Abstract
Caspases, aspartate-specific cysteine proteases, have fate-determining roles in many cellular processes including apoptosis, differentiation, neuronal remodeling, and inflammation (for review, see Yuan & Kroemer, 2010). There are a dozen caspases in humans alone, yet their individual contributions toward these phenotypes are not well understood. Thus, there has been considerable interest in activating individual caspases or using their activity to drive these processes in cells and animals. We envision that such experimental control of caspase activity can not only afford novel insights into fundamental biological problems but may also enable new models for disease and suggest possible routes to therapeutic intervention. In particular, localized, genetic, and small-molecule-controlled caspase activation has the potential to target the desired cell type in a tissue. Suppression of caspase activation is one of the hallmarks of cancer and thus there has been significant enthusiasm for generating selective small-molecule activators that could bypass upstream mutational events that prevent apoptosis. Here, we provide a practical guide that investigators have devised, using genetics or small molecules, to activate specific caspases in cells or animals. Additionally, we show genetically controlled activation of an executioner caspase to target the function of a defined group of neurons in the adult mammalian brain.
1. USE OF CHEMICAL-INDUCED DIMERIZERS TO ACTIVATE CASPASES
1.1. Controlling protein–protein interactions leads to selective activation of caspases
The elucidation of cell signaling has been empowered by chemical genetic approaches. In particular, protein-fragment complementation assays and chemical-induced dimerization (CID) have been very powerful approaches (Fig. 8.1A) (Fegan, White, Carlson, & Wagner, 2010; Remy & Michnick, 2007; Spencer, Wandless, Schreiber, & Crabtree, 1993). While the ability to control transcription-based circuits has been known for decades, only recently have signaling biologists learned to harness and engineer orthogonal circuits to selectively activate proteases. Proteolysis is an irreversible posttranslational-modification that directs the fate of cells in apoptosis, protein degradation, blood coagulation, and many other processes. Here, we discuss practical approaches that we and others have recently employed to selectively activate caspases using CID strategies.
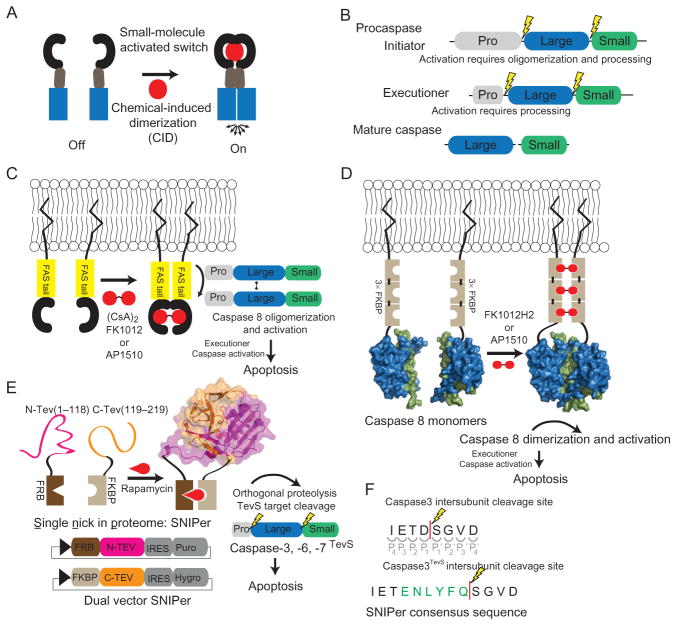
Figure 8.1.
Schematic overview of chemical genetic strategies for selective caspase activation. (A) Chemical-induced dimerization (CID) utilizes dimerization domains, dependent on cell-permeable small molecules, to control the proximity of signal transduction domains (blue). The chemical genetic strategy of CID creates an orthogonal circuit in the cell, thereby enabling the control of the ON/OFF state of signal transduction. (B) Procaspases are composed of three domains; a prodomain, a large subunit, and a small subunit. Initiators have a long prodomain and require both oligomerization and processing for activation. The executioners are predimerized and they have a short prodomain and proteolysis at the large–small domain junction is sufficient for activation. (C) Utilizing a CID-based approach, FAS tails are dimerized by the addition of the symmetric small molecule, AP1510 or FK1012, thereby reconstituting the caspase-8 (PDB ID 1QTN) activation scaffold and ultimately resulting in apoptosis. (D) Direct caspase-8 activation via CID. (E) The engineered split-tobacco etch viral (TEV) variant, single nick in proteome (SNIPer), used for inducible and selective cleavage of the executioner caspase isoforms (TEV structure PDB ID 1LVM). Each SNIPer half is expressed from a single plasmid that coexpresses an IRES-driven drug-resistance marker for stable cell line engineering. The addition of rapamycin causes the heterodimerization of FKBP and FRB, thereby rescuing TEV protease activity. (F) The TEV protease consensus sequence (green) is inserted into the caspase-processing site to generate a TevS allele, susceptible to SNIPer-mediated proteolysis.
There are two classes of apoptotic caspases: initiators (caspase-8, -9, -10) which are activated by oligomerization, and the primary executioners (caspase-3 and -7) that are activated by proteolysis mediated by upstream proteases (initiator caspases or granzyme B) (Fig. 8.1B). The purpose of this first section is to (1) briefly review previous approaches and research highlights that employed chemical genetic strategies used to activate caspases, primarily caspase-8, (2) summarize the progress that our laboratories have made in selectively turning on apoptosis via orthogonal executioner caspase activation, and (3) provide a detailed method for selective activation of executioner caspases employing this chemical genetic strategy.
1.2. Oligomerization strategies for selective caspase activation
1.2.1 Selective caspase-8 oligomerization and activation
The first target for CID activation of caspases was caspase-8, a classic initiator caspase involved in the extrinsic apoptosis pathway (Muzio, Stockwell, Stennicke, Salvesen, & Dixit, 1998; Yang, Chang, & Baltimore, 1998). CID activation of caspase-8 facilitated elegant experimentation of the induced-proximity hypothesis (Salvesen & Dixit, 1999), which is that the monomeric proenzyme is activated through dimerization. Previous studies have shown that heterologous expression of the inactive zymogens retained modest catalytic activity prior to their proteolytic processing. Initial tests of the caspase-8 cellular activation mechanism were described in two simultaneous papers that demonstrated the role of receptor oligomerization in caspase-8 activation (Belshaw, Spencer, Crabtree, & Schreiber, 1996; Spencer et al., 1996). This work showed that it was possible to activate procaspase-8 through CID of the intracellular FAS receptors using either a homodimerization analog of FK506, FK1012, a cyclosporin derivative (CsA)2 (Fig. 8.1C). A year later, the symmetric small molecule, AP1510, was used to oligomerize FAS tails (Amara et al., 1997). The CID approach was then applied directly to caspase-8. Competing caspase-8 chimeras, containing trimeric homodimerization domains demonstrated that oligomerization was sufficient for robust caspase-8 activation (Muzio et al., 1998; Yang et al., 1998) (Fig. 8.1D). Other investigators showed that conditional dimerization of known monomeric initiator caspases-8, -9, or 10 could result in activation yet the predimerized executioner caspase-3 was not activated by oligomerization (Chen, Orozco, Spencer, & Wang, 2002). CIDs have now been applied to several caspases and together constitute a collection of artificial death switches (Steller, 1998).
Further work showed that during activation, procaspase-8 both dimerizes and undergoes proteolysis (Chang, Xing, Capacio, Peter, & Yang, 2003; Hughes et al., 2009; Kang et al., 2008; Murphy, Creagh, & Martin, 2004; Pop, Fitzgerald, Green, & Salvesen, 2007; Sohn, Schulze-Osthoff, & Jänicke, 2005). Utilizing the unique specificity of tobacco etch viral (TEV) protease in mammalian cells and the previously described CID approaches for oligomerization, both methods of activation have been beautifully tested simultaneously (Oberst et al., 2010). Oberst et al. showed that dimerization alone in the absence of proteolysis of the intersubunit linker or prodomain failed to lead to activation, as detected by IETDase activity. Similarly, selective cleavage by TEV protease, in mutated linker regions with the TEV consensus sequences failed to activate the caspase-8. Only the combination of induced dimerization and proteolytic processing resulted in full activation of caspase-8. These experiments highlight the utility of engineered inducible and orthogonal activation strategies for the caspases, thereby permitting a more complete understanding of their function and regulation.
1.2.2 In vivo strategies for selective caspase activation
Inducible caspase activation has been utilized in several in vivo strategies that include conditional induction of apoptosis during development in C. elegans, mouse adipocytes, and a caspase-9-based artificial death switch for T-cell therapies. In order to reconstitute Ced-3/caspase-3 activity in C. elegans during development, individual subunits of caspase-3 chimeras were fused to antiparallel leucine-zippers and expressed by separate promoters, and selective apoptosis was observed only when promoters overlapped in cells (Chelur & Chalfie, 2007). Pajvani and colleagues demonstrated the power in reversible CID-based approaches in mice, upon removal of the dimerizer, thereby leading to the inactivation of the caspase-8 death switch in adipocytes; mice regained weight lost during treatment (Pajvani et al., 2005). Cell-based treatments for cancer and regenerative-based medicine hold promise, yet in the event of an adverse effect, elimination of the engineered cells would be critical. Toward this end, several artificial death switches, utilizing similar CID strategies discussed above, have demonstrated their usefulness in selectively killing engineered T-cells, demonstrated in mice and later in human patients (Di Stasi et al., 2011; Straathof et al., 2005).
1.3. Design of orthogonal and selective proteolysis for individual caspase activation
1.3.1 Inducible and selective proteolysis
We have recently shown one can selectively activate the executioner caspases through an orthogonal and selective CID approach of an initiating protease. This utilized a reengineered version of the split-TEV protease (Wehr et al., 2006), we call the SNIPer (single nick in proteome). The SNIPer was optimized by truncating the C-terminus of TEV and swapping the order of the heterodimerization domains of FKBP/FRB (Gray, Mahrus, & Wells, 2010). This new design greatly reduced background of TEV activity caused by small amounts of constitutive TEV dimer formation. The SNIPer leads to the rescue of the TEV activity by the addition of the cell-permeable small-molecule, rapamycin, which was shown using a genetically encoded TEV FRET reporter (Gray et al., 2010). Selective and exclusive proteolysis of each target caspase by TEV is achieved by inserting the TEV consensus cleavage motif into the endogenous caspase-processing sites (Fig. 8.1E). Once each caspase is activated, it is able to process endogenous targets, including wild-type caspases, but cannot activate the TEV-sensitive caspase alleles (caspaseTevS) since they are missing the caspase consensus site.
