Abstract
The ubiquitous water-borne Gram-negative bacterium Aeromonas salmonicida subsp. salmonicida is the causative agent of furunculosis, a worldwide disease in fish farms. Plasmids carrying antibiotic resistance genes have already been described for this bacterium. The aim of the present study was to identify and characterize additional multidrug resistance plasmids in A. salmonicida subsp. salmonicida. We sequenced the plasmids present in two multiple antibiotic-resistant isolates using high-throughput technologies. We also investigated 19 other isolates with various multidrug resistance profiles by genotyping PCR and assessed their resistance to tetracycline. We identified variants of the pAB5S9 and pSN254 plasmids that carry several antibiotic resistance genes and that have been previously reported in bacteria other than A. salmonicida subsp. salmonicida, which suggests a high level of interspecies exchange. Genotyping analyses and the antibiotic resistance profiles of the 19 other isolates support the idea that multiple versions of pAB5S9 and pSN254 exist in A. salmonicida subsp. salmonicida. We also identified variants of the pRAS3 plasmid. The present study revealed that A. salmonicida subsp. salmonicida harbors a wide variety of plasmids, which suggests that this ubiquitous bacterium may contribute to the spread of antibiotic resistance genes in the environment.
INTRODUCTION
The Gram-negative bacterium Aeromonas salmonicida subsp. salmonicida is an opportunistic fish pathogen (1). It is the etiological agent of furunculosis, a disease that especially affects salmonids in fish farms (2). While antibiotics are commonly used to treat A. salmonicida subsp. salmonicida infections, multidrug-resistant isolates have been frequently detected (3–5), preventing the effective treatment of furunculosis.
Many fully characterized plasmids from A. salmonicida subsp. salmonicida have provided antibiotic resistance to this species (2). All the known plasmids in A. salmonicida subsp. salmonicida harboring antibiotic resistance genes include at least a tetracycline resistance gene. The vast majority of the plasmids bearing antibiotic resistance genes confer multiple types of resistance to A. salmonicida subsp. salmonicida, including the large (167-kb) plasmid pAsa4, which provides resistance against chloramphenicol, spectinomycin, streptomycin, sulfonamides, tetracycline, mercury, and quaternary ammonium compounds (6). A plasmid bearing multiple resistance genes that is similar to the large pSN254 plasmid in Salmonella enterica (7) has been partially described in A. salmonicida subsp. salmonicida (3). This pSN254-like plasmid can be transferred via conjugation from A. salmonicida subsp. salmonicida to multiple receivers, including Escherichia coli, Edwardsiella tarda, and Aeromonas hydrophila (3).
Plasmid variants appear to be relatively frequent in A. salmonicida subsp. salmonicida. The best example is the pRAS3 plasmid. To date, two variants of this plasmid (pRAS3.1 and pRAS3.2) have been described (8). The differences between them are very subtle and consist of two additional repetition units in pRAS3.1, one that is 22 nucleotides in length and another that is 6 nucleotides in length. pAsal1B is another example of a plasmid variant. In this case, pAsal1B differs from the parental plasmid (pAsal1) by the presence of an AS5 insertion sequence (IS) and a fragment of the same IS (9).
Given these observations regarding plasmids in A. salmonicida subsp. salmonicida, it is possible that additional multidrug-resistant plasmids in this bacterium remain unidentified. Thus, we analyzed 78 A. salmonicida subsp. salmonicida isolates to identify those displaying multidrug resistance. By using next-generation sequencing methods, we detected pRAS3, pAB5S9, and pSN254 plasmid variants. We showed that A. salmonicida subsp. salmonicida displays marked heterogeneity in terms of the repertoire of plasmids bearing antibiotic resistance genes. This plasmid diversity may have major consequences in terms of treatments for furunculosis and highlights the significant flow of antibiotic resistance genes among different water-borne bacteria.
MATERIALS AND METHODS
Bacterial isolates and growth conditions.
The A. salmonicida subsp. salmonicida isolates used in the present study were the same as those analyzed by Trudel et al. (9) (see Table S1 in the supplemental material). They were grown on furunculosis agar (10) for 2 or 3 days at 18°C.
Antibiogram analyses.
One colony of each strain was resuspended in 2 ml of furunculosis broth, which was shaken at 200 rpm overnight at 18°C. The cultures were adjusted to an optical density (OD) at 595 nm of 0.9 and were spread on Mueller-Hinton agar medium (Oxoid, Canada) to determine their levels of resistance to the following antibiotics with the disc diffusion method: ampicillin (10 μg), chloramphenicol (30 μg), erythromycin (15 μg), gentamicin (10 μg), nalidixic acid (30 μg), sulfamethoxazole-trimethoprim (23.75 μg/1.25 μg), streptomycin (10 μg), tetracycline (5 μg), and trimethoprim (5 μg) (Becton Dickinson, Sparks, MD, USA). The plates were incubated at 18°C to prevent possible instability of the plasmids bearing antibiotic resistance genes. This kind of instability has already been described for pAsa5, a large plasmid bearing the type III secretion system (11–13). Bacterial growth after 48 h was recorded. Since there is, to our knowledge, no control chart for the diameter of the zones of antibiotic-driven growth inhibition at this temperature with this bacterium, antibiotic resistance was determined based on the relative sizes of the growth inhibition zones of the isolates compared to that of the sensitive strain 01-B526, which has been sequenced and has no antibiotic resistance genes (14), and to that of the resistant strain m15879-11 (see below) (data not shown). Every assay was performed at least in duplicate.
Assessment of the tetracycline MIC.
