Abstract
The study of a class of small non-coding RNA molecules, named microRNAs (miRNAs), has advanced our understanding of many of the fundamental processes of cancer biology and the molecular mechanisms underlying tumor initiation and progression. MiRNA research has become more and more attractive as evidence is emerging that miRNAs likely play important regulatory roles virtually in all essential bioprocesses. Looking at this field over the past decade it becomes evident that our understanding of miRNAs remains rather incomplete. As research continues to reveal the mechanisms underlying cancer therapy efficacy, it is clear that miRNAs contribute to responses to drug therapy and are themselves modified by drug therapy. One important area for miRNA research is to understand the functions of miRNAs and the relevant signaling pathways in the initiation, progression and drug-resistance of tumors to be able to design novel, effective targeted therapeutics that directly target pathologically essential miRNAs and/or their target genes. Another area of increasing importance is the use of miRNA signatures in the diagnosis and prognosis of various types of cancers. As the study of non-coding RNAs is increasingly more popular and important, it is without doubt that the next several years of miRNA research will provide more fascinating results.
Keywords: Cancer biology, Drug therapy, miRNAs, Nanoparticles, Oncogene, Tumor suppressor
Introduction
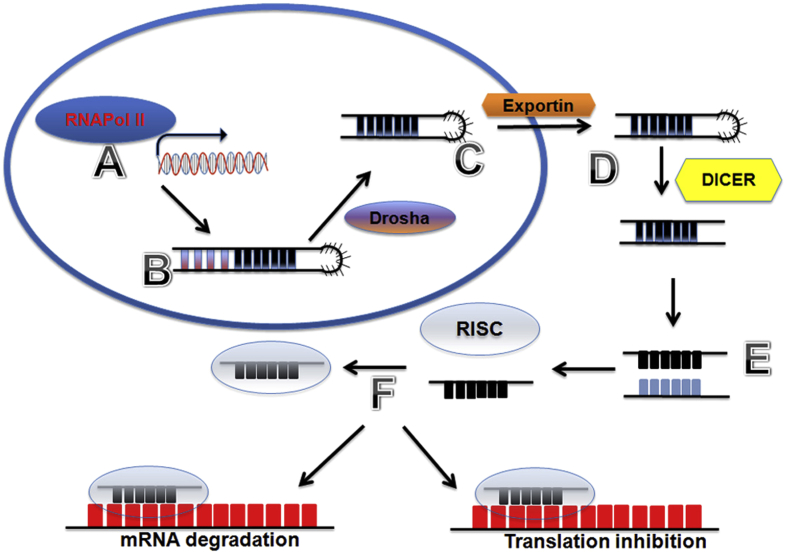
The discovery of lin-4 in Caenorhabditis elegans,1, 2, 3, 4, 5 the first microRNA (miRNA), led to the identification of several hundred other miRNA molecules. MiRNAs are short noncoding RNA molecules of roughly 19–24 nucleotides in length and are a large class, with more than 1,000 members, of small-regulatory RNAs in mammalian genomes. MiRNAs are initially processed as a primary miRNA (pri-miRNA) transcript by RNA polymerase II (or by RNA polymerase III in rare cases) and associated transcription factors. The pri-miRNAs are further processed by Drosha, a nuclear RNase III enzyme, to produce precursor miRNAs (pre-miRNAs). The pre-miRNAs are then transported out of the nucleus into the cytoplasm where they are further processed by the Dicer, another RNase III-familial endonuclease, to become ∼22 base-pairing (bp) miRNA duplexes. Mature miRNAs are then unwound from miRNA duplexes and loaded into the RNA-induced silencing complex (RISC). The RISC with the mature miRNAs can regulate gene expression by binding to the mRNA transcripts of target genes, usually at the 3′ untranslated regions (3′-UTRs). The miRNA-RISC complex can block translation of target mRNA into protein and/or induce degradation of target mRNA transcript (see Fig. 1).6, 7, 8
Figure 1.
Processing of microRNA. RNA polymerase II and appropriate transcription factors stimulate transcription of the microRNA gene (A) into a primary long transcript with a stem loop structure called a primary microRNA transcript (pri-miR). The primary transcript (B) is then processed by Drosha, an RNAase III enzyme, to produce a small precursor hairpin microRNA (pre-miR). The precursor microRNA (C) is then shuttled outside the nucleus by Exportin to the cytoplasm for further process. In the cytoplasm the precursor microRNA is then processed into a mature 19–24 nucleotide duplex (D) by another RNAase enzyme Dicer. Next, the duplex is split into a primary and secondary strand (E); then the primary strand is loaded into the RNA-induced silencing complex (RISC). Next the microRNA with RISC targets specific messenger RNA (mRNA) transcripts (F) at the seed region to induce either mRNA degradation (left) or block translation (right).
Binding of an animal miRNA to its target gene transcript relies mainly on a 6–8 nucleotide pair region of near perfect complementarity between the 5′ end (i.e., the ‘seed region’) of the miRNA and its target mRNA sequence. Although miRNAs usually bind to the 3′ UTRs of target transcripts, evidence is emerging that the target sites can be located outside of the 3′ UTRs, such as 5′ untranslated regions (5′-UTRs) or protein-coding regions. Interestingly, recent studies have demonstrated that allelic and sequence variants can i) create a new miRNA binding site where one did not exist before, ii) remove a miRNA binding site, or iii) change the affinity of a particular miRNA for a binding site.9, 10 Furthermore, as a single miRNA can regulate tens or even hundreds of different target genes while a single gene may be regulated by multiple miRNAs; the miRNA-target networks are pretty complex.
This review will focus on the functions of miRNAs in the pathogenesis of tumors and the potential clinical implications of miRNAs in the diagnosis, prognosis and therapy of cancers. First, it will outline the functions of miRNAs in cancer biology as tumor suppressors and/or oncogenes and how miRNAs can synergistically work together. Next, it will discuss miRNAs related to cancer therapy/treatment. Finally, it will focus on miRNAs as diagnostic and prognostic tools in the context of the tumor itself and circulating in blood plasma.
The regulations and functions of MiRNAs in cancers
For the past ten years the study of miRNAs in eukaryotic organisms has grown exponentially and it has been well established that these small RNAs are master regulators of gene expression and multiple other biological processes. MiRNAs were first identified in the mid-1990s with the discovery of Let-7 and Lin-4 in a worm (Caenorhabditis elegans),1, 2, 11 but it was not until 2000/2001 when it was determined that these miRNAs exist and are highly conserved in multiple eukaryotic organisms and mammalian species.3, 4, 5 This was a major finding in the field of miRNAs, showing not only that they exist in multiple organisms but also they are likely major regulators of biological processes in these organisms. The next landmark event came soon after in 2002 and 2003 when it was found that miRNAs were aberrantly expressed in cancer12, 13, 14 and subsequently several seminal studies were reported in 2005 that provided compelling evidence to demonstrate the importance of miRNAs in cancer.15, 16, 17 Since then, the study of miRNAs in cancer exploded dramatically and still represents a very important and challenging area of research (see examples in Table 1, Table 2).
Table 1.
