Abstract
Ethylene metabolism in higher plants is regulated by a wide array of endogenous and environmental factors. During most physiological processes, ethylene levels are mainly determined by a strict control of the rate-limiting biosynthetic steps responsible for the production of 1-aminocyclopropane-1-carboxylic acid (ACC) and its subsequent conversion to ethylene. Responsible for these reactions, the key enzymes ACC synthase and ACC oxidase are encoded by multigene families formed by members that can be differentially regulated at the transcription and post-translational levels by specific developmental and environmental signals. Among the wide variety of environmental cues controlling plant ethylene production, light quality, duration, and intensity have consistently been demonstrated to influence the metabolism of this plant hormone in diverse plant tissues, organs, and species. Although still not completely elucidated, the mechanisms underlying the interaction between light signal transduction and ethylene evolution appears to involve a complex network that includes central transcription factors connecting multiple signaling pathways, which can be reciprocally modulated by ethylene itself, other phytohormones, and specific light wavelengths. Accumulating evidence has indicated particular photoreceptors as essential mediators in light-induced signaling cascades affecting ethylene levels. Therefore, this review specifically focuses on discussing the current knowledge of the potential molecular mechanisms implicated in the light-induced responses affecting ethylene metabolism during the regulation of developmental and metabolic plant responses. Besides presenting the state of the art in this research field, some overlooked mechanisms and future directions to elucidate the exact nature of the light–ethylene interplay in higher plants will also be compiled and discussed.
Keywords: ACC, ACO, ACS, ethylene, light signaling, photoreceptors, phytochrome
INTRODUCTION
Light is one of the most influential and versatile environmental stimuli controlling plant life. It varies not only in quantity (fluence) but also in quality (wavelength), periodicity (photoperiod), and direction (unidirectional or diffuse). As photoautotroph and sessile organisms, most higher plants rely on sophisticated and plastic mechanisms to use light as both an energy source and abiotic signal to control decisive developmental adjustments. Not unexpectedly, during evolution plants have evolved a variety of photosensory systems that perceive the light environment and integrate this information into intrinsic developmental programs. Acting at the interface between the external and the internal plant environments, four main families of photoreceptors are responsible for the perception of light signals and their transduction through an array of gene expression modifications that will ultimately lead to adjustments in the plant growth and morphogenic patterns—a sequence of light-triggered processes collectively known as photomorphogenesis (Neff et al., 2000; Ma et al., 2001; Gyula et al., 2003; Franklin et al., 2005; Franklin and Quail, 2010).
During photomorphogenic responses the initial light cue is often translated into changes in the hormonal homeostasis in particular tissues or even throughout the entire plant. Hence, by modulating their hormonal status plants can rapidly adjust growth fitness to constantly changing environments (Li et al., 2004; Achard et al., 2006; Wolters and Jürgens, 2009). The early development of seedlings, which in nature is often skotomorphogenic, and the transition to photomorphogenic growth upon light exposure is one of the best-studied light-controlled processes in eudicotyledons, and it is tightly regulated by an intricate interplay between light signals and plant hormones. In fact, most of the current knowledge about light signaling pathways and ethylene biosynthesis, signal transduction, and response pathways has arisen from experimental approaches based on the etiolated seedling system. The simplicity of the light-induced responses in dark-grown seedlings made it ideal for quickly identifying photomorphogenic mutants in the model plant Arabidopsis thaliana (Liscum and Hangarter, 1993; Stepanova and Ecker, 2000; Li et al., 2004; Stepanova and Alonso, 2005; Chen and Chory, 2011; Boron and Vissenberg, 2014). In addition, the triple response phenotype of etiolated seedlings triggered by ethylene has also been proven as a simple and useful trait to screen for ethylene mutants in the same model plant (Bleecker et al., 1988; Guzman and Ecker, 1990; Lin et al., 2009).
Ethylene is an important growth regulator of numerous developmental aspects during plant life cycle (e.g., both vegetative and reproductive development, and responses to biotic and abiotic stresses), playing a key role in signaling pathways responsible for adaptive adjustments of plant’s fitness in a continuously fluctuating array of environmental signals. Moreover, ethylene biosynthesis is highly regulated by both developmental and external inputs, and stress-induced ethylene production can mediate multiple physiological and morphological responses involved in redirecting all required resources from standard growth to promote plant defense, resistance, resource forage, and/or “escape” mechanisms, including senescence, abscission, and plastic alterations in tissue/organ elongation and shoot–root ratios. However, the variable degree of plasticity observed in such ethylene-mediated responses seems to be dependent on the environmental challenge and species-specific features (Pierik et al., 2006, 2007; Lin et al., 2009; Wolters and Jürgens, 2009).
The development of photomorphogenic and hormonal mutant collections together with a considerable improvement in the experimental methods employed in molecular and genetics research over the last years have paved the way for elegant studies that have begun to shed light on the “black box” of how light signaling controls ethylene production to coordinate plant development. For example, it has been shown that light can modulate ethylene biosynthesis through particular photoreceptor-mediated pathways. Moreover, some transcription factors have been identified as potential integrators for light signaling and ethylene biosynthesis modulation. Furthermore, many regulators of ethylene biosynthesis at both transcriptional (Zhang et al., 2009; Wan et al., 2011; Xiao et al., 2013) and post-transcriptional levels (Wang et al., 2002, 2004; Chae et al., 2003; Liu and Zhang, 2004; Argueso et al., 2007; Joo et al., 2008; Prasad et al., 2010; Wan et al., 2011) have also been documented in the last decades. Besides controlling ethylene biosynthesis, light also influences ethylene signaling and, although not covered in this review, this is also an important aspect that has received increasing attention in the last decades.
Here we have compiled the major controlling mechanisms of both light signaling and ethylene biosynthesis and discussed the latest information on the potential signaling networks connecting both these pathways, with an emphasis on the emerging evidence of molecular mechanisms regulating the transcription and post-translational modifications of key participants in the ethylene biosynthetic pathway.
LIGHT PERCEPTION AND SIGNALING MECHANISMS
Light perception and signal transduction are responsible for modulating many processes throughout the plant life cycle and in diverse environmental contexts. Among the different wavelengths detected by the plant photosensory system, red light (RL), blue light (BL), UV-A and UV-B radiation are particularly informative and their perception involves four main families of information-transducing photoreceptors: the RL-absorbing phytochromes, the UV-A/BL-absorbing cryptochromes and phototropins, and the recently identified UVR8 receptors, which essentially perceive UV-B radiation. These signaling molecules provide the plants with information concerning various aspects of the light environment, thereby playing a vital role in plant survival and optimal growth (Franklin et al., 2005; Franklin and Quail, 2010; Rizzini et al., 2011). Detailed signaling mechanisms triggered by photoreceptors have been the focus of excellent reviews (Quail, 2002; Gyula et al., 2003; Chory, 2010; Chen and Chory, 2011); therefore, only general information about this theme will be provided here.
IDENTITY AND GENERAL FEATURES OF PLANT PHOTORECEPTORS
Phytochromes are the RL and far-red light (FRL) photoreceptors that play essential roles during plant photomorphogenesis (Chen and Chory, 2011). They are part of a chromoprotein multigene family, which in Arabidopsis is divided into photodegradable (phytochrome A—PHYA) and photostable (phytochrome B, C, D, and E—PHYB, PHYC, PHYD, and PHYE) types. These photoreceptors are formed by the association of one globular apoprotein and one chromophore which confers the properties of activation in response to the RL spectrum and photoreversibility upon FRL exposure (Wagner et al., 2005). The perception of RL is mediated by the chromophore (Wagner et al., 2005), which is synthesized in plastids and combined with the phytochrome apoproteins in the cytosol. Under dark conditions, the chromophore is maintained in a stable-inactive conformation (PHYPr) whereas the RL triggers the photoconversion of the inactive state PHYPr into the active form (PHYPfr), resulting in the phytochrome translocation from the cytoplasm into the nucleus (Ulijasz et al., 2010; Song et al., 2011; Figure 1). Despite the relatively well-conserved structure, PHYA and PHYB have different response patterns in terms of light stimulation and action mechanisms. PHYA-mediated responses are mainly triggered by FRL and this phytochrome does not show photoreversibility (Quail, 2002). Once in the active form, PHYAPfr associates with the proteins FAR RED ELONGATED HYPOCOTYL (FHR) and FHR-LIKE (FHL) and transiently accumulates in the nucleus (Kircher et al., 2002; Pfeiffer et al., 2012). On the other hand, PHYB is primarily activated by RL, presents R/FR photoreversibility, and accumulates in the nucleus for longer periods than PHYA (Gil et al., 2000). Furthermore, distinct members of the PHY family can display a certain degree of organ specificity in response to RL (Tepperman et al., 2004).
FIGURE 1.
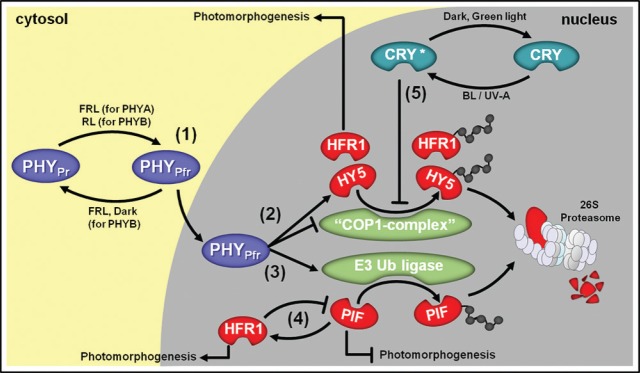
Simplified overview of light signal transduction via phytochromes (PHY) and cryptochromes (CRY) and their main interacting proteins. (1) Upon far-red light (FRL) or red light (RL) irradiation, cytosolic PHYAPr and PHYBPr, respectively, are converted into their active Pfr forms, which migrate to the nucleus. (2) In the cell nucleus, PHYPfr represses CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) E3-complex (“COP1 complex”), which is responsible for targeting positive components of the light signal transduction pathway such as LONG HYPOCOTYL 5 (HY5) and LONG HYPOCOTYL IN FAR-RED 1 (HFR1) to proteasomal degradation. The expression of genes coding for HY5 and HFR1 is also promoted by PHYPfr and these transcription factors are responsible for stimulating the expression of numerous photomorphogenesis-associated genes. (3) In parallel, PHYPfr activates an unidentified E3 Ubiquitin (Ub) ligase responsible for targeting light signal transduction repressor proteins such as PHYTOCHROME-INTERACTING FACTORS (PIFs) to degradation via 26S proteasome. (4) PIFs are also known to stimulate HFR1 and HFR1 blocks PIFs transcriptional activity. (5) Upon blue light (BL) or UV-A exposure, nuclear-localized CRY is converted into active CRY*, which also repress “COP1 complex” action.
The cryptochromes (CRY) are nuclear proteins that primarily respond to BL and UV-A radiation (Ahmad and Cashmore, 1997) and present an intricate interaction with phytochromes during the control of numerous photomorphogenic responses (Guo et al., 2001; Facella et al., 2008). In Arabidopsis, cryptochrome 1 (CRY1) is photostable and acts mainly at high light fluence (Yang et al., 2000; Herbel et al., 2013) whereas cryptochrome 2 (CRY2) is photodegradable and plays an important role at relatively lower light fluence (Lin et al., 1998; Guo et al., 2001; Gyula et al., 2003). CRY1/2 phosphorylation and subsequent molecular modifications are triggered soon after BL/UV-A exposure, facilitating the CRY interaction with other proteins (Kondoh et al., 2011). Such BL/UV-A-driven modifications in the CRY molecules can be reversed in complete darkness or upon exposure to green radiation (Sellaro et al., 2010).
In contrast, phototropins 1 and 2 are membrane-localized photoreceptors that undergo auto-phosphorylation in response to BL/UV-A radiation (Christie, 2007) and play an important role in many phototropic responses in plants (Kinoshita et al., 2001; Sakai et al., 2001). In the case of the UVR8 receptors, the perception of UV-B radiation occurs via tryptophan residues present at the protein homodimeric interface which leads to the monomerization and subsequent association of the UVR8 monomers with CONSTITUTIVE PHOTOMORPHOGENESIS 1 (COP1), which functions as an E3 ubiquitin ligase in multiple protein complexes and represents a master repressor during light signaling cascades. Such protein interaction will ultimately result in differential gene expression (Rizzini et al., 2011; Wu et al., 2012), and its potential outputs will be discussed later in the context of light signaling mechanisms controlling the transcription of genes involved in ethylene biosynthesis.
