Abstract
Plasmodium falciparum has the capacity to escape the actions of essentially all antimalarial drugs. ATP-binding cassette (ABC) transporter proteins are known to cause multidrug resistance in a large range of organisms, including the Apicomplexa parasites. P. falciparum genome analysis has revealed two genes coding for the multidrug resistance protein (MRP) type of ABC transporters: Pfmrp1, previously associated with decreased parasite drug susceptibility, and the poorly studied Pfmrp2. The role of Pfmrp2 polymorphisms in modulating sensitivity to antimalarial drugs has not been established. We herein report a comprehensive account of the Pfmrp2 genetic variability in 46 isolates from Thailand. A notably high frequency of 2.8 single nucleotide polymorphisms (SNPs)/kb was identified for this gene, including some novel SNPs. Additionally, we found that Pfmrp2 harbors a significant number of microindels, some previously not reported. We also investigated the potential association of the identified Pfmrp2 polymorphisms with altered in vitro susceptibility to several antimalarials used in artemisinin-based combination therapy and with parasite clearance time. Association analysis suggested Pfmrp2 polymorphisms modulate the parasite's in vitro response to quinoline antimalarials, including chloroquine, piperaquine, and mefloquine, and association with in vivo parasite clearance. In conclusion, our study reveals that the Pfmrp2 gene is the most diverse ABC transporter known in P. falciparum with a potential role in antimalarial drug resistance.
INTRODUCTION
Plasmodium falciparum, the lethal pathogen of human malaria, is notorious for its capacity to develop resistance to chemotherapy. From the first observations involving quinine (1) to the latest reports on emerging artemisinin (ART) resistance (2–4), the parasite has shown a resilient ability to evade the action of essentially every launched antimalarial drug, independently of the chemical structure involved. Such an evasive capacity raises the hypothesis that the parasite is presently developing broad-range resistance phenotypes, possibly akin to the extensive drug resistance witnessed in other infectious diseases, namely, tuberculosis (5). This is a legitimate concern, particularly because of the recent reports of decreased ART sensitivity of the parasite in Southeast Asia.
Conventional in vitro drug response and the parasite clearance rate (2, 3) are the main measurements to assess the resistance phenotype of the parasite, but these procedures involve demanding and time-consuming protocols. A practical alternative is the use of molecular sentinel tools, i.e., molecular markers based on genetic variations with a valuable predictive capacity in the identification of the resistance status of the analyzed infection. The development of such tools is dependent on the understanding of the drug resistance mechanisms and their associations with variants of particular genes. Such information can also provide key clues for the development of new evidence-based resistance-refractory antimalarials. Moreover, this type of information is of particular relevance at a moment when technology for point-of-care genetic analysis is starting to emerge, allowing a first glimpse of the possibility of personalized medicine in the future (6).
Transporter proteins belonging to the ATP-binding cassette (ABC) superfamily are well known to be involved in drug efflux, as they are associated with resistance in a large variety of phylogenetically different biological systems (7). These proteins are able to transport substrates across cell membranes against a concentration gradient, an action driven by ATP hydrolysis. In particular, the multidrug resistance-associated protein (MRP)-like subclass of ABC transporters is well known for transport of drugs out of cells, contributing to resistance as well as to the redox metabolism pathway (8). Since the antimalarial chloroquine (CQ), and potentially mefloquine (MQ), was reported to be transported by the human MRP1 and MRP4 proteins, it has been speculated that the putative P. falciparum MRP could have the same capacity and thus contribute to drug resistance (9, 10). Indeed, P. falciparum is presently known to contain in its genome two genes coding for MRP-like proteins, namely, Pfmrp1 (11) and Pfmrp2 (12). Both PfMRP1 (PlasmoDB gene ID PF3D7_0112200) and PfMRP2 (PlasmoDB gene ID PF3D7_1229100) proteins are localized in the cytoplasmic membrane of the parasite in the asexual stages (13). Additionally, it was observed that the loss of PfMRP1 drug transport capability resulted in an increased accumulation of antimalarials that was paralleled by an enhanced in vitro susceptibility to several antimalarial drugs, including CQ, quinine, and ART (14). A number of studies on Pfmrp1 diversity have associated single nucleotide polymorphisms (SNPs) in this gene to the parasite in vivo drug responses, manifested by the selection of specific SNPs upon treatment (15–17).
In contrast, our knowledge on Pfmrp2 is limited, and its biodiversity and possible involvement in antimalarial drug resistance are still to be disclosed. The localization of PfMRP2 in the plasma membrane suggests that this ABC transporter may be of relevance in the efflux of xenobiotics from the parasite cytoplasm, as previously reported for the structurally related PfMRP1 (14). Also, we recently showed that upon MQ exposure in vitro, different levels of Pfmrp2 transcription induction were observed between sensitive and less susceptible strains (18). Additionally, Pfmrp1 and Pfmrp2 genes have essentially opposite transcriptional patterns throughout the P. falciparum asexual intraerythrocytic cell cycle, so that each protein is expressed during different morphological stages (18, 19). This pattern further suggests a potential functional complementation between the two proteins that might be of relevance in complex phenotypes of the cell cycle stage-specific drug response. Lastly, recent microarray-based approaches have identified a P. falciparum 3D7 subvariant which carries a ca. 4-kb deletion at the Pfmrp2 5′ putative promoter that is associated with decreased susceptibility to CQ and MQ (20).
