Abstract
Burkholderia cenocepacia is notorious for causing respiratory tract infections in people with cystic fibrosis. Infections with this organism are particularly difficult to treat due to its high level of intrinsic resistance to most antibiotics. Multidrug resistance in B. cenocepacia can be ascribed to different mechanisms, including the activity of efflux pumps and biofilm formation. In the present study, the effects of deletion of the 16 operons encoding resistance-nodulation-cell division (RND)-type efflux pumps in B. cenocepacia strain J2315 were investigated by determining the MICs of various antibiotics and by investigating the antibiofilm effect of these antibiotics. Finally, the expression levels of selected RND genes in treated and untreated cultures were investigated using reverse transcriptase quantitative PCR (RT-qPCR). Our data indicate that the RND-3 and RND-4 efflux pumps are important for resistance to various antimicrobial drugs (including tobramycin and ciprofloxacin) in planktonic B. cenocepacia J2315 populations, while the RND-3, RND-8, and RND-9 efflux systems protect biofilm-grown cells against tobramycin. The RND-8 and RND-9 efflux pumps are not involved in ciprofloxacin resistance. Results from the RT-qPCR experiments on the wild-type strain B. cenocepacia J2315 suggest that there is little regulation at the level of mRNA expression for these efflux pumps under the conditions tested.
INTRODUCTION
Species belonging to the Burkholderia cepacia complex (Bcc), a cluster of phylogenetically closely related and phenotypically similar Gram-negative bacteria, can cause severe respiratory tract infections in people with cystic fibrosis (1). Although there are considerable regional differences, the majority of patients with cystic fibrosis worldwide are infected with either Burkholderia multivorans or Burkholderia cenocepacia (2). Infections with B. cenocepacia are particularly difficult to treat due to their high level of resistance against a wide range of antimicrobial agents (1, 3, 4). Contributing to this is the fact that Bcc strains, including B. cenocepacia strains, readily form biofilms on various biotic and abiotic surfaces (5). While the molecular mechanisms contributing to the decreased susceptibility of cells in a biofilm have not yet been completely elucidated, protection provided by matrix components, biofilm-specific protection against oxidative stress, and biofilm-specific expression of efflux pumps are thought to play an important role (6).
In the genome of B. cenocepacia strain J2315, a large number of efflux systems have been identified (7–10). The roles of some members of the resistance-nodulation-cell division (RND) efflux pump family have been investigated in more detail. The RND-3 (BCAL1674 to BCAL1676) and RND-4 (BCAL2820 to BCAL2822) efflux systems were shown to contribute to the intrinsic resistance of B. cenocepacia J2315 to various compounds and to mediate accumulation of quorum-sensing molecules in the growth medium (8). A transcriptomic analysis of RND-4 and RND-9 (BCAM1945 to BCAM1947) deletion mutants revealed that in B. cenocepacia, RND pumps also influence other phenotypic traits important for pathogenesis (9). Recently, we showed that the RND-4 efflux system is involved in the resistance to a new thiopyridine drug effective against B. cenocepacia (11). In addition, we reported on the identification of lifestyle-specific efflux pumps involved in B. cenocepacia tolerance to the disinfectant chlorhexidine (CHX) (12). Cells grown in biofilms were less tolerant to CHX in a mutant in which the RND-9 efflux pump was inactivated with respect to the wild-type strain, while planktonic cells were less tolerant in a mutant in which the RND-4 efflux pump was inactivated. In a double mutant in which both RND-4 and RND-9 efflux systems were inactivated, planktonic and sessile cells were hypersusceptible to CHX.