HUB TRANSCRIPTION FACTORS UNDER PHYTOCHROME CONTROL
Downstream of the RL perception, an increasing number of transcription factors has been identified as central integrators of light and multiple internal signals to optimize plant development. A key mechanism by which PHYs regulate gene expression is by modulating the protein stability of target transcription factors in the nucleus. One of the most important PHYPfr-mediated signaling pathways relies on the regulation of a multigene family of basic helix–loop–helix transcription factors known as PHYTOCHROME INTERACTION FACTOR (PIF). Accordingly, under inductive light conditions PHYAPfr and/or PHYBPfr migrates to the nucleus, where they can physically interact with PIF proteins and promote their ubiquitination and subsequent degradation via proteasome 26S (Figure 1). Under continuous darkness, PIF proteins inhibit the transcription of many genes associated with photomorphogenic responses; therefore, PHY activation and migration to the nucleus followed by its positive influence on PIF degradation represents a major point of transcriptional control of such genes under PIF-dependent inhibition (Ni et al., 1999; Khanna et al., 2004; Shen et al., 2007; Leivar et al., 2008; Lorrain et al., 2008; Chen and Chory, 2011). Besides playing a central role during the scoto-to-photomorphogenesis transition, PIFs are also involved in many developmental processes triggered by low R/FR conditions, such as shade avoidance responses, during which they control the transcription of genes coding for important proteins such as PIF3-LIKE 1 (PIL-1) and ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2 (ATHB2), and some components of the biosynthetic routes of plant hormones such as auxins, gibberellin, and ethylene (Martínez-Garcia et al., 2000; Leivar et al., 2009).
Active PHYAPfr and PHYBPfr are also known to increase the levels of LONG HYPOCOTYL 5 (HY5), which is another crucial transcription factor associated with light signaling and several photomorphogenic responses in plants. Hence, under continuous darkness HY5 undergoes very rapid destabilization mediated by COP1 (Gyula et al., 2003; Chory, 2010; Boron and Vissenberg, 2014). In fact, COP1 usually acts in association with other regulatory proteins (“COP1 complex”) promoting the ubiquitin-dependent degradation of both HY5 and PHYs, thereby representing an important element in the post-translational regulation of the PHY-dependent signaling cascades. In an intricate crosstalk with PHY, CRY also plays a fundamental role in the stabilization of HY5 since both CRY1 and CRY2 have been shown to increase the half-life of this protein by directly interacting with COP1 and promoting its destabilization or removal from the cell nucleus (Figure 1), which ultimately leads to an extended window of time for the stimulatory action of HY5 on the transcription of genes associated with photomorphogenic responses. Moreover, CRY also increases the half-life of PHYA by destabilizing the COP1 complex (Schwechheimer and Deng, 2000; Gyula et al., 2003; Chory, 2010; Fankhauser and Ulm, 2011; Boron and Vissenberg, 2014).
INFLUENCE OF DIVERSE LIGHT REGIMES ON ETHYLENE EVOLUTION
Light influence on ethylene evolution rate has been consistently demonstrated in several plants and can either stimulate or inhibit ethylene production depending on the tissue, organ, species, plant developmental phase, and the nature of light signal (Abeles et al., 1992; Corbineau et al., 1995; Halliday and Fankhauser, 2003; Kurepin et al., 2010). For example, European production of lilies can be hampered by long periods of low light, which induce higher production of ethylene, causing the abscission of developing flower buds (Abeles et al., 1992). On the other hand, potato plants treated with continuous light for faster tuber development displayed up to 15-fold increase in ethylene production, while ethylene levels rapidly dropped when these plants were transferred back to the 12 h light/dark photoperiod (Wheeler and Tibbitts, 1986). Despite the physiological response, most light-triggered adjustments in ethylene production seem to be mainly under PHY and CRY control (Vandenbussche et al., 2003; Kurepin et al., 2010).
PHYPfr-mediated signaling usually represses ethylene emission in several monocotyledons and eudicotyledons, as reported for etiolated seedlings of pea (Pisum sativum; Goeschl et al., 1967; Kang and Burg, 1972; Steed et al., 2004), light-grown seedlings of sorghum (Sorghum bicolor; Finlayson et al., 1998, 1999), coleoptiles and apical segments of etiolated rice (Oryza sativa) seedlings (Imaseki et al., 1971), light-grown leaves of both oat (Avena sativa; Corbineau et al., 1995) and wheat (Triticum aestivum; Jiao et al., 1987), and adult plants of both tobacco (Nicotiana tabacum; Pierik et al., 2004a) and Arabidopsis thaliana (Vandenbussche et al., 2003; Bours et al., 2014). Given its important role in RL perception and signaling, PHYB is the photoreceptor more closely associated with the negative regulation of ethylene levels in several plant models (Finlayson et al., 1998; Vandenbussche et al., 2003). In addition, the inhibitory effect of RL on ethylene emission depends on plant light exposure duration, radiation fluence (Imaseki et al., 1971; Pierik et al., 2004b; Kurepin et al., 2010), and some light-evoked adjustments on the plant circadian clock (Finlayson et al., 1998). However, in some plant systems PHYA also seems to play a prominent role in repressing ethylene production by direct action and/or via CRY1 repression, as reported for de-etiolating phyAphyB mutant pea seedlings (Foo et al., 2006).
CENTRAL METABOLIC INTERMEDIATES AND ENZYMES IN ETHYLENE BIOSYNTHESIS
Ethylene is a relatively simple unsaturated two-carbon gas which can be produced by numerous non-biological chemical reactions catalyzed by heat, oxidation, light, or ionizing radiation. In fact, non-biological model systems were systematically employed during the initial search for metabolic components and steps involved in ethylene production (Yang and Hoffman, 1984; Abeles et al., 1992). Such approaches led to the unexpected discovery that ethylene could be chemically generated from the amino acid methionine and its derivatives (Yang et al., 1966). Following this breakthrough, significant advances were achieved in ethylene research with biological systems, providing the basis of knowledge regarding the ethylene biosynthetic pathway (Yang and Hoffman, 1984; Abeles et al., 1992; Kende, 1993).
Ethylene biosynthesis in higher plants is now well characterized by a relatively simple metabolic pathway, which is, however, coordinated with some other equally important synthetic pathways involved in the plant metabolism regulation (e.g., polyamines). The identification of the intermediate components in ethylene biosynthesis allowed further elucidation of the two committed reactions in this pathway, which comprise the rate-limiting enzymes 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS, EC 4.4.1.14) and ACC oxidase (ACO, EC 1.4.3; Yang and Hoffman, 1984; Kende, 1993; Zarembinski and Theologis, 1994; Argueso et al., 2007; McClellan and Chang, 2008; Harpaz-Saad et al., 2012). Both ACS and ACO are encoded by multigene families whose members have been well characterized in some plant species and they are recognized as major players in ethylene biosynthetic regulation. However, as we will discuss in more detail below, the regulatory mechanisms of ethylene biosynthesis usually converge on the modulation of ACS proteins (Fluhr and Mattoo, 1996; Wang et al., 2002).
YANG CYCLE AND SYNTHETIC PATHWAYS ASSOCIATED WITH ETHYLENE PRODUCTION
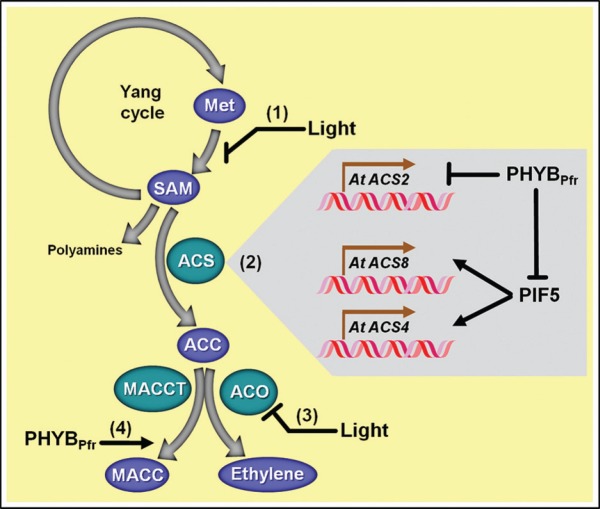
Ethylene biosynthesis in planta starts with the conversion of L-methionine to S-adenosyl methionine (AdoMet or SAM) in a reaction catalyzed by the enzyme L-methionine–S-adenosyltransferase (SAM synthetase, EC 2.5.1.6; Figure 2). The subsequent conversion of SAM to ACC is catalyzed by ACS and is generally considered the first committed and rate-limiting step in ethylene biosynthesis. The following crucial step in this pathway is catalyzed by ACO in a reaction that converts ACC to ethylene, CO2, HCN, and H2O (Adams and Yang, 1977, 1979; Lürssen et al., 1979; Yang and Hoffman, 1984; McKeon and Yang, 1987; Argueso et al., 2007; Lin et al., 2009). Behind this relatively linear sequence of few reactions hides a fundamental metabolic mechanism that prevents potential depletion of the methionine pools when a high rate of ethylene production is required. Such metabolic detail is significant because methionine, the biological precursor of ethylene, is a very scarce sulfur-containing amino acid and the amounts of sulfur available are usually limited in plants. In this sense, the methylthio-group from methionine needs to be recycled after SAM production. This is possible because the conversion of SAM to ACC by ACS also produces 5′-methylthioadenosine (MTA) as a by-product that retains the reduced methylthio-group and is readily recycled back to methionine through the Yang cycle. Therefore, the Yang cycle facilitates the occurrence of high rates of ethylene biosynthesis without influencing the steady-state levels of methionine pool (Yang and Hoffman, 1984; Miyazaki and Yang, 1987; Argueso et al., 2007; Zheng et al., 2013).
FIGURE 2.
Schematic representation of light-evoked impacts on ethylene biosynthetic pathway. (1) Light irradiation negatively impacts the conversion of methionine (Met) to S-adenosyl methionine (SAM). (2) Light also affects the transcription of particular 1-aminocyclopropane 1-carboxylic acid (ACC) synthases (ACS), responsible for converting SAM into ACC. In Arabidopsis, whereas ACS2 gene transcription is negatively controlled by PHYBPfr, ACS8/4 gene transcription is stimulated by PIF5, and, accordingly, PIF5 protein stability is negatively influenced by active phytochrome B (PHYBPfr). (3) Light is also known to inhibit the transcription and activity of ACC oxidase (ACO), whose activity converts ACC into ethylene. (4) PHYBPfr also stimulates the conversion of ACC into the non-volatile ACC metabolite 1-malonyl aminocyclopropane-1-carboxylic acid (MACC) via MACC transferase (MACCT). Biosynthetic enzymes and metabolic substrates or products are represented with green and blue ovals, respectively. Light-dependent transcriptional control of particular ACS genes is highlighted by the gray area of the figure.
Besides being the direct precursor of ethylene, ACC is also suggested as a cell-signaling molecule per se. ACC is a soluble molecule that seems to be translocated throughout different plant organs, and its translocation within flowers has been suggested to play an important role in floral senescence (Abeles et al., 1992; Harpaz-Saad et al., 2012; Yoon and Kieber, 2013a). Hence, it appears logical that most tissues can actively regulate the endogenous ACC levels during plant development. This function is mainly controlled by ACS and ACO; however, the cellular pool of ACC can be also modulated by other metabolic reactions leading to its conjugation (Yang and Hoffman, 1984; McKeon and Yang, 1987; Martin and Saftner, 1995; Peiser and Yang, 1998; Staswick and Tiryaki, 2004; Kombrink, 2012). Accordingly, ACC can be converted into ACC derivates, such as jasmonic acid-ACC (JA-ACC), γ-glutamyl-ACC (GACC), and 1-(malonylamino) cyclopropane-1-carboxylate (MACC). Both GACC and JA-ACC comprise minor moieties in the pool of ACC derivates, whose potential biological functions still remain poorly characterized. In this sense, the physiological significance of such derivates might be underestimated (Staswick and Tiryaki, 2004; Kombrink, 2012). Conversely, MACC formation seems to be a metabolic sink that allows depletion of ACC levels under certain conditions. This reaction is catalyzed by ACC-N-malonyltransferase (MACCT; Figure 2), an enzyme that displays increased activity during late stages of tomato fruit ripening (Yang and Hoffman, 1984). Some hypotheses suggest MACC as an end metabolite derived from ACC that can be easily accumulated (Yang and Hoffman, 1984; McKeon and Yang, 1987), while others consider MACC as a means for temporary storage of ACC in a non-reactive form which could be hydrolyzed back to ACC when needed for ethylene production (Hoffman et al., 1982; Jiao et al., 1986; Hanley et al., 1989).
LIGHT SIGNALING ON METABOLIC REGULATION OF THE ETHYLENE PRECURSOR LEVELS
One of the mechanisms involved in the PHYPfr-dependent regulation of ethylene production in plants precisely relies on regulating the abundance of the immediate ethylene precursor ACC. Although still not fully characterized at the molecular level, the active form PHYBPfr has been shown to promote the rapid conjugation of ACC into MACC during the light-induced seedling de-etiolation of wheat, thereby decreasing the internal ACC pool available for ethylene formation (Jiao et al., 1987; Figure 2). Interestingly, these events occurred without any modification in the extractable MACCT activity, indicating that light might have exerted its inductive effects by a mechanism other than increasing the MACCT levels (Jiao et al., 1987). Such mechanism could represent some potential post-translational control of MACCT yet unknown.
