Abstract
Metastasis remains the most common cause of death in most cancers, with limited therapies for combating disseminated disease. While the primary tumor microenvironment is an important regulator of cancer progression, it is less well understood how different tissue environments influence metastasis. We analyzed tumor-stroma interactions that modulate organ tropism of brain, bone and lung metastasis in xenograft models. We identified a number of potential modulators of site-specific metastasis, including cathepsin S as a regulator of breast-to-brain metastasis. High cathepsin S expression at the primary site correlated with decreased brain metastasis-free survival in breast cancer patients. Both macrophages and tumor cells produce cathepsin S, and only the combined depletion significantly reduced brain metastasis in vivo. Cathepsin S specifically mediates blood-brain barrier transmigration via proteolytic processing of the junctional adhesion molecule (JAM)-B. Pharmacological inhibition of cathepsin S significantly reduced experimental brain metastasis, supporting its consideration as a therapeutic target for this disease.
Introduction
Cancer cells in a primary tumor are adept at exploiting their local tissue microenvironment. By contrast, when metastatic cells leave these favorable surroundings, they must possess or acquire traits to enable survival and colonization of potentially hostile tissue environments1, 2. The obstacles that metastasizing tumor cells encounter vary between organs, and are influenced by non-cancerous stromal cells in the local microenvironment. For example, the blood-brain barrier, composed of endothelial cells, astrocytes and pericytes, presents a more formidable structure for tumor cells to penetrate, compared to the fenestrated capillaries in bone marrow2. Tumor cells that extravasate in different tissue microenvironments then encounter distinct cell types that can positively or negatively regulate metastatic outgrowth. Indeed, dissemination can occur to multiple organs, yet metastatic tumors typically grow in only one or a few sites3, indicating critical roles for the microenvironment in this process.
Multiple steps of the metastatic cascade require proteolytic activity. Most studies to date have focused on how proteases regulate angiogenesis and invasion in the primary tumor 4, 5. Proteases are organized into highly regulated networks in which their activity is controlled by endogenous inhibitors and interacting partners5. The mechanisms by which proteases regulate tumor progression are highly specific, including cleavage of cell-adhesion molecules6 and processing of growth factors, chemokines and kinases5-10, and are not only limited to protein degradation and matrix remodeling. We therefore investigated the expression of proteolytic effectors in different metastatic sites to determine if protease function is important in the rate-limiting steps of extravasation, seeding and outgrowth.
While previous studies profiling constituent cell types of different tumor microenvironments have been informative in identifying stromal gene signatures, often with prognostic value, they involved manipulation of the tumor to isolate individual cell types, and in most cases stromal cells alone were analyzed, without comparative expression data for the tumor cells11-16. Moreover, the focus has largely been on tumor-stroma interactions in the primary tumor, without consideration of the metastatic process. Here, we specifically examine the interplay between cancer cells and the microenvironment in intact xenograft tumors at distinct stages of metastasis using a dual species-specific microarray platform. We focused on three organ sites to which breast cancer commonly metastasizes: the brain, bone and lung17-19. We find that tumor cell-derived proteases and their inhibitors predominantly undergo stage-specific changes in expression during metastatic seeding and outgrowth in different organs, whereas stromal-derived genes are primarily regulated in a tissue-specific manner. From these analyses, we show that cathepsin S expression is regulated in both a cell type-specific and stage-dependent manner, and enhances breast-to-brain metastasis.
Results
Differential expression of proteases and protease inhibitors in different metastatic microenvironments
To investigate tumor-stroma interactions in different environments we used a mouse model for organ-specific experimental metastasis (Fig. 1a). In this model, three different metastatic variants of the human breast cancer cell line MDA-MB-231 (Refs. 20-22) were injected either intracardially or intravenously into immunocompromised mice, resulting in the development of brain, bone, or lung metastases. While previous studies focused on profiling tumor cell-specific expression in each of these cell line variants20-22, we have been able to additionally capture the stromal contribution by removing intact whole tumors at distinct stages of metastatic seeding and outgrowth in different organs, followed by expression analyses (Fig. 1a, Supplementary Fig. 1).
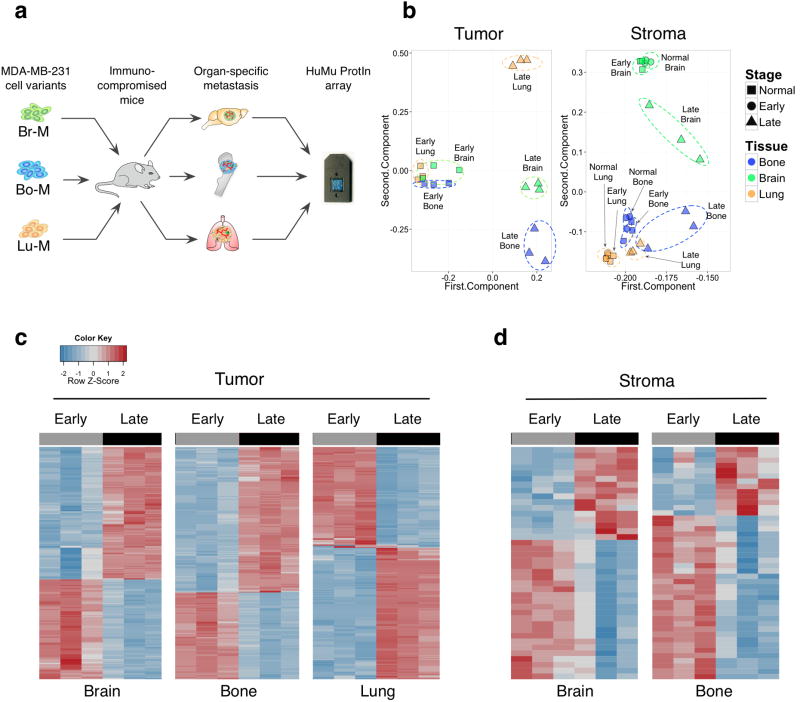
Figure 1. HuMu ProtIn array enables simultaneous acquisition of gene expression changes in tumor and stromal cells.
(a) Schematic of the experimental design employed to analyze tumor stroma interactions in different metastatic microenvironments using the dual species-specific HuMu ProtIn (Human/ Murine Proteases and Inhibitors) array in xenografted animals. Br-M = brain metastatic, Bo-M = bone metastatic, and Lu-M = lung metastatic variants of the MDA-MB-231 cell line. (b) Principal component analysis of the HuMu array data: the 1st and 2nd component are plotted on the x and y axes respectively. These two components together represent the largest sources of variation in the dataset. The first and second components represent 89.98% and 8.44% respectively of the variance in the tumor gene space, and 90.83% and 4.06% in the stromal gene space. This analysis revealed variation in tumor gene expression that was predominantly associated with differences between early- and late-stage metastasis. Meanwhile, variation in the stroma was associated with both stage and tissue. Dotted ellipses were drawn manually to indicate related data points within stage or organ. (c,d) Heatmaps of (c) tumor- and (d) stroma-derived genes that were differentially expressed between early and late metastases across different organ sites. The lung stroma did not show extensive differences between early and late stages (Supplementary Table 1g).
We used a platform that enabled simultaneous analysis of tumor and stromal gene expression; the “HuMu ProtIn” custom array (Hu= Human, Mu= Murine, Prot= Protease and In= Inhibitor), which surveys proteases, their endogenous inhibitors and interacting partners23. The uniqueness of this microarray is based on the species-specificity of the probe sets, with no cross-reactivity between human and mouse genes23. This platform thus allowed us to distinguish between mRNA expression changes in the tumor (human) and stromal (mouse) gene space in response to metastatic seeding and outgrowth (early- and late-stage metastases respectively, Supplementary Fig. 1), with the goal of identifying tumor-stroma interactions that modulate organ-specific metastasis. Each of the metastatic cell variants was transduced with a triple-fusion TK-GFP-Luc imaging vector, enabling non-invasive bioluminescence imaging (BLI) as a read-out of metastatic burden, as previously described24. Early- and late-stage metastases in each organ site were harvested based on BLI output, as described in the methods, corresponding to micrometastatic and macrometastatic disease respectively.
Principal component analysis (PCA) was used to evaluate global trends in proteolytic network expression across tissue and stage for both tumor cells and stromal cells (Fig. 1b). Analysis of tumor cell-specific gene expression revealed pronounced changes between early- and late-stage metastases across all three organs. Meanwhile, stromal genes were differentially expressed between early- and late-stage metastases in a tissue-dependent manner. The brain and bone stroma showed the most robust changes in gene expression as metastases progressed. Across all tissues, there were few changes in gene expression between the normal tissue (i.e. non-tumor burdened), and the stroma of early metastases (brain, bone = 0 genes, lung = 3 genes, Supplementary Table 1a). This could reflect the relatively low disease burden at the early stages, resulting in a minimal impact on the organ as a whole. Alternatively, expression changes in proteolytic genes in the stroma may not be as important in the earliest stages of metastatic extravasation and seeding, possibly due to a predominant role for tumor-supplied proteases or non-protease factors in the stroma.
Differential gene expression analyses revealed that many genes changed in tumor cells in the brain, bone and lung (242, 241 and 245 genes respectively) between early- and late-stage metastases (Fig. 1c, Supplementary Table 1b-d). By comparison, there were fewer stage-specific differentially expressed genes identified in the brain and bone stroma (40 and 44 genes respectively), and only one differentially expressed gene, haptoglobin, in the lung stroma (Fig. 1d, Supplementary Table 1e-g). A substantial proportion of tumor-derived genes were shared across all three metastatic sites (176 genes, Supplementary Fig. 2a), whereas few stromal genes were shared across >1 metastatic site (10 genes, Supplementary Fig. 2b). Rather, there were multiple tissue-specific proteases and inhibitors in the brain, bone and lung stroma (Supplementary Fig. 2c). Quantitative real-time PCR (qPCR) confirmed tissue-specific enrichment of representative candidates for each organ in normal tissue, and early- and late-stage metastases (Supplementary Fig. 2d). Representative proteases and protease inhibitors that exhibited stage-specific gene expression changes between early and late metastases were validated using immunostaining (Fig. 2a-e) and qPCR (Supplementary Fig. 3a-e). We also validated several genes identified in lung metastasis xenografts (Supplementary Table 1a) in the immunocompetent MMTV-PyMT model of breast-to-lung metastasis (Supplementary Fig. 3f, g).
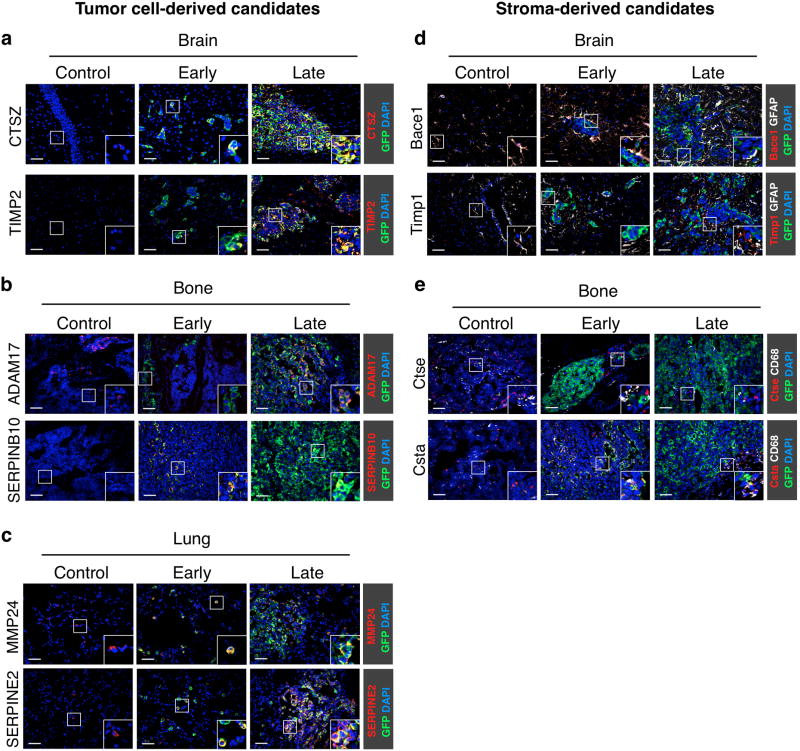
Figure 2. Independent validation of differentially expressed genes in experimental brain, bone and lung metastases.
