LETTER
Carbapenem resistance in Acinetobacter baumannii is a growing threat to the effective treatment of nosocomial infections. Acquired carbapenemases (e.g., OXA-23, OXA-24, and their variants) are a major source of carbapenem resistance (1). A. baumannii has a chromosomal carbapenem-hydrolyzing class D β-lactamase (CHDL) called OXA-51. There are nearly 100 clinical variants of OXA-51 documented (2–4), and mutations in blaOXA-51 are sometimes associated with an increase in the MIC values of carbapenems (5).
Starting with the blaOXA-51 gene from A. baumannii (GenBank accession number AJ309734), we synthesized the I129L and L167V variants of OXA-51. These substitutions occur in the active site and are associated with large increases in carbapenem MICs (4, 5). The mature form (residues 26 to 274) of all three genes were cloned into pET-24a, expressed, and purified (>95%) as described previously for OXA-24/40 (6). OXA-51 displays high Km and low kcat values for ampicillin (Table 1; methods are described in reference 7). OXA-51's affinity for carbapenems appears to be tighter (Km, 5 to 150 μM), although not as tight as that seen for OXA-23 or OXA-24/40 (doripenem Km, 10 to 30 nM) (7–9). Carbapenem Km values (and Ks values) were much lower for both mutants, suggesting greatly increased affinity that approaches or equals that seen with OXA-24/40 and OXA-23.
TABLE 1.
OXA-51 Km and kcat values for β-lactam substrates
| CHDL and antimicrobial | Km (μM)d | Ks (μM)a | kcat (s−1) | kcat/Km (μM−1 · s−1) |
|---|---|---|---|---|
| OXA-51 | ||||
| Ampicillin | >10,000b | >25 | ||
| Imipenem | 105 ± 3 | >79 | 0.660 ± 0.005 | 0.0063 ± 0.0002 |
| Doripenem | 4.7 ± 0.2 | >4.1 | 0.0730 ± 0.0009 | 0.016 ± 0.001 |
| Cefotaxime | NA | <0.02 | ||
| Ceftriaxone | NA | <0.02 | ||
| Aztreonam | NA | <0.02 | ||
| Cefoxitin | NA | <0.02 | ||
| OXA-51 I129L | ||||
| Ampicillin | 9,100 ± 900b | 160 ± 6 | 0.018 ± 0.002 | |
| Imipenem | <2 | 0.610 ± 0.094 | 0.330 ± 0.008 | 0.54 ± 0.08c |
| Doripenem | <2 | 0.110 ± 0.010 | 0.140 ± 0.003 | 1.4 ± 0.2c |
| Cefotaxime | NA | <0.02 | ||
| Ceftriaxone | NA | <0.02 | ||
| Aztreonam | NA | <0.02 | ||
| Cefoxitin | NA | <0.02 | ||
| OXA-51 L167V | ||||
| Ampicillin | 8,000 ± 600b | 470 ± 12 | 0.058 ± 0.004 | |
| Imipenem | <2 | 0.190 ± 0.040 | 0.150 ± 0.003 | 0.81 ± 0.20c |
| Doripenem | <2 | <0.05 | 0.032 ± 0.002 | >0.70c |
| Cefotaxime | NA | <0.02 | ||
| Ceftriaxone | NA | <0.02 | ||
| Aztreonam | NA | <0.02 | ||
| Cefoxitin | NA | <0.02 |
Ks values were determined using ampicillin as a reporter substrate (14).
For substrates with high Km values, a 0.2-mm path length cuvette was used.
kcat/Ks (μM−1 · s−1).
NA, not applicable.
To illuminate the structural basis of these results, we generated a model of OXA-51 using the PHYRE2 protein fold recognition server (Fig. 1) (10). The general fold of the predicted structure matches quite well OXA-24-doripenem (68% sequence identity) and shows especially strong overlap for active-site residues, including I129 (V130 in OXA-24/40) and L167 (L168). The δ methyl group of I129 is predicted to clash strongly with the hydroxyethyl group present on all carbapenems, explaining the relatively low affinity of those β-lactams for OXA-51 compared to that for OXA-24/40. Attempts to relieve this clash by rotation of the I129 side chain were unsuccessful, with further clashes forming between the δ methyl group and the side chain of L167 or between the γ2 methyl group of I129 and the doripenem hydroxyethyl group. Modeling the L167V mutation using PyMOL (11) relieves the former clash by providing space to accommodate the δ methyl group after rotating it toward V167. Modeling the I129L mutation eliminates the γ2 methyl group and thus allows L129 to rotate in the other direction without causing that group to clash with the hydroxyethyl moiety. The lower Km for both variants with respect to substrates containing α-hydroxyethyl groups can thus be explained by mutation-induced remodeling of the active site to better accommodate that group (2). The OXA-51 model also predicts the presence of the hydrophobic bridge found in other class D carbapenemases. The presence of a tryptophan in OXA-51 (W222) in place of the methionine found in the OXA-24/40 and OXA-23 bridge (M223 and M221) may further explain kinetic differences among CHDLs (12). The homologous W222 in the OXA-51 model shows much less conformational flexibility and therefore accounts for the very weak binding of ampicillin (Km, >10 mM compared to <500 μM for OXA-24/40 and OXA-23). Interestingly, there are known clinical variants of OXA-51-like enzymes with substitutions for W222 that are predicted to increase the flexibility at this position (e.g., W222G in OXA-79 and W222L in OXA-200) (13).