POST-TRANSLATIONAL MECHANISMS CONTROLLING ETHYLENE BIOSYNTHESIS
Both ACS and ACO family members are suggested to be at certain degree under post-translational regulation by proteasome-mediated degradation (Chen et al., 2005). However, particular attention has been devoted to unraveling the molecular structure and regulatory mechanisms of ACS proteins in Arabidopsis. For this reason, the following topics will mainly focus on discussing the central mechanisms involved in both transcriptional and post-translational control of ACS members in this plant model. Likewise, some evidence on ACS and ACO regulation in other species will be also comparatively discussed when opportune.
Briefly, ACS proteins belong to the α-superfamily of pyridoxal-5′-phosphate-dependent enzymes, which are represented in Arabidopsis by eight active members differentially regulated by several developmental and environmental signals. In addition to these active ACS enzymes, the Arabidopsis genome encodes an additional ACS (ACS1) that is catalytically inactive (Yamagami et al., 2003; Tsuchisaka and Theologis, 2004). Based primarily on the C-terminal sequence domains, ACS proteins are usually classified into three main groups which depict distinct regulatory features affecting the stability of the respective ACS proteins (Chae and Kieber, 2005; Yoshida et al., 2005). In Arabidopsis, type I ACS proteins (ACS2 and ACS6) have an extended C-terminus that contains phosphorylation sites (four conserved serine residues) targeted by mitogen-activated protein kinases (MPK) and calcium-dependent protein kinases (CDPK; Figure 3). Type II group includes isoforms (ACS4, ACS5, ACS8, and ACS9) with a shorter C-terminus that only contains a putative CDPK target site. Type III class is represented by one single member, ACS7, which has a very short C-terminal domain with no recognized phosphorylation sites (Tatsuki and Mori, 2001; Sebastià et al., 2004; Liu and Zhang, 2004; Chae and Kieber, 2005). Interestingly, this criterion of ACS classification has also been used for tomato ACS enzymes (Yoshida et al., 2005, 2006), indicating a general degree of structural conservation within ACS family. However, tomato encodes 10 LeACS isoenzymes with particular functional patterns (Yoshida et al., 2005, 2006).
FIGURE 3.
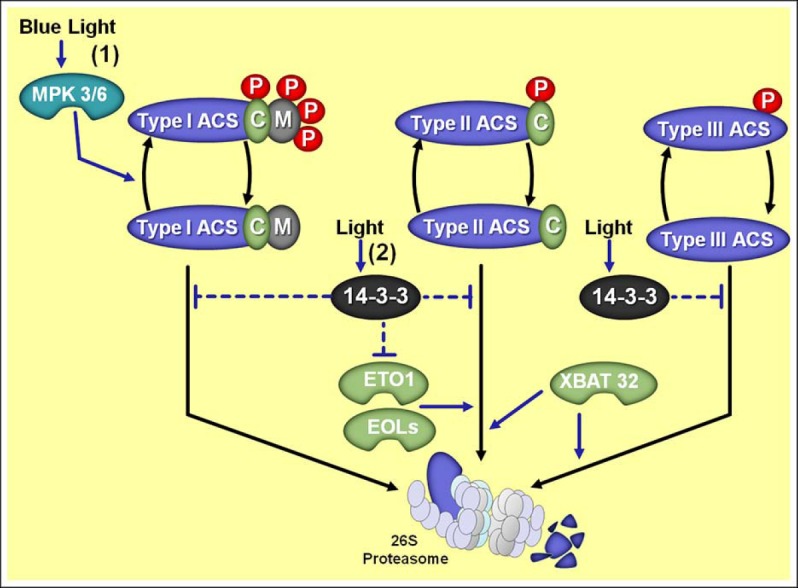
Overview of the distinct regulatory mechanisms controlling stability of type I, II, and III 1-aminocyclopropane 1-carboxylic acid synthases (ACS) and their regulation by light. (1) Blue light stimulates mitogen-activated protein kinases 3 and 6 (MPK 3/6) which in turn phosphorylate type I ACSs, thereby preserving ACSs from 26S proteasome-mediated degradation. (2) Light also stimulates 14-3-3 proteins which influence ACSs protein stabilization by both direct protein interaction and destabilization of ETHYLENE-OVERPRODUCER1 (ETO1) and ETO1/2-LIKE (EOL1/2) responsible for targeting type II ACS proteins for proteasomal degradation. Moreover, the RING-type E3 ligase XBAT32 affects both type II and III ACSs degradation. Protein phosphorylation is represented with red ovals and “P” letters. Phosphorylation sites targeted by calcium-dependent protein kinases (CDPK) are represented with green ovals and “C” letters. Phosphorylation sites targeted by MPK are represented with gray ovals and “M” letters. Note that type I ACS have both CDPK (“C”) and MPK (“M”) phosphorylation sites, type II ACS lacks only the MPK (“M”) phosphorylation site and type III ACS lacks both phosphorylation sites.
ACS PHOSPHORYLATION
Several experimental approaches have consistently suggested an important role for ACS phosphorylation and dephosphorylation during ethylene biosynthesis (Spanu et al., 1994; Tuomainen et al., 1997; Zhong and Burns, 2003), which regulates protein stability in an ACS isoform-dependent manner (Joo et al., 2008; Hahn and Harter, 2009; Figure 3). Genetic and biochemical studies have indicated CDPK and/or MPK as important molecules that cooperatively regulate stability of most ACS proteins and thus ethylene production, in response to developmental and environmental stimuli (Tatsuki and Mori, 2001; Liu and Zhang, 2004; Huang et al., 2013). As discussed previously, particular residues at the C-terminal domain of different isoforms of ACS represent putative target sites for phosphorylation, with MPK usually targeting three Ser residues that are distinct from the CDPK target site in the C-terminal extended region of a subset of ACS isoenzymes (Liu and Zhang, 2004; Sebastià et al., 2004; Joo et al., 2008; Huang et al., 2013).
Although very little information is available on CDPK signaling mechanisms during ACS regulation, it is widely accepted that the MPK3/6 module of MPK kinase (MKK) cascades plays relevant roles in the regulation of ethylene biosynthesis (Liu and Zhang, 2004; Joo et al., 2008; Xu et al., 2008; Hahn and Harter, 2009; Ju and Chang, 2012). Accordingly, the MPK3/6 module is mostly under control of MKK4, MKK5, and MKK9, constituting an important step in various signaling pathways involved in stress-induced responses in plants (Xu et al., 2008; Yoo et al., 2008; Beckers et al., 2009). For example, the stress-activated MKK4/5 signaling cascade can positively regulate ethylene biosynthesis by activating MPK6 (Liu and Zhang, 2004). In addition, ACS6 was initially identified as a potential substrate for MPK3/6 phosphorylation (Feilner et al., 2005) and further studies have confirmed that the MPK3/6 module is associated with enhanced stability of both type I ACS2 and ACS6 (Liu and Zhang, 2004; Joo et al., 2008; Figure 3). Interestingly, the autocatalytic ethylene production is often stress related and at least in Arabidopsis relies on post-translational regulation of type I ACSs by MPK3/6 cascade (Vandenbussche et al., 2012). Furthermore, the MKK9–MPK3/6 module is also suggested as a potential regulatory mechanism of ACS2/6 stabilization in some particular biological contexts (Popescu et al., 2009), supporting previous suggestions of the possible role of MKK9 and MPKs in ethylene biosynthesis control (Joo et al., 2008; Xu et al., 2008).
Therefore, ACS phosphorylation appears to be a major mechanism controlling the enzyme activity by increasing protein stability (Tsuchisaka and Theologis, 2004; Chae and Kieber, 2005; Argueso et al., 2007). Besides, tobacco plants under stressful conditions display a rapid increase in ethylene production mediated by an active NtMEK2 (MPKK)/SIPK (MPK) cascade that triggers a dramatic increase in ACS activity, which is followed by the transcriptional activation of a subgroup of ACS and ACO genes (Kim et al., 2003). As discussed in the following sections, recent data also suggest that phosphorylation might increase ACS affinity to directly interact with other proteins that influence ACS stability (Yoon and Kieber, 2013b).
ACS DEGRADATION
As previously discussed, the ubiquitin–26S proteasome has been linked to diverse functions in plants, including hormone signaling, photomorphogenesis, and stress-triggered responses (Yi and Deng, 2005; Stone and Callis, 2007; Vierstra, 2009; Santner and Estelle, 2010). In fact, many aspects of ethylene biosynthesis are also highly regulated at the post-transcriptional level by degradation of proteins controlled by the ubiquitin–26S proteasome system. Genetic studies in Arabidopsis have begun to reveal a number of ethylene biosynthetic enzymes which are targeted for proteasomal degradation, including the type II ACS proteins and the type III ACS7 (Wang et al., 2004; Yoshida et al., 2005; Christians et al., 2009; Lyzenga et al., 2012). Although not fully understood, ample evidence indicates that protein phosphorylation preserves ACS proteins from 26S proteasome-mediated degradation (Figure 3), providing a direct and rapid mechanism to change ethylene production (Spanu et al., 1994; Kelley and Estelle, 2012).
Details on post-translational regulation of ACS proteins have been derived from analyses of the ethylene-overproducing (eto1–3) mutants of Arabidopsis. Accordingly, dark-grown eto seedlings display the triple-response phenotype even in the absence of exogenous ethylene application and produces 10- to 40-fold more ethylene in the dark than the wild-type seedlings (Guzman and Ecker, 1990; Woeste et al., 1999; Chae et al., 2003; Wang et al., 2004). The increased ethylene biosynthesis in eto1 and eto2 mutations results from two different modifications at the C-terminal region of ACS5 isoform, while eto3 mutation is the result of a single amino acid change at the C-terminal region of ACS9 (Chae et al., 2003; Chae and Kieber, 2005). Such mutations were identified to cause disruptions within the TOE (for target of ETO1) domain of these ACS, which is recognized as a target site for ubiquitin–26S proteasome degradation. In fact, ETHYLENE-OVERPRODUCER1 (ETO1)/ETO1/2-LIKE (EOL1/2) genes code for CULLIN-3 E3 ubiquitin ligases which recognize and directly interact with the TOE domain of type II ACS proteins, targeting them for rapid degradation via 26S proteasome (Vogel et al., 1998; Chae et al., 2003; Wang et al., 2004; Yoshida et al., 2005, 2006; Christians et al., 2009; Figure 3). Additionally, the stability of both ACS7 (type III) and ACS4 (type II) was recently found to be turned over in a 26S proteasome-dependent manner through the participation of a RING-type E3 ligase named XBAT32 (Lyzenga et al., 2012), which also negatively modulates the abundance of ACS proteins and ethylene biosynthesis (Prasad and Stone, 2010; Prasad et al., 2010; Lyzenga et al., 2012). As the type III ACS7 lacks a C-terminal extension with the TOE sequence (Lyzenga et al., 2012), XBAT32 seems to represent a still unknown TOE-independent mechanism controlling ACS stability.
Adding an extra level of complexity in the ubiquitin–26S proteasome-dependent ACS stability control, recent findings revealed that ACS stability is also affected by ACS interaction with 14-3-3 proteins (Yoon and Kieber, 2013c; Figure 3). In this sense, 14-3-3 seems to interact with multiple isoforms from all three classes of ACS proteins, and this interaction increases the stability of the ACS proteins (Yoon and Kieber, 2013b). The molecular mechanism behind such regulation appears to involve the direct interaction of 14-3-3 with ACS proteins by decreasing their degradation by a still unknown ETO1/EOL-independent mechanism. These findings support the previous hypothesis that there is at least one further system acting to degrade type II ACS proteins in addition to the ETO1/EOLs, and the 14-3-3 proteins seem to antagonize this second degradation pathway (Lyzenga et al., 2012; Yoon and Kieber, 2013c). However, 14-3-3 proteins also seem to facilitate increased ACS stability by their interaction with the ETO1/EOLs E3 ligases which down-regulates their stability, thus increasing their degradation in an ubiquitin/proteasome-dependent manner and, consequently, decreasing the abundance of the ubiquitin ligases that target a subset of ACS proteins for degradation (Yoon and Kieber, 2013b).
In rice, the type II OsACS1 can interact with 14-3-3 proteins and the C-terminal domain of this enzyme was presumably phosphorylated by OsCDPK on a 14-3-3 recognition motif, suggesting that the phosphorylated OsACS1 may interact with rice 14-3-3 proteins, thus preventing ETO1 from binding to OsACS1 and induce this enzyme degradation (Yao et al., 2007). Therefore, such a regulatory mechanism might explain why after CDPK-dependent phosphorylation ACS proteins display increased protein stability (Yao et al., 2007; Yoon and Kieber, 2013c). Moreover, some evidence has indicated that 14-3-3 proteins interacted with ACO2 and ACO4 proteins in a yeast two-hybrid system, thus indicating that 14-3-3 proteins may regulate the ethylene biosynthesis pathway by modulating both ACS and ACO proteins (Huang et al., 2013).
