Abstract
Nitrofurantoin has been used for decades for the treatment of urinary tract infections (UTIs), but clinically significant resistance in Escherichia coli is uncommon. Nitrofurantoin concentrations in the gastrointestinal tract tend to be low, which might facilitate selection of nitrofurantoin-resistant (NIT-R) strains in the gut flora. We subjected two nitrofurantoin-susceptible intestinal E. coli strains (ST540-p and ST2747-p) to increasing nitrofurantoin concentrations under aerobic and anaerobic conditions. Whole-genome sequencing was performed for both susceptible isolates and selected mutants that exhibited the highest nitrofurantoin resistance levels aerobically (ST540-a and ST2747-a) and anaerobically (ST540-an and ST2747-an). ST540-a/ST540-an and ST2747-a (aerobic MICs of >64 μg/ml) harbored mutations in the known nitrofurantoin resistance determinants nfsA and/or nfsB, which encode oxygen-insensitive nitroreductases. ST2747-an showed reduced nitrofurantoin susceptibility (aerobic MIC of 32 μg/ml) and exhibited remarkable growth deficits but did not harbor nfsA/nfsB mutations. We identified a 12-nucleotide deletion in ribE, encoding lumazine synthase, an essential enzyme involved in the biosynthesis of flavin mononucleotide (FMN), which is an important cofactor for NfsA and NfsB. Complementing ST2747-an with a functional wild-type lumazine synthase restored nitrofurantoin susceptibility. Six NIT-R E. coli isolates (NRCI-1 to NRCI-6) from stools of UTI patients treated with nitrofurantoin, cefuroxime, or a fluoroquinolone harbored mutations in nfsA and/or nfsB but not ribE. Sequencing of the ribE gene in six intestinal and three urinary E. coli strains showing reduced nitrofurantoin susceptibility (MICs of 16 to 48 μg/ml) also did not identify any relevant mutations. NRCI-1, NRCI-2, and NRCI-5 exhibited up to 4-fold higher anaerobic MICs, compared to the mutants generated in vitro, presumably because of additional mutations in oxygen-sensitive nitroreductases.
INTRODUCTION
Nitrofurantoin belongs to the group of synthetic nitrofuran compounds and is an antibiotic that has been available for clinical use for more than 60 years (1). Upon oral administration, it is rapidly absorbed in the small intestine and is excreted by the kidneys into urine, where it reaches high therapeutic concentrations (200 μg/ml) (2). For this reason, nitrofurantoin is prescribed solely for the treatment of uncomplicated lower urinary tract infections (UTIs) (1). It is active against a wide spectrum of Gram-positive and Gram-negative bacteria, including most urinary tract pathogens (3). The antibacterial effect of nitrofurantoin is activated by reduction of the drug into toxic intermediate compounds (4). These intermediates can interact with protein and DNA, thereby inhibiting multiple bacterial enzymes involved in DNA, RNA, and protein synthesis and carbohydrate metabolism (1). The reduction of nitrofurantoin is mediated by nitroreductase systems of the bacterial host. In Escherichia coli, there are two types of nitroreductases, based on their sensitivities to oxygen, namely, type I (oxygen-insensitive) nitroreductases and type II (oxygen-sensitive) nitroreductases (5). Nitrofurantoin resistance in E. coli was shown previously to result primarily from stepwise mutations in nfsA and nfsB genes, encoding oxygen-insensitive nitroreductases (5, 6). While the majority of these changes constituted insertions of insertion sequence (IS) elements into the genes, deletions and missense mutations have also been described (5). Mutations in genes encoding oxygen-sensitive nitroreductases have not been described as yet.
Although nitrofurantoin has been used for decades, clinically significant resistance in E. coli remains uncommon (4). This might be ascribed to an associated fitness cost, especially in the presence of nitrofurantoin at high therapeutic concentrations in the urine (7). As mentioned earlier, most of the nitrofurantoin is absorbed in the small intestine; however, 6 to 13% of the drug reaches the colon (1, 8). Therefore, due to low concentrations of nitrofurantoin in the gastrointestinal tract, selection of nitrofurantoin-resistant (NIT-R) strains in this niche is highly probable. Additionally, low oxygen levels in the gastrointestinal tract might favor mutations in genes encoding oxygen-sensitive nitroreductases. NIT-R strains were found previously in the fecal flora with very low prevalence rates (0.6 to 2%) (9–11); however, these strains were not investigated molecularly. In this study, two nitrofurantoin-susceptible (NIT-S) intestinal E. coli strains were used as parent strains for in vitro generation of NIT-R mutants under aerobic and anaerobic growth conditions. Known and novel nitrofurantoin resistance determinants were identified by whole-genome sequencing and in vitro confirmational experiments, and the growth rates of these strains were assessed. Finally, we compared the resistance determinants and growth rates of the mutants generated in vitro with those of six clinical NIT-R E. coli strains recovered from the fecal flora of four UTI patients who were treated with nitrofurantoin, cefuroxime, ciprofloxacin, or norfloxacin.
MATERIALS AND METHODS
Isolation and characterization of E. coli strains from stool and urine samples.
Six NIT-R E. coli strains (MICs of ≥64 μg/ml) were isolated from stool samples from four UTI patients who were treated with nitrofurantoin, cefuroxime, ciprofloxacin, or norfloxacin. The recovery of these strains was performed as part of a large clinical trial investigating the emergence of antibiotic-resistant Enterobacteriaceae in the fecal flora of UTI patients after antibiotic treatment and the spread of these resistant strains to household members (A. Stewardson, N. Adriaensens, M. Godycki-Cwirko, H. Goossens, C. Lammens, S. Malhotra-Kumar, J. Vervoort, and S. Harbarth, unpublished data). Six intestinal and three urinary E. coli isolates with reduced nitrofurantoin susceptibility (MICs of <64 μg/ml) were also included in the study. The E. coli strains were identified by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry using the Microflex LT system (Bruker Daltonics, Bremen, Germany) with MALDI Biotyper 3.0 software (database version 5627). Routine susceptibility tests were performed by disk diffusion testing (disks from BD BBL, Franklin Lakes, NJ), according to the CLSI guidelines (12), for the following antibiotics: amoxicillin (plus clavulanate), piperacillin (plus tazobactam), cefotaxime (plus clavulanate), ceftazidime (plus clavulanate), cefepime, cefoxitin, aztreonam, ertapenem, meropenem, ciprofloxacin, fosfomycin, nitrofurantoin, and trimethoprim-sulfamethoxazole. The nitrofurantoin MICs of all investigated strains were determined by agar dilution, and breakpoints were applied according to CLSI guidelines (susceptible, ≤32 μg/ml; intermediate, 64 μg/ml; resistant, ≥128 μg/ml), utilizing a multipoint inoculator (Multipointelite; MAST Group Ltd., Bootle, United Kingdom) (12). The inoculated nitrofurantoin-containing Mueller-Hinton (MH) plates were incubated for 16 to 20 h at 37°C in an O2 incubator (Thermo Scientific, Waltham, MA) or Bugbox anaerobic workstation (Ruskinn Technology Ltd., Bridgend, United Kingdom), to determine the MICs under aerobic and anaerobic growth conditions, respectively.
