Summary
We analyzed proteomes of colon and rectal tumors previously characterized by the Cancer Genome Atlas (TCGA) and performed integrated proteogenomic analyses. Somatic variants displayed reduced protein abundance compared to germline variants. mRNA transcript abundance did not reliably predict protein abundance differences between tumors. Proteomics identified five proteomic subtypes in the TCGA cohort, two of which overlapped with the TCGA “MSI/CIMP” transcriptomic subtype, but had distinct mutation, methylation, and protein expression patterns associated with different clinical outcomes. Although copy number alterations showed strong cis- and trans-effects on mRNA abundance, relatively few of these extend to the protein level. Thus, proteomics data enabled prioritization of candidate driver genes. The chromosome 20q amplicon was associated with the largest global changes at both mRNA and protein levels; proteomics data highlighted potential 20q candidates including HNF4A, TOMM34 and SRC. Integrated proteogenomic analysis provides functional context to interpret genomic abnormalities and affords a new paradigm for understanding cancer biology.
Introduction
The Cancer Genome Atlas (TCGA) has characterized the genomic features of human cancers1–6 and thereby presents the challenge of explaining how genomic alterations drive cancers7. Because proteins link genotypes to phenotypes, the Clinical Proteomic Tumor Analysis Consortium (CPTAC) is performing proteomic analyses of TCGA tumor specimens for selected cancer types. Here we present the first integrated proteogenomic characterization of human cancer with an analysis of the TCGA colorectal cancer (CRC) specimens6.
The TCGA study affirmed well-established genomic features of CRC and described three transcriptional subtypes, 17 chromosomal regions of significant focal amplification and 28 regions of significant focal deletion, and linked genomic features of CRC to critical signaling pathways. The drivers underlying copy number alterations (CNAs) and transcriptional subtypes are largely unknown, and an integrative analysis of both genomic and proteomic data may provide a more comprehensive understanding of the information flow from DNA to protein to phenotype.
Identification of peptides, proteins and variant sequences
We performed liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based shotgun proteomic analyses on 95 TCGA tumor samples (Extended Data Fig. 1, Supplementary Methods 1–4), the clinical and pathological characteristics and TCGA datasets for which are summarized in Supplementary Table 1. Benchmark quality control (QC) samples from one basal and one luminal human breast tumor xenograft were analyzed in alternating order after every five CRC samples (Supplementary Methods 2).
We identified a total of 124,823 distinct peptides among the 95 samples, corresponding to 6,299,756 spectra in an assembly of 7,526 protein groups with a protein-level False Discovery Rate (FDR) of 2.64% (Supplementary Methods 5.1–5.2, Extended Data Fig. 2). To facilitate integration between genomic and proteomic data, a gene-level assembly of the peptides identified 7,211 genes.
A fundamental question in proteogenomics is which protein coding alterations are expressed at the protein level. Because standard database search approaches cannot identify variant pe3ptides from MS/MS data, we also performed database searches with customized sequence databases from matched RNA-Seq data for individual samples8, 9 (Supplementary Methods 5.3, Extended Data Fig. 3).
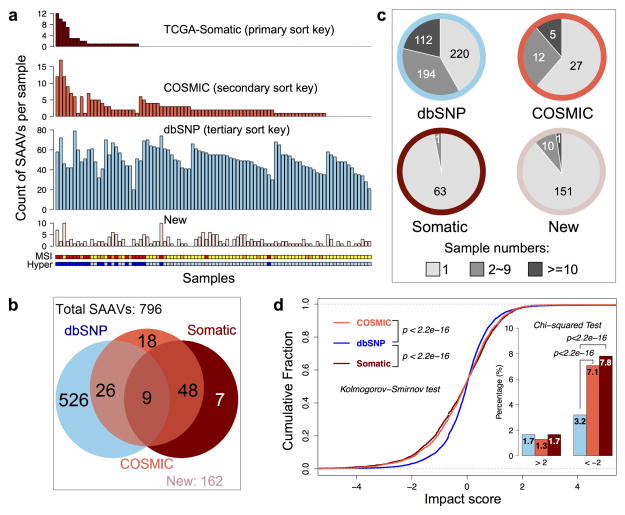
We identified 796 single amino acid variants (SAAVs) across all 86 tumors for which matched RNA-Seq data was available (Fig. 1a–b, Supplementary Table 2–3), among which 64 corresponded to somatic variants reported by TCGA and 101 were reported in the COSMIC database (i.e. COSMIC-supported variants). Of the remaining SAAVs, 526 were listed in the dbSNP database (i.e. dbSNP-supported variants) and are likely to be germline variants. The 162 previously unreported SAAVs might be explained by novel somatic or germline variants, RNA editing, or, in some cases, false discovery.
Figure 1. Summary of detected single amino acid variants (SAAVs) and the impact of single nucleotide variants (SNVs) on protein abundance.
a, The number of different types of SAAVs (TCGA-reported somatic variants, COSMIC-supported variants, dbSNP-supported variants, and new variants) in individual tumor samples. The samples are ordered by the number of detected somatic variants, then COSMIC-supported variants, and then dbSNP-supported variants. The Microsatellite instability (MSI) and hypermutation (Hyper) status are labeled below the bar charts for each sample (MSI-High: red, MSI-Low: orange, Microsatellite Stable: yellow; hypermutated: blue, non-hypermutated: sky blue; no data: grey). The number of somatic variants and COSMIC-supported variants were significantly higher in MSI-High and hypermutated tumors, whereas the other two types of SAAVs were randomly distributed across the data set. b, The total numbers for different types of SAAVs and their overlapping relations. All 796 detected SAAVs were annotated based on previous reports in dbSNP (left circle), COSMIC (middle circle), or TCGA-reported somatic variants (right circle), and their overlapping relations are shown in the Venn diagram. There are 162 SAAVs that have not been reported previously in these databases (new). c, Distribution of the frequency of occurrence (1 sample: light grey, 2–9 samples: grey, >=10 samples: dark grey) for different types of SAAVs. Border colors of the pie charts correspond to different SAAV types using the same color scheme as in (a). Whereas 58% of dbSNP-supported variants occurred in two or more samples, almost all somatic variants each occurred in only one sample. d, SNVs detected in RNA-Seq data were separated into three categories (dbSNP-supported, COSMIC-supported, and TCGA-Somatic). The impact of individual SNVs on protein abundance was calculated (see supplementary methods) and the impact scores for different categories of SNVs were plotted as cumulative fraction curves with two-sided p values from the Kolmogorov-Smirnov test labeled. The percentage of SNVs with an absolute impact score greater than 2 was also plotted as an inset, with p values from the Chi-squared test. Sample size for the dbSNP-supported, COSMIC-supported and TCGA-Somatic variants were 12184, 7492, and 3302, respectively.
The identified somatic variants were clearly enriched in the hypermutated samples, whereas the germline variants showed no association with hypermutation (Fig. 1a). Although 58% of the germline variants occurred in two or more samples, almost all somatic variants occurred in only one sample (Fig. 1c). The low identification rate for somatic variants may reflect relatively low sequence coverage in shotgun proteomics; however, somatic variants also might negatively impact protein abundance, possibly by reducing translational efficiency or protein stability10. Using the protein abundance quantification method described below and detailed in Supplementary Methods 5.4, we found that somatic variants exerted a significantly stronger negative impact on protein abundance than did dbSNP-supported variants (p<2.2 × 10-16, Kolmogorov-Smirnov (KS) test, Fig. 1d, Supplementary Methods 5.5). The percentage of variants with an impact score of less than −2 was doubled for somatic variants compared to dbSNP-supported variants (p<2.2 × 10-16, Chi-squared test, Fig. 1d).
Cancer-related variant proteins may serve as candidate protein biomarkers or therapeutic targets. The 108 somatic or COSMIC-supported protein variants mapped to 105 genes, including known cancer genes in the Cancer Gene Census database such as KRAS, CTNNB1, SF3B1, ALDH2, and FH. The list also included 14 targets of FDA-approved drugs or drugs in clinical trials4, such as ALDH2, HSD17B4, PARP1, P4HB, TST, GAK, SLC25A24, and SUPT16H. A subset of variant peptide sequences, including K-ras (G12D) were verified by targeted analyses of tumor lysates spiked with synthetic, isotope-labeled peptide standards (Supplementary Methods 6). One example is shown in Extended Data Fig. 4.