(a-e) Representative images of control (non tumor-burdened) tissue, early- and late-stage site-specific metastases (classified by BLI intensity as in Supplementary Fig. 1; n=3 samples for each stage and tissue) showing immunofluorescence staining of tumor- and stromal-derived proteases and protease inhibitors exhibiting stage-dependent expression changes in the HuMu ProtIn array. (a) Brain sections were stained with antibodies against the protease CTSZ (red) and the protease inhibitor TIMP2 (red) as representative candidates that were differentially expressed in tumor cells. (b) Bone sections were stained with antibodies against the protease ADAM17 (red) and the protease inhibitor SERPINB10 (red) to represent differentially expressed candidates in tumor cells in bone metastases. (c) Lung sections were stained with antibodies against the protease MMP24 (red) and the protease inhibitor SERPINE2 (red) to confirm stage-differential expression in tumor cells in lung metastases. (d) Staining for the stromal-derived protease Bace1 (red) and the protease inhibitor Timp1 (red) confirmed stage-specific expression changes in GFAP+ astrocytes (white). (e) Staining for the protease Ctse (red) and the protease inhibitor Csta (red) confirmed stage-specific stromal changes in bone metastasis. CD68+ macrophages (red) were identified as the predominant source for Csta in bone metastases. All sections were stained with GFP (green) to visualize tumor cells and DAPI as a nuclear counter stain. Scale bar indicates 50 μm.
We next asked whether expression changes in stromal cells in organ-specific metastases are a general response to tumor cell colonization of the respective tissue, or if they are specific to the metastatic cell variant used. In the models used here, bone metastases occasionally develop in animals inoculated with the brain-metastatic (Br-M) variant, and conversely brain metastases can be observed in mice inoculated with bone-metastatic (Bo-M) cells. This allowed us to compare stromal and tumor gene expression in the ‘matched’ (Br-M to brain, Bo-M to bone) and ‘mismatched’ (Br-M to bone, Bo-M to brain) samples. Interestingly, for the genes tested, we found that stromal expression changes depend on tumor-stroma interactions specific to the metastatic tumor cell variant (Supplementary Fig. 3a, b). By contrast, tumor gene expression in different metastatic variants responds to the same microenvironment in a similar manner, suggesting an important effect of the stroma on the tumor gene expression program (Supplementary Fig. 3c, d).
Cathepsin S is negatively associated with metastasis-free survival in patients with brain metastasis
While previous analyses have identified genes associated with site-specific metastases, few studies have uncovered genes that are concordantly or discordantly expressed in tumor cells and stroma. We therefore took advantage of the species specificity of the HuMu arrays to separately profile stroma- and tumor-derived genes in cross-species analyses. We identified genes for which both human and mouse homologs were significantly altered in each metastatic site (Fig. 3a, Supplementary Fig. 4a, b). Cathepsin S showed an intriguing expression pattern: while tumor-derived cathepsin S was high in early brain metastases and decreased in late-stage metastases, stromal cathepsin S displayed the inverse pattern, with higher expression in late-stage brain metastases versus early-stage. qPCR using species-specific probes for cathepsin S confirmed these data in an independent sample set (Fig. 3b). To distinguish between the cellular sources of cathepsin S, we will refer to tumor/ human CTSS in capitals, and stromal/ mouse Ctss in lower-case.
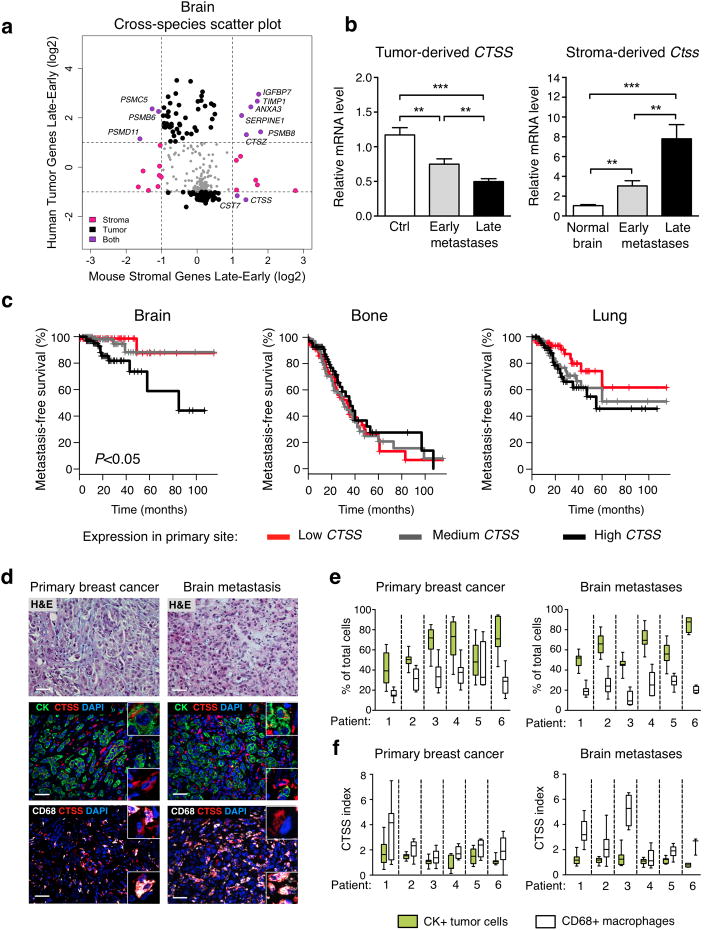
Figure 3. Cathepsin S shows highly regulated stage- and cell type-specific expression changes in experimental brain metastases, and cathepsin S expression in primary breast tumors is inversely correlated with brain metastasis-free survival in patients.
(a) Cross-species scatter plot shows log-fold expression changes in the tumor and stromal gene space in early- vs. late-stage brain metastases. Differentially expressed genes in either the stroma (mouse) or tumor (human) gene space are shown in pink or black respectively. Genes that are differentially expressed in both the stromal and tumor gene space are shown in purple. Grey dots represent homologs with either insufficient fold change or P values. Horizontal and vertical lines denote fold change cut-off for significance. (b) Expression of tumor-derived and stromal-derived cathepsin S (CTSS or Ctss respectively) in Br-M control (Ctrl; n=11 mice) cell line, normal brain (n=12 mice), and brain metastases (classified by BLI intensity; n=16 mice for early-stage and n=17 mice for late-stage metastases). mRNA expression is depicted relative to Ctrl for CTSS and relative to normal brain for Ctss. All assays were performed in triplicate and gene expression was normalized to Ubc (stromal genes), B2M (tumor cell-derived genes). (c) Metastasis-free survival (MFS) for breast cancer patients (GSE12276 data set) based on low, medium and high CTSS expression at the primary site. (d) Representative images of matched primary breast cancer and brain metastasis patient samples stained for CTSS (red) and CD68 (macrophages; white), or pan-cytokeratin (CK, tumor cells; green), and DAPI (nuclei; blue). Images are representative of specimens quantified in (e). Scale bar indicates 50 μm. (e) Quantification of proportions of tumor cells and macrophages, presented as the percentage of total DAPI+ cells, in matched primary breast cancer (n=6 samples) and brain metastases (n=6 samples). (f) Quantification of the CTSS index as a measure of relative CTSS levels in tumor cells and macrophages. Data are presented as bars + s.e.m. or as box plots (Boxes; values between the 25th and 75th percentile, whiskers; minimum and maximum values, horizontal line; median). P values were obtained using two-tailed unpaired t-test for (b) and a log-rank test for (c). **P<0.01, and ***P<0.001.
We investigated whether there were any associations between CTSS expression at the primary site and organ-specific metastasis-free survival (MFS) using a dataset of locally advanced primary breast cancer with clinical annotation20. Patients were separated into three equal tertiles of low, medium and high CTSS expression as described in the methods. Kaplan-Meier analysis was used to assess MFS for brain, bone and lung. Interestingly, the high CTSS expression group was associated with decreased MFS only for the brain, and not bone or lung (Fig. 3c). This was further evident in a complementary Cox proportional hazards model analysis (Supplementary Fig. 4c).
We similarly determined whether other tumor genes that were differentially expressed in the experimental model (Fig. 1c) were associated with differences in patient survival (Supplementary Table 2). In addition to CTSS, 26 other genes were significantly associated with brain MFS. Of these, 23 genes were negatively associated with brain MFS (Supplementary Fig. 4c, Supplementary Table 2a). 30 genes were associated with bone MFS, of which 6 genes were negatively associated (Supplementary Fig. 4d, Supplementary Table 2b). 59 genes were associated with lung MFS, of which 45 genes were negatively associated (Supplementary Fig. 4e, Supplementary Table 2c). Although tumor cells underwent largely congruent changes in gene expression from early- to late-stage metastases across the three organs (Supplementary Fig. 2a), only 20 of these genes were significantly associated with MFS at multiple sites, whereas the majority of genes were associated with tissue-specific MFS (Supplementary Fig. 4f, Supplementary Table 2a-c).
CST7 in brain metastasis, together with CTSS and SERPINA3 in bone metastasis, were the only genes that showed similar stage-dependent and cell type-specific expression changes to CTSS in brain metastasis (Fig. 3a, Supplementary Fig. 4a). Given that we did not observe an association of CTSS expression with patient bone MFS, and neither CST7 nor SERPINA3 expression associated with brain and bone MFS respectively (Supplementary Fig. 4a, b), we chose to further investigate the potential role of cathepsin S specifically in brain metastasis, a function not previously ascribed to this protease, or any cathepsin family member.
The patient expression data above was derived from whole tumor samples, thus precluding cell type-specific expression analyses. We therefore stained a set of patient samples of brain metastases, with matched primary breast tumors in a subset of cases (Supplementary Table 3). Across all samples (breast cancer and brain metastases), we found the major cell types contributing to the tumor mass were cytokeratin (CK)+ tumor cells and CD68+ macrophages, with a minor fraction representing CK-CD68- cells (Fig. 3d, e, Supplementary Fig. 5a-d). CTSS levels were highest in CD68+ macrophages, with expression also in CK+ tumor cells, albeit at lower levels than in macrophages, in both primary tumors and matched brain metastases (Fig. 3d, f, Supplementary Fig. 5a, b, e). CTSS expression in tumor cells was observed in all molecular subtypes of breast cancer analyzed (Fig. 3d, f, Supplementary Fig. 5a, b, e, Supplementary Table 3).