Examples of tumor-suppressor miRNAs.
| MicroRNA | Cancer typea | Function | Reference |
|---|---|---|---|
| miR-29b | AML | Represses Sp1 which resulted in c-KIT inhibition | 19, 20 |
| miR-34b/c | Lung cancer | A positive feedback between p53 and miR-34 mediates tumor suppression in human lung cancer | 21, 22 |
| miR-126 | Breast, lung, and colon cancers | Plays a critical tumor-suppressor role in tumor initiation and metastasis | 23, 24, 25, 26, 27 |
| miR-150 | AML | A critical tumor-suppressor gatekeeper in AML by targeting FLT3 and Myb | 28 |
| miR-155 | Breast cancer | Downregulates RAD51 and sensitizes cancer cells to irradiation | 29, 30 |
| miR-181a/b | AML | Their increased expression is associated with good prognosis and hinders tumor cell growth | 31, 32 |
| miR-375 | Breast cancer | Forced expression re-sensitizes cells to tamoxifen treatment | 33 |
| miR-494 | Lung cancer | Regulated by ERK1/2 it modulates proliferation and apoptosis response | 34 |
| miR-495 | AML; gastric cancer | Specifically down-regulated in MLL-rearranged AML; Shown to block migration and invasion | 35, 36 |
| miR-551a | Gastric cancer | Forced expression leads to a block in migration and invasion | 35 |
AML, Acute Myeloid Leukemia.
Table 2.
Examples of oncogenic miRNAs.
| MicroRNA | Cancer typea | Function | Reference |
|---|---|---|---|
| miR-9 | AML | Specifically overexpressed in MLL-rearranged AML and promotes leukemia progression | 37 |
| miR-17-92 | AML | Up-regulated in MLL-rearranged AML and targets p21 and RASSF2 | 38, 39, 40 |
| miR-21 | Breast cancer | Overexpression of miR-21 contributes to proliferation and metastasis | 41 |
| miR-27a | NSCLC | Promotes proliferation in NSCLC cells | 42 |
| miR-30a/c | RCC | Downregulation leads to increased expression of HIF2a | 43 |
| miR-126 | AML | Up-regulated in core-binding factor (CBF) leukemia | 44 |
| miR-181a/b | Breast, liver and colon cancers | Promote tumorigenesis and tumor progression | 45, 46, 47 |
| miR-196a | Gastric cancer | Promoted EMT, migration and invasion | 48 |
| miR-196b | AML | Upregulated in MLL-rearranged AML and targets Fas | 49 |
| miR-421 | Gastric cancer | Marker of circulating tumor cells | 50 |
AML, Acute Myeloid Leukemia; NSCLC, Non-Small Cell Lung Cancer; RCC, Renal Cell Carcinomas.
Furthermore, it is becoming evident that an emerging hallmark of cancer is the dysregulation of miRNAs, both in the tumor itself and in the surrounding microenvironment. Future work in this field will undoubtedly ensure its position in the new hallmarks of cancer.18 Dysregulated miRNAs can contribute to tumorigenesis by playing tumor-suppressive and/or oncogenic roles. As exampled in Table 1, many miRNAs, including miR-29b,19, 20 miR-34b/c,21, 22 miR-126,23, 24, 25, 26, 27 miR-150,28 miR-155,29, 30 miR-181a/b,31, 32 miR-375,33 miR-494,34 miR-49535, 36 and miR-551a,35 play critical tumor-suppressor roles in tumorigenesis and/or mark a good prognosis for patients. These miRNAs have shown to have potent antitumorigenic properties. However, there are also numerous oncogenic miRNAs including miR-9,37 miR-17-92,38, 39, 40 miR-21,41 miR-27a,42 miR-30a/c,43, miR-126,44 miR-181a/b,45, 46, 47 miR-196a,48 miR-196b,49 and miR-421.50 The intricate role of miRNAs in tumorigenesis appears to be simple yet it remains complicated. This can be attributed to several reasons briefly reviewed in the following section.
There is enormous complexity in miRNA-mediated gene regulation
First, a single miRNA can regulate up to hundreds of genes51, 52, 53, 54 (while prediction programs can suggest thousands of genes that are targets of a single miRNA) making it difficult to determine which genes are direct targets of a single miRNA.55, 56, 57, 58 Second, given the fact that a single mRNA target contains potential binding sites for multiple individual miRNAs, a single gene can be regulated simultaneously or sequentially by multiple miRNAs.55, 56, 57 Third, a single miRNA can regulate target mRNAs by both repressing translation and inducing mRNA degradation within mammalian species53, 54, 59, 60, 61, 62, 63 makes miRNA-mediated gene regulation even complicated.
The mechanism underlying translational repression is still unclear but it is known that the inhibition is determined by two potential criteria.6 The first criterion is whether mRNAs are present in an mRNA-protein complex (mRNP) for inhibition of initiation or in the form of large polysomes involved in elongation inhibition.6, 64, 65, 66, 67, 68 The second criterion is whether inhibited mRNAs contain internal ribosome entry site (IRES).6, 7, 65, 66, 68 Although the exact mechanisms leading to translational inhibition remain unclear, several hypothesizes have been proposed.6, 7, 8, 45, 46, 47 Notably, it was proposed that miRISC (miRNA loaded in RISC) may repress the elongation process, thus miRISC has a role in the promotion of early ribosome dissociation from mRNAs.6, 7, 8, 69 Another hypothesis is that the miRNA complex mediates repression through accumulation of target RNA transcripts in P-bodies (processing bodies) as P-bodies containing mRNA have been suggested to not be part of the translation process.7, 70, 71
Regulation of mRNA by miRNAs can also occur through mRNA degradation and actually, mRNA destabilization usually comprised the major component of repression in mammalian species.72 It has been shown that miRNAs with high degree of sequence complementation to target mRNA can induce the mRNA degradation processes.7 While several components of the complex are known to participate in regulation of mRNA degradation it appears that mechanisms including deadenylation, decapping and exonucleolytic digestion of mRNA are also involved.6, 7, 65, 66, 68, 73, 74, 75 The seed sequence similarity is thought to be important in the initial selection of mRNA targets where the number, type and position of mismatches of the miRNA/mRNA complex is critical.6, 7, 76, 77 Thus, the full downstream effects of a single miRNA may not be well understood until it is clear exactly how that miRNA is inhibiting its potential targets.
