Abstract
Over the past several years, single-nucleotide polymorphisms (SNPs) within the mprF open reading frame (ORF) have been proposed to be associated with a gain-of-function phenotype in terms of daptomycin (DAP) nonsusceptibility (referred to as daptomycin resistance [DAP-R] herein for ease of presentation) in Staphylococcus aureus. We investigated the frequencies of SNPs within the mprF ORF and the relationships of such SNPs to cross-resistance between DAP and cationic host defense peptides (HDPs). Thirty-five well-characterized, unique DAP-susceptible (DAP-S) and DAP-R methicillin-resistant S. aureus (MRSA) isolates of the clonal complex 5 genotype were used. In addition to mprF SNPs and DAP-HDP cross-resistance, several other key genotypic and phenotypic metrics often associated with DAP-R were delineated, as follows: (i) mprF expression, (ii) membrane phospholipid content, (iii) positive surface charge, (iv) DAP binding, and (v) cell wall thickness profiles. A number of DAP-S strains (MICs of ≤1 μg/ml) exhibited mprF SNPs, occasionally with high-level mprF sequence variation from the genotype reference strain. However, none of these SNPs were localized to well-chronicled mprF hot spot locations associated with DAP-R in S. aureus. In contrast, all 8 DAP-R isolates demonstrated SNPs within such known mprF hot spots. Moreover, only the DAP-R strains showed MprF gain-of-function phenotypes, enhanced mprF expression, higher survival against two prototypical HDPs, and reduced DAP binding. Although a heterogenous array of mprF SNPs were often found in DAP-S strains, only selected hot spot SNPs, combined with concurrent mprF dysregulation, were associated with the DAP-R phenotype.
INTRODUCTION
There have been many recent reports of clinical Staphylococcus aureus strains that have evolved in vitro daptomycin (DAP) resistance (DAP-R) (note that although the official term is daptomycin nonsusceptibility, we will utilize daptomycin resistance in this paper for ease of presentation) in the context of failing DAP treatment regimens, especially in endovascular infections (1–5). One of the key features of many DAP-R strains is the acquisition of one or more single-nucleotide polymorphisms (SNPs) in a relatively restricted cadre of genes, especially in mprF (multiple peptide resistance factor) (3, 6–8). The mprF gene is responsible for the synthesis and translocation (flipping) of the positively charged phospholipid (PL) lysyl-phosphatidylglycerol (L-PG) within its cell membrane (CM). This process contributes substantially to the net positive surface charge in S. aureus, particularly in adaptive responses to cationic antimicrobial agents (3, 7, 9). Although DAP is inherently an anionic molecule, its bactericidal activity is absolutely dependent on it undergoing extensive complexing with calcium, rendering the functional DAP complex positively charged. Therefore, genes such as mprF that affect surface charge are hypothesized to be integral to DAP-R, potentially via charge repulsive electrostatic mechanisms (10). In addition, recent data from our laboratories have shown that both clinical and in vitro-derived DAP-R S. aureus strains frequently exhibit cross-resistance with cationic host defense peptides (HDPs), such as those derived from polymorphonuclear leukocytes (PMNs) and platelets (8, 11, 12).
The cataloguing of mprF SNPs associated with DAP-R in S. aureus has principally emerged from studies of individual isogenic DAP-susceptible (DAP-S) and DAP-R strain pairs. However, such data have not been systematically evaluated in a population-based survey of mprF sequences from well-defined groups of S. aureus strains with differing DAP MICs. Thus, in the current study, we investigated the frequencies of mprF gain-of-function SNPs and their relationships to both DAP-R and cross-resistance to prototypical HDPs. We also compared these outcomes with several genotypic and phenotypic determinants previously linked to mprF-mediated DAP-R in S. aureus, including positive surface charge (3, 13), CM phospholipid content and asymmetry (9, 14), mprF transcription profiles (7, 15), DAP surface binding (16), and cell wall thickening (8).
MATERIALS AND METHODS
Bacterial strains.
The 27 methicillin-resistant S. aureus (MRSA) bloodstream study isolates were selected from a recently described overall strain collection of 47 MRSA strains (11, 17). Since these 47 strains were from distinct genotypic backgrounds, we focused only on isolates genotyped as clonal complex 5 (CC5; the most common clonotype in this collection) to maintain relative strain homogeneity. Sixteen of the 27 CC5 strains had DAP MICs of 0.25 to 0.5 μg/ml, while the remaining 11 strains had DAP MICs of 1 μg/ml; both groups are DAP-S by presumptive breakpoints (18). In addition, 8 DAP-R isolates (MICs of >2 μg/ml) previously genotyped as CC5 were randomly selected from our DAP-R strain collection. All of the DAP-S MRSA isolates were from bacteremic patients who had never received DAP and have been characterized previously (11, 17). The DAP-R MRSA isolates (MICs of >2 μg/ml) were from patients who had been treated with DAP and failed therapy. The latter strains have not been reported before.
All strains were grown in tryptic soy broth (TSB; Difco Laboratories, Detroit, MI), Mueller-Hinton broth (Difco Laboratories, Detroit, MI), or Luria broth (LB; Difco Laboratories) as indicated below, depending on the individual experiment. Liquid cultures were grown in Erlenmeyer flasks at 37°C with shaking (250 rpm) in a volume that was no greater than 10% of the flask volume. Preliminary studies showed that all MRSA isolates in this investigation had similar in vitro growth kinetics and growth yields (data not shown).