In this study, we aimed to determine whether RND efflux systems in B. cenocepacia J2315 play a lifestyle-specific role in resistance to antibiotics. To this end, we constructed 15 RND-deleted strains and evaluated the susceptibility of planktonic and sessile cells. In addition, we quantified the expression levels of genes encoding RND efflux systems in planktonic and sessile cells after exposure to antibiotics.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown in Luria-Bertani (LB) broth (Difco) or Mueller-Hinton (MH) broth (Difco), with shaking at 200 rpm, or on LB agar at 37°C.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| Burkholderia cenocepacia | ||
| J2315 (WT) | CF clinical isolate | BCCM/LMG Bacteria Collection |
| D1 | J2315 ΔBCAS0591–BCAS0593 | 8 |
| D2 | J2315 ΔBCAS0764–BCAS0766 | This study |
| D3 | J2315 ΔBCAL1672–BCAL1676 | 8 |
| D4 | J2315 ΔBCAL2820–BCAL2822 | 8 |
| D5 | J2315 ΔBCAL1778 | This study |
| D6–7 | J2315 ΔBCAL1079–BCAL1081 | This study |
| D8 | J2315 ΔBCAM0925–BCAM0927 | This study |
| D9 | J2315 ΔBCAM1945–BCAM1947 | 12 |
| D10 | J2315 ΔBCAM2549–BCAM2551 | This study |
| D11 | J2315 ΔBCAM0710–BCAM0712 | This study |
| D12 | J2315 ΔBCAM0433–BCAM0435 | This study |
| D13 | J2315 ΔBCAL1811–BCAL1813 | This study |
| D14 | J2315 ΔBCAS0582–BCAS0584 | This study |
| D15 | J2315 ΔBCAM1419–BCAM1421 | This study |
| D16 | J2315 ΔBCAL2134–BCAL2136 | This study |
| Escherichia coli | ||
| DH5α | F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK− mK+) supE44 thi-1 ΔgyrA96 relA1 | Laboratory stock |
| SY327 | araD Δ(lac pro) argE(Am) recA56 nalA λ pir; Rifr | 24 |
| Plasmids | ||
| pGEM-T Easy | Vector for PCR cloning, Ampr | Promega |
| pGP-ISceI-XCm | oriR6K, Tpr, Cmr, mob+, containing the ISceI restriction site | 15 |
| pRK2013 | oricolE1, RK2 derivative, Kanr mob+ tra+ | 25 |
| pDA-ISceI-SacB | pDA12 encoding the ISceI homing endonuclease | 15 |
CF, cystic fibrosis; Ampr, ampicillin resistance; Cmr, chloramphenicol resistance; Kanr, kanamycin resistance; Rifr, rifampin resistance; Tetr, tetracycline resistance; Tpr, trimethoprim resistance.
Construction of deletion mutants.
Manipulation of DNA was performed as described previously (13). Restriction enzymes and T4 DNA ligase were purchased from Promega and used following the manufacturer's instructions. PCRs were performed by using the MJ Mini personal thermal cycler (Bio-Rad). To amplify PCR products, HotStar Taq DNA polymerase and Q solution (Qiagen) were used according to the manufacturer's instructions. DNA fragments were cloned into pGEM-T Easy vector (Promega) and sequenced using the standard M13for and M13rev primers. The construction of mutant strains D1, D4, and D9 was described previously (8, 12). The D3 strain was constructed as previously described (8), but counterselection for the Escherichia coli helper and donor strains was done on ampicillin-containing medium. All B. cenocepacia mutant strains were constructed by using the method described by Flannagan et al. (14) and subsequently optimized by Hamad et al. (15). Briefly, the upstream and downstream DNA sequences (about 500 bp each) that flank the operon targeted for deletion were cloned into pGP-ISceI-XCm. This suicide plasmid contains a unique restriction site for the endonuclease ISceI. E. coli DH5α and E. coli SY327 cells were transformed by electroporation (13). Plasmids were mobilized into B. cenocepacia J2315 by triparental mating, as described previously (16), using E. coli DH5α carrying the helper plasmid pRK2013. Exconjugants were selected in the presence of trimethoprim (200 μg/ml), chloramphenicol (400 μg/ml), and ampicillin (200 μg/ml) as counterselection for E. coli helper (DH5α) and donor (SY327) strains. pDA-ISceI-SacB plasmid (encoding the ISceI endonuclease) was then introduced into the transformants by conjugation. This second step induces site-specific double-strand breaks and subsequent homologous recombination, resulting in exconjugants resistant to tetracycline and susceptible to trimethoprim. Colonies were selected on LB agar plates containing tetracycline (250 μg/ml) and ampicillin (200 μg/ml). The desired gene deletions were confirmed by PCR amplification using primers which anneal to sequences flanking the regions of homology (see Table S1 in the supplemental material), with the HotStar HiFidelity polymerase kit (Qiagen). The specific amplification conditions were optimized for each primer pair. Then, the deleted strains were cured from pDA-ISceI-SacB plasmid by culturing for several days in fresh LB medium at 37°C. Detection of cured mutants was achieved by growing B. cenocepacia on LB agar plates without salt and supplemented with 5% (wt/vol) sucrose and then screening the resulting colonies for loss of tetracycline resistance.
