Summary
Recently, we described the existence of the ubiquitin fold modifier1 (Ufm1) and its conjugation pathway in Leishmania donovani. We demonstrated the conjugation of Ufm1 to proteins such as mitochondrial trifunctional protein (MTP) that catalyzes β-oxidation of fatty acids in L. donovani. To elucidate the biological roles of the Ufm1-mediated modifications, we made an L. donovani Ufm1 null mutant (Ufm1−/−). Loss of Ufm1 and consequently absence of Ufm1 conjugation with MTP resulted in diminished acetyl-CoA, the end product of the β-oxidation in the Ufm1−/− amastigote stage. The Ufm1−/− mutants showed reduced survival in the amastigote stage in vitro and ex vivo in human macrophages. This survival was restored by re-expression of wild type Ufm1 with concomitant induction of acetyl-CoA but not by re-expressing the non-conjugatable Ufm1, indicating the essential nature of Ufm1 conjugation and β-oxidation. Both cell cycle analysis and ultrastructural studies of Ufm1−/− parasites confirmed the role of Ufm1 in amastigote growth. The defect in vitro growth of amstigotes in human macrophages was further substantiated by reduced survival. Therefore, these studies suggest the importance of Ufm1 in Leishmania pathogenesis with larger impact on other organisms and further provide an opportunity to test Ufm1−/− parasites as drug and vaccine targets.
Keywords: Ubiquitin fold modifier 1, Ufm1, Ufm1mediated protein conjugation, β-oxidation, acetyl-CoA, cell division, trypanosomatid, Leishmania donovani, mitochondrial trifunctional protein, MTP
Introduction
Leishmaniasis is a spectrum of diseases caused by protozoan parasites belonging to several different Leishmania species. These blood borne pathogens are currently prevalent in 88 countries around the World with an estimated 2 million new cases each year (Kaye and Aebischer, 2011). At present there are no effective vaccines against any of the clinical forms of leishmaniasis. Recent advances in genome sequencing ushered in post-genomic analysis of Leishmania parasites in terms of parasite biology in the sand fly vector and mammalian host, including host responses (Kaye and Scott, 2011). Yet a comprehensive model depicting pathogenesis associated with any form of leishmaniasis remains elusive, partly because of the complexity that underlies the numerous factors that control parasite virulence.
Protein modifications by ubiquitin and ubiquitin-like proteins (ubl) are widely described in eukaryotes and more recently discovered even in prokaryotic cells (Pearce et al., 2008; Hochstrasser, 2009). The modification of target proteins by a ubl involves covalent attachment of ubl to a substrate protein (Kerscher et al., 2006). The best-known consequence of ubl conjugation is the targeting of proteins for degradation by the proteasome (Hershko and Ciechanover, 1998). In addition to proteasomal targeting, conjugation by ubl has been shown to affect a broad range of functions including subcellular localization, endocytosis, membrane trafficking, protein kinase activation, DNA repair, chromatin dynamics and protein-protein interactions (Chen and Sun, 2009). Ubiquitin-fold modifier 1 (Ufm1) has recently been identified as a novel protein-conjugating system, displaying a similar tertiary structure to ubiquitin (Komatsu et al., 2004). Attachment of Ufm1 to its substrate proteins has been shown to follow enzymatic reactions commonly found in many ubl conjugation reactions. Ufm1 is synthesized as a precursor form and processed C terminally by two specific proteases, UfSP1 and UfSP2 (Kang et al., 2007). The processed Ufm1 is activated by the E1-like enzyme, Uba5, and then transferred to an E2 enzyme, Ufc1. Finally the Ufm1 is covalently conjugated to the substrate proteins via an E3-like enzyme Ufl1 (Tatsumi et al., 2010). So far the only host protein, C20orf116, with unknown function that is modified by Ufm1 has been shown in mouse studies (Tatsumi et al., 2010, Lemaire et al., 2011). Interestingly, this protein has been shown to be upregulated in beta cells in the islets of Langerhans and knock down of Ufm1 resulted in increased apoptosis under ER stress (Lemaire et al., 2011).
A number of ubls such as SUMO, NEDD8, HUB1 and Urm1 are conserved in most parasitic protozoa with exceptions such as Ufm1, FAT10, FUB1 and ISG15 that appear to be absent in Plasmodium (Ponder and Bogyo, 2007). Analyses of SUMO mediated modifications revealed novel targets such as metacaspase-3 and secretory proteins with RING finger domains in T. cruzi (Hashimoto et al., 2010; Bayona et al., 2011), in T. brucei (Liao et al., 2010) and Plasmodium (Issar et al., 2008) suggesting the importance of such modifications in these organisms. The conjugation functions of ufm1 in Leishmania or other trypanosomatid parasites remain unknown even though homologs of the necessary enzymes, involved in the mammalian Ufm1 conjugation, are also conserved in these organisms. Recently we described the existence of the Ufm1 pathway in Leishmania (Gannavaram et al., 2011). This pathway in Leishmania, including homologs of ubiquitin fold modifier1 (Ufm1), E1 like enzyme Uba5 and E2 like enzyme Ufc1 is localized in the mitochondria. Interestingly the protein targets for Ufm1 conjugation in Leishmania (Gannavaram et al., 2011) are distinct from the single protein identified so far as a target in mammalian cells (Tatsumi et al., 2010).
In this report, we have demonstrated the in vivo conjugation of L. donovani ufm1 to its target mitochondrial trifunctional protein. Deletion of Ufm1 affects this interaction and results in loss of β-oxidation of fatty acids. In addition, we also observed that disruption of Ufm1 alters specifically the growth of the intracellular form of the Leishmania parasite in vitro and ex vivo.
Results
Generation of Ufm1−/− null mutant in promastigotes
To investigate the in vivo functions of the Ufm1 mediated conjugations in L. donovani, we generated a L. donovani promastigote Ufm1 null mutant by homologous recombination (Fig. 1A). The promastigotes were transfected by electroporation and the two alleles of Ufm1 were replaced by sequential recombination with targeting constructs containing hygromycin and neomycin markers flanked by DNA fragments corresponding to 5′ and 3′ untranslated regions of LdUfm1. The Southern blot analysis showed complete loss of the coding region of the LdUfm1 in double knockout mutants when hybridized with 32P labeled Ufm1 coding sequence as a probe (Fig. 1B, lane denoted Ufm1−/− in LdUfm1 blot). Loss of LdUfm1 expression in the LdUfm1−/− parasite was confirmed by Western blot analysis using anti-Ufm1 antibody (Fig. 1C, Ufm1−/− lane). LdUfm1 expression was restored by transfecting the promastigote null mutant cells with the pXG-Phleo vector containing the coding sequence of either wild type or a Ufm1 truncated at the C-terminus including the conjugatable glycine residue (Ufm1−/−+Ufm1WT, Ufm1−/−+Ufm1ΔC, Fig. 1C). Immunoblotting with an anti-tubulin antibody revealed equal loading of protein (Fig 1C).
Fig. 1. Ufm1 gene disruption in the L. donovani promastigote genome.
A. Schematic diagram showing design of constructs for LdUfm1 gene disruption in the L. donovani genome. Neo construct is flanked on the 5′ and 3′ sides with LdUfm1 5′-UTR and 3′-UTR respectively. The positions of SalI restriction sites are indicated. B. Southern blot analysis of the genomic DNA of Leishmania wild-type (WT) and LdUfm1 gene-deleted (Ufm1−/−) promastigote parasites with LdUfm1 ORF, Hygromycin (Hyg) and Neomycin (Neo) as 32P-labelled probes. The genomic DNA from the parasites digested with the restriction enzyme SalI and was used in the analysis. C. Western blot analysis of the lysates of wild-type (WT) and LdUfm1 gene-deleted (Ufm1−/−) and parasites reexpressing either conjugatable (Ufm1−/−+Ufm1WT) or non-conjugatable Ufm1 (Ufm1−/−+Ufm1ΔC). The blots were probed with an anti-Ufm1 antibody (α-Ufm1) or an anti-α tubulin antibody (anti-α-tubulin) as a loading control.