The MICs of the strains to DAP were determined by standard Etest (AB Biodisk, Dalvagen, Sweden) on Mueller-Hinton agar plates, following the manufacturer's protocol (including calcium supplementations) (Difco Laboratories, Detroit, MI). DAP-R was defined as an Etest MIC of ≥2 μg/ml (19).
DNA isolation and mprF sequencing.
Genomic DNA was isolated from S. aureus using the method of Dyer and Iandolo (20). PCR amplification of the mprF ORFs was performed as previously described, using the primer pair mprF-F-bam and mprF-R-sph (15). DNA sequencing of the mprF ORFs was kindly performed at City of Hope, Duarte, CA. The mprF sequences from S. aureus MU50 (CC5) (GenBank accession number BA000017.4) were used as the consensus reference sequences to identify mprF SNPs among the study strains.
Host defense peptide susceptibility assays.
We studied prototypical HDPs from human PMNs and rabbit platelets. Human neutrophil peptide-1 (hNP-1), an archetypal α-defensin, was purchased from Peptide International (Louisville, KY). Thrombin-induced platelet microbicidal proteins (tPMPs) from fresh rabbit platelets were obtained as previously detailed; this preparation contains a mixture of tPMPs but is predominantly tPMP-1 (20–22). The bioactivity of these preparations (μg/ml equivalents) was determined by the standard Bacillus subtilis killing assay as outlined before (21, 22).
Standard MIC testing in nutrient broth may underestimate HDP activities (22, 23). Accordingly, in vitro bactericidal assays were carried out with hNP-1 and tPMPs using a well-characterized 2-h microdilution method in Eagle's minimal essential medium (3, 23). These assays were performed with either tPMPs (1 and 2 μg/ml bioactivity equivalents) or hNP-1 (10 and 20 μg/ml), using an initial inoculum of 5 × 103 CFU S. aureus cells (3, 23). These HDP concentrations were selected based on extensive pilot studies showing their inability to completely eradicate the starting inocula of the strain collections over the 2-h exposure period. Data were calculated and expressed as the relative percentage of surviving CFU (± standard deviation [SD]) of HDP-exposed versus HDP-unexposed cells, with the survival of each parental strain set at 100%. A minimum of three independent experiments was performed for each of the HDP assays.
Surface charge assays.
To quantify the relative positive cell surface charges in the strain sets, cytochrome c binding assays were performed as described before (24, 25). The binding of the highly cationic cytochrome c (pI = 10) (Sigma) was measured spectrophotometrically (optical density at 530 nm [OD530]), which quantifies the amount of this polycation remaining within the reaction mixture supernatants following exposure to the study strains for 30 min; the higher the amounts of residual cytochrome c in the supernates, the more relative positive surface charge exists (3, 8, 24, 26). A minimum of three independent experiments was performed for each evaluation.
RNA isolation and quantitative real-time PCR (qRT-PCR).
Four isolates from each of the three DAP MIC groups were randomly selected for characterizing mrpF expression profiles. To quantify the expression of the mprF gene, total RNA was isolated from the S. aureus cell pellets using the RNeasy kit (Qiagen, Valencia, CA) and the FastPrep FP120 instrument (BIO 101, Vista, CA) according to the manufacturer's recommended protocols. For RNA isolation, fresh overnight cultures of S. aureus strains were used to inoculate TSB to an OD600 of 0.1. Cell pellets were then obtained at either exponential (2 h) or late stationary (12 h) growth phases using previously described methods (27).
For qRT-PCR analyses, 2 μg of DNase-treated RNA was reverse transcribed using the SuperScript III first-strand synthesis kit (Invitrogen) according to the manufacturer's protocols. Quantification of cDNA levels was performed following the instructions of the Power SYBR green master mix kit (Applied Biosystems) on an ABI Prism 7000 sequence detection system (Applied Biosystems). The mprF and gyrB genes were detected using their respective specific primers as described before (27, 28). Fold changes in the expression levels of mprF were quantified in relation to the levels of gyrB.
CM phospholipid contents.
The three major S. aureus CM phospholipids (PLs) are L-PG, PG, and cardiolipin (CL) (9, 10, 29). To quantify the relative proportions of these three PLs in our strain sets, CM PLs were extracted from the selected S. aureus strains as described previously (26). The target PLs were separated and identified via two-dimensional thin-layer chromatography by their electrophoretic mobilities and ninhydrin staining properties (3, 26) and then removed from the plates and chemically quantified by spectrophotometer as described previously (3, 26). In addition, the proportion of synthesized L-PG which was translocated to the outer CM leaflet was quantified spectrophotometrically as detailed before, using the L-PG-targeting outer-CM-impermeable UV probe fluorescamine (14, 26). The latter fluorophore only binds to outer CM amine-containing PLs, such as L-PG. The composite data were expressed as the relative proportions (±SD) of the three PLs and the percentages of L-PG translocated to the outer CM, respectively. At least three independent experiments were performed to analyze the overall PL contents and L-PG translocations.