LIGHT SIGNALING AND THE POST-TRANSLATIONAL CONTROL OF ACS AND ACO PROTEINS
Recently, Yoon and Kieber (2013c) reported that etiolated seedlings of Arabidopsis submitted to light treatment present a rapid (within 2 h) increase in the levels ACS5 proteins without corresponding changes in ACS5 transcripts, suggesting that light stimuli act by increasing ACS5 stability in this plant system. Accordingly, light treatment triggers the opposite effect on EOL2 protein levels, which exhibit a significant and rapid reduction as soon as 2 h after light exposure. These authors have suggested that the mechanism by which 14-3-3 proteins control ACS stability could also be regulated by light stimuli; however, such hypothetical connection remains uncertain. Furthermore, some evidence indicates that the particular isoform SbACO2 from sorghum might show a light-regulated post-transcriptional regulation throughout the diurnal cycle. This suggestion was based on the observation that diurnal fluctuations in SbACO2 transcript abundance were translated into diel changes in enzymatic activity under unshaded environment, but not under simulated high-shade conditions (Finlayson et al., 1999). Additionally, recent findings have shown significant connections between MPK and light signaling pathways in plants which indicate the MKK3–MPK6 cascade being actively regulated by BL signaling at several levels (Sethi et al., 2014). Therefore, it is tempting to speculate that MKK3–MPK6-dependent cascades might possibly be implicated in the interplay between light signaling and ACS post-translational control (Figure 3).
TRANSCRIPTIONAL MECHANISMS CONTROLLING ETHYLENE BIOSYNTHESIS
Transcriptional regulation of ACS and ACO genes is one of the pivotal mechanisms controlling ethylene biosynthesis (Wan et al., 2011). As previously discussed, although regulatory mechanisms affecting ACS activity are generally considered the crucial regulatory point for ethylene production, increasing evidence has indicated that modulation of ACO expression can also represent a significant point of control for ethylene production under particular circumstances (Rudus et al., 2013; Xiao et al., 2013). Accordingly, a number of studies have reported that both ACS and ACO family members can be differentially expressed in diverse plant organ/tissues, distinct developmental phases, and in response to different environmental stimuli (Liang et al., 1992; Kende, 1993; Zarembinski and Theologis, 1994; Vahala et al., 1998; Wang et al., 2002; Argueso et al., 2007). Differences in the expression levels of each ACS and ACO genes might be an important means for adjusting differential ethylene production within a particular organ or plant tissue. For instance, the ACS multigene family of Arabidopsis has a prominent member-specific spatial regulation, suggesting a tissue-specific diversity of ethylene production (Tsuchisaka and Theologis, 2004). Likewise, members of the ACO gene family of tomato (Solanum lycopersicum) are also differentially regulated at the transcriptional level during climacteric fruit ripening, indicating distinct roles played by each ACO homolog during different tomato fruit developmental stages (Rudus et al., 2013; Xiao et al., 2013).
Additionally, both ACS and ACO expression can be directly and/or indirectly controlled by several signaling connections with other plant hormones such as auxin, brassinosteroid, and gibberellin (Guzman and Ecker, 1990; Joo et al., 2006). In this sense, auxin has been widely recognized as one of the most significant hormones controlling ethylene biosynthesis through transcriptional regulation of several ACS genes (Abel et al., 1995; Tsuchisaka and Theologis, 2004; Stepanova et al., 2007). In Arabidopsis, with the exception of ACS1/7/9 genes, the transcription of all other ACS members coding for functional enzymes are induced by auxin (Yamagami et al., 2003). In fact, the auxin–ethylene crosstalk often occurs via reciprocal regulation at biosynthetic level (Stepanova et al., 2007) with auxin apparently acting mostly via up-regulation of ACS4 transcription in Arabidopsis (Liang et al., 1992; Abel et al., 1995; Zhu and Guo, 2008). On the other hand, some data on the crosstalk between auxin and ethylene during transcriptional regulation of two ACO genes in rice (OsACO2/3) have indicated that auxin effects on ethylene biosynthesis are highly dependent of the ethylene status itself. For example, OsACO3 is induced by ethylene, but not in the presence of auxin, whereas OsACO2 is induced by auxin, but in a reduced extent when in the presence of ethylene (Chae et al., 2000). Hence, auxin induction of ACO transcripts is generally considered an indirect effect of auxin-stimulated ethylene production resulting from an increase in ACS activity (Peck and Kende, 1995). Hence, direct regulation of ACO transcription seems to be mainly induced by ethylene itself by an autocatalytic process that will be discussed in the next sections of this review.
ETHYLENE CONTROLLING ITS OWN BIOSYNTHESIS VIA ACS AND ACO TRANSCRIPTION
As mentioned previously, ethylene can modulate its own metabolism (Tsuchisaka et al., 2009) and this process usually occurs in an extremely flexible, developmentally and environmentally sensitive manner. Several studies have been reported on either positive or negative signaling feedback loops wherein ethylene regulates its own production (Sisler et al., 1985; Nakajima et al., 1990; Rottmann et al., 1991; Van Der Straeten et al., 1992; Woodson et al., 1992; Lincoln et al., 1993; Rodrigues-Pousada et al., 1999; Wang et al., 2002; Zhong and Burns, 2003). Therefore, depending on the plant tissue and the developmental/environmental context, ethylene can either restrain (auto-inhibition or system 1) or promote (autocatalysis or system 2) its own biosynthesis. Moreover, it has also been suggested that particular members of ACS gene family can play specific roles in these two systems of ethylene production in plants (Yang and Hoffman, 1984; Liu et al., 1985; Nakatsuka et al., 1998; Vandenbussche et al., 2012).
Negative feedback is particularly active during vegetative growth under non-stressful conditions and non-climacteric fruit development while autocatalytic ethylene production is usually related to stressful conditions, senescence and climacteric fruit ripening (McMurchie et al., 1972; Nakatsuka et al., 1998; Vandenbussche et al., 2012). During tomato fruit ripening different ACS and ACO genes are induced at particular stages of ripening. For example, LeACS2 and LeACS4 are mainly associated with autocatalytic ethylene production while LeACS6 is mostly related to the auto-inhibitory system responsible for the maintenance of basal ethylene production (Nakatsuka et al., 1998; Barry et al., 2000; Cara and Giovannoni, 2008).
In Arabidopsis, the auto-inhibitory system also operates through the tightly controlled activities of several ACS proteins, which generate the basal ethylene levels in a coordinated manner (Yoshii and Imaseki, 1982; Tsuchisaka et al., 2009). The transcription of ACS6 gene in this same species is generally associated with stress-induced conditions, in which ethylene autocatalysis plays an important role (Tsuchisaka et al., 2009; Vandenbussche et al., 2012). Furthermore, at least two ACO genes are ethylene inducible in Arabidopsis, suggesting that a feedback mechanism is in place to ensure that there will be no limitations to ethylene production once the precursor ACC is produced (Alonso et al., 2003; Zhong and Burns, 2003; Chen et al., 2005). Interestingly, the autocatalytic ethylene production in Arabidopsis is usually mediated by MPK cascade modules associated with stress-triggered signaling pathways. For example, besides post-translationally controlling ACS protein stability, the MPK3/6 module plays an important role in the ethylene autocatalytic pathway by inducing the transcription of the WRKY33 transcription factor which, in turn, directly interacts with ACS2/6 promoters (Smékalová et al., 2014). In addition, besides triggering ACS2/6 transcription up-regulation, the MKK9-MPK3/6 module also positively regulates the transcript abundance of several ERF (ETHYLENE RESPONSE FACTORS) genes (Xu et al., 2008; Yoo et al., 2008).
ERF or EREBP (ETHYLENE RESPONSE ELEMENT-BINDING PROTEIN) genes represent one of the largest families of transcription factors in the plant kingdom which regulates a diverse array of processes (including ethylene production) in response to multiple developmental and environmental cues (Smékalová et al., 2014). These plant-specific transcription factors function as trans-acting regulators at the last step of ethylene signaling pathway, and usually exhibit highly conserved motifs that includes AP2 (APETALA2)/ERF DNA-binding domain, RAV (related to ABI3/VP1), and DREB (dehydration-responsive element binding protein). These ERF domains can specifically interact with cis-elements in promoters of the ethylene-responsive genes, such as GCC box and/or dehydration-responsive elements/C-repeat (DRE/CRT) motifs, thus regulating the expression of different downstream genes (Ohme-Takagi and Shinshi, 1995; Park et al., 2001; Huang et al., 2004; Lin et al., 2009; Zhang et al., 2009; Kohli et al., 2013). In addition, some ERFs can induce the transcription of additional transcription factors, inducing a transcription-factor cascade that might facilitate the signal amplification and fine-tuning of signal output (Chen et al., 2005).
Several studies have already shown that ERFs participate in a feedback regulation of ethylene production by modulating the expression of ethylene biosynthesis genes during fruit ripening (Zhang et al., 2009; Sharma et al., 2010; Lee et al., 2012; Pirrello et al., 2012). For example, ectopic expression of the ERF.B3-SRDX dominant repressor in tomato caused reduction in ethylene biosynthesis associated with the down-regulation of ACS and ACO genes, which presented the GCC box and DRE/CRT motifs at their promoters (Liu et al., 2013; Pirrello et al., 2012). In addition, LeERF2/TERF2 activates ethylene biosynthesis by promoting the expression of ACS genes in tobacco and of both ACS and ACO genes in tomato (Zhang et al., 2009). On the other hand, the ERF repressor SlAP2a opposes tomato ripening by suppressing the expression of ACS2/4 and ACO1 genes, then causing down-regulation of ethylene biosynthesis (Chung et al., 2010). Interestingly, distinct MaERFs of banana (Musa acuminata) can be differentially expressed in a tissue-dependent manner during fruit ripening while different MaERFs appeared to regulate the transcription of particular ACS and/or ACO in either positive (MaERF9-induced activation of MaACS1) or negative (MaERF11-induced repression of MaACS1 and MaACO1) way (Xiao et al., 2013). Therefore, although the important role of multiple ERFs in the regulation of fruit ripening has been consistently described by several studies, little information is available about the signaling pathways involved in ERF regulation and the additional mechanisms modulating their target genes.
Furthermore, ERF proteins also play important roles in plant response to a variety of stress-related stimuli that involves modulation of ethylene synthesis (Xu et al., 2006; Wang et al., 2008; Hattori et al., 2009; Yaish et al., 2010). For instance, the ERF genes SUB1A and OsDERF1 of rice are, respectively, positive and negative regulators in drought response. Upon drought stress, the expression of both these ERFs is modulated, affecting in turn the ethylene production via alterations in the expression of ethylene biosynthesis genes (Xu et al., 2006; Fukao et al., 2011; Wan et al., 2011). Consequently, it has become clear that ERF transcription factors are the primary targets mediating stress-induced responses involving changes in ethylene signaling and/or biosynthesis. However, very little is known about the signaling mechanisms controlling ERF proteins in ethylene biosynthesis at the transcriptional level (Zhang et al., 2009), which highlights the great demand to identify transcription factors and other signaling elements involved in ethylene biosynthesis adjustments. The potential regulatory role of these transcription factors during the light-induced pathway leading to the regulation of ACS and/or ACO transcription will be discussed below.
LIGHT SIGNALING AND THE TRANSCRIPTIONAL CONTROL OF ACS AND ACO ENZYMES
PHYB is well known to module ethylene emission by repressing the transcription of genes encoding particular ACS isoenzymes (Figure 2). Illustrating this regulatory mechanism, mutations in phyA and phyB provide, respectively, increases on transcription of ACS1 in pea (Foo et al., 2006) and ACS2 in Arabidopsis (Bours et al., 2014). In fact, the genome-expression profiling of Arabidopsis revealed the light-regulated suppression of ACS, ACO, and SAM synthase transcripts (Ma et al., 2001). Accordingly, in this plant species the RL-evoked inhibition of ethylene emission not only depends on the repression of ACS2 gene transcription, but is also strongly associated with the PHYB-dependent degradation of PIF proteins, such as PIF4 and particularly PIF5. Interestingly, PIF1 and PIF3 biochemically interact with both PHYA and PHYB, whereas PIF4 and PIF5 are only degraded when associated with active PHYB and this PHYB-mediated degradation of PIF4 and PIF5 is considered to be an important mechanism responsible for controlling ethylene production in plant responses triggered by either darkness or low R/FR conditions, such as skotomorphogenic growth and shade avoidance responses (Lorrain et al., 2008; Shin et al., 2009).
As previously described, PIF5 protein is constitutively localized in the nucleus, where it exerts a repressive influence on the transcription of genes associated with photomorphogenic responses and presents a positive impact on genes associated with low R/FR responses, such as ACS4/8 (Figure 2). In agreement, Arabidopsis plants over-expressing PIF5 exhibited up to 300-fold increases in ACS4 transcripts even in the presence of light (Khanna et al., 2007). Although less prominent than observed for the ACS4 expression, PIF5 also positively influences ACS8 expression, which is the most abundantly expressed ACS gene in Arabidopsis and is the ACS gene most strongly controlled by endogenous ethylene and circadian clock (Yamagami et al., 2003; Thain et al., 2004). Due to such prominent impact on ethylene production, the PIF5 over-expression in Arabidopsis resulted in the triple response phenomenon characteristically triggered by excessive ethylene production. Moreover, PIF5 is also known to destabilize PHYB by promoting its degradation via proteasome 26S, and this PIF5-triggered degradation of PHYB might accentuate even more the ethylene production in conditions of high PIF5 protein abundance (Khanna et al., 2007).