Antimicrobial susceptibility testing for planktonic cells.
MICs were determined in duplicate according to the EUCAST broth microdilution protocol using flat-bottom 96-well microtiter plates (TPP; Trasadingen, Switzerland) and MH medium, as described previously (17, 18). Chloramphenicol and ciprofloxacin were obtained from Sigma-Aldrich (St. Louis, MO, USA), tobramycin from TCI Europe (Zwijndrecht, Belgium), minocycline from Certa (Braine-l'Alleud, Belgium), and meropenem from AstraZeneca (London, United Kingdom). The highest concentrations tested were 1,024 μg/ml (tobramycin), 128 μg/ml (meropenem, minocycline, and chloramphenicol), and 64 μg/ml (ciprofloxacin). The MIC was defined as the lowest concentration for which no significant difference in optical density (λ = 590 nm) was observed between the inoculated and blank wells after 24 h of incubation. All MIC determinations were performed in duplicate, and replicates never differed more than 2-fold. When a 2-fold difference was observed between replicates, the lowest concentration was recorded as the MIC.
Antimicrobial susceptibility testing for biofilms.
The antimicrobial susceptibility of sessile cells was determined as described previously (17). In brief, biofilms were grown for 24 h (4 h of adhesion and 20 h of biofilm formation) in MH broth and then exposed to antibiotics for 24 h in 0.9% (wt/vol) NaCl. Biofilms were grown on silicone discs (Q7-4735; Dow Corning, Midland, MI, USA) placed in the wells of a 24-well microtiter plate (TPP). After treatment, all discs were rinsed and transferred to 10 ml of MH broth. Sessile cells were removed from the discs by three cycles of vortexing (30 s) and sonication (30 s in a Branson 3510; Branson Ultrasonics Corp., Danbury, CT, USA), and the number of cells was determined using conventional plate count methods. At least six discs were investigated per condition. We used a Mann-Whitney test to determine whether differences in CFU per biofilm were significant (P < 0.05).
RNA extraction and RT-qPCR.
The expression of BCAL1675 (RND-3), BCAL2820 (RND-4), BCAM0925 (RND-8), BCAM0926 (RND-8), and BCAM1945 (RND-9) was measured by reverse transcriptase quantitative PCR (RT-qPCR) in treated and untreated sessile and planktonic cells. Biofilms were grown as described above and subsequently treated with tobramycin (8 μg/ml) or ciprofloxacin (2 μg/ml). To measure the expression in mature planktonic cells, an overnight culture in LB broth was diluted to an optical density of 0.1 (∼108 CFU/ml). After an additional 24 h of growth, cell suspensions with an optical density of 1 (∼109 CFU/ml) were also treated with tobramycin (8 μg/ml) or ciprofloxacin (2 μg/ml). The amount of antibiotic concentration used was high enough to have an effect on the number of CFU while being low enough to allow the recovery of sufficient cells for RNA extraction. After 24 h of treatment, RNA was extracted, as described previously (19) (n = 3), and cDNA was synthesized using the qScript cDNA SuperMix (Quanta Biosciences, Leuven, Belgium).
The RT-qPCR experiments were performed using the Perfecta SYBR green FastMix (Quanta Biosciences) on a Bio-Rad CFX96 real-time system C1000 thermal cycler, as described previously (18, 19). The primer concentration was 300 nM. The primer sequences are listed in Table S2 in the supplemental material. Three reference genes (BCAL2694, BCAS0175, and BCAM2784) were included to allow accurate normalization. These reference genes were previously found to be stably expressed (18). Statistical data analysis was performed using SPSS software, version 21 (SPSS, Chicago, IL, USA). A Mann-Whitney test was used to determine whether differences in expression were significant (P < 0.05).
Crystal violet staining.
Biofilm formation and crystal violet staining of the wild-type (WT) strain and mutants D3 and D4 were carried out as described previously (20). After staining, absorbance (590 nm) was measured using an Envision Xcite multilabel reader (PerkinElmer LAS, Boston, MA).