Combined depletion of cathepsin S in tumor and stromal cells reduces experimental brain metastasis
We investigated the stromal cell source of Ctss in the experimental brain metastasis model. Seeding and outgrowth of brain metastasis induced a stromal response characterized by accumulation of astrocytes and macrophages/microglia in metastatic lesions (Supplementary Fig. 1d). Detection of cathepsin S using an antibody that recognizes both mouse and human homologs, in combination with cell-type specific markers, identified macrophages as the predominant stromal cell type expressing Ctss in brain metastases and normal brain (Fig. 4a). We observed a gradual increase of Ctss expression in Iba1+ macrophages from normal brain to early- and late-stage metastases. CTSS expression was also detectable in tumor cells, though at lower levels than in macrophages, mirroring the patient analyses. At late stages, CTSS expression was undetectable in the majority of the tumor cells. We found a similar expression pattern in an immunocompetent brain metastasis model (Supplementary Fig. 3g). These data confirm the stage- and cell type-dependent expression changes at the protein level as predicted by the HuMu array.
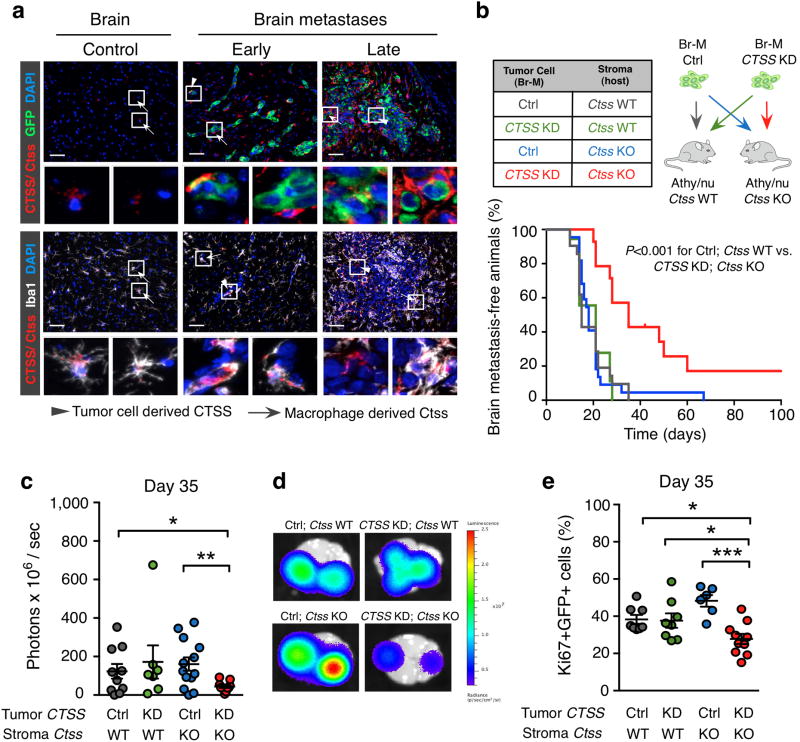
Figure 4. Macrophages are the predominant source of stromal-derived cathepsin S and only combined depletion of tumor- and stromal-derived cathepsin S reduces experimental brain metastasis.
(a) Representative images of normal brain, early- and late-stage brain metastasis (classified by BLI intensity) co-stained for Ctss/CTSS (red) and GFP (tumor cells; green) or Iba1 (macrophages/microglia; white). Tumor cell-derived CTSS is indicated by the arrowhead and macrophage-derived Ctss is indicated by the arrow. Images are representative of 5 independent specimens for each stage. (b) Kaplan-Meier curve shows the percentage of brain metastasis-free animals in the 4 experimental groups indicated in the table. Ctrl; Ctss WT (n=21 mice), CTSS KD; Ctss WT (n=16 mice), Ctrl; Ctss KO (n=22 mice), and CTSS KD; Ctss KO (n=12 mice). (c) Quantification of the ex vivo BLI intensity on day 35 after Br-M tumor cell inoculation. Ctrl; Ctss WT (n=10 mice), CTSS KD; Ctss WT (n=7 mice), Ctrl; Ctss KO (n=13 mice), and CTSS KD; Ctss KO (n=11 mice). (d) Representative ex vivo BLI images of the 4 experimental groups as shown in (c). (e) Quantification of tumor cell proliferation (percentage of Ki67+GFP+ cells) on day 35 after tumor cell inoculation. Ctrl; Ctss WT (n=8 mice), CTSS KD; Ctss WT (n=8 mice), Ctrl; Ctss KO (n=6 mice), and CTSS KD; Ctss KO (n=10 mice). Scale bar indicates 50 μm. Circles represent individual mice and horizontal lines represent the mean ± s.e.m. P values were obtained with Mantel-Cox log-rank test for MFS and with two-tailed unpaired t-test for numerical data. *P<0.05, **P<0.01 and ***P<0.001.
Given the reciprocal, cell type-specific expression pattern of cathepsin S, we sought to investigate if tumor and stromal sources play important, perhaps complementary roles in the seeding and outgrowth of experimental brain metastases. To address this, we performed short hairpin (sh)-RNA-mediated CTSS knockdown (KD) in Br-M cells, achieving a 90% reduction of CTSS expression at both the mRNA and protein level, and a corresponding reduction in secreted CTSS protein (Supplementary Fig. 6a-c). CTSS knockdown did not alter tumor cell proliferation in culture (Supplementary Fig. 6d). After backcrossing Ctss knockout (KO) mice25 into the Athy/nu background, we generated four experimental groups (shown in Fig. 4b), to analyze the effects of targeting tumor or stromal cathepsin S alone, or in combination, compared to the control group. Interestingly, only the combined removal of tumor and stromal cathepsin S significantly reduced brain metastasis incidence (Fig. 4b, Control (Ctrl); Ctss wild-type (WT) vs. CTSS KD; Ctss KO), whereas targeting either source separately had no effect. A separate cohort of mice for all four experimental groups was aged until day 35 post-tumor cell injection, which was selected as the time point by which all mice in the control group had developed brain metastases (Fig. 4b). Brain imaging at this endpoint revealed a 64% decrease in BLI signal only in the CTSS KD; Ctss KO group (Fig. 4c, d). Together, these results indicate that while there is a stage-dependency to cell type-specific cathepsin S expression, contributions from both cellular sources are apparently required to regulate brain metastasis.
Cathepsin S promotes transmigration of the blood-brain barrier by metastatic cells
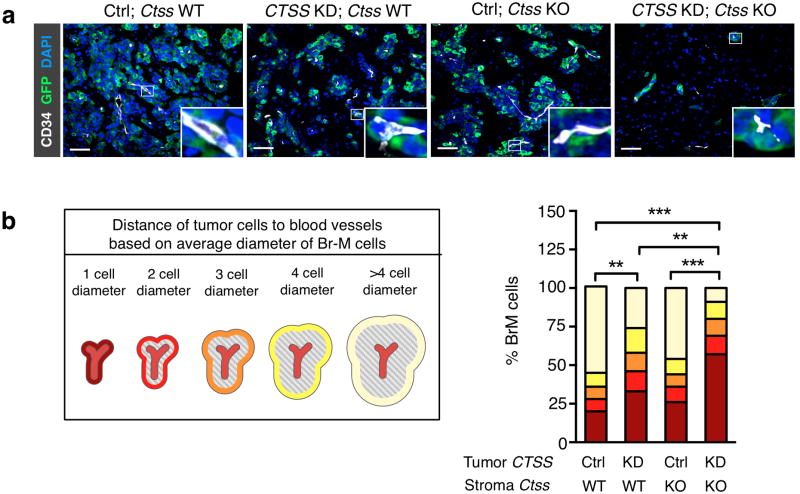
To gain insights into the mechanisms underlying impaired metastatic seeding and/ or outgrowth specifically in the CTSS KD; Ctss KO group, we next analyzed multiple tumorigenic processes at day 35. We found that the size and proliferation rate of brain metastases in CTSS KD; Ctss KO mice were significantly lower than any of the other groups (Fig. 4c-e), while there were no differences in apoptosis (Supplementary Fig. 6e). The small lesions which did develop in the CTSS KD; Ctss KO mice were closely apposed to the vasculature, with the majority of tumor cells being only 1 cell diameter from the vessel, and a pronounced reduction in growth away from blood vessels (Fig. 5a, b). Similarly, analysis of the GFP+ tumor cell area relative to the CD34+ blood vessel area confirmed this reduction (Supplementary Fig. 6f). Critically, Ctss deletion did not alter blood vessel density or permeability in the normal brain of non-tumor bearing animals (Supplementary Fig. 6g, h). These results are suggestive of either a potential defect in seeding of brain metastatic cells in the earliest stages, or a subsequent impairment in colonization, or perhaps deficiencies in both processes.
Figure 5. Cathepsin S deficiency in tumor cells and macrophages impairs metastatic seeding and outgrowth.
(a) Representative images of brain metastases (day 35) stained for GFP (tumor cells; green), the endothelial cell marker CD34 (white), and DAPI to visualize nuclei (blue). Scale bar indicates 50 μm. Images are representative of independent specimens quantified in (b). (b) GFP+ tumor cells were categorized based on their localization relative to blood vessels, defined as the distance of tumor cells from blood vessels (1 to >4 average tumor cell diameter), and the percentage of tumor cells in each defined area was quantified using Metamorph image analysis software. Ctrl; Ctss WT (n=4 mice), CTSS KD; Ctss WT (n=6 mice), Ctrl; Ctss KO (n=6 mice), and CTSS KD; Ctss KO (n=6 mice). Categorical data are plotted as stacked bars. P values were obtained with an ordinal Chi-square test for categorical data. **P<0.01 and ***P<0.001.
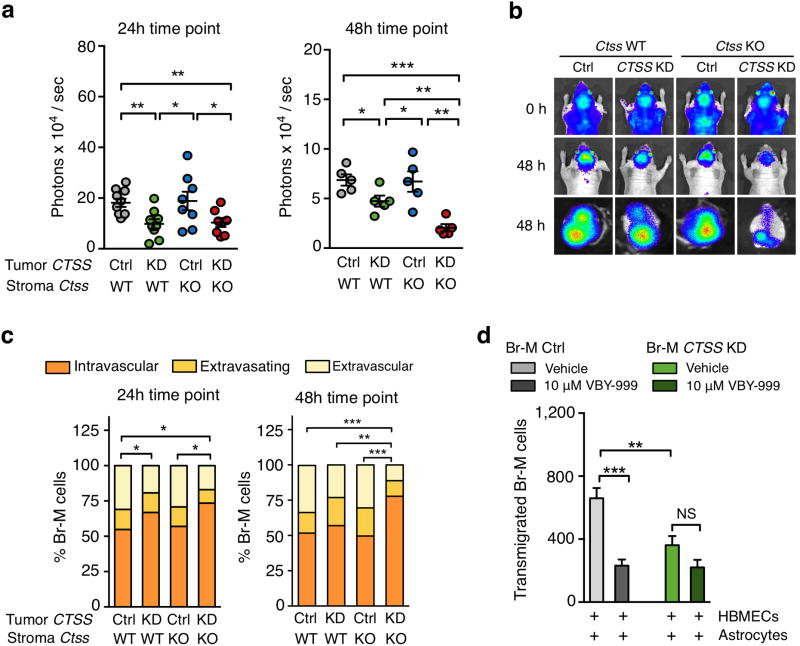
To investigate these possibilities, we assessed metastatic seeding across the four experimental groups. We examined the earliest steps of brain metastatic cell homing and survival26, specifically the first 48h. We found that 24h after CTSS KD tumor cell injection, BLI signal was reduced in both WT and Ctss KO mice, with a further decrease in BLI signal at 48h only in the CTSS KD; Ctss KO group (Fig. 6a, b). Similarly, analysis of the proportion of viable tumor cells still within the blood vessel lumen (intravascular), in the process of extravasating, or fully extravascular, revealed significant differences in the CTSS KD; WT group at 24h, and in the CTSS KD; Ctss KO group at both 24h and 48h (Fig. 6c).