Demonstration of mitochondrial trifunctional protein (MTP) as an in vivo target for Ufm1 conjugation in amstigotes
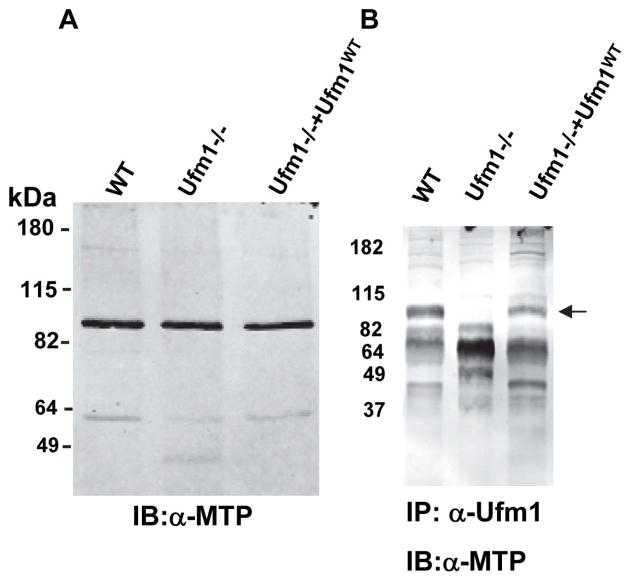
Previously we have identified proteins modified by Ufm1 conjugation in L. donovani amastigotes by mass spectrometry. One of the proteins identified was the mitochondrial trifunctional protein α-subunit (Gannavaram et al., 2011). The trifunctional protein involves three consecutive enzyme activities in the mitochondrial beta-oxidation of long-chain acyl-CoA esters: 2-enoyl-CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase and 3-ketoacyl-CoA thiolase. The mitochondrial β-oxidation produces acetyl-CoA and reduced NAD/FAD, with the acetyl-CoA feeding into the TCA cycle (Goetzman, 2011). In order to characterize the interaction between Ufm1 and the trifunctional protein in vivo in the amastigote forms, co-immunoprecipitation assay was performed with an anti-Ufm1 antibody and the blots were probed with an anti-human MTP antibody (Fig. 2B). To determine human MTP antibody will cross react with Leishmania MTP, we first demonstrated that anti-human peptide MTP antibody as well as anti-human full-length MTP antibody does react with Leishmania homolog of MTP in Leishmania amastigote lysates using an immunoblot (Fig. 2A; Fig. 1SA, respectively). Ufm1 does conjugate with MTP as revealed by the presence of a ~90kDa band corresponding to MTP in wild type Leishmania (Fig. 2B; WT) and in the parasite line re-expressing Ufm1 (Fig 2B. Ufm1−/−+Ufm1WT) but not in the Ufm1−/−, indicating the specificity of the molecular interaction between Ufm1 and MTP proteins (Fig 2B). Together, these results demonstrated that LdUfm1 can interact with mitochondrial trifunctional protein. This result is consistent with our previous observation that Ufm1 is predominantly found in the mitochondria of Leishmania and other trypanosomatids, as is the case with mitochondrial trifunctional protein (Gannavaram et al., 2011, Panigrahi et al., 2008).
Fig. 2. Validation of mitochondrial trifunctional protein as a target for conjugation by LdUfm1.
(A) Immunoblot analysis to show reactivity of anti-human MTP peptide antibodies with Leishmania donovani amastigote lysates. Protein lysates from wild type L. donovani cells (WT), Ufm1−/− or mutant amastigote parasites transfected with conjugatable Ufm1 (Ufm1−/−+Ufm1WT) were resolved on SDS-PAGE and the immunoblots were probed with anti-MTP antibodies. (B) Immunoblot analysis following immunoprecipitation reactions. Protein lysates from wild type L. donovani cells (WT), Ufm1−/− or mutant amastigote parasites transfected with conjugatable Ufm1 (Ufm1−/−+Ufm1WT) were used in co-immunoprecipitation reactions with α-Ufm1 antibodies and the eluates were resolved on SDS-PAGE and the immunoblots were probed with anti-human full length or peptide MTP antibodies. The bands corresponding to LdMTP are indicated with arrow mark.
Disruption of Ufm1 conjugation leads to reduced acetyl-CoA production in amstigotes
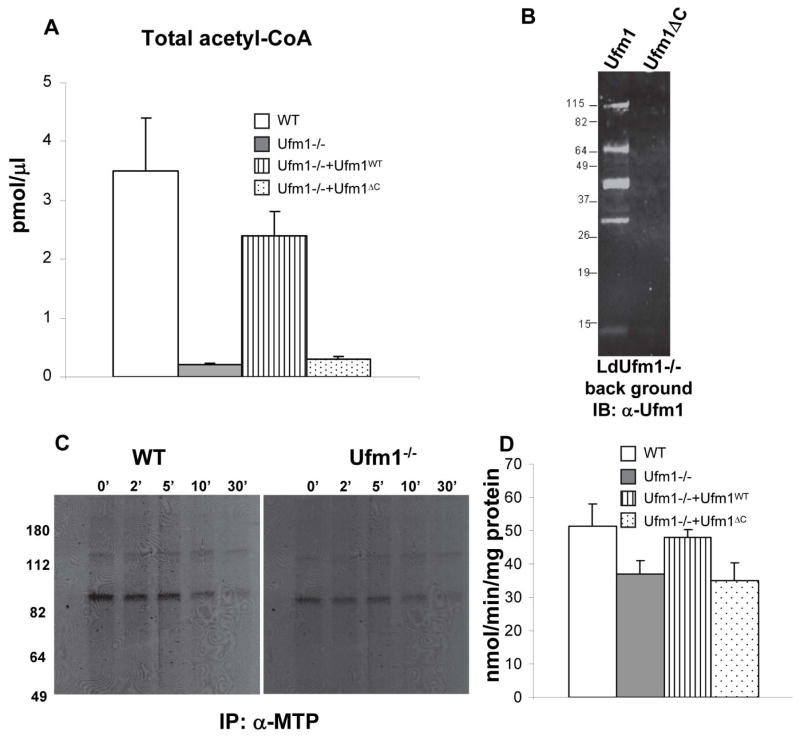
Mitochondrial trifunctional protein is a protein that catalyzes several reactions in beta oxidation leading to production of acetyl-CoA. Therefore any alterations in the conjugation of Ufm1 to the MTP might result in altered production of acetyl-CoA. To test this possibility, we determined the level of acetyl-CoA in Leishmania wild type cells, Ufm1−/− and in knock out cells re-expressing either conjugatable Ufm1 or non-conjugatable C-terminal deleted Ufm1. These experiments were performed with the axenic amastigotes. Results showed that Leishmania Ufm1−/− amastigotes produce significantly less acetyl-CoA compared to wild type cells (Fig. 3A WT vs Ufm1−/−). The acetyl-CoA levels are restored when a wild type Ufm1 is re-expressed in Ufm1−/− background (Fig. 3A Ufm1−/−+Ufm1WT) but not when a non-conjugatable Ufm1 is re-expressed (Fig. 3A Ufm1−/−+Ufm1ΔC). This showed that modification of MTP by Ufm1 is essential for its activities as revealed by the decreased acetyl-CoA synthesis when conjugation is blocked either in the knockout parasites or the mutant Ufm1 expressors. Restoration of Ufm1 conjugation in the cells reexpressing Ufm1WT in the knockout background was verified by the presence of Ufm1 conjugates that were purified under denaturing conditions using Ni-NTA agarose on an immunoblot of the purified conjugates with α-Ufm1 antibody (Fig. 3B Ufm1) and not in the cells lines re-expressing non conjugatable Ufm1 (Fig. 3B, Ufm1 delta C). The pattern of Ufm1 conjugates in the reexpressing cells was slightly different to that observed in the wild type demonstrated previously (Gannavaram et al, 2011). This might be because of the different Leishmania expression vectors (pKSNeo in Gannavaram et al., 2011 versus pXGPhleo in the current study) used in these experiments that rely on different regulatory sequences (A2 regulatory elements in pKSNeo and DHFR-TS elements in pXG-Phleo) driving the expression of the exogenous gene.
