Abstract
Campylobacter jejuni is a leading food-borne pathogen, and its antibiotic resistance is of serious concern to public health worldwide. C. jejuni is naturally competent for DNA transformation and freely takes up foreign DNA harboring genetic information responsible for antibiotic resistance. In this study, we demonstrate that C. jejuni transfers antibiotic resistance genes more frequently in biofilms than in planktonic cells by natural transformation.
TEXT
The increasing prevalence of antibiotic-resistant Campylobacter jejuni, a leading food-borne pathogen, is a serious public health problem worldwide (1), significantly compromising the efficacy of current antimicrobial chemotherapy for the treatment of human campylobacteriosis and resulting in adverse patient outcomes (2). The acquisition of antibiotic resistance genes by horizontal gene transfer plays a critical role in the dissemination of antibiotic resistance in pathogenic bacteria (3). Most mechanisms of horizontal gene transfer involve mobile genetic elements, such as plasmids, phages, and transposons, whereas natural transformation enables naturally competent bacteria to take up foreign DNA, even genomic DNA, from the environment (4). Unlike other enteric pathogens, such as Salmonella and pathogenic Escherichia coli, C. jejuni is naturally competent for DNA transformation; thus, C. jejuni can freely take up foreign DNA under normal growth conditions (5).
C. jejuni develops biofilms on various abiotic surfaces (6). Interestingly, Campylobacter spp. are frequently isolated from biofilms in various natural settings (7), suggesting that biofilm formation may help this fastidious bacterium survive in the environment. A biofilm is a microbial community encased in an extracellular matrix consisting of DNA, proteins, and polysaccharides (8). Although C. jejuni biofilms also contain extracellular DNA (eDNA) (9), little is known about the transfer of antibiotic resistance by natural transformation in the biofilms of this naturally competent pathogenic bacterium. In this study, we aimed to demonstrate the role of natural transformation in the spread of antibiotic resistance in C. jejuni biofilms.
C. jejuni strain NCTC 11168 was grown in Mueller-Hinton (MH) medium at 42°C under microaerobic conditions (5% O2, 10% CO2, 85% N2). To determine the transfer frequency of antibiotic resistance genes by natural transformation, we constructed two C. jejuni strains by integrating the aphA3 and cat genes, which confer resistance to kanamycin and chloramphenicol, respectively, into a noncoding region of the 23S rRNA gene clusters on the chromosome of C. jejuni, as described previously (10, 11). C. jejuni biofilms were prepared as described in our previous studies (12, 13). Briefly, overnight cultures of kanamycin-resistant (Kanr) and chloramphenicol-resistant (Cmr) C. jejuni strains on MH agar plates were resuspended in MH broth to an optical density at 600 nm (OD600) of 0.07. The bacterial suspension culture was grown at 42°C with shaking at 200 rpm for 5 h under microaerobic conditions. After dilution of the C. jejuni cultures by adding the same volume of fresh MH broth, equal volumes of two cultures were mixed and added to 24-well plates. After 24 h of incubation without agitation under microaerobic conditions, the supernatant was removed to determine the transmission frequency of antibiotic resistance genes in C. jejuni in a planktonic state. After washing the biofilms with phosphate-buffered saline (PBS) three times, fresh MH broth (1 ml) was added to each well containing biofilms. Since C. jejuni biofilms continuously shed bacterial cells (14), this method allows for the growth of C. jejuni cells that are released from biofilms without disruption of the biofilms (13). After 24 h of incubation, supernatants were collected to measure the gene transfer frequency in C. jejuni cells that were released from the biofilms. The level of biofilm formation was measured by using crystal violet staining (12). Occasionally, biofilms were prepared in the presence of DNase I (Sigma-Aldrich) at 37°C. Total bacterial counts were measured by serial dilution and growth on MH agar plates, and the count of the C. jejuni population that acquired double resistance to kanamycin and chloramphenicol by natural transformation was determined by growth on MH agar plates supplemented with kanamycin (30 μg · ml−1) and chloramphenicol (10 μg · ml−1).