In vitro generation of NIT-R mutants.
Two NIT-S E. coli strains (MICs of 4 to 16 μg/ml), of multilocus sequence types (STs) ST540 and ST2747 (ST540-p and ST2747-p, respectively) (as determined from whole-genome sequencing data), were recovered from stool samples from two UTI patients and were used as parent strains for in vitro generation of NIT-R mutants under aerobic and anaerobic growth conditions. A maximum of 3 selection rounds were implemented, to allow isolation of both single-step and multistep mutants. In each selection round, 1 × 108 to 2 × 108 CFU of the parent strains were spread homogeneously on MH agar plates supplemented with increasing nitrofurantoin concentrations (0.5 to 4 times the MIC for the parent strain) and were incubated for 24, 48, and 72 h at 37°C under aerobic (O2 incubator) and anaerobic (Bugbox) growth conditions. Resistant mutants were counted, and 10 to 20 isolates were subcultured on MH agar supplemented with nitrofurantoin, to confirm the resistance phenotype. Aerobic and anaerobic nitrofurantoin MICs were determined by agar dilution, and strains of interest were utilized for the next selection round.
Whole-genome sequencing to identify known and novel nitrofurantoin resistance determinants.
Whole-genome sequencing was performed (Pacific Biosciences, Menlo Park, CA) for ST540-p and ST2747-p and for the mutants generated in vitro that exhibited the highest nitrofurantoin resistance levels aerobically (ST540-a, with aerobic and anaerobic MICs of 256 and 32 μg/ml, respectively, and ST2747-a, with aerobic and anaerobic MICs of 64 and 16 μg/ml, respectively) and anaerobically (ST540-an, with aerobic and anaerobic MICs of 128 and 32 μg/ml, respectively, and ST2747-an, with aerobic and anaerobic MICs of 32 and 16 μg/ml, respectively). Genomic DNA was isolated (MasterPure complete DNA and RNA purification kit; Epicentre, Madison, WI), and DNA concentrations were measured (NanoDrop; Thermo Scientific). Library preparation and sequencing reactions were performed using a PacBio DNA template prep kit 2.0 (3 kb to 10 kb) and a PacBio DNA sequencing kit 2.0 with C2 chemistry. Sequence runs for six single-molecule real-time (SMRT) cells were performed with a PacBio RS II sequencer with a 1 by 180-min movie time SMRT cell. SMRT Analysis Portal 2.0 was used for filtering of the reads and subreads, with default parameters, and filtered data of ∼320 Mb for each cell per strain were considered for assembly. The genomes were assembled using the hierarchical genome assembly process (HGAP), which is available with the SMRT Analysis packages and was accessed through SMRT Analysis Portal 2.0 (13).
The genomes from ST540-p, ST540-a, and ST540-an and from ST2747-p, ST2747-a, and ST2747-an were compared and checked for genomic rearrangements using Mauve 2.3.1 (14). Single-nucleotide polymorphisms (SNPs) were identified by Show-SNPs, a script associated with MUMmer 3.23 (15). The presence of IS elements or mutations in nfsA and nfsB and other genomic changes that might potentially be in as yet unknown resistance determinants were confirmed by PCR sequencing using target-specific primer pairs (primer sequences available on request) (7).
Validation of novel nitrofurantoin resistance determinant.
The contribution of lumazine synthase to nitrofurantoin resistance was evaluated by complementing the lumazine synthase mutant generated in vitro (ST2747-an) with a functional wild-type lumazine synthase allele. A 559-bp fragment containing the coding sequence of wild-type ribE, including 65-bp upstream and 23-bp downstream regions (GenBank accession no. CP007392; bases 1080502 to 1081060) (13), was prepared by PCR amplification, and the PCR fragment was cloned in a pCR2.1 vector by Topo TA cloning (Invitrogen, Carlsbad, CA), according to the manufacturer's protocol. The resulting recombinant vector, LMZ-pCR2.1, was electrotransformed in ST2747-an, which was made electrocompetent by successive washing steps with an ice-cold 10% glycerol suspension at log phase. Transformants were selected on LB agar supplemented with kanamycin (50 μg/ml) and were screened for the presence of the correct insert by PCR sequencing using M13 primers. The presence of functional and/or mutated lumazine synthase was confirmed by PCR detection using an internal primer set that resulted in PCR fragments with a 12-nucleotide size difference (primer sequences for lumazine synthase primers available on request). Aerobic and anaerobic nitrofurantoin MICs of the complemented strain were determined to detect the restoration of nitrofurantoin susceptibility.
Growth rate measurements.
Growth rates of susceptible, resistant, and complemented mutant strains were measured in the absence and presence of nitrofurantoin. Each strain was analyzed in duplicate by adding 20 μl of a 0.5 McFarland standard suspension to 180 μl of LB broth, with final concentrations of nitrofurantoin ranging from 0 to 512 μg/ml, in a flat-bottomed 96-well plate (Greiner Bio-One, Monroe, NC). Automatic measurements of the optical density at 600 nm (OD600) were performed every 4 min for at least 21 h at 37°C, with a Multiskan GO microplate spectrophotometer (Thermo Scientific). The fitness of the strains, in terms of individual growth rate deficits, was evaluated by comparing the times of onset of growth, the slopes of the log phase, and the maximum OD600 values reached at fixed time points.
RESULTS
NIT-R mutants generated in vitro.
