Abstract
Rubella virus causes a relatively benign disease in most cases, although infection during pregnancy can result in serious birth defects. An effective vaccine has been available since the early 1970s and outbreaks typically do not occur among highly vaccinated (≥2 doses) populations. Nevertheless, considerable inter-individual variation in immune response to rubella immunization does exist, with single dose seroconversion rates ~95%. Understanding the mechanisms behind this variability may provide important insights into rubella immunity. In the current study, we examined associations between single nucleotide polymorphisms (SNPs) in selected cytokine, cytokine receptor, and innate/antiviral genes and immune responses following rubella vaccination in order to understand genetic influences on vaccine response. Our approach consisted of a discovery cohort of 887 subjects ages 11–22 at the time of enrollment and a replication cohort of 542 older adolescents and young adults (ages 18–40). Our data indicate that SNPs near the butyrophilin genes (BTN3A3/BTN2A1) and cytokine receptors (IL10RB/IFNAR1) are associated with variations in IFNγ secretion and that multiple SNPs in the PVR gene, as well as SNPs located in the ADAR gene, exhibit significant associations with rubella virus-specific IL-6 secretion. This information may be useful, not only in furthering our understanding immune responses to rubella vaccine, but also in identifying key pathways for targeted adjuvant use to boost immunity in those with weak or absent immunity following vaccination.
Keywords: Polymorphism, Genetic, Polymorphism, Single Nucleotide, Cytokines, Receptor, Cytokine, Immunity, Cellular, Measles-Mumps-Rubella Vaccine, MMR
Introduction
Rubella virus, a member of the Togaviridae family, typically causes a relatively benign illness, although arthritis and arthralgia have been observed in adults. Less frequent complications include both encephalitis and thrombocytopenia.(Reef and Plotkin 2013) Infection during pregnancy, especially early in pregnancy, can cause congenital rubella syndrome (CRS), leading to devastating fetal abnormalities. Since the advent of rubella vaccines and their widespread use in the 1970s, the number of cases of both rubella and CRS has dropped dramatically. Nevertheless, rubella remains a significant public health concern due to inadequate vaccine coverage(Metcalf et al. 2012), which leads to yearly outbreaks including a current nationwide outbreak in Japan with over 5,000 cases in 2013.
In the U.S., rubella vaccines are given as part of multi-component formulations that also include measles and mumps (MMR) or measles, mumps, and varicella (MMRV).(Jacobsen et al. 2009; Klein et al. 2010; Marin et al. 2010) These vaccines are highly effective, eliciting protective immunity in >95% of recipients after a single dose, and in >99% of recipients after two doses.(Reef and Plotkin 2013; Vesikari et al. 2012) Both humoral and cellular immune responses are elicited following immunization with rubella-containing vaccines and both contribute to protection against subsequent infection.(Dhiman et al. 2010b; Honeyman et al. 1974; Mitchell et al. 1999; Reef and Plotkin 2013; Vesikari and Buimovici-Klein 1975) At the population level, there is considerable inter-individual variation in immune responses to vaccines, including those containing rubella. We have previously identified a number of genetic polymorphisms associated with variations in rubella vaccine response.(Dhiman et al. 2010a; Haralambieva et al. 2010; Kennedy et al. 2010; Ovsyannikova et al. 2010a; Ovsyannikova et al. 2010b; Ovsyannikova et al. 2004, 2005; Ovsyannikova et al. 2009a; Ovsyannikova et al. 2009b; Ovsyannikova et al. 2010c)
Here we report a meta-analysis combining data from two independent cohorts in order to identify genetic polymorphisms associated with rubella virus-specific cellular immune responses.
Methods
Subject Recruitment and Demographics
The study set was a large sample of 1,429 healthy children, older adolescents, and healthy adults (age 11 to 40 years), consisting of two independent cohorts: a Rochester cohort and a San Diego cohort. The methods described herein are similar or identical to those published for our previous studies.(Dhiman et al. 2010a; Haralambieva et al. 2010; Kennedy et al. 2010; Ovsyannikova et al. 2010a; Ovsyannikova et al. 2010b; Ovsyannikova et al. 2004, 2005; Ovsyannikova et al. 2009a; Ovsyannikova et al. 2009b; Ovsyannikova et al. 2010c) Similarly, clinical and demographic characteristics of these cohorts have been previously reported. This study reports our findings among the Caucasian subset of each cohort.
The Rochester cohort comprised 1,145 individuals enrolled into three age-stratified cohorts of healthy, school-age children and young adults from all socioeconomic strata in Rochester, MN. Specifically, between December 2001 and August 2002, we enrolled 346 healthy children (age 12 to 18 years, cohort #1).(Ovsyannikova et al. 2004, 2005) Between December 2006-August 2007, we enrolled 440 healthy children (age 11 to 18 years, cohort #2), as previously published.(Haralambieva et al. 2010; Ovsyannikova et al. 2010a) In November 2008-September 2009, we enrolled 388 healthy children, enriched with African-American youth (age 11 to 22 years, cohort #3).(Ovsyannikova et al. 2011a; Ovsyannikova et al. 2012b) Parental consent was obtained for all participants and each subject had written records of having received two doses of measles-mumps-rubella (MMR, Merck) vaccine. Of the 1,145 individuals recruited into these three studies, 1,039 (90.7%) had sufficient samples and were successfully assayed for the measures of immune response. This cohort was predominantly of Caucasian descent, and after removing non-Caucasian individuals, the analysis set comprised 887 (85.4% of 1,039) participants.