Fig. 3. Physiological effects of Ufm1 deletion in L. donovani amastigotes.
(A) Total acetyl-CoA was measured from the amastigotes of L. donovani wild-type (WT), or Ufm1 knock out (Ufm1−/−) or mutants reexpressing either conjugatable (Ufm1−/−+Ufm1WT) or non-conjugatable Ufm1 (Ufm1−/−+Ufm1ΔC) proteins. (B). Immunoblot showing the restoration of Ufm1 conjugation in the mutant parasites reexpressing the conjugatable (Ufm1−/−+Ufm1WT) or non-conjugatable Ufm1 (Ufm1−/−+Ufm1ΔC) proteins purified using nickel-NTA resin and probed with α-Ufm1 antibodies. (C) Autoradiograms showing the 35S-met labeled mitochondrial trifunctional protein immunoprecipitated from the labeled L. donovani wild type and Ufm1−/− amastigotes at the time points indicated (0, 2, 5, 10 and 30 minutes) using α-MTP antibodies. (D) 3-hydroxyacyl-CoA dehydrogenase activity was measured using acetoacetyl-CoA as substrate from the amastigotes of the L. donovani wild-type (WT), or Ufm1 knock-out (Ufm1−/−) or mutants re-expressing either conjugatable (Ufm1−/−+Ufm1WT) or non-conjugatable Ufm1 (Ufm1−/−+Ufm1ΔC) proteins.
To test whether the Ufm1 interaction with MTP influences its activity by altering the stability of MTP, we first measured the steady state levels of MTP in Leishmania wild type, Ufm1−/− and the Ufm1 re-expressing amastigotes. The Leishmania wild type, Ufm1−/− and the Ufm1 re-expressing parasites all showed similar levels of MTP suggesting that the Ufm1 conjugation or lack thereof does not affect the stability of MTP (Fig. 2A). To further test whether Ufm1 deletion has any effect on the stability of MTP, we performed pulse chase experiments. Leishmania wild type or Ufm1−/− amastigotes were labeled with 35S-Met and the stability of the radioactively labeled MTP was monitored by immunoprecipitation reactions using anti-MTP antibody at different time points after labeling. Results showed that the stability of MTP is not affected in the Ufm1−/− parasites as indicated by the absence of degradation products or the lack of loss of radioactivity associated with MTP bands on the autoradiogram during chase (Fig 3C WT vs Ufm1−/−). These results suggest that for the acetyl-COA production interaction between Ufm1 and MTP is essential and loss of Ufm1 does not impact MTP stability.
To further test the effect of deletion of Ufm1 on the enzymatic activities of MTP, that comprises of 3-hydroxyacyl-CoA dehydrogenase, 2-enoyl-CoA hydratase and 3-ketoacyl-CoA thiolase, we measured the activity of the 3-hydroxyacyl-CoA dehydrogenase, one of the three activities associated with the MTP, in the mitochondria isolated from either wild type or Ufm1−/− and the Ufm1 re-expressing amastigotes. The results showed a significant reduction in the dehydrogenase activity in the Ufm1−/− compared to wild type amastigotes (Fig 3D p<0.05). This result suggests that the reduction in total acetyl-CoA observed in Ufm1−/− amastigotes is in part due to the reduction in the 3-hydroxyacyl-CoA dehydrogenase dehydrogenase activity in the absence of Ufm1 protein.
Loss of Ufm1 expression results in reduced parasite growth in vitro and ex vivo
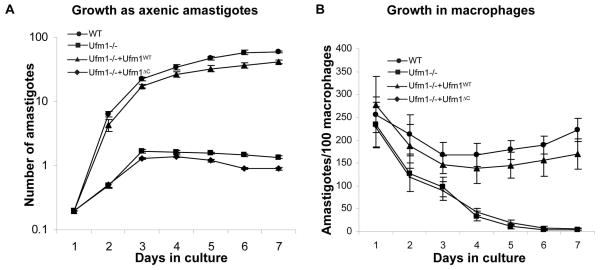
Since Ufm1-deficient L. donovani show defects in fatty acid metabolism in the amastigote stage of the parasite and the fatty acid metabolism is necessary for energy generation, we wanted to analyze what effect loss of Ufm1 has on growth in vitro as well as ex vivo in human macrophages. Survival of the axenic amastigotes was analyzed by counting viable cells over a period 7 days. The data showed that the Ufm1−/− failed to grow as amastigotes after 3 days in culture. Re-expression of wild type Ufm1 restored the growth. The growth was not restored when a C-terminal deleted Ufm1 was expressed in the Ufm1−/− background (Fig 4A). Next, we examined their growth in macrophages ex vivo. To this end, in vitro differentiated human macrophages were infected with stationary phase cultures of wild type, LdUfm1−/− and Ufm1 re-expressing promastigotes (Fig. 4B). The results at 6 h post-infection showed that the percentage of macrophages that are infected with the parasites was similar with all four cell types. These macrophage cultures were subsequently examined at 1, 2, 3, 4, 5, 6 and 7 days post-infection, and the percentage of infected macrophages was calculated. After 3 days the Ld Ufm1−/− and Ld Ufm1−/− with Ufm1ΔC expressing cells start to show significant decline in growth (Fig. 4B), whereas wild type control cells and add-back cells continue to grow inside macrophages (Fig. 4B). By day 6, Ld Ufm1−/− and Ld Ufm1−/− with Ufm1ΔC expressing cells were completely cleared from the macrophages. These results indicate that lack of Ufm1 conjugation with its cellular substrates could result in growth reduction in amastigotes.
Fig. 4. Effects of Ufm1 deletion on the survival of L. donovani amastigotes.
(A) The growth of L. donovani wild-type (WT), or Ufm1 knock out (Ufm1−/−) or mutants reexpressing either conjugatable (Ufm1−/−+Ufm1WT) or non-conjugatable Ufm1 (Ufm1−/−+Ufm1ΔC) proteins was monitored in axenic amastigote culture. (B) Human macrophages differentiated from monocytes were infected with stationary phase promastigote parasites from L. donovani wild type (WT), Ufm1 knock out (Ufm1−/−) or mutants reexpressing either conjugatable (Ufm1−/−+Ufm1WT) or non-conjugatable Ufm1 (Ufm1−/−+Ufm1ΔC) proteins for six hours (10:1 parasite-to-macrophage ratio) and the numbers of amastigotes in these cultures were determined over a period of 6 days by microscopic observation of Diff-quik reagent stained slides. The data are expressed as the number of amastigotes per 100 macrophages. Error bars indicate the standard deviation. The results are the mean of three independent experiments.
