Abstract
blaNDM-5 was found in Escherichia coli strain 0215 from a Chinese patient without travel history. Genomic sequencing and conjugation experiments were performed. Strain 0215 belonged to sequence type 167 (ST167) and had other resistance determinants, including blaTEM-135, blaCTX-M-14, and aac(6′)-Ib. blaNDM-5 was carried by a 47-kb self-transmissible IncX3 plasmid and was in a complex genetic context similar to that of blaNDM-1 on IncX3 plasmids. IncX3 plasmids might have emerged as a common vehicle mediating the spread of blaNDM.
TEXT
NDM (New Delhi metallo-β-lactamase) is a type of carbapenemase with the ability to hydrolyze all β-lactams except monobactams (1). NDM enzymes confer resistance to carbapenems, which have long served as reliable and potent agents against Gram-negative bacilli. Bacterial isolates producing NDM were first identified in 2008 (2) and have emerged worldwide, representing a serious challenge for clinical management and infection control. Most NDM-producing isolates belong to various species of Enterobacteriaceae or Acinetobacter.
Until now, 12 variants of NDM enzymes (NDM-1 to -12) have been discovered and assigned in the Lahey Clinic database (see http://www.lahey.org/Studies/other.asp#table 1). The NDM-5-encoding gene, blaNDM-5, was first identified in an Escherichia coli strain (EC045) from a patient in the United Kingdom who had a recent history of hospitalization in India (3). NDM-5 differs from NDM-1 in two amino acid substitutions (Val88Leu and Met154Leu) (3), which appear to confer increased resistance to carbapenems and broad-spectrum cephalosporins (3). The goal of this study was to characterize a Chinese E. coli strain carrying blaNDM-5.
TABLE 1.
E. coli of ST167 carrying blaCTX-M and/or blaNDM
| blaCTX-M | blaNDM | Location(s) | Host(s) | Source or reference |
|---|---|---|---|---|
| blaCTX-M-1 | Germany, The Netherlands | Cattle, swine, human | 27, 28 | |
| blaCTX-M-2 | Germany | Cattle | 27 | |
| blaCTX-M-3 | Germany | Swine | 27 | |
| blaCTX-M-9 | Mongolia | Wild bird | 29 | |
| blaCTX-M-14 | Spain | Human | 30, 31, MLST databasea | |
| blaCTX-M-14 | blaNDM-1 | China | Human | 19 |
| blaCTX-M-14 | blaNDM-5 | China | Human | This study |
| blaCTX-M-15 | blaNDM-7 | Franceb | Human | 20 |
| blaCTX-M-15 | Canada, Germany, Norway, Russia, Spain | Human, swine, turkey | 30, 32, 33, MLST database |
From a French patient who had recently traveled to Myanmar.
Strain 0215 was recovered from a routine screening rectal swab of a 75-year-old male patient with an acute exacerbation of chronic obstructive pulmonary disease in September 2013. The rectal swab was streaked directly onto a ChromID CARBA agar plate (bioMérieux, Lyon, France) selective for carbapenem-resistant Enterobacteriaceae. Strain 0215 was identified as E. coli by partially sequencing the 16S rRNA gene amplified with the universal primers 27F and 1492R (4). The MICs of imipenem, meropenem, amikacin, ceftazidime, and ciprofloxacin were determined using the microdilution broth method following the recommendations of the Clinical and Laboratory Standards Institute (5). Strain 0215 was resistant to imipenem (MIC, 512 μg/ml), meropenem (MIC, 256 μg/ml), ceftazidime (MIC, >256 μg/ml), and ciprofloxacin (MIC, 128 μg/ml) but was susceptible to amikacin (MIC, 16 μg/ml).