DAP binding assays.
To determine the relative profiles of whole-cell DAP binding to the selected S. aureus strains, bacteria were grown to an OD600 of 0.4 to 0.6 in LB supplemented with calcium (50 μg/ml) and then incubated with Bodipy-fluorescein-labeled DAP (Bodipy-DAP, 8 μg/ml; supplied courtesy of Cubist Pharmaceuticals, Lexington, MA) for 20 min at 37°C with shaking. Excess unincorporated label was removed by washing the cells three times in LB. In the final wash step, the cells were counterstained with DAPI (4′,6-diamidino-2-phenylindole; 2 μg/ml) DNA stain to visualize the cells (16) and then imaged using a DeltaVision deconvolution microscope (Applied Precision, Inc., Issaquah, WA) (16). The DAP-binding phenotype of the MRSA cells was quantified by the relative number of DAP-binding fluorescent spots/cell (more than 100 cells were queried for each strain).
Cell wall thickness.
The cell wall thicknesses of all study strains were compared by transmission electron microscopy (TEM) (8). The mean thickness (nm ± SD) of 100 cells was determined for the set of strains at a constant magnification of ×190,000 (model number 100CX; JEOL, Tokyo, Japan) using digital image capture and morphometric measurement (Advanced Microscopy Techniques version 54, Danvers, MA).
Statistical analysis.
The Kruskal-Wallis analysis of variance (ANOVA) test with the Tukey post hoc correction for multiple comparisons was utilized where indicated below. Significance was determined at a P value of <0.05.
RESULTS
SNPs within mprF genes.
A total of 35 S. aureus strains were subjected to mprF open reading frame (ORF) sequencing. Sequencing analyses of mprF genes revealed that 11/16 isolates with DAP MICs of ≤0.5 μg/ml and 6/11 isolates with DAP MICs of 1 μg/ml had mprF sequences identical to that of the S. aureus MU50 reference strain. In contrast, 5/16 isolates with DAP MICs of ≤0.5 μg/ml exhibited an amino acid substitution changing isoleucine at position 498 to asparagine (I498N) versus the MU50 sequence. In addition, analyses of 3/11 isolates with DAP MICs of 1 μg/ml had a distinct point mutation (Q692E; glutamine to glutamic acid). Interestingly, two strains in the latter group (MICs of 1 μg/ml) had 30 other amino acid substitutions within their mprF ORFs, suggesting high levels of heterogeneity within mprF ORFs in some S. aureus strains. All isolates with DAP MICs of ≥2 μg/ml had only one amino acid substitution, either at position 341 (L341S; leucine to serine) or 826 (L826F; leucine to phenylalanine), within their mprF ORFs. These latter SNPs have been well defined in the literature as high-frequency mprF mutational loci associated with DAP-R via gain-of-function phenotypes (9, 12, 13, 30, 31).
In vitro susceptibilities to HDPs.
In general, strain groups with higher DAP MICs exhibited significantly higher survival profiles when exposed in vitro to the two prototypical HDPs in this study (Table 1). Specifically, strains with DAP MICs of 1 μg/ml exhibited lower susceptibility to tPMP killing than strains with DAP MICs of ≤0.5 μg/ml (P < 0.01). In contrast, strains with DAP MICs of 1 μg/ml failed to show significantly reduced susceptibility to hNP-1 compared to those with DAP MICs of ≤0.5 μg/ml. Importantly, DAP-R isolates (DAP MICs of ≥2 μg/ml) exhibited significantly reduced killing by hNP-1 compared to strains in the other two MIC groups (P < 0.001).
TABLE 1.
Host defense peptide susceptibility among the study strain sets
| HDP (concn used)a | % Survival (mean ± SD) after 2-h exposure to HDP in group with DAP MIC of: |
P value |
||||
|---|---|---|---|---|---|---|
| ≤0.5 μg/ml | 1 μg/ml | ≥2 μg/ml | 0.5 vs 1 μg/ml | 1 vs 2 μg/ml | 0.5 vs 2 μg/ml | |
| tPMPs (2 μg/ml) | 10.1 ± 12.9 | 35.3 ± 24.9 | 43.1 ± 30.2 | <0.01 | NSb | <0.001 |
| tPMPs (1 μg/ml) | 19.1 ± 15.7 | 47.8 ± 26.2 | 59.0 ± 27.8 | <0.01 | NS | <0.001 |
| hNP-1 (20 μg/ml) | 17.2 ± 14.3 | 19.5 ± 15.9 | 29.1 ± 10.3 | NS | <0.05 | <0.001 |
| hNP-1 (10 μg/ml) | 31.7 ± 21.9 | 39.8 ± 22.3 | 65.4 ± 20.3 | NS | <0.05 | <0.001 |
tPMPs, thrombin-induced platelet microbicidal proteins; hNP-1, human neutrophil defensin-1.
NS, not significant.
Expression of mprF.