The MIC of tetracycline was assessed for 21 A. salmonicida subsp. salmonicida isolates (Table 1). The isolates were inoculated on furunculosis agar from frozen stocks and were grown at 18°C for 48 h before each experiment. Several colonies of each isolate were suspended in fresh LB medium (EMD Millipore, Canada). The optical densities (OD) of the bacterial suspensions were measured at 595 nm, the suspensions were diluted to 0.1 of the OD, and 3 × 107 bacteria (30 μl) were deposited in the wells of 48-well microplates. Tetracycline (Calbiochem) was serially diluted in LB medium, and an aliquot of each dilution was placed into a well of the 48-well microplates to obtain antibiotic concentrations ranging from 0 to 256 μg/ml in a final volume of 300 μl. The plates were incubated at 18°C for 48 h with shaking at 200 rpm in a Tecan Infinite F200 PRO microplate reader (Tecan, Morrisville, NC, USA). Growth was assessed during and at the end of the incubation period. Every assay was performed at least in duplicate.
TABLE 1.
Characteristics of multidrug-resistant A. salmonicida subsp. salmonicida isolates harboring a tetracycline resistance gene
| Isolatea | Antibiotic resistanceb | MIC (μg/ml) for tetracyclinec | Plasmids foundd |
|---|---|---|---|
| 01-B522 | SXT, TET, TMP | 128 | Cryptic, pAsal1, pAsa5, pAsa4-like |
| 07-9324 | STR, AMP, TET, CHL | 16 | Cryptic, pAsal1, pAsa5, pSN254-like |
| 07-7817 | STR, AMP, TET, CHL | 16 | Cryptic, pAsal1, pAsa5, pSN254-like |
| 08-2647 | SXT, STR, AMP, TET | 16 | Cryptic, pAsal1, pAsa5, pSN254-like |
| 07-7287 | STR, AMP, TET, CHL | 16 | Cryptic, pAsal1, pAsa5, pSN254-like |
| 08-2783 | STR, AMP, TET | 16 | Cryptic, pAsal1, pAsa5, pSN254-like |
| 08-4188 | STR, AMP, TET, CHL | 16 | Cryptic, pAsal1, pAsa5, pSN254-like |
| 2009-157K5 | STR, TET, ERY | 128 | Cryptic, pAsal1, pAsa5, pRas3.3 |
| 2010-47K18 | STR, TET, CHL | 128 | Cryptic, pAsal1, pAsa5, pRas3.4, pAB5S9-like |
| 2004-05MF26 | NAL, SXT, STR, AMP, TET, ERY, GEN, CHL | 16 | Cryptic, pAsal1, pAsa5, pSN254b |
| 2004-68K52 | NAL, SXT, STR, AMP, TET, CHL | 64 | Cryptic, pAsal1, pAsa5, pSN254-like |
| 2009-195K29 | STR, TET, ERY | 32 | Cryptic, pAsal1, pAsa5, pRas3.3 |
| 2009-144K3 | SXT, STR, TET, ERY, CHL | 256 | Cryptic, pAsal1, pAsa5, pRas3.3, pAB5S9b |
| M15448-11 | STR, AMP, TET, CHL | 32 | Cryptic, pAsal1, pAsa5 |
| M16474-11 | STR, AMP, TET, ERY, CHL | 32 | Cryptic, pAsal1, pAsa5 |
| M14481-11 | SXT, STR, AMP, TET, CHL | 16 | Cryptic, pAsal1, pAsa5, pSN254-like |
| M15879-11 | SXT, STR, AMP, TET, CHL | 16 | Cryptic, pAsal1, pAsa5, pSN254-like |
| M17739-11 | SXT, STR, AMP, TET, CHL | 16 | Cryptic, pAsal1, pAsa5, pSN254-like |
| M13732-11 | STR, AMP, TET, CHL | 16 | Cryptic, pAsal1, pAsa5, pSN254-like |
| M17053-11 | STR, AMP, TET, CHL | 16 | Cryptic, pAsal1, pAsa5, pSN254-like |
| M15469-11 | STR, AMP, TET, CHL | 16 | Cryptic, pAsal1, pAsa5, pSN254-like |
The origin and source of these isolates are given in Table S1 in the supplemental material.
NAL, nalidixic acid; SXT, sulfamethoxazole-trimethoprim; STR, streptomycin; AMP, ampicillin; TET, tetracycline; ERY, erythromycin; GEN, gentamicin; TMP, trimethoprim; CHL, chloramphenicol.
The MIC of the sensitive 01-B526 isolate was 0.5 μg/ml.
The three cryptic plasmids pAsa1, pAsa2, and pAsa3, as well as pAsal1 and pRAS3, were identified in these isolates in a previous study by Trudel et al. (9). The pAsa5, pAsa4-like, pAB5S9-like, and pSN254-like plasmids were found by PCR genotyping in the present study (see Table S2 in the supplemental material).
DNA extraction and genomic sequencing.
The total genomic DNA of the isolates was extracted using DNeasy blood and tissue kits (Qiagen, Canada). A 5-kb mate pair library was prepared using isolate 2009-144K3 and was sequenced on a GS-FLX+ instrument (Roche, Branford, CT, USA). A TruSeq shotgun library prepared with isolate 2004-05MF26 was sequenced using the Illumina MiSeq sequencing system (Illumina, San Diego, CA, USA). These two isolates were sequenced at the Plateforme d'Analyses Génomique of the Institut de Biologie Intégrative et des Systèmes (IBIS, Université Laval). The resulting reads of the 2009-144K3 isolate were de novo assembled with Newbler version 2.5.3 (15). In the case of isolate 2004-05MF26, the reads were filtered using Trimmomatic version 0.32 (16) according to the parameters suggested in the manual and were de novo assembled with Ray version 2.3.1 (17) at a k-mer length of 75.
Sequence analyses.
Contigs resulting from the de novo assemblies were first mapped on the chromosome and the A449 plasmid sequence using the CONTIGuator Web server (18) with default parameters. The large unmapped contigs were investigated by conducting BLASTx and BLASTn searches of the NCBI databases nr/nt. For the 2009-144K3 isolate, two contigs exhibited high degrees of identity with the pAB5S9 plasmid (19), while one contig exhibited a high degree of identity with the pRAS3 plasmid (8). The two pAB5S9-like contigs were assembled using Consed version 35 (20), and the junctions were verified by PCR amplifications. The resulting plasmid was named pAB5S9b. A third-party annotation was produced for the original pAB5S9 plasmid (GenBank accession number EF495198). Since the pRAS3-like plasmid was in a single contig, no subsequent assembly was required, and it was named pRAS3.3 (see Results and Discussion).