We screened the carbapenemase-encoding genes blaGES, blaKPC, blaIMP, blaNDM, blaOXA-48, and blaVIM for strain 0215 using PCR as described previously (6–9), and only blaNDM was detected. The complete coding sequence of blaNDM was amplified with an additional pair of primers (NDM-up and NDM-dw) (6), and sequencing revealed the presence of blaNDM-5. Since the discovery of blaNDM-5 in E. coli strain EC045, blaNDM-5 has also been found in a Klebsiella pneumoniae strain (MGR-K165, GenBank accession number KF220657) and three E. coli strains (GenBank accession numbers KJ150692, KF284078, and KF284079) (10) in India. Very recently, blaNDM-5 was also found in three E. coli strains in Algeria (11) and one E. coli strain in Spain (12). In addition, a recent report claimed that blaNDM-5 was also detected in another E. coli strain, KOEC3, in India (13). However, only 240 bp of the 813-bp blaNDM coding sequence are available; therefore, we were unable to confirm blaNDM-5 in this case. Nonetheless, blaNDM-5 has been found in different locations in India, including Lucknow (10) and Chennai (GenBank accession numbers KF220657 and KJ150692) and potentially in Goa (3) and Umiam (13), suggesting that blaNDM-5 might have been circulating in India. However, the patient from whom strain 0215 was recovered had no history of travel to Africa, Europe, or South Asia during his lifetime. To our knowledge, this is the first report of blaNDM-5 in China.
Genomic DNA of strain 0215 was prepared using the QIAamp DNA minikit (Qiagen, Hilden, Germany) and was subjected to paired-end whole-genomic sequencing with a 500-bp insert size and a ca. 140× coverage using the HiSeq 2500 Sequencer (Illumina, San Diego, CA) following the manufacturer's protocol at the Beijing Genomics Institute (Beijing, China). The reads were assembled to contigs using the Velvet program (14). A total of 7,354,784 clean reads and 661,930,560 clean bases were obtained from the genomic sequencing for strain 0215. The GC content was 50.66%. There is no reference genomic sequence of sequence type 167 (ST167) E. coli available, and ST167 is of the ST10 clonal complex. The genomic sequence of E. coli strain K-12 substrain MG1655 (GenBank accession number U00096), which belongs to ST10, was therefore selected as a reference for the further assembly and annotation of the genomic sequence of strain 0215 using the PAGIT and Prokka programs. PAGIT is a toolkit for ordering contigs, closing gaps, correcting sequence errors, and transferring annotation (15), and Prokka is a tool for annotating prokaryotic genomes (16). Reads were assembled to 243 contigs that were ≥100 bp in length.
Phylogenetic group (A, B1, B2, and D) typing, which was performed as described previously (17), assigned strain 0215 to group A. The sequence type (ST) of strain 0215 was assigned using the assembled genomic sequence to query the seven alleles of the multilocus sequence-typing scheme for E. coli (see http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) (18). Strain 0215 belonged to ST167. In contrast, the first E. coli strain carrying blaNDM-5 was of ST648 (3). E. coli strains of ST167 have been found carrying blaNDM-1 in China (19) and blaNDM-7 in a French patient after recent travel to Myanmar (20). In addition, ST167 E. coli strains have been found carrying a variety of extended-spectrum β-lactamase (ESBL) genes, including blaCTX-M-1, blaCTX-M-2, blaCTX-M-3, blaCTX-M-14, and blaCTX-M-15 in various countries (Table 1).
In addition to blaNDM-5, strain 0215 had multiple resistance genes, including blaTEM-135 (a non-ESBL variant of blaTEM-1), blaCTX-M-14 (an ESBL gene widely distributed in China), blaampC, aac(6′)-Ib, cmlA1 (encoding a chloramphenicol exporter), floR (encoding a florfenicol/chloramphenicol resistance protein), mph (encoding macrolide 2′-phosphotransferase I), and tet(A) (conferring resistance to tetracycline). Of note, the AmpC enzyme of strain 0215 is identical to the NCBI reference sequence WP_024176402 in amino acids. The same blaampC variant has also been found in blaCTX-M-15-carrying strain EC66, which belongs to the phylogenetic group A and the O101 type from India, but its sequence type is not available (21).