We generated mutants in vitro by plating two NIT-S intestinal E. coli strains (ST540-p and ST2747-p) on MH agar supplemented with nitrofurantoin at up to 32 μg/ml and incubating the strains under aerobic and anaerobic growth conditions. ST540-p had aerobic and anaerobic MICs of 16 and 4 μg/ml, respectively, while ST2747-p showed heteroresistance to nitrofurantoin, with aerobic and anaerobic MIC ranges of 4 to 16 and 2 to 8 μg/ml, respectively (Table 1). The mutants from the first selection round that were derived from ST540-p and ST2747-p had maximum aerobic and anaerobic MICs of 64 and 16 μg/ml (ST540-p mutants) and 16 and 4 μg/ml (ST2747-p mutants), respectively (see Fig. S1 in the supplemental material). A set of five mutants with different MICs (aerobic MICs of 16 to 64 μg/ml and anaerobic MICs of 4 to 16 μg/ml) were used as starting strains for the second selection round, on MH agar supplemented with nitrofurantoin at up to 128 μg/ml. These mutants, derived from first-round mutants of ST540-p and ST2747-p, had maximum aerobic and anaerobic MICs of 256 and 32 μg/ml (ST540-p mutants) and 32 and 16 μg/ml (ST2747-p mutants), respectively. Finally, a set of seven mutants from the second selection round with different MICs (aerobic MICs of 16 to 128 μg/ml and anaerobic MICs of 16 to 32 μg/ml) were used as starting strains for the third selection round, on MH agar supplemented with nitrofurantoin at up to 512 μg/ml. These mutants, derived from second-round mutants of ST540-p and ST2747-p, had maximum aerobic and anaerobic MICs of 256 and 32 μg/ml (ST540-p mutants) and 128 and 32 μg/ml (ST2747-p mutants), respectively.
TABLE 1.
Aerobic and anaerobic nitrofurantoin MICs and substitutions, insertions, and deletions in NfsA, NfsB, and lumazine synthase of NIT-S intestinal E. coli strains (ST540-p and ST2747-p), mutants that showed the highest nitrofurantoin resistance levels aerobically (ST540-a and ST2747-a) and anaerobically (ST540-an and ST2747-an), and a complemented mutant strain (ST2747-an/cm)
| Strain | Nitrofurantoin MIC (μg/ml) |
Substitutions, insertions, and deletionsa |
|||
|---|---|---|---|---|---|
| Aerobic | Anaerobic | NfsA | NfsB | Lumazine synthase | |
| ST540-p | 16 | 4 | WT | WT | WT |
| ST540-a | 256 | 32 | Ins of IS1 | Ins of IS1 | WT |
| ST540-an | 64 | 16 | Stop at position 100 | WT | WT |
| ST2747-p | 4–16 | 2–8 | WT | WT | WT |
| ST2747-a | 128 | 32 | Stop at position 20 | Stop at position 57 | WT |
| ST2747-an | 32 | 16 | WT | WT | Del of positions 131–134 |
| ST2747-an/cm | 16 | 8 | WT | WT | WT plus del of positions 131–134 |
WT, wild type; ins, insertion; del, deletion.
Both ST540-p and ST2747-p, together with their mutants that showed the highest nitrofurantoin MICs aerobically (ST540-a and ST2747-a) and anaerobically (ST540-an and ST2747-an), were subjected to whole-genome sequencing (13). Full genomes were compared to identify mutations in the known resistance determinants nfsA and nfsB, as well as to check for other genomic changes indicative of as-yet-undescribed nitrofurantoin resistance determinants. The aerobically isolated mutant ST540-a had aerobic and anaerobic MICs of 256 and 32 μg/ml, respectively, and both its nfsA and nfsB genes were interrupted by insertion of a mobile IS1 element, at nucleotide positions 579 and 561, respectively (Table 1). The anaerobically isolated mutant ST540-an had aerobic and anaerobic MICs of 64 and 16 μg/ml, respectively, and had a nonsense mutation in nfsA, leading to a premature stop in NfsA at position 100. The nfsB gene remained unaffected. The aerobically isolated mutant ST2747-a had aerobic and anaerobic MICs of 128 and 32 μg/ml, respectively, and both its nfsA and nfsB genes harbored a nonsense mutation, leading to premature stops in NfsA and NfsB at positions 20 and 57, respectively. The anaerobically isolated mutant ST2747-an had aerobic and anaerobic MICs of 32 and 16 μg/ml, respectively, but did not exhibit changes in nfsA or nfsB even though this mutant showed at least 2-fold increases in the MICs for nitrofurantoin, compared to its parent strain. Based on whole-genome sequencing data, we identified a 12-nucleotide deletion in ribE, a gene encoding 6,7-dimethyl-8-ribityllumazine synthase (lumazine synthase). Lumazine synthase is an essential enzyme involved in the riboflavin biosynthesis pathway. The 12-nucleotide deletion in ribE at nucleotide positions 391 to 402, which was confirmed in vitro by PCR and bidirectional sequencing, resulted in a four-amino acid loss (TKAG) in the active site of lumazine synthase, at positions 131 to 134 (Fig. 1) (16). In ST540-a, ST540-an, and ST2747-a, lumazine synthase remained unaffected. An overview of all other proteins with mutations in their nucleotide coding sequences, based on in silico comparative genome analysis of parent and mutant strains, is provided in Table S1 in the supplemental material.
FIG 1.

Lumazine synthase sequences for ST2747-p and ST2747-an. *, amino acid residues of the active site. The boxed area shows the deletion of amino acid positions 131 to 134 in ST2747-an.
Identification and validation of novel nitrofurantoin resistance determinant.
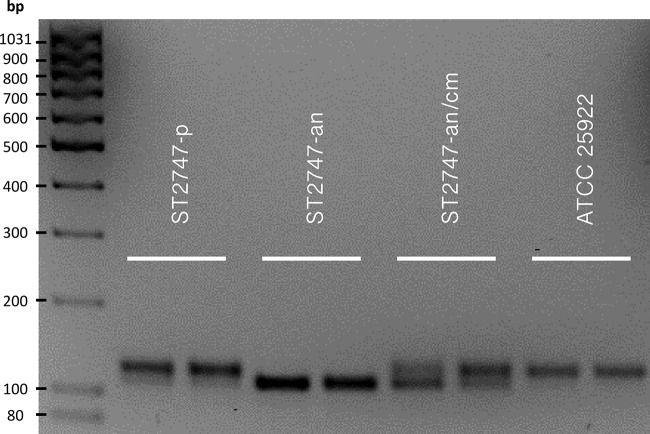
To confirm the role of the ribE deletion in mediating nitrofurantoin resistance in the mutant ST2747-an, we complemented this strain with a recombinant vector encoding a functional wild-type lumazine synthase (LMZ-pCR2.1), which would potentially restore nitrofurantoin susceptibility. Prior to susceptibility testing, we demonstrated the presence of both wild-type and mutated lumazine synthase in the complemented mutant strain (ST2747-an/cm) by PCR detection of ribE. Separation of the PCR products of ST2747-an/cm by agarose gel electrophoreses showed the presence of two fragments, with a 12-bp size difference (Fig. 2). MICs for nitrofurantoin were tested by agar dilution and are shown in Table 1. Introducing LMZ-pCR2.1 into ST2747-an resulted in 2-fold decreases of the aerobic and anaerobic nitrofurantoin MICs, from 32 to 16 μg/ml and from 16 to 8 μg/ml, respectively. These decreased MIC values were similar to those of the parent ST2747-p strain (Table 1).