RESULTS AND DISCUSSION
Deletion of specific RND efflux systems affects susceptibility of planktonic cells.
In the absence of antibiotics, the growth of the mutant strains was not affected, and doubling times and the maximum optical density reached during the stationary phase were similar for the mutants and the wild-type strain (data not shown). For the wild-type strain, the MICs were 8 and 256 μg/ml for ciprofloxacin and tobramycin, respectively; this is in agreement with previously reported data (17). For most mutants, the MICs for ciprofloxacin and tobramycin did not differ much from those observed for the wild-type strain, with the exception of strains D3 and D4 (Table 2), which were more susceptible to both ciprofloxacin and tobramycin. The increased susceptibility of the D4 mutant to ciprofloxacin and tobramycin was reported (8, 9). In addition, both strains showed increased susceptibility to minocycline (as did strain D16); strain D4 was also more sensitive to chloramphenicol than the wild-type strain (Table 2). These data strongly suggest that in planktonic B. cenocepacia J2315 cells, RND-3 and RND-4 efflux pumps are involved in the efflux of ciprofloxacin and tobramycin, while the other RND efflux systems do not play a major role in this process. As the D3 strain used in the present study was prepared by counterselecting the helper and donor E. coli strains on ampicillin-containing medium, the data regarding the involvement of the RND-3 efflux pump in aminoglycoside resistance are consistent with those reported by Hamad and collaborators (15) on B. cenocepacia K56-2. The RND-4 pump also seems to be involved in the efflux of minocycline and chloramphenicol in planktonic B. cenocepacia J2315 cells.
TABLE 2.
MICs of the various antibiotics for some of the B. cenocepacia strains tested
| Strain | MIC (μg/ml) fora: |
||||
|---|---|---|---|---|---|
| CIP | TOB | MIN | MER | CHL | |
| J2315 (WT) | 8 | 256 | 16 | 64 | 32 |
| D3 | 2 | 2 | —b | 32 | 16 |
| D4 | 2 | 128 | 4 | 64 | 8 |
| D16 | 8 | 256 | 4 | 32 | 16 |
CIP, ciprofloxacin; TOB, tobramycin; MIN, minocycline; MER, meropenem; CHL, chloramphenicol.
—, no clear minocycline breakpoint was observed for this mutant.
Inactivation of some RND efflux pumps increases the bactericidal effect of antibiotics against B. cenocepacia biofilm cells.
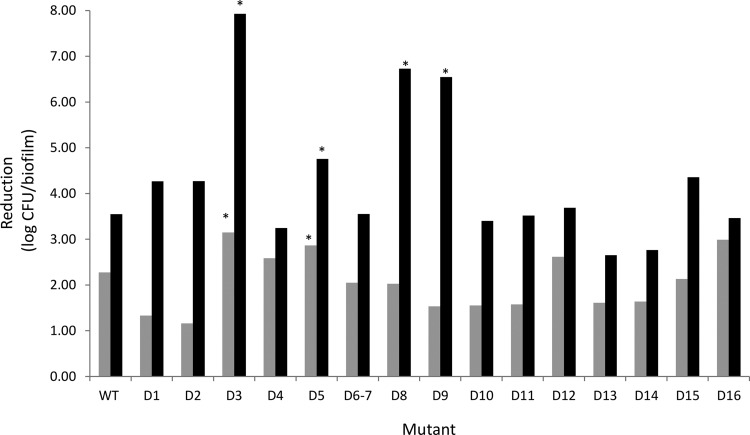
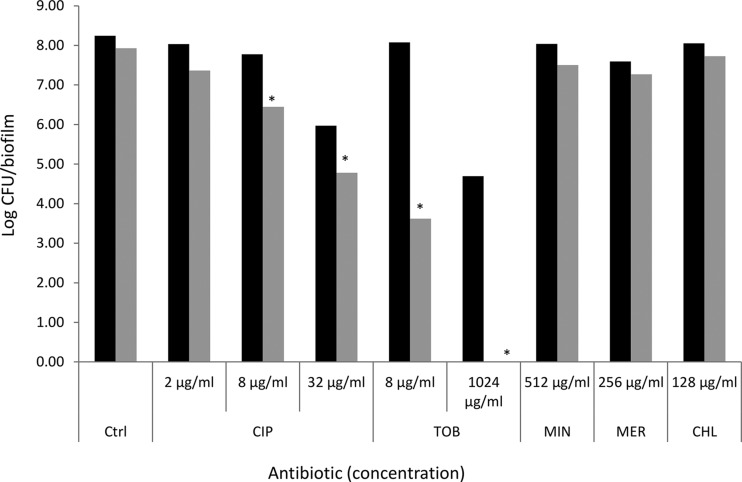
In the absence of antibiotics, biofilms formed by the wild-type strain and the various mutants contained approximately the same number of culturable cells (∼108 CFU/biofilm) (see Table S3 in the supplemental material). The lack of a difference in biofilm formation between the WT and D4 strains was confirmed with crystal violet staining; no difference in staining was observed between the WT (average absorbance, 0.33 ± 0.13) and D4 (average absorbance, 0.26 ± 0.20; P = 0.193) strains (see Fig. S1 in the supplemental material). The situation was less clear for strain D3, which showed an increased absorbance after crystal violet staining (average absorbance, 0.58 ± 0.16; P = 0.012). Combined, the data obtained