In the case of the 2004-05MF26 isolate, a single unmapped contig was found. This contig corresponded to a pSN254-like plasmid, which differed markedly from the pSN254 plasmid originally found in S. enterica (7). As such, it was named pSN254b (see Results and Discussion).
Annotations for the pAB5S9, pAB5S9b, and pRAS3.3 plasmids were performed manually. Open reading frames (ORFs) were found by a BLASTx search of the NCBI nonredundant database, and each ORF was verified. Given the size of the pSN254b plasmid, it was annotated using a custom Perl script. ORFs of ≥30 amino acids were found using the GETORF function of EMBOSS (21). Their products were then identified using a similarity search of a local formatted database with the heuristic fasta35 algorithm (22). The coordinates of each ORF-encoded protein were determined using a similarity search with tfasty35 (23) of the main sequence, which in this case was pSN254b. The search generated output files compatible with the Artemis genome viewer (24) and the Sequin annotation tool. Each ORF from the resulting annotation was manually verified.
The genomic maps and GC skews of the plasmids were generated using DNAPlotter version 1.10 (25) and were visualized using the genome viewer Artemis, version 15 (24).
PCR analyses.
The DNA templates, PCR mixtures, and program cycles were done as previously described (9). The PCR assays were performed at least twice, and suitable positive and negative controls were included in each assay.
The PCR primers used in the present study are listed in Table S2 in the supplemental material. We designed primers that were specific for each multidrug resistance plasmid (pAsa4, pAB5S9, pSN254). In the case of pAB5S9, two primer pairs detected pAB5S9 and pAB5S9b with the same amplicon size, while the third primer pair detected different amplicon sizes for the two plasmids (see Table S2). For pSN254, two primer pairs detected pSN254 from S. enterica and pSN254b from A. salmonicida subsp. salmonicida with the same amplicon size, while the third primer pair was specific to pSN254b (see Table S2).
The two regions in pRAS3-like plasmids which exhibit variations among repetition units were sequenced for all pRAS3-like positive isolates other than 2009-144K3 (i.e., for 2010-47K18, 2009-157K5, and 2009-195K29) using the Sanger method on a 3130XL apparatus (Applied Biosystems) at the Plateforme d'Analyses Génomique of the Institut de Biologie Intégrative et des Systèmes (IBIS, Université Laval).
Nucleotide sequence accession numbers.
The sequences of pAB5S9, pAB5S9b, pRAS3.3, and pSN254b were deposited in GenBank under the accession numbers BK008853, KJ909292, KJ909291, and KJ909290, respectively.
RESULTS AND DISCUSSION
We tested the antibiotic resistance of 78 A. salmonicida subsp. salmonicida isolates, of which 30 were resistant to at least two antibiotics (Table S1). To investigate the putative presence of new plasmids carrying multidrug resistance genes, we used high-throughput methods to determine the sequences of the 2009-144K3 and 2004-05MF26 isolates, which were resistant to a large number of antibiotics but displayed differences in their antibiotic resistance profiles (Table 1). These bacteria were isolated from New Brunswick and Quebec, Canada, respectively.
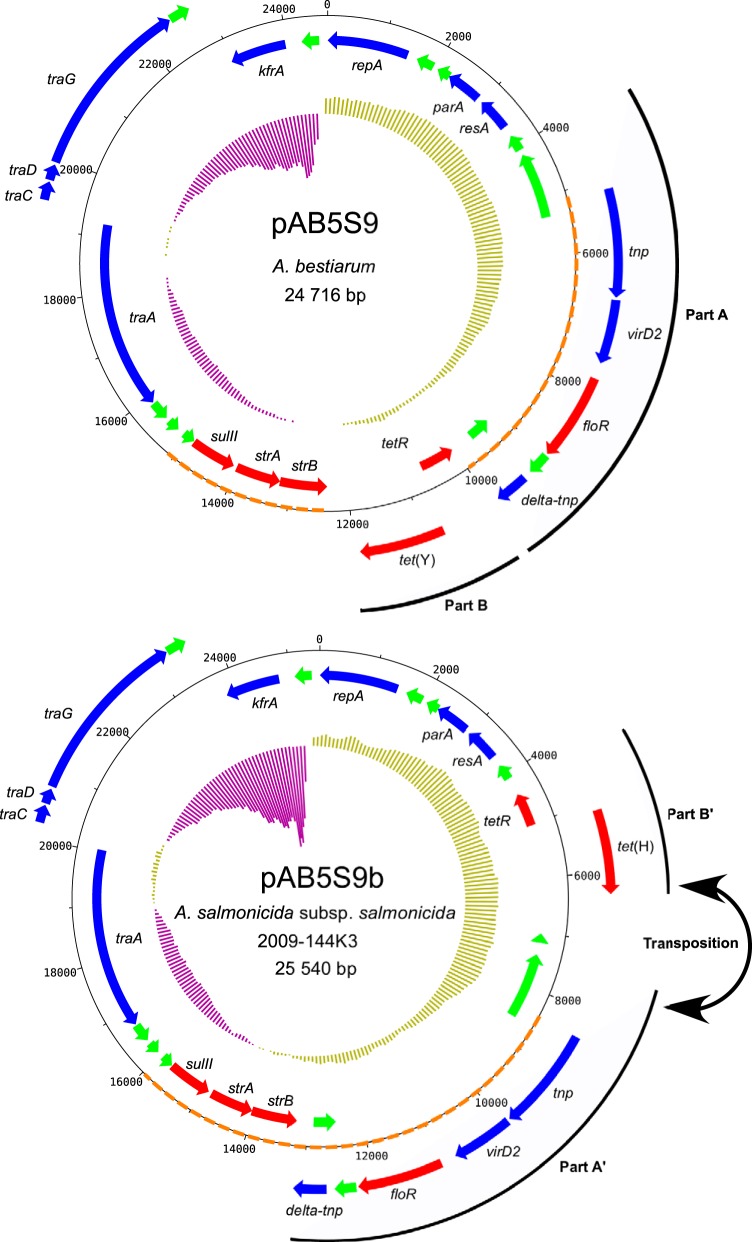
The sequencing of the A. salmonicida subsp. salmonicida 2009-144K3 isolate identified a large contig that shared no identity with the chromosome or known plasmid sequences associated with A. salmonicida subsp. salmonicida. Further analyses revealed a high level of homology with pAB5S9, a previously described plasmid from Aeromonas bestiarum (19), which carries multiple antibiotic resistance genes. pAB5S9b, the 2009-144K3 variant, exhibited a highly conserved gene distribution and gene order with its homolog except for a transposition of two regions (Fig. 1). One of these regions (Fig. 1, part A) had a complete putatively functional transposase and a truncated transposase (delta-tnp). These transposases have been reported to be part of an ISCR2 element in A. bestiarum (19). The other region (Fig. 1, part B) contained two genes involved in tetracycline resistance. Surprisingly, one of these genes in the pAB5S9 plasmid, tet(Y), did not appear to have a homologous gene in the pAB5S9b plasmid. However, another gene involved in tetracycline resistance, tet(H), was found at the same position. Moreover, the pAB5S9b variant contained genes that should confer resistance to sulfonamide (sulII), streptomycin (strA and strB), florfenicol (floR), and chloramphenicol (floR).