Conjugation experiments were carried out in broth using azide-resistant E. coli strain J53 as the recipient. Transconjugants were selected on plates containing 4 μg/ml meropenem plus 150 μg/ml sodium azide. The presence of blaNDM in transconjugants was confirmed using PCR, and enterobacterial repetitive intergenic consensus (ERIC)-PCR (22) was used for further distinguishing transconjugants from the donor strain. blaNDM-5 was transferred to E. coli recipient J53, although none of the other resistance genes listed above were cotransferred with blaNDM-5. Therefore, blaNDM-5 was located on a self-transmissible plasmid, designated pNDM5_0215 here, in strain 0215. However, PCR-based replicon typing (PBRT) (23) failed to assign pNDM5_0215 to a replicon type, as all replicon-typing PCRs were negative. Genomic sequencing of strain 0215 revealed a 34-kb contig, which is identical to an IncX3 plasmid (pNDM_MGR194) carrying blaNDM-5 in K. pneumoniae strain MGR-K165. Therefore, the sequence of pNDM_MGR194 (GenBank accession number KF220657) was used as reference for assembling pNDM5_0215, which was completely circularized by filling the gaps using long-range PCR (Fermentas, Burlington, Canada; Fig. 1) and Sanger sequencing using an ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA) at the Beijing Genomics Institute. Indeed, pNDM5_0215 was a 47-kb IncX3 plasmid and had only two minor differences from pNDM_MGR194, the absence of one copy of a 22-bp iteron that is involved in regulating plasmid replication and the presence of an additional copy of the insertion sequence IS5 on pNDM5_0215 (Fig. 1). Otherwise, the two plasmids were identical. IncX plasmids are narrow-host-range plasmids of Enterobacteriaceae and include at least five subtypes, IncX1 to IncX5 (24). A few IncX3 plasmids carrying ESBL and/or carbapenemase genes from various species of Enterobacteriaceae found in several countries have been completely sequenced (Table 2).
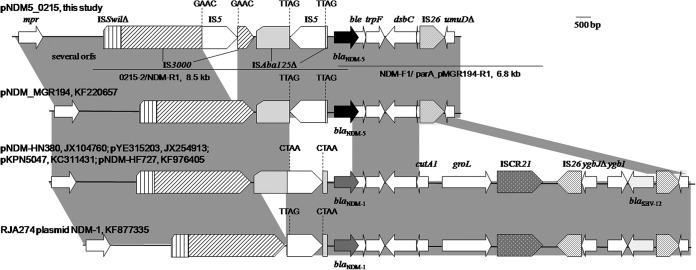
FIG 1.
Genetic contexts of blaNDM-1 and blaNDM-5 on IncX3 plasmids. Plasmid names of each structure and their GenBank accession numbers are listed. The identical regions are highlighted in gray. The 4-bp flanking sequences of each IS5 are shown. Between mpr and ISSwil, there are several ORFs of unknown function, which are indicated here by “several orfs.” Two long-range PCR products used to fill in the gap of pNDM5_0215 are shown by lines, and primer names and amplicon sizes are indicated. Primers NDM-F1 and NDM-R1 are from reference 6, while primers 0215-2 (TGGTGCTGGTTATCTGTGCT) and parA_pMGR194-R1 (CCGTTATCTGTCCGCTTTTC) were newly designed.
TABLE 2.
IncX3 plasmids with a complete sequence available in GenBank
| Plasmid | bla gene carried | Host species | Location | GenBank accession no. | Reference or source |
|---|---|---|---|---|---|
| pIncX-SHV | blaSHV-11 | K. pneumoniae | Italy | JN247852 | 34 |
| pKP13d | blaKPC-2 | K. pneumoniae | Brazil | CP003997 | 35 |
| pKPC-NY79 | blaKPC-2 | K. pneumoniae | China | JX104759 | 36 |
| pKpS90 | blaKPC-2, blaSHV-12 | K. pneumoniae | France | JX461340 | 37 |
| pNDM-HN380 | blaNDM-1, blaSHV-12 | K. pneumoniae | China | JX104760 | 38 |
| pYE315203 | blaNDM-1, blaSHV-12 | Citrobacter freundii | China | JX254913 | |
| pKPN5047 | blaNDM-1, blaSHV-12 | K. pneumoniae | China | KC311431 | |
| RJA274 plasmid NDM-1 | blaNDM-1, blaSHV-12 | Raoultella planticola | China | KF877335 | |
| pNDM-HF727 | blaNDM-1, blaSHV-12 | Enterobacter cloacae | China | KF976405 | 36 |
| pNDM_MGR194 | blaNDM-5 | K. pneumoniae | India | KF220657 | |
| pNDM5-0215 | blaNDM-5 | E. coli | China | This study |
Although blaNDM-5 has been identified in several cases, its genetic context was largely uninvestigated. Nonetheless, the available complete sequences of pNDM5_0215 and pNDM_MGR194 allowed us to reveal the genetic context of blaNDM-5. On the two IncX3 plasmids, blaNDM-5 is in the same genetic context, except there is an insertion of an additional copy of IS5 into IS3000 on pNDM5_0215, as evidenced by the presence of the characteristic 4-bp direct target repeats (Fig. 1). In such a context, blaNDM-5 is adjacent to an incomplete ISAba125, which is interrupted by the insertion of IS5 and is also truncated by the insertion of IS3000 at its left end upstream. A zinc metalloproteinase-encoding gene, mpr, lies further upstream of blaNDM-5. Several putative open reading frames (ORFs) of unknown function and a truncated ISSwil are present between mpr and IS3000. Downstream of blaNDM-5, there are ble (mediating bleomycin resistance), trpF (encoding a phosphoribosylanthranilate isomerase), dsbC (encoding an oxidoreductase), a remnant of ctuA1 (encoding an ion-tolerant protein) that is truncated by the insertion of IS26, and a truncated umuD gene (encoding a mutagenesis protein) (Fig. 1).