with plate counts and crystal violet staining suggest that deletion of some RND efflux pumps does not affect B. cenocepacia biofilm formation, while the deletion of others may lead to increased production of biofilm matrix and/or the production of an altered matrix. This is in contrast with the findings of earlier studies in which the deletion of efflux pumps in Salmonella enterica serovar Typhimurium (21, 22) or Escherichia coli (23) negatively affected biofilm formation. More research is required to elucidate the mechanisms behind these differences in crystal violet staining. Treatment of the WT biofilm with ciprofloxacin (32 μg/ml) or tobramycin (1,024 μg/ml) led to significant reductions in the number of sessile cells recovered (log reduction, 2.28 and 3.55, respectively; P < 0.05). Similarly, treatment of all mutant biofilms with ciprofloxacin (32 μg/ml) led to a significant (P < 0.05) reduction in sessile cell numbers, with log reductions ranging from 1.16 (strain D2) to 3.15 (strain D3) (Fig. 1; see also Table S3 in the supplemental material). Similarly, treatment with tobramycin (1,024 μg/ml) led to a significant (P < 0.05) decrease in the number of CFU/biofilm for all mutants investigated, with log reductions ranging from 2.65 (strain D13) to 7.93 (strain D3) (Fig. 1; Table S3). For the latter strain, treatment with 1,024 μg/ml of tobramycin led to complete eradication of the biofilm, while this treatment led to near eradication of biofilms formed by strains D8 and D9. We subsequently focused on biofilms formed by strain D3 (which showed the highest reductions when exposed to high concentrations of tobramycin and ciprofloxacin) and exposed these biofilms to different concentrations of these antibiotics, as well as to minocycline (512 μg/ml), meropenem (256 μg/ml), and chloramphenicol (128 μg/ml) (Fig. 2). While no significant differences were observed between the number of CFU in a wild-type biofilm and those in a biofilm formed by strain D3 following treatment with ciprofloxacin (2 μg/ml), minocycline, meropenem, or chloramphenicol, sessile cells from strain D3 were hypersusceptible to intermediate concentrations of ciprofloxacin (8 μg/ml) and to low (8 μg/ml) and high (1,024 μg/ml) concentrations of tobramycin (P < 0.05). Together, these data indicate that the RND-3 efflux system is important for the protection of sessile B. cenocepacia J2315 cells against ciprofloxacin and tobramycin but not against minocycline, meropenem, or chloramphenicol. Our data also suggest that RND-8 and RND-9 efflux systems play important roles in the protection of sessile cells against tobramycin.
FIG 1.
Reduction in number of CFU/biofilm after treatment of B. cenocepacia biofilms with ciprofloxacin (32 μg/ml) (gray bars) or tobramycin (1,024 μg/ml) (black bars). Data are expressed as log reduction in CFU/biofilm (compared to the untreated biofilm). For the WT and all mutant biofilms investigated, the treated biofilms always contained significantly fewer CFU than the corresponding untreated control (P < 0.05). *, significantly more reduction than observed for the WT strain (P < 0.05).