FIG 1.
Maps of pAB5S9 and pAB5S9b. Genes are represented by arrows. Genes on the outside are transcribed clockwise, whereas genes on the inside are transcribed counterclockwise. Red, green, and blue arrows represent genes coding for antibiotic resistance, hypothetical proteins, and other functions, respectively. The inner ring represents the GC skew calculated by DNAPlotter, with a window and step size of 10,000 bp and 100 bp, respectively. Parts A and B in pAB5S9 are transposed in pAB5S9b and are indicated as parts A′ and B′. The orange dotted lines represent the SXT resistance regions in pAB5S9 and pAB5S9b.
A region in the original pAB5S9 (19) shares a high degree of homology with a part of the SXT resistance element of Vibrio cholerae that contains the floR, strB, strA, and sulII genes (26). The region is cut in half in pAB5S9 by the tetR and tet(Y) genes (19). Interestingly, the SXT resistance region in pAB5S9b in A. salmonicida subsp. salmonicida is not altered (Fig. 1), suggesting that this plasmid might be closer to an ancestral SXT-like element than the original pAB5S9.
pAB5S9b is 824 bp larger than the pAB5S9 plasmid in A. bestiarum. This difference is mainly due to an insertion between parts A′ and B′ of the variant in A. salmonicida subsp. salmonicida. The insertion bears a single ORF that encodes a short hypothetical protein. An ortholog of this ORF has been annotated only in Acinetobacter baumannii (GenBank accession number EXS20171), an opportunistic Gram-negative human pathogen.
The pAB5S9 plasmid was first discovered in France (19). Our study identified, for the first time, a plasmid in the pAB5S9 family isolated in North America from a different Aeromonas species. This discovery showed that the distribution of pAB5S9 may be geographically independent and may be involved in the propagation of antibiotic resistance in many regions of the world.
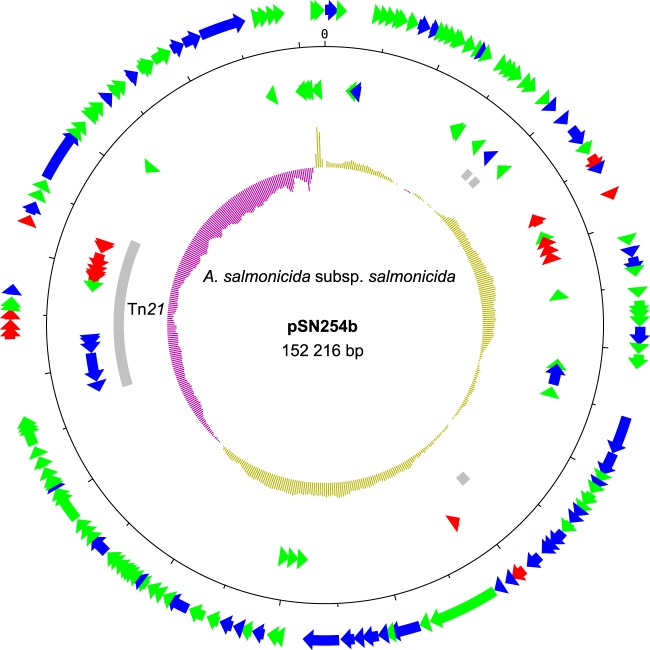
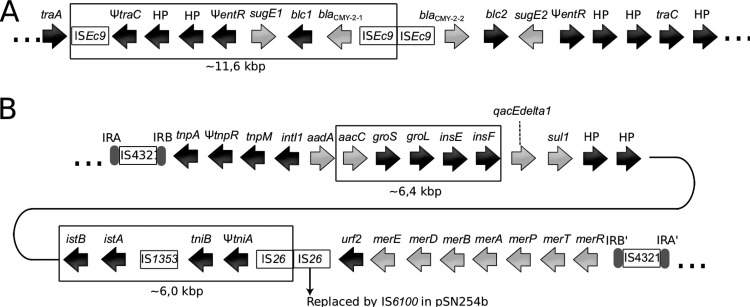
The pSN254 variant of the 2004-05MF26 isolate differed significantly from the original plasmid and was named pSN254b (Fig. 2). The variant in A. salmonicida subsp. salmonicida was smaller than the replicon identified in S. enterica by approximately 24 kbp. Three sections in pSN254b were missing; one was approximately 11.6 kbp, one was 6.4 kpb, and one was 6.0 kbp. The 11.6-kbp section corresponded to a duplicated fragment previously reported in S. enterica (7) that contains genes encoding hypothetical proteins, some of which are involved in multidrug resistance, including proteins for quaternary ammonium compound resistance (SugE1) and the CMY-2 beta-lactamase proteins (Fig. 3A). This section in the pSN254 plasmid in S. enterica is bordered by ISEc9, suggesting that it may be a putative duplicated composite transposon. The pSN254b variant contains a single copy of this IS. However, we cannot discriminate between the possible insertion of this putative transposon in pSN254 or its deletion from pSN254b by conservative transposition. This deletion is interesting, because it was previously reported in another plasmid, pAR060302, which is in the same incompatibility plasmid group (IncA/C) (27). This suggests that the two plasmids share a high degree of homology.