Quantification of protein abundance
To quantify protein abundance, we used spectral counts, which are the total number of MS/MS spectra acquired for peptides from a given protein11 (Supplementary Methods 5.4, Supplementary Table 4). Analysis of data from benchmark QC samples demonstrated platform reproducibility throughout the analyses and enabled evaluation of data normalization methods (Extended Data Fig. 5a–b). Based on the minimal spectral count requirement established using the QC data set (Extended Data Fig. 5c), 3,899 genes with a protein-level FDR of 0.43% were used to compare relative protein abundance across tumor samples.
mRNA abundance does not reliably predict protein abundance
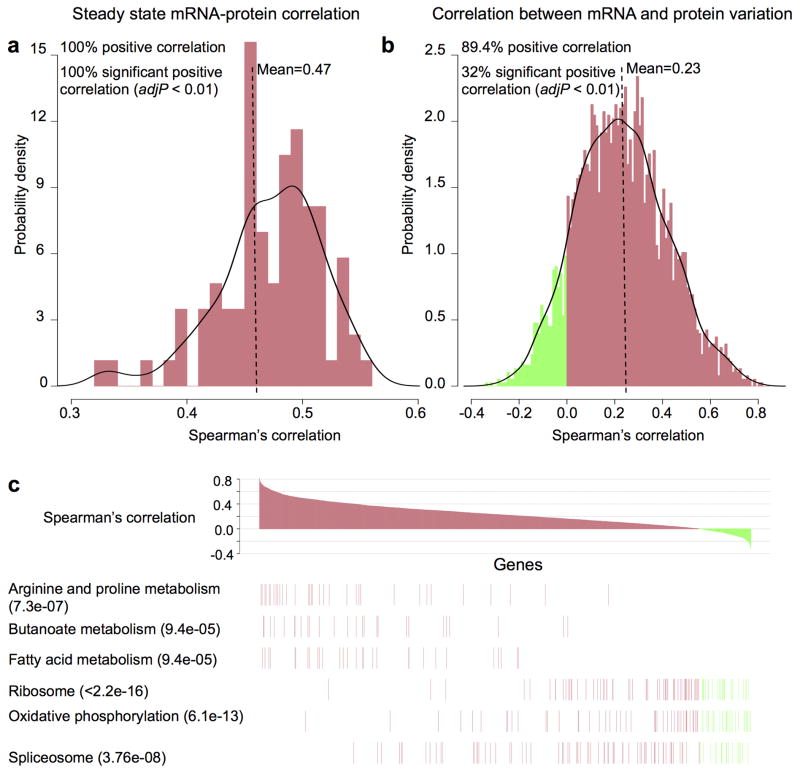
The matched proteomic and RNA-Seq measurements from the TCGA CRC tumors allowed the first global analysis of transcript-protein relationships in a large human tumor cohort (Supplementary Methods 7). First, we compared the steady state mRNA and protein abundance for each gene within individual samples (Supplementary Methods 7.2–7.3, Extended Data Fig. 6a). All samples showed significant positive mRNA-protein correlation (multiple-test adjusted p value < 0.01, Spearman’s correlation coefficient) and the average correlation between steady state mRNA and protein abundance in individual samples was 0.47 (Fig. 2a), which is comparable to previous reports in multi-cellular organisms12.
Figure 2. Correlations between mRNA and protein abundance in TCGA tumors.
a, Steady state mRNA and protein abundance were positively correlated in all 86 samples (multiple-test adjusted p value < 0.01) with a mean Spearman’s correlation coefficient of 0.47. b, mRNA and protein variation were positively correlated for most (89.4%) mRNA-protein pairs across the 87 samples, but only 32% showed significant correlation (multiple-test adjusted p value < 0.01), with a mean Spearman’s correlation coefficient of 0.23. c, mRNA and protein levels displayed dramatically different correlation for genes involved in different biological processes. Genes encoding intermediary metabolism functions showed high mRNA-protein correlations, whereas genes involved in oxidative phosphorylation, RNA splicing and ribosome components showed low or negative correlations. Multiple-test adjusted two-sided p-values from the Kolmogorov-Smirnov test were provided in the parentheses following the KEGG pathway names. Red and green in the figures indicate positive- and negative-correlations, respectively.
Next, we examined the concordance between mRNA and protein variation of individual genes across the 87 tumors for which 3,764 genes had both mRNA and protein measurements suitable for relative abundance comparison (Supplementary Methods 7.2, 7.4). Although 89% of the genes showed a positive mRNA-protein correlation, only 32% had statistically significant correlations (Fig. 2b). The average Spearman’s correlation between mRNA and protein variation was 0.23, which was comparable to reported values for yeast, mouse and human cell lines13–15.
To test whether the concordance between protein and mRNA variation is related to the biological function of the gene product, we performed KEGG enrichment analysis (Supplementary Methods 7.5, Supplementary Table 5). Genes involved in several metabolic processes showed concordant mRNA and protein variation, whereas other gene classes showed low or even negative concordance in mRNA and protein variation (Figure 2c). We also found that genes with stable mRNA and stable protein tend to have higher mRNA-protein correlation than those with unstable mRNA and unstable protein (p = 5.27 × 10-6, two-sided Wilcoxon rank-sum test, Supplementary Methods 7.6, Extended Data Fig. 6b). mRNA measurements thus are poor predictors of protein abundance variations and both biological functions of the gene products and mRNA and protein stability may govern mRNA-protein correlation.
Impact of copy number alterations on mRNA and protein abundance
The TCGA study identified 17 regions of significant focal amplification and 28 regions of significant focal deletion. We examined the impact of CNAs on mRNA and protein abundance, including both cis-effects on the abundance of genes in the same loci and trans-effects on the abundance of genes at other loci in the genome (Supplementary Methods 8).
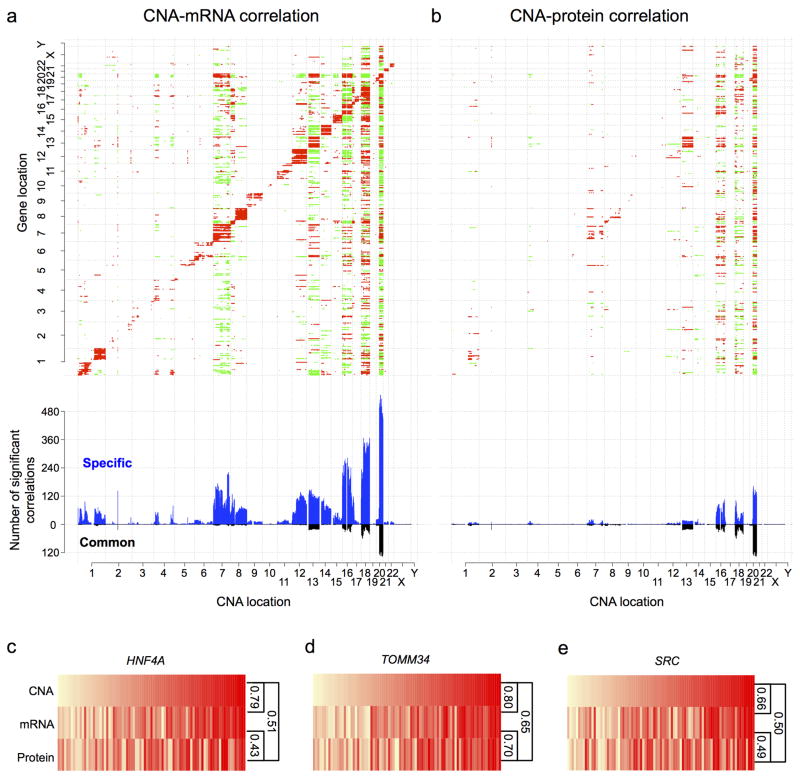
For all 23,125 genes with a CNA measurement in the TCGA data set, we calculated Spearman’s correlation with mRNA and protein abundance, respectively for the 3,764 genes with both mRNA and protein measurements (Supplementary Methods 8.1). Examination of the matrix visualizing significant CNA-mRNA correlations (multiple-test adjusted p value < 0.01) revealed strong positive correlations along the diagonal (Fig. 3a), suggesting strong cis-effects of CNAs on mRNA abundance. Most of the diagonal signals corresponded to previously reported arm-level changes6. In contrast, the diagonal pattern was much weaker for CNA-protein correlations (Fig. 3b).