Ultrastructural studies indicate growth defects in Ufm1−/−amastigotes
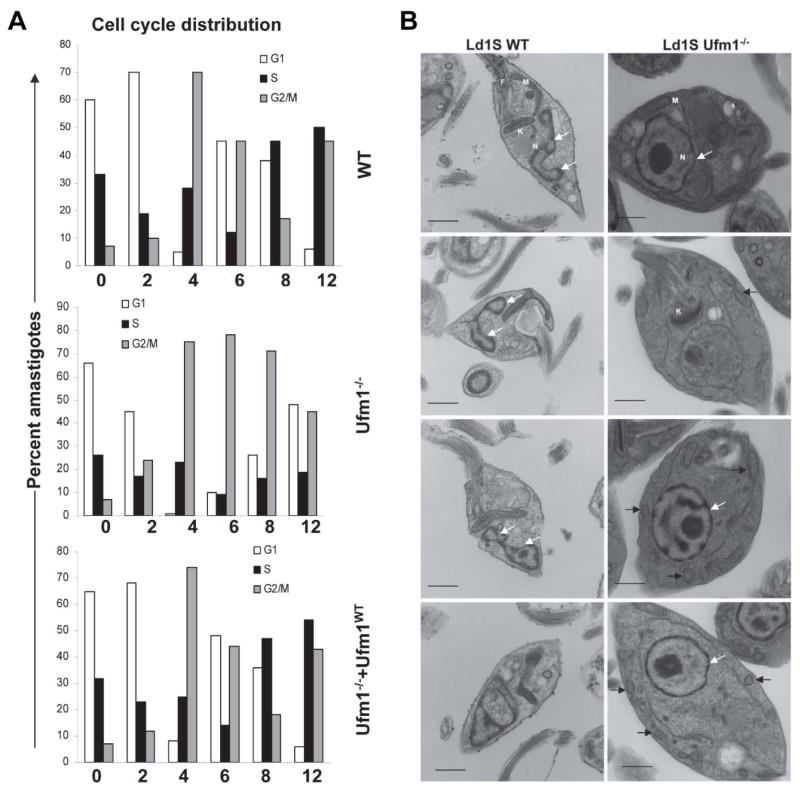
To analyze the cause of the growth arrest of axenic LdUfm1−/− amastigote cells, axenic amastigote cultures at the 24- time point were subjected to cell cycle analysis. The cells were synchronized at the G1-S boundary by hydroxy urea (HU) treatment prior to cell cycle analysis. Before the HU treatment, the characteristic profile of phase distribution observed in all the cell lines was: Ld1S: G0-G1, 53.1±0.9%; S, 25.0±1.27%; G2-M, 21.7±1.7%; Ld1SUfm1−/−: G0-G1, 58.4±0.7%; S, 15.1±5.4%; G2-M, 26.5±6.0%; Ld1SUfm1−/−+Ufm1WT: G0/G1, 49.52±3.75%; S, 38.29±3.9%; G2-M, 12.28±6.45%. The progression of the cell cycle phases was dissected by flow cytometry at 2 hour intervals after releasing the HU block. Approximately 90% of wild type cells, Ufm1−/− and the Ufm1 re-expressing cells were arrested in G1-S boundary by the overnight HU treatment. Upon releasing the HU block, the G2-M peak reached a maximum by 4 hours in WT, Ufm1−/− and Ufm1−/−+Ufm1WT in a synchronized manner. However, the subsequent progression of the cell cycle from G2-M to G1 was significantly delayed in Ufm1−/− cells, compared with wild type cells (Fig. 5A; 48% vs 5%, respectively). Re-expression of Ufm1 in the Ufm1−/− cells restored cell cycle progression (Fig. 5A; 5%). Cell cycle analysis also showed that the arrest in growth of the LdUfm1−/− axenic amastigotes occurred with a significant accumulation of cells in the G2-M phase (starting from 6 hr after HU release) indicating delay in their transit through this phase (Fig. 5A). The wild type and Ufm1 re-expressing cells undergo multiple DNA duplication events while the Ufm1−/− cells are unable to enter a DNA synthesis phase as evidenced by the DNA content (Fig. 5A). These results strongly suggest that, in addition to its essential role in the modification of MTP and thus β-oxidation of fatty acids, Ufm1 is also involved in Leishmania cell division through as yet unidentified substrate proteins.
Fig. 5. Cell cycle and ultrastuctural analysis of L. donovani wild type and Ufm1−/− axenic amastigotes.
(A). The log-phase amastigote cells (~1×107/ml) were synchronized at the G1-S border with 200 μg hydroxyurea for 12 hours. DNA content was measured after staining with propidium iodide (PI) and the cell cycle phases were analyzed by flow cytometry at 2 hour intervals up to 12 hours. The G1, S and G2-M distributions in various cell cycle phases at different time intervals were calculated from the actual data using MOD-FIT software. The values shown are means ± s.d. (B) Electron micrographs of L. donovani wild type and Ufm1−/− axenic amastigotes. One hundred images from both cell types were analyzed and a representative samples are shown. F, flagellum, M mitochondria, N nucleus, K kinetoplast, arrows point to mitochondria. White arrows show the nuclei and the black arrows indicate mitochondria.
To further investigate into the defect of amastigote cell cycle due to lack of Ufm1 we analyzed morphological abnormalities in the Ufm1−/− amastigotes compared to wild type cells by electron microscopy. In wild type cells, the nucleus undergoing division can be seen where as in the Ufm1−/− cells, such dividing nuclei were absent (Fig 5B). In wild type cells, mitochondria showed typical morphology where as in Ufm1−/− several fragmented mitochondria were apparent (Fig. 5B top and bottom panels, indicated by arrows).
Discussion
Ubiquitin-like protein modifiers (Ubls) regulate a broad range of biological functions including endocytosis, membrane trafficking, protein kinase activation, DNA repair and chromatin dynamics in eukaryotic cells (Hochstrasser, 2009). Recently we demonstrated the existence of Ufm1 and its conjugation pathway in L. donovani (Gannavaram et. al. 2011). In this report we generated Ufm1 knockout mutant parasites, LdUfm1−/−, to characterize the function of the Ufm1 pathway and its role in Leishmania pathogenesis. LdUfm1−/− parasites grow as promastigotes but are growth defective in the amastigote stage, suggesting the importance of LdUfm1 in the virulent form of the parasite. This is consistent with our earlier observations that Ufm1 mediated modifications are predominant only in amastigote stages. In that study we showed there were more Ufm1 conjugated proteins in the amastigote stage than the promastigote stage and also that alterations in Ufm1 conjugation by overexpression of mutant forms of Ufm1 selectively affect the growth of amastigotes (Gannavaram et al., 2011).
Ubls such as Ubiquitin, SUMO and NEDD have been identified in medically important parasitic protozoa are being investigated in these organisms (Hashimoto et al., 2010; Artavanis-Tsakonas et al., 2010). Recently, Ubl targets that are unique to parasitic protozoa have been described. For instance, several proteins including metacaspase-3, thymidine hydroxylase and histone acetyl transferase were found to be potential sumoylation targets in Trypanosoma cruzi (Bayona et al., 2011). Arguably, sumoylation of thymidine hydroxylase and histone acetyl transferase, DNA and chromatin modification proteins, might represent regulation of transcriptional activity in these parasites. However, metacaspases have been implicated in programmed cell death pathways in plants, yeasts and protozoan parasites including Leishmania and in T. brucei metacaspases have been shown to be associated with endosomes. (Helms et al., 2006, Lee et al., 2007, Meslin et al., 2011). Therefore modification of metacaspase by SUMO might represent novel functions in the protozoan parasites such as T. cruzi. Previously, we showed by mass spectrometry that there are several potential targets for LdUfm1 (Gannavaram et. al. 2011). Therefore, deletion of Ufm1 in L. donovani would facilitate investigation of Ufm1 function in L. donovani with respect to its targets.