TEV cleavage sequences were introduced into the two major processing sites in caspases-3, -6, and -7. Selective cleavage of the site between the prodomain and the large subunit did not lead to activation, but all three caspases were activated upon SNIPer-mediated proteolysis between the large and small subunit. When these alleles were stably transfected into human 293 cells, only activation of caspases-3 and -7 lead to apoptosis, whereas caspase-6 did not, therefore suggesting it is not a typical executioner. The executioner caspases were shown to be ubiquitinated and degraded by the ubiquitin proteasome system after activation, and proteasome inhibitors greatly accelerate SNIPer-mediated caspaseTevS-driven apoptosis. Interestingly, caspase-6TevS activation by SNIPer is lethal in the presence of proteasome inhibitors.
1.3.2 Generating SNIPer substrates: TEVs alleles
In order to generate SNIPer-cleavable substrates, we employ an insertion strategy using standard molecular biology techniques. Briefly, we replace the P1 residue of the endogenous cleavage site with the six residues required for TEV recognition and cleavage (ENLYFQ↓). This insertion yields an orthogonal substrate (i.e., no longer cleavable by caspases) because the substrate lacks the required aspartic residue followed by a small amino acid (G/S/A). We also verify the P1′ residue as TEV prefers G/S at P1′ and depending on the endogenous sequence, this may also need to be mutated. A variety of PCR techniques can be used to insert the six residues required for TEV cleavage, such as standard overlap extension PCR or kunkel mutagenesis for simultaneous insertions (Tonikian, Zhang, Boone, & Sidhu, 2007). We always test our constructs using transient transfections prior to making stable cell lines.
1.4. Cell engineering
Direct caspase dimerization or orthogonal upstream protease activation both require substantial cellular engineering. The requirements for each strategy vary and each has its own set of advantages and disadvantages. Here, we discuss in detail our approach to cellular engineering stable cell lines that express the SNIPer and subsequent screening for caspaseTevS-sensitive proteolysis. It should be noted that since the SNIPer is a highly selective and orthogonal mechanism capable of inducing caspase processing, the target substrate does not need to be a caspase but the processing site needs to be accessible to TEV protease. Our first approach consisted of engineering cells to constitutively express the SNIPer using traditional MMLV retrovirus transduction (Ory, Neugeboren, & Mulligan, 1996). However, we recently have had better success using a lentiviral-based approach that enables for a wider range of engineered target cell lines (Cooray, Howe, & Thrasher, 2012).
1.4.1 Protocol for SNIPer stable cell line engineering (inducible and selective proteolysis)
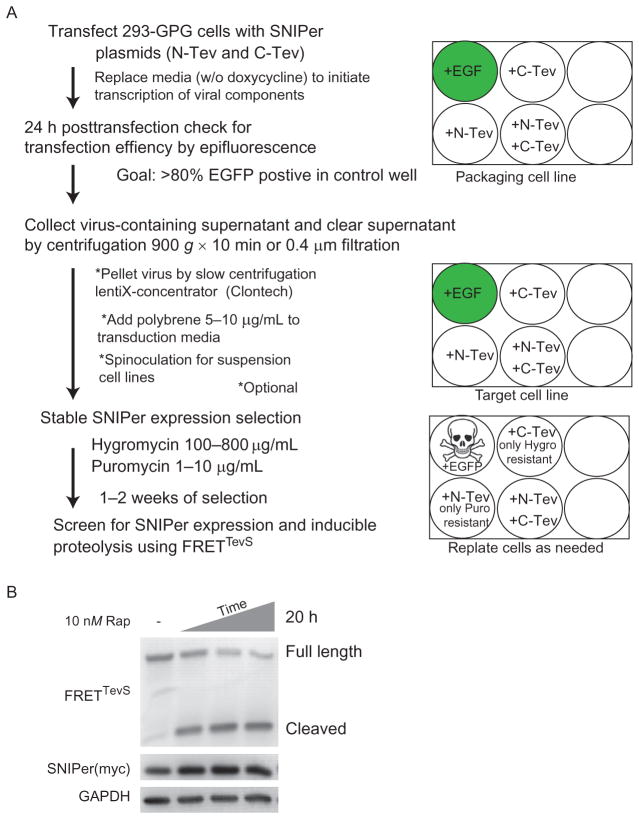
Our strategy for stable expression of both halves of SNIPer has been described previously (Gray et al., 2010). The purpose of this section is to highlight some specific details, technical challenges, and potential solutions that can be anticipated during cellular engineering with the SNIPer (Fig. 8.2).
Figure 8.2.
Cell engineering overview. (A) Summary of the protocol used for virus production and stable cell line engineering of SNIPer expressing cells. (B) A representative Western immunoblot of SNIPer engineered cells, transfected with FRETTevS reporter, shows selective proteolysis of the reporter over time.
Culture HEK293/GPG cells in DMEM supplemented with 10% FBS, nonessential amino acids, sodium pyruvate, L-glutamine, penicillin–streptomycin, 300 μg/mL G418, 2 μg/mL puromycin, and 1 μg/mL doxycycline.
A modified protocol for lipofectamine2000 (Life Technologies) is used to transfect GPG cells. We find robust transfection efficiency using only ¼ of the recommended amount of lipid mixture and that the decreased amount also minimizes cellular toxicity. We co-transfect both N- and C-terminal SNIPer plasmids together to increase successful generation of double-positive clones. The media is replaced after 5 h with fresh DMEM without doxycycline and antibiotics. Transfection efficiency is visually inspected at 24 h posttransfection by examining a control well that has been transfected in parallel with a fluorescent protein (EGFP or mCherry). We generally observe >80% transfection efficiency.
Viral-containing supernatant is collected at 48 and 72 h posttransfection.
The viral supernatant is clarified of cellular debris by either centrifugation at 900 × g for 10 min or passage through a SFCA 0.4 μm filter. Optional: virus concentration by Clontech Lenti-X concentrator.
Cleared supernatant is added to previously plated adherent target cell lines (at 60–70% confluency) and incubated overnight. Polybrene increases effective viral titers by neutralizing the charge on the cell surface. Appropriate concentrations of polybrene should be screened on each new cell types. Typically, we use 8 μg/mL for HEK293 and HeLa cells. Standard spinoculation protocols may be employed when targeting suspension cell lines.
We start selection 24 h post target cell transduction. At this point, puromycin and hygromycin are simultaneously added to select for double-positive cells, those that have integrated both halves of the SNIPer. Each half of the SNIPer also coexpresses via internal ribosomal entrysite (IRES)-dependent translation of anantibiotic resistance marker (Fig. 8.1E). Duals election usually takes ~14 days and cells are replated as necessary.
-
Tips on successful selections
Adjusting the growing density of the cells aids in the successful recovery of cells difficult to transduce. Drug selection is also very dependent upon cell density and rate of cell division. We always use a nontransduced cell line treated with selection antibiotics to ensure that all nontransduced cells are eliminated.
In most cases, heterogeneous pools of SNIPer positive cells are sufficient for further study and transfection with caspaseTevS isoforms. The SNIPer positive cell line can be used as a parental cell line for further transduction with caspaseTevS isoforms.
Typical ranges for selections when using puromycin and hygromycin are 1–10 μg/mL and 100–800 μg/mL, respectively. The effective concentration is cell-type specific. To ensure proper selection, we perform serial selections, first with puromycin since it is fast acting and then with the slower acting hygromycin. When working with a new cell line, we utilize fluorescent proteins with puromycin and hygromycin resistance to get immediate and observable feedback on transduction efficiency and progression of selection.
1.4.2 Assessing SNIPer-mediated proteolysis and rapamycin side effects
In addition to binding the SNIPer, rapamycin also binds to endogenous FK506-binding protein (FKBP) forming a complex that binds the FRB domain of mTOR and results in mTOR inhibition. We, and others, have shown that this problem is reduced by minimizing the dose of rapamycin (typically 5–10 nM needed to activate the SNIPer) and in combination the over-expression of the FKBP and FRB from the SNIPer, acts as a buffer to compete with endogenous FK506 binding. Nonetheless, we recommend measuring the level of mTOR inhibition by monitoring the phosphorylation state of the mTOR substrate S6 kinase via Western immunoblotting. We typically monitor this using antibodies for p70 S6 kinase and phoso-p70 S6 kinase (Cell Signaling Technology, #9202 and #9205). One should expect levels of mTOR inhibition in the presence of SNIPer to vary depending on the particular cell line used and the SNIPer expression level.
As a first screen of SNIPer function on a TevS target substrate, it is convenient to test it in transiently transfected cells. We typically use a molar ratio of 6:1 SNIPer halves to substrateTevS. This ratio yields the most consistent results for substrate cleavage. Once substrate cleavage is observed to be rapamycin dependent, then one should move to stably transfected cells to ensure more homogeneity in the cell population.
Western immunoblotting is our standard technique for tracking cleavage of engineered TEV cleavage site containing-substrates and expression of the SNIPer (each half is N-terminally myc tagged). It should be noted that FRB fusions are regularly observed to be less stable than FKBP fusions (Edwards & Wandless, 2007). The kinetics of cleavage by the SNIPer varies depending on the substrate being studied and the readout. We initially test for substrate cleavage following 8 h of rapamycin treatment, but we observe rapamycin-dependent cleavage in as little as 1 h after its addition with certain substrates.