Figure 6. Cathepsin S mediates blood-brain barrier transmigration of brain metastatic cells.
(a) Quantification of BLI intensity at the indicated time points relative to BLI signal immediately after tumor cell inoculation. Ctrl; Ctss WT (n=10 mice), CTSS KD; Ctss WT (n=9 mice), Ctrl; Ctss KO (n=8 mice), and CTSS KD; Ctss KO (n=8 mice) for the 24h time point, and n=5 for each group for the 48h time point. (b) Representative BLI images in the 4 experimental groups immediately (0h) and 48h after tumor cell injection in vivo (top panels) and ex vivo (lower panel). Images are representative of the number of mice as in (a). (c) Tumor cells were categorized based on their localization relative to the vasculature defined as intravascular, extravasating and extravascular, and the percentage of viable tumor cells in each category was quantified at the indicated time points. Ctrl; Ctss WT (n=4 mice), CTSS KD; Ctss WT (n=3 mice), Ctrl; Ctss KO (n=3 mice), and CTSS KD; Ctss KO (n=4 mice) for the 24h time point, and n=4 mice for each group for the 48h time point. (d) Quantification of the number of transmigrated Br-M Ctrl and CTSS KD cells in the presence or absence of the cathepsin S-specific inhibitor VBY-999 through an in vitro BBB formed with human brain microvascular endothelial cells (HBMEC) in co-culture with astrocytes. 200 fields of view (FOV) were analyzed per sample. n=25, 15, 22, 15 samples were analyzed in three independent experiments. Circles represent individual mice and horizontal lines represent the mean ± s.e.m. for numerical data shown in (a). Graphs represent mean + s.e.m in (d). Categorical data are plotted as stacked bars. P values were obtained with two-tailed unpaired t-test for numerical data and with an ordinal Chi-square test for categorical data. NS = not significant, *P<0.05, **P<0.01, and ***P<0.001.
Given the initial reduction in tumor cell extravasation in CTSS KD; Ctss WT mice (Fig. 6a, c), although the incidence of detectable brain metastasis was ultimately not affected (Fig. 4b), we assessed subsequent metastatic colony outgrowth. While there was an initial trend towards delayed growth in the CTSS KD; Ctss WT cohort, brain metastases ultimately grew to the same extent as controls (Supplementary Fig. 7a). By contrast, tumor growth kinetics in the CTSS KD; Ctss KO group did not recover over the same period (Supplementary Fig. 7a). This suggests that tumor- and stromal-derived cathepsin S show some functional redundancy during seeding and outgrowth, and the impact of each cellular source is most likely regulated by differential expression levels at distinct stages. Additionally, tumor cell derived-CTSS may be important for the initial steps of blood-brain barrier (BBB) transmigration and extravasation into the brain, whereas stromal-supplied Ctss is subsequently involved in supporting tumor cell survival to successfully form brain micrometastases, and only their combined depletion impairs both metastatic seeding and outgrowth. A similar finding was reported in a colorectal carcinoma model, where depletion of both tumor and stromal cathepsin S was required to slow tumor growth27.
Cathepsin S promotes BBB transmigration via junctional protein cleavage
The BBB is a selective barrier between the systemic circulation and the brain, which is formed by specialized endothelial cells, pericytes and astrocytes28. While the BBB restricts the entry of most macromolecules, it is not an impenetrable barrier to transmigration of metastasizing cancer cells. We therefore examined the potential role of tumor cell-supplied CTSS in BBB penetration, by using an in vitro BBB assay29. We performed genetic or pharmacological depletion of CTSS in Br-M cells via shRNA-mediated knockdown or a cathepsin S-specific inhibitor VBY-999 respectively, which did not affect Br-M cell viability (Supplementary Fig. 8a). CTSS depletion did not affect the ability of Br-M cells to cross a BBB formed by human umbilical endothelial vein cells (HUVECs) and astrocytes (Supplementary Fig. 8b). By contrast, when human brain microvascular endothelial cells (HBMECs) were used instead of HUVECs, this presented a stronger barrier to Br-M transmigration, which was impaired by 55-65% via genetic or pharmacological depletion of CTSS respectively (Fig. 6d). HBMEC monolayers (without astrocytes) also significantly decreased CTSS KD Br-M cell transmigration (Supplementary Fig. 8b), whereas transmigration across HUVECs or astrocytes alone was not altered by CTSS depletion (Supplementary Fig. 8b).
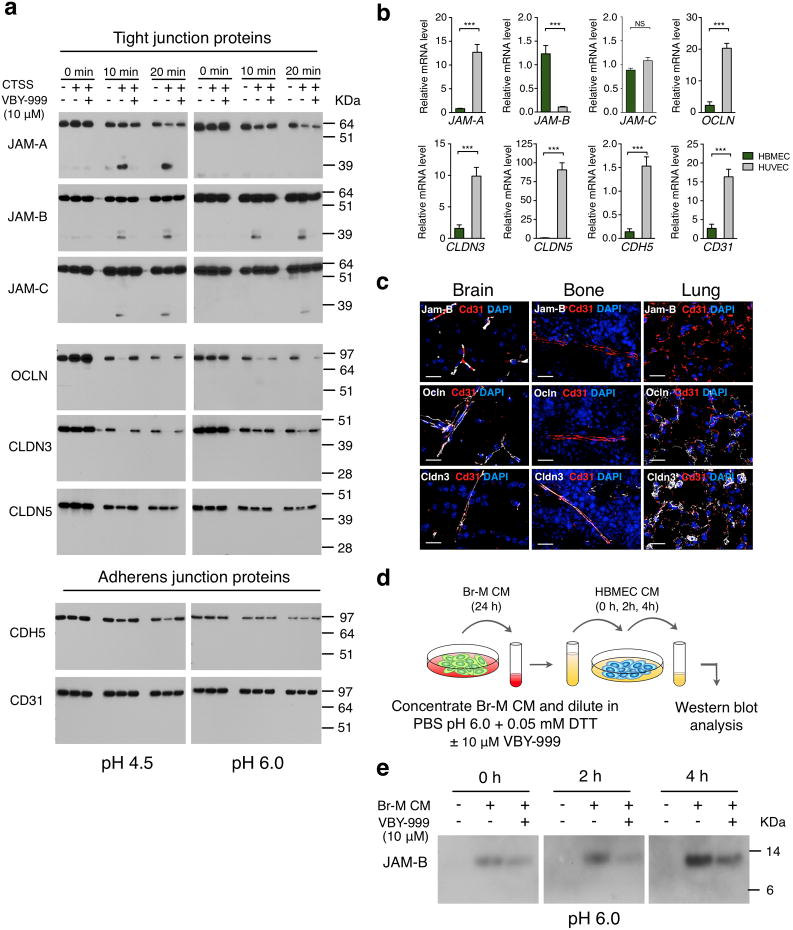
Tight and adherens junctions between adjacent cells are critical for maintaining BBB integrity, and are composed of different proteins including junctional adhesion molecules (JAMs), occludin, claudins and cadherins28, 30, 31. Therefore, we investigated whether any of these proteins represented potential CTSS substrates. We performed biochemical cleavage assays using recombinant CTSS and recombinant proteins for each of the potential substrates, under similar conditions to those previously reported for E-cadherin cleavage by CTSS6. CTSS efficiently cleaved the JAM family members JAM-A, -B and -C at pH 4.5, the acidic pH of the lysosome, and maintained robust cleavage of JAM-B specifically at pH 6.0, the acidified pericellular pH measured in solid tumors32. Importantly, cathepsin S retains activity at neutral pH33. JAM cleavage was inhibited by VBY-999 in all cases (Fig. 7a). Occludin and Claudin (CLDN)-3 were also cleaved by CTSS, whereas CLDN5 and the adherens junction proteins CD31 and CDH5 (VE-cadherin) were unaffected (Fig. 7a). We examined expression of these genes in HBMECs and HUVECS, and found that only JAM-B was significantly higher in HBMECs (Fig. 7b). Tissue-specific enrichment of junctional protein genes was confirmed by querying published datasets34 (Supplementary Fig. 8c). Staining of several junctional proteins further revealed tissue-specific expression for Jam-B, which was detectable only in brain, and not bone or lung, while Occludin and Cldn3 showed a broader expression pattern (Fig. 7c).
Figure 7. Cathepsin S cleaves tight junction proteins that regulate blood-brain barrier integrity.
(a) Western blot analysis of CTSS-mediated cleavage of recombinant tight junction proteins (junctional adhesion molecules (JAM)-A, -B and -C, occludin (OCLN), claudins (CLDN)-3 and-5), and adherens junction proteins cadherin 5 (CDH5) and CD31 for the indicated time points at pH 4.5 and pH 6.0 in the presence or absence of the cathepsin S-specific inhibitor VBY-999. VBY-999 was used at 10 μM, a concentration that efficiently inhibits cathepsin S. (b) mRNA expression of the tight junction and adherens junction molecules in HUVECs and HBMECs (n=9 samples for each cell line). All assays were run in triplicate and gene expression was normalized to B2M. Expression is depicted relative to expression in HBMECs. (c) Representative images of control brain, bone, and lung sections stained for the tight junction proteins Jam-B, Ocln or Cldn 3 (white), with CD31 (red) to visualize blood vessels. DAPI was used as a nuclear counterstain. Images are representative of 3 independent specimens. (d) Schematic of the cell-based cleavage assay. (e) Western blot analysis showing increased JAM-B in HBMEC-conditioned media (CM) after incubation with Br-M cell CM for the indicated time points. Addition of the cathepsin S specific inhibitor VBY-999 (10 μM) resulted in reduced accumulation of JAM-B in HBMEC CM at the indicated time points. Incubation with PBS pH 6.0, 0.05 mM DTT served as a control for baseline JAM-B shedding of HBMEC. Scale bar indicates 20 μm. Graphs represent mean + s.e.m. P values were obtained using two-tailed unpaired t-test. NS = not significant, ***P<0.001. Each western blot shows the representative result of three independent experiments. Uncropped images of blots are shown in Supplementary Fig. 9.
As the effects of CTSS depletion or inhibition on Br-M transmigration were only observed when HBMECs were used in the BBB assay, and given the organ-specificity of Jam-B expression (Fig. 7c), we reasoned that JAM-B might be the most relevant substrate in this context. We aimed to identify the putative cleavage location for CTSS in JAM-B. We compared fragment sizes of each cleavage product that was detectable with JAM-A, -B, or -C specific antibodies to fragments that contain the immunoglobulin (Ig)G1 domain, which is linked to recombinant JAM proteins (Supplementary Fig. 8d). The molecular weight of JAM cleavage products and pH dependence of JAM processing by CTSS suggests that all 3 family members share a similar but not fully conserved CTSS cleavage site that is localized close to the transmembrane domain. Alignment of the amino acid sequence of the JAM family members identified the putative cleavage site for CTSS (Supplementary Fig. 8e), which is consistent with CTSS specificity preferences previously identified in biochemical studies35-37. Cleavage in this region likely leads to shedding of the JAM extracellular domain, thereby disrupting cell-cell adhesion. We performed cell-based assays to determine if tumor cell-secreted CTSS mediates removal of JAM-B from the HBMEC surface (Fig. 7d). Indeed, incubation of HBMECs with Br-M cell-conditioned media (CM) led to a CTSS-mediated accumulation of JAM-B in HBMEC CM after 2-4h. The effect was decreased by addition of VBY-999 (Fig. 7e, Supplementary Fig. 8f). These results are consistent with impaired BBB transmigration in vitro and in vivo when CTSS is targeted, as CTSS-mediated cleavage of the JAMB extracellular domain would be expected to disrupt tight junction integrity, thereby allowing tumor cells to breach the BBB.