Figure 3. Effects of copy number alterations (CNAs) on mRNA and protein abundance.
a,b, The top panels show copy number-abundance correlation matrices for mRNA abundance (a) and protein abundance (b) with significant positive and negative correlations (multiple-test adjusted p value < 0.01, Spearman’s correlation coefficient) indicated by red and green colors, respectively, and genes ordered by chromosomal location on both x and y-axes. The bottom panels show the frequency of mRNAs/proteins associated with a particular copy number alteration, where blue and black bars represent associations specific to mRNA/protein or common to both mRNA and protein, respectively. c–e, HNF4A, TOMM34 and SRC showed significant CNA-mRNA, mRNA-protein, and CNA-protein correlations (Spearman’s correlation coefficient). The color grade from light yellow to red indicates relatively low-level to high-level CNA, relative mRNA abundance or relative protein abundance among the 85 samples, which were ordered by copy number data.
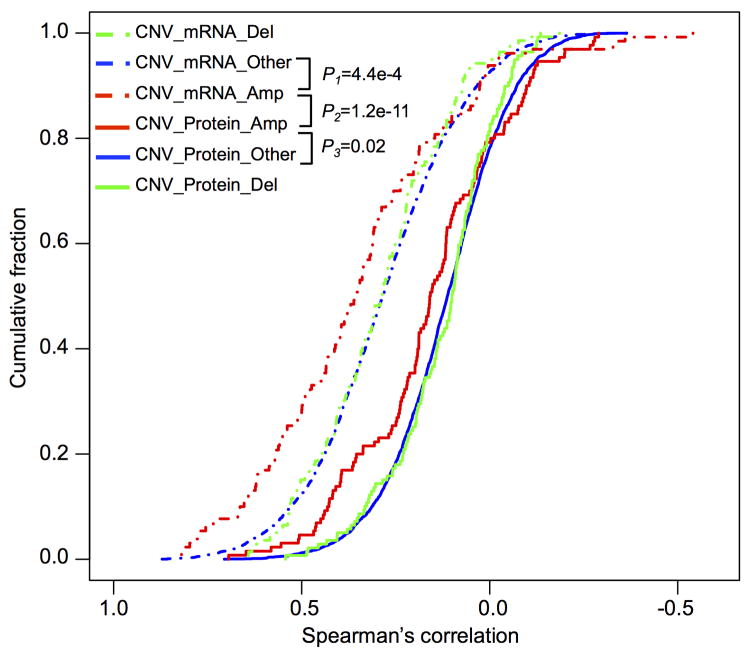
To further investigate the cis-effects of CNAs, we separated all genes with CNA, mRNA, and protein measurements into those in focal amplification regions, focal deletion regions, and non-focal regions (i.e., chromosomal regions without focal amplification or deletion). As shown in Extended Data Figure 7, CNA-mRNA correlations were significantly higher than CNA-protein correlations for genes in all three groups (p<1.0 × 10-10, KS test). Moreover, genes in the focal amplification regions showed significantly higher CNA-mRNA and CNA-protein correlations than genes in the non-focal regions (p=4.4 × 10-4 and 0.02, respectively, KS test). However, the same trend was not observed for genes in the focal deletion regions. Therefore, focal amplifications have the strongest cis-effects on both mRNA and protein abundance, suggesting that selection for high protein abundance may drive CNA in regions of focal amplification. On the other hand, many CNA-driven mRNA level increases do not translate into increased abundance of the corresponding proteins.
Figure 3a–b also revealed multiple trans-acting CNA hot spots, defined as chromosomal loci whose amplification is significantly associated with abundance changes of many transcripts or proteins at other loci. Chromosomes 20q, 18, 16, 13 and 7 contained the five strongest hot spots driving global mRNA abundance variation. These hot spots also were strongest at the protein level. Most hot spot-related transcript changes did not propagate to the protein level, presumably reflecting buffering of protein abundance by post-transcriptional regulation16, 17. Notably, many hot spot-associated protein level alterations occurred in the absence of corresponding mRNA alterations, suggesting that the same trans-acting hot spot may exert independent effects at both the transcriptome and proteome levels.
The 20q amplification was associated with the largest global changes in both mRNA and protein levels in this univariate analysis. The same conclusion was reached with a regularized multivariate regression analysis method, remMap18 (Supplementary Methods 8.2, Supplementary Table 6–9). These data highlight the importance of 20q amplification in CRC, which has not been well documented in previous studies. Among the 79 genes in the 20q region with quantifiable protein measurements, 67 (85%) showed significant CNA-mRNA correlation, but only 40 (51%) showed significant CNA-protein correlation (multiple-test adjusted p value < 0.01, Spearman’s correlation coefficient, Supplementary Table 10).
Because significant CNA-protein correlations identify amplified sequences that translate to high protein abundance, proteomic measurements can help prioritize genes in amplified regions for further examination. Of particular interest among the 40 genes is HNF4A (Fig. 3c), a candidate driver gene nominated by TCGA for the 20q13.12 focal amplification peak6. HNF4α is a transcription factor with a key role in normal gastrointestinal development19 and is increasingly being linked to CRC20. However, there are contradictory reports on whether HNF4α acts as an oncogene or a tumor suppressor gene in CRC20. Upon reanalysis of the HNF4A shRNA knockdown data for CRC cell lines from the Achilles project21, we found that the dependency of CRC cells on HNF4α correlated significantly with the amplification level of HNF4A (Supplementary Methods 8.3, Extended Data Fig. 8), which may partially explain the contradictory roles reported for HNF4α in CRC. Other interesting candidates included TOMM34 (Fig. 3d), which is over-expressed frequently in CRC tumors and is involved in the growth of CRC cells22, and SRC (Fig. 3e), which encodes a non-receptor tyrosine kinase implicated in several human cancers including CRC23.
Proteomic subtypes of CRC
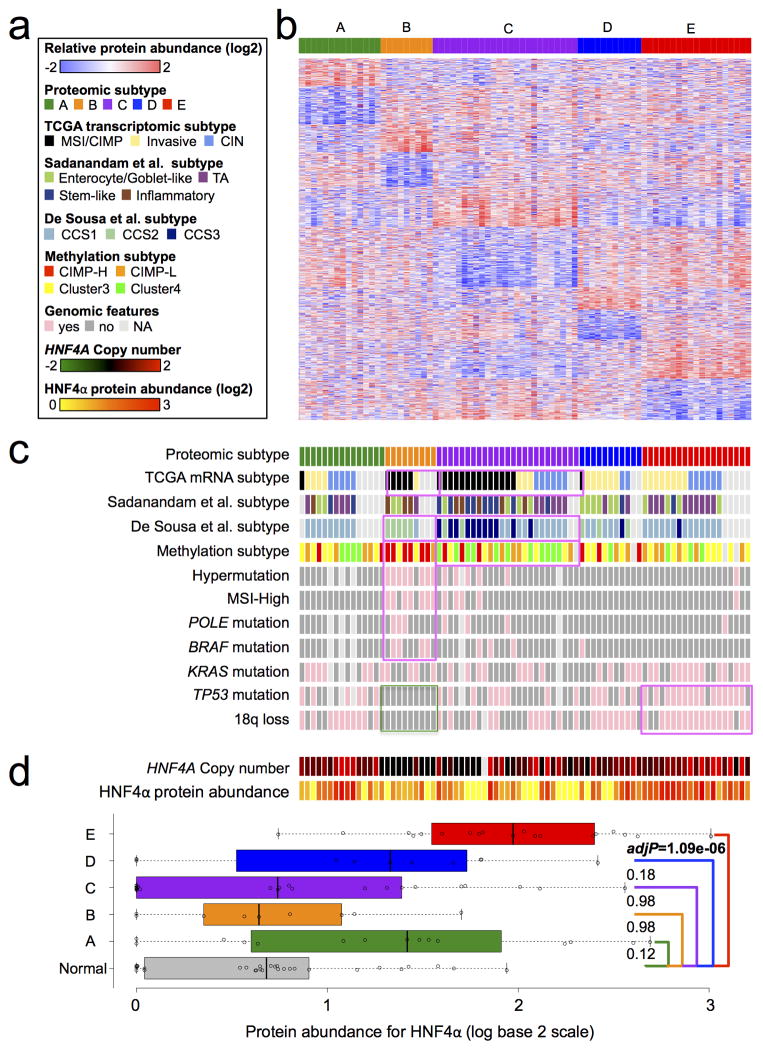
The TCGA study reported three transcriptomic subtypes of CRC, designated “MSI/CIMP” (microsatellite instability/CpG island methylator phenotype), “Invasive”, and “CIN” (chromosomal instability). Given the limited correlation between mRNA and protein levels, we asked whether CRC subtypes can be better represented with proteomics data. Using the Consensus Clustering24 method (Supplementary Methods 9.1–9.2, Extended Data Fig. 9), we identified five major proteomic subtypes in this tumor cohort, with 15, 9, 25, 11, and 19 cases in subtypes A through E, respectively (Fig. 4a–b).