We focused here on the study of Pfmrp2 complete open reading frame (ORF) sequence diversity in a set of adapted parasites from an ART resistance focus at the Thai-Burma border, disclosing a previously unknown natural variation of this gene. Taking advantage of available data on in vitro antimalarial 50% inhibitory concentrations (IC50s) and parasite clearance times (PCTs) for this set of samples (21, 22), we explored the possible association of the polymorphisms found with modulation of drug sensitivity.
MATERIALS AND METHODS
P. falciparum field isolates.
Forty-six culture-adapted P. falciparum field isolates, originally adapted at Karolinska Institutet (21), were enrolled in this study. All were isolated from clinical cases of uncomplicated malaria diagnosed and managed at the Shoklo Malaria Unit in the Mae Sot District, Tak Province, Thailand, where parasite clearance time data were collected. The chosen parasite set of infections represented a relatively homogeneous and specific population, in line with the objectives of obtaining a better understanding of the specific influence of the Pfmrp2 variation in drug responses.
The study was ethically cleared by the relevant institutions, and the blood samples were obtained upon informed consent provided in the local language (21).
Pfmrp2 molecular analysis.
The Pfmrp2 gene (6,327 bp on the 3D7 laboratory strain) was PCR amplified from genomic DNA, extracted from culture-adapted Thai strains (21) in 8 fragments, in order to cover the entire ORF. Amplification and sequencing primers are described in Table S1 in the supplemental material. The PCR amplifications were performed in a total volume of 50 μl with 0.2 μM each forward and reverse primer, 0.2 mM deoxynucleoside triphosphates, 2.5 mM MgCl2, and 2 U GoTaq DNA polymerase (Promega Biotech AB). The thermal cycle program was performed with a starting DNA denaturation temperature of 94°C for 2 min, followed by three touchdown cycles: 10 cycles of 93°C for 15 s, 57°C for 30 s, and 72°C for 90 s; 25 cycles of 93°C for 15 s, 55°C for 30 s, and 72°C for 90 s; followed by 10 cycles of 93°C for 15 s, 53°C for 30 s, and 72°C for 90 s. Fragments that did not amplify were subjected to a nested amplification with the primers described in Table S1 in the supplemental material and the same thermal cycle program. The resulting PCR products were confirmed through agarose gel electrophoresis for specificity and a low presence of unspecific amplification. The direct sequencing of the amplicons was outsourced to Macrogen Inc. (Seoul, South Korea).
Analysis of the sequence chromatograms.
Due to the complexity of the gene, sequence chromatogram files were analyzed by using two bioinformatic approaches. Sequence chromatogram files were analyzed by base calls using phred version 0.020425.c (23) and aligned to the P. falciparum 3D7 genome reference sequence (PlasmoDB gene ID PF3D7_1229100) by using the alignment program ssaha2 version 2.5.1 (24). The program was configured to report only the best alignment for each match. A custom Perl script was used to call single nucleotide polymorphisms and polymorphic microindels (MIs) from the alignments. A position was called polymorphic if either the position had more than one read to support the nucleotide difference or the quality score of the position was 20 or higher. The Tablet program was used to visualize alignments (25). The other approach to analyze the sequence chromatogram files was with the Staden package (26). To identify sequence polymorphisms, the consensus sequence for each strain was compared with the corresponding gene sequence of the Plasmodium falciparum 3D7 genome reference sequence by using MUSCLE (27) and manually inspected for alignment errors.
In addition to the 3D7 genome reference sequence, five other reference and published parasite lines sequences were downloaded: Dd2 (http://www.ncbi.nlm.nih.gov/nuccore/AASM00000000), Hb3 (http://www.ncbi.nlm.nih.gov/nuccore/AANS00000000), UGT5.1 (http://www.ncbi.nlm.nih.gov/nuccore/AMYP00000000), RAJ116 (http://www.ncbi.nlm.nih.gov/nuccore/ACBR00000000), and IGH-CR14 (http://www.ncbi.nlm.nih.gov/nuccore/ACBS00000000). The Pfmrp2 gene sequence was extracted and used in the alignments for sequence comparisons with our set of samples.
Determined antimalarial in vitro 50% inhibitory concentrations and parasite clearance times.
The 46 Thai isolates were previously phenotyped for the in vitro IC50s of ART, dihydroartemisinin (DHA), MQ, lumefantrine (LUM), piperaquine (PPQ), and CQ. Briefly, the IC50s were determined by culturing the parasite (synchronized at ring stage with 0.05% parasitemia and 1.5% hematocrit) in the presence of serial dilutions of the aforementioned antimalarial drugs. After 72 h of incubation at 37°C under restricted O2 conditions, the cells were lysed and analyzed by the histidine-rich protein 2-based double-site sandwich enzyme-linked immunosorbent assay (28). Further detailed information can be found in the original reports (21, 22).
In this set of Thai isolates, the patient parasite clearance times ranged from 6 to 132 h. A high parasite clearance rate was detected in 19 patients, with PCTs from 6 to 36 h, and 27 patients had a low parasite clearance rate, with PCTs between 90 and 132 h. In vitro drug susceptibility and PCT phenotype data and Pfmrp2 genotype associations were tested by using a one-way analysis of variance (ANOVA) with SPSS statistics software.
Topology of the PfMRP2 protein.