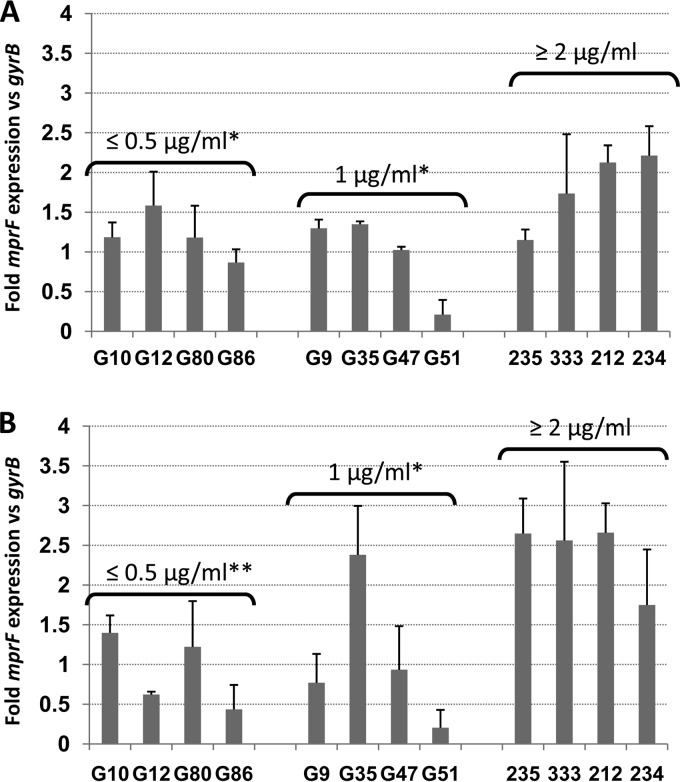
As demonstrated by the results in Fig. 1A and B, isolates with DAP MICs of ≥2 μg/ml exhibited significantly higher mean fold mprF expression levels than the two DAP-S groups of isolates during both exponential and stationary growth phases. However, there were no significant differences in the mean fold levels of mprF expression between strains with DAP MICs of ≤0.5 and those with DAP MICs of 1 μg/ml, irrespective of the growth phase queried.
FIG 1.
Transcription of mprF genes among the study strains during exponential growth phase (A) and stationary phase (B). Total cellular RNA samples from the strains grown in Trypticase soy broth were isolated at 2 h (exponential growth phase) or 12 h (stationary phase) postinoculation and subjected to qRT-PCR analyses. Means ± SDs are shown. *, P < 0.05 versus results for group with DAP MICs of ≥2 μg/ml; **, P < 0.01 versus results for group with DAP MICs of ≥2 μg/ml.
PL content and translocation of L-PG.
The proportion of total L-PG within the overall PL content was significantly increased (∼2-fold) in isolates with DAP MICs of ≥2 μg/ml compared to its proportions in strains in the other two MIC groups (Table 2). This profile was mainly observed relative to increases in net L-PG synthesis rather than enhanced translocation (flipping) profiles. As expected, in addition to the enhanced L-PG synthesis phenotype, the DAP-R group displayed a substantial compensatory reduction in the amount of CM PG compared to the amounts in the other two DAP-S isolate groups.
TABLE 2.
Phospholipid profiles among the three DAP MIC groups of MRSA isolatesa
| DAP MIC strain group | Inner L-PG | Outer L-PG | Proportion (mean % ± SD) among overall PL content |
||
|---|---|---|---|---|---|
| L-PG | PG | CL | |||
| ≤0.5 μg/ml | 11.2 ± 1.0† | 2.0 ± 1.3 | 13.2 ± 1.3† | 81.5 ± 2.2† | 5.3 ± 1.3* |
| 1 μg/ml | 13.6 ± 4.1† | 2.3 ± 1.0 | 15.9 ± 4.8† | 75.5 ± 8.3† | 8.7 ± 5.2 |
| ≥2 μg/ml | 22.0 ± 5.3 | 2.5 ± 0.4 | 24.5 ± 5.6 | 68.0 ± 11.5 | 7.5 ± 6.0 |
L-PG, lysyl-phosphatidylglycerol; PG, phosphatidylglycerol; CL, cardiolipin; *, P < 0.05 versus results for group with DAP MICs of ≥2 μg/ml; †, P < 0.01 versus results for group with DAP MICs of ≥2 μg/ml.
Bodipy-DAP whole-cell binding.