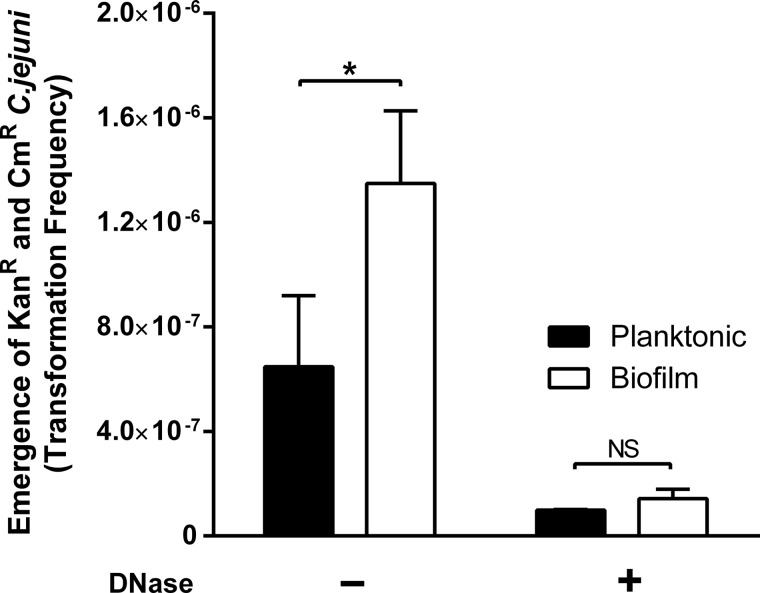
Co-culture of two singly resistant (Kanr or Cmr) C. jejuni strains resulted in the emergence of doubly resistant (Kanr and Cmr) C. jejuni colonies by natural transformation (Fig. 1). The presence of the resistance genes (aphA3 and cat) in the doubly resistant colonies was confirmed by PCR (data not shown). The frequency of the development of a doubly resistant C. jejuni population was higher in biofilms than in planktonic cells by about 6.5-fold (Fig. 1). Given the absence of plasmids, prophages, and other mobile genetic elements from the genome of C. jejuni NCTC 11168 (15), the transmission of the chromosomally encoded resistance genes is possible only by natural transformation. To confirm the role of eDNA in the spread of antibiotic resistance, we treated biofilms with DNase I. If the transfer of resistance results from natural transformation, DNase treatment will degrade eDNA and affect the transfer frequency. Interestingly, DNase treatment substantially reduced the frequency of the emergence of doubly resistant C. jejuni in biofilms and in planktonic cells. In addition, the frequencies were reduced to similar levels between biofilms and planktonic cells. Presumably, DNase I treatment reduced the availability of free eDNA in biofilms and in planktonic cells and minimized the differences. In addition, we observed that DNase I treatment decreased the level of biofilm formation of C. jejuni in a concentration-dependent manner (Fig. 2). Svensson et al. also reported that C. jejuni biofilms contain a DNase I-sensitive component (9). In Pseudomonas aeruginosa, eDNA is known to stabilize biofilm structures, particularly in the early stage of biofilm development (16).
FIG 1.
Frequency of emergence of doubly resistant C. jejuni isolates by natural transformation. The frequency of development of doubly resistant (Kanr and Cmr) C. jejuni colonies was calculated by dividing the bacterial count on MH agar plates supplemented with kanamycin and chloramphenicol by the total bacterial count. One hundred units of DNase I was added to each biofilm sample. The results are shown as the means and standard deviations (SD) of triplicate samples in a single experiment. The experiment was repeated three times. Statistical significance (*, P < 0.05) was calculated with the Student t test. NS, nonsignificant.
FIG 2.
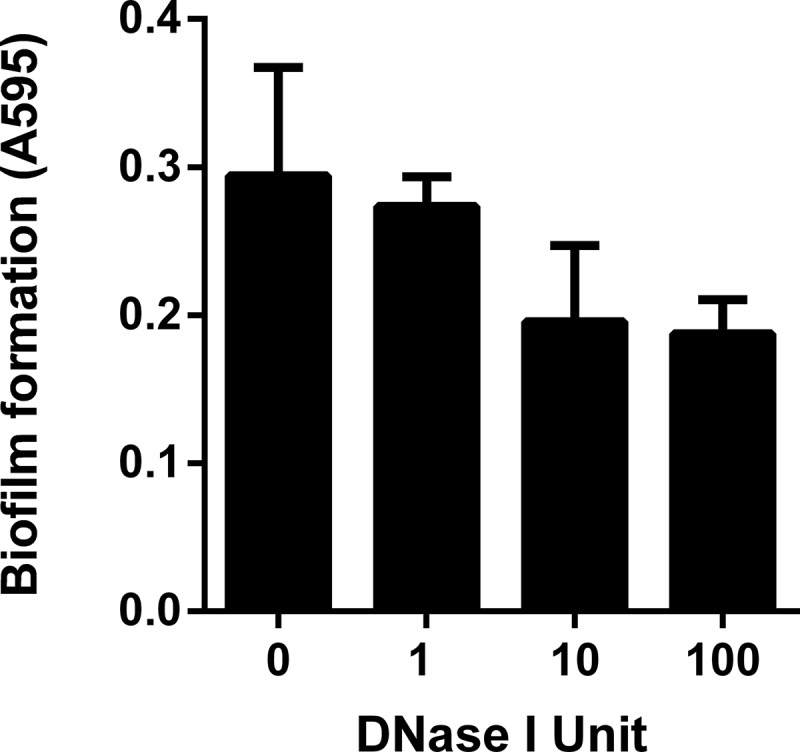
Decreased biofilm formation by DNase I treatment. Biofilms were grown for 24 h under microaerobic conditions. The results are shown as the means and SD of triplicate samples in a single experiment. The experiment was repeated three times.