1.4.3 Considerations when employing SNIPer-mediated proteolysis
When designing a SNIPer-mediated research project, it is important to be aware of some of its limitations. As is common with any protein-engineering strategy that introduces conditional control of an enzyme, background activity, for example, TEV protease activity in the absence of rapamycin, presents a challenge and needs to be thoroughly considered when looking at the overall signaling pathway. An additional consideration is the partial loss of catalytic activity of engineered variants. It has been shown that split-TEV constructs suffer from both constitutive background and low maximal activity level (Wehr et al., 2006; Williams, Puhl, & Ikeda, 2009). The SNIPer was designed to minimize background proteolysis as this was essential for selective caspaseTevS activation. For other substrates, where complete cleavage of the substrate is necessary, there are additional challenges with the SNIPer. First, the endogenous substrate will compete with the function of the engineered TevS allele, thereby masking the functional consequence of substrateTevS cleavage. Second, the SNIPer has only about ~30% the native activity of intact full-length TEV. In some cases, the transcriptional expression rate of the full-length substrate may exceed the catalytic rate of the SNIPer. If complete cleavage of the substrate is required, then other approaches may need to be considered.
1.4.4 Next-generation SNIPer-based stable cell line creation (lentiviral-based engineering and single-vector versions)
The adaptation of the SNIPer to a lentiviral-based backbone enables the creation of a wider range of cell lines with higher titers, including nondividing cells and cell lines more resistant to traditional retroviral delivery of transgenes. A single-vector version with bicistronic expression of the SNIPer has been constructed and varying iterations are currently being tested for robust and reliable inducible proteolysis. We will make these available upon request once validation is completed (C. W. Morgan & J. A. Wells unpublished).
1.5. Monitoring cell death phenotypes
Following the successful observation of rapamycin-induced cleavage of the targeted protein in cells using Western immunoblotting, the phenotype of this cleavage event can be characterized. This volume offers many detailed protocols, and in addition, Galluzzi et al. reviewed the latest techniques for monitoring cell death (Galluzzi et al., 2009). Assay selection should be matched with the specific cell line and available equipment for characterization. Often, the most convenient method for apoptosis is to measure cell viability following activation by standard methods such as Cell Titer Glo (Promega), MTT assay, or propidium iodide staining. Depending on the cell type, concentration of rapamycin, expression levels of the split-TEV halves, caspaseTevS allele, the time to death, and homogeneity of the cells will need to be screened. Generally, with caspase-3TevS we initially observe a decrease in viability starting at 3 h following activation. Once an initial dose response and corresponding time course have been completed, more detailed characterization of the engineered cell line is suggested using TUNEL and Annexin V staining, DNA laddering, and caspase-3 and PARP cleavage. Together, these phenotypic observations constitute hallmarks of apoptosis.
1.6. Conclusions and future questions
Harnessing CID strategies has expanded our ability to test different activation modes for apoptotic caspases, oligomerization, and processing. Previous research on selective activation on caspases has given us crucial insight into the process that cells use to dismantle themselves during apoptosis. The chemical genetic tools outlined above give researchers an unprecedented tool to selectively induce apoptosis in a controlled circuit that conveniently feeds into the endogenous apoptotic pathway. Further engineering for inducible and selective activation needs to be undertaken for the inflammatory caspases-1, -4, -5, -11, and the remaining apoptotic caspase-2. The ability to specifically control caspase signal transduction through selective activation could play important roles in future cell-based therapies in the form of death switches. The conditional control of caspase activation will certainly be important for basic science research, as we uncover more details about the role of proteolysis in controlling cell fate decisions and the cellular mechanisms that antagonizes caspase activation. The chemical genetic strategies outlined above enable researchers highly selective control to turn on caspase-mediated signaling pathways.
2. USE OF Cre-LoxP AND A SELF-ACTIVATING CASPASE-3TevS FOR CONDITIONAL APOPTOSIS OF NEURONS
2.1. Introduction
Unlike the cells in most other tissues, neurons that are born during development are primarily the same neurons that will make up the nervous system for the remainder of an organism’s life. Neurons are postmitotic cells and do not readily regenerate, meaning that any lesions incurred during an organism’s life will be permanent. Moreover, neurons are remarkably heterogeneous, anatomically, molecularly, and functionally. In order to meaningfully study the function of such diverse neuronal cell types in anatomically distinct regions, we need a means of performing targeted lesions and other manipulations to assess their function. We have utilized the caspase-3TevS to specifically ablate progesterone receptor (PR)-expressing neurons in the ventrolateral region of the ventromedial hypothalamus (VMHvl) in mice, and showed that this region is critical for sexual behavior in both sexes and aggression in males.
2.2. Viral-mediated neuronal ablation
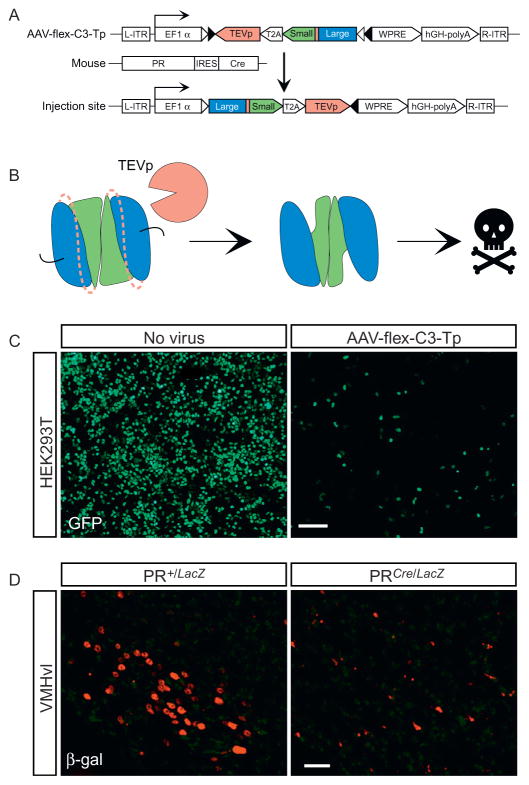
We have encoded the caspase-3TevS described in Section 1, and a full-length TEV protease, in a single adeno-associated virus (referred to as AAV-flex-taCasp3-TEVp) such that expression of both transgenes and enzymatic activation of caspase-3TevS is dependent on Cre recombinase. In this review, we refer to this virus as AAV-flex-C3-Tp. Cre-loxP is a bipartite genetic system that allows for reproducible, inducible targeting of any molecularly defined cell type. We injected this virus into the VMHvl of PR-IRES-Cre mice and were able to ablate, on average, ≥ 95% of PR neurons while leaving neighboring Cre-nonexpressing cells intact (Fig. 8.3) (Yang et al., 2013). This system is highly modular and applicable not just to PR neurons, but, in principle, any molecularly distinct neuronal population or other Cre-expressing cell type in the body. Indeed, we have successfully used this virus to ablate other distinct neuronal populations engineered to express Cre recombinase in vivo (E. K. Unger & N. M. Shah unpublished observations).
Figure 8.3.
Utilizing AAV-flex-C3-Tp. (A) Strategy to ablate PR-expressing cells in vivo. (B) Within Cre+ cells, the TEV protease will cleave its consensus recognition sequence (dashed lines) between the large and small subunits of the caspase-3 dimer, activating it and leading to apoptosis. (C) The AAV-flex-C3-Tp encoding plasmid was transfected into HEK293T cells expressing the fusion protein Cre-GFP. Cell death was evaluated after 1 week. (D) AAV-flex-C3-Tp was stereotaxically targeted to the ventrolateral region of the VMH (VMHvl) of adult PR+/LacZ and PRCre/LacZ mice which harbor the transgenes nuclear LacZ (PRLacZ allele) or Cre recombinase (PRCre allele) inserted into the PR locus by homologous recombination. Cell death was evaluated after 4 weeks. Scale bars represent 100 μm (C) and 25 μm (D). (C) and (D) are reproduced from Yang et al. (2013).
2.2.1 Self-activating caspase-3TevS transgene
Caspase-3 was modified to encode the TEV protease consensus recognition sequence, caspase-3TevS, as described above. Our virus coexpresses caspase-3TevS and TEV protease linked by a T2A sequence. T2A is a short, self-cleaving peptide that allows multiple proteins to be expressed from the same transgene (Provost, Rhee, & Leach, 2007). T2A has been shown to be more efficient than an IRES for the expression of multiple products and is effective in neurons (Tang et al., 2009). It is also smaller and thus easier to package within the AAV genome.
2.2.2 Cre-loxP
Our strategy uses the bipartite Cre-loxP system wherein Cre recombinase recognizes and recombines two short palindromic loxP sites eliminating the intervening sequence (Orban, Chui, & Marth, 1992; Sauer, 1987; Sauer & Henderson, 1988). In our strategy, Cre has been knocked-in to the PR locus to generate the PR-IRES-Cre mouse strain, such that expression of Cre mirrors the endogenous expression of PR (Yang et al., 2013). The virally encoded caspase-3TevS and TEV protease are flanked by loxP sites such that in PR-expressing cells, both proteins will be expressed and induce apoptosis. Although AAV can infect cells indiscriminately, only Cre-expressing cells will switch on functional expression of the encoded transgene, thereby enabling genetically targeted ablation of the desired cell type exclusively without bystander toxicity to Cre-nonexpressing cells.
2.2.3 Flex
Caspase-3 is an executioner caspase, and leaky expression of the enzyme even at low levels can elicit apoptosis. We therefore wished to restrict caspase-3 expression exclusively to cells expressing Cre recombinase. Accordingly, we employed a strategy (flex switch) that utilizes flanking heterologous loxP sites to encode the desired transgene, caspase-3TevS-T2A-TEV protease in our case, in an orientation reversed to that of the promoter driving expression of the transgene (Atasoy, Aponte, Su, & Sternson, 2008; Schnütgen et al., 2003; Sohal, Zhang, Yizhar, & Deisseroth, 2009). This strategy ensures that the transgene cannot be expressed except in the presence of Cre recombinase, whose activity irreversibly switches the transgene on to the same strand as the promoter. In our case, the flex switch restricts coexpression of caspase-3TevS and TEV protease to Cre-expressing cells.
2.2.4 AAV
We chose to encode the caspase-3TevS-T2A-TEV protease transgene in an AAV because it is the best option for our in vivo application. First, it is not a retrovirus and thus only requires BSL-1. Second, it has extremely low immunogenicity so there is less worry of an immune response. Third, it has a very broad tropism, which can be made still broader with different serotypes—there are currently 11 in use today with “preferences” for different cell types (Taymans et al., 2007). Fourth, it can be produced at very high titer (1012–1013), and there are several commercial facilities dedicated to producing high-titer AAV for a nominal fee. A potential drawback of using AAV is that its genome is small and it can only accept 4–5 kb DNA inserts. However, this was not an issue for our studies because our transgenic insert is within this range.