In July 2005-September 2006, we enrolled an additional 1,076 healthy older adolescents and healthy adults (age 18 to 40 years) from armed forces personnel in San Diego, CA.(Kennedy et al. 2009; Ovsyannikova et al. 2011b) As members of the U.S. military, they represent a cross section of the U.S. population with proven vaccine-induced immunity to MMR. Of the 1,076 subjects recruited into this study, 997 (92.7%) subjects had provided consent for use of their samples and data in other studies, and had sufficient samples which were successfully assayed for rubella neutralizing antibody levels. Of those subjects, 542 (54.4%) were of Caucasian descent and were included in this study. The Institutional Review Boards of the Mayo Clinic and the Naval Health Research Center (NHRC) approved the study, and written informed consent was obtained from each subject, from the parents of all children who participated in the study, as well as written assent from age-appropriate participants.
Rubella virus-specific cytokine secretion
The levels of secreted cytokines following stimulation of PBMCs with live rubella virus were measured, as previously described by our group.(Dhiman et al. 2010b; Ovsyannikova et al. 2009b) Briefly, 2 × 105 PBMCs were stimulated with the W-Therien strain of rubella virus (a gift from Dr. Teryl Frey, Georgia State University, Atlanta, GA) with optimized MOI and incubation times depending on the specific cytokine measured. For the measurement of IL-2, IL-6, and IFN-γ, PBMCs were stimulated with an MOI of 5. The levels of secreted TNF-α were determined after viral stimulation with an MOI of 0.05. The supernatants were removed post stimulation at 24 hrs. for IL-6, 48 hrs. IFN-γ, eight days for IL-2 and TNF-α, and all samples were stored at −80 °C until assayed. Cytokine levels were quantified using BD OptEIA™ Human ELISA kits, and absorbance levels were measured using a Molecular Devices SpectraMax 340PC.
SNP selection and candidate SNP genotyping methods
The SNP selection and genotyping methods described herein are similar or identical to those published for our previous genetic association studies.(Haralambieva et al. 2010; Ovsyannikova et al. 2010a; Ovsyannikova et al. 2010b) The replication effort included follow-up on 299 previously identified (Rochester cohort) genetic associations (p<0.05) between SNPs in immune response genes (cytokine and cytokine receptor genes, Toll-like receptor genes, vitamin D and vitamin A receptor family genes, antiviral effector and other innate genes) and immune measures after rubella vaccination. In addition, another 571 candidate tagSNPs selected from eight newly discovered viral receptor and attachment factor genes (MOG, PVR, PVRL1, PVRL2, PVRL3, PVRL4, BTN2A1, BTN3A1) were identified using the tagSNP selection approach(Carlson et al. 2004) based on linkage disequilibrium (LD) and included on the genotyping panel. Genotypes for 102 of these SNPs were already available for the replication (San Diego) cohort from a previously performed genome-wide SNP genotyping using the Illumina Infinium HumanHap550 or HumanHap650Y BeadChip arrays. All previously discovered significant SNPs not present on the Illumina 550/650 arrays (n=197 SNPs) were genotyped using a custom Illumina GoldenGate 768-plex genotyping assay. The selection criteria included: SNPs with validation data; successful predictive genotyping scores for Illumina GoldenGate assays; a minor allele frequency (MAF) ≥0.05; and a pairwise linkage disequilibrium (LD) threshold of r2 ≥ 0.90. SNP-specific deviation from Hardy-Weinberg Equilibrium (HWE) was tested and we excluded any SNP that displayed violations of HWE (p<0.001). The SNP list was further post-processed and refined using the SNPPicker program (Sicotte et al. 2011) in order to accommodate a set of Illumina platform constraints and to pick tagSNPs optimally across the multiple population groups. A total of 768 SNPs (including 197 replication SNPs not previously genotyped in the San Diego cohort, and 571 SNPs selected from eight new candidate genes) were selected based on this approach.
Our genotyping methods have been previously described.(Dhiman et al. 2007) Genomic DNA was extracted from blood using the Puregene extraction kit (Gentra Systems Inc., Minneapolis, MN). DNA was quantified using the Picogreen method (Molecular Probes). The 768 SNPs were genotyped using a custom-designed 768-plex Illumina GoldenGate™ assay (Illumina Inc., San Diego, CA) following the manufacturer’s instructions. The BeadStudio 2 software was used to call genotypes. Corriel Trio DNA samples (Mother: 1347-02 NA11875, Father: 1347-01 NA10859, Daughter: 1347-08 NA11875) and two other genomic DNA controls were used for quality control and to test for genotyping reproducibility. All data were transferred electronically to SAS for further analysis. The genotyping quality using the Illumina GoldenGate assay was high, with a total genotyping success rate >99.7% in both cohorts and 96–100% reproducibility rate for controls and replicate samples. For the Caucasian-only analyses, we used data on 555 SNPs from 887 Rochester subjects and 565 SNPs from 542 San Diego subjects.