Expression of miRNAs can be regulated at both transcriptional and post-transcriptional levels
Given the essential functions of miRNAs in maintaining virtually all important biological processes, including development, differentiation and apoptosis, the stringent control of miRNAs, at both transcriptional and post-transcriptional levels, is critical.78 Numerous studies have analyzed the regulation of miRNAs at the transcription sites, including direct binding by transcription factors, hyper/hypomethylation of miRNA promoters, and changes in histone assembly and chromatin packing.79, 80, 81, 82, 83, 84, 85
However, fewer studies have looked at the regulation of miRNAs at downstream processing steps. It is important to identify other cofactors existing in or cooperating with the Drosha or Dicer complexes and how they assist the Drosha or Dicer complexes in regulating the maturation or degradation of miRNAs. We recently have shown that in a subtype of acute myeloid leukemia (AML) that carries chromosomal translocations involving the mixed lineage leukemia (MLL) gene, the primary transcription of miR-150 is significantly up-regulated by MLL-fusion proteins but its maturation process is substantially inhibited by the MYC/LIN28 functional axis; as a result, the level of mature miR-150 is significantly down-regulated and thereby expression of its critical target genes such as FLT3 and MYB is significantly up-regulated, which ultimately lead to cell transformation and leukemogenesis.28
The function of miRNAs can be tissue-specific
A single miRNA may play distinct roles in different tissues. For instance, the miR-181 family has traditionally been described in several solid malignancies, such as breast, liver and colon cancer, as an oncomiR (defined as a miRNA that acts as an oncogene to promote tumorigenesis and tumor progression)45, 46, 47; however, in AML this family acts as tumor suppressor.31, 32 Similarly, miR-126 has been previously reported as a tumor-suppressor gene in various types of solid tumors including breast, lung, and colon cancers, etc23, 24, 25, 26, 27; we recently found that miR-126 functions as an oncogene in the development of leukemias, particularly core-binding factor (CBF) leukemias bearing t(8;21) or inv(16)/t(16;16).44 This concept is now widely accepted in the scientific community and is in part attributed to the ability of miRNAs to target different genes in different tissues. For example, the miR-17-92 cluster, initially transcribed as a polycistronic transcript to later produce 6 mature miRNA transcripts, is overexpressed in lymphoma and MLL-rearranged AML and plays an important oncogenic role likely through repressing expression of its tumor-suppressor target genes such as Pten, Rassf2, and p2116, 38, 39, 40, 86, 87; in contrast, in breast cancer the miR-17-92 cluster functions as a tumor suppressor likely through negatively regulating oncogenic target genes involved in cell proliferation, cellular invasion, and tumor metastasis such as AIB1 and CCND1.88, 89, 90, 91
It is possible that different target genes are co-expressed with the miRNA regulators and thereby are regulated by the miRNAs in distinct types of cells or tissues. Another possibility is that the regulatory miRNAs are recruited into different target transcripts in distinct types of cells or tissues. It has been shown over the past several years that there are multiple RNA-binding proteins that assist in processing of miRNAs. For example, HnRNP A1, a member of nucleo-cytoplasmic shuttling proteins previously implicated in mRNA metabolism, was shown to specifically bind to pri-miR-18a and facilitated Drosha-mediated processing, thus acting as a key factor in regulation of miR-18a processing.6, 7, 92, 93 These messenger ribonucleoprotein complexes (mRNPs) were critical in the inhibition of miRNA targets. Thus, this might suggest that general RNA binding proteins have an auxiliary function in the regulation of miRNAs and probably may help to preferentially inhibit different target genes in distinct types of cells or tissues.
Individual miRNAs can regulate both oncogenes and tumor suppressors simultaneously
Interestingly, a single miRNA can simultaneously regulate both tumor suppressive and oncogenic target genes within a single cancer. This was recently described in our paper from Li et al (2012).49 In this paper we showed that miR-196b was upregulated in MLL-rearranged AML and resulted in repression of the tumor suppressor and pro-apoptotic genes such as FAS; surprisingly, we found that miR-196b also targets HOXA9/MEIS1, characterized oncogenes that are co-upregulated with miR-196b by MLL-fusion proteins.49 In normal hematopoiesis, miR-196b likely helps to fine-tune expression of Hox genes and Meis1 in primitive hematopoietic stem cells (HSCs) and to remove their residual transcripts in committed progenitor cells to allow further differentiation of the cells; when miR-196b is down-regulated, Fas becomes up-regulated to maintain homeostasis of the cells.49 In MLL-fusion-mediated leukemogenesis, aberrant up-regulation of miR-196b by MLL fusions results in the persistent repression of expression of its tumor-suppressor targets (e.g. FAS), along with the residual high level of HOXA9/MEIS1; as a result, aberrant overexpression of miR-196b inhibits differentiation, disrupts cell homeostasis and promotes cell proliferation via inhibiting apoptosis, which eventually results in cell transformation and leukemogenesis.49 This means, despite what was described previously, that a miRNA may not simply be described as a tumor-suppressor or oncomiR as the scientific community has done in the past. This also implies that tumor initiation and development, contributed by aberrant regulation of miRNAs, might be more complex than previously thought and has important implications for using miRNAs as a therapeutic avenue.
MiRNAs mediate drug response and themselves are modulated in therapy
As miRNAs were being linked to several hallmarks of cancer in tumor cells, there was a hypothesis that miRNA expression could be altered by cancer therapy and associated with drug response. This hypothesis is supported by two lines of evidence that are not mutually exclusive: differential expression of miRNAs in tumor cells before treatment has been associated with response to chemotherapy; changes in miRNA expression have been observed in cancer cells following treatment with effective therapy. It is highly likely that these observations are linked together within a single cancer (see Fig. 2). For example, it was found within lung cancer a group of miRNAs were regulated by epithelial growth factor receptor (EGFR) and hepatocyte growth factor receptor (MET).94 Both EGFR and MET are overexpressed in lung cancer driving tumorigenesis and, interestingly, the investigators found that these miRNAs bestowed resistance to tyrosine kinase therapy.94 Specifically, they found that gefitinib (a tyrosine kinase inhibitor) treatment inhibited miR-221, miR-222, miR-30b and miR-30c, all positively regulated by EGFR and MET, and resulted in increased apoptosis in lung cancer cell lines.94 Furthermore, increased expression of miR-30c, miR-221 and miR-222 in gefitinib-responsive cells attenuated sensitivity and knockdown of these miRNAs bestowed sensitivity to gefitinib in normally gefitinib-insensitive cells.94
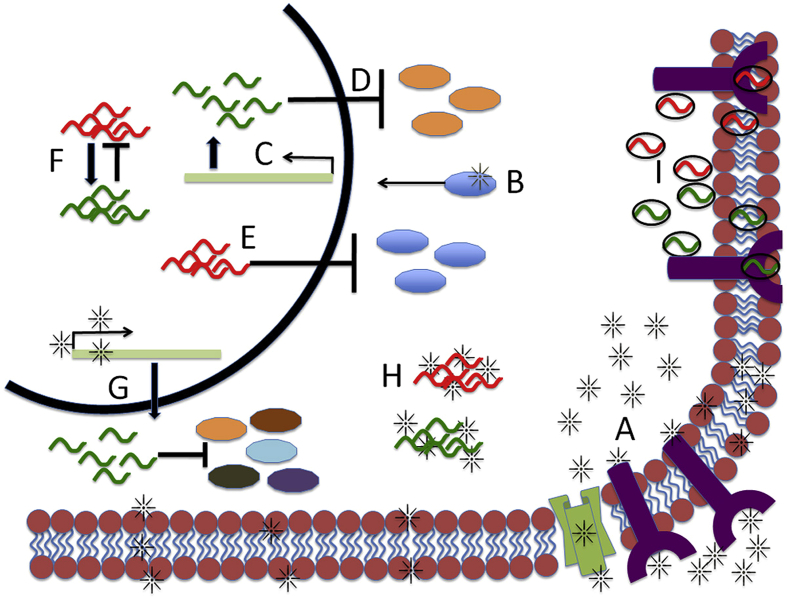
Figure 2.
MicroRNAs and cancer therapy. There are multiple ways which microRNAs can be affected by drug therapy. Drugs can either through the cell membrane or bind to receptors or cellular channels (A) to enter the cell. Once inside drugs can bind to protein targets or transcription factors (B) to affect miRNA expression (C) or conversely block the activation of protein targets and prevent that target from activating or blocking a miRNA. By driving miRNA expression this can now lead to inhibition of oncogenes (D) or tumor suppressors (E). Furthermore, miRNAs could potentially regulate each other (F) meaning drugs can have multiple effects on miRNAs. Another possibility is that the drug can directly bind to the regulatory region of miRNAs either inhibiting or inducing expression (G), which can then lead to decrease of miRNA-target genes. Drugs can also potentially bind to miRNAs themselves or to miRNA binding partners (H) leading to a change in miRNA function. Finally miRNAs themselves can be drugs either as modified nucleic acids or as oligos or antisense oligos and then packaged into either viruses or microvesicles and macrovesicles. These miRNAs can enter the cell freely to inhibit downstream targets or potentially bind to cellular receptors (I). Thus, by changing miRNA expression it could change the ability of a cell to respond to drugs either by activating resistance or bestowing sensitivity through a multitude of mechanisms. These are some of the most common mechanisms that miRNAs can be affect by drugs or act as a drug themselves.