Furthermore, it has become clear that an integrated, multi-hormonal network is a common signaling mechanism involved in ethylene biosynthesis control (Chae et al., 2000). For example, besides acting as repressors of gibberellins (GA) responses, DELLA proteins can also physically interact with PIF proteins, thereby inhibiting PIF responses via a PHY-independent mechanism. Such a DELLA-dependent PIF suppression mainly takes place under complete darkness, when both these proteins are at abundant levels. Thus, the influence of GA on the transcriptional regulation of some ethylene-related genes, such as ACS8, seems to be associated with the DELLA destabilization and consequent repression of PIF degradation. Another overlap between ethylene and GA consists of the fact that active PHYBPfr represses active GA biosynthesis, which in turn decreases ethylene emission via DELLA-dependent and independent pathways (Keller et al., 2011). Accordingly, low R/FR conditions promote DELLA degradation, leading to increased PIF-related responses (de Lucas et al., 2008) which include the stimulation of ethylene biosynthesis. Moreover, light responses triggered by low fluence radiation, such as shade avoidance, are well known to involve both auxins and ethylene, during which an intensive crosstalk between these phytohormones and PHYB is suggested (Vandenbussche et al., 2003; Millenaar et al., 2009; Pierik et al., 2009). For example, Arabidopsis mutants exhibiting low auxin sensitivity, such as auxin resistant 2 (axr2) and (axr1–3), usually present phenotypical similarities to wild-type plants maintained under complete darkness or low R/FR radiation. Treatment of these mutants with ethylene has been shown to revert this phenotype, suggesting that low R/FR leads to increased auxin levels, and these elevated auxin levels are responsible for stimulating ethylene production under these conditions. Additionally, auxin and low R/FR apparently share the same signaling pathways to increase ACS6/8 transcript abundance (Vandenbussche et al., 2003). However, some contrasting results obtained with low R/FR-treated Arabidopsis plants (Millenaar et al., 2009; Pierik et al., 2009) have indicated that signaling interactions among PHYs, auxins, and ethylene during low-light induced responses can be highly plastic and dependent on specific experimental conditions (i.e., plant material obtained by distinct growth methods, different periods of exposure to the light treatment).
Adding even more complexity to the crosstalk between light signaling and regulation of ethylene biosynthesis in plants, it is currently known that LONG HYPOCOTYL IN FAR-RED1 (HFR1) is particularly relevant during low F/FR responses. Accordingly, HFR1 is a nuclear protein structurally similar to PIF3 (Fairchild et al., 2000), which is phosphorylated by COP1 and usually marked for degradation under complete darkness (Figure 1). However, in the presence of light, especially under BL- or FRL-rich radiation, this protein remains much more stable due to the presence of active PHYA and CRYs. Some FRL-induced responses that are associated with modifications in ethylene emission (e.g., repression of ACS8 transcription) are apparently influenced by the biochemical interaction between PHYA-HFR1 or via the heterodimerization of HFR1 with PIF3 (Fairchild et al., 2000). However, it is also possible that such FRL-induced responses associated with modifications in ethylene evolution occurs through a more indirect pathway involving the HFR1-induced repression of genes and/or enzymes of other plant hormone biosynthetic pathways, such as GA and auxin (Sessa et al., 2005). Recent work has also suggested that PIFs and COP1 complexes synergistically repress photomorphogenesis in the dark, indicating that PIF proteins might inhibit HY5 by direct and indirect mechanisms (Xu et al., 2014). Furthermore, COP1 was reported as capable of physically interact with PIF3-LIKE1 (PIL1) and promote PIL1 degradation via the 26S proteasome, whereas PHYB physically interacts with PIL1 and enhances PIL1 protein accumulation upon RL irradiation, possibly via suppressing the COP1–PIL1 association (Jang et al., 2010; Luo et al., 2014).
Since PIF and COP1/HY5-mediated pathways are generally considered the two main light signaling branches downstream of the photoreceptors (Figure 1), it is likely that these components participate as central integrators between signaling pathways mediated by light and hormones cues (Yamamoto et al., 1998; Lau and Deng, 2010). In fact, some studies have consistently indicated PIF and COP/HY5 as hub steps of convergence and integration of signaling information mediated by both light and hormones. For example, such a PIF/COP/HY5-mediated network appears to control the opposite effects induced by ethylene on hypocotyl elongation of etiolated Arabidopsis seedlings when grown under darkness (elongation promotion) or light exposure (elongation repression; Feng et al., 2008; Liang et al., 2012). Interestingly, both these signaling steps are located downstream of ETHYLENE INSENSITIVE 3 (EIN3) during ethylene-regulated hypocotyl development in dark-grown seedlings (Ang and Deng, 1994; An et al., 2010). The transcription factors EIN3 and EIN3-LIKE 1 (EIL1) are intermediates in the ethylene signaling transduction responsible for the subsequent activation of ERF genes during multiple physiological processes (An et al., 2010; Liang et al., 2012). Moreover, COP1 appears to positively regulate the accumulation of EIN3 protein through a yet non-identified regulatory mechanism (Zhong et al., 2009) while COP1 affects the transcription of EIN3 downstream genes such as ERF1 (Solano et al., 1998; Liang et al., 2012).
CONCLUSION
Light has been acknowledged for some time as an important environmental cue capable of modulating ethylene biosynthesis in higher plants. While substantial progress has been made in identifying the metabolic components and enzymatic steps involved in ethylene biosynthetic route, our current understanding of how ethylene production is controlled by light signaling pathways is still very limited. Over the past decades, the identification of several mutants affected in particular aspects of ethylene production and/or photomorphogenic responses has facilitated significant advances in both research fields. As a result, some important breakthroughs on mechanistically explaining how light perception and signal transduction can modulate ethylene biosynthesis have been recently achieved, particularly in terms of the light-evoked changes in transcriptional and post-translational regulation of ACS and ACO enzymes.
Besides the direct effects of light-associated proteins on particular ethylene biosynthetic elements, we might also remain open-minded to conceive relatively more complex interconnection nodes, in which other plant hormones act in between the light perception and the actual modification of ethylene biosynthetic steps. Exciting new models of light–ethylene interaction networks might probably emerge when the combinatory influence of distinct photoreceptors (e.g., phytochromes and cryptochromes) or different environmental stimuli (e.g., light and biotic or abiotic stresses) started to be considered. Moreover, our knowledge about the mechanistic interplays between light and ethylene production might become even more complex as the research currently performed mainly in few plant models (e.g., Arabidopsis, tomato, pea) is extended to a wider range of species. Given the profuse influence of both light and ethylene on plant development, growth, and metabolism, studying the interplay between these stimuli is unarguably a promising research field for years to come.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ACKNOWLEDGMENTS
Work in the authors’ laboratory has been supported by Fundação de Amparo a Pesquisa do Estado de São Paulo (grant no. 2013/18056-2, São Paulo Research Foundation – FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
REFERENCES
- Abel S., Nguyen M. D., Chow W., Theologis A. (1995). ACS4, a primary indoleacetic acid-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsisthaliana. Structural characterization, expression in Escherichia coli, and expression characteristics in response to auxin. J. Biol. Chem. 270, 19093–19099. 10.1074/jbc.270.32.19093 [DOI] [PubMed] [Google Scholar]
- Abeles F. B., Morgan P. W., Salveit M. E. (1992). Ethylene in Plant Biology. San Diego: Academic Press. [Google Scholar]
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., et al. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94. 10.1126/science.1118642 [DOI] [PubMed] [Google Scholar]
- Adams D. O., Yang S. F. (1977). Methionine metabolism in apple tissue: implication of S-adenosylmethionine as an intermediate in the conversion of methionine to ethylene. Plant Physiol. 60, 892–896. 10.1104/pp.60.6.892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D. O., Yang S. F. (1979). Ethylene biosynthesis: identification of aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Natl. Acad. Sci. U.S.A. 76, 170–174. 10.1073/pnas.76.1.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M., Cashmore A. R. (1997). The blue-light receptor cryptochrome 1 shows functional dependence on phytochrome A or phytochrome B in Arabidopsis thaliana. Plant J. 11, 421–427. 10.1046/j.1365-313X.1997.11030421.x [DOI] [PubMed] [Google Scholar]
- Alonso J. M., Stepanova A. N., Solano R., Wisman E., Ferrari S., Ausubel F. M., et al. (2003). Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 100, 2992–2997. 10.1073/pnas.0438070100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F., Zhao Q., Ji Y., Li W., Jiang Z., Yu X., et al. (2010). Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22, 2384–2401. 10.1105/tpc.110.076588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang L. H., Deng X. W. (1994). Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6, 613–628. 10.1105/tpc.6.5.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso C. T., Hansen M., Kieber J. J. (2007). Regulation of ethylene biosynthesis. J. Plant Growth Regul. 26, 92–105 10.1007/s00344-007-0013-5 [DOI] [Google Scholar]
- Barry C. S., Llop-Tous M. I., Grierson D. (2000). The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol. 123, 979–986. 10.1104/pp.123.3.979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers G. J. M., Jaskiewicz M., Liu Y., Underwood W. R., He S. Y., Zhang S., et al. (2009). Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21, 944–953. 10.1105/tpc.108.062158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker A. B., Estelle M. A., Somerville C., Kende H. (1988). Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241, 1086–1089. 10.1126/science.241.4869.1086 [DOI] [PubMed] [Google Scholar]
- Boron A. K., Vissenberg K. (2014). The Arabidopsis thaliana hypocotyl, a model to identify and study control mechanisms of cellular expansion. Plant Cell Rep. 33, 697–706. 10.1007/s00299-014-1591-x [DOI] [PubMed] [Google Scholar]
- Bours R., van Zanten M., Pierik R., Bouwmeester H., van der Krol A. (2014). Antiphase light and temperature cycles affect PHYTOCHROME B-controlled ethylene sensitivity and biosynthesis, limiting leaf movement and growth of Arabidopsis. Plant Physiol. 163, 882–895. 10.1104/pp.113.221648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cara B., Giovannoni J. J. (2008). Molecular biology of ethylene during tomato fruit development and maturation. Plant Sci. 175, 106–113 10.1016/j.plantsci.2008.03.021 [DOI] [Google Scholar]
- Chae H. S., Cho Y. G., Park M. Y., Lee M. C., Eun M. Y., Kang B. G., et al. (2000). Hormonal cross-talk between auxin and ethylene differentially regulates the expression of two members of the 1-aminocyclopropane-1-carboxylate oxidase gene family in rice (Oryza sativa L.). Plant Cell Physiol. 41, 354–362. 10.1093/pcp/41.3.354 [DOI] [PubMed] [Google Scholar]
- Chae H. S., Faure F., Kieber J. J. (2003). The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell 15, 545–559. 10.1105/tpc.006882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae H. S., Kieber J. J. (2005). Eto Brute? Role of ACS turnover in regulating ethylene biosynthesis. Trends Plant Sci. 10, 291–296. 10.1016/j.tplants.2005.04.006 [DOI] [PubMed] [Google Scholar]
- Chen M., Chory J. (2011). Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 21, 664–671. 10.1016/j.tcb.2011.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. F., Etheridge N., Schaller G. E. (2005). Ethylene signal transduction. Ann. Bot. 95, 901–915. 10.1093/aob/mci100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J. (2010). Light signal transduction: an infinite spectrum of possibilities. Plant J. 61, 982–991. 10.1111/j.1365-313X.2009.04105.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians M. J., Gingerich D. J., Hansen M., Binder B. M., Kieber J. J., Vierstra R. D. (2009). The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J. 57, 332–345. 10.1111/j.1365-313X.2008.03693.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie J. M. (2007). Phototropin blue-light receptors. Annu. Rev. Plant Biol. 58, 21–45. 10.1146/annurev.arplant.58.032806.103951 [DOI] [PubMed] [Google Scholar]