Figure 4. Proteomic subtypes of colon and rectal cancers, associated genomic features, and relative abundance of HNF4α.
a, Figure legends for b, c and d. b, Identification of five proteomic subtypes. Tumors are displayed as columns, grouped by proteomic subtypes as indicated by different colors. Proteins used for the subtype classification are displayed as rows. The heat map presents relative abundance of the proteins (logarithmic scale in base 2) in the 90 tumor cohort. c, Association of proteomic subtypes with major colorectal cancer-associated genomic alterations and previously published transcriptomic and methylation subtypes. Subtypes significantly overlapped with a transcriptomic or methylation subtype are highlighted by pink boxes. Both proteomic subtypes B and C showed significant overlap with the TCGA MSI/CIMP subtype. In addition, they showed significant overlap with the CCS2 and CCS3 subtypes in the De Sousa et al. classification, respectively. Proteomic subtype B significantly overlapped with the TCGA CIMP-H methylation subtype, whereas subtype C significantly overlapped with a non-methylation subtype (TCGA cluster 4 methylation subtype). Subtypes over-represented with a specific genomic alteration are also highlighted by pink boxes. The green box highlights the absence of TP53 mutations and 18q loss in subtype B. d, The top panel shows HNF4A copy number and relative abundance of HNF4α in the five subtypes; the bottom panel compares relative abundance of HNF4α in the five subtypes to that in normal colon samples, respectively, and the adjP values are based on the two-sided Wilcoxon rank-sum test followed by multiple-test adjustment.
We tested the association between the subtype classification and established genomic and epigenomic features of CRC using Fisher’s exact test (Fig. 4c, Supplementary Table 11). Almost all hypermutated and MSI-high tumors were included in subtypes B and C, as well as tumors with POLE and BRAF mutations. However, statistically significant association with these features was only observed for subtype B (multiple-test adjusted p value < 0.05). Moreover, whereas subtype B was significantly associated with the TCGA CIMP-H (CIMP-high) methylation subtype, subtype C was significantly associated with a non-CIMP subtype (cluster 4). Another unique feature of subtype B was the lack of TP53 mutations and chromosome 18q loss. These results clearly established the association between proteomic subtype B and MSI-High and CIMP, but suggest that subtype C might have different biological underpinnings.
The remaining three subtypes were associated with CIN, another well-accepted genetic property of CRC. In particular, subtype E was significantly associated with both TP53 mutations and 18q loss, genomic features frequently associated with CIN tumors25. Interestingly, subtype E was also associated with HNF4A amplification and relatively higher abundance of HNF4α protein (Fig. 4d). HNF4α abundance was significantly higher in subtype E tumors compared to normal colon samples (multiple-test adjusted p value = 1.09 × 10-6, two-sided Wilcoxon rank-sum test); however, significant up-regulation of HNF4α was not observed for other subtypes (Supplementary Methods 10). This result, together with our reanalysis of shRNA knockdown data from the Achilles project (Extended Data Fig. 8), suggest that HNF4α dependency might be particularly associated with the subset of tumors or cells with HNF4A amplification.
We also examined the association between the subtype classification and clinical features and found only that stage II tumors were significantly enriched in subtype C (multiple-test adjusted p value < 0.05, Supplementary Table 11). Supervised statistical analyses at the individual protein level for 13 clinical and genomic features also identified few, if any significant protein effects of these features, except for hypermutation status, MSI status, and 18q loss (Supplementary Table 12), suggesting that the proteomic subtypes identified by the unsupervised clustering analysis captured the major proteome variations across the tumors.
Next, we compared the proteomic subtype classification with the TCGA transcriptional subtype classification for the 62 samples that had both subtype labels. Proteomic subtypes B and C both showed significant association with the TCGA subtype “MSI/CIMP” (Fig. 4b, Supplementary Table 11); however, they differ considerably at genomic, epigenomic, and proteomic levels (Fig. 4b). We also examined alternative classifications of the TCGA samples based on two recently published transcriptomic subtype classifiers26, 27. Proteomic subtype C, but not subtype B, showed enriched overlap with the “Stem-like” subtype described by Sadanandam et al.27 and the “CCS3” subtype described by De Sousa et al.26. Interestingly, tumors with “Stem-like” and “CCS3” classifications both have poor prognosis, which suggests that proteome subtype C also may be associated with poor prognosis. Therefore, the ability to distinguish subtype B from C through proteomics data is important, because MSI-High tumors typically have better prognosis25.
Protein signatures and networks associated with proteomic subtypes
To better understand the biology underlying the proteomic subtypes, we identified protein signatures for each subtype by supervised comparison of protein abundance in that subtype against all others; we also required signature proteins for a subtype to be significantly different in abundance compared to normal colon samples from 30 individuals analyzed on the same proteome analysis platform (Supplementary Methods 10, Supplementary Table 13–14). As shown in Extended Data Figure 10a, all CRC subtypes displayed more than 2,000 (>60%) significant protein abundance differences compared to normal colon. Although a full validation of the proteomic subtypes and protein signatures for the subtypes will require proteomic profiling data from an independent tumor cohort, a low cross-validation error rate of 3.8% demonstrated good generalizability of the subtypes and their signature proteins (Supplementary Methods 11).
We performed Gene Ontology enrichment analysis for the subtype signatures using WebGestalt28 (Supplementary Methods 11, Supplementary Table 15). Genes involved in “response to wounding” were significantly enriched in the up-signature of subtype C (multiple-test adjusted p value < 2.2 × 10-16, Fisher’s exact test). The wound-response gene signature is a powerful predictor of poor clinical outcome in patients with early stage breast cancers29. This result further links our subtype C to poor prognosis.
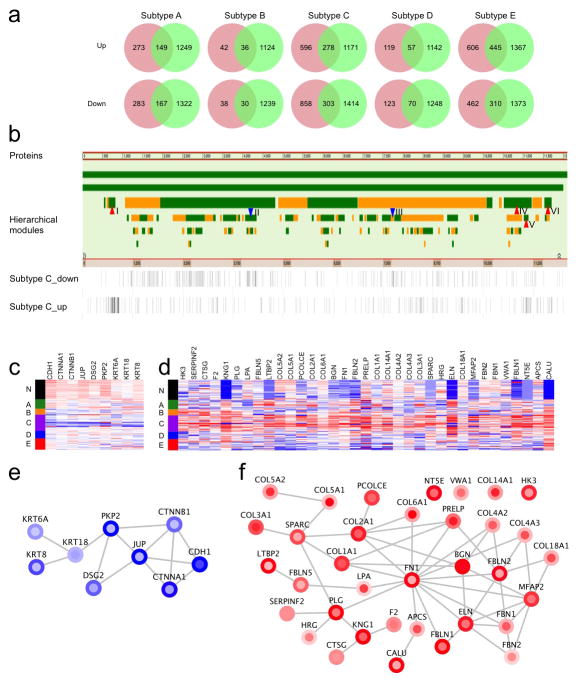
To better understand the functional networks underlying this subtype with potential clinical importance, we uploaded the up- and down-signatures of subtype C to NetGestalt30 for enriched protein-protein interaction network module analysis. Four network modules were enriched with genes in the up-signature for subtype C, whereas two modules were enriched with genes in the down-signature (multiple-test adjusted p value < 0.01, Fisher’s exact test, Extended Data Fig. 10b). Notably, the down-signature enriched module (III) included the E-cadherin (CDH1)-β-catenin (CTNNB1)-α-catenin (CTNNA1) complex (Extended Data Fig. 10c, 10e). E-cadherin, the most under-expressed protein in the sub-network, suppresses invasion in lobular breast carcinoma31 and is a switch for the epithelial-to-mesenchymal transition (EMT), which is associated with poor prognosis in colon cancer32. Other components of the module were desmosomal proteins (PKP2, JUP and DSG2) and cytokeratins (KRT18, KRT6A and KRT8). Reduction in both desmosome formation and cytokeratin expression is associated with EMT33. Moreover, proteins in the most significantly up-regulated network module (Extended Data Fig. 10d, 10f) included collagens (COL1A1 and COL3A1) and extracellular matrix glycoproteins (FN1, BGN, FBN1, and FBN2) that also are markers of EMT34, 35. These data strengthen the association of subtype C with poor prognosis and relate it to EMT activation.