An alternative approach to delivering the caspase-3TevS-T2A-TEV protease transgene would be to generate a mouse strain harboring this transgene in a Cre-dependent manner. However, promoter sequences that drive transgene expression in a spatiotemporally appropriate manner in the adult VMHvl remain to be defined, and our viral approach circumvents this caveat. Our approach does necessitate, however, that the virus must be injected manually into every experimental animal. Viruses provide the added convenience of being much faster and less resource intensive to generate, produce, and maintain than mouse lines.
2.3. Protocol: Cloning the caspase-3TevS-T2A-TEVp construct and AAV plasmid expression
-
Assembling the caspase-3TevS-T2A-TEVp construct
The caspase-3TevS allele described above (see Section 1.3) was stitched to a T2A-TEVp sequence using overlapping PCR. The resulting caspase-3TevS-T2A-TEVp transgene was cloned in an orientation reverse to that of the ubiquitous promoter, EF1α, into the pAAV-EF1α-DIO-WPRE-pA plasmid (Sohal et al., 2009). This plasmid is currently available from Addgene.
-
Tips for growing AAV plasmids
Subcloning in AAV vectors can be difficult because the two encoded inverted terminal repeats (ITRs) often render the plasmid unstable. It is therefore desirable to perform as many subcloning steps as possible in a standard plasmid lacking ITRs prior to moving the transgene into the AAV vector.
When transforming, be sure to use high-competency cells that have been specifically designed for unstable sequences. We use MAX Efficiency® Stbl2™ high-competency cells.
Plate these cells on standard LB+ampicillin (amp; 100 μg/mL) plates and grow 16–20 h at 30 °C.
Pick a colony for a starter culture and grow it for 8–10 h at 30 °C in LB with amp (75 μg/mL). It is best to pick several colonies from which to make starter cultures and then expand only the best growing one for the final large-scale maxi-prep culture.
The large-scale culture should be grown in 1:1 LB:2×YT with amp (75 μg/mL) at 30 °C for 16–20 h. Because these cells are grown at 30 °C rather than 37 °C, they will expand much more slowly. These are also low-copy number plasmids meaning that the yield will be quite low. It may be better to split the starter culture into several larger cultures rather than simply growing a single culture for a longer period of time.
We use the Endo-Free Plasmid Maxi Kit from Qiagen to purify the plasmid DNA. In our hands, the yield from these columns is ~300 μg/L culture.
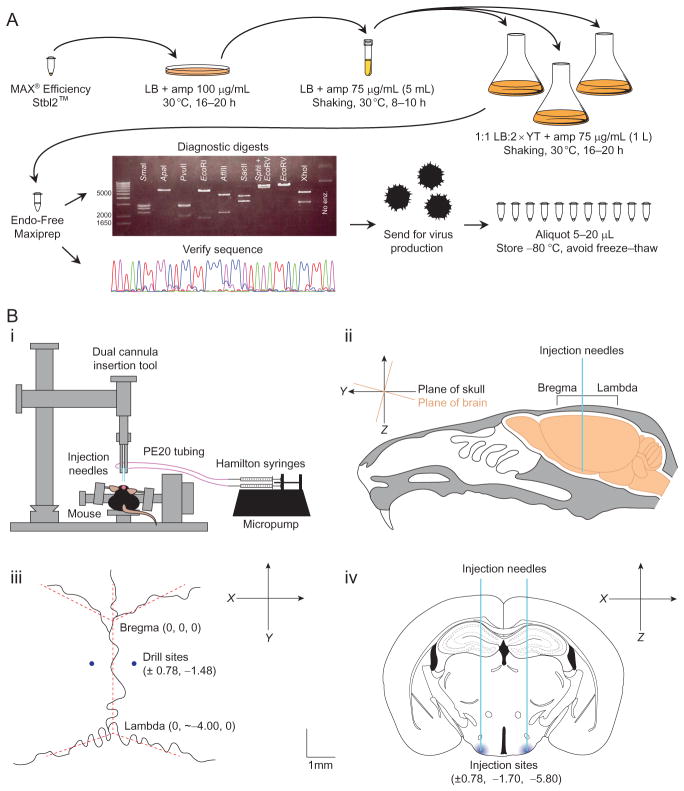
Diagnostic digests of the purified plasmid DNA are essential to exclude recombination events triggered by the ITRs. We digested the AAV-flex-C3-Tp plasmid with eight different enzymes to ensure each element was present and of the expected size (Fig. 8.4A). Some viral cores even require specific digests before they will initiate virus production from the plasmid DNA.
Verifying the sequence of the plasmid is also very important, including all subcloning boundaries and the ITRs. Because of the repeats, sequencing will often fail just inside the ITRs, but it is important to verify that they are present and that the flanking sequences are intact. We sequenced the AAV-flex-C3-Tp plasmid with nine different primers (Table 8.1) that each covered 700–900 bp in an overlapping manner to give the entire sequence of the insert and ITRs.
Most AAV’s are produced in dedicated cores. Many viral cores have a minimum requirement for the plasmid you provide them, typically 300–500 μg of plasmid DNA.
Once the virus is made, it should be aliquoted (typically 5–20 μL) and frozen at −80 °C. Once the aliquot is thawed, it should not be refrozen as freeze–thaw cycles reduce the titer of the virus significantly. Thawed aliquots should remain on ice or at 4 °C, and we typically use each aliquot within 1 week of thawing.
Figure 8.4.
Overview of AAV virus production and injection. (A). Summary of the protocol used for AAV plasmid growth and verification for virus production. Maxipreps originating from the same colony were pooled for diagnostics. (B) Schematic of virus injection. (i) Stereotax and micropump setup: injection needles are connected to Hamilton syringes by PE20 tubing. The syringes are depressed steadily and simultaneously by a micropump. (ii) Orientation of the brain within the cranial cavity. Note the 15° slant of the skull relative to the brain which is common in inbred strains. (iii) Cranial sutures with bregma and lambda marked. (iv) Coronal section through the VMHvl where we injected the virus. Note that the y coordinate is different from the drilling site in (iii) because of the 15° slant. Scale bar represents 1 mm (iii and iv). Figure (iv) was modified from Paxinos Brain Atlas (Paxinos & Franklin, 2004).
Table 8.1.
List of primers used to sequence the AAV-flex-C3-Tp
| Sequence | Orientation | Position | Reads through |
|---|---|---|---|
| CCT CTG ACT TGA GCG TCG AT | Forward | 7107 | Into left ITR |
| ACA CGA CAT CAC TTT CCC AG | Reverse | 314 | Into left ITR |
| TTC TCA AGC CTC AGA CAG TGG | Forward | 1413 | lox/lox, TEVpro, T2A, small |
| AAT CAT GTC CCT GCC GTC GAT C | Forward | 2035 | T2A, small, TEV, large |
| AGA GGG GAT CGT TGT AGA AGT C | Reverse | 2766 | TEV, small, T2A, TEVpro |
| AAA GCA GCG TAT CCA C | Reverse | 3393 | lox/lox, large, TEV, small |
| TAG AAG GAC ACC TAG TCA GA | Reverse | 4053 | pA, WPRE, lox/lox large |
| TCA AGC GAT TCT CCT GCC TC | Forward | 4282 | pA, into right ITR |
| TAC TAT GGT TGC TTT GAC GT | Reverse | 4703 | Into right ITR |
2.4. Protocol: Injecting AAV-flex-C3-Tp into the adult brain
AAV particles, like other viruses and drugs, can be stereotaxically injected into a specific brain region, and the particular characteristics of the virus or chemical determine the subsequent rate and extent of diffusion within the injected area. The spread of AAV particles is limited to 0.5–2 mm from the tip of the injection needle, depending on the volume injected and the size of the needle. The timing of the surgery naturally dictates the onset of the ablation of the desired neuronal subset, offering temporal control, although expression of the virally encoded, Cre-dependent transgene is not immediate, and animals need time to recover from the surgery. Some tissue damage is unavoidable, but this can be reduced by using a smaller needle or pulled glass pipette. There can be significant differences in the tropism of different AAV serotypes for different cell types, including different neuronal populations, and it is important to utilize a serotype that affords maximal infection of the target cells. The advantages of viral delivery—flexibility in encoding transgenes, temporal and anatomical precision—make it ideal for ablating specific cell types in specific regions.
-
Surgery
Anesthetize the animal. The most common ketamine-based cocktails such as Ketamine/Xylazine/Acepromazine do not induce surgical-plane anesthesia, so addition of an inhalational anesthetic such as 0.5% isoflurane is preferable. If inhalational anesthesia is used as the primary modality of anesthesia, additional analgesics will be required.
Shave and sterilize the scalp.
We use a stereotaxic alignment system from Kopf (model 1900) because it allows the precision in head-angle alignment necessary for our deep brain areas. Small deviations in alignment or rotation will be exaggerated in deeper areas. A deflection of only a 100 μm will miss the target region completely. This model allows for the precise measurement and adjustment of the rotation of the skull in all planes such that variation can be accounted for between individual mice (Fig. 8.4Bi).
Mount the mouse on the bite bar and secure the ear bars as indicated in the instruction manual. Many inbred strains including 129 and B6 have a 15° slant to their skull as compared to wild-caught mice and thus it is advisable to use an angled bite bar. It is also advisable to use nonrupture ear bars as these are more humane.
Make an incision that is roughly 1 cm long along the top of the scalp. It should be as small as possible (to reduce pain and healing time for the animal) while still allowing for good visualization of skull landmarks (bregma and lambda) (Fig. 8.4Bii–iii).
Align the instrument to bregma and position and level the skull according to the instruction manual. Bregma is not always well-defined because of the nonlinear cranial sutures (Fig. 8.4Biii), and visualizing lines that most closely approximate the sutures in situ to construct an ideal bregma is recommended (dashed lines in Fig. 8.4Biii). To minimize variability in generating such an ideal bregma, we also recommend that the user maintain a reasonably fixed angle at the intersection of the lines that approximate the sutures. If using the double pressure gauge tool (alignment indicator tool) from Kopf Instruments to level the skull, be sure not to press too hard. The mouse skull is easily deformed, and this will lead to mistargeting.