In summary, eDNA is a major structural component of C. jejuni biofilms and plays an important role in the transfer of antibiotic resistance in C. jejuni by natural transformation. This study presents the first empirical demonstration that naturally competent C. jejuni is highly capable of taking up antibiotic resistance genes in biofilms. Since biofilms are a common environment for microorganisms in natural settings, biofilms appear to be an important reservoir that generates antibiotic-resistant C. jejuni under normal growth conditions even without antibiotic selective pressure.
ACKNOWLEDGMENTS
This study is supported by NSERC Discovery Grant 401843-2012-RGPIN and by intramural funding from the University of Alberta.
We thank Lynn McMullen and Michael Gänzle (University of Alberta) for sharing their laboratory facilities.
Footnotes
Published ahead of print 29 September 2014
REFERENCES
- 1.Engberg J, Aarestrup FM, Taylor DE, Gerner-Smidt P, Nachamkin I. 2001. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 7:24–34. 10.3201/eid0701.010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helms M, Simonsen J, Olsen KE, Molbak K. 2005. Adverse health events associated with antimicrobial drug resistance in Campylobacter species: a registry-based cohort study. J. Infect. Dis. 191:1050–1055. 10.1086/428453. [DOI] [PubMed] [Google Scholar]
- 3.Juhas M. 18 July 2013. Horizontal gene transfer in human pathogens. Crit. Rev. Microbiol. 10.3109/1040841X.2013.804031. [DOI] [PubMed] [Google Scholar]
- 4.Seitz P, Blokesch M. 2013. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol. Rev. 37:336–363. 10.1111/j.1574-6976.2012.00353.x. [DOI] [PubMed] [Google Scholar]
- 5.Jeon B, Muraoka W, Sahin O, Zhang Q. 2008. Role of Cj1211 in natural transformation and transfer of antibiotic resistance determinants in Campylobacter jejuni. Antimicrob. Agents Chemother. 52:2699–2708. 10.1128/AAC.01607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalmokoff M, Lanthier P, Tremblay TL, Foss M, Lau PC, Sanders G, Austin J, Kelly J, Szymanski CM. 2006. Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J. Bacteriol. 188:4312–4320. 10.1128/JB.01975-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maal-Bared R, Bartlett KH, Bowie WR, Hall ER. 2012. Campylobacter spp. distribution in biofilms on different surfaces in an agricultural watershed (Elk Creek, British Columbia): using biofilms to monitor for Campylobacter. Int. J. Hyg. Environ. Health 215:270–278. 10.1016/j.ijheh.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Nadell CD, Xavier JB, Foster KR. 2009. The sociobiology of biofilms. FEMS Microbiol. Rev. 33:206–224. 10.1111/j.1574-6976.2008.00150.x. [DOI] [PubMed] [Google Scholar]
- 9.Svensson SL, Davis LM, MacKichan JK, Allan BJ, Pajaniappan M, Thompson SA, Gaynor EC. 2009. The CprS sensor kinase of the zoonotic pathogen Campylobacter jejuni influences biofilm formation and is required for optimal chick colonization. Mol. Microbiol. 71:253–272. 10.1111/j.1365-2958.2008.06534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang S, Zhang Q, Ryu S, Jeon B. 2012. Transcriptional regulation of the CmeABC multidrug efflux pump and the KatA catalase by CosR in Campylobacter jejuni. J. Bacteriol. 194:6883–6891. 10.1128/JB.01636-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlyshev AV, Wren BW. 2005. Development and application of an insertional system for gene delivery and expression in Campylobacter jejuni. Appl. Environ. Microbiol. 71:4004–4013. 10.1128/AEM.71.7.4004-4013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oh E, Jeon B. 2014. Role of alkyl hydroperoxide reductase (AhpC) in the biofilm formation of Campylobacter jejuni. PLoS One 9:e87312. 10.1371/journal.pone.0087312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bae J, Jeon B. 2013. Increased emergence of fluoroquinolone-resistant Campylobacter jejuni in biofilm. Antimicrob. Agents Chemother. 57:5195–5196. 10.1128/AAC.00995-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reuter M, Mallett A, Pearson BM, van Vliet AH. 2010. Biofilm formation by Campylobacter jejuni is increased under aerobic conditions. Appl. Environ. Microbiol. 76:2122–2128. 10.1128/AEM.01878-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668. 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 16.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]