Such a context of blaNDM-5 on IncX3 plasmids is highly similar to those of blaNDM-1 on several other IncX3 plasmids (Fig. 1). All contexts of blaNDM-1 and blaNDM-5 on IncX3 plasmids are bounded by mpr at one end and by ΔumuD at the other. The highly similar contexts suggest that blaNDM-5 might have evolved from blaNDM-1 via point mutations in the mpr-ΔumuD region of an IncX3 plasmid. Of note, an IS26-bounded region containing blaSHV-12, ygbI (a putative dehydrogenase gene), and a truncated ygbJ (a putative DEOR-type transcriptional regulator gene) is present downstream of blaNDM-1 on IncX3 plasmids (Fig. 1). It is therefore reasonable to propose that the absence of groL, ISCR21, and the region containing blaSHV-12 from the context of blaNDM-5 might be explained by the insertion of an addition of IS26 into cutA1 and the following homologous recombination between two copies of IS26, which resulted in the deletion. The IncX3 plasmids carrying blaNDM-1 or blaNDM-5 also had nearly identical backbones, further supporting the idea that blaNDM-5 might have emerged on IncX3 plasmids. A few blaNDM-carrying IncX3 plasmids have been found in China but at various locations, suggesting that a common IncX3 plasmid carrying blaNDM is circulating in various Enterobacteriaceae species in China. However, IncX3 plasmids carrying blaNDM are not restricted to China but also have been identified in Germany (blaNDM-7, in an E. coli isolate from a Yemeni patient) (25), India (blaNDM-5, pNDM_MGR194), and the United Arab Emirates (blaNDM-1) (26), suggesting IncX3 plasmids might have emerged as a common platform mediating the spread of blaNDM. In addition, two nearly identical blaNDM-5-carrying plasmids, pNDM5_0215 and pNDM_MGR194, were found in China and India separately. This might suggest unrecognized international transfers of the same IncX3 plasmid or, less likely, the independent emergence of blaNDM-5 on the same plasmid in each country. The association of IncX3 plasmids and blaNDM variants and the epidemiology of IncX3 plasmids in Enterobacteriaceae species warrant more studies.
Nucleotide sequence accession number.
Reads of the 0215 genomic sequence have been deposited into the NCBI database under BioProject identification no. PRJNA244631 and submission identification no. SUB492451.
ACKNOWLEDGMENT
This work was partially supported by a grant from the National Natural Science Foundation of China (project no. 81222025).