Cathepsin S inhibition reduces experimental brain metastasis formation
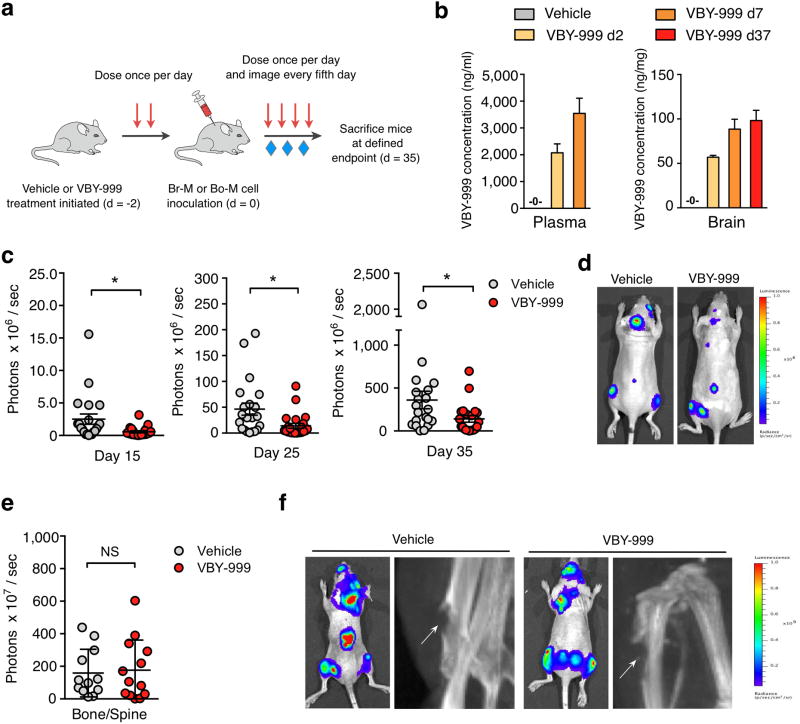
We next examined whether pharmacological inhibition of cathepsin S reduced metastatic seeding and colonization in a preclinical prevention trial (Fig. 8a). Mice were treated with VBY-999 for 2 days to inhibit cathepsin S activity prior to Br-M cell inoculation, and were then continuously treated with VBY-999 until the trial endpoint of 35 days post-tumor cell inoculation. Pharmacokinetic analysis showed that VBY-999 levels in the plasma were significantly above the required concentration for target inhibition at the time of Br-M cell inoculation, and confirmed that VBY-999 efficiently crosses the BBB with stable concentrations in the brain throughout the trial (Fig. 8b). Interestingly, BLI signal was reduced by 65-77% at different time points during the trial and at the 35 day endpoint (Fig. 8c, d).
Figure 8. Pharmacological inhibition of cathepsin S reduces brain metastasis formation in a preclinical trial.
(a) Schematic of the prevention trial experimental design. (b) Quantification of VBY-999 concentrations in plasma and brain tissue at the indicated time points after treatment started (n=3 mice for each group). (c) Quantification of BLI intensity in the head region at the indicated time points after Br-M cell inoculation. n=20 mice for vehicle group (5% dextrose in water (D5W)) and n=21 mice for VBY-999 treatment group (100 mg/kg/day). The BLI signal in the VBY-999 versus control group is 77, 70 and 65% reduction at each of the three successive time points indicated. (d) Representative BLI images at the trial endpoint, d35 after Br-M cell inoculation. Images are representative of the number of mice as in (c). (e) Quantification of BLI intensity at d35 after Bo-M tumor cell inoculation in the bone and spine region. Vehicle (n=12 mice) and VBY-999 (n=13 mice). (f) Representative BLI and X-ray images at day 35 after Bo-M cell inoculation. Arrows indicate osteolytic lesions. Images are representative of the number of mice as in (e). Bars represent mean + s.e.m. for (b), circles represent individual mice and horizontal lines represent the mean ± s.e.m for (c, e). P values were obtained using two-tailed unpaired t-test. NS = not significant, *P<0.05.
Initiation of VBY-999 treatment in fully established, end-stage brain metastases did not alter tumor burden (Supplementary Fig. 7b, c), indicating that targeting this enzyme is most critical in seeding and early outgrowth. We investigated the organ specificity of cathepsin S inhibition by assessing bone metastasis in prevention trials, using two different approaches. First, as bone and spine metastases can develop in the Br-M model, we assessed whether these lesions were affected by VBY-999. There was no significant difference between the treatment groups, which was supported by the finding that genetic depletion of cathepsin S did not affect bone metastasis formation (Supplementary Fig. 7d, e). Similarly, a VBY-999 prevention trial specifically in the bone metastasis model showed no change in BLI output or development of osteolytic metastases (Fig. 8e, f, Supplementary Fig. 7f). These data are consistent with our finding that CTSS expression levels in patients only correlated with brain MFS, and not bone MFS. In sum, cathepsin S inhibition is efficient in substantially and specifically reducing brain metastasis if cathepsin S activity is blocked throughout the disease course.
Discussion
Here we identify a brain metastasis-promoting role for cathepsin S. Using gene expression screens of distinct metastatic microenvironments in animal models, and analyses of patient samples, we uncovered a specific association between high cathepsin S levels and the development of brain metastases. Interestingly, cathepsin S was regulated in both a cell type-specific and stage-dependent manner, with high expression in tumor cells in early stage-metastases, which is subsequently downregulated concomitant with increased macrophage-specific expression in late-stage lesions. This pattern of expression shares intriguing similarities with lung metastatic niche development, whereby tenascin C (TNC) is initially provided by the tumor cells to support extravasation into the lung. TNC production later switches to infiltrating stromal cells as lung metastases expand38. Cathepsin S also shows cell type-specific expression in normal tissues, and is predominantly produced by immune and stromal cells6, 39, 40. Induction of cathepsin S expression in tumor cells of epithelial origin may indicate a type of ‘leukocytic mimicry’ whereby metastatic tumor cells could implement immune cell-like expression programs that enhance mobilization and cell motility. Indeed, similarities between the behavior of metastatic tumor cells and leukocytes have been noted41, which could be particularly advantageous during BBB transmigration2.
Considering that extravasation into the brain parenchyma is a rate-limiting step in brain metastasis30, 42, 43, we further investigated the contribution of cathepsin S to this specific stage. We identified JAM-B cleavage by cathepsin S as an important mechanism by which BBB transmigration of brain-metastatic cells is enhanced. Consistently, genetic or pharmacological targeting of cathepsin S significantly impaired experimental brain metastasis. We found that both tumor- and stroma-derived cathepsin S are important for metastatic seeding and outgrowth, and infer that this relates to a requirement for high levels of the enzyme overall, rather than distinct functions in these two cell types. Thus, while the levels of tumor-CTSS are highest at the step of metastatic cell extravasation, there is nonetheless stromal-Ctss already expressed at this stage, albeit at much lower levels than in later stages. Similarly, while stromal cells become the predominant source of cathepsin S during colonization, there is still some tumor-CTSS present. Therefore, both sources need to be depleted, otherwise there is some degree of functional compensation from the cell type that is not targeted. Given that our analysis of breast cancer patients revealed a significant association between high cathepsin S levels in the primary site and reduced brain MFS, it may be instructive to specifically consider this patient population for any future therapeutic intervention to reduce brain metastasis by cathepsin S inhibition.
The proteolytic gene expression screens performed herein resulted in the identification of numerous differentially expressed genes that are regulated in a stage- or tissue-specific manner in different metastatic microenvironments. By querying whether expression of these genes in primary breast cancer patients was associated with MFS in brain, bone or lung, we were able to apply an additional filter that allowed subsequent restriction of the gene lists to only those that showed a significant correlation with survival, which represent interesting targets for future investigation. Moreover, determining expression levels of these genes at the primary site may be useful as prognostic markers to evaluate the risk for patients to develop tissue-specific metastasis. As such, the identification of proteases in different metastatic microenvironments, many of which can be pharmacologically inhibited45-49, reveals an abundance of potential targets for metastatic disease.
Materials and Methods
Mice
All animal studies were approved by the Institutional Animal Care and Use Committee of Memorial Sloan-Kettering Cancer Center (MSKCC). Athymic/nude mice were purchased from NCI Frederick or bred within the MSKCC animal facility. The cathepsin S knockout mouse line (Ctss KO) was generated as described previously25 and backcrossed for 6 generations to the Athymic/nude background. NOD/SCID mice were purchased from Charles River Laboratories. MMTV-PyMT50 immunocompetent transgenic mice (FVB/n) were bred within the MSKCC animal facility.
Cell lines
Brain- (Br-M), bone- (Bo-M) and lung- (Lu-M) metastatic variants20-22 of the human breast cancer cell line MDA-MB-231 (denoted ‘parental’) were provided by J.M. and labeled with a triple-imaging vector (TK-GFP-Luc; TGL)24 to allow for non-invasive in vivo imaging of tumor growth over time. The MDA-MB-231 variants were cultured in DMEM + 10% FBS. Mouse Br-M variants were derived from the TS1 cell line51, which was previously isolated from a MMTV-PyMT mammary tumor, through sequential rounds of in vitro and in vivo selection for homing to the brain. These are denoted PyMT-BrM cells. Human umbilical vein endothelial cells (HUVEC) were purchased from the ATCC. Human brain microvascular endothelial cells (HBMEC) and human astrocytes (HA) were purchased from Sciencell. HUVEC and HBMEC were cultured on gelatin coated cell culture dishes, and HA on poly-L-lysine coated cell culture dishes in endothelial cell media (ECM, Sciencell) + 10% FBS supplemented with endothelial cell growth factors (ECGF).
Generation of experimental brain, bone and lung metastases
For brain and bone metastasis generation in xenografted mice, 1×104 brain-metastatic cells (Br-M) or 1×105 bone-metastatic cells (Bo-M) were inoculated into the left cardiac ventricle of 6-8 week old female Athymic/nude mice. For lung metastasis generation, 1×105 lung-metastatic cells (Lu-M) were injected into the lateral tail vein of 6-8 week old female NOD/SCID mice. For brain metastasis generation in immunocompetent mice, 1×105 PyMT-BrM cells were inoculated into the left cardiac ventricle of 6-8 week old female FVB/n mice. Early- and late-stage metastases were defined by bioluminescence intensity (BLI) at the time of tissue harvest for samples used in the microarray analysis and the independent sample set used for validation. Brain metastases that had a BLI output between 4.3×106 to 4.2×107 photons / sec were classified as early-stage metastases and were collected between 3-4 weeks after tumor cell inoculation. Late-stage brain metastases had a BLI output between 1.6×108 to 6.4×108 photons / sec and were collected between 5-8 weeks after tumor cell inoculation. Histological and morphometric analyses of these different stages showed that early-stage brain metastases are comprised of clusters of ∼50-200 cells, and can be considered similar to ‘micrometastases’, and late-stage metastases consist of clusters of ∼5,000-15,000 cells, corresponding to ‘macrometastases’. Representative images of the different stages are shown in Supplementary Figure 1d. Early-stage bone metastases were defined by a BLI intensity that ranged between 6.3×106 to 1.1×108 photons / sec and were harvested 3 weeks after tumor cell inoculation. Late-stage bone metastases showed a minimal BLI intensity of 8×108 photons / sec and a maximal BLI intensity of 2.5×109 photons / sec and were harvested 5 weeks after tumor cell inoculation. Histological and morphometric analyses of bone metastases showed that early-stage lesions are comprised of clusters of ∼50-200 cells, and late-stage metastatic clusters consist of ∼2,000-10,000 cells. Representative images of the different stages are shown in Supplementary Figure 1e. The generation of ‘mismatched’ samples (Br-M in bone or Bo-M in brain) followed the same criteria for early- and late-stage metastasis. For experiments shown in Supplementary Figure 3a-e, stromal-derived candidates were chosen to represent genes that show expression changes in an organ-specific manner (see Supplementary Fig. 2b, Supplementary Table 1e-g). Tumor-derived genes were selected to represent genes with expression changes confined to only one organ (see Supplementary Fig. 2a, Supplementary Table 1b-d). Early-stage lung metastases were harvested 48h after tumor cell inoculation. The BLI intensity at this time point ranged between 2.1×106 to 1.7×107 photons / sec. Late-stage lung metastases were harvested 5 weeks after tumor cell inoculation with an average BLI intensity between 8.1×108 to 3×109 photons / sec. Histological and morphometric analyses of lung metastases showed that early-stage lesions are comprised of cells diffusely present throughout the lung (∼2,000-4,000 cells per sectional plane, per entire lung), and late-stage metastatic clusters consist of ∼1,000-5,000 cells. Representative images of the different stages are shown in Supplementary Figure 1f. Late-stage lung metastases from the spontaneous MMTV-PyMT breast-to-lung metastasis model were harvested from 14 week-old female PyMT mice.