Statistical Analyses
Subject demographics were summarized separately for the combined Rochester cohorts and the San Diego cohort. Summaries comprised counts and percentages for categorical variables and medians and 25th and 75th percentiles for continuous variables. A principal components analysis was performed to confirm racial groupings using data available from prior genome-wide analyses. Because non-Caucasian samples comprised few individuals, and genetic and phenotypic differences among racial groups can bias association studies, we focused on the Caucasian participants in this analysis.
Linear mixed effects models were used to assess associations between the various cellular immune response measures and SNP genotypes within the Rochester and San Diego cohorts. Our assessments of the cellular immunity measures were measured in multiple wells, some with and some without stimulation with rubella virus. We included the results from each assay well in our analyses, and accounted for within-subject effects by modeling an unstructured covariance matrix within the linear mixed effects regression models. In these analyses, we assumed an additive genetic model and tested for differential SNP associations on the stimulation effect through a per-SNP genotype variable representing the number of minor alleles carried. In order to ensure that the cellular immune response measures conformed to linear models assumptions, an inverse normal transformation was applied to each phenotype prior to analysis. Individual SNP associations were tested for significance after adjustment for potential differences due to gender, age at enrollment, immunization age, and time from last immunization to enrollment. After testing for associations between SNPs and phenotypes within the two cohorts, a fixed effects meta-analysis was performed for each SNP and for each phenotype.(S and K 2008) Pooled estimates were obtained and tested for significance, and tests for heterogeneity of effect between cohorts were obtained. Q-values, which estimate the probability that a p-value reflects a false positive finding, were computed separately for the p-values from the different phenotypes using the methods of Storey et al. (Storey 2002, 2003) for SNPs that did not display evidence of significant heterogeneity between cohorts (p>0.10). SNPs with meta-analysis p-values below 0.01 were considered to be of potential interest, and SNPs with q-values below 0.1 were considered to be significantly associated with the corresponding phenotype.
Results
Because of the different mix of races in each cohort, and the effect of race on immune outcome, we confined this report to the Caucasian subset of each cohort (Table 1), as this was the largest racial subset available for each cohort. We initially tested secretion of four cytokines (IL-2, IL-6, IFNγ, TNFα) in response to in vitro rubella virus stimulation in our immunized subjects. Both IL-2 and TNFα secretion were largely undetectable in San Diego subjects and therefore were not included in this report. The Rochester cohort (n=887) had a median IL-6 response of 3,629.3 pg/ml (IQR= 3,083.5 – 4,002.4) and the San Diego cohort (n=542) had a median IL-6 response of 4,128.9 pg/ml (IQR = 3,514.5 – 4,816.3). This difference in IL-6 secretion was significant (p<0.0001). IFNγ secretion had a median value of 6.3 pg/ml (IQR= 1.7 – 19.7) in the Rochester cohort and a median value of −1.8 pg/ml (IQR= −6.4 – −2.9) in the San Diego cohort. These negative values likely indicate that rubella virus exerts a suppressive effect on basal IFNg levels in most of the San Diego cohort subjects. The differential IFNγ response was also significant (p< 0.0001).
Table 1.
Subject Demographics
| Rochester Cohort (n=887) | San Diego Cohort (n=542) | |
|---|---|---|
|
| ||
| Median age in years at enrollment | 15 (13 – 17) | 23 (22 – 27) |
|
| ||
| Median age in years at most recent vaccination | 10 (5 – 12) | 19 (18 – 21) |
|
| ||
| Years since most recent vaccination | 6.4 (4.6 – 8.5) | 3.0 (2.2 – 3.9) |
|
| ||
| Gender | ||
|
Male Female |
487 (55%) 400 (45%) |
394 (73%) 148 (27%) |
Individually, the two cohorts are fairly small for genetic association studies and may have lacked sufficient power to detect the small effects typically expected from individual SNPs. This is readily apparent in the San Diego cohort p-values (Tables 2 and 3). With this in mind, we performed a meta-analysis using data from both cohorts. The genotyping panel contained SNPs located within or near a variety of cytokine and cytokine receptor genes, as well as innate and antiviral genes (Supplemental Table 1). We identified a number of SNPs that are potentially associated with variations in IFNγ secretion levels. While not meeting thresholds set to control false discovery, the most significant SNPs (p< 0.01) are shown in Table 2. These included: two intergenic SNPs located between the IL10RB and IFNAR1 genes; a SNP in TOP2A, encoding a DNA topoisomerase; and another intergenic SNP located between two butyrophilin genes, BTN3A3 and BTN2A1.
Table 2.