FIG 2.
Susceptibility of wild-type biofilms (black bars) and biofilms formed by the D3 mutant (gray bars) when exposed to antibiotics in various concentrations [data are expressed as log(CFU/biofilm)]. CIP, ciprofloxacin; TOB, tobramycin; MIN, minocycline; MER, meropenem; CHL, chloramphenicol. *, significantly more reduction than observed for the WT strain (P < 0.05).
Analysis of gene expression suggests there is little regulation at the mRNA level.
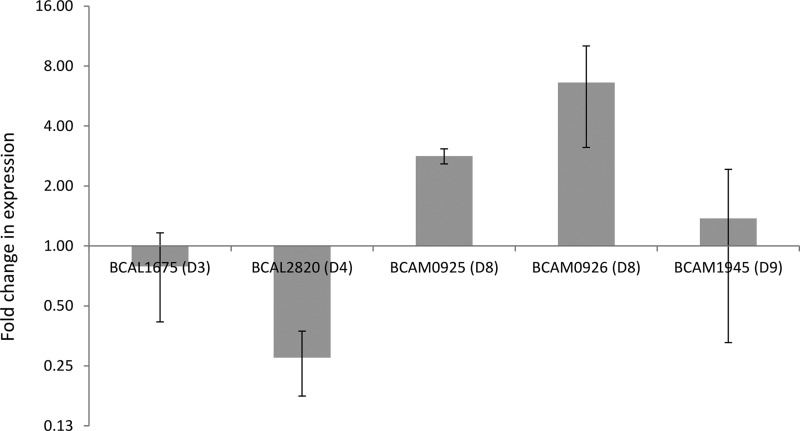
To further explore the role of certain RND efflux systems in B. cenocepacia drug resistance, we isolated mRNA from planktonic (stationary phase) and sessile cultures exposed to ciprofloxacin (2 μg/ml) or tobramycin (8 μg/ml) and measured the expression of genes encoding parts of the RND-3 (BCAL1675), RND-4 (BCAL2820), RND-8 (BCAM0925 and BCAM0926), and RND-9 (BCAM1945) efflux systems. When we compared the expression of these genes between untreated sessile and planktonic cultures, we noted a moderate overexpression of both components of the RND-8 efflux system and a moderate downregulation of BCAL2820 (RND-4) in biofilms (Fig. 3). The expression levels of BCAL1675 (RND-3) and BCAM1945 (RND-9) were similar in sessile and planktonic cells (Fig. 3).
FIG 3.
Average relative expression of genes encoding components of B. cenocepacia J2315 RND efflux systems in untreated biofilms compared to expression in untreated planktonic cells. Error bars represent standard errors of the means (n = 3).
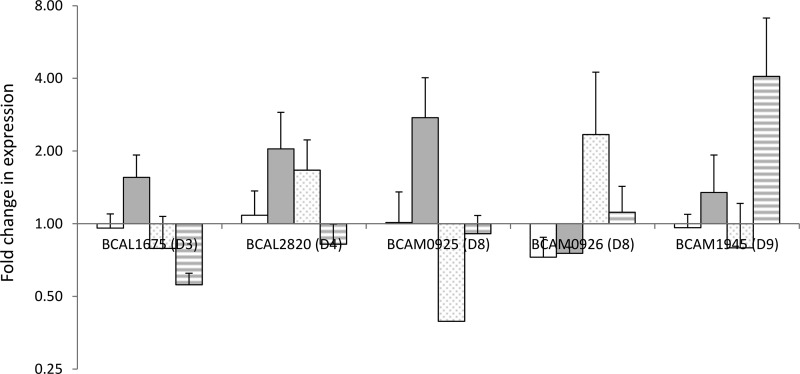
When we compared the expression levels in cultures treated with ciprofloxacin or tobramycin versus untreated cultures, we noticed small changes in expression for BCAL1675 (RND-3) and BCAL2820 (RND-4) (Fig. 4). The picture for components of the RND-8 efflux system is confusing, as BCAM0925 is only upregulated in planktonic cultures treated with tobramycin, while BCAM0926 is only upregulated in biofilms treated with ciprofloxacin. BCAM1945 (RND-9) shows a >4-fold upregulation in tobramycin-treated biofilms compared to untreated biofilms (Fig. 4). However, none of those differences are statistically significant (P > 0.05). These data suggest that, overall, there is little regulation at the level of mRNA expression for these efflux pumps under the conditions tested. For ciprofloxacin, this might be due to the relatively low concentration used, but this explanation is unlikely for tobramycin, as the concentration used (8 μg/ml) has an obvious effect on biofilms of B. cenocepacia strain D3 and is above the MIC (2 μg/ml) for this mutant.