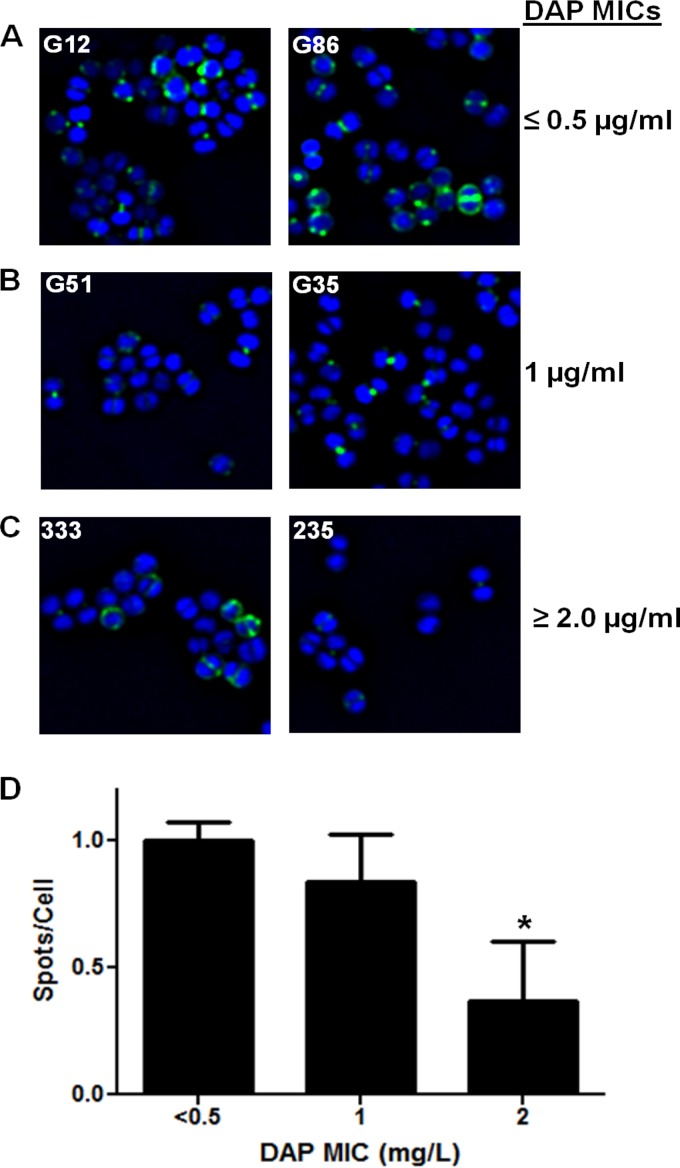
As shown by the results in Fig. 2A, B, and C, isolates in the two DAP-S MIC groups bound significantly more Bodipy-DAP than DAP-R strains (P < 0.05). In agreement with a previous publication (16), the DAP binding in DAP-S cells occurred focally and preferentially on the staphylococcal surface, especially near the septal division plane. When quantified by the mean number of fluorescent DAP spots/cell, this confirmed the significantly decreased surface binding observed among the DAP-R isolates (Fig. 2D).
FIG 2.
Representative binding of fluorescent DAP (Bodipy-DAP) to S. aureus bacteria of different DAP MIC groups. S. aureus strains from groups with DAP MICs of ≤0.5 μg/ml (A), 1 μg/ml (B), and ≥0.5 μg/ml (C) were stained with Bodipy-labeled DAP. (D) Intensity of fluorescence from cells stained with Bodipy-DAP. Mean results ± SD are shown. *, P < 0.05 versus results for DAP-S groups.
Net surface charge.
Cytochrome c binding analyses of all 35 strains revealed that the DAP-R isolates had significantly enhanced mean positive surface charge compared to both groups of DAP-S isolates (P = 0.0029 versus strains with DAP MICs of 1 μg/ml and P = 0.0034 versus strains with DAP MICs of ≤0.5 μg/ml) (see Table S1 in the supplemental material). There were no significant differences in relative surface charge between isolates in the two DAP-S MIC groups (P = 0.57).
Cell wall thickness.
The cell wall thicknesses determined by TEM did not differ significantly among the three MIC groups of study strains (see Table S1 in the supplemental material). The average cell wall thicknesses for the groups with DAP MICs of ≤0.5, 1, and ≥2 μg/ml were 25.0 ± 2.3, 25.7 ± 2.4, and 24.8 ± 3.7 nm, respectively.
DISCUSSION
In the present investigation, we studied a total of 35 well-characterized, unique DAP-S and DAP-R MRSA strains to evaluate potential interrelationships among the frequencies of SNPs within mprF, DAP-R phenotype, and HDP cross-resistance phenotypes. A number of interesting findings emerged from this investigation.
First, mprF sequencing revealed that there are distinct SNPs within the mprF ORFs in a substantial proportion of DAP-S strains of the CC5 genotype (i.e., groups with DAP MICs of ≤0.5 or 1 μg/ml). Thus, there are mprF sequence variations not only in the DAP-R S. aureus strains, as previously published (3, 7, 8, 13), but also in selected DAP-S strains of this genotypic complex. However, the two SNPs found in the DAP-S strains, I498N and Q692E, have never been described before in association with enhanced L-PG synthesis/translocation or the DAP-R phenotype. The five DAP-S strains with the I498N SNP (DAP MICs of ≤0.5 μg/ml) and the three DAP-S strains with the Q692E SNP (DAP MICs of 1 μg/ml group) did not show any significant difference in CM PL contents, surface positive charge, or in vitro HDP susceptibilities compared to these characteristics in the DAP-S strains without such SNPs. These data imply that these mutations are not in essential locations within mprF that dictate changes in MprF structure or function or translate to modifications of CM PL profiles. Surprisingly, in addition to these latter SNPs, two strains in the group with DAP MICs of 1 μg/ml had 30 different amino acid substitutions within their MprF proteins, connoting a high frequency of mprF sequence heterogeneity in some DAP-S S. aureus strains. This hypermutational mprF heterogeneity, however, did not yield enhanced MprF functionality (i.e., in L-PG synthesis or translocation). In contrast, unlike the DAP-S strains, all isolates with DAP MICs of ≥2 μg/ml had mprF SNPs which have been well characterized as being associated with the DAP-R phenotype (8, 9, 11, 28, 29). In line with previous studies (12–14), the DAP-R strain group exhibited excess CM synthesis of L-PG, enhanced surface positive charge, and a reduced DAP-binding phenotype. These sequencing data, in combination with the PL profiling data described above, indicate that whereas mprF SNPs and, even, frequent mprF heterogeneities were found in some DAP-S isolates, SNPs within hot spot loci of the mprF ORF are required for the MprF gain-of-function phenotypes.