SNPs associated with variations in IFNγ response to Rubella
| Rochester Cohort | San Diego Cohort | Meta | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP IDa | Chrb | Genec | Location | MAFd | Genotype | Ne | Median (IQR)f |
Cohort p-value |
MAFd | Genotype | Ne | Median (IQR)f |
Cohort p-value |
p-valueg | q-valueh |
| rs471692 | 17 | TOP2A | Intron | 20.18 | GG | 521 | 6.6 (1.9,20.8) | 0.150 | 19.13 | GG | 309 | −1.3 (−5.7,3.2) | 0.002 | 0.0007 | 0.237 |
| GA | 277 | 6.0 (1.5,18.2) | GA | 147 | −2.7 (−7.4,1.2) | ||||||||||
| AA | 31 | 5.0 (1.1,13.1) | AA | 17 | −1.7 (−7.1,1.4) | ||||||||||
|
| |||||||||||||||
| rs7278931 | 21 | IL10RB/IFNAR1 | Intergenic | 17.45 | GG | 555 | 5.9 (1.6,17.6) | 0.011 | 18.50 | GG | 310 | −1.9 (−6.3,2) | 0.040 | 0.0018 | 0.315 |
| GA | 231 | 6.6 (2,21.7) | GA | 143 | −2 (−6.5,3.2) | ||||||||||
| AA | 24 | 8.8 (2.9,35.9) | AA | 15 | 2.6 (−10.5,4.2) | ||||||||||
|
| |||||||||||||||
| rs8134731 | 21 | IL10RB/IFNAR1 | Intergenic | 18.15 | AA | 561 | 6 (1.6,18) | 0.053 | 18.82 | AA | 312 | −1.9 (−6.3,2) | 0.053 | 0.0069 | 0.650 |
| AG | 238 | 7.1 (2,22.1) | AG | 147 | −2.0 (−6.5,3.3) | ||||||||||
| GG | 30 | 8.8 (2.9,33.6) | GG | 15 | 2.6 (−10.5,4.2) | ||||||||||
|
| |||||||||||||||
| rs10484440 | 6 | BTN3A3/BTN2A1 | Intergenic | 13.00 | AA | 614 | 6.6 (1.7,21.8) | 0.071 | 13.03 | AA | 357 | −2 (−6.5,2.8) | 0.055 | 0.0091 | 0.650 |
| AG | 202 | 5.9 (1.9,16.6) | AG | 111 | −1.7 (−6.6,2.5) | ||||||||||
| GG | 7 | 0.0 (−0.6,5.3) | GG | 5 | −2.5 (−4.4, −.8) | ||||||||||
rs SNP identification number,
Chromosomal location,
Gene or genetic region containing the indicated SNP,
Minor Allele Frequency
Number of subjects with a given genotype,
Median outcome measurement for each genotype group. Results expressed as pg/ml. The interquartile range (IQR) is shown in parentheses,
P-values were adjusted for demographic and clinical variables as well as inflation of significance described in the Methods section,
Q-values computed using the methods of Storey et al from the distribution of p-values from SNPs whose heterogeneity tests did not reach significance.
Table 3.
SNPs associated with variations in IL-6 response to Rubella.
| Rochester Cohort | San Diego Cohort | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP IDa | Chrb | Genec Location |
MAFd | Genotype | Ne | Median (IQR)f |
Cohort p-value |
MAFd | Genotype | Ne | Median (IQR)f |
Cohort p-value |
Meta p-valueg |
q-valueh |
| rs474247 | 1 | TNFRSF1B | 19.5 | GG | 539 | 3,615 (3,099–4,041) | 4.2 × 10–6 | 23.1 | GG | 278 | 4,152 (3,539–4,909) | 0.2771 | 4.9 × 10–6 | 0.001 |
| Intron | GA | 258 | 3,637 (3,120–3,969) | GA | 167 | 4,050 (3,364–4,674) | ||||||||
| AA | 31 | 3,694 (3,025–3,985) | AA | 28 | 4,096 (3,411–4,715) | |||||||||
|
| ||||||||||||||
| rs9393725 | 6 | BTN1A1/BTN2A1 | 1.6 | AA | 803 | 3,634 (3,097–4,008) | 0.0008 | 1.5 | AA | 459 | 4,141 (3,491–4,820) | 0.0806 | 0.00014 | 0.014 |
| Intergenic | AC | 26 | 3,469 (2,997–3,797) | AC | 14 | 3,829 (3,559–4,702) | ||||||||
| CC | 0 | (,) | CC | 0 | (−) | |||||||||
|
| ||||||||||||||
| rs7260482 | 19 | PVR/LOC147710 | 25.8 | AA | 452 | 3,665 (3,131–3,997) | 0.0004 | 28.8 | AA | 240 | 4,130 (3,503–4,824) | 0.1717 | 0.00017 | 0.014 |
| Intergenic | AC | 313 | 3,547 (3,025–4,052) | AC | 195 | 4,181 (3,548–4,876) | ||||||||