To predict the transmembranes (TM) in the ABC transporter PfMRP2, we used two different software programs. The most-used program was TMHMM server v. 2.0 (transMembrane hidden Markov model; http://www.cbs.dtu.dk/services/TMHMM/). The other program used was the OCTOPUS software (29), which uses a combination of hidden Markov models and artificial neural networks. The topology predictor program OCTOPUS, in particular, is the first to integrate modeling of reentrant, membrane dip, and TM hairpin regions in the topological grammar. It performs a homology search by using BLAST to create a sequence profile used as the input to a set of neural networks that predict both the preference for each residue to be located in a TM, interface, close loop, or globular loop environment and to be inside or outside the membrane. These predictions are used as input for a two-track hidden Markov model, which uses them to calculate the most likely topology. To get a reliable model, we merged the data of the two transmembrane predictive software programs. Table 1 and Fig. S1 in the supplemental material show the membrane-spanning domains and trans-membrane prediction of PfMRP2, and Fig. 1 shows a PfMRP2 two-dimensional (2D) representation with the localization of the analyzed SNPs and microindels.
TABLE 1.
Membrane-spanning domains and transmembrane predictions for PfMRP2
| MSD | TM | Predicted TM domaina (aa span) based on: |
|||
|---|---|---|---|---|---|
| Previously published (13) | OCTOPUS | TMHMM | O/T merged | ||
| MSD1 | 1 | 133–155 | 133–157 | 132–155 | 134–155 |
| 2 | 180–202 | 178–199 | 180–203 | 180–199 | |
| 3 | 402–424 | 401–422 | 396–418 | 401–418 | |
| 4 | 434–456 | 425–449 | 428–454 | 428–448 | |
| 5 | 517–539 | 511–531 | 511–533 | 512–531 | |
| 6 | 554–575 | 544–564 | 548–565 | 551–562 | |
| — | 787–807 | — | — | ||
| MSD2 | 7 | — | 1397–1419 | 1398–1426 | 1400–1417 |
| 8 | 1430–1452 | 1445–1472 | 1446–1471 | 1448–1470 | |
| 1473–1495 | — | 1485–1506 | — | ||
| 9 | 1510–1532 | 1524–1545 | 1526–1546 | 1528–1545 | |
| 10 | 1553–1570 | 1547–1564 | 1547–1565 | 1547–1564 | |
| 11 | 1574–1593 | 1629–1658 | 1629–1651 | 1631–1651 | |
| 12 | 1654–1676 | 1667–1689 | 1671–1693 | 1671–1689 | |
The TM segments shown are those reported from a previous study (13) or predicted in this study by using the OCTOPUS and TMHMM software. To obtain a reliable model, we supported the information for the TM segments obtained by merging the OCTOPUS/TMHMM probabilities (i.e., data in the O/T merged column). TM domain amino acid ranges shown in bold had the greatest differences from the data obtained from reference 13 or via analysis using the OCTOPUS and TMHMM software programs. TM column represents the transmembrane number for the O/T merged model. Dashes (—) represent TM segments.
FIG 1.
2D representation of PfMRP2, with the location of the analyzed SNPs and microindels. The structure depicted refers to the reference P. falciparum 3D7 clone sequence, comprising 2,108 aa. Note that the numbering of amino acids follows the 3D7 reference genomic data, independently of the microindel variation observed in the studied Thai set of parasites.
Statistical analysis.
To evaluate associations between the genetic polymorphisms found in Pfmrp2 and in vitro drug resistance, a one-way ANOVA was performed to compare the mean IC50s of ART, DHA, MQ, LUM, PPQ, and CQ. This analysis was complemented with chi-square analysis to verify the presence of differences in the parasite clearance rates that were high (<36 h) and low (>90 h). The SPSS package (IBM SPSS Statistics v. 19) was used to conduct all statistical analyses, and results were considered significant when P was <0.05.
Nucleotide sequence accession numbers.
The Pfmrp2 consensus sequences from the 46 Thai parasites were submitted to GenBank (accession numbers KM213123 to KM213168).
RESULTS
Analysis of the primary sequence organization of PfMRP2.
The predicted two-dimensional structure of PfMRP2 suggested a classical ABC transporter protein, with 12 TM domains, distributed in two membrane-spanning domains (MSDs), and two nucleotide-binding domains (NBDs). The data support previously proposed models (13) of PfMRP2 as a transporter with 2 MSDs of 6 units each (prediction probability range, 0.9 to 1). However, according to our analysis, we detected some differences in TM7 and TM11 predictions when we used the merged data from TMHMM and OCTOPUS (Table 1; see also Fig. S1 in the supplemental material). In our work, we identified TM7 between amino acids (aa) 1400 and 1417 which were not detected in previous analysis (13), while T11 was repositioned. These observed divergences did not interfere with the analysis or interpretation of our data, since none of the subsequently identified SNPs were located in these regions.
In spite of its large size, PfMRP2 is predicted to represent a short MRP-type protein, as it did not show the extra 5 TM N-terminal domains present in the long MRPs, such as the human MRPs 1, 2, 3, 6, and 7. This was largely compensated by a large (∼850-aa) NBD1-containing cytoplasmic loop. Also notable was the unusually distal localization of the NBD2 ABC signature plus Walker B motif, which was only 17 aa from the protein C-terminal.
Pfmrp2 polymorphisms.
The Pfmrp2 ORF was sequenced for 46 P. falciparum strains that originated from the Thai-Burma border. The full sequence was determined for 37/46 isolates, and on average 93% of the gene was covered for the remaining 9 isolates. A total of 12 nonsynonymous and 6 synonymous SNPs were identified, in comparison with the 3D7 reference sequence. Among these SNPs, we found novel SNPs not previously annotated within PlasmoDB (http://plasmodb.org) nor observed in recently performed genome surveys (30) or in other available genome sequencing data (31) (Table 2). The proportions of the analyzed SNPs in this set of isolates varied from a single event (1/46) to situations of monomorphism (46/46).
TABLE 2.