Discussion
Our proteomic characterization of the genomically-annotated TCGA colon tumors illustrates the power of integrated proteogenomic analysis. The data demonstrate that protein abundance cannot be reliably predicted from DNA or RNA-level measurements. mRNA and protein levels were modestly correlated, as earlier cell and animal model studies suggest36, but over two thirds of these correlations were not statistically significant in the TCGA tumor set. Although most CNAs in CRC drive mRNA abundance changes, relatively few translated to consistent changes in protein abundance.
Genomic and proteomic technologies provide reinforcing data. RNA-Seq data facilitates the discovery of variant proteins, which could serve as possible biomarker candidates or therapeutic targets. Combined mRNA and protein profiling data can identify potentially relevant genes in amplified chromosomal regions. This approach, which revealed the importance of chromosome 20q amplification and provided new insights into the role of HNF4α in CRC, can be broadly extended to understand roles of CNAs in other cancers. Proteomics identified CRC subtypes similar to those detectable by transcriptome profiles, but further captured features not detectable in transcript profiles. The separation of the TCGA “MSI/CIMP” subtype into distinct proteotypes illustrates the unique potential of proteomics-based subtyping. After validation in independent cohorts, protein subtype signatures could be directly translated into laboratory tests for tumor classification. Integrated proteogenomic analysis, as demonstrated in this study, will enable new advances in cancer biology, diagnostics and therapeutics.
Methods Summary
All tumor samples for the current study were obtained through the TCGA Biospecimen Core Resource (BCR) as described previously6. No other selection criteria other than availability were applied for this study. Patient-derived xenograft tumors from established Basal and Luminal-B breast cancer intrinsic subtypes 37, 38 were raised subcutaneously in 8 week old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice (Jackson Labs, Bar Harbor, Maine) as previously described39, 40. Normal colon biopsies were obtained from screening colonoscopies performed between July 2006 and October 2010 under Vanderbilt University IRB approval #061096.
Tissue proteins were extracted and tryptic peptide digests were analyzed by multidimensional liquid chromatography-tandem mass spectrometry. Xenograft QC samples were run after every 5 colorectal tumor samples. Raw data were processed for peptide identification by database and spectral library searching and identified peptides were assembled as proteins and mapped to gene identifiers for proteogenomic comparisons. Quantitative proteomic comparisons were based on spectral count data. Detailed descriptions of the samples, LC-MS/MS analysis, and data analysis methods can be found in Supplementary Methods. All of the primary mass spectrometry data on TCGA tumor samples are deposited at the CPTAC Data Coordinating Center as raw and mzML files and complete protein assembly datasets for public access (https://cptac-data-portal.georgetown.edu).
Extended Data
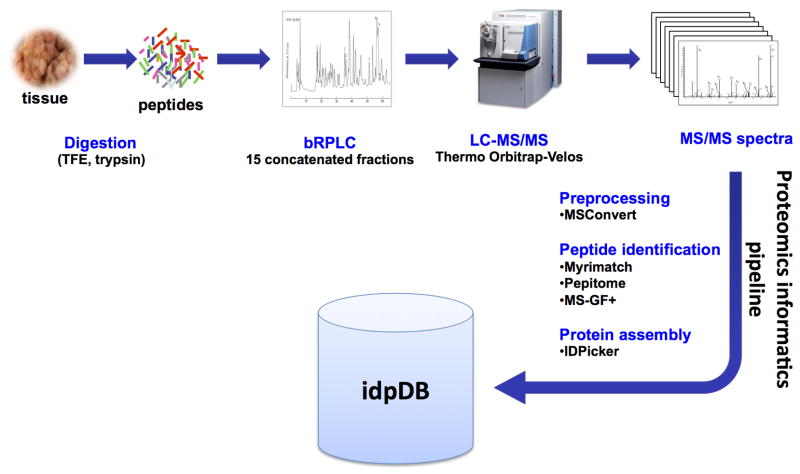
Extended Data Figure 1. Mass spectrometry (MS)-based proteomics workflow.
Protein was extracted from frozen tumor tissue and used to generate tryptic digests. The resulting tryptic peptides were fractionated using off-line basic-reverse phase high-pressure liquid chromatography (bRPLC). Collected fractions were pooled and used for reverse phase HPLC in-line with a Thermo Orbitrap-Velos MS instrument. Raw data was processed by MSConvert and then used for database and spectral library searching using three different search engines (Myrimatch, Pepitome and MS-GF+). Identified peptides were assembled using IDPicker 3 with selected filters as described in the methods. IDPicker 3 stores its protein assemblies for a specified set of filters in the idpDB format. These SQLite databases associate spectra with peptides, peptides with proteins, and LC-MS/MS experiments with a hierarchy of experiments.
Extended Data Figure 2. Relaxing the false discovery rate (FDR) of peptide-spectrum match (PSM) for high-confident proteins increases spectral counts.
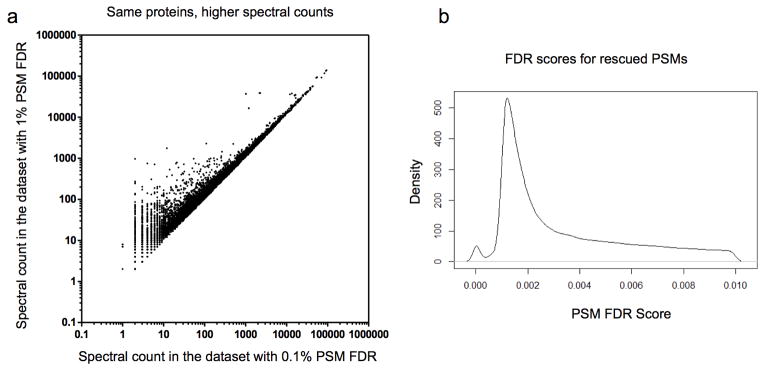
To increase spectral counts and improve statistical comparisons, we first created a protein assembly that maximized the number of proteins identified (at 0.1% PSM FDR) and then relaxed the PSM FDR to 1% exclusively for the set of confidently identified proteins. This strategy led to increased spectral counts from 4,896,831 to 6,299,756, a 29% increase. a, Spectral count plot of all 7,526 confidently identified proteins demonstrates the increase in the absolute number of spectra identified for each protein, but no decrease for any of the proteins. Each dot in the figure represents one of the 7,526 proteins; x-axis and y-axis represent the spectral counts obtained in the data sets with 0.1% and 1% PSM FDR, respectively, both plotted on a log scale. b, Density plot showing the distribution of PSM FDR scores for all rescued PSMs. Rescued PSMs are of high quality with a median PSM FDR score of less than 0.2%, indicating the maintained integrity of the data set.
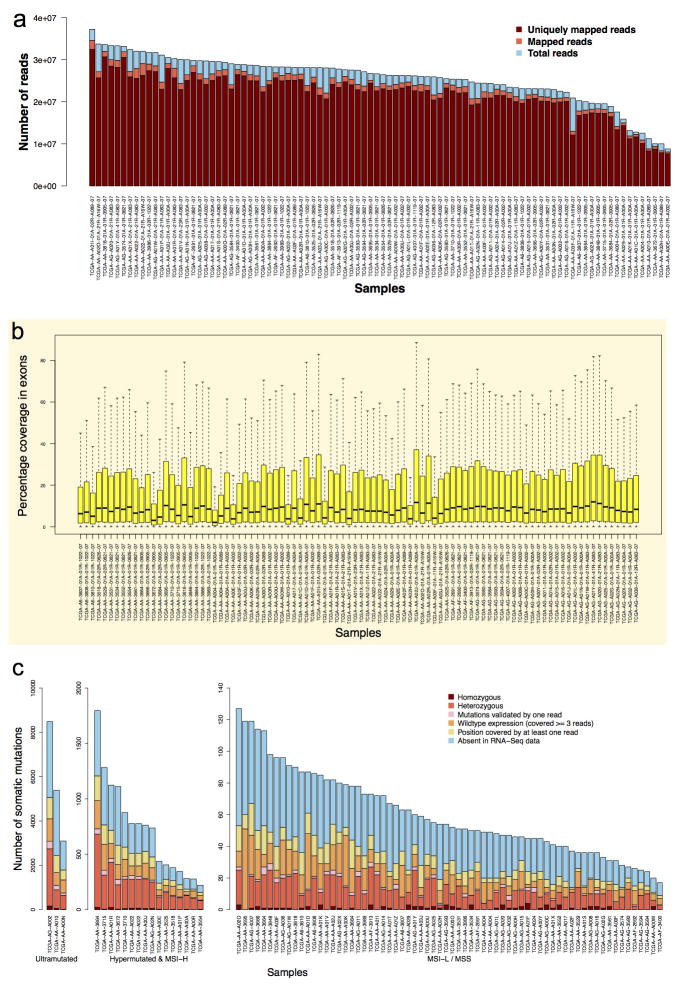
Extended Data Figure 3. Reads mapping, exon coverage and missense somatic variants in RNA-Seq data.
a, Summary of total RNA-Seq read counts and mapping results for individual samples. b, Distribution of percentage sequence coverage in exons for individual samples. Among all 228,157 exons, 76% were expressed, but only 64% had an average coverage greater than 1. Exons with no coverage were not included in the box plots. c, Number of missense somatic variants detected by RNA-Seq in individual samples. Approximately 54% of the mutation positions were covered by RNA-Seq reads and only 43% were covered by three or more reads.