| CC | 55 | 3,458 (3,099–3,881) | CC | 35 | 3,698 (3,175–4,282) | |||||||||
|
| ||||||||||||||
| rs2229857 | 1 | ADAR | 28.2 | GG | 442 | 3,617 (3,081–4,003) | 0.0003 | 30.4 | GG | 235 | 4,031 (3,448–4,783) | 0.0003 | 0.00026 | 0.016 |
| Coding | GA | 308 | 3,634 (3,120–4,027) | GA | 189 | 4,267 (3,533–4,914) | ||||||||
| AA | 78 | 3,744 (3,099–3,975) | AA | 49 | 4,076 (3,541–4,611) | |||||||||
|
| ||||||||||||||
| rs9616 | 1 | ADAR | 28.3 | TT | 429 | 3,654 (3,135–4,034) | 0.0001 | 28.6 | TT | 245 | 4,198 (3,574–4,828) | 0.0003 | 0.00038 | 0.019 |
| UTR | TA | 326 | 3,605 (3,065–3,964) | TA | 185 | 4,002 (3,376–4,803) | ||||||||
| AA | 69 | 3,642 (2,981–3,978) | AA | 43 | 4,127 (3,530–4,880) | |||||||||
|
| ||||||||||||||
| rs10410651 | 19 | PVR | 25.3 | AA | 465 | 3,665 (3,127–3,999) | 0.0015 | 28.8 | AA | 241 | 4,133 (3,509–4,828) | 0.1467 | 0.00046 | 0.019 |
| Intron | AG | 307 | 3,561 (3,024–4,063) | AG | 197 | 4,174 (3,548–4,847) | ||||||||
| GG | 56 | 3,458 (3,105–3,874) | GG | 35 | 3,698 (3,175–4,282) | |||||||||
|
| ||||||||||||||
| rs203709 | 19 | PVR | 25.5 | AA | 464 | 3,663 (3,127–3,997) | 0.002 | 28.9 | AA | 240 | 4,130 (3,503–4,860) | 0.1405 | 0.00059 | 0.021 |
| Intron | AT | 307 | 3,554 (3,010–4,063) | AT | 198 | 4,178 (3,548–4,847) | ||||||||
| TT | 57 | 3,458 (3,110–3,881) | TT | 35 | 3,698 (3,175–4,282) | |||||||||
|
| ||||||||||||||
| rs7250339 | 19 | PVR/LOC147710 | 24.1 | AA | 470 | 3,650 (3,130–3,994) | 0.0058 | 26.2 | AA | 262 | 4,164 (3,533–4,905) | 0.0531 | 0.0008 | 0.025 |
| Intergenic | AG | 300 | 3,572 (3,044–4,047) | AG | 183 | 4,151 (3,503–4,813) | ||||||||
| GG | 47 | 3,396 (3,058–3,900) | GG | 28 | 3,633 (3,152–4,259) | |||||||||
|
| ||||||||||||||
| rs1466435 | 19 | BCAM/PVRL2 | 5.2 | CC | 748 | 3,633 (3,092–4,006) | 0.004 | 3.2 | CC | 441 | 4,117 (3,497–4,787) | 0.1661 | 0.0014 | 0.037 |
| Intergenic | CG | 76 | 3,620 (3,124–3,989) | CG | 32 | 4,299 (3,541–5,051) | ||||||||
| GG | 5 | 3,353 (3,077–3,834) | GG | 0 | (-) | |||||||||
|
| ||||||||||||||
| rs28483039 | 19 | PVR | 23.8 | GG | 482 | 3,658 (3,127–3,999) | 0.0103 | 26.2 | GG | 262 | 4,164 (3,533–4,905) | 0.0531 | 0.0015 | 0.037 |
| Intron | GA | 299 | 3,576 (3,025–4,061) | GA | 183 | 4,151 (3,503–4,813) | ||||||||
| AA | 48 | 3,364 (3,063–3,891) | AA | 28 | 3,633 (3,151–4,259) | |||||||||
|
| ||||||||||||||
| rs1127317 | 1 | ADAR | 27.9 | AA | 446 | 3,625 (3,084–4,021) | 0.0021 | 29.8 | AA | 238 | 4,041 (3,448–4,711) | 0.3959 | 0.0018 | 0.039 |
| UTR | AC | 306 | 3,634 (3,125–4,013) | AC | 187 | 4,276 (3,533–4,914) | ||||||||
| CC | 77 | 3,694 (3,024–3,975) | CC | 47 | 4,076 (3,541–4,611) | |||||||||
|
| ||||||||||||||
| rs7104562 | 11 | PVRL1 | 2.5 | AA | 788 | 3,631 (3,079–4,004) | 0.0006 | 2.3 | AA | 452 | 4,129 (3,500–4,810) | 0.9782 | 0.0019 | 0.039 |
| Intron | AG | 41 | 3,637 (3,247–3,963) | AG | 21 | 4,155 (3,418–5,043) | ||||||||
| GG | 0 | (,) | GG | 0 | (-) | |||||||||
|
| ||||||||||||||
| rs2770150 | 9 | ASTN2/TLR4 | 27.7 | AA | 427 | 3,634 (3,135–4,001) | 0.0094 | 24.8 | AA | 270 | 4,191 (3,450–4,830) | 0.1888 | 0.0035 | 0.069 |
| Intergenic | AG | 342 | 3,625 (3,039–4,004) | AG | 167 | 4,076 (3,584–4,771) | ||||||||
| GG | 59 | 3,640 (3,024–4,121) | GG | 36 | 3,859 (3,226–4,801) | |||||||||
|
| ||||||||||||||
