemother. 68:1697–1706. 10.1093/jac/dkt092. [DOI] [PubMed] [Google Scholar]
- 5.Ross JE, Mendes RE, Flamm RK, Jones RN. 2012. Abstr. 52nd Intersci. Conf. Antimicrob. Agents Chemother., abstr C2-139. [Google Scholar]
- 6.Mendes RE, Deshpande LM, Castanheira M, Dipersio J, Saubolle M, Jones RN. 2008. First report of cfr-mediated resistance to linezolid in human staphylococcal clinical isolates recovered in the United States. Antimicrob. Agents Chemother. 52:2244–2246. 10.1128/AAC.00231-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flamm RK, Farrell DJ, Mendes RE, Ross JE, Sader HS, Jones RN. 2012. LEADER surveillance program results for 2010: an activity and spectrum analysis of linezolid using 6801 clinical isolates from the United States (61 medical centers). Diagn. Microbiol. Infect. Dis. 74:54–61. 10.1016/j.diagmicrobio.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 8.Farrell DJ, Mendes RE, Ross JE, Sader HS, Jones RN. 2011. LEADER Program results for 2009: an activity and spectrum analysis of linezolid using 6,414 clinical isolates from 56 medical centers in the United States. Antimicrob. Agents Chemother. 55:3684–3690. 10.1128/AAC.01729-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locke JB, Zuill DE, Sahm DF, Deane J, Denys GA, Shaw KJ. 2013. Abstr. 23rd Eur. Congr. Clin. Microbiol. Infect. Dis., abstr O 449. [Google Scholar]
- 10.Ross JE, Deshpande LM, Castanheira M, Jones RN. 2011. Abstr. 111th Gen. Meet. Am. Soc. Microbiol., abstr A-4. [Google Scholar]
- 11.Streit JM, Ross JE, Mendes RE, Flamm RK, Jones RN, Hogan PA. 2013. Abstr. IDWeek 2013 Infect. Dis. Soc. Am., abstr 737. [Google Scholar]
- 12.Farrell DJ, Mendes RE, Ross JE, Jones RN. 2009. Linezolid surveillance program results for 2008 (LEADER Program for 2008). Diagn. Microbiol. Infect. Dis. 65:392–403. 10.1016/j.diagmicrobio.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Mendes RE, Flamm RK, Hogan PA, Ross JE, Jones RN. 2014. Summary of linezolid activity and resistance mechanisms detected during the 2012 LEADER Surveillance Program for the United States. Antimicrob. Agents Chemother. 58:1243–1247. 10.1128/AAC.02112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw KJ, Barbachyn MR. 2011. The oxazolidinones: past, present, and future. Ann. N. Y. Acad. Sci. 1241:48–70. 10.1111/j.1749-6632.2011.06330.x. [DOI] [PubMed] [Google Scholar]
- 15.Long KS, Vester B. 2012. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob. Agents Chemother. 56:603–612. 10.1128/AAC.05702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Im WB, Choi SH, Park JY, Finn J, Yoon SH. 2011. Discovery of torezolid as a novel 5-hydroxymethyl-oxazolidinone antibacterial agent. Eur. J. Med. Chem. 46:1027–1039. 10.1016/j.ejmech.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 17.Locke JB, Finn J, Hilgers M, Morales G, Rahawi S, Kedar GC, Picazo JJ, Im W, Shaw KJ, Stein JL. 2010. Structure-activity relationships of diverse oxazolidinones for linezolid-resistant Staphylococcus aureus strains possessing the cfr methyltransferase gene or ribosomal mutations. Antimicrob. Agents Chemother. 54:5337–5343. 10.1128/AAC.00663-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaw KJ, Poppe S, Schaadt R, Brown-Driver V, Finn J, Pillar CM, Shinabarger D, Zurenko G. 2008. In vitro activity of TR-700, the antibacterial moiety of the prodrug TR-701, against linezolid-resistant strains. Antimicrob. Agents Chemother. 52:4442–4447. 10.1128/AAC.00859-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Locke JB, Zuill DE, Scharn CR, Deane J, Sahm DF, Jenkins SG, Goering RV, Shaw KJ. 2013. Abstr. 53rd Intersci. Conf. Antimicrob. Agents Chemother., abstr C1-517. [Google Scholar]
- 20.Deane J, Opiela C, Shah D, Shaw K, Locke J, Sahm D. 2013. Abstr. 53rd Intersci. Conf. Antimicrob. Agents Chemother., abstr C2-090. [Google Scholar]
- 21.Locke JB, Hilgers M, Shaw KJ. 2009. Novel ribosomal mutations in Staphylococcus aureus strains identified through selection with the oxazolidinones linezolid and torezolid (TR-700). Antimicrob. Agents Chemother. 53:5265–5274. 10.1128/AAC.00871-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CLSI. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—9th ed, vol 32, no. 2 CLSI document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 23.Schaadt R, Sweeney D, Shinabarger D, Zurenko G. 2009. The in vitro activity of TR-700, the active ingredient of the antibacterial prodrug TR-701, a novel oxazolidinone antibacterial agent. Antimicrob. Agents Chemother. 53:3236–3239. 10.1128/AAC.00228-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goering RV, Ribot EM, Gerner-Smidt P. 2011. Pulsed field gel electrophoresis: laboratory and epidemiologic considerations for interpretation of data, p 167–177 In Persing DH, Tenover FC, Nolte FS, Hayden RT, van BelkumA. (ed), Molecular microbiology: diagnostic principles and practice, 2nd ed. ASM Press, Washington, DC. [Google Scholar]
- 25.Locke JB, Rahawi S, Lamarre J, Mankin AS, Shaw KJ. 2012. Genetic environment and stability of cfr in methicillin-resistant Staphylococcus aureus CM05. Antimicrob. Agents Chemother. 56:332–340. 10.1128/AAC.05420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schenk S, Laddaga RA. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133–138. [DOI] [PubMed] [Google Scholar]
- 27.Locke JB, Morales G, Hilgers M, Kedar GC, Rahawi S, Picazo JJ, Shaw KJ, Stein JL. 2010. Elevated linezolid resistance in clinical cfr-positive Staphylococcus aureus isolates is associated with co-occurring mutations in ribosomal protein L3. Antimicrob. Agents Chemother. 54:5352–5355. 10.1128/AAC.00714-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karlyshev AV, Pallen MJ, Wren BW. 2000. Single-primer PCR procedure for rapid identification of transposon insertion sites. Biotechniques 28:1078, 1080, 1082. [DOI] [PubMed] [Google Scholar]
- 29.Kehrenberg C, Schwarz S. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50:1156–1163. 10.1128/AAC.50.4.1156-1163.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mendes RE, Deshpande LM, Bonilla HF, Schwarz S, Huband MD, Jones RN, Quinn JP. 2013. Dissemination of a pSCFS3-like cfr-carrying plasmid in Staphylococcus aureus and Staphylococcus epidermidis clinical isolates recovered from hospitals in Ohio. Antimicrob. Agents Chemother. 57:2923–2928. 10.1128/AAC.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaMarre JM, Locke JB, Shaw KJ, Mankin AS. 2011. Low fitness cost of the multidrug resistance gene cfr. Antimicrob. Agents Chemother. 55:3714–3719. 10.1128/AAC.00153-11. [DOI] [PMC free article] [PubMed] [Google Scholar]