Second, we have previously shown that some DAP-R S. aureus isolates express more mprF transcripts over a standard in vitro growth cycle than their DAP-S parental strains (7, 8). As shown by the results in Fig. 1, DAP-R isolates (DAP MICs of ≥2 μg/ml) showed higher levels of mprF transcription than isolates from the two DAP-S groups during both exponential and stationary phases, indicating that the DAP-R isolates used in this study express more net mprF transcripts in their mutated form (L341S or L826F).
Third, recently published studies by our group and others have suggested that DAP-R S. aureus strains commonly exhibit cross-resistance to certain cationic HDPs, especially those associated with defense against endovascular infections, i.e., tPMPs from platelets and hNP-1 from neutrophils (4, 9, 10, 29). In agreement with these prior investigations (12), the present strain groups having higher DAP MICs showed significantly reduced susceptibilities to these two prototypical HDPs (Table 1). In particular, DAP-R isolates exhibited the highest levels of resistance to these two HDPs. These data indicate that only hot spot SNPs (L341S and L826F) within mprF are correlated with the DAP-R and HDP cross-resistance phenotypes.
Fourth, in previous publications (10, 12), the latter two SNPs have been associated with excess production of L-PG. In agreement with these reports (12–14), all the DAP-R isolates in this study with either the L341S or the L826F SNP showed excess synthesis of L-PG. Excess synthesis and translocation of L-PG are believed to contribute to the relative positive surface charge in S. aureus; thus, as expected, the increased L-PG synthesis profiles of our DAP-R isolates versus those of the DAP-S isolates paralleled such surface charge analyses. The observation that only inner L-PG synthesis and not outer leaflet translocation was considerably increased in the DAP-R strains should mainly be viewed in the context of the concomitant reduction in PG content. Since PG serves as a putative initial docking site for DAP within the staphylococcal CM, reduced levels of PG would be associated with greater DAP-R (28, 32). In addition, DAP molecules which cross the outer CM could also theoretically be charge repulsed at the level of the inner CM in DAP-R strains. The observation that such DAP-R isolates bound significantly less Bodipy-DAP than DAP-S strains is likely due to a composite of reduced CM DAP docking sites and inner-CM charge repulsion mechanisms.
Last, in addition to the above-described concepts of hot spot mutations in mprF, dysregulation of mprF expression, and DAP/HDP cross-resistance, we investigated cell wall thickness phenotypes in our strain sets. Prior investigations from our laboratory and others have documented a common (albeit not universal) association between a thickened cell wall phenotype and DAP/HDP resistance in S. aureus (8, 12, 13). The mechanistic relationship between DAP-R and a thickened cell wall phenotype is hypothesized to rest in either a mechanical barrier to DAP penetration and/or affinity trapping of calcium-DAP within the cell wall structure. However, in the present study, the cell wall thicknesses of DAP-R isolates were not significantly different from those of isolates in the two DAP-S groups. This observation further underscores the importance of mutant mprF-associated electrostatic perturbations as one mechanism of DAP-R phenotypes.
It is important to recognize the limitations of the present studies. Our data are somewhat restricted in that only a single clonal complex (CC5) genotype was interrogated. It will be important to repeat such analyses with other relevant genotype strains commonly causing clinical syndromes, such as CC8, CC22, and CC30 (33–35). Moreover, we did not query other gene perturbations that may influence mprF which also have been linked to the DAP-R phenotype, including yycFG, dltA, and rpoB (36). Furthermore, we studied a relatively small MRSA strain sample size. Nonetheless, the results of the current studies reveal novel insights regarding the multiple functions of mprF heterogeneity in antibiotic and host defense avoidance by MRSA.
In conclusion, our data suggest the following: (i) there are individual point mutations (SNPs), as well as high levels of heterogeneity within mprF ORFs in some DAP-S S. aureus strains; (ii) however, only selected hot spot point mutations resulting in mprF dysregulation appear to yield the MprF gain-of-function phenotype (e.g., enhanced L-PG synthesis); (iii) in turn, the latter phenotype appears to be linked to surface positive charge modifications; and (iv) these genotypic and phenotypic abnormalities are observed predominantly in DAP-R isolates and are associated with HDP cross-resistance.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from the National Institutes of Health (AI-39108 to A.S.B. and AI-111661 to M.R.Y.), the American Heart Association (12BGIA11780035 to S.-J.Y.), and the National Institute of Child Health and Human Development (U54 HD071600-01 to G.S.).
Footnotes
Published ahead of print 6 October 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03422-14.