SNPs identified in Pfmrp2 by sequencing the Thai isolates
| Chromosome positiona | Nucleotide position | Nucleotide change | Amino acid position | Amino acid change | Frequency (proportion) | Location |
|---|---|---|---|---|---|---|
| 1198644 | 561 | T → A | 187 | Synonymous | 0.02 (1/46) | TM2 |
| 1198610*# | 595 | C → G | 199 | L → V | 1.00 (46/46) | TM2 |
| 1198321 | 884 | C → G | 295 | T → R | 0.02 (1/46) | loop |
| 1198275 | 930 | G → T | 310 | L → F | 0.02 (1/46) | loop |
| 1198224 | 981 | C → T | 327 | Synonymous | 0.02 (1/46) | loop |
| 1197428*§ | 1777 | A → G | 593 | N → D | 0.07 (3/46) | NBD1 |
| 1197341* | 1864 | A → G | 622 | N → D | 0.22 (10/46) | NBD1 |
| 1197064*§# | 2141 | A → T | 714 | K → I | 1.00 (46/46) | NBD1 |
| 1196808*§ | 2397 | C → T | 799 | Synonymous | 0.39 (16/41) | NBD1 |
| 1195791*§ | 3414 | T → C | 1138 | Synonymous | 0.20 (9/44) | NBD1 |
| 1194626*# | 4579 | T → A | 1527 | S → T | 1.00 (46/46) | TM9 |
| 1194614*# | 4591 | C → A | 1531 | L → I | 1.00 (46/46) | TM9 |
| 1194093 | 5112 | T → C | 1704 | Synonymous | 0.02 (1/46) | NBD2 |
| 1193954 | 5251 | A → T | 1751 | N → Y | 0.09 (3/34) | NBD2 |
| 1193947§ | 5258 | T → C | 1753 | F → S | 0.07 (3/46) | NBD2 |
| 1193882 | 5323 | A → G | 1775 | I → V | 0.14 (5/36) | NBD2 |
| 1193861 | 5344 | A → T | 1782 | M → L | 0.02 (1/46) | NBD2 |
| 1193820 | 5385 | A → T | 1795 | Synonymous | 0.08 (3/37) | NBD2 |
The only three nonsynonymous SNPs located in the predicted TM domains of the transporter, L199V (TM2), S1527T (TM9), and L1531I (TM9), were found to be monomorphic among the studied isolates, suggesting that inheritance was from a common ancestor (Table 2).
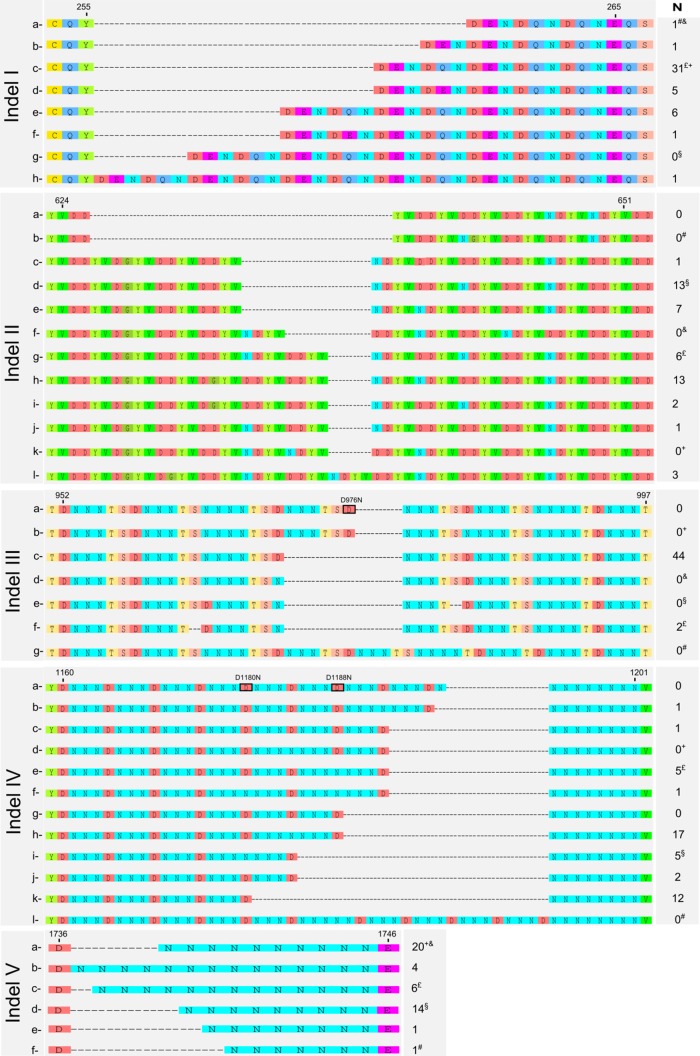
We also found a significant number of MIs, based on the presence of tandem repeats (Fig. 1 and 2), relative to the 3D7 reference sequence. This is an unusual observation among P. falciparum ABC transporters. The identified MIs comprise 5 regions of the gene (herein referred as MI-I to MI-V) and lead to a notable length variation of the coded protein (2,109 to 2,135 aa). Within the MI regions, we also found variations in single nucleotides, troubling a forthright sequence alignment. Three of these variations (D976N, D1180N, and D1188N) are annotated as SNPs within PlasmoDB. Due to the changeability of possible alignments, allocating an amino acid position becomes biased, and therefore we decided not to include them as definitive SNPs but instead as residues of variability within the MIs (Fig. 2).
FIG 2.