FIG 2.
Map of pSN254b. Genes are represented by arrows. Genes on the outside are transcribed clockwise, whereas genes on the inside are transcribed counterclockwise. Red, green, and blue arrows represent genes coding for antibiotic resistance, hypothetical proteins, and other functions, respectively. Gray rectangles represent mobile elements, such as transposons and IS. The inner ring represents the GC skew calculated by DNAPlotter with a window and step size of 20,000 bp and 300 bp, respectively. Each tick mark represents a 5,000-bp step.
FIG 3.
Differences between pSN254 and pSN254b (A) Putative composite transposon. The genes and the IS in the box are present in the pSN254 of S. enterica but are absent from the pSN254b of A. salmonicida subsp. salmonicida. (B) Transposon Tn21. The genes and the IS in the boxes are present in the pSN254 of S. enterica but are absent from the pSN254b of A. salmonicida subsp. salmonicida. Light gray arrows indicate genes coding for antibiotic and mercury resistance.
Interestingly, the 6.4-kbp and 6.0-kbp fragments, which are present in pSN254 but absent in pSN254b, are located in the transposon Tn21 (Fig. 3B), which is also present in the large plasmid pAsa4 (6). The first cassette located immediately downstream from the attI site is aadA, which was previously identified in this transposon (28) and is known to confer resistance to streptomycin and spectinomycin by adenylylation (29). However, five other cassettes follow the aadA cassette in the pSN254 plasmid of S. enterica. These cassettes consist of the 6.4-kbp section and are not present in the pSN254b plasmid of A. salmonicida subsp. salmonicida.
The other section absent from pSN254b (6.0 kbp) corresponds to the cassettes for the istB and istA genes, the insertion sequence IS1353, and the tni module, which encodes an ATPase and a transposase. Another interesting characteristic of the A. salmonicida subsp. salmonicida Tn21 is the replacement of IS26 by IS6100, which is present in a wide range of hosts (30). Overall, pSN254b provides resistance to florfenicol, chloramphenicol, tetracycline, streptomycin, spectinomycin, sulfonamide, quaternary ammonium compounds, beta-lactam antibiotics, and mercury.
pSN254b is related to pSN254, a blaCMY-2 plasmid, and shares a typical backbone with other plasmids from the A/C incompatibility plasmid group (IncA/C). The majority of the genes with known functions codes for the replication/maintenance of the plasmid and for a type IV conjugative transfer system (7). Putative hypothetical protein-coding genes make up approximately half of the gene repertoire of the plasmid, suggesting that additional functions for this plasmid may be discovered in the future.
Variations among blaCMY-2 plasmids have been reported (27), and it has been proposed that, if a blaCMY-2 plasmid were discovered in A. salmonicida, it would be the first time that the spread of a blaCMY-2 plasmid was associated with a marine disease.
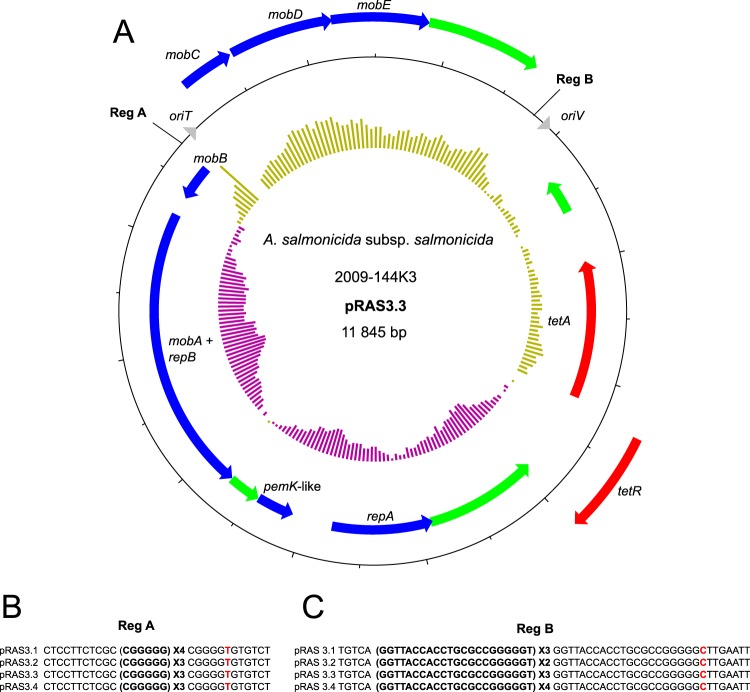
Two variants of pRAS3, an IncQ-related plasmid that carries genes for tetracycline resistance, have been described (8). They display minor differences, mainly in repetition units at two sites. We identified two additional variants, pRAS3.3 and pRAS3.4. They differed in terms of the repetition units located at the same sites in pRAS3.1 and pRAS3.2 (Fig. 4). Four isolates which are known to bear pRAS3 (9) were included in our analysis. In addition to 2009-144K3, these isolates included 2010-47K18, 2009-157K5, and 2009-195K29. Of these four isolates, three bore pRAS3.3, and one bore pRAS3.4 (Table 1). Tandem repeated sequences ended with a highly similar repeated unit but had a point mutation at the last nucleotide for the two repetition spots. The repetition spot identified as region A (Reg A) ended with a repeated unit with a transversion (Fig. 4B), while the repeated unit of Reg B had a transition (Fig. 4C). In these two cases, the repetitions were close to the regulation sequence involved in mobilization or replication (oriT or oriV, respectively). We propose that these repeated sequences may be involved in the regulation of plasmid transfer or copy number, as previously suggested for IncQ plasmids (8, 31, 32), but further studies are required to confirm this.