Previous studies to test the effect of a ubl conjugation to a substrate protein typically relied on site directed mutagenesis of the specific lysine residue involved in covalent linkage with the particular ubl. An approach to identification of the target lysine includes the observation that the diglycine moiety from the ubl remains on a ubiquitinated lysine residue after trypsin digestion. Therefore a monoclonal antibody that enriches for peptides containing lysine residues modified by diglycine has been utilized to enrich ubiquitinated peptides (Xu et al., 2010). Still, identification of the ubiquitination site typically requires high protein coverage and is complicated by the difficulty of confirming that the identified proteins are indeed ubiquitinated. Given the difficulty in taking such an approach, Ufm1 knock out mutants in Leishmania represent an opportunity to investigate Ufm1 function in parasitic protozoa.
All eukaryotic cells are equipped to degrade a wide array of fatty acid species. The task is shared by mitochondria and peroxisomes with the latter generally handling the unusual species and mitochondria everything else. The two organelles functionally interact through transport of vesicles (Braschi et al., 2010). Many fatty acids are truncated in peroxisomes before degradation is completed in the mitochondria. The peroxisomal pathway produces acetyl-CoA, which is funneled into synthetic pathways such as isoprenoid synthesis but no energy is produced (Kovacs et al., 2007). The mitochondrial pathway produces acetyl-CoA and reduced NAD/FAD, with the acetyl-CoA progressing into the TCA cycle to yield additional reducing equivalents that are converted to ATP. Genome analyses suggested that Leishmania is capable of oxidizing fatty acids via βoxidation in two separate cellular compartments the glycosomes and the mitochondria. Homologs of enzymes involved in β-oxidation of fatty acids have been identified in Leishmania (Opperdoes and Szikora, 2006). Several acyl-CoA dehydrogenases with specificities for very long, long, intermediate and short/branched fatty acids were also found with a mitochondrial transit peptide indicating that Leishmania mitochondria are capable of utilizing a variety of fatty acids (Opperdoes and Michels, 2008). Importantly, amastigotes have been shown to contain greater activities than promastigotes of the enzymes that catalyse β-oxidation of fatty acids in L. mexicana, suggesting that βoxidation of fatty acids is relatively less important in promastigotes where glycolysis is the main source of metabolism and more important in the intracellular stage (Coombs et al., 1982). Further, a recent study that examined differentiation of L. donovani from promastigote to amastigote revealed the dynamics of the metabolome associated with the differentiation (Rosenzweig et al., 2008). During differentiation, the parasites shifted from glucose to fatty acid oxidation as the main source of metabolic energy. Consistent with this an increase in β-oxidation capacity including a ~13-fold increase in the expression of the long chain fatty acyl CoA synthetase, the rate-limiting enzyme of _βoxidation, up regulation of TCA cycle enzymes, mitochondrial respiratory chain, and oxidative phosphorylation proteins were observed during differentiation into amastigotes. In addition, differentiating parasites also up-regulated gluconeogenesis, producing sugars from glycerol and amino acids (Rosenzweig et al., 2008). These observations are consistent with our result that Ufm1 conjugation to MTP to modulate β-oxidation of fatty acids is pertinent in the amastigote stage. Further, in Leishmania amastigotes, Ufm1, a mitochondrial protein, modifies the mitochondrial trifunctional protein. This protein catalyzes the β-oxidation of fatty acids, culminating in the production of acetyl-CoA. The Ufm1−/− parasites were analyzed for their capacity to synthesize acetyl-CoA. Results showed that in the mutant parasites the interaction between Ufm1 and the Leishmania homolog of MTP is lost and as a result have reduced levels of acetyl-CoA, indicating the importance of Ufm1 conjugation in this process. To further explore the mechanism underlying reduction in acetyl-CoA production, we measured one of the three activities comprising of MTP and found that it was significantly reduced in Ufm1−/− parasites (Fig 3D). The alteration in the dehydrogenase activity in the Ufm1−/− amastigotes was not as drastic as to explain the reduction in acetyl-CoA, a product of the cumulative activities of the MTP (Fig 3A). It is likely that in the absence of Ufm1 the downstream activities such as hydratase and thiolase are also affected that might result in the severe reduction in the end product acetyl-CoA as observed in Fig. 3A. Future studies are necessary to measure the downstream activities to determine the relative contribution of the deficiencies to the acetyl-CoA levels observed.
Mammalian cells have multiple routes of energy acquisition. Particularly, knockout mice have been created for 12 fatty acid oxidation genes, including four of the acyl-CoA dehydrogenases, both subunits of trifunctional protein, short/medium-chain hydroxyacyl-CoA dehydrogenase, and two enzymes required for oxidation of polyunsaturated fatty acids indicating level of redundancy in mammalian metabolism (Goetzman, 2011). However, Leishmania amastigotes survive in the phagolysosomal compartment where limited nutrient availability imposes metabolic constraints on the parasites. Therefore parasite specific optimizations may be necessary and it may be argued that Ufm1 conjugation to MTP represents such optimization. Since Ufm1 conjugation to MTP results in the generation of acetyl-CoA due to β-oxidation of long chain fatty acids and acetyl-COA is needed in the TCA cycle to generate energy i.e. ATP, it is possible that alteration of Ufm1-MTP conjugation could lead to energy deficit that could lead to defect in cell cycle and growth as observed in the present study. Further, morphological changes such as lack of nuclear division and mitochondrial fragmentation observed due to lack of Ufm1 could be directly correlated with deficit in Ufm1-MTP conjugation or conjugation with as yet unidentified substrate proteins in amastigotes since we have previously shown that Ufm1 can conjugate to multiple proteins in amastigote stages.
In conclusion, our study demonstrate for the first time that Ufm1 mediates protein modifications and its role in Leishmania pathogenesis that have not been described in other organisms. Parasite stage specific modifications by ubls appear to be a common paradigm in parasites. Studies of Ubl mediated modifications in T. cruzi, T. brucei and L. donovani show evidence to this effect. Studies of Ufm1 mediated modifications in parasitic protozoa are likely to lead to a deeper understanding of their contribution to parasite pathogenesis. Further, since Ufm1 deficiency does not affect promastigote stages, other parts of the Ufm1 conjugation system such as Uba5 and Ufc1 may also be targets for deletion by genetic approaches. Such mutants would further help in revealing the functions of these proteins. It is of particular importance that growth of Ufm1 knock-out mutant is attenuated in the amastigote stages, as such stage specific attenuation makes it a promising candidate to be tested as a vaccine. In addition, the studies described here with Ufm1 in Leishmania could help in the understanding of its function in other organisms.
Experimental procedures
Plasmids and parasite cultures
Leishmania donovani promastigotes (strain 1S, WHO designation: MHOM/SD/62/1S) were grown in M199 medium containing 10% heat inactivated fetal bovine serum. Promastigotes were transfected by electroporation and selected for growth in medium containing Geneticin (G418) up to 100μg/ml or Hygromycin up to 50μg/ml or both. These drug-resistant cells were used in all subsequent experiments. Axenic amastigotes of wild type or the mutant lines were generated following a published protocol (Debrabant et al., 2004). Such amastigotes have been demonstrated to have gene expression signatures consistent with the tissue derived amastigotes (Duncan et al, 2001).The Leishmania expression plasmid pXG-Phleo (Freedman and Beverley, 1993) was used to express either the full-length or where indicated, the mutant forms of L. donovani Ufm1, in transfected Leishmania parasites. For this purpose, the full-length gene encoding L. donovani Ufm1 was amplified with oligos based on the L. infantum putative Ufm1 sequence. The point mutation G98A was introduced by PCR to generate Ufm1 mutants. These oligos introduced 6His and HA tags at the N′ terminus of the fusion protein and contained BamHI restriction sites on either end. The BamHI insert was subcloned into the BamHI site of pXG-Phleo, resulting in plasmid constructs that were used to overexpress wild type or mutant Ufm1 in Leishmania transfectants.