- Chung M. Y., Vrebalov J., Alba R., Lee J., McQuinn R., Chung J. D., et al. (2010). A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 64, 936–947. 10.1111/j.1365-313X.2010.04384.x [DOI] [PubMed] [Google Scholar]
- Corbineau F., Rudnicki R. M., Goszczynska D. M., Come D. (1995). The effect of light quality on ethylene production in leaves of oat seedlings (Avena sativa L.). Environ. Exp. Bot. 35, 227–233 10.1016/0098-8472(94)00052-7 [DOI] [Google Scholar]
- de Lucas M., Daviere J. M., Rodriguez-Falcon M., Pontin M., Iglesias-Pedraz J. M., Lorrain S., et al. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484. 10.1038/nature06520 [DOI] [PubMed] [Google Scholar]
- Facella P., Lopez L., Carbone F., Galbraith D. W., Giuliano G., Perrotta G. (2008). Diurnal and circadian rhythms in the tomato transcriptome and their modulation by cryptochrome photoreceptors. PLoS ONE 3:e2798. 10.1371/journal.pone.0002798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild C. D., Schumaker M. A., Quail P. H. (2000). HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 14, 2377–2391. 10.1101/gad.828000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Ulm R. (2011). Light-regulated interactions with SPA proteins underlie cryptochrome-mediated gene expression. Genes Dev. 25, 1004–1009. 10.1101/gad.2053911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feilner T., Hultschig C., Lee J., Meyer S., Immink R. G., Koenig A., et al. (2005). High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol. Cell. Proteomics 4, 1558–1568. 10.1074/mcp.M500007-MCP200 [DOI] [PubMed] [Google Scholar]
- Feng S., Martinez C., Gusmaroli G., Wang Y., Zhou J., Wang F., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479. 10.1038/nature06448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlayson S. A., Jung I. J., Mullet J. E., Morgan P. W. (1999). The mechanism of rhythmic ethylene production in sorghum. The role of phytochrome B and simulated shading. Plant Physiol. 119, 1083–1089. 10.1104/pp.119.3.1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlayson S. A., Lee I. J., Morgan P. W. (1998). Phytochrome B and the regulation of circadian ethylene production in sorghum. Plant Physiol. 116, 17–25 10.1104/pp.116.1.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluhr R., Mattoo A. K. (1996). Ethylene-biosynthesis and perception. Crit. Rev. Plant Sci. 15, 479–523. [Google Scholar]
- Foo E., Ross J. J., Davies N. W., Reid J. B., Weller J. L. (2006). A role for ethylene in the phytochrome-mediated control of vegetative development. Plant J. 46, 911–921. 10.1111/j.1365-313X.2006.02754.x [DOI] [PubMed] [Google Scholar]
- Franklin K. A., Larner V. S., Whitelam G. C. (2005). The signal transducing photoreceptors of plants. Int. J. Dev. Biol. 49, 653–664. 10.1387/ijdb.051989kf [DOI] [PubMed] [Google Scholar]
- Franklin K. A., Quail P. H. (2010). Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61, 11–24. 10.1093/jxb/erp304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T., Yeung E., Bailey-Serres J. (2011). The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 23, 412–427. 10.1105/tpc.110.080325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil P., Kircher S., Adam E., Bury E., Kozma-Bognar L., Schäfer E., et al. (2000). Photocontrol of subcellular partitioning of phytochrome-B:GFP fusion protein in tobacco seedlings. Plant J. 22, 135–145. 10.1046/j.1365-313x.2000.00730.x [DOI] [PubMed] [Google Scholar]
- Goeschl J. D., Pratt H. P., Bonner B. A. (1967). An effect of light on the production of ethylene and the growth of the plumular portion of etiolated pea seedlings. Plant Physiol. 42, 1077–1080. 10.1104/pp.42.8.1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Mockler T., Duong H., Lin C. (2001). SUB1, an Arabidopsis Ca2+-binding protein involved in cryptochrome and phytochrome co-action. Science 291, 487–490. 10.1126/science.291.5503.487 [DOI] [PubMed] [Google Scholar]
- Guzman P., Ecker J. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2, 513–523. 10.1105/tpc.2.6.513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyula P., Schäfer E., Nagy F. (2003). Light perception and signalling in higher plants. Curr. Opin. Plant Biol. 6, 446–452 10.1016/S1369-5266(03)00082-7 [DOI] [PubMed] [Google Scholar]
- Hahn A., Harter K. (2009). Mitogen-activated protein kinase cascades and ethylene: signaling, biosynthesis, or both? Plant Physiol. 149, 1207–1210. 10.1104/pp.108.132241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday K. J., Fankhauser C. (2003). Phytochrome-hormonal signalling networks. New Phytol. 157, 449–463 10.1046/j.1469-8137.2003.00689.x [DOI] [PubMed] [Google Scholar]
- Hanley K. M., Meir S., Bramlage W. J. (1989). Activity of ageing carnation flower parts and the effects of 1-(malonylamino)cyclopropane-1-carboxylic acid-induced ethylene. Plant Physiol. 91, 1126–1130. 10.1104/pp.91.3.1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpaz-Saad S., Yoon G. M., Mattoo A. K., Kieber J. J. (2012). The formation of ACC and competition between polyamines and ethylene for SAM. Annu. Plant Rev. 44, 53–81 10.1002/9781118223086.ch3 [DOI] [Google Scholar]
- Herbel V., Orth C., Wenzel R., Ahmad M., Bittl R., Batschauer A. (2013). Lifetimes of Arabidopsis cryptochrome signaling states in vivo. Plant J. 74, 583–592. 10.1111/tpj.12144 [DOI] [PubMed] [Google Scholar]
- Hattori Y., Nagai K., Furukawa S., Song X.-J., Kawano R., Sakakibara H., et al. (2009). The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460, 1026–1030. 10.1038/nature08258 [DOI] [PubMed] [Google Scholar]
- Hoffman N. E., Yang S. F., McKeon T. (1982). Identification and metabolism of 1-(malonylamino)cyclopropane-1-carboxylic acid as a major conjugate of 1-aminocyclopropane-1-carboxylic acid, an ethylene precursor in higher plants. Biochem. Biophys. Res. Commun. 104, 765–770 10.1016/0006-291X(82)90703-3 [DOI] [PubMed] [Google Scholar]
- Huang S.-J., Chang C.-L., Wang P.-H., Tsai M.-C., Hsu P.-H., Chang I.-F. (2013). A type III ACC synthase, ACS7, is involved in root gravitropism in Arabidopsis thaliana. J. Exp. Bot. 64, 4343–4360. 10.1093/jxb/ert241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Zhang Z., Zhang X., Zhang H., Huang D., Huang R. (2004). Tomato TERF1 modulates ethylene response and enhances osmotic stress tolerance by activating expression of downstream genes. FEBS Lett. 573, 110–116. 10.1016/j.febslet.2004.07.064 [DOI] [PubMed] [Google Scholar]
- Imaseki H., Pjon C.-J., Furuya M. (1971). Phytochrome action in Oryza sativa L. – red and far red reversible effect on the production of ethylene in excised coleoptiles. Plant Physiol. 48, 241–244. 10.1104/pp.48.3.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.-C., Henriques R., Seo H. S., Nagatani A., Chua N.-H. (2010). Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22, 2370–2383. 10.1105/tpc.109.072520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X. Z., Philosoph-Hadas S., Su L. Y., Yang S. F. (1986). The conversion of 1-(malonylamino)cyclopropane-1-carboxylic acid to 1-aminocyclopropane-1-carboxylic acid in plant tissues. Plant Physiol. 81, 637–641. 10.1104/pp.81.2.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X. Z., Yip W. K., Yang S. F. (1987). The effect of light and phytochrome on 1-aminocyclopropane-1-carboxylic acid metabolism in etiolated wheat seedling leaves. Plant Physiol. 85, 643–647. 10.1104/pp.85.3.643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo S., Liu Y., Lueth A., Zhang S. (2008). MPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J. 54, 129–140. 10.1111/j.1365-313X.2008.03404.x [DOI] [PubMed] [Google Scholar]
- Joo S., Seo Y. S., Kim S. M., Hong D. K., Park K. Y., Kim W. T. (2006). Brassinosteroid induction of AtACS4 encoding an auxin responsive 1-aminocyclopropane-1-carboxylate synthase 4 in Arabidopsis seedlings. Physiol. Plant. 126, 592–604 10.1111/j.1399-3054.2005.00602.x [DOI] [Google Scholar]
- Ju C., Chang C. (2012). Advances in ethylene signalling: protein complexes at the endoplasmic reticulum membrane. AoB Plants 2012:pls031 10.1093/aobpla/pls031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B. G., Burg S. P. (1972). Involvement of ethylene in phytochrome-mediated carotenoid synthesis. Plant Physiol. 49, 631–633. 10.1104/pp.49.4.631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M. M., Jaillais Y., Pedmale U. V., Moreno J. E., Chory J., Ballar C. L. (2011). Cryptochrome 1 and phytochrome B control shade-avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J. 67, 195–207. 10.1111/j.1365-313X.2011.04598.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D. R., Estelle M. (2012). Ubiquitin-mediated control of plant hormone signaling. Plant Physiol. 160, 47–55. 10.1104/pp.112.200527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H. (1993). Ethylene biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 283–307 10.1146/annurev.pp.44.060193.001435 [DOI] [Google Scholar]
- Khanna R., Huq E., Kikis E. A., Al-Sady B., Lanzatella C., Quail P. H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix–loop–helix transcription factors. Plant Cell 16, 3033–3044. 10.1105/tpc.104.025643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Shen Y., Marion C. M., Tsuchisaka A., Theologis A., Schäfer E., et al. (2007). The basic helix–loop–helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19, 3915–3929. 10.1105/tpc.107.051508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Yi H., Choi G., Shin B., Song P.-S., Choi G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15, 2399–2407. 10.1105/tpc.014498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Doi M., Suetsugu N., Kagawa T., Wada M., Shimazaki K. (2001). Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414, 656–660. 10.1038/414656a [DOI] [PubMed] [Google Scholar]
- Kircher S., Gil P., Kozma-Bognár L., Fejes E., Speth V., Husselstein-Muller T., et al. (2002). Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14, 1541–1555. 10.1105/tpc.001156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli A., Sreenivasulu N., Lakshmanan P., Kumar P. P. (2013). The phytohormone crosstalk paradigm takes center stage in understanding how plants respond to abiotic stresses. Plant Cell Rep. 32, 945–957. 10.1007/s00299-013-1461-y [DOI] [PubMed] [Google Scholar]
- Kombrink E. (2012). Chemical and genetic exploration of jasmonate biosynthesis and signaling paths. Planta 236, 1351–1366. 10.1007/s00425-012-1705-z [DOI] [PubMed] [Google Scholar]
- Kondoh M., Shiraishi C., Müller P., Ahmad M., Hitomi K., Getzoff E. D., et al. (2011). Light-induced conformational changes in full-length Arabidopsis thaliana cryptochrome. J. Mol. Biol. 413, 128–137. 10.1016/j.jmb.2011.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepin L. V., Walton L. J., Yeung E. C., Chinnappa C. C., Reid D. M. (2010). The interaction of light irradiance with ethylene in regulating growth of Helianthus annuus shoot tissues. Plant Growth Regul. 62, 43–50 10.1007/s10725-010-9483-8 [DOI] [Google Scholar]
- Lau O. S., Deng X. W. (2010). Plant hormone signaling lightens up: integrators of light and hormones. Curr. Opin. Plant Biol. 13, 571–577. 10.1016/j.pbi.2010.07.001 [DOI] [PubMed] [Google Scholar]
- Lee J. M., Joung J.-G., McQuinn R., Chung M.-Y., Fei Z., Tieman D., et al. (2012). Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J. 70, 191–204. 10.1111/j.1365-313X.2011.04863.x [DOI] [PubMed] [Google Scholar]
- Leivar P., Monte E., Al-Sady B., Carle C., Storer A., Alonso J. M., et al. (2008). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20, 337–352. 10.1105/tpc.107.052142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J. M., Monte E., Calderon R. H., Liu T. L., Quail P. H. (2009). Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21, 3535–3553. 10.1105/tpc.109.070672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Johnson P., Stepanova A., Alonso J. M., Ecker J. R. (2004). Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev. Cell 7, 193–204. 10.1016/j.devcel.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Liang X., Abel S., Keller J. A., Shen N. F., Theologis A. (1992). The 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 89, 11046–11050. 10.1073/pnas.89.22.11046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Wang H., Mao L., Hu Y., Dong T., Zhang Y., et al. (2012). Involvement of COP1 in ethylene- and light-regulated hypocotyl elongation. Planta 236, 1791–1802. 10.1007/s00425-012-1730-y [DOI] [PubMed] [Google Scholar]