FIG 2.
PCR detection of wild-type and/or mutated ribE in ST2747-p, ST2747-an, and ST2747-an/cm. ATCC 25922 was included as a positive control for wild-type ribE. The PCR fragments were separated, together with a MassRuler low range DNA ladder (Thermo Scientific), on a 2% agarose gel for 1.5 h at 7.5 V/cm with 0.5× Tris-borate-EDTA buffer (40 mM Tris-HCl, 40 mM borate, 1 mM EDTA). The upper band (119 bp) corresponded to the wild-type ribE of ST2747-p and the lower band (107 bp) to the mutated ribE of ST2747-an.
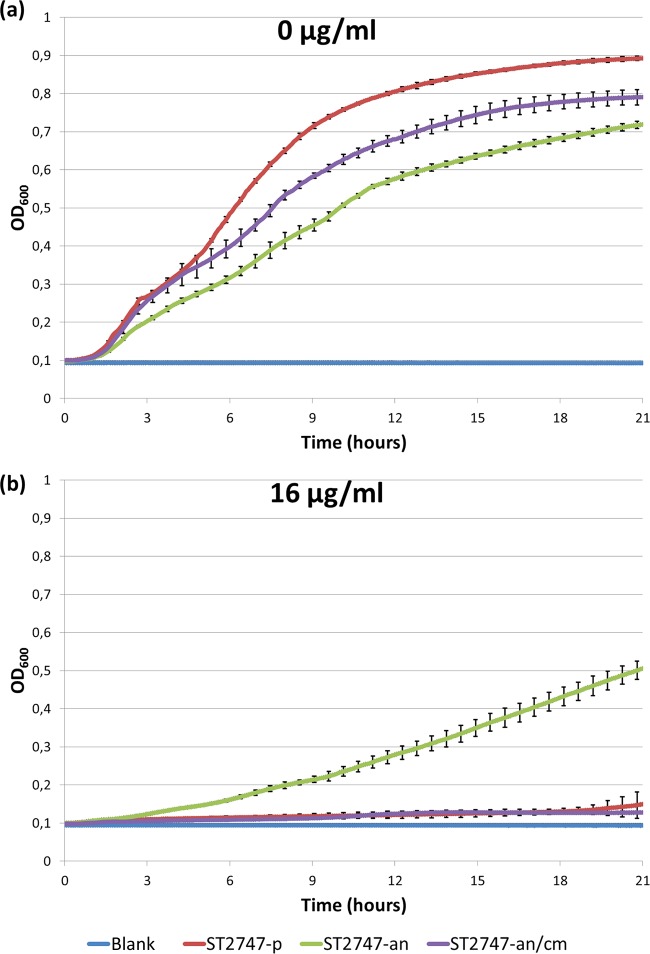
Colonies formed by the ST2747-an mutant after 16 to 20 h of incubation on MH agar were smaller than those of ST2747-p, indicating a potential fitness cost associated with the ribE deletion. On the other hand, colonies formed by the complemented ST2747-an/cm mutant were similar in size to those of S2747-p. To quantify differences in growth rates of the strains, we performed automated OD600 measurements in the absence and presence of nitrofurantoin (Fig. 3). In the absence of nitrofurantoin, the onset of growth of ST2747-an was clearly delayed and the growth rate was remarkably lower during the log phase, compared to ST2747-p, with an OD600 difference of ±0.2 after 21 h of incubation at 37°C. In contrast, the onset of growth of ST2747-an/cm was identical to that of ST2747-p. The growth rate of ST2747-an/cm remained similar for the first 5 h without significant changes, followed by a slight decrease that ultimately led to an OD600 difference of ±0.1 after 21 h of incubation. In the presence of 16 μg/ml nitrofurantoin, ST2747-an was still able to grow, at a lower growth rate, while both ST2747-p and ST2747-an/cm failed to grow.
FIG 3.
Growth rate measurements (OD600 values) for ST2747-p, ST2747-an, and ST2747-an/cm, in the absence (a) and presence (b) of 16 μg/ml nitrofurantoin.
NIT-R clinical E. coli strains.
Routine disk diffusion susceptibility testing enabled selection of NIT-R clinical E. coli strains (nitrofurantoin zone diameters of ≤14 mm) recovered from stool samples from UTI patients. In total, six unique NIT-R E. coli strains could be recovered from four UTI patients treated with nitrofurantoin, cefuroxime, ciprofloxacin, or norfloxacin. Aerobic and anaerobic nitrofurantoin MICs were determined by agar dilution and ranged from 64 to 256 μg/ml and from 16 to 128 μg/ml, respectively (Table 2). We sequenced the complete coding sequences of the previously identified resistance determinants nfsA and nfsB and the novel resistance determinant ribE. All six isolates were found to show at least one mutation in nfsA (Table 2). While we observed missense mutations alone in two isolates (NRCI-1 and NRCI-3), the other four isolates also carried nonsense mutations, leading to a premature stop in NfsA at either position 139 (NRCI-6) or position 147 (NRCI-2, NRCI-4, and NRCI-5). Mutations in nfsB were identified in all isolates except one (NRCI-2). For three isolates, these mutations were missense mutations, either exclusively (NRCI-1 and NRCI-6) or in combination with a nonsense mutation (NRCI-3) leading to a premature stop in NfsB at position 138. For the remaining two isolates (NRCI-4 and NRCI-5), the nfsB gene was disrupted by insertion of a mobile IS26 element at nucleotide position 249; however, no mutations in ribE were observed. We also sequenced the ribE gene in six intestinal and three urinary E. coli strains that showed reduced nitrofurantoin susceptibility (zone diameters of ≤14 mm and MICs of 16 to 48 μg/ml). In total, four synonymous substitutions (T108C, C267T, T345G, and C429T) in ribE were detected in these strains. In addition to nitrofurantoin resistance, the NRCI-1 to NRCI-6 strains exhibited resistance to ciprofloxacin and trimethoprim-sulfamethoxazole, with zone diameters of ≤15 mm and ≤10 mm, respectively. Additionally, NRCI-1 and NRCI-2 showed extended-spectrum β-lactamase (ESBL) production by the combination disk method (ceftazidime and cefotaxime zone diameters of ≤22 mm and ≤27 mm, respectively, with zone diameter increases of ≥5 mm when the drugs were combined with clavulanate).