Microindels identified in the Pfmrp2 by sequencing the Thai isolates and aligning the sequences with publicly available sequencing data. The 5 microindels identified were subcategorized as MI variants of type a (the 3D7 reference genomic sequence type); the other types are designated b to l for the variants found among the Thai isolates (N) and the publicly available genomes. Symbols for publicly available genomes: £, Dd2; &, Hb3; #, UGT5.1; §, RAJ116; +, IGH-CR14. Highlighted black boxes denote amino acid positions that are described in the PlasmoDB database (http://www.plasmodb.org) as single nucleotide polymorphisms.
To additionally support the observation of such complex Pfmrp2 MI regions, publicly available sequence data from 5 distinct parasite lines (Dd2, Hb3, UGT5.1, RAJ116, and IGH-CR14) were aligned with the set of Thai isolates under study. The resulting analysis confirmed the presence of the 5 MIs determined in our set of samples, further increasing the number of variants in each MI region (Fig. 2). All MI regions were located outside the TM domains.
Genotype/phenotype associations.
The isolates under study were previously shown to have a relatively large range of IC values for the tested antimalarial drugs, with median IC50s (maximum to minimum) of 7.4 nM (1.2 to 19.5 nM) for ART, 1.2 nM (0.3 to 5.3 nM) for DHA, 92.5 nM (16.5 to 270.8 nM) for MQ, 11.9 nM (2.0 to 40.8 nM) for LUM, and 39.4 nM (13.8 to 108.2 nM) for PPQ; all strains were highly resistant to CQ, with IC50s of >450 nM (454 to 1,027 nM) (21, 22).
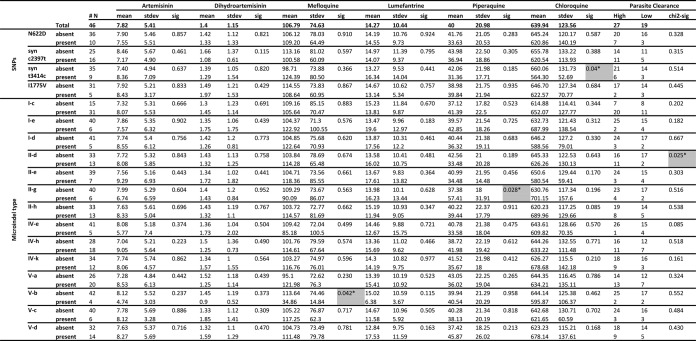
One-way analysis of variance revealed an association between the synonymous SNP coding for an asparagine at position 1138 (codon change of GAT → GAC at nucleotide position 3414) and the parasite response to CQ (P = 0.040), with an average IC50 for the t3414 variant of 660 nM and for the c3414 variant of 564.3 nM (Fig. 3). No other significant association was found within the identified SNPs.
FIG 3.
SNPs and microindels and their associations with IC50 values and parasite clearance rates. We used one-way ANOVA to compare associations between IC50s and the presence (+) or absence (−) of the SNP or microindel type. Chi-square analysis was used to compare associations between high (<36-h) or low (>90-h) parasite clearance rates and the presence (+) or absence (−) of the SNP or microindel type. *, P < 0.05 (significant findings are also highlighted in gray, for additional emphasis. #, the number of samples bearing each genotype. Results represent only the analysis for genotypes with allele frequencies of >8%.
The 5 MIs identified were subcategorized into MI variants that were type a, such as the 3D7 reference genomic sequence (Fig. 2). With this classification approach, associative trends were revealed for MI-II-g type and PPQ IC50s (P = 0.028), with mean IC50 of 37.4 nM in the absence of the polymorphism and of 57.4 nM in isolates containing this type of polymorphism. The MI-V-b type showed a trend of association with MQ susceptibility (P = 0.042) that encompassed an increased number of asparagines (N) from 10 (mean IC50 of 113.6 nM) to 14 (mean IC50 of 34.9 nM) (Fig. 3).
Interestingly, we observed the PCT phenotype to be influenced by MI polymorphisms. For MI-II, the presence of the type II-a allele was significantly more frequent in those parasites with high clearance rates (P = 0.025) (Fig. 3).
DISCUSSION
The PfMRP2 ABC transporter has been found to be located in the plasma membrane (13). Conceptually, its localization and predicted function suggest this ABC transporter is of potential relevance in the efflux of xenobiotics from the parasite cytoplasm, as reported for the structurally related PfMRP1 (14).
The present work shows that Pfmrp2 harbors frequent and complex natural variations, including SNPs and MIs based on tandem repeats. The accumulation of these polymorphisms gives rise to a significant level of diversity in this protein, more extensive than that documented for any of the two previously well-investigated ABC transporters in P. falciparum, Pgh-1 and PfMRP1. This high diversity implies the existence of PfMRP2 natural variants with different sizes and possible conformations and consequent varied transporting capacities.
Compared with the first characterization of this gene by Mu et al., in which 97 isolates from geographically distinct regions were analyzed, our investigations revealed a notably higher frequency of SNP/kb (2.8 versus 1.3 SNP/kb) (33). Of note, since the main goal of our work was to understand the influence of the Pfmrp2 gene in drug susceptibility and parasite clearance, the set of parasites used in our study was expected to represent a relatively homogeneous sample. As such, and as in most other similar types of studies, the observed polymorphism frequencies should not be taken as representative of Southeast Asian P. falciparum populations as a whole. On the other hand, we expect most of the observed polymorphisms to be relatively common (i.e., >1% frequency) in the targeted region. Also such a low prevalence can rapidly increase in time upon drug pressure. Such events can be relevant in the context of this work, due to the notorious tendency for Southeast Asia parasite populations to develop resistance.