| rs2228145 | 1 | IL6R | 37.5 | AA | 323 | 3,611 (3,120–3,999) | 0.0069 | 39.8 | AA | 178 | 4,159 (3,574–4,847) | 0.3192 | 0.0043 | 0.077 |
| Coding | AC | 394 | 3,629 (3,066–4,023) | AC | 211 | 4,076 (3,460–4,888) | ||||||||
| CC | 111 | 3,670 (3,065–3,992) | CC | 84 | 4,006 (3,366–4,676) | |||||||||
|
| ||||||||||||||
| rs3852861 | 19 | PVRL2 | 39.4 | CC | 295 | 3,681 (3,245–4,030) | 0.0043 | 40.7 | CC | 167 | 3,980 (3,435–4,828) | 0.602 | 0.0053 | 0.090 |
| Intron | CA | 408 | 3,554 (3,038–4,003) | CA | 230 | 4,209 (3,539–4,820) | ||||||||
| AA | 126 | 3,594 (3,127–3,987) | AA | 75 | 4,155 (3,455–4,683) | |||||||||
|
| ||||||||||||||
| rs962859 | 21 | IL10RB | 42.7 | AA | 257 | 3,683 (3,183–4,052) | 0.0023 | 42.9 | AA | 165 | 4,042 (3,425–4,720) | 0.8688 | 0.0063 | 0.100 |
| Intron | AC | 424 | 3,611 (3,048–4,002) | AC | 200 | 4,168 (3,539–4,927) | ||||||||
| CC | 135 | 3,540 (3,090.5–3,889) | CC | 106 | 4,154 (3,522–4,683) | |||||||||
|
| ||||||||||||||
| rs12416901 | 11 | PVRL1 | 2.8 | AA | 785 | 3,631 (3,081–4,004) | 0.0063 | 2.2 | AA | 453 | 4,127 (3,497–4,807) | 0.8519 | 0.01 | 0.146 |
| Intron | AG | 44 | 3,574 (3,221–3,932) | AG | 20 | 4,258 (3,476–5,084) | ||||||||
| GG | 0 | (,) | GG | 0 | (-) | |||||||||
rs SNP identification number,
Chromosomal location,
Gene or genetic region containing the indicated SNP,
Minor Allele Frequency
Number of subjects with a given genotype,
Median outcome measurement for each genotype group. Results expressed as pg/ml. The interquartile range (IQR) is shown in parentheses,
P-values were adjusted for demographic and clinical variables as well as inflation of significance described in the Methods section.
Q-values computed using the methods of Storey et al from the distribution of p-values from SNPs whose heterogeneity tests did not reach significance.
Furthermore, we identified multiple significant SNP:IL-6 response associations. The most significant SNP associations (p< 0.01) are shown in Table 3. All but one of these met our false-positive threshold (q-value<0.10). These SNPs are located in the following genes: ADAR, encoding the RNA-specific adenosine deaminase; IL10RB; IL6R; PVR, the poliovirus receptor; PVRL1 and PVRL2, the poliovirus receptor-related proteins that also mediate herpesvirus entry; and TNFRSF1B, encoding the p75 TNFα receptor. The list also includes: five SNPs located in or near the PVR gene (Figure 1), which encodes the poliovirus receptor; an intergenic SNP located in between BCAM, the basal cell adhesion molecule gene and PVRL2; as well as a single SNP near the TLR4 and astrotactin 2 genes.
Figure 1. Locus-zoom plot of selected region of Chromosome 19.
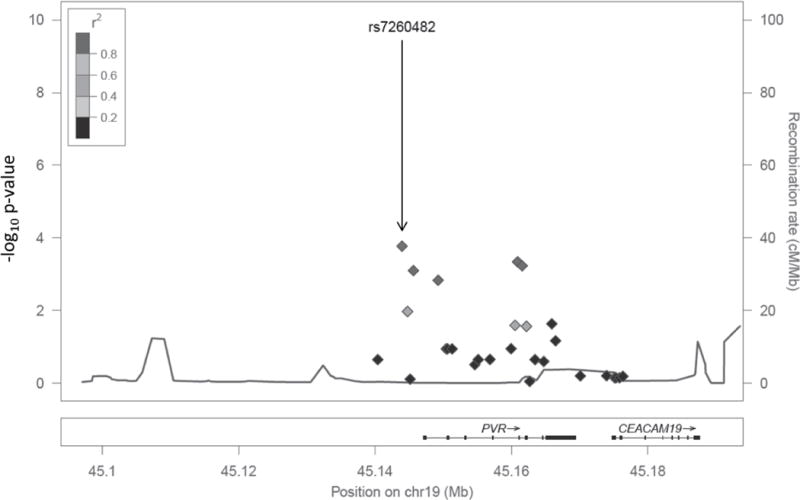
A genetic region encompassing the PVR and CEACAM19 genes contained multiple SNPs significantly associated with rubella-specific IL-6 secretion following vaccination (Table 3). P-value is depicted on the left Y-axis. Each SNP is marked by the solid diamonds and LD (with respect to rs72604S2) is indicated by the grey color of the diamond (There are no SNPs with r2 between 0.6 – 0.8 or between 0.2 – 0.4). The recombination rate is mapped as the grey line at the bottom of the plot and on the right Y-axis. Underneath the plot each gene and its chromosomal position is indicated. Thick segments of each gene indicate exons, while the arrows indicate the direction of transcription.