REFERENCES
- 1.Bayer AS, Schneider T, Sahl H-G. 2013. Mechanisms of daptomycin resistance in Staphylococcus aureus: role of the cell membrane and cell wall. Ann. N. Y. Acad. Sci. 1277:139–158. 10.1111/j.1749-6632.2012.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayden MK, Rezai K, Hayes RA, Lolans K, Quinn JP, Weinstein RA. 2005. Development of daptomycin resistance in vivo in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5285–5287. 10.1128/JCM.43.10.5285-5287.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones T, Yeaman MR, Sakoulas G, Yang SJ, Proctor RA, Sahl HG, Schrenzel J, Xiong YQ, Bayer AS. 2008. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 52:269–278. 10.1128/AAC.00719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murthy MH, Olson ME, Wickert RE, Fey PD, Jalali Z. 2008. Daptomycin non-susceptible methicillin-resistant Staphylococcus aureus USA 300 isolate. J. Med. Microbiol. 57:1036–1038. 10.1099/jmm.0.2008/000588-0. [DOI] [PubMed] [Google Scholar]
- 5.Skiest DJ. 2006. Treatment failure resulting from resistance of Staphylococcus aureus to daptomycin. J. Clin. Microbiol. 44:655–656. 10.1128/JCM.44.2.655-656.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mishra NN, Yang S-J, Chen L, Muller C, Saleh-Mghir A, Kuhn S, Peschel A, Yeaman MR, Nast CC, Kreiswirth BN, Crémieux A-C, Bayer AS. 2013. Emergence of daptomycin resistance in daptomycin-naïve rabbits with methicillin-resistant Staphylococcus aureus prosthetic joint infection is associated with resistance to host defense cationic peptides and mprF polymorphisms. PLoS One 8:e71151. 10.1371/journal.pone.0071151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang SJ, Xiong YQ, Dunman PM, Schrenzel J, Francois P, Peschel A, Bayer AS. 2009. Regulation of mprF in daptomycin-nonsusceptible Staphylococcus aureus. Antimicrob. Agents Chemother. 53:2636–2637. 10.1128/AAC.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang S-J, Nast CC, Mishra NN, Yeaman MR, Fey PD, Bayer AS. 2010. Cell wall thickening is not a universal accompaniment of the daptomycin nonsusceptibility phenotype in Staphylococcus aureus: evidence for multiple resistance mechanisms. Antimicrob. Agents Chemother. 54:3079–3085. 10.1128/AAC.00122-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst C, Staubitz P, Mishra NN, Yang SJ, Hornig G, Kalbacher H, Bayer AS, Kraus D, Peschel A. 2009. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 5:e1000660. 10.1371/journal.ppat.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, Kalbacher H, Nieuwenhuizen WF, Jung G, Tarkowski A, van Kessel KPM, van Strijp JAG. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with L-lysine. J. Exp. Med. 193:1067–1076. 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishra NN, Bayer AS, Moise PA, Yeaman MR, Sakoulas G. 2012. Reduced susceptibility to host-defense cationic peptides and daptomycin coemerge in methicillin-resistant Staphylococcus aureus from daptomycin-naive bacteremic patients. J. Infect. Dis. 206:1160–1167. 10.1093/infdis/jis482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishra NN, McKinnell J, Yeaman MR, Rubio A, Nast CC, Chen L, Kreiswirth BN, Bayer AS. 2011. In vitro cross-resistance to daptomycin and host defense cationic antimicrobial peptides in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 55:4012–4018. 10.1128/AAC.00223-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang S-J, Mishra NN, Rubio A, Bayer AS. 2013. Causal role of single nucleotide polymorphisms within the mprF gene of Staphylococcus aureus in daptomycin resistance. Antimicrob. Agents Chemother. 57:5658–5664. 10.1128/AAC.01184-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mishra NN, Yang SJ, Sawa A, Rubio A, Nast CC, Yeaman MR, Bayer AS. 2009. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus (MRSA). Antimicrob. Agents Chemother. 53:2312–2318. 10.1128/AAC.01682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang SJ, Kreiswirth BN, Sakoulas G, Yeaman MR, Xiong YQ, Sawa A, Bayer AS. 2009. Enhanced expression of dltABCD is associated with development of daptomycin nonsusceptibility in a clinical endocarditis isolate of Staphylococcus aureus. J. Infect. Dis. 200:1916–1920. 10.1086/648473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhand A, Bayer AS, Pogliano J, Yang SJ, Bolaris M, Nizet V, Wang G, Sakoulas G. 2011. Use of anti-staphylococcal β-lactams to increase daptomycin activity in eradicating persistent bacteremia due to methicillin-resistant Staphylococcus aureus (MRSA): role of enhanced daptomycin binding. Clin. Infect. Dis. 53:158–163. 10.1093/cid/cir340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moise PA, Forrest A, Bayer AS, Xiong YQ, Yeaman MR, Sakoulas G. 2010. Factors influencing time to vancomycin-induced clearance of nonendocarditis methicillin-resistant Staphylococcus aureus bacteremia: role of platelet microbicidal protein killing and agr genotypes. J. Infect. Dis. 201:233–240. 10.1086/649429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial susceptibility testing; 20th informational supplement, CLSI document M100-S22. CLSI, Wayne, PA. [Google Scholar]