Our data are suggestive of an involvement of this gene in modulating parasite drug sensitivity to at least some of the drugs tested. In the particular case of CQ, and acknowledging that all the analyzed Thai isolates carry the pfcrt 76T allele (21), we observed an association between the parasite sensitivity and a synonymous SNP coding for an asparagine at aa 1138 (GAT → GAC). Explaining the effect of a synonymous mutation in a phenotype has been a challenge in other areas of drug transport proteins. The presently most favored hypothesis is based on preferential codon usage. In P. falciparum, the two possible triplets that code for the asparagine amino acid have very distinct frequencies. Codon usage analysis describes the triplet GAU appearing at a greater frequency (55%) than the triplet GAC (only 9%) (34). The use of alternative codons with different levels of intracellular availability in terms of loaded tRNA molecules is thought to influence translation rates. This hypothesis has been experimentally put forward through investigations of the synonymous t3434c SNP in the human P-glycoprotein, which is known to influence the exposure of some orally administered drugs (35). Indeed, it has been demonstrated that the nucleotide change disturbs the pace of the mRNA translation process, leading to changes in the conformational dynamics of the nascent protein toward its final three-dimensional shape (36, 37). Such a mechanistic hypothesis is worthy of investigation in the context of P. falciparum biology.
As documented for other ABC transporters in P. falciparum (32), PfMRP2 has divergent insertions throughout its protein sequence, which normally occur within protein domain linkers. Although already observed in other P. falciparum ABC transporters, namely, Pgh-1, the number and extension of polymorphisms of these structures in PfMRP2 are unprecedented (Fig. 1 and 2). All MI regions found are localized outside the TM domains. Their exclusion from such protein regions is probably related to the inability of the transmembrane domains to accommodate this type of polymorphism, as the polymorphisms most likely alter the intramembrane α-helix structure, leading to functionally deleterious distortions in the three-dimensional structure of the protein. Considering their location, the extension of these polymorphisms is expected to have a nonnegligible effect in the protein structure and potentially in its function. Unfortunately, in contrast with our previous work on the ABC transporter Pgh-1 (32), the large number of variable insertions/deletions impaired the development of a meaningful homology structure model to validate its impact on the protein's function.
Suggestively, associative trends were revealed between variations in the MI-II region and the IC50s of PPQ, a 4-aminoquinoline structurally related to CQ. These data are consistent with the fact that CQ and PPQ show a degree of cross-resistance (22). An increase of asparagines from 10 to 14 at the MI-V region was also found associated with increased MQ susceptibility. It is conceivable that these variations alter the steady-state conformations of the cytoplasmic regions of the protein and with that, the interaction with the antimalarial drugs, affecting their efflux out of the parasite. The proposed potential role of PfMRP2 in the drug response to CQ and MQ is further supported by the recent DNA microarray-based observations that associated Pfmrp2 promoter polymorphisms with the parasite CQ and MQ responses in vitro (20).
Recently, Okombo et al. (38) found associations between variation in the size of the PfMRP2 MI-III and the in vitro parasite responses to lumefantrine. Since this MI region was not polymorphic in our analyzed set of isolates (all have the DNNNTS sequence deleted), our study does not confirm or deny the previously reported observation. It is possible that this might be related to the fact that, although the present study includes a substantial number of laboratory-adapted strains, the herein-revealed biodiversity of this gene will demand future studies with larger sample sizes for definitive determination of the role of PfMRP2 in the parasite response to antimalarial drugs.
We are reporting for the first time a likely influence of the variable MI regions on the parasite clearance time upon ACT treatment (artesunate-mefloquine). Although these data are preliminary, they suggest a potential role of PfMRP2 in the capacity for the parasite to withstand clinical levels of artemisinin (and possibly mefloquine). It is possible that some of these variants involve fitness costs that become evident upon exposure to a stressor, like the aforementioned drugs. Though a change in fitness characteristics by itself may not be considered a resistance response (39), our data point to the potential contribution of PfMRP2 in the capacity of the parasite in evading the action of these powerful but short-term drugs.
The likely influence of PfMRP2 on the parasite response to multiple drugs mirrors that reported for PfMRP1 and its proposed role in CQ, artemisinin, quinine, mefloquine, and lumefantrine (14, 16, 40, 41). The present report adds to several independent lines of evidence by our investigators and others, suggesting the importance of MRP-like proteins in parasite drug responses, highlighting the genetic variances found herein with their potential use as molecular sentinel tools.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by project grants from the Swedish Development Cooperation Agency, Department for Research Cooperation (SWE-2007-174 and SWE-2009-165). M.I.V. and N.S.O. are recipients of postdoctoral fellowship from Fundação para a Ciência e Tecnologia (FCT)/Ministerio da Ciência e Ensino Superior, Portugal, MCES (SFRH/BPD/76614/2011 and UMINHO/BPD/15/2014, respectively). The Shoklo Malaria Research Unit is part of the Mahidol Oxford University Tropical Medicine Research Unit and is supported by the Wellcome Trust of Great Britain.
We thank Zbynek Bozdech and Margarida Saraiva for helpful discussions and for critically reading the manuscript.
Footnotes
Published ahead of print 29 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03337-14.