Meanwhile, the effect of metabolism or chemotherapy-based diet on miRNA expression and subsequent tumorigenesis has also bee investigated. For example, Mandal et al found that the bioactive component of fish oil, docosahexaenoic acid (DHA), inhibited miR-21, a protumorigenic miRNA.41 Another group found the diet/microbe-derived short chain fatty acid butyrate, a known histone deacetylase (HDAC) inhibitor used in cancer treatment particularly in colon cancer, blocked tumorigenesis through inhibiting miR-106b expression and subsequently promoting expression of p21, a direct target of miR-106b.95 Similarly, it was reported that butyrate resulted in decreased expression of the oncogenic miRNA cluster miR-17-92; they found this similar result was observed in other HDAC inhibitors such as trichostatin A (TSA) and suberoylanilide hydroxamic acid (SAHA).96 Several of these inhibitors are clinically approved for the treatment of a few cancers and are currently being evaluated for other cancers, which provide a novel avenue for research.
Together these results show that miRNAs both modulate response to chemotherapy and are themselves modulated by chemotherapy. These effects can be direct or through indirect as are the examples of diet-induced miRNA changes. The new wave of cancer therapy is focusing on drugs that specifically target miRNAs as well as complementary signaling pathways to synergize with the routine use of chemotherapy in cancer patients.
MiRNA-based cancer therapy
Due to the central role of miRNAs in cancer initiation and progression, they have been a source of interest for several years, specifically whether these miRNAs can be targeted or not.97 As already described, it has been observed that several drugs can alter miRNA expression. Moreover, if certain cancers are particularly “addicted” to particular miRNAs (named oncomiR addiction) then targeting specific miRNAs selectively should minimize off-target toxicity.98 Investigators are interested in designing inhibitors for oncogenic miRNAs and mimics for tumor-suppressor miRNAs that can act alone or synergistically with currently approved treatments (see Fig. 2).99
In the past ten years several methods of design of miRNA inhibitors have been designed. One method of miRNA inhibition has been the use of modified nucleic acids that can bind and inhibit the mature miRNAs.99 An example of this is the anti-miR which can have various chemical modifications that enhance function and increase stability such as locked nucleic acids (LNAs),99, 100, 101 peptic nucleic acids, phosphorothioate modifications, 2′O-Me and 2′-fluoro substitutions and morpholinos.59, 99, 100, 101, 102, 103, 104, 105 Such miRNA inhibitors have been tested successfully in multiple experiments and are currently being evaluated for clinical purposes.101, 105 In contrast, nude miRNA mimics can be packaged directly with delivery vehicles for therapy.106
Another active area with miRNA research has been the delivery of miRNA inhibitors or miRNA mimics to target host regions. One effective method is the use of engineered viruses that induce miRNA inhibition through the expression of transcripts complementary to mature miRNA sequences. These viral delivery methods have had significant success in laboratory studies but have limited effectiveness in patients due to concerns about off-target effects of the viruses such as immunogenicity and chromosomal incorporations.107 Another limitation is the inability to specifically target the virus to the tumor and thereby generating toxicity as several of these miRNAs control host processes important for issues outside of the tumor.107, 108, 109 Another mechanism for the delivery of miRNA-based therapies is the use of nanoparticles.110, 111 The nanoparticle delivery method appears attractive because it avoids several of the concerns used for viral delivery systems.107 The nanoparticle delivery method has been shown to hold great potential. For example, Su et al found that lipid nanoparticles that contained 2′fluoro-modified anti-miR-122 significantly inhibited tumor growth.110 Another method recently developed was the use of biodegradable polymer nanoparticles containing anti-miRNAs to inhibit miR-155 in a mouse model of pre-B-cell lymphoma.111 Nanoparticles conjugated with targeting molecules for specific binding have also been designed and tested. For example, Huang et al designed transferring-conjugated anionic lipopolyplex nanoparticles carrying miR-29b and showed their specific binding to AML cells and significant anti-leukemia activities in vitro and in vivo.106
MiRNAs as biomarkers for diagnosis and prognosis in cancers
Currently, there are dozens of clinical trials that are assessing the correlation between miRNA expression and cancer diagnosis and prognosis. Several of these trials have been started in the past five years (www.clinicaltrials.gov) while new ones are being designed. Due to the pleiotropic effects of miRNAs they have been an attractive avenue for patient diagnosis evaluation and prognosis. Also, miRNAs, attributed to their size, are highly stable and resistant to RNAses and thus have a higher level of stability than mRNA.
MiRNAs in cancer tissues
Expression profiles of many miRNAs derived from tumor tissues have been shown to be useful in diagnosis and prognosis of the patients. For example, Lu et al demonstrated that miRNA expression profiles can be used to precisely classify various types of cancers, and are superior to mRNA expression profiles in classification of poorly differentiated tumors.15 MiR-181 family has been shown to provide an independent prognostic assessment in both cytogenetically normal and abnormal acute myeloid leukemia patients.31, 32, 112, 113 Similarly, a study from Yang et al showed that seven miRNAs, miR-15b*, miR-23a, miR-133a, miR-150*, miR-197, miR-497 and miR-548b-5p, were significantly decreased in the serum of patients with advanced stage (grade II–IV) astrocytomas, and the miRNA-based signature could accurately distinguish between normal vs. cancer patients.114
Circulating MiRNAs in serum or plasma
Furthermore, several studies have identified stable miRNAs in human serum or plasma and have seen that there are distinctive patterns of expression of these miRNAs in these patients.56, 57, 115, 116, 117, 118, 119, 120, 121 Circulating miRNAs have also been shown to be able to serve as diagnostic/prognostic indicators. For example, Chen et al identified a comprehensive miRNA signature in the serum of normal patients comprising roughly 190 miRNAs; they further demonstrated that cancer patients with solid malignancies had a completely different miRNA expression signature.122 Differential expression of circulating miRNAs has since been seen in patients of many types of cancers in the past several years including multiple myeloma, nasopharyngeal carcinoma, gastric cancer, prostate cancer, breast cancer, colon cancer, pancreatic cancer, diffuse large B-cell lymphoma, squamous cell carcinoma, lung cancer, ovarian cancer and several others48, 50, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142, 143, 144, 145, 146, 147, 148, 149, 150, 151, 152, 153, 154, 155, 156, 157, 158, 159, 160, 161 (reviewed further in Ref.149).