Discussion
Immune responses to antigenic stimuli, either infection or vaccination, are influenced, in part, by host genetics. Our work has focused on the role of genetic polymorphisms on immune responses to viral vaccines. We have previously identified a number of genetic variants (mainly HLA alleles and SNPs) associated with variations in cellular and/or humoral immunity to rubella vaccine. (Dhiman et al. 2010a; Dhiman et al. 2008; Haralambieva et al. 2010; Jacobson et al. 2009; Kennedy et al. 2010; Ovsyannikova et al. 2010b; Ovsyannikova et al. 2004, 2005; Ovsyannikova et al. 2009a; Ovsyannikova et al. 2006; Ovsyannikova et al. 2009b; Ovsyannikova et al. 2010c; Pankratz et al. 2010) Our current study design incorporates several important elements: the use of SNP data to define race/ethnicity, and the inclusion of two separate cohorts for SNP discovery and replication. Our study also has several limitations that include: the chance for rubella exposure and/or disease in our San Diego cohort; relatively small cohort sizes (especially when compared to cancer gene association studies with tens of thousands of subjects); and response outcomes that reflect complex immunologic processes controlled by multiple genes and pathways, where individual SNPs have minor contributions to the spectrum of immune response. These limitations are especially evident in the San Diego cohort-specific data, as demonstrated by the larger p-values observed for both the IFNγ and IL-6 associations. Potential confounding variables were the notable differences in age, time since vaccination, and gender composition between our two cohorts, and these were accounted for in our statistical analyses. In spite of these limitations, our meta-analysis did identify a number of statistically significant genetic associations across two independent cohorts.
We identified four SNPs significantly associated with variations in IFNγ response in both of our cohorts. Three of the SNPs are intergenic and one is intronic; it is possible that they are not the causal variant, but are in close LD to the causal variant. However, most of the variations found with each genotype are extremely small (1–2 pg/ml) and the majority of the San Diego cohort had minimal to negative values (indicating that in vitro stimulation with rubella virus suppressed IFNγ secretion), making the biological relevance unclear. The one exception is rs10484440 where, in the Rochester cohort, there was no IFNγ response (median IFNγ = 0 pg/ml) for individuals homozygous for the G allele. Homozygous A and heterozygous individuals had median IFNγ secretion levels of ~ 6pg/ml. Two of the SNPs were located near the IL10RB gene and genetic polymorphisms in this region have also been associated with chronic hepatitis B infection,(Romporn et al. 2013) cytokine responses to smallpox vaccine,(Ovsyannikova et al. 2012a) HIV infection outcomes,(Shrestha et al. 2010) immune responses following MMR vaccination,(Dhiman et al. 2008) and malaria susceptibility.(Khor et al. 2007)
Among the SNPs associated with IL-6 secretion after rubella virus stimulation, one was located within the TNFRSF1B gene, three SNPs were located in the ADAR gene, and five SNPs were located in or near the PVR gene. Each of the genetic associations demonstrated stronger signals in the Rochester cohort than in the San Diego cohort. Several factors may contribute to this: better vaccination dates in the Rochester cohort medical records, which allowed us to more accurately capture time since last vaccination; the smaller size of the San Diego cohort; and the potential impact of undocumented, chance rubella exposure or additional vaccinations in the San Diego cohort. This is especially apparent with rs474247 in TNFRSF1B, with a Rochester cohort p-value of 4.2 × 10−6 and a San Diego cohort p-value of 0.2771. In a previous study, we reported cytokine and cytokine receptor SNPs associated with rubella-specific cytokine secretion in a subset (n=738) of the Rochester cohort. In that study, we identified multiple TNFRSF1B SNPs associated with variations in IL-6 secretion.(Dhiman et al. 2010a) In the meta-analysis, the A allele of rs474247, an intronic SNP in TNFRSF1B, was associated with an allele-dose dependent increase in IL-6 secretion. Binding of TNFα to TNFR1 and TNFR2 leads to activation of NF-kB and pro-inflammatory responses, including IL-6 production. Polymorphisms affecting TNFRSF1B expression and/or function can lead to variations in TNFR pathway activation, leading to alterations in pro-inflammatory cytokine secretion.(Till et al. 2005)
ADAR encodes an RNA-specific adenosine deaminase involved in pre-mRNA splicing, RNA stability and other RNA structure-related activities. The two ADAR-specific SNPs associated with IL-6 response were among the most significant SNPs in the Rochester cohort and were the only SNPs with strong evidence of association in the San Diego cohort. ADAR has been shown to exert antiviral properties by RNA editing of a number of viral RNA genomes including: vesicular stomatitis virus, measles virus, hepatitis C virus, hepatitis delta virus, and HIV. Two of these three SNPs (rs2229857, rs1127317) were also associated with variations in cytokine secretion after measles stimulation.(Haralambieva et al. 2011) The non-synonymous SNP, rs2229857 encoding a K>R change, is also associated with sustained response to IFN therapy for chronic HCV infection.(Hwang et al. 2006; Welzel et al. 2009)