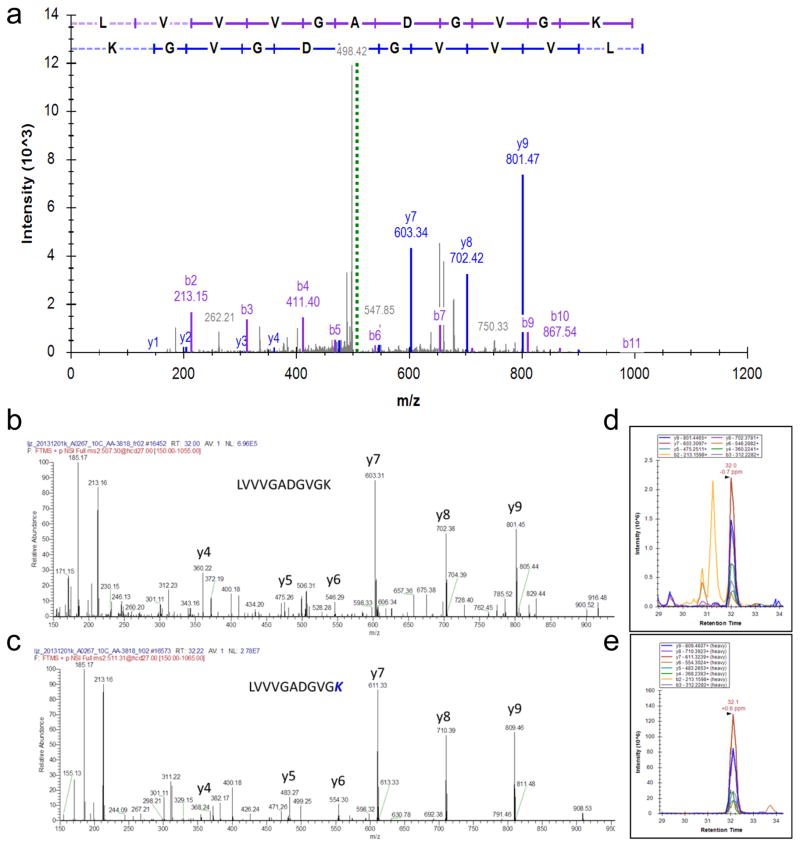
Extended Data Figure 4. PRM (Parallel reaction monitoring) validation results.
a, PRM data for the variant sequence LVVVGADGVGK (KRAS G12D in TCGA-AA-3818). b, PRM data for the variant sequence LVVVGADGVGK (KRAS G12D in TCGA-AG-A00Y). c, PRM data for the variant sequence TPVLFDVYEIK (ANXA11 I278V in TCGA-AF-3400). d, PRM data for the variant sequence DLEDLFFK (SRSF9 Y35F in TCGA-AA-A01P). Single amino acid variants (SAAVs) identified in the TCGA shotgun data set were validated using PRM analyses. Three distinct SAAVs in four TCGA samples were selected for validation. The TCGA samples were freshly prepared in the same manner as the original samples analyzed by shotgun proteomics. Each sample was spiked with 12.5 fmol/μL of a mixture of all isotopically labeled peptides. Using an inclusion list containing the precursor m/z values representing both unlabeled (endogenous) and labeled peptides, each fraction was analyzed by PRM for the variant peptides. For each variant shown in a–d, the top MS/MS spectra display represents the spectrum identified in the initial shotgun analyses of the TCGA samples. The two annotated spectra shown below the original spectra represent the MS/MS of the unlabeled endogenous variant peptide and the spiked respective labeled peptide in the PRM analysis of the TCGA sample, respectively. The chromatographic traces show the overlapping transitions and retention time of both the endogenous and labeled variant peptide, respectively.
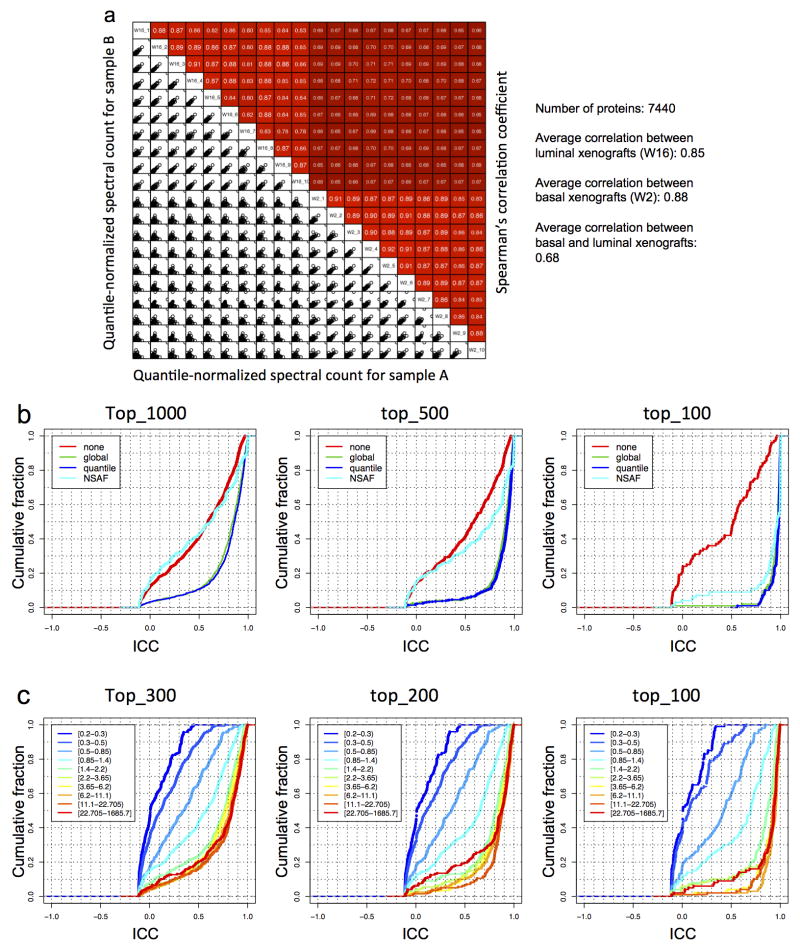
Extended Data Figure 5. Platform evaluation and analysis method selection using quality control (QC) samples.
a, The lower-left half (uncolored) depicts pair-wise scatter plots of the samples, with x- and y-axes representing quantile-normalized spectral counts for samples in corresponding columns and rows, respectively. The upper-right half (red colored) depicts pair-wise Spearman’s correlation coefficients for the same comparisons. b, For each normalization method (none, global, quantile, and NSAF), we calculated the intraclass correlation coefficients (ICCs) for individual proteins in the QC data set. The analysis was done for the top 1000, 500, or 100 proteins with the largest variance and the cumulative fraction curves were plotted. In most scenarios, quantile normalization generated slightly higher ICC scores than global normalization, and both methods clearly outperformed the NSAF normalization. c, We sorted all proteins in the QC data set based on their total spectral counts and then divided the proteins into 10 bins with equal number of proteins. Average spectral count ranges for each bin are shown in the brackets in the legend box. For each bin, we calculated the ICCs for individual proteins in the bin. The analysis was done for the top 300, 200, or 100 proteins with the largest variance in each bin. The cumulative fraction curves were plotted. Protein bins with spectral counts less than 1.4 showed clearly lower ICC scores, whereas the ICC score curves started to converge when the average spectral count was greater than 1.4.
Extended Data Figure 6. Extended data for mRNA-protein correlation analysis.