New function of MiRNAs other than targeting mRNAs
Increasing evidence shows that both normal and tumor cells (especially circulating tumor cells (CTCs)162, 163 in terms of solid tumors) can package miRNAs in exosomes (10–100 nm in diameter), microvesicles (100–1000 nm), membrane microparticles (50–80 nm) and apoptotic bodies (50–500 nm) which appear to be specifically-packaged miRNAs.164 These miRNAs can control functions in both closely neighboring cells and distantly located cells. This provides a solid basis to study miRNAs in CTCs and in extracellular vesicles. Further, with new research it has been discovered that miRNAs can bind to proteins, challenging the current dogma of miRNAs acting solely on target mRNAs.165 Thus, miRNAs packed in these extracellular vesicles can have diverse effects outside of mRNA binding. This thought was very recently supported when it was discovered that miRNAs (miR-21 and miR-29a) within exosomes can bind to toll-like receptors (TLRs) and could activate immune cells as ligands for these receptors.165 Although this might have been hypothesized with previous knowledge that single viral RNAs can be ligands for these receptors,166, 167 these findings have never been shown before in miRNAs and open up an entirely undiscovered and novel field looking at the function of miRNAs as ligands. Additionally, this could mean that miRNAs could be potential ligands for the large class of orphan receptors where no ligand has been identified to date168, 169 or they could be ligands for a myriad of other receptors.
Conclusions
As we look at the last decade of miRNA research in cancer biology and the incredible work that has been accomplished, it is clear that there is still much to be discovered. The future work in the next ten years will be focused on understanding how miRNAs regulate target genes in cancer initiation, progression, metastasis, relapse, and drug response and resistance, which will provide a more comprehensive mechanistic analysis of these noncoding RNAs. Furthermore, more noncoding RNAs are constantly being studied and are also an increasing area of focus. Future work will undoubtedly focus on the co-interaction and regulation of noncoding RNAs and how they together contribute to disease. Once we have gained this understanding, it will allow researchers to design better miRNA inhibitors or miRNA mimics that will guide the creation of new and more effective drug therapies. However, there are still lots of challenges to be solved before we applied miRNA-based therapies into the clinic to treat cancer patients.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
We would like to dedicate this review to Dr. Janet Rowley whose inspiration, mentorship and critique was instrumental to the design of this review. This work was supported in part by the National Institutes of Health (NIH) R01CA127277 and R01CA182528 (to JC), F31 CA171702 (to CP), the Howard Hughes Med-into-Grad Translational Research Program (to CP), American Cancer Society (ACS) Research Scholar grant (to JC) and the Spastic Paralysis Foundation of the Illinois, Eastern Iowa Branch of Kiwanis International (to JC).
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. Dec 3 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. Dec 3 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. Oct 26 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 4.Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. Oct 26 2001;294(5543):862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 5.Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. Oct 26 2001;294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 6.Wahid F., Shehzad A., Khan T., Kim Y.Y. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochimica et Biophysica Acta (BBA) – Molecular Cell Research. 2010;1803(11):1231–1243. doi: 10.1016/j.bbamcr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Carthew R.W., Sontheimer E.J. Origins and mechanisms of miRNAs and siRNAs. Cell. 20 Feb 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alemán L.M., Doench J., Sharp P.A. Comparison of siRNA-induced off-target RNA and protein effects. RNA. March 1 2007 doi: 10.1261/rna.352507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abelson J.F., Kwan K.Y., O'Roak B.J. Sequence variants in SLITRK1 are associated with Tourette's Syndrome. Science. October 14, 2005;310(5746):317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 10.Yu Z., Li Z., Jolicoeur N. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res. 2007;35(13):4535–4541. doi: 10.1093/nar/gkm480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moss E.G., Lee R.C., Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. Mar 7 1997;88(5):637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 12.Calin G.A., Dumitru C.D., Shimizu M. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. Nov 26 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McManus M.T. MicroRNAs and cancer. Semin Cancer Biol. Aug 2003;13(4):253–258. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 14.Michael M.Z., OC S.M., van Holst Pellekaan N.G., Young G.P., James R.J. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. Oct 2003;1(12):882–891. [PubMed] [Google Scholar]
- 15.Lu J., Getz G., Miska E.A. MicroRNA expression profiles classify human cancers. Nature. Jun 9 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 16.He L., Thomson J.M., Hemann M.T. A microRNA polycistron as a potential human oncogene. Nature. Jun 9 2005;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Donnell K., Wentzel E., Zeller K., Dang C., Mendell J. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. Mar 4 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Garzon R., Liu S., Fabbri M. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. Jun 18 2009;113(25):6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu S., Wu L.C., Pang J. Sp1/NFkappaB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell. Apr 13 2010;17(4):333–347. doi: 10.1016/j.ccr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He L., He X., Lim L.P. A microRNA component of the p53 tumour suppressor network. Nature. Jun 28 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okada N., Lin C.P., Ribeiro M.C. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev. Mar 1 2014;28(5):438–450. doi: 10.1101/gad.233585.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tavazoie S.F., Alarcon C., Oskarsson T. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. Jan 10 2008;451(7175):147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J., Du Y.Y., Lin Y.F. The cell growth suppressor, mir-126, targets IRS-1. Biochem Biophys Res Commun. Dec 5 2008;377(1):136–140. doi: 10.1016/j.bbrc.2008.09.089. [DOI] [PubMed] [Google Scholar]
- 25.Liu B., Peng X.C., Zheng X.L., Wang J., Qin Y.W. MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer. Feb 13 2009 doi: 10.1016/j.lungcan.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Crawford M., Brawner E., Batte K. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun. Sep 5 2008;373(4):607–612. doi: 10.1016/j.bbrc.2008.06.090. [DOI] [PubMed] [Google Scholar]
- 27.Guo C., Sah J.F., Beard L., Willson J.K., Markowitz S.D., Guda K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. Nov 2008;47(11):939–946. doi: 10.1002/gcc.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang X., Huang H., Li Z. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. Oct 16 2012;22(4):524–535. doi: 10.1016/j.ccr.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gasparini P., Lovat F., Fassan M. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc Natl Acad Sci USA. March 10 2014 doi: 10.1073/pnas.1402604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rokah O.H., Granot G., Ovcharenko A. Downregulation of miR-31, miR-155, and miR-564 in chronic myeloid leukemia cells. PLoS ONE. 2012;7(4):e35501. doi: 10.1371/journal.pone.0035501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z., Huang H., Li Y. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood. Mar 8 2012;119(10):2314–2324. doi: 10.1182/blood-2011-10-386235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwind S., Maharry K., Radmacher M.D. Prognostic significance of expression of a single microRNA, miR-181a, in cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. Dec 20 2010;28(36):5257–5264. doi: 10.1200/JCO.2010.29.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward A., Balwierz A., Zhang J.D. Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene. Feb 28 2013;32(9):1173–1182. doi: 10.1038/onc.2012.128. [DOI] [PubMed] [Google Scholar]