For tissue isolation, mice were anesthetized with 10 mg/ml ketamine; 1 mg/ml xylazine and retro-orbitally injected with 15 mg/ml luciferin. Mice were then intracardially perfused with PBS. Tumor-burdened tissue was identified by the presence of BLI signal for brain and bone metastases. For lung metastases, part of the left lung lobe was collected. Snap frozen samples were collected for RNA and protein isolation and tissues were fixed in 4% paraformaldehyde (PFA) for histology.
Microarray analysis
For microarray analysis, all samples were prepared and processed by the Genomics Core Facility at MSKCC. RNA was isolated using Trizol (Invitrogen) and the quality was assessed on an Agilent Bioanalyzer. Total RNA was reverse transcribed and labeled using the Genechip 3′ IVT Express Kit (Affymetrix). The resulting cRNA was hybridized to HuMu ProtIn chips (Affymetrix). All bioinformatics analyses were completed in R using the Bioconductor suite52. The ‘affy’ package53 was used for robust multi-array average normalization followed by quantile normalization. Mouse and human samples and probes were normalized separately. With the exception of the cross-species scatterplots, all subsequent bioinformatics analyses regarded the tumor and stroma separately.
The ‘limma’ package54 was used to identify differentially expressed genes, defined by a fold change cutoff of ± 2 and a false discovery rate of 10%. Tissue-specific genes were identified by the intersection of pairwise comparisons: e.g. lung stroma-specific genes were identified by the intersection of genes significantly enriched in lung vs. bone and genes significantly enriched in lung vs. brain. Stage-specific genes were identified in a tissue-specific manner comparing early- and late-stage metastases.
Principal component analysis (PCA) was completed using the covariance matrix in the ‘princomp’ function in R. Homologs for mouse and human genes were identified using the HomoloGene Database through the NCBI (http://www.ncbi.nlm.nih.gov/homologene). Homolog pairs were plotted with mouse/stroma tissue-specific, early vs. late, fold change on the x axis, and human/tumor tissue-specific, early vs. late, fold change on the y axis.
External datasets and survival analysis
For gene expression analysis of mouse endothelial cells, raw data from GSE47067 34 was imported into R and normalized as above. For patient analysis, normalized gene expression data was downloaded from the GEO (GSE12276) 20. Each gene was mean centered and scaled by standard deviation. Patients were split into tertiles (lower 33%, middle 33%, upper 33%) of CTSS gene expression for Kaplan-Meier survival analysis. The scaled, continuous CTSS gene expression was used for Hazard Ratio (HR) calculation. Similar analyses were completed for genes in Supplementary Figure 4c-e. Survival analysis was completed using the ‘survival’ package in R55. Hazard ratios were determined utilizing the ‘coxph’ function from the ‘survival’ package. Nominal P values are reported for HR significance in Supplementary Table 2 with a significance cutoff of 0.05 used to identify genes significantly associated with metastasis-free survival. P values were generated using the log-rank statistic for Kaplan-Meier analysis and Wald's test for the Hazard Ratio analysis.
Clinical samples
Primary breast tumors and brain metastases were provided by E.B., P.K.B., and W.C.H.. This research was approved by MSKCC Institutional Review Board (IRB) protocol 06-107 and Dana-Farber/Harvard Cancer Center (DF/HCC) protocol 10-454. All participants provided written consent. Information about the clinical samples can be found in Supplementary Table 3.
Generation of CTSS knockdown lines
Five shRNA sequences targeting CTSS were obtained from the RNAi Codex and RNAi Consortium. shRNA sequences were inserted into the targeting hairpin sequence for the pRetroSuper vector. Correct insertion into the vector was verified by digestion and sequencing of the vector. Plasmids with the correct shRNA targeting sequence were transfected into H29 viral packaging cells. Viral particles were concentrated from the H29 cell supernatant, added to the target cells in the presence of polybrene and cells were selected with puromycin. The following shRNA sequences were tested for their knockdown efficiency: CTSS-1 shRNA; 5′ – CTAAATTAACCTAAACGTA -3′, CTSS-2 shRNA; 5′ – CAGAGAAATATCACATATA -3′, CTSS-3 shRNA; 5′ – GATAAAGTTTGCTAAGTAA -3′, CTSS-4 shRNA; 5′ – CTAAATTAACCTAAACGT -3′, and CTSS-5 shRNA; 5′ – CTTTCCAGTACATCATTGA -3′. CTSS-3 shRNA was used for subsequent experiments to target CTSS with 90% knockdown (KD) efficiency. A non-targeting shRNA (5′ - CGCCATAAATATAACTTTA -3′) was used as a control.
Targeting tumor- and stroma-derived cathepsin S in vivo
1×104 Br-M cells (Br-M CTSS KD or Br-M Ctrl) were inoculated into the left ventricle of 6-8 week old female Athymic/nude or Ctss KO Athymic/nude mice. Metastasis formation was monitored once weekly by BLI using a Xenogen IVIS-200 Optical In Vivo Imaging System to determine metastasis incidence in the four experimental groups shown in the table in Figure 4b. Lesions that reached the cut-off of 1×106 photons / sec in a defined region in the head area were scored positively for brain metastasis in the incidence curve presented in Figure 4b. In addition, numerical values of the increase in BLI intensity present the kinetics of tumor progression (Supplementary Fig. 7a). An independent cohort of mice was injected with Br-M cells as described above and sacrificed at day 35 after tumor cell inoculation for subsequent analysis of proliferation, apoptosis, angiogenesis, and metastatic outgrowth.
For in vivo extravasation experiments, Athymic/nude or Ctss KO Athymic/nude mice were inoculated with 5×105 Br-M Ctrl or Br-M CTSS KD cells. BLI intensity was monitored at 0h, 24h and 48h after tumor cell inoculation and the BLI intensity was plotted relative to the BLI intensity immediately after tumor cell inoculation (0h time point). Analyses of BLI intensity were performed blinded to the group allocation. No statistical method was used to predetermine sample size.
Identification of cathepsin S inhibitor VBY-999
VBY-999 was provided by Virobay Inc., Menlo Park, CA and is part of an extensive structure-based drug discovery program. VBY-999 is a covalent reversible inhibitor with an electrophilic nitrile warhead. The detailed chemical synthesis and structure of compounds in the structural series including VBY-999 can be found in issued US Patent 7,547,701. Recombinant purified human and mouse cathepsin S were used to assess potency of VBY-999 and determine inhibition constants. Activity on the peptide substrate Z-Leu-Arg-AMC was determined in vitro by measuring hydrolysis of the substrate with spectrofluorimetric quantitation of AMC. VBY-999 was preincubated with cathepsin S for 15 min at room temperature (25°C) after which the substrate was added to initiate the 30 min reaction. Assay incubation buffer included 25 mM CH3COONa, pH 4.5, 2.5 mM DTT, and 0.05 M NaCl. Appropriate reaction conditions and peptide substrates for other cysteine and serine proteases were utilized to screen for selectivity of VBY-999 for cathepsin S. VBY-999 has an inhibition constant Ki(app) = 290 pM on the purified human cathepsin S enzyme, and > 3000-fold selectivity versus the related cathepsins K, L, B, and F. Potency on the closely related cathepsins K, L, and F was Ki(app) ≥ 3 μM, with potency on cathepsin B Ki(app) = 700 nM. Potency on mouse cathepsin S enzyme was verified on mouse cathepsin S purified enzyme. VBY-999 has an inhibition constant Ki(app) = 690 pM on mouse cathepsin S. No measurable inhibition was detected for any other cysteine, serine or aspartyl proteases tested.
VBY-999 inhibitor preclinical trial
For administration to mice, VBY-999 was formulated in a nanoparticle-based suspension formulation and further diluted in 5% dextrose in water (D5W) at a concentration of 10 mg/ml. Subcutaneous dosing of VBY-999 provided a dosing formulation and route that allowed high and sustained plasma concentrations of the drug to be achieved, which was confirmed using a bioanalytical LC-MS/MS method after 2 and 7 days of treatment (Fig. 8b). This results in full inhibition of the enzyme target for the duration of the trial, following once-daily dosing. In order to determine if VBY-999 had sufficient penetration of the CNS to be available for cathepsin S inhibition in the brain and at the blood-brain barrier, and to confirm that concentrations in the brain remain stable throughout the duration of the trial, VBY-999 concentration was determined by LC-MS/MS at day 2, day 7, and day 37 after treatment initiation (Fig. 8b). These data indicate that VBY-999 levels in the plasma and brain were significantly above the required concentration for target inhibition at the time of tumor cell inoculation and remain stable throughout the 37-day treatment schedule at a level sufficiently greater than the enzyme inhibition constant, and are thus expected to effectively inhibit cathepsin S activity. For the prevention trials, mice were randomly assigned into vehicle and VBY-999 treatment groups and treatment was started two days before tumor cell inoculation (d= -2). Mice were dosed with 100 mg/kg VBY-999 or vehicle (D5W) by subcutaneous injection once daily. At day 0, Athymic/nude mice were inoculated with 1×104 Br-M Ctrl cells or 1×105 Bo-M Ctrl cells. Metastasis formation was monitored every fifth day by bioluminescence imaging using a Xenogen IVIS-200 Optical In Vivo Imaging System during the trial period from day 0 to day 35 after tumor cell inoculation. For the Bo-M trial, mice were subjected to X-ray analysis at d35 after tumor cell inoculation using a SPECT-CT scanner (X-SPECT). For the regression trial, mice were stratified into vehicle and VBY-999 treatment groups at d27 after tumor cell inoculation to achieve equal average BLI intensity at the time of treatment start at d28. Mice were dosed daily with either vehicle or VBY-999 (100 mg/kg) for 7 days and metastasis growth was monitored by BLI imaging at d32 and d35. Analyses of BLI intensity were performed blinded to the group allocation. No statistical method was used to predetermine sample size.
RNA isolation, cDNA synthesis and quantitative real-time PCR
RNA was isolated with Trizol, DNase treated, and 0.2 μg of RNA was used for cDNA synthesis. Details about the Taqman assays can be found in Supplementary Table 4. All species-specific Taqman assays were chosen based on their location in the mRNA sequence that allows for maximal discrimination between mouse and human transcripts. For each Taqman assay, species specificity was tested by qPCR using mouse or human samples as controls.