FIG 4.
Map of pRAS3.3 and differences among variants. A. The map of pRAS3.3 in the 2009-144K3 isolate. Genes are represented by arrows. Genes on the outside are transcribed clockwise, whereas genes on the inside are transcribed counterclockwise. Red, green, and blue arrows represent genes coding for antibiotic resistance, hypothetical proteins, and other functions, respectively. The inner ring represents the GC skew calculated by DNAPlotter with a window and step size of 1,000 bp and 50 bp, respectively. Each tick mark represents a 500-bp step. (B and C) Representation of the differences in the number of repeat units between all pRAS3 variants for the region (Reg) A and Reg B spots. The red nucleotides are point mutations in the last repetition unit, either a transversion (B) or a transition (C).
Like pRAS3.1 and pRAS3.2, pRAS3.3 has the same two ORFs that code for an active toxin-antitoxin system related to the PemK/Mazf family (32, 33).
After identifying pAB5S9b and pSN254b, we PCR genotyped the multidrug-resistant A. salmonicida subsp. salmonicida isolates harboring a tetracycline resistance gene in order to identify other isolates that potentially bear these plasmids or pAsa4. Only the tetracycline-resistant isolates were tested, because pAB5S9b, pSN254b, pAsa4, and pRAS3 shared resistance to this antibiotic. In addition to 2009-144K3 and 2004-05MF26, 19 isolates were tested (Table 1). The genotyping primers were designed to detect pAB5S9 and pSN254. Other primers were also used to detect the presence of pAsa4 and pAsa5 (see Table S1 in the supplemental material). When combined with data from our previous study on small plasmids from the same isolates (9), the genotyping provided a more complete view of the plasmid profiles.
As expected for pathogenic isolates, they all displayed positive PCR results for pAsa5 (Table 1), which encodes the type III secretion system essential for the virulence of A. salmonicida subsp. salmonicida (34). One isolate (01-B522) was positive for pAsa4. However, this isolate displayed less antibiotic resistance than expected for a bacterium bearing pAsa4 (6). Given this, the pAsa4 plasmid in the 01-B522 isolate is likely a variant of the pAsa4 plasmid (i.e., pAsa4-like) in the A449 reference strain. Further analyses of this plasmid will be required once its complete sequence has been determined in order to evaluate its relationship to pAsa4.
In addition to 2009-144K3, only the 2010-47K18 isolate contained a pAB5S9 plasmid (Table 1). The three pAB5S9 primer pairs gave positive PCR signals for 2010-47K18, suggesting that this isolate bears a pAB5S9b plasmid. However, the 2010-47K18 isolate was not resistant to sulfamethoxazole, unlike 2009-144K3, whose resistance is provided by the sulII gene on pAB5S9 and pAB5S9b. As such, the pAB5S9b-like plasmid in 2010-47K18 was probably another variant of the pAB5S9 family. Interestingly, these two isolates harboring the pAB5S9 variants also each bore a pRAS3 plasmid. The two plasmid types carried resistance genes for tetracycline. As expected, these isolates, as well as the isolate containing the pAsa4-like plasmid and an isolate likely bearing an unknown plasmid (see below), had the highest tetracycline MICs.
Lastly, in addition to 2004-05MF26, 13 isolates exhibited positive PCR signals for all of the pSN254 primer pairs (Table 1 and data not shown. This is surprising, since these isolates had four different antibiotic profiles with respect to sulfamethoxazole, streptomycin, ampicillin, tetracycline, and chloramphenicol resistance. These results indicated that multiple variants of pSN254 (pSN254-like) may exist in various populations of A. salmonicida subsp. salmonicida. Further analyses are required to confirm this possibility.
Only four of the isolates did not bear one of the multidrug-resistance plasmids (pAB5S9, pSN254, pAsa4). Isolates 2009-157K5 and 2009-195K29 displayed resistance to tetracycline, erythromycin, and streptomycin. Since these two isolates possessed a pRAS3 plasmid, which confers tetracycline resistance, and since resistance to erythromycin and streptomycin is frequently due to point mutations on chromosomal genes (35), it is plausible that these isolates contained no other plasmids than those found by genotyping and plasmid profiling (9). However, the MIC of tetracycline for 2009-157K5 was higher than that for 2009-195K29. This may be due to a mutation in the pRAS3 plasmid of one of the isolates that increases its copy number, the expression of a tet gene, or another parameter related to the level of resistance to tetracycline. Another scenario is that 2009-157K5 bears another yet-unidentified plasmid that also codes for tetracycline resistance. The two other isolates that lacked a known multidrug resistance plasmid (M15448-11 and M16474-11) displayed potentially chromosome-encoded resistance to streptomycin and erythromycin (35) as well as resistance to ampicillin, tetracycline, and chloramphenicol. This suggests that these isolates may possess other multidrug resistance plasmids that have not yet been identified.
Overall, the present study showed that A. salmonicida subsp. salmonicida harbors a high number of plasmid variants. Two such variants (pAB5S9 and pSN254) have been identified in other bacterial genera or species, including S. enterica, a well-documented human pathogen. Moreover, pSN254 plasmids have been shown to be transferable by conjugation from A. salmonicida subsp. salmonicida to E. coli, A. hydrophila, and E. tarda (3). In fact, DNA transfer between nonhuman and human pathogenic bacteria is probably frequent under some conditions. It is possible that the genomic rearrangement activity seen in A. salmonicida subsp. salmonicida (36), coupled with a DNA flow between this bacterium and human pathogens, plays a role in spreading antibiotic resistance among human pathogens.