- 34.Romano G., Acunzo M., Garofalo M. MiR-494 is regulated by ERK1/2 and modulates TRAIL-induced apoptosis in non–small-cell lung cancer through BIM down-regulation. Proc Natl Acad Sci USA. Oct 9 2012;109(41):16570–16575. doi: 10.1073/pnas.1207917109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z., Cao Y., Jie Z. miR-495 and miR-551a inhibit the migration and invasion of human gastric cancer cells by directly interacting with PRL-3. Cancer Lett. Oct 1 2012;323(1):41–47. doi: 10.1016/j.canlet.2012.03.029. [DOI] [PubMed] [Google Scholar]
- 36.Jiang X., Huang H., Li Z. miR-495 is a tumor-suppressor microRNA down-regulated in MLL-rearranged leukemia. Proc Natl Acad Sci USA. Nov 20 2012;109(47):19397–19402. doi: 10.1073/pnas.1217519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen P., Price C., Li Z. miR-9 is an essential oncogenic microRNA specifically overexpressed in mixed lineage leukemia–rearranged leukemia. Proc Natl Acad Sci USA. July 9 2013;110(28):11511–11516. doi: 10.1073/pnas.1310144110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mi S., Li Z., Chen P. Aberrant overexpression and function of the miR-17-92 cluster in MLL-rearranged acute leukemia. Proc Natl Acad Sci USA. Feb 23 2010;107(8):3710–3715. doi: 10.1073/pnas.0914900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z., Luo R.T., Mi S. Consistent deregulation of gene expression between human and murine MLL rearrangement leukemias. Cancer Res. Feb 1 2009;69(3):1109–1116. doi: 10.1158/0008-5472.CAN-08-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong P., Iwasaki M., Somervaille T.C. The miR-17-92 microRNA polycistron regulates MLL leukemia stem cell potential by modulating p21 expression. Cancer Res. May 1 2010;70(9):3833–3842. doi: 10.1158/0008-5472.CAN-09-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandal C.C., Ghosh-Choudhury T., Dey N., Choudhury G.G., Ghosh-Choudhury N. miR-21 is targeted by omega-3 polyunsaturated fatty acid to regulate breast tumor CSF-1 expression. Carcinogenesis. Oct 2012;33(10):1897–1908. doi: 10.1093/carcin/bgs198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Acunzo M., Romano G., Palmieri D. Cross-talk between MET and EGFR in non-small cell lung cancer involves miR-27a and Sprouty2. Proc Natl Acad Sci USA. May 21, 2013;110(21):8573–8578. doi: 10.1073/pnas.1302107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moch H., Lukamowicz-Rajska M. miR-30c-2-3p and miR-30a-3p: new pieces of the jigsaw puzzle in HIF2α regulation. Cancer Discov. January 1, 2014;4(1):22–24. doi: 10.1158/2159-8290.CD-13-0897. [DOI] [PubMed] [Google Scholar]
- 44.Li Z., Lu J., Sun M. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci USA. Oct 7 2008;105(40):15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y., Yu Y., Tsuyada A. Transforming growth factor-beta regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene. Mar 24 2011;30(12):1470–1480. doi: 10.1038/onc.2010.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang B., Hsu S.H., Majumder S. TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene. Mar 25 2010;29(12):1787–1797. doi: 10.1038/onc.2009.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei Z., Cui L., Mei Z., Liu M., Zhang D. miR-181a mediates metabolic shift in colon cancer cells via the PTEN/AKT pathway. FEBS Lett. May 2 2014;588(9):1773–1779. doi: 10.1016/j.febslet.2014.03.037. [DOI] [PubMed] [Google Scholar]
- 48.Tsai K.W., Liao Y.L., Wu C.W. Aberrant expression of miR-196a in gastric cancers and correlation with recurrence. Genes Chromosomes Cancer. Apr 2012;51(4):394–401. doi: 10.1002/gcc.21924. [DOI] [PubMed] [Google Scholar]
- 49.Li Z., Huang H., Chen P. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat Commun. 2012;2:688. doi: 10.1038/ncomms1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou H., Xiao B., Zhou F. MiR-421 is a functional marker of circulating tumor cells in gastric cancer patients. Biomarkers: biochemical indicators of exposure, response, and susceptibility to chemicals. Mar 2012;17(2):104–110. doi: 10.3109/1354750X.2011.614961. [DOI] [PubMed] [Google Scholar]
- 51.Pillai R.S., Bhattacharyya S.N., Artus C.G. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. Sep 2 2005;309(5740):1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 52.Valencia-Sanchez M.A., Liu J., Hannon G.J., Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. Mar 1 2006;20(5):515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 53.Bagga S., Bracht J., Hunter S. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. Aug 26 2005;122(4):553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 54.Lim L.P., Lau N.C., Garrett-Engele P. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. Feb 17 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 55.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. Jan 23 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 56.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. Jan 23 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cullen B.R. Transcription and processing of human microRNA precursors. Mol Cell. Dec 22 2004;16(6):861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 58.Ambros V. The functions of animal microRNAs. Nature. Sep 16 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 59.Krutzfeldt J., Rajewsky N., Braich R. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. Dec 1 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 60.Jackson A.L., Bartz S.R., Schelter J. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. Jun 2003;21(6):635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 61.Sood P., Krek A., Zavolan M., Macino G., Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci USA. Feb 13 2006 doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He H., Jazdzewski K., Li W. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. Dec 27 2005;102(52):19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Farh K.K., Grimson A., Jan C. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. Dec 16 2005;310(5755):1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 64.Humphreys D.T., Westman B.J., Martin D.I., Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA. Nov 22 2005;102(47):16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kiriakidou M., Tan G.S., Lamprinaki S., De Planell-Saguer M., Nelson P.T., Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. Jun 15 2007;129(6):1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 66.Mathonnet G., Fabian M.R., Svitkin Y.V. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. Sep 21 2007;317(5845):1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 67.Thermann R., Hentze M.W. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. Jun 14 2007;447(7146):875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 68.Ding X.C., Grosshans H. Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. EMBO J. Feb 4 2009;28(3):213–222. doi: 10.1038/emboj.2008.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Petersen C.P., Bordeleau M.E., Pelletier J., Sharp P.A. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. Feb 17 2006;21(4):533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 70.Eulalio A., Huntzinger E., Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. Apr 2008;15(4):346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 71.Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. Jul 15 2006;20(14):1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. Sep 4 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Giraldez A.J., M Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. Apr 2006;312(5770):75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 74.Wu L., Fan J., Belasco J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA. Mar 14 2006;103(11):4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wakiyama M., Takimoto K., Ohara O., Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. Aug 1 2007;21(15):1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seggerson K., Tang L., Moss E.G. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol. Mar 15 2002;243(2):215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- 77.Maroney P.A., Yu Y., Fisher J., Nilsen T.W. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat Struct Mol Biol. Dec 2006;13(12):1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- 78.Siomi H., Siomi M.C. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. May 14 2010;38(3):323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 79.Pekarsky Y., S U., Cimmino A., Palamarchuk A., Efanov A., Maximov V., Volinia S., Alder H., Liu C.G., Rassenti L., Calin G.A., Hagan J.P., Kipps T., Croce C.M. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66(24):11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 80.Iliopoulos D., Jaeger S.A., Hirsch H.A., Bulyk M.L., Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. Aug 27 2010;39(4):493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mavrakis K.J., Van Der Meulen J., Wolfe A.L. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL) Nat Genet. Jul 2011;43(7):673–678. doi: 10.1038/ng.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mavrakis K.J., Leslie C.S., Wendel H.G. Cooperative control of tumor suppressor genes by a network of oncogenic microRNAs. Cell Cycle. Sep 1 2011;10(17):2845–2849. doi: 10.4161/cc.10.17.16959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Garzon R., Calin G.A., Croce C.M. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 84.Nana-Sinkam S.P., Croce C.M. Non-coding RNAs in cancer initiation and progression and as novel biomarkers. Mol Oncol. Dec 2011;5(6):483–491. doi: 10.1016/j.molonc.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Visone R., Veronese A., Balatti V., Croce C.M. MiR-181b: new perspective to evaluate disease progression in chronic lymphocytic leukemia. Oncotarget. Feb 2012;3(2):195–202. doi: 10.18632/oncotarget.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mu P., Han Y.C., Betel D. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. Dec 15 2009;23(24):2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Olive V., Bennett M.J., Walker J.C. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. Dec 15 2009;23(24):2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shi M., Guo N. MicroRNA expression and its implications for the diagnosis and therapeutic strategies of breast cancer. Cancer Treat Rev. Jan 24 2009 doi: 10.1016/j.ctrv.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 89.Hossain A., Kuo M.T., Saunders G.F. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. Nov 2006;26(21):8191–8201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu Z., Wang C., Wang M. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol. Aug 11 2008;182(3):509–517. doi: 10.1083/jcb.200801079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu Z., Willmarth N.E., Zhou J. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc Natl Acad Sci USA. May 4 2010;107(18):8231–8236. doi: 10.1073/pnas.1002080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guil S., Caceres J.F. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. Jul 2007;14(7):591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 93.Eiring A.M., Harb J.G., Neviani P. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. Mar 5 2010;140(5):652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Garofalo M., Romano G., Di Leva G. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med. Jan 2012;18(1):74–82. doi: 10.1038/nm.2577. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 95.Hu S., Dong T.S., Dalal S.R. The microbe-derived short chain fatty acid butyrate targets miRNA-dependent p21 gene expression in human colon cancer. PLoS One. 2011;6(1):e16221. doi: 10.1371/journal.pone.0016221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Humphreys K.J., Cobiac L., Le Leu R.K., Van der Hoek M.B., Michael M.Z. Histone deacetylase inhibition in colorectal cancer cells reveals competing roles for members of the oncogenic miR-17-92 cluster. Mol Carcinog. Jun 2013;52(6):459–474. doi: 10.1002/mc.21879. [DOI] [PubMed] [Google Scholar]
- 97.Watashi K., Yeung M.L., Starost M.F., Hosmane R.S., Jeang K.T. Identification of small molecules that suppress microRNA function and reverse tumorigenesis. J Biol Chem. Aug 6 2010;285(32):24707–24716. doi: 10.1074/jbc.M109.062976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cheng C.J., Slack F.J. The duality of oncomiR addiction in the maintenance and treatment of cancer. Cancer J. May–Jun 2012;18(3):232–237. doi: 10.1097/PPO.0b013e318258b75b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lennox K.A., Behlke M.A. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. Dec 2011;18(12):1111–1120. doi: 10.1038/gt.2011.100. [DOI] [PubMed] [Google Scholar]
- 100.Elmen J., Lindow M., Schutz S. LNA-mediated microRNA silencing in non-human primates. Nature. Apr 17 2008;452(7189):896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 101.Stenvang J., Silahtaroglu A.N., Lindow M., Elmen J., Kauppinen S. The utility of LNA in microRNA-based cancer diagnostics and therapeutics. Semin Cancer Biol. Apr 2008;18(2):89–102. doi: 10.1016/j.semcancer.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 102.Lu Y., Xiao J., Lin H. A single anti-microRNA antisense oligodeoxyribonucleotide (AMO) targeting multiple microRNAs offers an improved approach for microRNA interference. Nucleic Acids Res. Jan 9 2009 doi: 10.1093/nar/gkn1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wolfrum C., Shi S., Jayaprakash K.N. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. Oct 2007;25(10):1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 104.Stenvang J., Petri A., Lindow M., Obad S., Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3(1):1. doi: 10.1186/1758-907X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sheridan C. Gene therapy finds its niche. Nat Biotechnol. Feb 2011;29(2):121–128. doi: 10.1038/nbt.1769. [DOI] [PubMed] [Google Scholar]
- 106.Huang X., Schwind S., Yu B. Targeted delivery of microRNA-29b by transferrin-conjugated anionic lipopolyplex nanoparticles: a novel therapeutic strategy in acute myeloid leukemia. Clin Cancer Res. May 1 2013;19(9):2355–2367. doi: 10.1158/1078-0432.CCR-12-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thomas C.E., Ehrhardt A., Kay M.A. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. May 2003;4(5):346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 108.Haraguchi T., Ozaki Y., Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. Apr 2009;37(6):e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Greish K. Enhanced permeability and retention of macromolecular drugs in solid tumors: a royal gate for targeted anticancer nanomedicines. J Drug Target. Aug–Sep 2007;15(7–8):457–464. doi: 10.1080/10611860701539584. [DOI] [PubMed] [Google Scholar]
- 110.Su J., Baigude H., McCarroll J., Rana T.M. Silencing microRNA by interfering nanoparticles in mice. Nucleic Acids Res. Mar 2011;39(6):e38. doi: 10.1093/nar/gkq1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Babar I.A., Cheng C.J., Booth C.J. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci USA. June 26 2012;109(26):E1695–E1704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marcucci G., Radmacher M.D., Maharry K. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. May 1 2008;358(18):1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 113.Marcucci G., Maharry K., Radmacher M.D. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B Study. J Clin Oncol. Nov 1 2008;26(31):5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang C., Wang C., Chen X. Identification of seven serum microRNAs from a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int J Cancer. Jan 1 2013;132(1):116–127. doi: 10.1002/ijc.27657. [DOI] [PubMed] [Google Scholar]
- 115.Leung A.K., Sharp P.A. microRNAs: a safeguard against turmoil? Cell. Aug 24 2007;130(4):581–585. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 116.Vasudevan S., Tong Y., Steitz J.A. Switching from repression to activation: microRNAs can up-regulate translation. Science. Dec 21 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 117.Volinia S., Calin G.A., Liu C.G. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. Feb 14 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. Jun 13 2003;113(6):673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 119.McManus M.T., Sharp P.A. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet. Oct 2002;3(10):737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 120.Croce C.M. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. Oct 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fabbri M., Croce C.M. Role of microRNAs in lymphoid biology and disease. Curr Opin Hematol. Jul 2011;18(4):266–272. doi: 10.1097/MOH.0b013e3283476012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen X., Ba Y., Ma L. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. Oct 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 123.Gilad S., Meiri E., Yogev Y. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3(9):e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lawrie C.H., Gal S., Dunlop H.M. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. May 2008;141(5):672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 125.Mitchell P.S., Parkin R.K., Kroh E.M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. Jul 29 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang K., Zhang S., Marzolf B. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA. Mar 17 2009;106(11):4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lodes M.J., Caraballo M., Suciu D., Munro S., Kumar A., Anderson B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS One. 2009;4(7):e6229. doi: 10.1371/journal.pone.0006229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Brase J.C., Johannes M., Schlomm T. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. Feb 1 2011;128(3):608–616. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- 129.Tanaka M., Oikawa K., Takanashi M. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS One. 2009;4(5):e5532. doi: 10.1371/journal.pone.0005532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang J., Chen J., Chang P. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Philadelphia, Pa.) Sep 2009;2(9):807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ho A.S., Huang X., Cao H. Circulating miR-210 as a novel hypoxia marker in pancreatic cancer. Transl Oncol. Apr 2010;3(2):109–113. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kong X., Du Y., Wang G. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis. Dig Dis Sci. Feb 2011;56(2):602–609. doi: 10.1007/s10620-010-1285-3. [DOI] [PubMed] [Google Scholar]