Footnotes
Published ahead of print 22 September 2014
REFERENCES
- 1.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602. 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054. 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hornsey M, Phee L, Wareham DW. 2011. A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 55:5952–5954. 10.1128/AAC.05108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrant E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, NY. [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; twenty-third informational supplement. M100-S23 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 6.Zong Z, Zhang X. 2013. blaNDM-1-carrying Acinetobacter johnsonii detected in hospital sewage. J. Antimicrob. Chemother. 68:1007–1010. 10.1093/jac/dks505. [DOI] [PubMed] [Google Scholar]
- 7.Mendes RE, Kiyota KA, Monteiro J, Castanheira M, Andrade SS, Gales AC, Pignatari AC, Tufik S. 2007. Rapid detection and identification of metallo-β-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. J. Clin. Microbiol. 45:544–547. 10.1128/JCM.01728-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:622–632. 10.1128/AAC.44.3.622-632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford PA, Bratu S, Urban C, Visalli M, Mariano N, Landman D, Rahal JJ, Brooks S, Cebular S, Quale J. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin. Infect. Dis. 39:55–60. 10.1086/421495. [DOI] [PubMed] [Google Scholar]
- 10.Rahman M, Shukla SK, Prasad KN, Ovejero CM, Pati BK, Tripathi A, Singh A, Srivastava AK, Gonzalez-Zorn B. 2014. Prevalence and molecular characterisation of New Delhi metallo-β-lactamases NDM-1, NDM-5, NDM-6 and NDM-7 in multidrug-resistant Enterobacteriaceae from India. Int. J. Antimicrob. Agents 44:30–37. 10.1016/j.ijantimicag.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Sassi A, Loucif L, Gupta SK, Dekhil M, Chettibi H, Rolain JM. 2014. NDM-5 carbapenemase-encoding gene in multidrug-resistant clinical isolates of Escherichia coli from Algeria. Antimicrob. Agents Chemother. 58:5606–5608. 10.1128/AAC.02818-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sole Guiu M, Pitart C, Roman A, Moreno A, Roca I, Vila J, Marco F. 2014. Molecular characterisation of NDM-5 in an Escherichia coli isolate from a non-traveller patient in Spain, poster P1097 24th Eur. Congr. Clin. Microbiol. Infect. Dis., Barcelona, Spain, 10 to 13 May 2014. [Google Scholar]
- 13.Ghatak S, Singha A, Sen A, Guha C, Ahuja A, Bhattacharjee U, Das S, Pradhan NR, Puro K, Jana C, Dey TK, Prashantkumar KL, Das A, Shakuntala I, Biswas U, Jana PS. 2013. Detection of New Delhi metallo-β-lactamase and extended-spectrum β-lactamase genes in Escherichia coli isolated from mastitic milk samples. Transbound. Emerg. Dis. 60:385–389. 10.1111/tbed.12119. [DOI] [PubMed] [Google Scholar]
- 14.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558. 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151. 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Lou D, Xu Y, Shang Y, Li D, Huang X, Li Y, Hu L, Wang L, Yu F. 2013. First identification of coexistence of blaNDM-1 and blaCMY-42 among Escherichia coli ST167 clinical isolates. BMC Microbiol. 13:282. 10.1186/1471-2180-13-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuzon G, Bonnin RA, Nordmann P. 2013. First identification of novel NDM carbapenemase, NDM-7, in Escherichia coli in France. PLoS One 8:e61322. 10.1371/journal.pone.0061322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swain MT, Tsai IJ, Assefa SA, Newbold C, Berriman M, Otto TD. 2012. A post-assembly genome-improvement toolkit (PAGIT) to obtain annotated genomes from contigs. Nat. Protoc. 7:1260–1284. 10.1038/nprot.2012.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 21.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Canica MM, Park YJ, Lavigne JP, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61:273–281. 10.1093/jac/dkm464. [DOI] [PubMed] [Google Scholar]
- 22.Versalovic J, Koeuth T, Lupski JR. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823–6831. 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228. 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 24.Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. 2012. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68:43–50. 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Gottig S, Hamprecht AG, Christ S, Kempf VA, Wichelhaus TA. 2013. Detection of NDM-7 in Germany, a new variant of the New Delhi metallo-β-lactamase with increased carbapenemase activity. J. Antimicrob. Chemother. 68:1737–1740. 10.1093/jac/dkt088. [DOI] [PubMed] [Google Scholar]