FIG 4.
Average relative expression of genes encoding components of B. cenocepacia J2315 RND efflux systems in treated cultures compared to expression in untreated cultures. Error bars represent standard errors of the means (n = 3). White bars, planktonic cultures treated with ciprofloxacin (2 μg/ml); gray bars, planktonic cultures treated with tobramycin (8 μg/ml); dotted bars, biofilms treated with ciprofloxacin (2 μg/ml); striped bars, biofilms treated with tobramycin (8 μg/ml).
Conclusions.
The B. cenocepacia RND-3 efflux pump appears to be crucial for protecting planktonic and sessile cells against ciprofloxacin and tobramycin. Our data suggest that the RND-4 efflux pump is important for efflux of ciprofloxacin, tobramycin, minocycline, and chloramphenicol in planktonic cells but not in biofilms. Finally, the RND-8 and RND-9 efflux systems are required for protection against tobramycin activity only in biofilms but not in planktonic cultures. Contrary to our expectations, no significant differences in gene expression were noted for components belonging to the RND-3, RND-4, RND-8, and RND-9 efflux pumps when treated wild-type cultures were compared with untreated cultures. This suggests that the lifestyle-specific activity of these pumps is not (or at least not exclusively) regulated at the transcriptional level.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Italian Cystic Fibrosis Research Foundation (FFC project 10/2012 [G.R.], adopted by Associazione Trentina FC onlus in ricordo di Zaira Tutino, Gruppo di Sostegno FFC di Palermo–in ricordo di Elisa Pepe; Delegazione FFC di Imola), by the Fund for Scientific Research–Flanders (FWO-Vlaanderen), and by the Interuniversity Attraction Poles Programme initiated by the Belgian Science Policy Office.
We thank Lisa Slachmuylders and Sanne Kiekens for excellent technical assistance.
Footnotes
Published ahead of print 29 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03800-14.
REFERENCES
- 1.Mahenthiralingam E, Urban TA, Goldberg JB. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144–156. 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 2.LiPuma JJ. 2010. The changing microbial epidemiology in cystic fibrosis. Clin. Microbiol. Rev. 23:299–323. 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peeters E, Nelis HJ, Coenye T. 2008. Evaluation of the efficacy of disinfection procedures against Burkholderia cenocepacia biofilms. J. Hosp. Infect. 70:361–368. 10.1016/j.jhin.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 4.Drevinek P, Mahenthiralingam E. 2010. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin. Microbiol. Infect. 16:821–830. 10.1111/j.1469-0691.2010.03237.x. [DOI] [PubMed] [Google Scholar]
- 5.Coenye T. 2010. Social interactions in the Burkholderia cepacia complex: biofilms and quorum sensing. Future Microbiol. 5:1087–1099. 10.2217/fmb.10.68. [DOI] [PubMed] [Google Scholar]
- 6.Van Acker H, Van Dijck P, Coenye T. 2014. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol. 22:326–333. 10.1016/j.tim.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Guglierame P, Pasca MR, De Rossi E, Buroni S, Arrigo P, Manina G, Riccardi G. 2006. Efflux pump genes of the resistance-nodulation-division family in Burkholderia cenocepacia genome. BMC Microbiol. 6:66. 10.1186/1471-2180-6-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buroni S, Pasca MR, Flannagan RS, Bazzini S, Milano A, Bertani I, Venturi V, Valvano MA, Riccardi G. 2009. Assessment of three resistance-nodulation-cell division drug efflux transporters of Burkholderia cenocepacia in intrinsic antibiotic resistance. BMC Microbiol. 9:200. 10.1186/1471-2180-9-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bazzini S, Udine C, Sass A, Pasca MR, Longo F, Emiliani G, Fondi M, Perrin E, Decorosi F, Viti C, Giovannetti L, Leoni L, Fani R, Riccardi G, Mahenthiralingam E, Buroni S. 2011. Deciphering the role of RND efflux transporters in Burkholderia cenocepacia. PLoS One 6:e18902. 10.1371/journal.pone.0018902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perrin E, Fondi M, Papaleo MC, Maida I, Emiliani G, Buroni S, Pasca MR, Riccardi G, Fani R. 2013. A census of RND superfamily proteins in the Burkholderia genus. Future Microbiol. 8:923–937. 10.2217/fmb.13.50. [DOI] [PubMed] [Google Scholar]