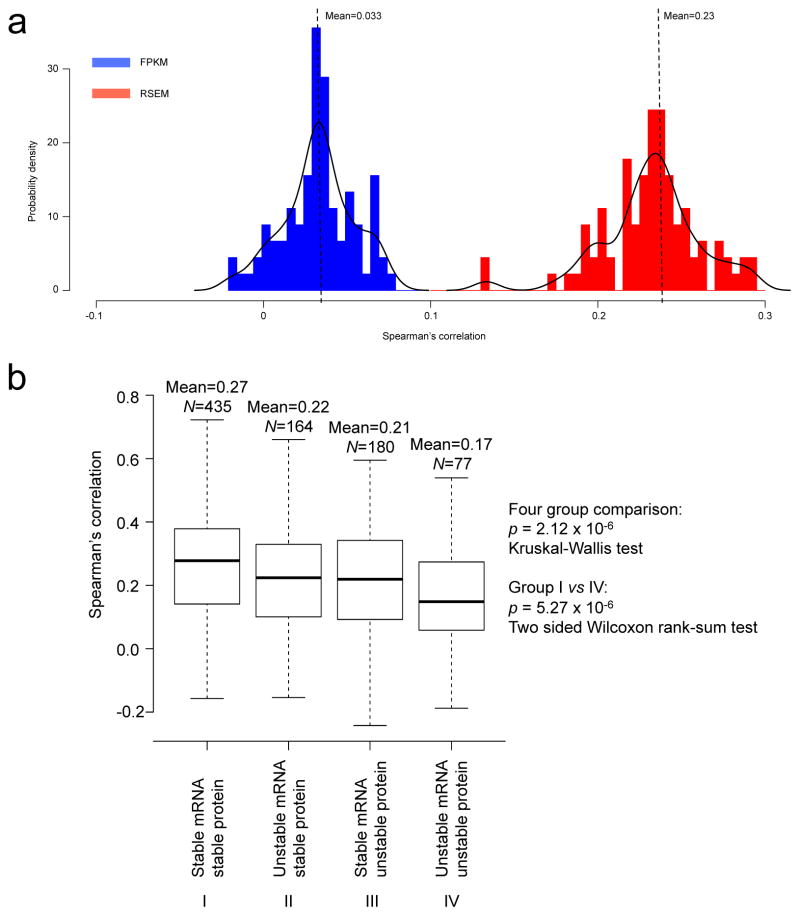
a, Evaluation of the length-bias in different RNA-Seq-based gene abundance estimation methods. The plot shows the distribution of correlation between gene length and estimated transcript abundance based on FPKM (Fragments Per Kilobase of exon per Million fragments mapped, blue curve) and RSEM (RNA-Seq Expectation Maximization, red curve), respectively. FPKM measure is independent of gene length, whereas the RSEM measure strongly correlates with gene length. b, Relationship between mRNA-protein correlation and the stability of the molecules. Human genes were separated into four categories based on the mRNA and protein half-lives of their mouse orthologs: stable mRNA/stable protein; stable mRNA/unstable protein, unstable mRNA/stable protein, and unstable mRNA/unstable protein. Distribution of mRNA-protein correlations for genes in each category was plotted in the box plots. Genes with stable mRNA and stable protein showed relatively higher mRNA-protein correlation whereas those with unstable mRNA and unstable protein showed relatively lower mRNA-protein correlation. Only common genes in both our study and the mouse study were included in the analysis. The total number of genes in each category (N) is labeled in the figure. The p value indicating correlation difference among the four categories was calculated based on the Kruskal-Wallis non-parametric ANOVA test. The p value indicating correlation difference between the stable mRNA/stable protein group and the unstable mRNA/unstable protein group was calculated based on the two-sided Wilcoxon rank-sum test.
Extended Data Figure 7. mRNA and protein-level cis-effect of copy number alterations (CNAs) in focal amplification, focal deletion and non-focal regions.
The figure plots cumulative fraction curves of CNA-mRNA (dashed lines) and CNA-protein (solid lines) expression correlations for genes in the focal amplification regions (red), focal deletion regions (green), and non-focal regions (blue), respectively. Focal amplification regions were defined in the TCGA study. Any chromosomal regions outside the focal amplification and deletion regions were considered as non-focal regions. CNA-mRNA correlations were significantly higher than CNA-protein correlations for genes in any of the three groups. Moreover, genes in the focal amplification regions showed the highest level of CNA-mRNA and CNA-protein correlations among the three groups of genes. P values were based on the two-sided kolmogorov-smirnov test.
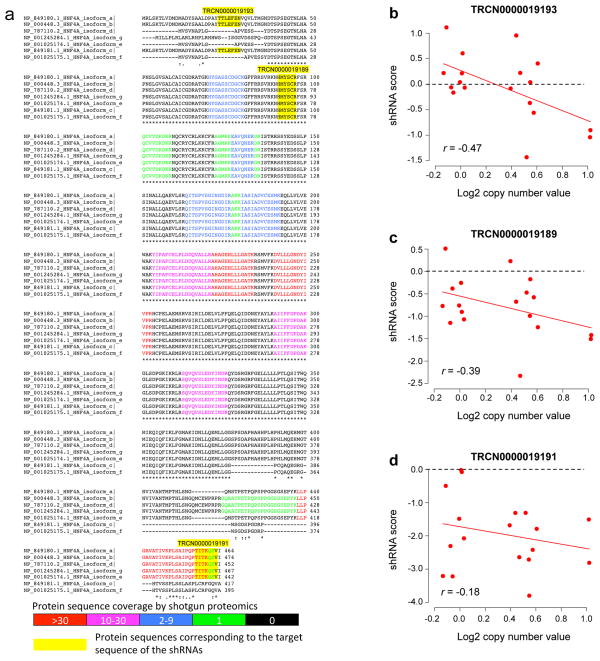
Extended Data Figure 8. HNF4α isoforms and the effect of HNF4A shRNA on the proliferation of colon cancer cells.
a, Multiple sequence alignment of the HNF4α isoforms, with peptides detected by shotgun proteomics and sequences corresponding to the shRNA target sequences highlighted. Different colors of the letters indicate different levels of sequence coverage in the shotgun proteomics study, as indicated by the color scale bar. Yellow boxes highlight sequences corresponding to the shRNA target sequences. TRCN0000019193 specifically targets P1 promoter-driven isoforms, whereas the other two target both types of isoforms. b–d, The P1- HNF4α specific shRNA showed mixed impacts (b), whereas shRNAs simultaneously targeting both P1- and P2- HNF4α showed a primarily negative impact on cell proliferation (c,d). Moreover, a stronger negative impact was associated with increased copy number, both for the P1- HNF4α specific shRNA (p=0.04, Spearman’s correlation [r]) and for all shRNAs (p=0.01, Spearman’s correlation p-values for individual shRNAs summarized by the Fisher’s combined probability test).
Extended Data Figure 9. Consensus matrices, the empirical cumulative distribution function (CDF) plot and core sample identification.
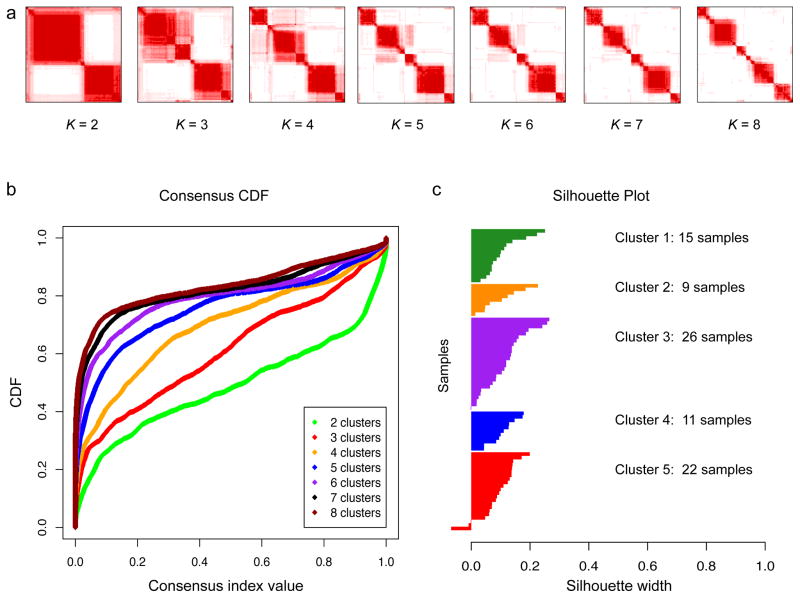
a, Consensus matrices of the 90 CRC samples for k = 2 to k = 8. The consensus matrices show the robustness of the discovered clusters to sampling variability (resampling 80% samples) for cluster numbers k = 2 to 8. In each consensus matrix, both the rows and the columns were indexed with the same sample order and samples belonging to the same cluster frequently are adjacent to each other. For each pair of samples, a consensus index, which is the percentage of times they belong to the same cluster during 1,000 runs of the clustering algorithm based on resampling was calculated. The consensus index for each pair of samples was represented by color gradient from white (0%) to red (100%) in the consensus matrix. b, CDF plots corresponding to the consensus matrices for k = 2 to k = 8. This plot shows the cumulative distribution of the entries of the consensus matrices within the 0–1 range. Skew toward 0 and 1 indicates good clustering. As k increases, the area under the CDF is hypothesized to increase markedly until k reaches the ktrue. In this case, 7 was considered as ktrue because the change of the area under the CDF was close to zero when k increased from 7 to 8. c, Silhouette plot for core sample identification. For each sample (y-axis), the silhouette width (x-axis) compares its similarity to its assigned class and to any other classes. Samples with higher similarity to their assigned class than to any other classes will get positive silhouette width score and be selected as core samples.