-
Choosing coordinates
Mouse brain atlas-based coordinates are very accurate, with small mouse-to-mouse variability, at least for standard inbred laboratory strains that we use such as C57Bl/6J (Lein et al., 2007; Paxinos & Franklin, 2004). However, because of how the cranial surface is angled relative to the brain, the final coordinates of the needle tip may not match the coordinates where the holes are drilled in the skull (Fig. 8.4Bii–iv). This tilt should be taken into consideration when establishing coordinates. For the VMHvl, we drill at ±0.78, −1.48 (x,y) and inject at ±0.78, −1.70, −5.80 (x,y,z, bregma=0,0,0). To ensure an accurate set of coordinates, each new region should be tested several times before injecting the experimental virus.
We usually establish our initial coordinates by injecting trypan blue, a dye that labels dead cells. For these studies, we euthanize the mouse with a lethal dose of anesthesia just prior to lowering the injection needles. The brain is dissected immediately, cut into slices thin enough to visualize the needle tracks, and placed in a dish of cold PBS to be viewed on a dissecting microscope.
These coordinates are subsequently refined by a more precise means involving the injection of a fluorescent-tagged reporter (such as cholera toxin B) or an AAV constitutively expressing a fluorescent reporter protein, and careful histological processing, to ensure accuracy. This is often a reiterative process, with the results of each stereotaxic injection trial leading to further refinement of the injection coordinates.
-
Injecting the virus
Assuming coordinates have been chosen, drill holes in the skull above the injection sites (Fig. 8.4Bii).
Lower the needles slowly (1–2 mm/min) to the correct z coordinate. Once there, create a space 50–100 μm above and below the target site, by moving the needles up and down a few times slowly to generate a small pocket for the virus solution to fill.
We use 33G needles (~200 μm outer diameter) coupled to Hamilton syringes via PE20 tubing. Our syringes are mounted on a micro-pump to maintain an even flow rate during infusion (Fig. 8.4Bi). If more precision is needed, pulled glass pipettes can be substituted for steel needles; however, glass pipettes are more fragile and are potentially deflected by fiber tracts within the brain.
Infuse the virus slowly, 60–100 nL/min. The volume can be anywhere from 50 to 1000 nL depending on the titer and efficacy of the virus. Larger volumes will spread farther. It is not recommended that you exceed 1 μL as this will potentially damage the target site.
Allow an additional 5 min for the virus to diffuse away from the needle tips before pulling the needles back up. Retract the needles at the same rate they were lowered.
-
Finishing
Close the wound with skin glue (vetbond, dermabond, or liquid bandaid). You may also use sutures, but in most cases this is not necessary. Wound clips are not recommended for the scalp because the mice will pull these out.
Be sure to use an appropriate analgesic. If an opiate-based analgesic is used (e.g., buprenorphine) monitor the mouse’s respiration until it returns to normal before administering the drug, as opiates are a respiratory depressant.
Wait 1–4 weeks before analyzing the mouse to allow adequate recovery time and for expression of the virus to peak. A time course should be performed for each neuronal population to establish a timepoint at which maximal cell loss has occurred. We wait 3–4 weeks after injecting AAV-flex-C3-Tp prior to behavioral testing.
2.5. The future of viral-mediated ablation in the brain
While stroke, cancer, trauma, and neurodegenerative victims have provided invaluable insights into localizing brain function, genetically targeted ablations of specific neuronal populations will enable high-resolution functional mapping of the neural circuits that encode behavior in health and disease. The electrolytic or surgical lesions of the past have offered us insight into regional functionalization of the brain, but now we have the means to determine the function of any group of adult neurons or model neurodegenerative disorders with remarkable accuracy. These genetically targeted studies will provide us with more precise understanding of the function of discrete neuronal populations and a testing ground for therapeutic interventions. As we mentioned earlier, our strategy is modular and can, in principle, be applied to any cell type in the brain or other organ system. We are also developing means of ablating even more specific neuronal populations with enhanced temporal control in the form of light- and ligand-activated caspases.
3. SMALL-MOLECULE ACTIVATORS OF CASPASES
3.1. Introduction
Allosteric modulation of enzymes with unnatural small molecules is an attractive way of modifying enzymatic activities by often offering greater selectivity over targeting conserved active sites. Allosteric control also allows the possibility to turn enzymes on, and there have been important recent discoveries in this area for kinases, deacetylases, dehydrogenases, phosphatases, and nucleases (for review, see Zorn & Wells, 2010). Upregulation of apoptotic signaling is common in precancerous cells and silencing of apoptosis is a hallmark of cancer cells. This apoptotic suppression is usually achieved by making lesions in signaling pathways upstream of the caspases, such as p53 (Lowe & Sherr, 2003). Thus, there has been a significant effort to activate executioner caspase with small molecules in hopes of differentially killing cancer cells.
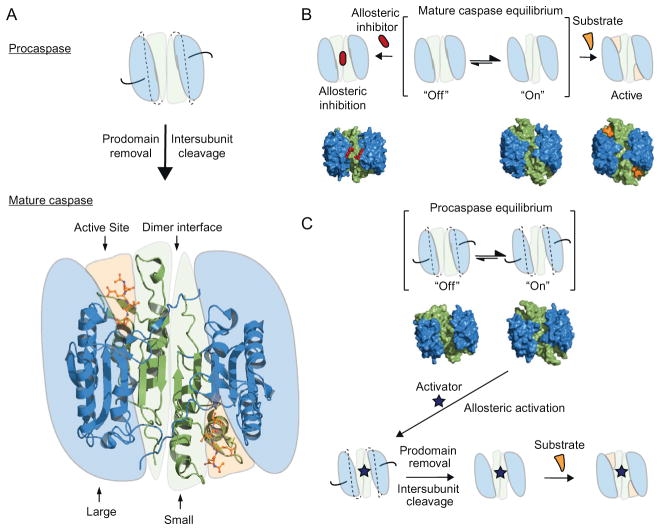
The caspases are expressed as inactive proenzymes, or zymogens, and their activation is under tight regulation in cells due to their fate-determining activity. For example, the zymogenicity (defined as the mature enzyme kcat/Km divided by the proenzyme kcat/Km) of procaspase-3 is greater than 107, making it the most inactive protease zymogen yet reported (Pop et al., 2007; Stennicke & Salvesen, 2000; Zorn, Wolan, Agard, & Wells, 2012). Upon proteolysis by upstream proteases, the N-terminal prodomain of the procaspase is removed, and the intersubunit linker is cut, the later being crucial to activation (Fig. 8.1B). This proteolytic activation leads to a dimer of heterodimers consisting of a large and small subunit, as illustrated by the crystal structure of mature caspase-7 (Fig. 8.5A) (Chai et al., 2001; Riedl et al., 2001; Wei et al., 2000). In this active state, the executioner caspases exist in a conformational equilibrium between an “off” and “on” state, but predominantly in the on-state to allow substrate binding in the active site (Fig. 8.5B). Interestingly, mature caspase-1 is predominantly in the off-state until driven to the on-state by binding substrate (Gao, Sidhu, & Wells, 2009). In the “off” state, the active caspase can be trapped in a nonfunctional conformation by allosteric inhibitors that bind to the dimer interface, as shown in previous studies of caspase-1, -5, and -7 (Gao & Wells, 2012; Hardy, Lam, Nguyen, O’Brien, & Wells, 2004; Scheer, Romanowski, & Wells, 2006). These results also raise the question as to whether small molecules can potentially be found to activate the pro-enzyme (Fig. 8.5C). More recently, our group demonstrated the feasibility of activating procaspase-3 and -7. Antibody fragments were selected for binding the on-state and could stimulate procaspase-3 activity by 1000-fold (Thomsen, Koerber, & Wells, 2013). These results also highlight the enormous barrier of activation of procaspases and suggest that finding robust allosteric activators of procaspase-3 will be challenging.
Figure 8.5.
Mechanism of caspase regulation. (A) Executioner procaspase maturation requires proteolysis of the N-terminal prodomain (black lines) and cleavage of the inter-subunit linker (dashed lines) between the large and small subunits. The crystal structure of mature caspase-7 is shown (PDB ID 1F1J) highlighting the dimer interface, the large and small subunits and the Ac-DEVD-CHO peptide occupying the active site (shown in sticks). (B) Mechanism of allosteric inhibition of caspases, showing the hotspot for allosteric binding located at the dimer interface. The surface representation of experimentally determined X-ray structures of caspase-7 is shown below the cartoons. From left to right, the allosteric-site ligand-bound structure of caspase-7 complexed with DICA, the ligand-free apo caspase-7 structure in an “on” conformation, and the active-site ligand-bound structure of caspase-7 in complex with the Ac-DEVD-CHO peptide (PDB ID 1SHJ, 1K86, 1F1J, respectively). (C) Mechanism of allosteric activation of procaspases. The brackets denote the open and closed active-site equilibrium, with the surface of the X-ray structures of procaspase-3 in the “on” and “off” states (PDB ID 4JR0 and 4JQY, respectively). Upon binding of a hypothetical allosteric activator, the proenzyme is locked into an “on” conformation, allowing possible proteolytic cleavage of the prodomain and intersubunit linkers.
3.2. Small-molecule activators of caspases
At present, four small-molecule compounds have been reported to activate procaspases in vitro and to induce cell death in cell culture. Neither of the binding sites for these small molecules have been structurally defined, nor have their stoichiometry of binding been determined. Interestingly, the mechanisms for the two that have been most extensively characterized are not through traditional allosteric means, that is, stoichiometric binding of a small molecule to the protein. Here, we review the discovery of each of these four compounds and provide practical suggestions for assays that may be useful for future discovery efforts.