Collection of conditioned media, protein isolation and western blotting
Conditioned media (CM) from Br-M cell lines was generated by incubating confluent cell layers in serum-free DMEM media for 24h. Collected CM was passed through 0.22 μm filters to remove cellular debris. For western blotting, CM was concentrated by centrifugation in Centrifugal Filter Units (Millipore). Proteins from cells or snap-frozen tissue were isolated through lysis or homogenization in RIPA lysis buffer (Pierce) with 1× complete Mini protease inhibitor cocktail (Roche). Membranes were probed with antibodies as indicated in Supplementary Table 5 and detected with HRP-conjugated secondary antibodies using chemoluminescence detection (Millipore). Bands from western blots were quantified in the dynamic range using the gel analysis module in ImageJ software.
Generation of Serpina3n antibody
Peptides targeting murine Serpina3n were determined via alignment of the protein sequence for serpina3n against mouse Serpin a3 family members, Serpina3b, c, f, g, k, and m, as well as human SERPINA3. From this alignment, divergent regions were located and peptides were chosen that corresponded to regions 373-396 (a3n-no1), 225-248 (a3n-no2), and 398-418 (a3n-no3) of Serpina3n. 10-14mg of each peptide was synthesized by the Pocono Rabbit Farm and Laboratory, with 2mg of each peptide conjugated to KLH and 2mg of each peptide conjugated to BSA. The KLH-conjugated peptides were used to generate an immune response in Armenian hamsters and BALB/c mice by the Monoclonal Antibody Core Facility at MSKCC. Serum from hamsters and mice was tested via ELISA using the BSA-conjugated peptides in Nunc Maxisorp ELISA plates. The best responding hamster to all three peptides was used for fusion, and positive colonies were screened by ELISA and for response to each peptide. Clones were screened by immunohistochemistry and immunofluorescence for ability to recognize murine Serpina3n in mouse tissue, and colony 13H5 (which responded to peptide a3n-no1) was selected for subcloning.
Immunocytochemistry
For immunocytochemistry, cells were cultured on glass coverslips and fixed in 4% PFA in 0.1M phosphate buffer for 20 min at room temperature. Cells were permeabilized in PBS with 0.25% Triton X-100 for 10 min. Cells were blocked in 0.5% PNB (phosphate-NaCl buffer) in PBS for at least 1h at room temperature, followed by incubation with goat anti-human CTSS primary antibody diluted 1:100 in 0.25% PNB overnight at 4°C. Donkey anti-goat Alexa568 secondary antibody (Molecular Probes) was used at a dilution of 1:500 in 0.25% PNB for 1h at room temperature.
Tissue preparation and immunostaining
Tissue for frozen histology was fixed in 4% PFA overnight, and brain and lung samples were subsequently transferred into 30% sucrose until the tissue was fully equilibrated. Bone samples were decalcified in 0.5 M EDTA, pH 8.0 for 14 days. All tissues were then embedded in OCT (Tissue-Tek) and 10 μm or 20 μm cryostat tissue sections were used for all subsequent analyses. For immunofluorescence, frozen sections were thawed and dried at room temperature and rehydrated. For the standard staining protocol, tissue sections were blocked in 0.5% PNB in PBS for at least 1h at room temperature, followed by incubation with primary antibody in 0.25% PNB overnight at 4°C. Primary antibody information and dilutions are listed in Supplementary Table 5. Fluorophore-conjugated secondary antibodies (Molecular Probes) were used at a dilution of 1:500 in 0.25% PNB for 1h at room temperature.
Paraffin-embedded sections were processed using a Ventana automated staining device. The automated deparaffinization/ rehydration, citrate buffer-based antigen retrieval, and blocking of unspecific protein binding and endogenous peroxidase was followed by incubation with mouse anti-human CD68 (Dako) primary antibody and goat anti-human CTSS (R&D Systems) or mouse anti-human CK (Dako) and goat anti-human CTSS (R&D Systems) overnight at 4°C. Sections were incubated with donkey-anti mouse HRP labeled secondary antibody (Jackson Immunoresearch, 1:200) in 0.25% PNB buffer in PBS for 1.5h followed by incubation with Alexa488 labeled tyramide (Invitrogen) at a 1:200 dilution in amplification buffer for 8 min. Following the signal amplification reaction, donkey anti-goat Alexa568 (Molecular Probes) was used at a dilution of 1:500 in 0.25% PNB for 1h at room temperature. Frozen sections that were used for Jam-B, Cldn3 and Ocln staining were processed using a Ventana automated staining device. The automated rehydration, citrate buffer-based antigen retrieval, and blocking of unspecific protein binding and endogenous peroxidase steps were followed by incubation with rat anti-mouse Jam-B (Pierce) primary antibody and goat anti-mouse Cd31 (R&D Systems), rabbit anti-mouse Cldn3 (Invitrogen) and goat anti-mouse Cd31, or rabbit anti-mouse Ocln (Invitrogen) and goat anti-mouse Cd31 overnight at 4°C. Sections were incubated with donkey anti-rat or donkey anti-rabbit biotin labeled secondary antibody (Vector, 1:200) in PBS + 0.03% Tween for 1.5h followed by incubation with Streptavidin-Cy5 (Invitrogen, 1:200) PBS + 0.03% Tween for 20 min. Donkey anti-goat Alexa568 (Molecular Probes) was used at a dilution of 1:500 in 0.25% PNB for 1h at room temperature. Apoptotic cells were stained via terminal dUTP nick end labeling (TUNEL) following the manufacturer's instructions (Trevigen), with the modification of using Streptavidin-Cy5 (Invitrogen; 1:200) instead of Streptavidin-FITC.
Microscopy and image analysis
Tissue sections and cells on coverslips were visualized under a Carl Zeiss Axioimager Z1 microscope equipped with an ApoTome.2 and a TissueGnostics stage to allow for automated image acquisition. The analysis of proliferation and apoptosis were performed using TissueQuest analysis software (TissueGnostics) as previously described56. All parameters of metastatic outgrowth and angiogenesis were quantitated using MetaMorph software (Molecular Devices). Briefly, vasculature was visualized by Texas Red Lectin (Vector Laboratories) injections or by staining of the endothelial cell marker CD34. Tumor cells were detected by their expression of the GFP reporter. The area covered by CD34 and GFP staining was quantified. To determine the number of tumor cells that are present within an area of 1 – >4 average tumor cell diameter, the blood vessel area was dilated by 1 – 4 average tumor cell diameter with an increment of 1 tumor cell diameter and the number of tumor cells in each area was determined. Tumor cells that were localized outside an area of 4 average tumor cell diameter were defined as >4 tumor cell diameter away from CD34+ blood vessels as illustrated in Figure 5b. Vessel density was quantified as the area covered by Texas Red Lectin relative to the area covered by DAPI.
To histologically quantify the percentage of intravascular, extravasating or extravasated tumor cells (Fig. 6c), brain sections were stained for TUNEL+ cells to exclude non-viable tumor cells from the analysis. Brain sections were automatically acquired using TissueQuest software (TissueGnostics), which used a z-stack (5 images above and below the focal plane, 0.3 μm steps, 20× objective) to generate a maximal intensity projection (MIP) image of each acquired brain area. Tumor cells were detected via cell tracker green (Invitrogen) and vasculature was visualized by Texas Red Lectin (Vector Laboratories) injections. Tumor cells were counted manually and their localization relative to the vasculature was determined.
For analysis of human samples, 5-10 fields of view were acquired using a 20× objective (total magnification 200×) and a Zeiss Apotome to ensure cells were in the same optical section. The number of CK+ tumor cells and CD68+ macrophages, and their relative CTSS intensities (CTSS index) was evaluated using CellProfiler 2.0 software. A CellProfiler module was generated that allowed for the detection of tumor cells and macrophages based on their DAPI and CK signal, or DAPI and CD68 signal, respectively. The CTSS signal intensity was measured in the whole cell population (DAPI+) and associated with a specific cell type (macrophages or tumor cells), and the proportion of CTSS signal associated with CK+ tumor cells or CD68+ macrophages was calculated relative to the overall CTSS signal intensity in all DAPI+ cells. Analyses of histological assessment were performed blinded to the group allocation.
Measurement of vessel permeability
6-8 week old Athymic/nude mice were injected with Evan's blue dye (30 mg/kg) into the tail vein. 30 mins after injection, mice were anesthetized and perfused with acidified fixative (1% PFA in 0.05 mM citrate buffer, pH 3.5). 30 mg of brain tissue was incubated in 500 μl formamide (Sigma) to extract Evan's blue at 60°C overnight. Absorbance was measured at 610 nm and 740 nm on a spectraMax 340pc plate reader (Molecular Devices).
In vitro blood-brain barrier transmigration assays
In vitro blood-brain barrier (BBB) transmigration assays were performed as previously described29. The artificial BBB was formed with either HUVECs or HBMECs (20,000 cells / well) in co-culture with HA cells (100,000 cells / well) for 3 days on Transwell-inserts with 3 μm fluoroblok membranes. Cell-tracker green (CMFDA)-labeled Br-M Ctrl or Br-M CTSS KD cells (20,000 cells / well) were allowed to transmigrate for 18h through the artificial BBB towards a FBS gradient, in the presence or absence of VBY-999 (10 μM). Tumor cell transmigration through empty inserts (coated with gelatin and poly-L-lysine) or inserts coated with HUVECs, HBMECs or HAs alone were used to determine the baseline migratory potential and the contribution of the single cell types to BBB formation. The number of transmigrated tumor cells was quantified by analyzing 200 fields of views (FOVs) that were acquired with a 20× objective (200× total magnification) using TissueQuest analysis software (TissueGnostics). Analysis was performed blinded to the group allocation.
In vitro and cell-based cleavage assays
Recombinant inactive CTSS was obtained from R&D Systems. CTSS was activated at 50 ng/μl in 50 mM sodium acetate, 5 mM DTT, 0.25 M NaCl (pH 4.5) for 1.5h at 37°C. For the in vitro cleavage reaction, activated CTSS was incubated with recombinant proteins in the presence or absence of the cathepsin S inhibitor VBY-999 (10 μM) for 0, 10 or 20 min in 50 mM sodium acetate, 5 mM DTT, 0.25 M NaCl at pH 4.5 and pH 6.0. Details about the recombinant proteins used in the in vitro cleavage assay can be found in Supplementary Table 6. The cleavage reaction was stopped by adding SDS sample buffer and reducing agent (Invitrogen) and the samples were boiled at 95°C for 5 min. Aliquots were subjected to western blot analysis as described above. Information about the antibodies can be found in Supplementary Table 5. All experiments were repeated independently at least three times.
For cell-based cleavage assays, HBMECs were grown to 100% confluence in a 10 cm plate. Conditioned media from Br-M cells was collected as described above. 200 μl of concentrated Br-M CM (collected from two 10 cm plates of confluent Br-M cells) was diluted in 6.5 ml PBS pH 6.0 + 0.05 mM DTT for each 10 cm plate of HBMECs. The cleavage reaction was performed in the presence or absence of VBY-999 (10 μM) for 0h, 2h, and 4h. PBS pH 6.0 + 0.05 mM DTT was used as a control. The supernatant from HBMEC cell layers was collected after the indicated time points, concentrated and subjected to western blot analysis as described above.
Proliferation assays
Cell growth rate was determined using an MTT cell proliferation kit (Roche). Briefly, cells were plated in triplicate in 96-well plates (2.5 × 103 for Br-M Ctrl and Br-M CTSS KD cells) in the presence or absence of 0.1 – 100μm VBY-999. 10 μl of MTT labeling reagent was added to each well and then incubated for 4h at 37°C, followed by the addition of 100 μl MTT solubilization reagent overnight. Absorbance was measured at 595 nm and 750 nm on a spectraMax 340pc plate reader (Molecular Devices).