TABLE 2.
Disk diffusion results, aerobic and anaerobic nitrofurantoin MICs, and substitutions and insertions in NfsA, NfsB, and lumazine synthase of NIT-R clinical E. coli strains
| Strain | Disk diffusion resulta |
Nitrofurantoin MIC (μg/ml) |
Substitution(s) or insertionb |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMX | AMC | PIP | TZP | CTX | CTX-CLAc | CAZ | CAZ-CLAc | FEP | FOX | ATM | ETP | MEM | CIP | FOF | F/M | SXT | Aerobic | Anaerobic | NfsA | NfsB | Lumazine synthase | |
| NRCI-1 | R | I | R | I | R | + | R | + | R | I | R | S | S | R | S | R | R | 256 | 64 | Q72K | G66D, I75M | WT |
| NRCI-2 | R | I | R | I | R | + | R | + | I | S | R | S | S | R | S | R | R | 256 | 64 | R15L, T117I, stop at position 147, L157F, D187G | WT | WT |
| NRCI-3 | R | S | R | S | S | − | S | − | S | S | S | S | S | R | S | R | R | 64 | 16 | R15C, T117I, D187G | I75M, stop at position 138 | WT |
| NRCI-4 | R | S | R | S | S | − | S | − | S | S | S | S | S | R | S | R | R | 64 | 16 | T117I, stop at position 147, L157F, D187G | Ins of IS26 | WT |
| NRCI-5 | R | R | R | R | S | − | R | − | S | S | R | S | S | R | S | R | R | 256 | 128 | T117I, stop at position 147, L157F, D187G | Ins of IS26 | WT |
| NRCI-6 | R | I | R | S | S | − | S | − | S | S | S | S | S | R | S | R | R | 64 | 32 | A138G, stop at position 139 | R121C | WT |
R, resistant; I, intermediate; S, susceptible; AMX, amoxicillin; AMC, amoxicillin-clavulanate; PIP, piperacillin; TZP, piperacillin-tazobactam; CTX, cefotaxime; CTX-CLA, cefotaxime-clavulanate; CAZ, ceftazidime; CAZ-CLA, ceftazidime-clavulanate; FEP, cefepime; FOX, cefoxitin; ATM, aztreonam; ETP, ertapenem; MEM, meropenem; CIP, ciprofloxacin; FOF, fosfomycin; F/M, nitrofurantoin; SXT, trimethoprim-sulfamethoxazole.
WT, wild type; ins, insertion.
+, ≥5-mm zone diameter increase; −, <5-mm zone diameter increase.
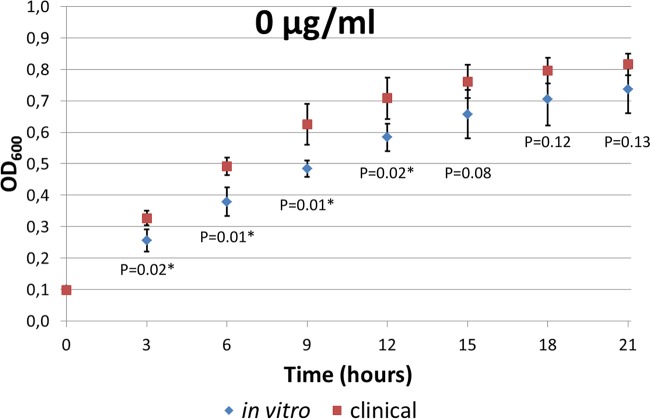
The growth rates of the NIT-R clinical isolates were assessed by automated measurements in the absence and presence of nitrofurantoin and were compared with those of the mutants generated in vitro (see Fig. S2 in the supplemental material). Generally, the resistant clinical isolates tended to be more fit than the mutants generated in vitro in the absence of nitrofurantoin, exhibiting steeper early log phases and reaching significantly higher OD600 values (P < 0.02, Student's t test) at up to 12 h of incubation (Fig. 4). In the stationary phase, the OD600 values exhibited by the clinical isolates were no longer significantly higher. In the presence of nitrofurantoin, the growth curves of the resistant clinical isolates did not differ significantly from those of the mutants generated in vitro (data not shown). When the isolates were examined individually, it was remarkable that, even though NRCI-1, NRCI-2, and NRCI-5 had aerobic MICs of 256 μg/ml, only NRCI-5 was able to grow during the growth rate experiments in the presence of 128 μg/ml nitrofurantoin. In the absence of nitrofurantoin, all three isolates showed good growth. The mutant ST540-a generated in vitro, with an aerobic MIC of 256 μg/ml, was also able to grow in the presence of 128 μg/ml nitrofurantoin; however, it reached the lowest OD600 values in the absence of nitrofurantoin, with an OD600 difference from its parent strain ST540-p of almost 0.3 after 21 h of incubation.
FIG 4.
Comparison of growth rates (OD600 values) for mutants generated in vitro versus NIT-R clinical isolates, in the absence of nitrofurantoin. Error bars, 95% confidence intervals. *, significant difference (P < 0.02, Student's t test).