The marked heterogeneity in the composition and distribution of plasmids in A. salmonicida subsp. salmonicida, as revealed in the present study, confirmed the importance of studying the genome of this bacterium. Our results suggest that multiple still-uncharacterized variants of pSN254, pAB5S9, pRAS3, and pAsa4 exist and indicate that additional multidrug resistance plasmids may be identified in the future. High-throughput sequencing technologies will likely increase documented cases of DNA transfer between A. salmonicida subsp. salmonicida and other bacteria. Given that A. salmonicida subsp. salmonicida is a ubiquitous bacterium in aquatic environments (37), and considering the constantly growing number of multidrug resistance plasmids that are being found in this bacterium, it is reasonable to consider A. salmonicida subsp. salmonicida a potential reservoir of antibiotic resistance genes that can be transferred to human pathogens.
This study revealed additional elements of a growing worldwide problem, which is the prevalence of multidrug-resistant bacteria. It is now clear that pollution of environments by human activities, such as contamination by heavy metals, has contributed to the selection for multidrug-resistant bacteria (38). It is also known that the use of antibiotics in veterinary medicine has led to an increase in antibiotic levels in water (39). Our study provides a better understanding of A. salmonicida subsp. salmonicida, which is a model for bacteria in environments, such as fish farms, that have been significantly altered by human activities. We hope that this will prompt more research on this issue, given its health and economic impacts.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Laboratoire de bactériologie clinique, Faculté de médecine vétérinaire (Université de Montréal, Montreal, QC, Canada), the Félix d'Hérelle Reference Center (Département de biochimie, de microbiologie et de bio-informatique, Université Laval, Quebec City, QC, Canada), the Quebec Ministère de l'Agriculture, des Pêcheries et de l'Alimentation, and the Aquatic Animal Health Department, Fisheries and Oceans Canada (NB, Canada), and we thank J. Frey (University of Bern, Switzerland) for the A. salmonicida subsp. salmonicida isolates.
M.V.T. received scholarships under the CREATE program of Ressources Aquatiques Québec (RAQ). K.H.T. and S.D.-D. received Alexander Graham Bell Canada Graduate Scholarships from the Natural Sciences and Engineering Research Council of Canada (NSERC). R.K.D. received a scholarship under the Canadian Francophony Scholarship Program (CFSP). This project was funded by an NSERC Discovery grant to S.J.C., a grant from the Société de recherche et de développement en aquaculture continentale (SORDAC) to S.J.C. and N.D., and a grant from the RAQ to M.F.
Footnotes
Published ahead of print 29 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03730-14.
REFERENCES
- 1.Derome N, Boutin S, Llewellyn M, Gauthier J. Opportunistic pathogens of fish. In Hurst CJ. (ed), The Rasputin effect: when commensals and symbionts become parasitic, in press Springer, New York, NY. [Google Scholar]
- 2.Dallaire-Dufresne S, Tanaka KH, Trudel MV, Lafaille A, Charette SJ. 2014. Virulence, genomic features, and plasticity of Aeromonas salmonicida subsp. salmonicida, the causative agent of fish furunculosis. Vet. Microbiol. 169:1–7. 10.1016/j.vetmic.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 3.McIntosh D, Cunningham M, Ji B, Fekete FA, Parry EM, Clark SE, Zalinger ZB, Gilg IC, Danner GR, Johnson KA, Beattie M, Ritchie R. 2008. Transferable, multiple antibiotic and mercury resistance in Atlantic Canadian isolates of Aeromonas salmonicida subsp. salmonicida is associated with carriage of an IncA/C plasmid similar to the Salmonella enterica plasmid pSN254. J. Antimicrob. Chemother. 61:1221–1228. 10.1093/jac/dkn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sandaa RA, Enger O. 1994. Transfer in marine sediments of the naturally occurring plasmid pRAS1 encoding multiple antibiotic resistance. Appl. Environ. Microbiol. 60:4234–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sørum H, L'Abée-Lund TM, Solberg A, Wold A. 2003. Integron-containing IncU R plasmids pRAS1 and pAr-32 from the fish pathogen Aeromonas salmonicida. Antimicrob. Agents Chemother. 47:1285–1290. 10.1128/AAC.47.4.1285-1290.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reith ME, Singh RK, Curtis B, Boyd JM, Bouevitch A, Kimball J, Munholland J, Murphy C, Sarty D, Williams J, Nash JH, Johnson SC, Brown LL. 2008. The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics 9:427. 10.1186/1471-2164-9-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welch TJ, Fricke WF, McDermott PF, White DG, Rosso M-L, Rasko DA, Mammel MK, Eppinger M, Rosovitz MJ, Wagner D, Rahalison L, LeClerc JE, Hinshaw JM, Lindler LE, Cebula TA, Carniel E, Ravel J. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2:e309. 10.1371/journal.pone.0000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.L'Abée-Lund TM, Sørum H. 2002. A global non-conjugative Tet C plasmid, pRAS3, from Aeromonas salmonicida. Plasmid 47:172–181. 10.1016/S0147-619X(02)00001-X. [DOI] [PubMed] [Google Scholar]
- 9.Trudel MV, Tanaka KH, Filion G, Daher RK, Frenette M, Charette SJ. 2013. Insertion sequence AS5 (IS AS5) is involved in the genomic plasticity of Aeromonas salmonicida. Mob. Genet. Elements 3:e25640. 10.4161/mge.25640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hänninen ML, Hirvelä-Koski V. 1997. Molecular and phenotypic methods for the characterization of atypical Aeromonas salmonicida. Vet. Microbiol. 56:147–158. 10.1016/S0378-1135(96)01339-9. [DOI] [PubMed] [Google Scholar]
- 11.Daher RK, Filion G, Tan SGE, Dallaire-Dufresne S, Paquet VE, Charette SJ. 2011. Alteration of virulence factors and rearrangement of pAsa5 plasmid caused by the growth of Aeromonas salmonicida in stressful conditions. Vet. Microbiol. 152:353–360. 10.1016/j.vetmic.2011.04.034. [DOI] [PubMed] [Google Scholar]