3.2.1 PAC-1
PAC-1, or procaspase-activating compound 1, was a small molecule discovered by the Hergenrother group as being capable of activating procaspase-3 in vitro and inducing apoptosis in various cell lines (Fig. 8.6A) (Putt et al., 2006). This compound was discovered by performing a high-throughput screen for 20,500 diverse small molecules, and looking for procaspase-3 activation by monitoring cleavage of a peptidic substrate (acetyl Asp-Glu-Val-Asp-p-nitroanilide (Ac-DEVD-pNa) at 200 μM). The compound PAC-1 activated procaspase-3 up to fourfold over background after 12 h of incubation with an EC50 of 0.22 and 4.7 μM for procaspase-3 and -7, respectively (Peterson et al., 2009). This activation is small considering the 107-fold change in activity for full activation of the proenzyme (Zorn et al., 2012). Nonetheless, PAC-1 was shown to induce cell death in several cancer cell lines in a manner proportional to the cellular concentration of procaspase-3, and was ineffective in MCF-7 cells that lack procaspase-3, although another group was unable to repeat this result (Denault et al., 2007). PAC-1 was also shown to significantly delay the progression of tumors in mice (Putt et al., 2006). The group later showed that PAC-1 is a zinc chelator agent that leads to activation of procaspase-3 by relieving the zinc inhibition of caspase-3 from their buffers that were missing EDTA (Peterson et al., 2009). These findings are consistent with previous studies showing that zinc inhibits caspase-3 with an IC50 of 0.1 μM (Perry et al., 1997). They also reported that the zinc-chelation mechanism was responsible for the cell death observed in cell culture leading to altered intracellular Ca2+ concentration, indicative of endoplasmic reticulum stress-induced apoptosis (West et al., 2012). In summary, PAC-1 may not be a direct activator of procaspase, but led to interesting discoveries about the role of metal chelation in caspase activation and initiation of cell death. Whether the cellular activity is a direct effect of chelating zinc from caspase-3 remains challenging to prove, given the many roles of zinc in cells.
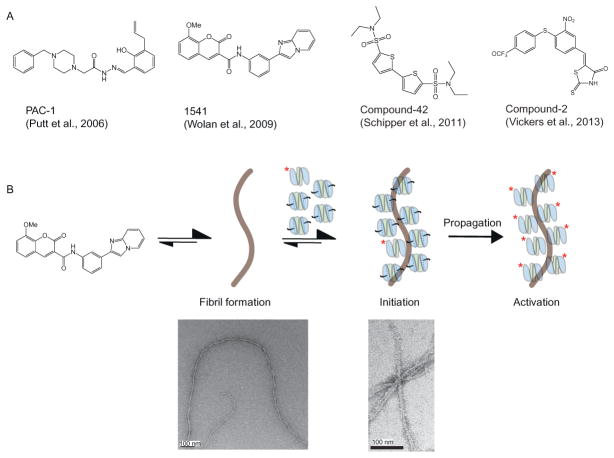
Figure 8.6.
Procaspase activation by small molecules. (A) Four small molecules have been reported to act as allosteric activators of procaspases; PAC-1 (Putt et al., 2006), 1541 (Wolan, Zorn, Gray, & Wells, 2009), compound-42 (Schipper, MacKenzie, Sharma, & Clark, 2011), and compound-2 (Vickers et al., 2013). (B) Mechanism of procaspase activation by 1541 nanofibrils. A small amount of active caspase (labeled with asterisk) is already present in E. coli-purified procaspase samples. The nanofibrils serve as a scaffold for colocalization of active caspase with the proenzyme, leading to activation. Figure (B) is adapted with permission from Zorn, Wille, Wolan, and Wells, (2011). Copyright (2011) American Chemical Society.
3.2.2 Compound-1541
In a separate HTS of over 62,000 compounds, our lab discovered a compound called 1541 (Fig. 8.6A), that lead to specific activation of procaspases-3 and -6 in vitro but not procaspase-7 (Wolan et al., 2009). Compound-1541 induces rapid and complete apoptosis with an EC50 ~2 μM. The apoptosis is minimally affected by deletion of caspase-8, a driver of the extrinsic pathway, and apoptosis proceeds without significant release of cytochrome C, a marker of the intrinsic pathway. These data were consistent with direct activation of caspase-3. Moreover, the rate of cell death is slowed in MCF-7 cells lacking caspase-3. However, we were unable to determine the structure of the compound bound to either the mature or zymogen form of caspase-3. Nor were we able to obtain clear biophysical evidence for stoichiometric binding, for example, by surface plasmon resonance (SPR) or isothermal titration calorimetry (ITC). Further characterization of the compounds by electron microcopy (EM) revealed the surprising finding that 1541 and its active analogs self-assemble into nanofibrils in solution (Fig. 8.5B) (Zorn et al., 2011). Using various biochemical and biophysical methods, we conclusively found that these nanofibrils colocalize caspase-3 with procaspase-3 and promote activation in vitro (Zorn et al., 2012).
Globular aggregates of small molecules are known to act as inhibitors and can be diagnosed by testing for detergent sensitivity, β-lactamase inhibition, and sensitivity to bovine serum albumin (BSA) (Coan & Shoichet, 2008; Feng et al., 2007; Seidler, McGovern, Doman, & Shoichet, 2003). However, we obtained mixed results by performing these tests on 1541 (Zorn et al., 2011). For example, detergents such as Triton or CHAPS did not disrupt procaspase-3 activation, nor did 1541 inhibit β-lactamase activity. However, the addition of BSA into the assay prevented procaspase-3 activation. A careful characterization of 1541 in various conditions (e.g., buffer, temperature, concentration, etc.) led us to observe higher molecular weight species using dynamic light scattering (DLS). Spin-down assays showed coprecipitation of the nanofibrils with procaspase-3. Moreover, sequestering procaspase-3 from the nanofibrils using a dialysis bag containing the pro-enzyme prevented activation. We hypothesize that unlike typical amorphous aggregates, that are in free equilibrium between monomer and aggregate, the nanofibrils have fewer ends available for monomer–nanofibrils exchange. This makes it extremely slow for the compound to reform fibrils, making it very unlikely for the compound to enter the dialysis bag and to reform fibrils on the inside of the membrane during the incubation period.
We found that activation in vitro depends on the presence of a small amount of active caspase to initiate proenzyme cleavage. For example, adding as little as 0.01 equivalent of the covalent caspase-3 inhibitor (Ac-DEVD-cmk) to procaspase-3 in our assays prevented procaspase activation by 1541. We believe this contaminant comes from tiny amounts of mature enzyme that is processed from the proenzyme, perhaps by endogenous E. coli proteases or by the procaspase itself during expression in bacteria. There is also a pronounced lag in the activation process in vitro, as is typical for the activation of proenzymes, in general. This lag is not due to assembly of the fibrils; we observe them forming instantly upon dilution into aqueous solution, as monitored by EM or DLS. Moreover, the inclusion of even a 1% stoichiometric amount of active caspase-3 shortens the lag period by 2.5-fold (Zorn et al., 2012). Taken together, our experiments show that 1541 nanofibrils colocalize caspase-3 with procaspase-3 in vitro, leading to activation of the proenzyme in a similar manner as a firecracker fuse. The spark of activation is provided by a small amount of active caspase that is colocalized to the fibril. Additional structural characterization of the nanofibrils with procaspase-3 will be important to understand the assembly and activation mechanism further.
Many questions remain to be answered regarding the cellular mechanism for 1541. How does 1541 induce apoptosis in cell culture with such potency? Our preliminary studies indicate that the fibrils are responsible for inducing apoptosis in cell culture (O. Julien & J. A. Wells, unpublished results), but the exact molecular mechanism of cell death is currently under investigation. Are the nanofibrils activating procaspases in cells in the same way they do in vitro? There are some intriguing similarities, both at the functional and structural levels, between 1541 nanofibrils and some amyloid fibrils that induce apoptosis such as Aβ (Zorn et al., 2011). The ease of making 1541 and the simplicity of forming these fibrils make it a potentially important model system for understanding cell death induced by such fibril-forming molecules.
3.2.3 Compound-42
Another small molecule, named compound-42 (Fig. 8.6A), was recently discovered by the Clark group that was capable of enhancing procaspase-3 activation by 27-fold (Schipper et al., 2011). Again, this is a small change considering the zymogenicity of the procaspase-3. The compound was discovered using computational docking screen of 62 compounds that targeted the allosteric inhibitory site located at the dimer interface of procaspase-3 (Hardy et al., 2004). They hypothesized that stabilizing the same interface on procaspase-3 could lead to allosteric activation of the zymogen. Based on their docking score, 13 compounds were tested in a procaspase-3 activation assay in vitro. One compound, compound-42, was found to increase procaspase-3 activity by 27-fold at 400 μM after 1 h incubation with the proenzyme. The authors hypothesized that compound-42 activates procaspase-3 by releasing the intersubunit linker from the dimer interface allowing the enzyme to auto-process this linker in trans. The group measured a KD of 4 μM for compound-42 to bind mature caspase-3 by ITC, but could not determine the binding affinity of compound-42 to procaspase-3. No cellular activity was reported.
3.2.4 Compound-2
Earlier this year, the Wolan group discovered a new compound that was capable of promoting the maturation of executioner procaspase-3 and -7 (Vickers et al., 2013). They used a clever fluorescence-polarization-based screen. This utilized a newly designed canonical DEVD-aldehyde peptide recognition motif, and a 5(6)-carboxyfluorescein at the N-terminus linked to a 3 × 6-aminohexanoic acid. The main advantage of this new probe is that it binds and inhibits any contaminating mature caspase-3 and -7, in a reversible covalent manner that may be present or produced during the activation assay, and does not get consumed like standard caspase probes that are based on the release of a fluorophore. Using HTS, they identified a small molecule, compound-2 (Fig. 8.6A), that the activated executioner procaspases in vitro with an EC50 of ~1–5 μM. Importantly, compound-2 and its analogs are capable of inducing apoptosis in cell culture with the same low μM EC50. These values are very similar to 1541. Further studies will be needed to confirm the stoichiometry and site of binding, as well as the mechanism of inducing apoptosis in cells.
3.3. A practical guide to avoid aggregating small molecules: The case of procaspase activators
3.3.1 Detergent in vitro
The use of small amounts of detergent during the high-throughput screen is the first step toward avoiding false positive results caused by small molecules (Fig. 8.7). This did not preclude the discovery of 1541 as the screen included 0.1% CHAPS to avoid standard aggregators. Vickers et al. also included some detergent in their assay buffer to discourage small-molecule aggregation. Once hits are found, it is useful to test the effect of increasing the detergent concentration on the enzymatic activity of the target of interest. If a decrease in compound potency is observed, aggregation is a strong possibility.
Figure 8.7.
Useful tests to ensure compounds are acting through a stoichiometric binding mechanism.