- 19.Boucher HW, Sakoulas G. 2007. Antimicrobial resistance: perspectives on daptomycin resistance, with emphasis on resistance in Staphylococcus aureus. Clin. Infect. Dis. 45:601–608. 10.1086/520655. [DOI] [PubMed] [Google Scholar]
- 20.Yeaman MR, Sullam PM, Dazin PF, Bayer AS. 1994. Platelet microbicidal protein alone and in combination with antibiotics reduces Staphylococcus aureus adherence to platelets in vitro. Infect. Immun. 62:3416–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeaman MR, Bayer AS, Koo S-P, Foss W, Sullam PM. 1998. Platelet microbicidal proteins and neutrophil defensin disrupt the Staphylococcus aureus cytoplasmic membrane by distinct mechanisms of action. J. Clin. Invest. 101:178–187. 10.1172/JCI562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeaman MR, Puentes SM, Norman DC, Bayer AS. 1992. Partial characterization and staphylocidal activity of thrombin-induced platelet microbicidal protein. Infect. Immun. 60:1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong YQ, Mukhopadhyay K, Yeaman MR, Adler-Moore J, Bayer AS. 2005. Functional interrelationships between cell membrane and cell wall in antimicrobial peptide-mediated killing of Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3114–3121. 10.1128/AAC.49.8.3114-3121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meehl M, Herbert S, Gotz F, Cheung A. 2007. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:2679–2689. 10.1128/AAC.00209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Gotz F. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405–8410. 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 26.Mukhopadhyay K, Whitmire W, Xiong YQ, Molden J, Jones T, Peschel A, Staubitz P, Adler-Moore J, McNamara PJ, Proctor RA, Yeaman MR, Bayer AS. 2007. In vitro susceptibility of Staphylococcus aureus to thrombin-induced platelet microbicidal protein-1 (tPMP-1) is influenced by cell membrane phospholipid composition and asymmetry. Microbiology 153:1187–1197. 10.1099/mic.0.2006/003111-0. [DOI] [PubMed] [Google Scholar]
- 27.Yang S-J, Bayer AS, Mishra NN, Meehl M, Ledala N, Yeaman MR, Xiong YQ, Cheung AL. 2012. The Staphylococcus aureus two-component regulatory system, GraRS, senses and confers resistance to selected cationic antimicrobial peptides. Infect. Immun. 80:74–81. 10.1128/IAI.05669-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertsche U, Weidenmaier C, Kuehner D, Yang S-J, Baur S, Wanner S, Francois P, Schrenzel J, Yeaman MR, Bayer AS. 2011. Correlation of daptomycin resistance in a clinical Staphylococcus aureus strain with increased cell wall teichoic acid production and D-alanylation. Antimicrob. Agents Chemother. 55:3922–3928. 10.1128/AAC.01226-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oku Y, Kurokawa K, Ichihashi N, Sekimizu K. 2004. Characterization of the Staphylococcus aureus mprF gene, involved in lysinylation of phosphatidylglycerol. Microbiology 150:45–51. 10.1099/mic.0.26706-0. [DOI] [PubMed] [Google Scholar]
- 30.Mehta S, Cuirolo AX, Plata KB, Riosa S, Silverman JA, Rubio A, Rosato RR, Rosato AE. 2012. VraSR two-component regulatory system contributes to mprF-mediated decreased susceptibility to daptomycin in in vivo-selected clinical strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 56:92–102. 10.1128/AAC.00432-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra NN, Bayer AS. 2013. Correlation of cell membrane lipid profiles with daptomycin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 57:1082–1085. 10.1128/AAC.02182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kilelee E, Pokorny A, Yeaman MR, Bayer AS. 2010. Lysyl-phosphatidylglycerol attenuates membrane perturbation rather than surface association of the cationic antimicrobial peptide 6W-RP-1 in a model membrane system: implications for daptomycin resistance. Antimicrob. Agents Chemother. 54:4476–4479. 10.1128/AAC.00191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferry T, Bes M, Dauwalder O, Meugnier H, Lina G, Forey F, Vandenesch F, Etienne J. 2006. Toxin gene content of the Lyon methicillin-resistant Staphylococcus aureus clone compared with that of other pandemic clones. J. Clin. Microbiol. 44:2642–2644. 10.1128/JCM.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lozano C, Porres-Osante N, Benito D, Zarazaga M, Torres C, Porres-Osante N, Rojo-Bezares B, Sáenz Y, Torres C, Crettaz J, Olarte I. 2013. Changes in genetic lineages, resistance, and virulence in clinical methicillin-resistant Staphylococcus aureus in a Spanish hospital. J. Infect. Chemother. 19:233–242. 10.1007/s10156-012-0486-4. [DOI] [PubMed] [Google Scholar]
- 35.Miller CE, Batra R, Cooper BS, Patel AK, Klein J, Otter JA, Kypraios T, French GL, Tosas O, Edgeworth JD. 2012. An association between bacterial genotype combined with a high-vancomycin minimum inhibitory concentration and risk of endocarditis in methicillin-resistant Staphylococcus aureus bloodstream infection. Clin. Infect. Dis. 54:591–600. 10.1093/cid/cir858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedman L, Alder JD, Silverman JA. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:2137–2145. 10.1128/AAC.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.