FIG 1.
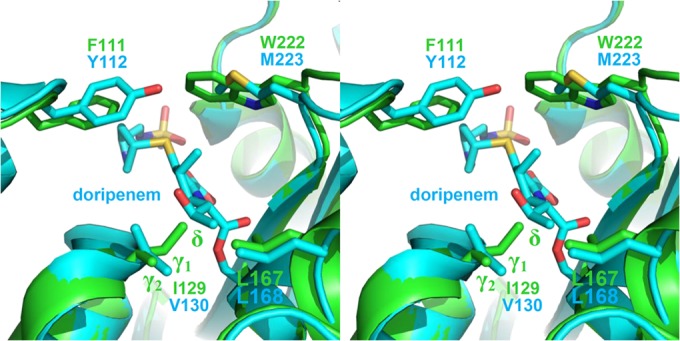
Stereodiagram of OXA-24/40-doripenem (cyan) aligned with a model of OXA-51 (green). The sequence of the mature OXA-51 enzyme (residues 26 to 274) was submitted to the PHYRE2 server. The resulting Protein Data Bank file was aligned with the structure of OXA-24/40-doripenem (Protein Data Bank accession no. 3PAE) using PyMOL.
In conclusion, it appears that selective pressure caused by treatment with carbapenems is leading to the emergence of another dangerous mechanism of resistance to those key “last-resort” drugs.
Footnotes
Published ahead of print 25 August 2014
REFERENCES
- 1.Limansky AS, Mussi MA, Viale AM. 2002. Loss of a 29-kilodalton outer membrane protein in Acinetobacter baumannii is associated with imipenem resistance. J. Clin. Microbiol. 40:4776–4778. 10.1128/JCM.40.12.4776-4778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans BA, Amyes SG. 2014. OXA β-lactamases. Clin. Microbiol. Rev. 27:241–263. 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown S, Amyes SG. 2005. The sequences of seven class D β-lactamases isolated from carbapenem-resistant Acinetobacter baumannii from four continents. Clin. Microbiol. Infect. 11:326–329. 10.1111/j.1469-0691.2005.01096.x. [DOI] [PubMed] [Google Scholar]
- 4.Chen TL, Lee YT, Kuo SC, Hsueh PR, Chang FY, Siu LK, Ko WC, Fung CP. 2010. Emergence and distribution of plasmids bearing the blaOXA-51-like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in Taiwan. Antimicrob. Agents Chemother. 54:4575–4581. 10.1128/AAC.00764-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zander E, Chmielarczyk A, Heczko P, Seifert H, Higgins PG. 2013. Conversion of OXA-66 into OXA-82 in clinical Acinetobacter baumannii isolates and association with altered carbapenem susceptibility. J. Antimicrob. Chemother. 68:308–311. 10.1093/jac/dks382. [DOI] [PubMed] [Google Scholar]
- 6.Schneider KD, Ortega CJ, Renck NA, Bonomo RA, Powers RA, Leonard DA. 2011. Structures of the class D carbapenemase OXA-24 from Acinetobacter baumannii in complex with doripenem. J. Mol. Biol. 406:583–594. 10.1016/j.jmb.2010.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaitany KC, Klinger NV, June CM, Ramey ME, Bonomo RA, Powers RA, Leonard DA. 2013. Structures of the class D carbapenemases OXA-23 and OXA-146: mechanistic basis of activity against carbapenems, extended-spectrum cephalosporins, and aztreonam. Antimicrob. Agents Chemother. 57:4848–4855. 10.1128/AAC.00762-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith CA, Antunes NT, Stewart NK, Toth M, Kumarasiri M, Chang M, Mobashery S, Vakulenko SB. 2013. Structural basis for carbapenemase activity of the OXA-23 β-lactamase from Acinetobacter baumannii. Chem. Biol. 20:1107–1115. 10.1016/j.chembiol.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santillana E, Beceiro A, Bou G, Romero A. 2007. Crystal structure of the carbapenemase OXA-24 reveals insights into the mechanism of carbapenem hydrolysis. Proc. Natl. Acad. Sci. U. S. A. 104:5354–5359. 10.1073/pnas.0607557104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371. 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 11.Schrödinger LLC. 2014. The PyMOL Molecular Graphics System, version 1.3. Schrödinger, LLC, Cambridge, MA. [Google Scholar]
- 12.June CM, Vallier BC, Bonomo RA, Leonard DA, Powers RA. 2014. The structural origins of oxacillinase specificity in class D β-lactamases. Antimicrob. Agents Chemother. 58:333–341. 10.1128/AAC.01483-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zander E, Nemec A, Seifert H, Higgins PG. 2012. Association between β-lactamase-encoding bla(OXA-51) variants and DiversiLab rep-PCR-based typing of Acinetobacter baumannii isolates. J. Clin. Microbiol. 50:1900–1904. 10.1128/JCM.06462-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Y, Prusoff WH. 1973. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22:3099–3108. 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]