- Lin C., Yang H., Guo H., Mockler T., Chen J., Cashmore A. R. (1998). Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. U.S.A. 95, 2686–2690. 10.1073/pnas.95.5.2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z. F., Zhong S. L., Grierson D. (2009). Recent advances in ethylene research. J. Exp. Bot. 60, 3311–3336. 10.1093/jxb/erp204 [DOI] [PubMed] [Google Scholar]
- Lincoln J. E., Campbell A. D., Oetiker J., Rottmann W. H., Oeller P. W., Shen N. F., et al. (1993). LE-ACS4, a fruit ripening and wound-induced 1-aminocyclopropane-1-carboxylate synthase gene of tomato (Lycopersicum esculentum). Expression in Escherichia coli, structural characterization, expression characteristics, and phylogenetic analysis. J. Biol. Chem. 268, 19422–19430. [PubMed] [Google Scholar]
- Liscum E., Hangarter R. P. (1993). Light-stimulated apical hook opening in wild-type Arabidopsis thaliana seedlings. Plant Physiol. 101, 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Pirrello J., Kesari R., Mila I., Roustan J.-P., Li Z., et al. (2013). A dominant repressor version of the tomato Sl-ERF.B3 gene confers ethylene hypersensitivity via feedback regulation of ethylene signaling and response components. Plant J. 76, 406–419. 10.1111/tpj.12305 [DOI] [PubMed] [Google Scholar]
- Liu Y., Hoffman N. E., Yang S. F. (1985). Promotion by ethylene of the capability to convert 1-aminocyclopropane-1-carboxylic acid to ethylene in preclimacteric tomato and cantaloupe fruit. Plant Physiol. 77, 407–411. 10.1104/pp.77.2.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16, 3386–3399. 10.1105/tpc.104.026609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P. D., Whitelam G. C., Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323. 10.1111/j.1365-313X.2007.03341.x [DOI] [PubMed] [Google Scholar]
- Luo Q., Lian H.-L., He S.-B., Li L., Jia K.-P., Yang H.-Q. (2014). COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell 26, 2441–2456. 10.1105/tpc.113.121657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lürssen K., Naumann K., Schroder R. (1979). 1-aminocyclopropane-1-carboxylic acid. An intermediate in ethylene biosynthesis in higher plants. Z. Pflanzenphysiol. 92, 285–294 10.1016/S0044-328X(79)80011-2 [DOI] [Google Scholar]
- Lyzenga W. J., Booth J. K., Stone S. L. (2012). The Arabidopsis RING-type E3 ligase XBAT32 mediates the proteasomal degradation of the ethylene biosynthetic enzyme, 1-aminocyclopropane-1-carboxylate synthase 7. Plant J. 71, 23–34. 10.1111/j.1365-313X.2012.04965.x [DOI] [PubMed] [Google Scholar]
- Ma L., Li J., Qu L., Hager J., Chen Z., Zhao H., et al. (2001). Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13, 2589–2607. 10.1105/tpc.13.12.2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. N., Saftner R. A. (1995). Purification and characterization of 1-aminocyclopropanecarboxylic acid N-malonyltransferase from tomato fruit. Plant Physiol. 108, 1241–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Garcia J. F., Huq E., Quail P. H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863. 10.1126/science.288.5467.859 [DOI] [PubMed] [Google Scholar]
- McClellan C. A., Chang C. (2008). The role of protein turnover in ethylene biosynthesis and response. Plant Sci. 175, 24–31. 10.1016/j.plantsci.2008.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon T. A., Yang S.-F. (1987). “Biosynthesis and metabolism of ethylene,” in Plant Hormones and Their Role in Plant Growth and Development, ed. Davies P. J. (Dordrecht: Kluwer Academic Publishers; ), 94–112 10.1007/978-94-009-3585-3_6 [DOI] [Google Scholar]
- McMurchie E. J., McGlasson W. B., Eaks I. L. (1972). Treatment of fruit with propylene gives information about the biogenesis of ethylene. Nature 237, 235–236. 10.1038/237235a0 [DOI] [PubMed] [Google Scholar]
- Millenaar F. F., van Zanten M., Cox M. C. H., Pierik R., Voesenek L. A. C. J., Peeters A. J. M. (2009). Differential petiole growth in Arabidopsis thaliana: photocontrol and hormonal regulation. New Phytol. 184, 141–152. 10.1111/j.1469-8137.2009.02921.x [DOI] [PubMed] [Google Scholar]
- Miyazaki J. H., Yang S. F. (1987). The methionine salvage pathway in relation to ethylene and polyamine biosynthesis. Physiol. Plant. 69, 366–370 10.1111/j.1399-3054.1987.tb04302.x [DOI] [Google Scholar]
- Nakajima N., Mori H., Yamazaki K., Imaseki H. (1990). Molecular cloning and sequence of a complementary DNA encoding 1-aminocyclopropane-1-carboxylate synthase induced by tissue wounding. Plant Cell Physiol. 31, 1021–1029. [Google Scholar]
- Nakatsuka A., Murachi S., Okunishi H., Shiomi S., Nakano R., Kubo Y., et al. (1998). Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant Physiol. 118, 1295–1305. 10.1104/pp.118.4.1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M. M., Fankhauser C., Chory J. (2000). Light: an indicator of time and place. Genes Dev. 14, 257–271. 10.1101/gad.14.3.257 [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J. M., Quail P. H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400, 781–784. 10.1038/23500 [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M., Shinshi H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182. 10.1105/tpc.7.2.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. M., Park C. J., Lee S. B., Ham B. K., Shin R., Paek K. H. (2001). Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13, 1035–1046. 10.1105/tpc.13.5.1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck S. C., Kende H. (1995). Sequential induction of the ethylene biosynthetic enzymes by indole-3-acetic acid in etiolated peas. Plant Mol. Biol. 28, 293–301. 10.1007/BF00020248 [DOI] [PubMed] [Google Scholar]
- Peiser G., Yang S. F. (1998). Evidence for 1-(malonylamino)-cyclopropane-1-carboxylic acid being the major conjugate of aminocyclopropane-1-carboxylic acid in tomato fruit. Plant Physiol. 116, 1527–1532. 10.1104/pp.116.4.1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A., Nagel M.-K., Popp C., Wüst F., Bindics J., Viczián A., et al. (2012). Interaction with plant transcription factors can mediate nuclear import of phytochrome B. Proc. Natl. Acad. Sci. U.S.A. 109, 5892–5897. 10.1073/pnas.1120764109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R., Whitelam G. C., Voesenek L. A. C. J., de Kroon H., Visser E. J. W. (2004a). Canopy studies on ethylene-insensitive tobacco identify ethylene as a novel element in blue light and plant–plant signalling. Plant J. 38, 310–319. 10.1111/j.1365-313X.2004.02044.x [DOI] [PubMed] [Google Scholar]
- Pierik R., Cuppens M. L. C., Voesenek L. A. C. J., Visser E. J. W. (2004b). Interactions between ethylene and gibberellins in phytochrome-mediated shade avoidance responses in tobacco. Plant Physiol. 136, 2928–2936. 10.1104/pp.104.045120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R., Djakovic-Petrovic T., Keuskamp D. H., de Wit M., Voesenek L. A. C. J. (2009). Auxin and ethylene regulate elongation responses to neighbor proximity signals independent of gibberellin and DELLA proteins in Arabidopsis. Plant Physiol. 149, 1701–1712. 10.1104/pp.108.133496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R., Sasidharan R., Voesenek L. A. C. J. (2007). Growth control by ethylene: adjusting phenotypes to the environment. J. Plant Growth Regul. 26, 188–200 10.1007/s00344-006-0124-4 [DOI] [Google Scholar]
- Pierik R., Tholen D., Poorter H., Visser E. J. W., Voesenek L. A. C. J. (2006). The Janus face of ethylene: growth inhibition and stimulation. Trends Plant Sci. 11, 176–183. 10.1016/j.tplants.2006.02.006 [DOI] [PubMed] [Google Scholar]
- Pirrello J., Prasad B. C. N., Zhang W., Chen K., Mila I., Zouine M., et al. (2012). Functional analysis and binding affinity of tomato Ethylene Response Factors provide insight on the molecular bases of plant differential responses to ethylene. BMC Plant Biol. 12:190. 10.1186/1471-2229-12-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu S., Popescu G. V., Bachan S., Zhang Z., Gerstein M., Snyder M., et al. (2009). MPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23, 80–92. 10.1101/gad.1740009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad M. E., Schofield A., Lyzenga W., Liu H., Stone S. L. (2010). Arabidopsis RING E3 ligase XBAT32 regulates lateral root production through its role in ethylene biosynthesis. Plant Physiol. 153, 1587–1596 10.1104/pp.110.156976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad M. E., Stone S. L. (2010). Further analysis of XBAT32, an Arabidopsis RING E3 ligase, involved in ethylene biosynthesis. Plant Signal. Behav. 5, 1425–1429. 10.4161/psb.5.11.13294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P. H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell. Biol. 3, 85–93. 10.1038/nrm728 [DOI] [PubMed] [Google Scholar]
- Rizzini L., Favory J.-J., Cloix C., Faggionato D., O’Hara A., Kaiserli E., et al. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332, 103–106. 10.1126/science.1200660 [DOI] [PubMed] [Google Scholar]
- Rodrigues-Pousada R., Van Caeneghem W., Chauvaux N., Van Onckelen H., Van Montagu M., Van Der Straeten D. (1999). Hormonal cross-talk regulates the Arabidopsis thaliana 1-aminocyclopropane-1-carboxylate synthase gene 1 in a developmental and tissue-dependent manner. Physiol. Plant. 105, 312–320 10.1034/j.1399-3054.1999.105217.x [DOI] [Google Scholar]
- Rottmann W. H., Peter G. F., Oeller P. W., Keller J. A., Shen N. F., Nagy B. P., et al. (1991). 1-aminocyclopropane-1-carboxylate synthase in tomato is encoded by a multigene family whose transcription is induced during fruit and floral senescence. J. Mol. Biol. 222, 937–961 10.1016/0022-2836(91)90587-V [DOI] [PubMed] [Google Scholar]
- Rudus I., Sasiak M., Kepczynski J. (2013). Regulation of ethylene biosynthesis at the level of 1-aminocyclopropane-1-carboxylate oxidase (ACO) gene. Acta Physiol. Plant. 35, 295–307 10.1007/s11738-012-1096-6 [DOI] [Google Scholar]
- Sakai T., Kagawa T., Kasahara M., Swartz T. E., Christie J. M., Briggs W. R., et al. (2001). Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. U.S.A. 98, 6969–6974. 10.1073/pnas.101137598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A., Estelle M. (2010). The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. 61, 1029–1040. 10.1111/j.1365-313X.2010.04112.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C., Deng X. W. (2000). The COP/DET/FUS proteins-regulators of eukaryotic growth and development. Semin. Cell Dev. Biol. 11, 495–503. 10.1006/scdb.2000.0203 [DOI] [PubMed] [Google Scholar]
- Sebastià C. H., Hardin S. C., Clouse S. D., Kieber J. J., Huber S. C. (2004). Identification of a new motif for CDPK phosphorylation in vitro that suggests ACC synthase may be a CDPK substrate. Arch. Biochem. Biophys. 428, 81–91. 10.1016/j.abb.2004.04.025 [DOI] [PubMed] [Google Scholar]
- Sellaro R., Crepy M., Trupkin S. A., Karayekov E., Buchovsky A. S., Rossi C., et al. (2010). Cryptochrome as a sensor of the blue/green ratio of natural radiation in Arabidopsis. Plant Physiol. 154, 401–409. 10.1104/pp.110.160820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G., Carabelli M., Sassi M., Ciolfi A., Possenti M., Mittempergher F., et al. (2005). A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 19, 2811–2815. 10.1101/gad.364005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi V., Raghuram B., Sinha A. K., Chattopadhyay S. (2014). A mitogen-activated protein kinase cascade module, MKK3-MPK6 and MYC2, is involved in blue light-mediated seedling development in Arabidopsis. Plant Cell 26, 3343–3357. 10.1105/tpc.114.128702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M. K., Kumar R., Solanke A. U., Sharma R., Tyagi A. K., Sharma A. K. (2010). Identification, phylogeny, and transcript profiling of ERF family genes during development and abiotic stress treatments in tomato. Mol. Genet. Genomics 284, 455–475. 10.1007/s00438-010-0580-1 [DOI] [PubMed] [Google Scholar]
- Shen Y., Khanna R., Carle C. M., Quail P. H. (2007). Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145, 1043–1051. 10.1104/pp.107.105601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I. S., Bae G., Lee C. H., et al. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl Acad. Sci. U.S.A. 106, 7660–7665. 10.1073/pnas.0812219106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisler E. C., Goren R., Huberman M. (1985). Effect of 2,5-norbornadiene on abscission and ethylene production in citrus leaf explants. Physiol. Plant. 63, 114–120 10.1111/j.1399-3054.1985.tb02828.x [DOI] [Google Scholar]
- Smékalová V., Doskocilová A., Komis G., Šamaj J. (2014). Crosstalk between secondary messengers, hormones and MPK modules during abiotic stress signalling in plants. Biotechnol. Adv. 32, 2–11. 10.1016/j.biotechadv.2013.07.009 [DOI] [PubMed] [Google Scholar]