Data presentation and statistical analysis
Data are presented as means with standard error (s.e.m.) or as statistical scatter plots using GraphPad Prism Pro5. Numeric data were analyzed using unpaired two-tailed Student's t-test unless otherwise noted. Kaplan-Meier survival curves, heatmaps and scatterplots were generated in R v 2.15.2 using the base R graphics, ‘gplots’ or ‘ggplot2’ packages. P values were generated using the Log-Rank statistic for Kaplan-Meier Analysis and Wald's test for the Hazard Ratio. P <0.05 was considered statistically significant. All code used to analyze the data and generate the plots is available at: https://bitbucket.org/bowmanr/joycelab-humu-brain-met-ctss.
Accession numbers
Data sets generated in this study are available online at NCBI GEO, Accession Number GSE47930. The accession numbers for the previously published data sets that were reanalyzed in this study are available online at NCBI GEO, and Accession Numbers GSE12276 20 and GSE47067 34.
Supplementary Material
Acknowledgments
We thank K. Simpson and X. Chen for excellent technical support, and members of the Joyce lab for insightful discussion. We thank L. Akkari, O. Olson and D. Yan for reading the manuscript. We are grateful to P. Bos for advice on the BBB assays and experimental brain metastasis model. We thank the MSKCC Core Facilities of Genomics, Molecular Cytology, Small Animal Imaging and Monoclonal Antibody Production for technical assistance. We thank H. Chapman (UCSF) for providing cathepsin S knockout mice. This research was supported by the following: US National Cancer Institute program grants of the Integrative Cancer Biology Program (CA148967; J.A.J., C.L.) and Tumor Microenvironment Network (CA126518; J.A.J., J.M.), the Health Research Science Board of New York, and the Alan and Sandra Gerry Metastasis Research Initiative (J.A.J.), Deutsche Forschungsgemeinschaft (SE2234/1-1; L.S.), US National Cancer Institute F31 fellowship CA167863 and Gerstner Sloan Kettering graduate program (R.L.B.), US National Cancer Institute F32 fellowship CA130329 (S.D.M.), and Canadian Institutes of Health Research (D.F.Q.).
Footnotes
Contributions: L.S., S.D.M. and J.A.J. designed experiments. L.S., S.D.M., D.F.Q. and B.T.E. performed experiments and analyzed data. R.L.B., F.R. and C.L. performed computational analyses. E.B., P.K.B., W.C.H., L.J.H. and J.M. provided patient samples or reagents. J.A.J., L.S. and R.L.B. wrote the manuscript. All authors edited or commented on the manuscript. J.A.J. conceived and supervised the study.
References
- 1.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 3.Goss PE, Chambers AF. Does tumour dormancy offer a therapeutic target? Nat Rev Cancer. 2010;10:871–877. doi: 10.1038/nrc2933. [DOI] [PubMed] [Google Scholar]
- 4.Affara NI, Andreu P, Coussens LM. Delineating protease functions during cancer development. Methods Mol Biol. 2009;539:1–32. doi: 10.1007/978-1-60327-003-8_1. [DOI] [PubMed] [Google Scholar]
- 5.Mason SD, Joyce JA. Proteolytic networks in cancer. Trends Cell Biol. 2011;21:228–237. doi: 10.1016/j.tcb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gocheva V, et al. Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 2006;20:543–556. doi: 10.1101/gad.1407406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy G. The ADAMs: signalling scissors in the tumour microenvironment. Nat Rev Cancer. 2008;8:929–941. doi: 10.1038/nrc2459. [DOI] [PubMed] [Google Scholar]
- 8.Butler GS, Overall CM. Proteomic identification of multitasking proteins in unexpected locations complicates drug targeting. Nat Rev Drug Discov. 2009;8:935–948. doi: 10.1038/nrd2945. [DOI] [PubMed] [Google Scholar]
- 9.Lopez-Otin C, Hunter T. The regulatory crosstalk between kinases and proteases in cancer. Nat Rev Cancer. 2010;10:278–292. doi: 10.1038/nrc2823. [DOI] [PubMed] [Google Scholar]
- 10.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allinen M, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Finak G, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 13.Erez N, Truitt M, Olson P, Arron ST, Hanahan D. Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-kappaB-dependent manner. Cancer Cell. 2010;17:135–147. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- 14.Katz AM, et al. Astrocyte-specific expression patterns associated with the PDGF-induced glioma microenvironment. PLoS One. 2012;7:e32453. doi: 10.1371/journal.pone.0032453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seaman S, et al. Genes that distinguish physiological and pathological angiogenesis. Cancer Cell. 2007;11:539–554. doi: 10.1016/j.ccr.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ojalvo LS, Whittaker CA, Condeelis JS, Pollard JW. Gene expression analysis of macrophages that facilitate tumor invasion supports a role for Wnt-signaling in mediating their activity in primary mammary tumors. J Immunol. 2010;184:702–712. doi: 10.4049/jimmunol.0902360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989;8:98–101. [PubMed] [Google Scholar]
- 18.Disibio G, French SW. Metastatic patterns of cancers: results from a large autopsy study. Arch Pathol Lab Med. 2008;132:931–939. doi: 10.5858/2008-132-931-MPOCRF. [DOI] [PubMed] [Google Scholar]
- 19.Lee YT. Breast carcinoma: pattern of metastasis at autopsy. J Surg Oncol. 1983;23:175–180. doi: 10.1002/jso.2930230311. [DOI] [PubMed] [Google Scholar]
- 20.Bos PD, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang Y, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 22.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz DR, et al. Hu/Mu ProtIn oligonucleotide microarray: dual-species array for profiling protease and protease inhibitor gene expression in tumors and their microenvironment. Mol Cancer Res. 2007;5:443–454. doi: 10.1158/1541-7786.MCR-06-0337. [DOI] [PubMed] [Google Scholar]
- 24.Ponomarev V, et al. A novel triple-modality reporter gene for whole-body fluorescent, bioluminescent, and nuclear noninvasive imaging. Eur J Nucl Med Mol Imaging. 2004;31:740–751. doi: 10.1007/s00259-003-1441-5. [DOI] [PubMed] [Google Scholar]
- 25.Shi GP, et al. Cathepsin S required for normal MHC class II peptide loading and germinal center development. Immunity. 1999;10:197–206. doi: 10.1016/s1074-7613(00)80020-5. [DOI] [PubMed] [Google Scholar]
- 26.Lorger M, Felding-Habermann B. Capturing changes in the brain microenvironment during initial steps of breast cancer brain metastasis. Am J Pathol. 2010;176:2958–2971. doi: 10.2353/ajpath.2010.090838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Small DM, et al. Cathepsin S from both tumor and tumor-associated cells promote cancer growth and neovascularization. Int J Cancer. 2013;133:2102–2112. doi: 10.1002/ijc.28238. [DOI] [PubMed] [Google Scholar]
- 28.Abbott NJ, Ronnback L, Hansson E. Astrocyte-endothelial interactions at the blood-brain barrier. Nat Rev Neurosci. 2006;7:41–53. doi: 10.1038/nrn1824. [DOI] [PubMed] [Google Scholar]
- 29.Eugenin EA, Berman JW. Chemokine-dependent mechanisms of leukocyte trafficking across a model of the blood-brain barrier. Methods. 2003;29:351–361. doi: 10.1016/s1046-2023(02)00359-6. [DOI] [PubMed] [Google Scholar]
- 30.Jia W, Martin TA, Zhang G, Jiang WG. Junctional adhesion molecules in cerebral endothelial tight junction and brain metastasis. Anticancer Res. 2013;33:2353–2359. [PubMed] [Google Scholar]
- 31.Liu WY, Wang ZB, Zhang LC, Wei X, Li L. Tight junction in blood-brain barrier: an overview of structure, regulation, and regulator substances. CNS Neurosci Ther. 2012;18:609–615. doi: 10.1111/j.1755-5949.2012.00340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallagher FA, et al. Magnetic resonance imaging of pH in vivo using hyperpolarized 13C-labelled bicarbonate. Nature. 2008;453:940–943. doi: 10.1038/nature07017. [DOI] [PubMed] [Google Scholar]
- 33.Kirschke H, Wiederanders B, Bromme D, Rinne A. Cathepsin S from bovine spleen. Purification, distribution, intracellular localization and action on proteins. Biochem J. 1989;264:467–473. doi: 10.1042/bj2640467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nolan DJ, et al. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev Cell. 2013;26:204–219. doi: 10.1016/j.devcel.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choe Y, et al. Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J Biol Chem. 2006;281:12824–12832. doi: 10.1074/jbc.M513331200. [DOI] [PubMed] [Google Scholar]
- 36.Oliveira M, et al. Improvement of cathepsin S detection using a designed FRET peptide based on putative natural substrates. Peptides. 2010;31:562–567. doi: 10.1016/j.peptides.2009.12.027. [DOI] [PubMed] [Google Scholar]
- 37.Biniossek ML, Nagler DK, Becker-Pauly C, Schilling O. Proteomic identification of protease cleavage sites characterizes prime and non-prime specificity of cysteine cathepsins B, L, and S. J Proteome Res. 2011;10:5363–5373. doi: 10.1021/pr200621z. [DOI] [PubMed] [Google Scholar]
- 38.Oskarsson T, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chapman HA. Endosomal proteases in antigen presentation. Curr Opin Immunol. 2006;18:78–84. doi: 10.1016/j.coi.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 40.Gupta S, Singh RK, Dastidar S, Ray A. Cysteine cathepsin S as an immunomodulatory target: present and future trends. Expert Opin Ther Targets. 2008;12:291–299. doi: 10.1517/14728222.12.3.291. [DOI] [PubMed] [Google Scholar]
- 41.Madsen CD, Sahai E. Cancer dissemination--lessons from leukocytes. Dev Cell. 2010;19:13–26. doi: 10.1016/j.devcel.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 42.Steeg PS, Camphausen KA, Smith QR. Brain metastases as preventive and therapeutic targets. Nat Rev Cancer. 2011;11:352–363. doi: 10.1038/nrc3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eichler AF, et al. The biology of brain metastases-translation to new therapies. Nat Rev Clin Oncol. 2011;8:344–356. doi: 10.1038/nrclinonc.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park ES, et al. Cross-species hybridization of microarrays for studying tumor transcriptome of brain metastasis. Proc Natl Acad Sci U S A. 2011;108:17456–17461. doi: 10.1073/pnas.1114210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turk B. Targeting proteases: successes, failures and future prospects. Nat Rev Drug Discov. 2006;5:785–799. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- 46.Palermo C, Joyce JA. Cysteine cathepsin proteases as pharmacological targets in cancer. Trends Pharmacol Sci. 2008;29:22–28. doi: 10.1016/j.tips.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 47.Drag M, Salvesen GS. Emerging principles in protease-based drug discovery. Nat Rev Drug Discov. 2010;9:690–701. doi: 10.1038/nrd3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fingleton B. Matrix metalloproteinases as valid clinical targets. Curr Pharm Des. 2007;13:333–346. doi: 10.2174/138161207779313551. [DOI] [PubMed] [Google Scholar]
- 49.Seidah NG, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov. 2012;11:367–383. doi: 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 50.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shree T, et al. Macrophages and cathepsin proteases blunt chemotherapeutic response in breast cancer. Genes Dev. 2011;25:2465–2479. doi: 10.1101/gad.180331.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 54.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry RA, Huber W, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 55.Therneau T. A Package for Survival Analysis in S. R package version 2.37-7. 2014 http://CRAN.R-project.org/package=survival.
- 56.Gocheva V, et al. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 2010;24:241–255. doi: 10.1101/gad.1874010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.