- 12.Stuber K, Burr SE, Braun M, Wahli T, Frey J. 2003. Type III secretion genes in Aeromonas salmonicida subsp. salmonicida are located on a large thermolabile virulence plasmid. J. Clin. Microbiol. 41:3854–3856. 10.1128/JCM.41.8.3854-3856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka KH, Dallaire-Dufresne S, Daher RK, Frenette M, Charette SJ. 2012. An insertion sequence-dependent plasmid rearrangement in Aeromonas salmonicida causes the loss of the type three secretion system. PLoS One 7:e33725. 10.1371/journal.pone.0033725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charette SJ, Brochu F, Boyle B, Filion G, Tanaka KH, Derome N. 2012. Draft genome sequence of the virulent strain 01-B526 of the fish pathogen Aeromonas salmonicida. J. Bacteriol. 194:722–723. 10.1128/JB.06276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y-J, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Ho CH, Irzyk GP, Jando SC, Alenquer MLI, Jarvie TP, Jirage KB, Kim J-B, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380. 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boisvert S, Laviolette F, Corbeil J. 2010. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J. Comput. Biol. 17:1519–1533. 10.1089/cmb.2009.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galardini M, Biondi EG, Bazzicalupo M, Mengoni A. 2011. CONTIGuator: a bacterial genomes finishing tool for structural insights on draft genomes. Source Code Biol. Med. 6:11. 10.1186/1751-0473-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon L, Cloeckaert A, Doublet B, Schwarz S, Bouju-Albert A, Ganière J-P, Le Bris H, Le Flèche-Matéos A, Giraud E. 2008. Complete sequence of the floR-carrying multiresistance plasmid pAB5S9 from freshwater Aeromonas bestiarum. J. Antimicrob. Chemother. 62:65–71. 10.1093/jac/dkn166. [DOI] [PubMed] [Google Scholar]
- 20.Gordon D, Green P. 2013. Consed: a graphical editor for next-generation sequencing. Bioinformatics 29:2936–2937. 10.1093/bioinformatics/btt515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277. 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 22.Pearson WR, Lipman DJ. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U. S. A. 85:2444–2448. 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearson WR, Wood T, Zhang Z, Miller W. 1997. Comparison of DNA sequences with protein sequences. Genomics 46:24–36. 10.1006/geno.1997.4995. [DOI] [PubMed] [Google Scholar]
- 24.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 25.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. 2009. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25:119–120. 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beaber JW, Hochhut B, Waldor MK. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184:4259–4269. 10.1128/JB.184.15.4259-4269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Call DR, Singer RS, Meng D, Broschat SL, Orfe LH, Anderson JM, Herndon DR, Kappmeyer LS, Daniels JB, Besser TE. 2010. blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob. Agents Chemother. 54:590–596. 10.1128/AAC.00055-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liebert CA, Hall RM, Summers AO. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandvang D. 1999. Novel streptomycin and spectinomycin resistance gene as a gene cassette within a class 1 integron isolated from Escherichia coli. Antimicrob. Agents Chemother. 43:3036–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dogra C, Raina V, Pal R, Suar M, Lal S, Gartemann K-H, Holliger C, van der Meer JR, Lal R. 2004. Organization of lin genes and IS6100 among different strains of hexachlorocyclohexane-degrading Sphingomonas paucimobilis: evidence for horizontal gene transfer. J. Bacteriol. 186:2225–2235. 10.1128/JB.186.8.2225-2235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gardner MN, Deane SM, Rawlings DE. 2001. Isolation of a new broad-host-range IncQ-like plasmid, pTC-F14, from the acidophilic bacterium Acidithiobacillus caldus and analysis of the plasmid replicon. J. Bacteriol. 183:3303–3309. 10.1128/JB.183.11.3303-3309.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loftie-Eaton W, Rawlings DE. 2009. Comparative biology of two natural variants of the IncQ-2 family plasmids, pRAS3.1 and pRAS3.2. J. Bacteriol. 191:6436–6446. 10.1128/JB.00864-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loftie-Eaton W, Rawlings DE. 2010. Evolutionary competitiveness of two natural variants of the IncQ-like plasmids, pRAS3.1 and pRAS3.2. J. Bacteriol. 192:6182–6190. 10.1128/JB.00176-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dacanay A, Knickle L, Solanky KS, Boyd JM, Walter JA, Brown LL, Johnson SC, Reith M. 2006. Contribution of the type III secretion system (TTSS) to virulence of Aeromonas salmonicida subsp. salmonicida. Microbiology 152:1847–1856. 10.1099/mic.0.28768-0. [DOI] [PubMed] [Google Scholar]
- 35.Coculescu BI. 2009. Antimicrobial resistance induced by genetic changes. J. Med. Life 2:114–123. [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka KH, Frenette M, Charette SJ. 2013. IS-mediated loss of virulence by Aeromonas salmonicida: a tangible piece of an evolutionary puzzle. Mob. Genet. Elements 3:e23498. 10.4161/mge.23498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janda JM, Abbott SL. 2010. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23:35–73. 10.1128/CMR.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martínez JL. 2008. Antibiotics and antibiotic resistance genes in natural environments. Science 321:365–367. 10.1126/science.1159483. [DOI] [PubMed] [Google Scholar]
- 39.Tello A, Austin B, Telfer TC. 2012. Selective pressure of antibiotic pollution on bacteria of importance to public health. Environ. Health Perspect. 120:1100–1106. 10.1289/ehp.1104650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.