- Solano R., Stepanova A., Chao Q., Ecker J. R. (1998). Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE FACTOR1. Genes Dev. 12, 3703–3714. 10.1101/gad.12.23.3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Psakis G., Lang C., Mailliet J., Gärtner W., Hughes J., et al. (2011). Two ground state isoforms and a chromophore D-ring photoflip triggering extensive intramolecular changes in a canonical phytochrome. Proc. Natl. Acad. Sci. U.S.A. 108, 3842–3847. 10.1073/pnas.1013377108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu P., Grosskopf D. G., Felix G., Boller T. (1994). The apparent turnover of 1-aminocyclopropane-1-carboxylate synthase in tomato cells is regulated by protein phosphorylation and dephosphorylation. Plant Physiol. 106, 529–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P. S., Tiryaki I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16, 2117–2127. 10.1105/tpc.104.023549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed C. L., Taylor L. K., Harrison M. A. (2004). Red light regulation of ethylene biosynthesis and gravitropism in etiolated pea stems. Plant Growth Regul. 43, 117–125. 10.1023/B:GROW.0000040116.10016.c3 [DOI] [PubMed] [Google Scholar]
- Stepanova A. N., Alonso J. M. (2005). Ethylene signalling and response pathway: a unique signalling cascade with a multitude of inputs and outputs. Physiol. Plant. 123, 195–206 10.1111/j.1399-3054.2005.00447.x [DOI] [Google Scholar]
- Stepanova A. N., Ecker J. R. (2000). Ethylene signaling: from mutants to molecules. Curr. Opin. Plant Biol. 3, 353–360 10.1016/S1369-5266(00)00096-0 [DOI] [PubMed] [Google Scholar]
- Stepanova A. N., Yun J., Likhacheva A. V., Alonso J. M. (2007). Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19, 2169–2185. 10.1105/tpc.107.052068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S. L., Callis J. (2007). Ubiquitin ligases mediate growth and development by promoting protein death. Curr. Opin. Plant Biol. 10, 624–632. 10.1016/j.pbi.2007.07.010 [DOI] [PubMed] [Google Scholar]
- Tatsuki M., Mori H. (2001). Phosphorylation of tomato 1-aminocyclopropane-1-carboxylic acid synthase, LE-ACS2, at the C-terminal region. J. Biol. Chem. 276, 28051–28057. 10.1074/jbc.M101543200 [DOI] [PubMed] [Google Scholar]
- Tepperman J. M., Hudson M. E., Khanna R., Zhu T., Chang S. H., Wang X., et al. (2004). Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red light-regulated gene expression during seedling de-etiolation. Plant J. 38, 725–739. 10.1111/j.1365-313X.2004.02084.x [DOI] [PubMed] [Google Scholar]
- Thain S. C., Vandenbussche F., Laarhoven L. J. J., Dowson-Day M. J., Wang Z. Y., Tobin E. M., et al. (2004). Circadian rhythms of ethylene emission in Arabidopsis. Plant Physiol. 136, 3751–3761. 10.1104/pp.104.042523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A., Theologis A. (2004). Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiol. 136, 2982–3000. 10.1104/pp.104.049999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A., Yu G. X., Jin H. L., Alonso J. M., Ecker J. R., Zhang X. M., et al. (2009). A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics 183, 979–1003. 10.1534/genetics.109.107102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomainen J., Betz C., Kangasjärvi J., Ernst D., Yin Z.-H., Langebartels C., et al. (1997). Ozone induction of ethylene emission in tomato plants: regulation by differential accumulation of transcripts for biosynthesis enzymes. Plant J. 12, 1151–1162 10.1046/j.1365-313X.1997.12051151.x [DOI] [Google Scholar]
- Ulijasz A. T., Cornilescu G., Cornilescu C. C., Zhang J., Rivera M., Markley J. L., et al. (2010). Structural basis for the photoconversion of a phytochrome to the activated Pfr form. Nature 463, 250–254. 10.1038/nature08671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahala J., Schalgnhaufer C. D., Pell E. J. (1998). Induction of ACC synthase cDNA by ozone in light-grown Arabidopsis thaliana leaves. Physiol. Plant. 103, 45–50 10.1034/j.1399-3054.1998.1030106.x [DOI] [Google Scholar]
- Vandenbussche F., Vaseva I., Vissenberg K., Van Der Straeten D. (2012). Ethylene in vegetative development: a tale with a riddle. New Phytol. 194, 895–909. 10.1111/j.1469-8137.2012.04100.x [DOI] [PubMed] [Google Scholar]
- Vandenbussche F., Vriezen W. H., Smalle J., Laarhoven L. J., Harren F. J., Van Der Straeten D. (2003). Ethylene and auxin control the Arabidopsis response to decreased light intensity. Plant Physiol. 133, 517–527. 10.1104/pp.103.022665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Straeten D., Rodrigues-Pousada R. A., Villarroel R., Hanley S., Goodman H., Van Montagu M. (1992). Cloning, genetic mapping, and expression analysis of an Arabidopsis thaliana gene that encodes 1-aminocyclopropane-1-carboxylate. Proc. Natl. Acad. Sci. U.S.A. 89, 9969–9973 10.1073/pnas.89.20.9969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R. D. (2009). The ubiquitin–26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10, 385–397. 10.1038/nrm2688 [DOI] [PubMed] [Google Scholar]
- Vogel J. P., Woeste K. W., Theologis A., Kieber J. J. (1998). Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc. Natl. Acad. Sci. U.S.A. 95, 4766–4771. 10.1073/pnas.95.8.4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J. R., Brunzelle J. S., Forest K. T., Vierstra R. D. (2005). A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438, 325–331. 10.1038/nature04118 [DOI] [PubMed] [Google Scholar]
- Wan L., Zhang J., Zhang H., Zhang Z., Quan R., Zhou S., et al. (2011). Transcriptional activation of OsDERF1 in OsERF3 and OsAP2-39 negatively modulates ethylene synthesis and drought tolerance in rice. PLoS ONE 6:e25216. 10.1371/journal.pone.0025216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. L., Li H., Ecker J. R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14, S131–S151. 10.1105/tpc.001768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. L.-C., Yoshida H., Lurin C., Ecker J. R. (2004). Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428, 945–950. 10.1038/nature02516 [DOI] [PubMed] [Google Scholar]
- Wang Q., Guan Y., Wu Y., Chen H., Chen F., Chu C. (2008). Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol. Biol. 67, 589–602. 10.1007/s11103-008-9340-6 [DOI] [PubMed] [Google Scholar]
- Wheeler R. M., Tibbitts T. W. (1986). Growth and tuberization of potato (Solanum tuberosum L.) under continuous light. Plant Physiol. 80, 801–804. 10.1104/pp.80.3.801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeste K. E., Ye C., Kieber J. J. (1999). Two Arabidopsis mutants that overproduce ethylene are affected in the post-transcriptional regulation of 1-aminocyclopropane-1-carboxylic acid synthase. Plant Physiol. 119, 521–530. 10.1104/pp.119.2.521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters H., Jürgens G. (2009). Survival of the flexible: hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 10, 305–317. 10.1038/nrg2558 [DOI] [PubMed] [Google Scholar]
- Woodson W. R., Park K. Y., Drory A., Larsen P. B., Wang H. (1992). Expression of ethylene biosynthetic pathway transcripts in senescing carnation flowers. Plant Physiol. 99, 526–532. 10.1104/pp.99.2.526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Hu Q., Yan Z., Chen W., Yan C., Huang X., et al. (2012). Structural basis of ultraviolet-B perception by UVR8. Nature 484, 214–219. 10.1038/nature10931 [DOI] [PubMed] [Google Scholar]
- Xiao Y.-Y., Chen J.-Y., Kuang J.-F., Shan W., Xie H., Jiang Y.-M., et al. (2013). Banana ethylene response factors are involved in fruit ripening through their interactions with ethylene biosynthesis genes. J. Exp. Bot. 64, 2499–2510. 10.1093/jxb/ert108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Xu X., Fukao T., Canlas P., Maghirang-Rodriguez R., Heuer S., et al. (2006). Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442, 705–708. 10.1038/nature04920 [DOI] [PubMed] [Google Scholar]
- Xu J., Li Y., Wang Y., Liu H., Lei L., Yang H., et al. (2008). Activation of MPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 283, 26996–27006. 10.1074/jbc.M801392200 [DOI] [PubMed] [Google Scholar]
- Xu X., Paik I., Zhu L., Bu Q., Huang X., Deng X. W., et al. (2014). PHYTOCHROME INTERACTION FACTOR1 enhance the E3 ligase activity of CONSTITUTIVE PHOTOMORPHOGENIC1 to synergistically repress photomorphogenesis in Arabidopsis. Plant Cell 26, 1992–2006. 10.1105/tpc.114.125591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaish M. W., El-Kereamy A., Zhu T., Beatty P. H., Good A. G., Bi Y. M., et al. (2010). The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 6:e1001098. 10.1371/journal.pgen.1001098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagami T., Tsuchisaka A., Yamada K., Haddon W. F., Harden L. A., Theologis A. (2003). Biochemical diversity among the 1-aminocyclopropane-1-carboxylate synthase isoenzymes encoded by the Arabidopsis gene family. J. Biol. Chem. 278, 49102–49112. 10.1074/jbc.M308297200 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y. Y., Matsui M., Ang L. H., Deng X. W. (1998). Role of a COP1 interactive protein in mediating light-regulated gene expression in Arabidopsis. Plant Cell 10, 1083–1094. 10.1105/tpc.10.7.1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. Q., Wu Y. J., Tang R. H., Liu D., Liu Y., Cashmore A. R. (2000). The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103, 815–827 10.1016/S0092-8674(00)00184-7 [DOI] [PubMed] [Google Scholar]
- Yang S. F., Hoffman N. E. (1984). Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 35, 155–189 10.1146/annurev.pp.35.060184.001103 [DOI] [Google Scholar]
- Yang S. F., Ku H. S., Pratt H. K. (1966). Ethylene production from methionine as mediated by flavin mononucleotide and light. Biochem. Biophys. Res. Commun. 24, 729–743 10.1016/0006-291X(66)90387-1 [DOI] [PubMed] [Google Scholar]
- Yao Y., Du Y., Jiang L., Liu J. Y. (2007). Interaction between ACC synthase 1 and 14-3-3 proteins in rice: a new insight. Biochemistry 72, 1003–1007. 10.1134/S000629790709012X [DOI] [PubMed] [Google Scholar]
- Yi C., Deng X. W. (2005). COP1—from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15, 618–625. 10.1016/j.tcb.2005.09.007 [DOI] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y. H., Tena G., Xiong Y., Sheen J. (2008). Dual control of nuclear EIN3 by bifurcate MPK cascades in C2H4 signalling. Nature 451, 789–795. 10.1038/nature06543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon G. M., Kieber J. J. (2013a). 1-Aminocyclopropane-1-carboxylic acid as a signalling molecule in plants. AoB Plants 5:plt017 10.1093/aobpla/plt017 [DOI] [Google Scholar]
- Yoon G. M., Kieber J. J. (2013b). 14-3-3 regulates 1-aminocyclopropane-1-carboxylate synthase protein turnover in Arabidopsis. Plant Cell 25, 1016–1028. 10.1105/tpc.113.110106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon G. M., Kieber J. J. (2013c). ACC synthase and its cognate E3 ligase are inversely regulated by light. Plant Signal. Behav. 8:e26478. 10.4161/psb.26478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Nagata M., Saito K., Wang K. L., Ecker J. R. (2005). Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol. 5: 14. 10.1186/1471-2229-5-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Wang K. L., Chang C. M., Mori K., Uchida E., Ecker J. R. (2006). The ACC synthase TOE sequence is required for interaction with ETO1 family proteins and destabilization of target proteins. Plant Mol. Biol. 62, 427–437. 10.1007/s11103-006-9029-7 [DOI] [PubMed] [Google Scholar]
- Yoshii H., Imaseki H. (1982). Regulation of auxin-induced ethylene biosynthesis. Repression of inductive formation of 1-aminocyclopropane-1-carboxylate synthase by ethylene. Plant Cell Physiol. 23, 639–649. [Google Scholar]
- Zarembinski T. I., Theologis A. (1994). Ethylene biosynthesis and action: a case of conservation. Plant Mol. Biol. 26, 1579–1597. 10.1007/BF00016491 [DOI] [PubMed] [Google Scholar]
- Zhang Z. J., Zhang H. W., Quan R. D., Wang X. C., Huang R. F. (2009). Transcriptional regulation of ethylene response factor LeERF2 in the expression of ethylene biosynthesis genes controls ethylene production in tomato and tobacco. Plant Physiol. 150, 365–377. 10.1104/pp.109.135830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Guo Y., Novák O., Dai X., Zhao Y., Ljung K., et al. (2013). Coordination of auxin and ethylene biosynthesis by the aminotransferase VAS 1. Nat. Chem. Biol. 9, 244–248. 10.1038/nchembio.1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G. V., Burns J. K. (2003). Profiling ethylene-regulated gene expression in Arabidopsis thaliana by microarray analysis. Plant Mol. Biol. 53, 117–131. 10.1023/B:PLAN.0000009270.81977.ef [DOI] [PubMed] [Google Scholar]
- Zhong S., Zhao M., Shi T., Shi H., An F., Zhao Q., et al. (2009). EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc. Natl. Acad. Sci. U.S.A. 106, 21431–21436. 10.1073/pnas.0907670106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Guo H. (2008). Genetic basis of ethylene perception and signal transduction in Arabidopsis. J. Integr. Plant Biol. 50, 808–815. 10.1111/j.1744-7909.2008.00710.x [DOI] [PubMed] [Google Scholar]