DISCUSSION
In this study, we generated NIT-R mutants in vitro through stepwise plating of susceptible strains (ST540-p and ST2747-p) on MH plates with increasing nitrofurantoin concentrations, under aerobic and anaerobic growth conditions. This ultimately resulted in the selection of the highly NIT-R strains ST540-a (aerobic and anaerobic MICs of 256 and 32 μg/ml, respectively) and ST2747-a (aerobic and anaerobic MICs of 128 and 32 μg/ml, respectively). Both resistant strains were generated by successive selection rounds performed exclusively under aerobic growth conditions. Remarkably, only one selection round was necessary for ST540-p to become intermediate resistant (aerobic MIC of 64 μg/ml), while ST2747-p remained susceptible (aerobic MIC of 32 μg/ml) after two selection rounds (see Fig. S1 in the supplemental material) (12). It was only after the third selection round that ST2747-p achieved a resistant MIC of 128 μg/ml under aerobic growth conditions. There are multiple possible explanations for why the emergence of nitrofurantoin resistance occurred more rapidly in ST540-p. First, ST540-p might have higher intrinsic mutability due to the presence of certain mobile elements that are transposed under the influence of environmental factors such as the presence of antibiotics (17, 18). Indeed, in ST540-a, both nfsA and nfsB genes were disrupted by insertion of an IS1 mobile element (Table 1), indicating that, under the influence of nitrofurantoin, IS1 mobile elements contribute to increased mutability. Additionally, ST540-p might have had preexisting low-level nitrofurantoin resistance mutations that facilitated the acquisition of additional mutations when the cells were placed under selection pressure with nitrofurantoin. Several such strains were described previously (6); they exhibited wild-type resistance to nitrofurantoin but could be mutated to high levels of nitrofurantoin resistance in a single step. Sequence analysis of one of those strains revealed that it had a wild-type nfsA gene and a nonsense mutation in nfsB, leading to a premature stop in NfsB at position 142 (5). We also isolated mutants that were generated by successive selection rounds performed exclusively under strict anaerobic growth conditions. These isolates, ST540-an and ST2747-an, exhibited aerobic and anaerobic MICs of 64 and 16 μg/ml (ST540-an) and 32 and 16 μg/ml (ST2747-an), respectively, which were much lower resistance levels than those generated under aerobic growth conditions. It is possible that an anaerobic environment plays a protective role in the emergence of nitrofurantoin resistance, indicating that growth conditions might be important determinants of bacterial mutability of (18). Moreover, all mutants generated in vitro showed anaerobic MIC values that were 2-fold to 8-fold lower than the aerobic MIC values. These results corroborate a previous study (7) and can probably be attributed to the oxygen-sensitive nitroreductase system, which becomes active in the absence of oxygen and hence compensates for the (partial) loss of activity of the oxygen-insensitive nitroreductase system (19, 20). In addition, another study using single- and double-knockout E. coli mutants targeting nfsA and nfsB showed that, at low oxygen levels, the mutants were still able to reduce the nitro compound 7-nitrocoumarin-3-carboxylic acid (7NCCA), indicating that NfsA and NfsB are not required for nitroreduction under anaerobic growth conditions (21).
In ST2747-an, mutations in nfsA or nfsB could not be detected, although a 2-fold MIC increase was observed. Whole-genome sequencing of the strain indicated a 12-nucleotide deletion in ribE, a gene encoding lumazine synthase. The ribE gene product consists of a 16.2-kDa subunit that builds up lumazine synthase, a hollow icosahedral capsid of 60 subunits with a total mass of about 1 MDa (16). The active site is located close to the inner surface of the capsid, and substrates need to access the active site by penetrating the capsid via narrow channels (22, 23). Lumazine synthase is an essential enzyme involved in the riboflavin biosynthesis pathway, where it catalyzes the following chemical reaction: 1-deoxy-l-glycerotetrulose-4-phosphate + 5-amino-6-(d-ribitylamino)uracil ⇄ 6,7-dimethyl-8-(d-ribityl)lumazine + 2 H2O + phosphate. Riboflavin, also known as vitamin B2, is mandatory for the production of flavin mononucleotide (FMN), which is an important cofactor of NfsA and NfsB (5). FMN is tightly associated with NfsA and NfsB, where it serves as a mediator for electron transfer from NAD(P)H to various substrates for nitroreduction (24). Enzymatic assays with 7NCCA demonstrated that both nitroreductases were greatly dependent on FMN for nitroreduction to proceed (21). The 12-nucleotide deletion in ribE resulted in a four-amino acid loss (TKAG) in the active site of lumazine synthase, at positions 131 to 134 (Fig. 1). This region is highly conserved among lumazine synthases of very diverse bacteria, such as Bacillus subtilis, Photobacterium leiognathi, and Haemophilus influenzae (16), suggesting that deletions within this region have major implications for the functionality of the enzyme. This might have ultimately resulted in the inability (or reduced ability) of ST2747-an to synthesize FMN, thereby hindering nitroreduction by both Nfs enzymes, particularly NfsB.
The involvement of lumazine synthase in the development of nitrofurantoin resistance was assessed by complementing the ST2747-an mutant with LMZ-pCR2.1, which encoded a functional wild-type lumazine synthase. This resulted in the restoration of nitrofurantoin susceptibility, which was demonstrated by 2-fold decreases of the aerobic and anaerobic MICs from 32 to 16 μg/ml and from 16 and 8 μg/ml, respectively, similar to the MIC values of the parent ST2747-p strain. These results confirm that the reduced nitrofurantoin susceptibility is caused by the TKAG deletion in lumazine synthase. Additionally, the impact of the deletion in lumazine synthase on bacterial growth rate was measured. We found clearly delayed growth onset and a 20% decrease in the bacterial growth of ST2747-an after 21 h of incubation in the absence of nitrofurantoin, compared to the parent strain (Fig. 3). A growth deficit is not entirely unexpected. Gram-negative bacteria are unable to take up riboflavin from the external environment (25), which causes them to be completely dependent on endogenous biosynthesis. As mentioned earlier, lumazine synthase is required for the production of riboflavin. Riboflavin itself does not have any biological activity; however, it is the precursor of FMN and flavin adenine dinucleotide (FAD). Both cofactors are mostly noncovalently bound not only to NfsA and NfsB but also to numerous other flavoproteins, which makes them essential components for cellular metabolism (26). These flavoproteins enable redox reactions involving one- and two-electron transfer processes and catalyze the dehydrogenation of metabolites, as well as oxidation and dehydroxylation reactions (27). Interestingly, the growth rate could be partially restored in the complemented mutant ST2747-an/cm, as the growth onset was not delayed and the rate remained similar to that of ST2747-p for the first 5 h of incubation. At the end of the experiment, ST2747-an/cm showed 10% increased bacterial growth, compared to ST2747-an. As expected, in the presence of 16 μg/ml nitrofurantoin, ST2747-an was still able to grow at a lower growth rate, while both ST2747-p and ST2747-an/cm failed to grow.