- 133.Li A., Omura N., Hong S.M. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. Jul 1 2010;70(13):5226–5237. doi: 10.1158/0008-5472.CAN-09-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Resnick K.E., Alder H., Hagan J.P., Richardson D.L., Croce C.M., Cohn D.E. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. Jan 2009;112(1):55–59. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 135.Heneghan H.M., Miller N., Lowery A.J., Sweeney K.J., Kerin M.J. MicroRNAs as Novel Biomarkers for Breast Cancer. J Oncol. 2009;2009:950201. doi: 10.1155/2010/950201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhu W., Qin W., Atasoy U., Sauter E.R. Circulating microRNAs in breast cancer and healthy subjects. BMC Research Notes. 2009;2:89. doi: 10.1186/1756-0500-2-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Roth C., Rack B., Muller V., Janni W., Pantel K., Schwarzenbach H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12(6):R90. doi: 10.1186/bcr2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Asaga S., Kuo C., Nguyen T., Terpenning M., Giuliano A.E., Hoon D.S. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. Jan 2011;57(1):84–91. doi: 10.1373/clinchem.2010.151845. [DOI] [PubMed] [Google Scholar]
- 139.Ng E.K., Chong W.W., Jin H. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. Oct 2009;58(10):1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 140.Huang Z., Huang D., Ni S., Peng Z., Sheng W., Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. Jul 1 2010;127(1):118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 141.Tsujiura M., Ichikawa D., Komatsu S. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. Mar 30 2010;102(7):1174–1179. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu R., Zhang C., Hu Z. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. Mar 2011;47(5):784–791. doi: 10.1016/j.ejca.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 143.Zhang C., Wang C., Chen X. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem. Dec 2010;56(12):1871–1879. doi: 10.1373/clinchem.2010.147553. [DOI] [PubMed] [Google Scholar]
- 144.Komatsu S., Ichikawa D., Takeshita H. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. Jun 28 2011;105(1):104–111. doi: 10.1038/bjc.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hu Z., Chen X., Zhao Y. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. Apr 1 2010;28(10):1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 146.Foss K.M., Sima C., Ugolini D., Neri M., Allen K.E., Weiss G.J. miR-1254 and miR-574-5p: serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J Thorac Oncol: Official Publication of the International Association for the Study of Lung Cancer. Mar 2011;6(3):482–488. doi: 10.1097/JTO.0b013e318208c785. [DOI] [PubMed] [Google Scholar]
- 147.Liu X.G., Zhu W.Y., Huang Y.Y. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol. Jun 2012;29(2):618–626. doi: 10.1007/s12032-011-9923-y. [DOI] [PubMed] [Google Scholar]
- 148.Wei J., Gao W., Zhu C.J. Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chin J Cancer. Jun 2011;30(6):407–414. doi: 10.5732/cjc.010.10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Weiland M., Gao X.H., Zhou L., Mi Q.S. Small RNAs have a large impact: circulating microRNAs as biomarkers for human diseases. RNA Biol. Jun 2012;9(6):850–859. doi: 10.4161/rna.20378. [DOI] [PubMed] [Google Scholar]
- 150.Wulfken L.M., Moritz R., Ohlmann C. MicroRNAs in renal cell carcinoma: diagnostic implications of serum miR-1233 levels. PLoS One. 2011;6(9):e25787. doi: 10.1371/journal.pone.0025787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.van Schooneveld E., Wouters M.C., Van der Auwera I. Expression profiling of cancerous and normal breast tissues identifies microRNAs that are differentially expressed in serum from patients with (metastatic) breast cancer and healthy volunteers. Breast Cancer Res. 2012;14(1):R34. doi: 10.1186/bcr3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yang M., Chen J., Su F. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.LaConti J.J., Shivapurkar N., Preet A. Tissue and serum microRNAs in the Kras(G12D) transgenic animal model and in patients with pancreatic cancer. PLoS One. 2011;6(6):e20687. doi: 10.1371/journal.pone.0020687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wong C.M., Wong C.C., Lee J.M., Fan D.N., Au S.L., Ng I.O. Sequential alterations of microRNA expression in hepatocellular carcinoma development and venous metastasis. Hepatology. May 2012;55(5):1453–1461. doi: 10.1002/hep.25512. [DOI] [PubMed] [Google Scholar]
- 155.Kim D.J., Linnstaedt S., Palma J. Plasma components affect accuracy of circulating cancer-related microRNA quantitation. J Mol Diagn. Jan 2012;14(1):71–80. doi: 10.1016/j.jmoldx.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang T., Lv M., Shen S. Cell-free microRNA expression profiles in malignant effusion associated with patient survival in non-small cell lung cancer. PLoS One. 2012;7(8):e43268. doi: 10.1371/journal.pone.0043268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gao W., Li J.Z., Ho W.K., Chan J.Y., Wong T.S. Biomarkers for use in monitoring responses of nasopharyngeal carcinoma cells to ionizing radiation. Sensors (Basel, Switzerland) 2012;12(7):8832–8846. doi: 10.3390/s120708832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Akiyoshi S., Fukagawa T., Ueo H. Clinical significance of miR-144-ZFX axis in disseminated tumour cells in bone marrow in gastric cancer cases. Br J Cancer. Oct 9 2012;107(8):1345–1353. doi: 10.1038/bjc.2012.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jones C.I., Zabolotskaya M.V., King A.J. Identification of circulating microRNAs as diagnostic biomarkers for use in multiple myeloma. Br J Cancer. Dec 4 2012;107(12):1987–1996. doi: 10.1038/bjc.2012.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang Z., Han J., Cui Y., Fan K., Zhou X. Circulating microRNA-21 as noninvasive predictive biomarker for response in cancer immunotherapy. Med Hypotheses. Jul 2013;81(1):41–43. doi: 10.1016/j.mehy.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 161.Wong K.F., Xu Z., Chen J., Lee N.P., Luk J.M. Circulating markers for prognosis of hepatocellular carcinoma. Expert Opin Med Diagn. Jul 2013;7(4):319–329. doi: 10.1517/17530059.2013.795146. [DOI] [PubMed] [Google Scholar]
- 162.Wong T.S., Liu X.B., Wong B.Y., Ng R.W., Yuen A.P., Wei W.I. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clin Cancer Res. May 1 2008;14(9):2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 163.Lin S.C., Liu C.J., Lin J.A., Chiang W.F., Hung P.S., Chang K.W. miR-24 up-regulation in oral carcinoma: positive association from clinical and in vitro analysis. Oral Oncol. Mar 2010;46(3):204–208. doi: 10.1016/j.oraloncology.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 164.Liu C.J., Kao S.Y., Tu H.F., Tsai M.M., Chang K.W., Lin S.C. Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral Dis. May 2010;16(4):360–364. doi: 10.1111/j.1601-0825.2009.01646.x. [DOI] [PubMed] [Google Scholar]
- 165.Fabbri M., Paone A., Calore F. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. Jul 31 2012;109(31):E2110–2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lund J.M., Alexopoulou L., Sato A. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci USA. Apr 13 2004;101(15):5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Heil F., Hemmi H., Hochrein H. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. Mar 5 2004;303(5663):1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 168.Fabbri M. TLRs as miRNA receptors. Cancer Res. Dec 15 2012;72(24):6333–6337. doi: 10.1158/0008-5472.CAN-12-3229. [DOI] [PubMed] [Google Scholar]
- 169.Fabbri M., Paone A., Calore F., Galli R., Croce C.M. A new role for microRNAs, as ligands of Toll-like receptors. RNA Biol. Feb 2013;10(2):169–174. doi: 10.4161/rna.23144. [DOI] [PMC free article] [PubMed] [Google Scholar]