- 26.Sonnevend A, Al Baloushi A, Ghazawi A, Hashmey R, Girgis S, Hamadeh MB, Al Haj M, Pal T. 2013. Emergence and spread of NDM-1 producer Enterobacteriaceae with contribution of IncX3 plasmids in the United Arab Emirates. J. Med. Microbiol. 62:1044–1050. 10.1099/jmm.0.059014-0. [DOI] [PubMed] [Google Scholar]
- 27.Schink AK, Kadlec K, Kaspar H, Mankertz J, Schwarz S. 2013. Analysis of extended-spectrum-β-lactamase-producing Escherichia coli isolates collected in the GERM-Vet monitoring programme. J. Antimicrob. Chemother. 68:1741–1749. 10.1093/jac/dkt123. [DOI] [PubMed] [Google Scholar]
- 28.Leverstein-van Hall MA, Dierikx CM, Cohen Stuart J, Voets GM, van den Munckhof MP, van Essen-Zandbergen A, Platteel T, Fluit AC, van de Sande-Bruinsma N, Scharinga J, Bonten MJ, Mevius DJ, National ESBL Surveillance Group 2011. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 17:873–880. 10.1111/j.1469-0691.2011.03497.x. [DOI] [PubMed] [Google Scholar]
- 29.Guenther S, Aschenbrenner K, Stamm I, Bethe A, Semmler T, Stubbe A, Stubbe M, Batsajkhan N, Glupczynski Y, Wieler LH, Ewers C. 2012. Comparable high rates of extended-spectrum-β-lactamase-producing Escherichia coli in birds of prey from Germany and Mongolia. PLoS One 7:e53039. 10.1371/journal.pone.0053039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oteo J, Diestra K, Juan C, Bautista V, Novais A, Perez-Vazquez M, Moya B, Miro E, Coque TM, Oliver A, Canton R, Navarro F, Campos J. 2009. Extended-spectrum β-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int. J. Antimicrob. Agents 34:173–176. 10.1016/j.ijantimicag.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 31.Valverde A, Canton R, Garcillan-Barcia MP, Novais A, Galan JC, Alvarado A, de la Cruz F, Baquero F, Coque TM. 2009. Spread of blaCTX-M-14 is driven mainly by IncK plasmids disseminated among Escherichia coli phylogroups A, B1, and D in Spain. Antimicrob. Agents Chemother. 53:5204–5212. 10.1128/AAC.01706-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer J, Rodriguez I, Baumann B, Guiral E, Beutin L, Schroeter A, Kaesbohrer A, Pfeifer Y, Helmuth R, Guerra B. 28 July 2014. blaCTX-M-15-carrying Escherichia coli and Salmonella isolates from livestock and food in Germany. J. Antimicrob. Chemother. 10.1093/jac/dku270. [DOI] [PubMed] [Google Scholar]
- 33.Naseer U, Haldorsen B, Tofteland S, Hegstad K, Scheutz F, Simonsen GS, Sundsfjord A, Norwegian ESBL Study Group 2009. Molecular characterization of CTX-M-15-producing clinical isolates of Escherichia coli reveals the spread of multidrug-resistant ST131 (O25:H4) and ST964 (O102:H6) strains in Norway. APMIS 117:526–536. 10.1111/j.1600-0463.2009.02465.x. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Fernandez A, Villa L, Carta C, Venditti C, Giordano A, Venditti M, Mancini C, Carattoli A. 2012. Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob. Agents Chemother. 56:2143–2145. 10.1128/AAC.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramos PI, Picao RC, Almeida LG, Lima NC, Girardello R, Vivan AC, Xavier DE, Barcellos FG, Pelisson M, Vespero EC, Medigue C, Vasconcelos AT, Gales AC, Nicolas MF. 2014. Comparative analysis of the complete genome of KPC-2-producing Klebsiella pneumoniae Kp13 reveals remarkable genome plasticity and a wide repertoire of virulence and resistance mechanisms. BMC Genomics 15:54. 10.1186/1471-2164-15-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho PL, Cheung YY, Lo WU, Li Z, Chow KH, Lin CH, Chan JF, Cheng VC. 2013. Molecular characterization of an atypical IncX3 plasmid pKPC-NY79 carrying blaKPC-2 in a Klebsiella pneumoniae. Curr. Microbiol. 67:493–498. 10.1007/s00284-013-0398-2. [DOI] [PubMed] [Google Scholar]
- 37.Kassis-Chikhani N, Frangeul L, Drieux L, Sengelin C, Jarlier V, Brisse S, Arlet G, Decre D. 2013. Complete nucleotide sequence of the first KPC-2- and SHV-12-encoding IncX plasmid, pKpS90, from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 57:618–620. 10.1128/AAC.01712-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho P, Li Z, Lo W, Cheung Y, Lin C, Sham P. 2012. Identification and characterization of a novel incompatibility group X3 plasmid carrying blaNDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg. Microb. Infect. 1:e39. 10.1038/emi.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]