Table S1 in the supplemental material gives an overview of proteins carrying mutations, based on in silico comparative genome analysis, including two other proteins that are indirectly involved in FMN biosynthesis. In ST540-a and ST540-an, we observed a potential missense mutation in araB, which encodes ribulokinase, an enzyme that participates in pentose and glucuronate interconversions. It catalyzes the phosphorylation of l-ribulose to l-ribulose-5-phosphate, which is an essential step in the metabolism of the carbon source l-arabinose (28). In the pentose phosphate pathway, l-ribulose-5-phosphate can be converted into d-ribulose-5-phosphate, a substrate for riboflavin metabolism, ultimately leading to the production of FMN. Additionally, we described a potential missense mutation in rbsR, encoding a ribose operon repressor, in ST540-an and ST2747-an. The repressor is a LacI-type transcriptional repressor of the ribose rbsDACBK operon. The RbsABC proteins constitute an ATP-binding cassette (ABC)-type high-affinity transporter for the carbon source d-ribose, while RbsD and RbsK are responsible for the conversion of d-ribose into d-ribose-5-phosphate in the pentose phosphate pathway (29). d-Ribose-5-phosphate can be easily converted into d-ribulose-5-phosphate and hence can be utilized for FMN production. A mutation in the inducer recognition site of RbsR might lead to an inability of d-ribose to induce transcription of the rbsDACBK operon. However, these mutations were not investigated further and need to be confirmed by in vitro analysis.
Routine analysis of strains recovered from stool samples from UTI patients resulted in the detection of six NIT-R clinical E. coli strains. These isolates had similar aerobic MICs, compared to the mutants generated in vitro (64 to 256 μg/ml); for some clinical isolates (NRCI-1, NRCI-2, and NRCI-5), however, the anaerobic MICs were up to 4-fold higher, reaching values of 128 μg/ml, compared with 32 μg/ml for the mutants generated in vitro (Table 2). In accordance with previous data, these clinical isolates might have acquired mutations in the oxygen-sensitive nitroreductase genes or other uncharacterized genes (7). When the types of mutations observed in nfsA and nfsB were compared, it was remarkable that all NIT-R clinical isolates had missense mutations in one or both genes, while such mutations were completely absent in the mutants generated in vitro. These mutations might have accumulated gradually over a longer time period. One of the mutations in NRCI-3 (R15C) was characterized previously (5) and represents a loss-of-function mutation. Similar to the in vitro mutants with aerobic MICs of ≥64 μg/ml, most NIT-R clinical isolates had nonsense mutations or large insertions of an IS element in nfsA and/or nfsB, with the exception of NRCI-1, which harbored only missense mutations in both genes. Furthermore, no mutations could be observed in ribE in the clinical isolates. Since the deletion in ribE was associated with reduced nitrofurantoin susceptibility (MICs of 32 μg/ml), we sequenced the ribE gene in six intestinal and three urinary E. coli isolates, with nitrofurantoin zone diameters of ≤14 mm and MICs of 16 to 48 μg/ml. Apart from a few synonymous substitutions, the ribE gene was largely unaffected in these strains. These results, combined with the growth rate deficit associated with the ribE deletion, which is indicative of a fitness cost, might underlie the lack of recovery of such mutants in the community. Interestingly, steeper early log phases and significantly higher OD600 values (P < 0.02, Student's t test) at up to 12 h of incubation in the absence of nitrofurantoin were observed among the resistant clinical isolates, compared to the mutants generated in vitro (Fig. 4). These clinical strains might have improved their biological costs of resistance through the acquisition of compensatory mutations (30), although a recent study of NIT-R clinical strains showed a lack of clonal spread of such isolates, reiterating the high fitness costs associated with nitrofurantoin resistance in E. coli (31). Presumably, the six NIT-R clinical E. coli strains were recovered from the fecal flora of the UTI patients because they showed coresistance to the antibiotics these patients were given, such as ciprofloxacin and cefuroxime.
The individual growth assessment method used to assess fitness costs in this study was not without limitations. Direct in vitro competition testing followed by plating for determination of viable counts might have been a more accurate method for fitness estimations (32); however, the minor differences in MICs between the competing strains precluded the use of a direct competition assay. Finally, while negative in vitro competition experiments do not exclude the possibility of fitness costs under in vivo conditions, positive in vitro results (such as ours) do allow a reasonable assumption that the resistance would engender a fitness deficit in the bacteria under natural conditions, as described previously (33). In conclusion, we identified a novel nitrofurantoin resistance determinant and clearly showed the growth deficits or presumptive fitness costs engendered by this and previously identified nitrofurantoin resistance determinants, thus providing a basis for the lack of widespread resistance to this unusual antibiotic in E. coli.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the Institute of Primary Care & Public Health at the Cardiff University School of Medicine for providing the NIT-S and NIT-R urinary E. coli isolates.
We have no conflicts of interest to declare.
J.V. and A.S. are supported by funding from the European Community (SATURN network contract FP7-Health-2009-Single Stage-241796). B.B.X. is supported by University of Antwerp research funds (BOF-DOCPRO grant 2012-27450) awarded to S.M.-K.
Footnotes
Published ahead of print 22 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03952-14.