REFERENCES
- 1.Rodrigues Coura J. 1987. Memoir of the memorias. Development of malaria hematozoa resistant to quinine. By Arthur Neiva, 1910. Mem. Inst. Oswaldo Cruz 82:303–309 (In Portugese.) [PubMed] [Google Scholar]
- 2.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 361:455–467. 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM. 2008. Evidence of artemisinin-resistant malaria in western Cambodia. N. Engl. J. Med. 359:2619–2620. 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 4.Takala-Harrison S, Clark TG, Jacob CG, Cummings MP, Miotto O, Dondorp AM, Fukuda MM, Nosten F, Noedl H, Imwong M, Bethell D, Se Y, Lon C, Tyner SD, Saunders DL, Socheat D, Ariey F, Phyo AP, Starzengruber P, Fuehrer HP, Swoboda P, Stepniewska K, Flegg J, Arze C, Cerqueira GC, Silva JC, Ricklefs SM, Porcella SF, Stephens RM, Adams M, Kenefic LJ, Campino S, Auburn S, Macinnis B, Kwiatkowski DP, Su XZ, White NJ, Ringwald P, Plowe CV. 2013. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc. Natl. Acad. Sci. U. S. A. 110:240–245. 10.1073/pnas.1211205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes KI. 2012. Antimalarial drugs and the control and elimination of malaria, p 1–17 In Staines HM, Krishna S. (ed), Treatment and prevention of malaria. Springer, Basel, Switzerland. [Google Scholar]
- 6.Gil JP. 2013. Malaria pharmacogenomics: return to the future. Pharmacogenomics 14:707–710. 10.2217/pgs.13.41. [DOI] [PubMed] [Google Scholar]
- 7.Klaassen CD, Aleksunes LM. 2010. Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol. Rev. 62:1–96. 10.1124/pr.109.002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haimeur A, Conseil G, Deeley RG, Cole SP. 2004. The MRP-related and BCRP/ABCG2 multidrug resistance proteins: biology, substrate specificity and regulation. Curr. Drug Metab. 5:21–53. 10.2174/1389200043489199. [DOI] [PubMed] [Google Scholar]
- 9.Vezmar M, Georges E. 1998. Direct binding of chloroquine to the multidrug resistance protein (MRP): possible role for MRP in chloroquine drug transport and resistance in tumor cells. Biochem. Pharmacol. 56:733–742. 10.1016/S0006-2952(98)00217-2. [DOI] [PubMed] [Google Scholar]
- 10.Wu CP, Klokouzas A, Hladky SB, Ambudkar SV, Barrand MA. 2005. Interactions of mefloquine with ABC proteins, MRP1 (ABCC1) and MRP4 (ABCC4) that are present in human red cell membranes. Biochem. Pharmacol. 70:500–510. 10.1016/j.bcp.2005.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klokouzas A, Tiffert T, van Schalkwyk D, Wu CP, van Veen HW, Barrand MA, Hladky SB. 2004. Plasmodium falciparum expresses a multidrug resistance-associated protein. Biochem. Biophys. Res. Commun. 321:197–201. 10.1016/j.bbrc.2004.06.135. [DOI] [PubMed] [Google Scholar]
- 12.Bozdech Z, Ginsburg H. 2004. Antioxidant defense in Plasmodium falciparum: data mining of the transcriptome. Malar. J. 3:23. 10.1186/1475-2875-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kavishe RA, van den Heuvel JM, van de Vegte-Bolmer M, Luty AJ, Russel FG, Koenderink JB. 2009. Localization of the ATP-binding cassette (ABC) transport proteins PfMRP1, PfMRP2, and PfMDR5 at the Plasmodium falciparum plasma membrane. Malar. J. 8:205. 10.1186/1475-2875-8-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raj DK, Mu J, Jiang H, Kabat J, Singh S, Sullivan M, Fay MP, McCutchan TF, Su XZ. 2009. Disruption of a Plasmodium falciparum multidrug resistance-associated protein (PfMRP) alters its fitness and transport of antimalarial drugs and glutathione. J. Biol. Chem. 284:7687–7696. 10.1074/jbc.M806944200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahlstrom S, Ferreira PE, Veiga MI, Sedighi N, Wiklund L, Martensson A, Farnert A, Sisowath C, Osorio L, Darban H, Andersson B, Kaneko A, Conseil G, Bjorkman A, Gil JP. 2009. Plasmodium falciparum multidrug resistance protein 1 and artemisinin-based combination therapy in Africa. J. Infect. Dis. 200:1456–1464. 10.1086/606009. [DOI] [PubMed] [Google Scholar]
- 16.Dahlstrom S, Veiga MI, Martensson A, Bjorkman A, Gil JP. 2009. Polymorphism in PfMRP1 (Plasmodium falciparum multidrug resistance protein 1) amino acid 1466 associated with resistance to sulfadoxine-pyrimethamine treatment. Antimicrob. Agents Chemother. 53:2553–2556. 10.1128/AAC.00091-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ursing J, Zakeri S, Gil JP, Bjorkman A. 2006. Quinoline resistance associated polymorphisms in the pfcrt, pfmdr1 and pfmrp genes of Plasmodium falciparum in Iran. Acta Trop. 97:352–356. 10.1016/j.actatropica.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Veiga MI, Ferreira PE, Schmidt BA, Ribacke U, Bjorkman A, Tichopad A, Gil JP. 2010. Antimalarial exposure delays Plasmodium falciparum intra-erythrocytic cycle and drives drug transporter genes expression. PLoS One 5:e12408. 10.1371/journal.pone.0012408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1:e5. 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mok S, Liong KY, Lim EH, Huang X, Zhu L, Preiser PR, Bozdech Z. 2013. Structural polymorphism in the promoter of pfmrp2 confers Plasmodium falciparum tolerance to quinoline drugs. Mol. Microbiol. 91:918–934. 10.1111/mmi.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veiga MI, Ferreira PE, Jornhagen L, Malmberg M, Kone A, Schmidt BA, Petzold M, Bjorkman A, Nosten F, Gil JP. 2011. Novel polymorphisms in Plasmodium falciparum ABC transporter genes are associated with major ACT antimalarial drug resistance. PLoS One 6:e20212. 10.1371/journal.pone.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veiga MI, Ferreira PE, Malmberg M, Jornhagen L, Bjorkman A, Nosten F, Gil JP. 2012. pfmdr1 Amplification is related to increased Plasmodium falciparum in vitro sensitivity to the bisquinoline piperaquine. Antimicrob. Agents Chemother. 56:3615–3619. 10.1128/AAC.06350-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ewing B, Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186–194. [PubMed] [Google Scholar]