Immunoprecipitation analysis
For immunoprecipitation analysis, 1×108 Leishmania amastigotes were lysed in 1 ml of NET buffer (150mM NaCl, 1mM EDTA, 10mM Tris–HCl, pH 7.5, 1% Nonidet P-40 with protease inhibitor cocktail) and the lysate was centrifuged at 12000 rpm for 20 min at 4°C to collect the supernatant. Five μl of anti-Ufm1 antibodies were added to 500μl to the lysate and the mixture was incubated under constant rotation overnight at 4°C. The complexes were immunoprecipitated with 25μl of Protein-A Sepharose beads by incubation under constant rotation at 4°C for 1hr. The precipitated complexes were washed five times with ice-cold NET buffer and eluted by boiling for 5 min in SDS sample buffer in the presence of β-mercaptoethanol. The supernatant was subjected to SDS–PAGE and analyzed by immunoblots with anti-MTP antibodies (Novus Biologicals).
For purification of 6xHis-tagged proteins under denaturing conditions, 2×108 transfectant Leishmania axenic amastigotes were lysed in 1 ml of denaturing lysis buffer (6M Gu-HCl, 0.1M NaH2PO4, and 0.01M Tris-HCl, pH 8.0, 5mM imidazole with 20mM N-ethylmaleimide) and the lysate was sonicated for 10 seconds and then centrifuged at 12000 rpm for 20 min at room temperature to remove debris. To the supernatant, 30 μl of Ni-NTA Agarose (Qiagen) was added and the mixture was incubated under constant rotation for 4 hours at room temperature. The precipitated material was washed five times with denaturing wash buffer (6M Gu-HCl, 0.1M NaH2PO4, and 0.01M Tris-HCl, pH 8.0) followed by buffer containing 8M urea, 0.1M NaH2PO4, 0.1% Triton-X100 and 0.01M Tris-HCl, pH 6.3. The complexes were eluted in elution buffer (0.1M NaH2PO4, 30% glycerol, 5% SDS, 0.2M imidazole and 0.15M Tris-HCl, pH 6.7). The resulting lysates were subjected to SDS-PAGE and analyzed by immunoblots with anti-Ufm1 antibodies.
35S-Met labeling and immunoprecipitation
35S-Methionine-labeled proteins were synthesized using axenic amastigotes according the methods described previously (Debrabant et al., 2002). 4×108 cells were labeled for 2 minutes and cells were collected after defined intervals of time. The cell lysates were incubated at 4°C for 6 h with the anti-human MTP antibodies. 40 μl of ProteinA-Sepharose beads were then added to each reaction and allowed to equilibrate for 1 h. The beads were washed 3 times (5 min each) in the immunoprecipitation buffer containing Tris pH 8.0, NaCl 100 mM, NP-40 0.5%, MgCl2 5 mM and DTT 1 mM. The bound proteins were eluted with SDS-PAGE sample buffer, analyzed by SDS-PAGE and fluorography.
Enzymatic activity
The activity of 3-hydroxyacyl-CoA dehydrogenase was measured in mitochondria isolated from the Leishmania amastigotes following the procedure published previously (Wanders et al., 1990). The activity was measured by using acetoacetyl-CoA as a substrate in an activity buffer containing 50mM MES (pH 6.16), 100mM potassium phosphate, 01mM DTT, 0.1% triton X-100, 0.1mM NADH and the decrease in absorbance at 340nm was measured.
Macrophage infection
Human elutriated monocytes were resuspended at 1.8×105 cells/ml in RPMI medium containing 10% FBS and macrophage colony-stimulating factor (20 ng/ml, ProSpec, Israel), plated at 0.5 ml/well on eight-chamber Lab-Tek tissue-culture slides (Miles Laboratories) and incubated for 9 days for differentiation into macrophages. The macrophage infection experiments were performed essentially as described earlier (Gannavaram et al., 2011).
Generation of a targeting construct for deletion of the LdUfm1 gene
The drug resistance markers hygromycin and neomycin were used to obtain LdUfm1−/−. To generate the targeting construct, a 920bp fragment from the 5′ region and an 823bp fragment from the 3′ region flanking the LdUfm1 open reading frame were amplified by PCR using L. donovani genomic DNA. The primers used to amplify 5′ flanking fragment included restriction sites HindIII and SpeI. Similarly, the primers added NotI and XbaI sites to the 3′ flanking fragment. The drug resistance markers hygromycin and neomycin were amplified with primers that add SpeI and NotI to the open reading frame. These DNA fragments were subcloned into the pCR2.1-Topo vector and the nucleotide sequence was determined to ensure fidelity. The plasmid containing the 5′ flanking fragment was digested with HindIII/SpeI, gel purified and ligated into a similarly digested plasmid containing either hygromycin or neomycin. The resultant plasmids, containing both the 5′flanking region and the drug resistance markers were digested with NotI/XbaI and the 3′flanking fragment isolated by NotI/XbaI digestion was ligated in to these sites. The authenticity of the final plasmid was confirmed by DNA sequencing. For the purpose of transfection, the targeting construct was prepared by digestion with HindIII/XbaI, which cuts out a linear fragment containing the Ufm1 5′ flanking sequence, the hygromycin/neomycin genes and the Ufm1 3′ flanking sequence. The fragment was gel purified and used in transfection.
Complementation of LdUfm1 in LdUfm1−/− parasites
To restore Ufm1 expression in the LdUfm1−/− parasites, the LdpUfm1 ORF was first PCR amplified using a LdUfm1 containing plasmid as template and the following common forward primer:5′-GGGGATCCATGTACCCATACGACGTCCCTGAC-3′, and reverse primers: 5′-GGGGATCCTCAACCGACGCGATCACGCGGAATCAGTCGGATCTC-3′ or 5′-GGGGATCCTCAGACGCGATCACGCGGAATCAGTCGGATCTC-3′ that amplify a wild type Ufm1 coding sequence or a ΔC variant ending in a C-terminal valine residue in two independent amplification reactions. The amplified product was subcloned into the pCR2.1-TOPO cloning vector. The fidelity of the cloned sequence was verified by nucleotide sequencing. The BamHI insert was ligated into the BamHI site of the pXG-Phleo vector (Freedman and Beverley, 1993) and the recombinant plasmids, pXG-Phleo-LdUfm1WT or pXG-Phleo-LdUfm1ΔC were transfected into the LdUfm1−/− promastigotes as described previously (Gannavaram et al., 2008). Transfected promastigotes were selected with minimal dose of phleomycin (10 mg/ml).
Isolation of genomic DNA and Southern blot analysis
Total genomic DNA was isolated from promastigotes with the Wizard genomic DNA purification kit (Promega Biosciences), following the method suggested by the manufacturer. The DNA was digested with restriction endonuclease SalI and separated on 1% agarose gels. Southern blot analysis of the resolved DNA was done as described previously using a 32P-labelled LdUfm1coding sequence as a probe (Selvapandiyan et al., 2004).