REFERENCES
- 1.Shah RR, Wade G. 1989. Reappraisal of the risk/benefit of nitrofurantoin: review of toxicity and efficacy. Adverse Drug React. Acute Poisoning Rev. 8:183–201. [PubMed] [Google Scholar]
- 2.Conklin JD. 1978. The pharmacokinetics of nitrofurantoin and its related bioavailability. Antibiot. Chemother. 25:233–252. [DOI] [PubMed] [Google Scholar]
- 3.Guay DR. 2001. An update on the role of nitrofurans in the management of urinary tract infections. Drugs 61:353–364. 10.2165/00003495-200161030-00004. [DOI] [PubMed] [Google Scholar]
- 4.McOsker CC, Fitzpatrick PM. 1994. Nitrofurantoin: mechanism of action and implications for resistance development in common uropathogens. J. Antimicrob. Chemother. 33(Suppl A):23–30. [DOI] [PubMed] [Google Scholar]
- 5.Whiteway J, Koziarz P, Veall J, Sandhu N, Kumar P, Hoecher B, Lambert IB. 1998. Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J. Bacteriol. 180:5529–5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCalla DR, Kaiser C, Green MH. 1978. Genetics of nitrofurazone resistance in Escherichia coli. J. Bacteriol. 133:10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandegren L, Lindqvist A, Kahlmeter G, Andersson DI. 2008. Nitrofurantoin resistance mechanism and fitness cost in Escherichia coli. J. Antimicrob. Chemother. 62:495–503. 10.1093/jac/dkn222. [DOI] [PubMed] [Google Scholar]
- 8.Rafii F, Hansen EB., Jr 1998. Isolation of nitrofurantoin-resistant mutants of nitroreductase-producing Clostridium sp. strains from the human intestinal tract. Antimicrob. Agents Chemother. 42:1121–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calva JJ, Sifuentes-Osornio J, Ceron C. 1996. Antimicrobial resistance in fecal flora: longitudinal community-based surveillance of children from urban Mexico. Antimicrob. Agents Chemother. 40:1699–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.London N, Nijsten R, van der Bogaard A, Stobberingh E. 1994. Carriage of antibiotic-resistant Escherichia coli by healthy volunteers during a 15-week period. Infection 22:187–192. 10.1007/BF01716700. [DOI] [PubMed] [Google Scholar]
- 11.Stamey TA, Condy M, Mihara G. 1977. Prophylactic efficacy of nitrofurantoin macrocrystals and trimethoprim-sulfamethoxazole in urinary infections: biologic effects on the vaginal and rectal flora. N. Engl. J. Med. 296:780–783. [DOI] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 13.Xavier BB, Vervoort J, Stewardson A, Adriaenssens N, Coenen S, Harbarth S, Goossens H, Malhotra-Kumar S. 2014. Complete genome sequences of nitrofurantoin-sensitive and -resistant Escherichia coli ST540 and ST2747 strains. Genome Announc. 2:e00239-14. 10.1128/genomeA.00239-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403. 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mortl S, Fischer M, Richter G, Tack J, Weinkauf S, Bacher A. 1996. Biosynthesis of riboflavin: lumazine synthase of Escherichia coli. J. Biol. Chem. 271:33201–33207. [DOI] [PubMed] [Google Scholar]
- 17.Kidwell MG, Lisch DR. 2000. Transposable elements and host genome evolution. Trends Ecol. Evol. 15:95–99. 10.1016/S0169-5347(99)01817-0. [DOI] [PubMed] [Google Scholar]
- 18.Martinez JL, Baquero F. 2000. Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 44:1771–1777. 10.1128/AAC.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson FJ, Mason RP, Hovsepian J, Holtzman JL. 1979. Oxygen-sensitive and -insensitive nitroreduction by Escherichia coli and rat hepatic microsomes. J. Biol. Chem. 254:4009–4014. [PubMed] [Google Scholar]
- 20.Marques de Oliveira I, Bonatto D, Pêgas Henriques JA. 2010. Nitroreductases: enzymes with environmental, biotechnological and clinical importance, p 1008–1019 In Mendez-Vilas A. (ed), Current research, technology and education topics in applied microbiology and microbial biotechnology, vol 2 Formatex Research Center, Badajoz, Spain. [Google Scholar]
- 21.Mercier C, Chalansonnet V, Orenga S, Gilbert C. 2013. Characteristics of major Escherichia coli reductases involved in aerobic nitro and azo reduction. J. Appl. Microbiol. 115:1012–1022. 10.1111/jam.12294. [DOI] [PubMed] [Google Scholar]
- 22.Ladenstein R, Schneider M, Huber R, Bartunik HD, Wilson K, Schott K, Bacher A. 1988. Heavy riboflavin synthase from Bacillus subtilis: crystal structure analysis of the icosahedral β60 capsid at 3.3 Å resolution. J. Mol. Biol. 203:1045–1070. [DOI] [PubMed] [Google Scholar]
- 23.Ritsert K, Huber R, Turk D, Ladenstein R, Schmidt-Base K, Bacher A. 1995. Studies on the lumazine synthase/riboflavin synthase complex of Bacillus subtilis: crystal structure analysis of reconstituted, icosahedral β-subunit capsids with bound substrate analogue inhibitor at 2.4 Å resolution. J. Mol. Biol. 253:151–167. 10.1006/jmbi.1995.0542. [DOI] [PubMed] [Google Scholar]
- 24.Zenno S, Koike H, Kumar AN, Jayaraman R, Tanokura M, Saigo K. 1996. Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi flavin oxidoreductase. J. Bacteriol. 178:4508–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer M, Bacher A. 2008. Biosynthesis of vitamin B2: structure and mechanism of riboflavin synthase. Arch. Biochem. Biophys. 474:252–265. 10.1016/j.abb.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Vogl C, Grill S, Schilling O, Stulke J, Mack M, Stolz J. 2007. Characterization of riboflavin (vitamin B2) transport proteins from Bacillus subtilis and Corynebacterium glutamicum. J. Bacteriol. 189:7367–7375. 10.1128/JB.00590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraaije MW, Mattevi A. 2000. Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem. Sci. 25:126–132. 10.1016/S0968-0004(99)01533-9. [DOI] [PubMed] [Google Scholar]
- 28.Lee N, Bendet I. 1967. Crystalline l-ribulokinase from Escherichia coli. J. Biol. Chem. 242:2043–2050. [PubMed] [Google Scholar]
- 29.Shimada T, Kori A, Ishihama A. 2013. Involvement of the ribose operon repressor RbsR in regulation of purine nucleotide synthesis in Escherichia coli. FEMS Microbiol. Lett. 344:159–165. 10.1111/1574-6968.12172. [DOI] [PubMed] [Google Scholar]
- 30.Andersson DI, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8:260–271. 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 31.Poulsen HO, Johansson A, Granholm S, Kahlmeter G, Sundqvist M. 2013. High genetic diversity of nitrofurantoin- or mecillinam-resistant Escherichia coli indicates low propensity for clonal spread. J. Antimicrob. Chemother. 68:1974–1977. 10.1093/jac/dkt159. [DOI] [PubMed] [Google Scholar]
- 32.Van Heirstraeten L, Coenen S, Lammens C, Hens N, Goossens H, Malhotra-Kumar S. 2012. Antimicrobial drug use and macrolide-resistant Streptococcus pyogenes, Belgium. Emerg. Infect. Dis. 18:1515–1518. 10.3201/eid1809.120049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andersson DI, Levin BR. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2:489–493. 10.1016/S1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.