- 24.Ning Z, Cox AJ, Mullikin JC. 2001. SSAHA: a fast search method for large DNA databases. Genome Res. 11:1725–1729. 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D. 2010. Tablet: next generation sequence assembly visualization. Bioinformatics 26:401–402. 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staden R. 1996. The Staden sequence analysis package. Mol. Biotechnol. 5:233–241. 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- 27.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noedl H, Bronnert J, Yingyuen K, Attlmayr B, Kollaritsch H, Fukuda M. 2005. Simple histidine-rich protein 2 double-site sandwich enzyme-linked immunosorbent assay for use in malaria drug sensitivity testing. Antimicrob. Agents Chemother. 49:3575–3577. 10.1128/AAC.49.8.3575-3577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viklund H, Elofsson A. 2008. OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 24:1662–1668. 10.1093/bioinformatics/btn221. [DOI] [PubMed] [Google Scholar]
- 30.Manske M, Miotto O, Campino S, Auburn S, Almagro-Garcia J, Maslen G, O'Brien J, Djimde A, Doumbo O, Zongo I, Ouedraogo JB, Michon P, Mueller I, Siba P, Nzila A, Borrmann S, Kiara SM, Marsh K, Jiang H, Su XZ, Amaratunga C, Fairhurst R, Socheat D, Nosten F, Imwong M, White NJ, Sanders M, Anastasi E, Alcock D, Drury E, Oyola S, Quail MA, Turner DJ, Ruano-Rubio V, Jyothi D, Amenga-Etego L, Hubbart C, Jeffreys A, Rowlands K, Sutherland C, Roper C, Mangano V, Modiano D, Tan JC, Ferdig MT, Amambua-Ngwa A, Conway DJ, Takala-Harrison S, Plowe CV, Rayner JC, Rockett KA, Clark TG, Newbold CI, Berriman M, MacInnis B, Kwiatkowski DP. 2012. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature 487:375–379. 10.1038/nature11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson T, Campino SG, Auburn S, Assefa SA, Polley SD, Manske M, MacInnis B, Rockett KA, Maslen GL, Sanders M, Quail MA, Chiodini PL, Kwiatkowski DP, Clark TG, Sutherland CJ. 2011. Drug-resistant genotypes and multi-clonality in Plasmodium falciparum analysed by direct genome sequencing from peripheral blood of malaria patients. PLoS One 6:e23204. 10.1371/journal.pone.0023204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferreira PE, Holmgren G, Veiga MI, Uhlen P, Kaneko A, Gil JP. 2011. PfMDR1: mechanisms of transport modulation by functional polymorphisms. PLoS One 6:e23875. 10.1371/journal.pone.0023875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mu J, Ferdig MT, Feng X, Joy DA, Duan J, Furuya T, Subramanian G, Aravind L, Cooper RA, Wootton JC, Xiong M, Su XZ. 2003. Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol. Microbiol. 49:977–989. 10.1046/j.1365-2958.2003.03627.x. [DOI] [PubMed] [Google Scholar]
- 34.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2013. GenBank. Nucleic Acids Res. 41:D36–D42. 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffmeyer S, Burk O, von Richter O, Arnold HP, Brockmoller J, Johne A, Cascorbi I, Gerloff T, Roots I, Eichelbaum M, Brinkmann U. 2000. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc. Natl. Acad. Sci. U. S. A. 97:3473–3478. 10.1073/pnas.97.7.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fung KL, Gottesman MM. 2009. A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. Biochim. Biophys. Acta 1794:860–871. 10.1016/j.bbapap.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. 2007. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315:525–528. 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 38.Okombo J, Abdi AI, Kiara SM, Mwai L, Pole L, Sutherland CJ, Nzila A, Ochola-Oyier LI. 2013. Repeat polymorphisms in the low-complexity regions of Plasmodium falciparum ABC transporters and associations with in vitro antimalarial responses. Antimicrob. Agents Chemother. 57:6196–6204. 10.1128/AAC.01465-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Froberg G, Ferreira PE, Martensson A, Ali A, Bjorkman A, Gil JP. 2013. Assessing the cost-benefit effect of a Plasmodium falciparum drug resistance mutation on parasite growth in vitro. Antimicrob. Agents Chemother. 57:887–892. 10.1128/AAC.00950-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hao M, Jia D, Li Q, He Y, Yuan L, Xu S, Chen K, Wu J, Shen L, Sun L, Zhao H, Yang Z, Cui L. 2013. In vitro sensitivities of Plasmodium falciparum isolates from the China-Myanmar border to piperaquine and association with polymorphisms in candidate genes. Antimicrob. Agents Chemother. 57:1723–1729. 10.1128/AAC.02306-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Briolant S, Henry M, Oeuvray C, Amalvict R, Baret E, Didillon E, Rogier C, Pradines B. 2010. Absence of association between piperaquine in vitro responses and polymorphisms in the pfcrt, pfmdr1, pfmrp, and pfnhe genes in Plasmodium falciparum. Antimicrob. Agents Chemother. 54:3537–3544. 10.1128/AAC.00183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.