Measurement of acetyl-CoA
Acetyl-CoA measurements were made using the PicoProbe acetyl CoA assay kit (Abcam) according to the protocol suggested by the manufacturer. The amastigote cell pellets (2×109 cells) after 24 hours of differentiation under axenic culture conditions were frozen on dry ice with 2 μl 1N perchloric acid for the purpose of deproteinization. The homogenate was centrifuged at 10,000Xg for 5 minutes. The supernatant was neutralized with 3M KHCO3, adding 1μl aliquots/10 μl supernatant until bubble evolution ceased. After incubation on ice for 5 minutes, the supernatant was centrifuged at 10,000xg for 2 minutes to pellet KClO4. The supernatant was used in acetyl-CoA estimation. To correct for background created by free CoASH and succ-CoA in samples, 10 μl of CoASH Quencher was added to each background sample to quench free CoA, then 2 μl of Quench Remover was added. Fluorescence was measured (Ex/Em :535/589nm) with a plate reader and the concentration of acetyl-CoA (pmol) was measured using a standard curve.
Electron microscopy
Pelleted cells were fixed with 2.5% glutaraldehyde, 0.5mM MgCl2, 0.25mM CaCl2, 0.12M sodium cacodylate buffer pH 7.2 at room temp for 20 min. then on ice for 40 min. After buffer washes cells were post-fixed for 1 hour on ice in 1% osmium tetroxide, 0.12M sodium cacodylate buffer pH 7.2. Samples were block stained in 1% uranyl acetate, dehydrated in graded ethanol solutions, and embedded in EMbed-812 (Electron Microscopy Sciences, Hatfield PA). Thin sections were stained with uranyl acetate and lead citrate and examined on a JEM 1400 electron microscope ( JEOL USA, Peabody MA) with an AMT XR-60 digital camera (Advanced Microscopy Techniques Corporation, Woburn MA).
Cell cycle analysis
For flow cytometry analysis, cells were fixed as described (Dvorak, 1993) with slight modifications. Briefly, 1×107 cells from Ld1sWT, LdUfm1−/− or LdUfm1−/−+Ufm1cultures (asynchronous) were centrifuged at 8000 rpm for 3 minutes, washed with cold PBS and resuspended in 50 μl phosphate-buffered saline (pH 7.2). The cell suspension was mixed with 150 μl of fixative solution (1% Triton X-100, 40 mM citric acid, 20 mM sodium phosphate, 200 mM sucrose) and incubated at room temperature for 5 minutes. Finally, 350 μl of diluent buffer (125 mM MgCl2 in PBS) was added and the samples were stored at 4°C until further use. The fixed cells were treated with 50 μg RNase (5 mg/ml in 0.2 M sodium phosphate buffer, pH 7.0) for 3 hours at 37°C. Then, 50 μg/ml propidium iodide (5 mg/ml in 1.12% sodium citrate) was added and the tubes were incubated at 25°C for 1 hour. The samples were left overnight for equilibration at 4°C. The samples were analyzed on a FACS Canto (Becton Dickinson), and the proportions of G1, S and G2-M populations were determined using Flowjo software. Around 20,000 events were collected for each sample. For synchronization experiments, cells were maintained in exponential growth phase (≥107 cells/ml). Leishmania cells (1×107cells/ml) were centrifuged and transferred into fresh M-199 containing 200 μg/ml hydroxyurea and incubated at 25°C for 12 hours. The cells were washed twice with PBS and resuspended in fresh M-199 containing 10% FCS without hydroxyurea. Aliquots were taken at regular time intervals and the samples were processed for FACS analysis after addition of propidium iodide as described above.
Supplementary Material
References
- Artavanis-Tsakonas K, Weihofen WA, Antos JM, Coleman BI, Comeaux CA, Duraisingh MT, Gaudet R, Ploegh HL. Characterization and structural studies of the Plasmodium falciparum ubiquitin and Nedd8 hydrolase UCHL3. J Biol Chem. 2010;285:6857–66. doi: 10.1074/jbc.M109.072405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson HJ, Babbitt PC, Sajid M. The global cysteine peptidase landscape in parasites. Trends Parasitol. 2009;25:573–581. doi: 10.1016/j.pt.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayona JC, Nakayasu ES, Laverriere M, Aguilar C, Sobreira TJ, Choi H, Nesvizhskii AI, Almeida IC, Cazzulo JJ, Alvarez VE. SUMOylation pathway in Trypanosoma cruzi: functional characterization and proteomic analysis of target proteins. Mol Cell Proteomics. 2011;10:M110 007369. doi: 10.1074/mcp.M110.007369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braschi E, Goyon V, Zunino R, Mohanty A, Xu L, McBride HM. Vps35 mediates vesicle transport between the mitochondria and peroxisomes. Curr Biol. 2010;20:1310–1315. doi: 10.1016/j.cub.2010.05.066. [DOI] [PubMed] [Google Scholar]
- Braschi E, McBride HM. Mitochondria and the culture of the Borg: understanding the integration of mitochondrial function within the reticulum, the cell, and the organism. Bioessays. 2010;32:958–966. doi: 10.1002/bies.201000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Chung DW, Ponts N, Cervantes S, Le Roch KG. Post-translational modifications in Plasmodium: more than you think! Mol Biochem Parasitol. 2009;168:123–134. doi: 10.1016/j.molbiopara.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Coombs GH, Craft JA, Hart DT. A comparative study of Leishmania mexicana amastigotes and promastigotes. Enzyme activities and subcellular locations. Mol Biochem Parasitol. 1982;5:199–211. doi: 10.1016/0166-6851(82)90021-4. [DOI] [PubMed] [Google Scholar]
- Debrabant A, Joshi MB, Pimenta PF, Dwyer DM. Generation of Leishmania donovani axenic amastigotes: their growth and biological characteristics. Int J Parasitol. 2004;34:205–217. doi: 10.1016/j.ijpara.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Debrabant A, Lee N, Pogue GP, Dwyer DM, Nakhasi HL. Expression of calreticulin P-domain results in impairment of secretory pathway in Leishmania donovani and reduced parasite survival in macrophages. Int J Parasitol. 2002;32:1423–1434. doi: 10.1016/s0020-7519(02)00134-0. [DOI] [PubMed] [Google Scholar]
- Donnelly S, Dalton JP, Robinson MW. How pathogen-derived cysteine proteases modulate host immune responses. Adv Exp Med Biol. 2011;712:192–207. doi: 10.1007/978-1-4419-8414-2_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan R, Alvarez R, Jaffe CL, Wiese M, Klutch M, Shakarian A, Dwyer D, Nakhasi HL. Early response gene expression during differentiation of cultured Leishmania donovani. Parasitol Res. 2001;87:897–906. doi: 10.1007/s004360100464. [DOI] [PubMed] [Google Scholar]
- Dvorak JA. Analysis of the DNA of parasitic protozoa by flow cytometry. Methods Mol Biol. 1993;21:191–204. doi: 10.1385/0-89603-239-6:191. [DOI] [PubMed] [Google Scholar]
- Freedman DJ, Beverley SM. Two more independent selectable markers for stable transfection of Leishmania. Mol Biochem Parasitol. 1993;62:37–44. doi: 10.1016/0166-6851(93)90175-w. [DOI] [PubMed] [Google Scholar]
- Gannavaram S, Sharma P, Duncan RC, Salotra P, Nakhasi HL. Mitochondrial associated ubiquitin fold modifier-1 mediated protein conjugation in Leishmania donovani. PLoS One. 2011;6:e16156. doi: 10.1371/journal.pone.0016156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannavaram S, Vedvyas C, Debrabant A. Conservation of the pro-apoptotic nuclease activity of endonuclease G in unicellular trypanosomatid parasites. J Cell Sci. 2008;121:99–109. doi: 10.1242/jcs.014050. [DOI] [PubMed] [Google Scholar]