Extended Data Figure 10. Network analysis of the subtype signature proteins.
a, The number of signature proteins for each subtype. For a given subtype, the red circle represents proteins that were different in abundance between the subtype and all other subtypes, the green circle represents proteins that were different in abundance between the subtype and normal colon tissues. The intersection between red and green circles contains the signature proteins for each subtype. b, Visualizing subtype C signature proteins in NetGestalt. Proteins in the iRef protein-protein interaction network are placed in a linear order together with the hierarchical modular organization of the network. Alternating bar colors (green and orange) are used to distinguish neighboring modules. Proteins in the up- and down-signatures of subtype C were visualized as two separate tracks below the network modules, where each bar represents a protein. These proteins are not randomly distributed in the network. Highlighted by red or blue arrows are four Network modules (I, IV, V, VI) significantly enriched with up-signature proteins and two modules (II and III) significantly enriched with down-signature proteins (adjusted p value < 0.01). c–d, Heat maps depicting relative abundance of down- and up-signature proteins of subtype C in modules III and I, respectively. Tumors are displayed as rows, grouped by normal controls (N) and proteomic subtypes (A–E) as indicated by different side bar colors. Proteins are displayed as columns. e–f, Network diagrams depicting the interaction of down- and up-signature proteins of subtype C in modules III and I, respectively. Node and node-border colors represent relatively higher or lower abundance in the subtype compared to other subtypes and normal colon tissues, respectively. Red and blue in the heat maps and the network diagrams represent relatively higher or lower abundance, respectively.
Supplementary Material
Acknowledgments
This work was supported by National Cancer Institute (NCI) CPTAC awards U24CA159988, U24CA160035, and U24CA160034, by NCI SPORE award P50CA095103 and NCI Cancer Center Support Grant P30CA068485, by National Institutes of Health grant GM088822 and by contract 13XS029 from Leidos Biomedical Research, Inc. Genomics data for this study were generated by The Cancer Genome Atlas pilot project established by the NCI and the National Human Genome Research Institute. Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at http://cancergenome.nih.gov/.
Footnotes
Supplementary Information is linked to the online version of this paper at www.nature.com/nature.
Author Contributions B.Z., R.J.C.S., D.L.T., L.J.Z. and D.C.L. designed the proteomic analysis experiments, data analysis workflow, and proteomic-genomic data comparisons. K.F.S., L.J.Z., R.J.C.S. and D.C.L. directed and performed proteomic analysis of colon tumor and quality control samples. J.W., X.W., J.Z., Q.L., Z.S., P.W., S.W., R.J.C.S. and B.Z. performed proteomic-genomic data analyses. M.C.C., S.K., R.J.C.S. and D.L.T. performed analyses of mass spectrometry data and adapted algorithms and software for data analysis. S.R.D., R.R.T and M.J.C.E. developed and prepared breast xenografts used as quality control samples. S.A.C, K.F.S. and D.C.L. designed strategy for quality control analyses. R.J.C.S., C.R.K, R.C.R, and H.R. coordinated acquisition, distribution and quality control evaluation of TCGA tumor samples. B.Z., J.W., R.J.C.S., R.J.C. and D.C.L. interpreted data in context of colon cancer biology. B.Z., R.J.C.S. and D.C.L. wrote the manuscript.
All of the primary mass spectrometry data on TCGA tumor samples are deposited at the CPTAC Data Coordinating Center as raw and mzML files for public access (https://cptac-data-portal.georgetown.edu).
The authors declare no competing financial interests.
Reprints and permissions information is available at www.nature.com/reprints.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Kandoth C, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.TCGA. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.TCGA. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.TCGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Zhang B. customProDB: an R package to generate customized protein databases from RNA-Seq data for proteomics search. Bioinformatics. 2013;29:3235–3237. doi: 10.1093/bioinformatics/btt543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, et al. Protein identification using customized protein sequence databases derived from RNA-Seq data. J Proteome Res. 2012;11:1009–1017. doi: 10.1021/pr200766z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim WK, et al. Identification and selective degradation of neopeptide-containing truncated mutant proteins in the tumors with high microsatellite instability. Clin Cancer Res. 2013;19:3369–3382. doi: 10.1158/1078-0432.CCR-13-0684. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 12.de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foss EJ, et al. Genetic variation shapes protein networks mainly through non-transcriptional mechanisms. PLoS Biol. 2011;9:e1001144. doi: 10.1371/journal.pbio.1001144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghazalpour A, et al. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 2011;7:e1001393. doi: 10.1371/journal.pgen.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gry M, et al. Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genomics. 2009;10:365. doi: 10.1186/1471-2164-10-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foss EJ, et al. Genetic basis of proteome variation in yeast. Nat Genet. 2007;39:1369–1375. doi: 10.1038/ng.2007.22. [DOI] [PubMed] [Google Scholar]
- 17.Fu J, et al. System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nat Genet. 2009;41:166–167. doi: 10.1038/ng.308. [DOI] [PubMed] [Google Scholar]
- 18.Peng J, et al. Regularized Multivariate Regression for Identifying Master Predictors with Application to Integrative Genomics Study of Breast Cancer. Annals of Applied Statistics. 2010;4:53–77. doi: 10.1214/09-AOAS271SUPP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garrison WD, et al. Hepatocyte nuclear factor 4alpha is essential for embryonic development of the mouse colon. Gastroenterology. 2006;130:1207–1220. doi: 10.1053/j.gastro.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chellappa K, Robertson GR, Sladek FM. HNF4alpha: a new biomarker in colon cancer? Biomarkers in medicine. 2012;6:297–300. doi: 10.2217/bmm.12.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheung HW, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci U S A. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimokawa T, et al. Identification of TOMM34, which shows elevated expression in the majority of human colon cancers, as a novel drug target. Int J Oncol. 2006;29:381–386. [PubMed] [Google Scholar]
- 23.Irby RB, et al. Activating SRC mutation in a subset of advanced human colon cancers. Nat Genet. 1999;21:187–190. doi: 10.1038/5971. [DOI] [PubMed] [Google Scholar]
- 24.Monti S, Tamayo P, Mesirov J, Golub TR. Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Machine Learning. 2003;52:91–118. [Google Scholar]
- 25.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 26.De Sousa EMF, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19:614–618. doi: 10.1038/nm.3174. [DOI] [PubMed] [Google Scholar]
- 27.Sadanandam A, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang HY, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi Z, Wang J, Zhang B. NetGestalt: integrating multidimensional omics data over biological networks. Nat Methods. 2013;10:597–598. doi: 10.1038/nmeth.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 32.Loboda A, et al. EMT is the dominant program in human colon cancer. BMC Med Genomics. 2011;4:9. doi: 10.1186/1755-8794-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geiger T, Sabanay H, Kravchenko-Balasha N, Geiger B, Levitzki A. Anomalous features of EMT during keratinocyte transformation. PLoS One. 2008;3:e1574. doi: 10.1371/journal.pone.0001574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiemer AK, Takeuchi K, Quinlan MP. Identification of genes involved in epithelial-mesenchymal transition and tumor progression. Oncogene. 2001;20:6679–6688. doi: 10.1038/sj.onc.1204872. [DOI] [PubMed] [Google Scholar]
- 35.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119:1429–1437. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 39.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li S, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4:1116–1130. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.