- 11.Scoffone VC, Spadaro F, Udine C, Makarov V, Fondi M, Fani R, De Rossi E, Riccardi G, Buroni S. 2014. Mechanism of resistance to an antitubercular 2-thiopyridine derivative that is also active against Burkholderia cenocepacia. Antimicrob. Agents Chemother. 58:2415–2417. 10.1128/AAC.02438-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coenye T, Van Acker H, Peeters E, Sass A, Buroni S, Riccardi G, Mahenthiralingam E. 2011. Molecular mechanisms of chlorhexidine tolerance in Burkholderia cenocepacia biofilms. Antimicrob. Agents Chemother. 55:1912–1919. 10.1128/AAC.01571-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 14.Flannagan RS, Linn T, Valvano MA. 2008. A system for the construction of targeted unmarked gene deletions in the genus Burkholderia. Environ. Microbiol. 10:1652–1660. 10.1111/j.1462-2920.2008.01576.x. [DOI] [PubMed] [Google Scholar]
- 15.Hamad MA, Skeldon AM, Valvano MA. 2010. Construction of aminoglycoside-sensitive Burkholderia cenocepacia strains for use in studies of intracellular bacteria with the gentamicin protection assay. Appl. Environ. Microbiol. 76:3170–3176. 10.1128/AEM.03024-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig FF, Coote JG, Parton R, Freer JH, Gilmour NJ. 1989. A plasmid which can be transferred between Escherichia coli and Pasteurella haemolytica by electroporation and conjugation. J. Gen. Microbiol. 135:2885–2890. [DOI] [PubMed] [Google Scholar]
- 17.Peeters E, Nelis HJ, Coenye T. 2009. In vitro activity of ceftazidime, ciprofloxacin, meropenem, minocycline, tobramycin and trimethoprim/sulfamethoxazole against planktonic and sessile Burkholderia cepacia complex bacteria. J. Antimicrob. Chemother. 64:801–809. 10.1093/jac/dkp253. [DOI] [PubMed] [Google Scholar]
- 18.Van Acker H, Sass A, Bazzini S, De Roy K, Udine C, Messiaen T, Riccardi G, Boon N, Nelis HJ, Mahenthiralingam E, Coenye T. 2013. Biofilm-grown Burkholderia cepacia complex cells survive antibiotic treatment by avoiding production of reactive oxygen species. PLoS One 8:e58943. 10.1371/journal.pone.0058943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Acker H, Sass A, Dhondt I, Nelis HJ, Coenye T. 2014. Involvement of toxin–antitoxin modules in Burkholderia cenocepacia biofilm persistence. Pathog. Dis. 71:326–335. 10.1111/2049-632X.12177. [DOI] [PubMed] [Google Scholar]
- 20.Peeters E, Nelis H, Coenye T. 2008. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 72:157–165. 10.1016/j.mimet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Kvist M, Hancock V, Klemm P. 2008. Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl. Environ. Microbiol. 74:7376–7782. 10.1128/AEM.01310-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baugh S, Phillips CR, Ekanayaka AS, Piddock LJ, Webber MA. 2014. Inhibition of multidrug efflux as a strategy to prevent biofilm formation. J. Antimicrob. Chemother. 69:673–681. 10.1093/jac/dkt420. [DOI] [PubMed] [Google Scholar]
- 23.Matsumura K, Furukawa S, Ogihara H, Morinaga Y. 2011. Roles of multidrug efflux pumps on the biofilm formation of Escherichia coli K-12. Biocontrol Sci. 16:69–72. 10.4265/bio.16.69. [DOI] [PubMed] [Google Scholar]
- 24.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652. 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.