- Goetzman ES. Modeling disorders of fatty acid metabolism in the mouse. Prog Mol Biol Transl Sci. 2011;100:389–417. doi: 10.1016/B978-0-12-384878-9.00010-8. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Murata E, Aoki T. Secretory protein with RING finger domain (SPRING) specific to Trypanosoma cruzi is directed, as a ubiquitin ligase related protein, to the nucleus of host cells. Cell Microbiol. 2010;12:19–30. doi: 10.1111/j.1462-5822.2009.01375.x. [DOI] [PubMed] [Google Scholar]
- Helms MJ, Ambit A, Appleton P, Tetley L, Coombs GH, Mottram JC. Bloodstream form Trypanosoma brucei depend upon multiple metacaspases associated with RAB11-positive endosomes. J Cell Sci. 2006;119:1105–17. doi: 10.1242/jcs.02809. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issar N, Roux E, Mattei D, Scherf A. Identification of a novel post-translational modification in Plasmodium falciparum: protein sumoylation in different cellular compartments. Cell Microbiol. 2008;10:1999–2011. doi: 10.1111/j.1462-5822.2008.01183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SH, Kim GR, Seong M, Baek SH, Seol JH, Bang OS, Ovaa H, Tatsumi K, Komatsu M, Tanaka K, Chung CH. Two novel ubiquitin-fold modifier 1 (Ufm1)-specific proteases, UfSP1 and UfSP2. J Biol Chem. 2007;282:5256–5262. doi: 10.1074/jbc.M610590200. [DOI] [PubMed] [Google Scholar]
- Kaye PM, Aebischer T. Visceral leishmaniasis: immunology and prospects for a vaccine. Clin Microbiol Infect. 2011;17:1462–1470. doi: 10.1111/j.1469-0691.2011.03610.x. [DOI] [PubMed] [Google Scholar]
- Kedzierski L, Zhu Y, Handman E. Leishmania vaccines: progress and problems. Parasitology. 2006;133(Suppl):S87–112. doi: 10.1017/S0031182006001831. [DOI] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Chiba T, Tatsumi K, Iemura S, Tanida I, Okazaki N, Ueno T, Kominami E, Natsume T, Tanaka K. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J. 2004;23:1977–1986. doi: 10.1038/sj.emboj.7600205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahav T, Sivam D, Volpin H, Ronen M, Tsigankov P, Green A, Holland N, Kuzyk M, Borchers C, Zilberstein D, Myler PJ. Multiple levels of gene regulation mediate differentiation of the intracellular pathogen Leishmania. FASEB J. 2011;25:515–525. doi: 10.1096/fj.10-157529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Gannavaram S, Selvapandiyan A, Debrabant A. Characterization of metacaspases with trypsin-like activity and their putative role in programmed cell death in the protozoan parasite Leishmania. Eukaryot Cell. 2007;6:1745–57. doi: 10.1128/EC.00123-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire K, Moura RF, Granvik M, Igoillo-Esteve M, Hohmeier HE, Hendrickx N, Newgard CB, Waelkens E, Cnop M, Schuit F. Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PLoS One. 2011;6:e18517. doi: 10.1371/journal.pone.0018517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Wang T, Fan K, Tu X. The small ubiquitin-like modifier (SUMO) is essential in cell cycle regulation in Trypanosoma brucei. Exp Cell Res. 2010 doi: 10.1016/j.yexcr.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Listovsky T, Brandeis M, Zilberstein D. Leishmania express a functional Cdc20 homologue. Biochem Biophys Res Commun. 2011;408:71–77. doi: 10.1016/j.bbrc.2011.03.118. [DOI] [PubMed] [Google Scholar]
- McConville MJ, Naderer T. Metabolic pathways required for the intracellular survival of Leishmania. Annu Rev Microbiol. 2011;65:543–561. doi: 10.1146/annurev-micro-090110-102913. [DOI] [PubMed] [Google Scholar]
- Meslin B, Beavogui AH, Fasel N, Picot S. Plasmodium falciparum metacaspase PfMCA-1 triggers a z-VAD-fmk inhibitable protease to promote cell death. PLoS One. 2011;6:e23867. doi: 10.1371/journal.pone.0023867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Opperdoes FR, Coombs GH. Metabolism of Leishmania: proven and predicted. Trends Parasitol. 2007;23:149–158. doi: 10.1016/j.pt.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Panigrahi AK, Zikova A, Dalley RA, Acestor N, Ogata Y, Anupama A, Myler PJ, Stuart KD. Mitochondrial complexes in Trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol Cell Proteomics. 2008;7:534–545. doi: 10.1074/mcp.M700430-MCP200. [DOI] [PubMed] [Google Scholar]
- Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder EL, Bogyo M. Ubiquitin-like modifiers and their deconjugating enzymes in medically important parasitic protozoa. Eukaryot Cell. 2007;6:1943–1952. doi: 10.1128/EC.00282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig D, Smith D, Opperdoes F, Stern S, Olafson RW, Zilberstein D. Retooling Leishmania metabolism: from sand fly gut to human macrophage. FASEB J. 2008;22:590–602. doi: 10.1096/fj.07-9254com. [DOI] [PubMed] [Google Scholar]
- Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvapandiyan A, Debrabant A, Duncan R, Muller J, Salotra P, Sreenivas G, Salisbury JL, Nakhasi HL. Centrin gene disruption impairs stage-specific basal body duplication and cell cycle progression in Leishmania. J Biol Chem. 2004;279:25703–25710. doi: 10.1074/jbc.M402794200. [DOI] [PubMed] [Google Scholar]
- Selvapandiyan A, Dey R, Gannavaram S, Lakhal-Naouar I, Duncan R, Salotra P, Nakhasi HL. Immunity to visceral leishmaniasis using genetically defined live-attenuated parasites. J Trop Med. 2012;2012:631460. doi: 10.1155/2012/631460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi K, Sou YS, Tada N, Nakamura E, Iemura S, Natsume T, Kang SH, Chung CH, Kasahara M, Kominami E, Yamamoto M, Tanaka K, Komatsu M. A novel type of E3 ligase for the Ufm1 conjugation system. J Biol Chem. 2010;285:5417–5427. doi: 10.1074/jbc.M109.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi K, Sou YS, Tada N, Nakamura E, Iemura SI, Natsume T, Kang SH, Chung CH, Kasahara M, Kominami E, Yamamoto M, Tanaka K, Komatsu M. A novel type of E3-ligase for the Ufm1-conjugation system. J Biol Chem. 2009 doi: 10.1074/jbc.M109.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi K, Yamamoto-Mukai H, Shimizu R, Waguri S, Sou YS, Sakamoto A, Taya C, Shitara H, Hara T, Chung CH, Tanaka K, Yamamoto M, Komatsu M. The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nat Commun. 2011;2:181. doi: 10.1038/ncomms1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders RJ, IJIst L, van Gennip AH, Jakobs C, de Jager JP, Dorland L, van Sprang FJ, Duran M. Long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency: identification of a new inborn error of mitochondrial fatty acid beta-oxidation. J Inherit Metab Dis. 1990;13:311–314. doi: 10.1007/BF01799383. [DOI] [PubMed] [Google Scholar]
- Williams RA, Woods KL, Juliano L, Mottram JC, Coombs GH. Characterization of unusual families of ATG8-like proteins and ATG12 in the protozoan parasite Leishmania major. Autophagy. 2009;5:159–172. doi: 10.4161/auto.5.2.7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xirodimas DP, Sundqvist A, Nakamura A, Shen L, Botting C, Hay RT. Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep. 2008;9:280–286. doi: 10.1038/embor.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WW, Charest H, Ghedin E, Matlashewski G. Identification and overexpression of the A2 amastigote-specific protein in Leishmania donovani. Mol Biochem Parasitol. 1996;78:79–90. doi: 10.1016/s0166-6851(96)02612-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.