3.3.2 Dynamic light scattering
In our hands, the gold standard test for detecting small-molecule aggregators is DLS. If performed cautiously, this test is capable of detecting all forms of aggregation (e.g., colloid particles, micelles, fibrils, etc.). We recommend all buffer solutions to be filtered at 0.22 μm before adding the compound of interest. Measurements should be made in the same time frame used for the enzyme and cellular assays. Low and high concentration of compounds should also be tested at various temperatures. Measurements should be performed in triplicate. Finally, it is useful to test a known aggregator as positive control (e.g., like tetra-iodophenolphthalein) and a known well-behaved small molecule as negative control (e.g., coumarins).
3.3.3 Nonspecific enzyme inhibition
Various assays can be used to detect the formation of small-molecule aggregates in vitro. For example, Shoichet and coworkers developed a β-lactamase inhibition assay based on the high sensitivity of this enzyme to be inhibited by colloidal particles (McGovern, Caselli, Grigorieff, & Shoichet, 2002). While useful for colloidal particles, 1541 nanofibrils did not inhibit β-lactamase. Another simple assay is to test if BSA perturbs the enzymatic activity caused by the compound of interest (Coan & Shoichet, 2007).
3.3.4 Transmission electron microscopy
Transmission electron microscopy is a powerful and relatively easy method to verify the presence of aggregators. However, it is important to remember that a negative result here does not rule out the presence of aggregation, since not all aggregators can be effectively detected using this method. This method was especially useful for observing 1541 nanofibrils.
3.3.5 Stoichiometry of binding
Various techniques allow the determination of the stoichiometry of binding of a compound of interest to its target. One can use ITC, X-ray crystallography, SPR, or nuclear magnetic resonance (NMR) spectroscopy. In addition, X-ray crystallography and NMR spectroscopy are powerful tools to determine the binding location of the small molecule on its target. As with EM, a single negative result does not reveal much, but one should be worried if all fails.
3.3.6 Detergent in cell culture
An easy way to test the presence of small-molecule aggregation is to use small amounts of detergent in cell culture (Owen, Doak, Wassam, Shoichet, & Shoichet, 2012). Specifically, Tween-80 is able to prevent small-molecule aggregation and has been shown by Owen and coworkers to have negligible toxicity in cell culture at low concentration (i.e., less than 0.1%). Recently, we have had success using this assay to block 1541 activity (O. Julien and J. A. Wells, data to be published).
3.3.7 Use of active-site titrants to rule out the involvement of mature caspases in proenzyme activation
Activation by 1541 requires a small amount of mature caspase, which can be tested for and blocked by addition of substoichiometric amounts of DEVD-chloromethyl ketone. We found mature caspase contaminants are unavoidable when using E. coli to express the procaspase. Such contaminants are even present if one uses the “noncleavable” D3A mutant. It is possible that alternate cleavage sites by host proteases are the cause of this mature enzyme contamination. Indeed, proteolysis at D169 has been shown to occur when the normal cleavage site D175 is unavailable (MacKenzie et al., 2013). In our hands, such spurious activation in E. coli expression systems can be reduced by using shorter expression times, but it is impossible to eliminate completely. Thus, the safest technique is to inactivate trace levels of mature caspase molecules in proenzyme preparations by adding as little as 0.01–0.05 equivalent of a covalent and irreversible caspase inhibitor (e.g., Ac-DEVD-chloromethyl ketone).
3.4. Conclusions
The most thoroughly characterized procaspase activators, PAC-1 and 1541, were shown to activate the proenzyme through unexpected mechanisms. PAC-1 is believed to remove zinc inhibition, and 1541 through nanofibril formation and activation by colocalization of mature and inactive caspase. These are still very useful tools and have provided important new insights about mechanisms of activation of caspases. Direct activation of executioner caspases remains a big challenge due to the high zymogenicity barrier of the proenzyme (Zorn et al., 2012). It may be easier to target procaspase-7 instead of procaspase-3 because of its lower zymogen activation barrier (Thomsen et al., 2013). It may even be more achievable to target the initiator caspases since they have even lower barriers and compounds that induce dimerization could potentially be found. Finding new drugs here would have a big impact, but many challenges remain.
Acknowledgments
We are grateful to Dan Gray, Julie Zorn, and Dennis Wolan for their pioneering work on Sections 1 and 3, and Cindy Yang and Dan Gray for their work on Section 2. We also thank Allison Doak, Jason Porter, Justin Rettenmaier, Cheryl Tajon, Nathan Thomsen, for their thoughtful discussions and careful reading of the chapter. C. W. M. is supported by a NSF Graduate Fellowship. O. J. is supported by a Banting Fellowship from the Government of Canada and the Canadian Institutes of Health Research. E. K. U. is supported by NIH grant F31NS078959. N. M. S. is supported by The Ellison Medical Foundation and the NIH (grants DP1MH099900, R01NS049488, and R01NS083872). J. A. W. is supported by NIH (grants R01 CA136779 and R01 GM081051).
References
- Amara JF, Clackson T, Rivera VM, Guo T, Keenan T, Natesan S, et al. A versatile synthetic dimerizer for the regulation of protein-protein interactions. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:10618–10623. doi: 10.1073/pnas.94.20.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. The Journal of Neuroscience. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw PJ, Spencer DM, Crabtree GR, Schreiber SL. Controlling programmed cell death with a cyclophilin-cyclosporin-based chemical inducer of dimerization. Chemistry & Biology. 1996;3:731–738. doi: 10.1016/s1074-5521(96)90249-5. [DOI] [PubMed] [Google Scholar]
- Chai J, Wu Q, Shiozaki E, Srinivasula SM, Alnemri ES, Shi Y. Crystal structure of a procaspase-7 zymogen: Mechanisms of activation and substrate binding. Cell. 2001;107:399–407. doi: 10.1016/s0092-8674(01)00544-x. [DOI] [PubMed] [Google Scholar]
- Chang DW, Xing Z, Capacio VL, Peter ME, Yang X. Interdimer processing mechanism of procaspase-8 activation. The EMBO Journal. 2003;22:4132–4142. doi: 10.1093/emboj/cdg414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelur DS, Chalfie M. Targeted cell killing by reconstituted caspases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2283–2288. doi: 10.1073/pnas.0610877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Orozco A, Spencer DM, Wang J. Activation of initiator caspases through a stable dimeric intermediate. The Journal of Biological Chemistry. 2002;277:50761–50767. doi: 10.1074/jbc.M210356200. [DOI] [PubMed] [Google Scholar]
- Coan KE, Shoichet BK. Stability and equilibria of promiscuous aggregates in high protein milieus. Molecular Biosystems. 2007;3:208–213. doi: 10.1039/b616314a. [DOI] [PubMed] [Google Scholar]
- Coan KE, Shoichet BK. Stoichiometry and physical chemistry of promiscuous aggregate-based inhibitors. Journal of the American Chemical Society. 2008;130:9606–9612. doi: 10.1021/ja802977h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooray S, Howe SJ, Thrasher AJ. Retrovirus and lentivirus vector design and methods of cell conditioning. Methods in Enzymology. 2012;507:29–57. doi: 10.1016/B978-0-12-386509-0.00003-X. [DOI] [PubMed] [Google Scholar]
- Denault JB, Drag M, Salvesen GS, Alves J, Heidt AB, Deveraux Q, et al. Small molecules not direct activators of caspases. Nature Chemical Biology. 2007;3:519. doi: 10.1038/nchembio0907-519. author reply 520. [DOI] [PubMed] [Google Scholar]
- Di Stasi A, Tey SK, Dotti G, Fujita Y, Kennedy-Nasser A, Martinez C, et al. Inducible apoptosis as a safety switch for adoptive cell therapy. The New England Journal of Medicine. 2011;365:1673–1683. doi: 10.1056/NEJMoa1106152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SR, Wandless TJ. The rapamycin-binding domain of the protein kinase mammalian target of rapamycin is a destabilizing domain. The Journal of Biological Chemistry. 2007;282:13395–13401. doi: 10.1074/jbc.M700498200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fegan A, White B, Carlson JC, Wagner CR. Chemically controlled protein assembly: Techniques and applications. Chemical Reviews. 2010;110:3315–3336. doi: 10.1021/cr8002888. [DOI] [PubMed] [Google Scholar]
- Feng BY, Simeonov A, Jadhav A, Babaoglu K, Inglese J, Shoichet BK, et al. A high-throughput screen for aggregation-based inhibition in a large compound library. Journal of Medicinal Chemistry. 2007;50:2385–2390. doi: 10.1021/jm061317y. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Aaronson SA, Abrams J, Alnemri ES, Andrews DW, Baehrecke EH, et al. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death and Differentiation. 2009;16:1093–1107. doi: 10.1038/cdd.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Sidhu SS, Wells JA. Two-state selection of conformation-specific antibodies. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3071–3076. doi: 10.1073/pnas.0812952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wells JA. Identification of specific tethered inhibitors for caspase-5. Chemical Biology & Drug Design. 2012;79:209–215. doi: 10.1111/j.1747-0285.2011.01261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray DC, Mahrus S, Wells JA. Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell. 2010;142:637–646. doi: 10.1016/j.cell.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy JA, Lam J, Nguyen JT, O’Brien T, Wells JA. Discovery of an allosteric site in the caspases. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12461–12466. doi: 10.1073/pnas.0404781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MA, Harper N, Butterworth M, Cain K, Cohen GM, MacFarlane M. Reconstitution of the death-inducing signaling complex reveals a substrate switch that determines CD95-mediated death or survival. Molecular Cell. 2009;35:265–279. doi: 10.1016/j.molcel.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Kang TB, Oh GS, Scandella E, Bolinger B, Ludewig B, Kovalenko A, et al. Mutation of a self-processing site in caspase-8 compromises its apoptotic but not its non-apoptotic functions in bacterial artificial chromosome-transgenic mice. Journal of Immunology. 2008;181:2522–2532. doi: 10.4049/jimmunol.181.4.2522. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: Progress and puzzles. Current Opinion in Genetics & Development. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- MacKenzie SH, Schipper JL, England EJ, Thomas ME, 3rd, Blackburn K, Swartz P, et al. Lengthening the intersubunit linker of procaspase 3 leads to constitutive activation. Biochemistry. 2013;52:6219–6231. doi: 10.1021/bi400793s. [DOI] [PubMed] [Google Scholar]
- McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. Journal of Medicinal Chemistry. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- Murphy BM, Creagh EM, Martin SJ. Interchain proteolysis, in the absence of a dimerization stimulus, can initiate apoptosis-associated caspase-8 activation. The Journal of Biological Chemistry. 2004;279:36916–36922. doi: 10.1074/jbc.M402039200. [DOI] [PubMed] [Google Scholar]
- Muzio M, Stockwell BR, Stennicke HR, Salvesen GS, Dixit VM. An induced proximity model for caspase-8 activation. The Journal of Biological Chemistry. 1998;273:2926–2930. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- Oberst A, Pop C, Tremblay AG, Blais V, Denault JB, Salvesen GS, et al. Inducible dimerization and inducible cleavage reveal a requirement for both processes in caspase-8 activation. The Journal of Biological Chemistry. 2010;285:16632–16642. doi: 10.1074/jbc.M109.095083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban PC, Chui D, Marth JD. Tissue- and site-specific DNA recombination in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:6861–6865. doi: 10.1073/pnas.89.15.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen SC, Doak AK, Wassam P, Shoichet MS, Shoichet BK. Colloidal aggregation affects the efficacy of anticancer drugs in cell culture. ACS Chemical Biology. 2012;7:1429–1435. doi: 10.1021/cb300189b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajvani UB, Trujillo ME, Combs TP, Iyengar P, Jelicks L, Roth KA, et al. Fat apoptosis through targeted activation of caspase 8: A new mouse model of inducible and reversible lipoatrophy. Nature Medicine. 2005;11:797–803. doi: 10.1038/nm1262. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates. San Diego, CA: Elsevier Academic Press; 2004. [Google Scholar]
- Perry DK, Smyth MJ, Stennicke HR, Salvesen GS, Duriez P, Poirier GG, et al. Zinc is a potent inhibitor of the apoptotic protease, caspase-3. A novel target for zinc in the inhibition of apoptosis. The Journal of Biological Chemistry. 1997;272:18530–18533. doi: 10.1074/jbc.272.30.18530. [DOI] [PubMed] [Google Scholar]
- Peterson QP, Goode DR, West DC, Ramsey KN, Lee JJ, Hergenrother PJ. PAC-1 activates procaspase-3 in vitro through relief of zinc-mediated inhibition. Journal of Molecular Biology. 2009;388:144–158. doi: 10.1016/j.jmb.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop C, Fitzgerald P, Green DR, Salvesen GS. Role of proteolysis in caspase-8 activation and stabilization. Biochemistry. 2007;46:4398–4407. doi: 10.1021/bi602623b. [DOI] [PubMed] [Google Scholar]
- Provost E, Rhee J, Leach SD. Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos. Genesis. 2007;45:625–629. doi: 10.1002/dvg.20338. [DOI] [PubMed] [Google Scholar]
- Putt KS, Chen GW, Pearson JM, Sandhorst JS, Hoagland MS, Kwon JT, et al. Small-molecule activation of procaspase-3 to caspase-3 as a personalized anticancer strategy. Nature Chemical Biology. 2006;2:543–550. doi: 10.1038/nchembio814. [DOI] [PubMed] [Google Scholar]
- Remy I, Michnick SW. Application of protein-fragment complementation assays in cell biology. BioTechniques. 2007;42:137, 139, 141. doi: 10.2144/000112396. passim. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Fuentes-Prior P, Renatus M, Kairies N, Krapp S, Huber R, et al. Structural basis for the activation of human procaspase-7. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:14790–14795. doi: 10.1073/pnas.221580098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvesen GS, Dixit VM. Caspase activation: The induced-proximity model. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:10964–10967. doi: 10.1073/pnas.96.20.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B. Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Molecular and Cellular Biology. 1987;7:2087–2096. doi: 10.1128/mcb.7.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer JM, Romanowski MJ, Wells JA. A common allosteric site and mechanism in caspases. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7595–7600. doi: 10.1073/pnas.0602571103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schipper JL, MacKenzie SH, Sharma A, Clark AC. A bifunctional allosteric site in the dimer interface of procaspase-3. Biophysical Chemistry. 2011;159:100–109. doi: 10.1016/j.bpc.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnütgen F, Doerflinger N, Calléja C, Wendling O, Chambon P, Ghyselinck NB. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nature Biotechnology. 2003;21:562–565. doi: 10.1038/nbt811. [DOI] [PubMed] [Google Scholar]
- Seidler J, McGovern SL, Doman TN, Shoichet BK. Identification and prediction of promiscuous aggregating inhibitors among known drugs. Journal of Medicinal Chemistry. 2003;46:4477–4486. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn D, Schulze-Osthoff K, Jänicke RU. Caspase-8 can be activated by interchain proteolysis without receptor-triggered dimerization during drug-induced apoptosis. The Journal of Biological Chemistry. 2005;280:5267–5273. doi: 10.1074/jbc.M408585200. [DOI] [PubMed] [Google Scholar]
- Spencer DM, Belshaw PJ, Chen L, Ho SN, Randazzo F, Crabtree GR, et al. Functional analysis of Fas signaling in vivo using synthetic inducers of dimerization. Current Biology. 1996;6:839–847. doi: 10.1016/s0960-9822(02)00607-3. [DOI] [PubMed] [Google Scholar]
- Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- Steller H. Artificial death switches: Induction of apoptosis by chemically induced caspase multimerization. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5421–5422. doi: 10.1073/pnas.95.10.5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennicke HR, Salvesen GS. Caspases—Controlling intracellular signals by protease zymogen activation. Biochimica and Biophysica Acta. 2000;1477:299–306. doi: 10.1016/s0167-4838(99)00281-2. [DOI] [PubMed] [Google Scholar]
- Straathof KC, Pulé MA, Yotnda P, Dotti G, Vanin EF, Brenner MK, et al. An inducible caspase 9 safety switch for T-cell therapy. Blood. 2005;105:4247–4254. doi: 10.1182/blood-2004-11-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Ehrlich I, Wölff SB, Michalski AM, Wolfl S, Hasan MT, et al. Faithful expression of multiple proteins via 2A-peptide self-processing: A versatile and reliable method for manipulating brain circuits. The Journal of Neuroscience. 2009;29:8621–8629. doi: 10.1523/JNEUROSCI.0359-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taymans JM, Vandenberghe LH, Haute CV, Thiry I, Deroose CM, Mortelmans L, et al. Comparative analysis of adeno-associated viral vector sero-types 1, 2, 5, 7, and 8 in mouse brain. Human Gene Therapy. 2007;18:195–206. doi: 10.1089/hum.2006.178. [DOI] [PubMed] [Google Scholar]
- Thomsen ND, Koerber JT, Wells JA. Structural snapshots reveal distinct mechanisms of procaspase-3 and -7 activation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8477–8482. doi: 10.1073/pnas.1306759110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonikian R, Zhang Y, Boone C, Sidhu SS. Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries. Nature Protocols. 2007;2:1368–1386. doi: 10.1038/nprot.2007.151. [DOI] [PubMed] [Google Scholar]
- Vickers CJ, González-Páez GE, Umotoy JC, Cayanan-Garrett C, Brown SJ, Wolan DW. Small-molecule procaspase activators identified using fluorescence polarization. Chembiochem. 2013;14:1419–1422. doi: 10.1002/cbic.201300315. [DOI] [PubMed] [Google Scholar]
- Wehr MC, Laage R, Bolz U, Fischer TM, Grünewald S, Scheek S, et al. Monitoring regulated protein-protein interactions using split TEV. Nature Methods. 2006;3:985–993. doi: 10.1038/nmeth967. [DOI] [PubMed] [Google Scholar]
- Wei Y, Fox T, Chambers SP, Sintchak J, Coll JT, Golec JM, et al. The structures of caspases-1, -3, -7 and -8 reveal the basis for substrate and inhibitor selectivity. Chemistry & Biology. 2000;7:423–432. doi: 10.1016/s1074-5521(00)00123-x. [DOI] [PubMed] [Google Scholar]
- West DC, Qin Y, Peterson QP, Thomas DL, Palchaudhuri R, Morrison KC, et al. Differential effects of procaspase-3 activating compounds in the induction of cancer cell death. Molecular Pharmaceutics. 2012;9:1425–1434. doi: 10.1021/mp200673n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DJ, Puhl HL, 3rd, Ikeda SR. Rapid modification of proteins using a rapamycin-inducible tobacco etch virus protease system. PLoS One. 2009;4:e7474. doi: 10.1371/journal.pone.0007474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolan DW, Zorn JA, Gray DC, Wells JA. Small-molecule activators of a proenzyme. Science. 2009;326:853–858. doi: 10.1126/science.1177585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Chang HY, Baltimore D. Autoproteolytic activation of pro-caspases by oligomerization. Molecular Cell. 1998;1:319–325. doi: 10.1016/s1097-2765(00)80032-5. [DOI] [PubMed] [Google Scholar]
- Yang CF, Chiang MC, Gray DC, Prabhakaran M, Alvarado M, Juntti SA, et al. Sexually dimorphic neurons in the ventromedial hypothalamus govern mating in both sexes and aggression in males. Cell. 2013;153:896–909. doi: 10.1016/j.cell.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Kroemer G. Alternative cell death mechanisms in development and beyond. Genes & Development. 2010;24:2592–2602. doi: 10.1101/gad.1984410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn JA, Wells JA. Turning enzymes ON with small molecules. Nature Chemical Biology. 2010;6:179–188. doi: 10.1038/nchembio.318. [DOI] [PubMed] [Google Scholar]
- Zorn JA, Wille H, Wolan DW, Wells JA. Self-assembling small molecules form nanofibrils that bind procaspase-3 to promote activation. Journal of the American Chemical Society. 2011;133:19630–19633. doi: 10.1021/ja208350u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn JA, Wolan DW, Agard NJ, Wells JA. Fibrils colocalize caspase-3 with procaspase-3 to foster maturation. The Journal of Biological Chemistry. 2012;287:33781–33795. doi: 10.1074/jbc.M112.386128. [DOI] [PMC free article] [PubMed] [Google Scholar]