The second genetic region with multiple significant SNP associations was found on chromosome 19 near the PVR and PVRL2 genes (Figure 1). PVR encodes for CD155, a transmembrane glycoprotein that mediates NK cell adhesion and effector function.(Sakisaka and Takai 2004) Multiple T cell and NK cell receptors (TIGIT, DNAM-1, CD96) interact with PVR and its related proteins and these interactions modulate T and NK cell activity.(Bottino et al. 2003; Fuchs et al. 2004; Yu et al. 2009) In fact, PVR and related proteins interact with a wide variety of ligands and participate in multiple immunologic functions and serve as cellular receptors for poliovirus, rhinovirus, and reovirus.(Xu and Jin 2010) SNPs rs3852861 and rs1466435 are located in or near the neighboring PVRL2 gene that encodes for a component of adherens junctions and serves as a cellular receptor for herpes simplex virus and pseudorabies virus. As illustrated in Figure 1, multiple SNPs located within these genetic regions were associated with differential IL-6 production. The presence of so many SNPs associated with immune response makes the TNFRSF1B and PVR regions excellent candidates for a fine mapping effort to assess correlations between these and other regional SNPs. This effort will be necessary to narrow down the possible causal variants. For example, rs203709 (Table 2) is in close LD (r2 = 0.959) with another PVR SNP (rs7255066) exhibiting moderately significant associations with multiple sclerosis in a large collaborative GWAS involving over 9,000 European individuals with multiple sclerosis.(Sawcer et al. 2011)
The differential evidence for genotype-phenotype associations between the two cohorts emphasizes the need to examine multiple populations in order to identify genetic regions expected to influence immune response. Furthermore, it highlights the importance of fine mapping studies that inform the selection of likely causal SNPs for targeted experiments designed to discover the underlying biology behind the variation in immune response. Our data so far highlight the importance of genetic association studies focused on vaccine response and suggest that genetic control of rubella vaccine-induced immunity may be mediated, in part, by novel genes and gene families such as PVR and butyrophilin genes. These findings, when combined with detailed studies elucidating the mechanisms behind identified genotype-phenotype associations, can then pave the way for potentially novel immunostimulatory therapies to improve viral vaccine efficacy and treat viral infections.
Supplementary Material
Acknowledgments
We appreciate the study subjects and their families’ willingness to participate in our study. We thank the Mayo Clinic nurses and study coordinators for their efforts in subject recruitment, Dr. Julie M. Cunningham and the Mayo Medical Genome Facility Genotyping Core for their assistance with genotyping efforts. We also thank Megan M. O’Byrne, Nathaniel D. Warner for their contributions to the statistical analyses. Research reported in this publication was supported by the National Institute of Allergy And Infectious Diseases of the National Institutes of Health under Award Number R37 AI048793, which recently received a MERIT award. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Ethical Standards
All experiments described in this study comply with current, applicable US laws.
Conflict of Interest Statement: Dr. Poland is the chair of a Safety Evaluation Committee for novel non-rubella investigational vaccine trials being conducted by Merck Research Laboratories. Dr. Poland offers consultative advice on vaccine development to Merck & Co. Inc., CSL Biotherapies, Avianax, Sanofi Pasteur, Dynavax, Novartis Vaccines and Therapeutics, PAXVAX Inc, and Emergent Biosolutions. Drs. Poland and Ovsyannikova hold two patents related to vaccinia and measles peptide research. These activities have been reviewed by the Mayo Clinic Conflict of Interest Review Board and are conducted in compliance with Mayo Clinic Conflict of Interest policies. This research has been reviewed by the Mayo Clinic Conflict of Interest Review Board and was conducted in compliance with Mayo Clinic Conflict of Interest policies. The other authors do not have any conflicts of interest.
References
- 2013 Nationwide rubella epidemic–Japan. MMWR. Morbidity and mortality weekly report. 2013;62:457–62. [PMC free article] [PubMed] [Google Scholar]
- Bottino C, Castriconi R, Pende D, Rivera P, Nanni M, Carnemolla B, Cantoni C, Grassi J, Marcenaro S, Reymond N, Vitale M, Moretta L, Lopez M, Moretta A. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. The Journal of experimental medicine. 2003;198:557–67. doi: 10.1084/jem.20030788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. American journal of human genetics. 2004;74:106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman N, Haralambieva IH, Kennedy RB, Vierkant RA, O’Byrne MM, Ovsyannikova IG, Jacobson RM, Poland GA. SNP/haplotype associations in cytokine and cytokine receptor genes and immunity to rubella vaccine. Immunogenetics. 2010a;62:197–210. doi: 10.1007/s00251-010-0423-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman N, Haralambieva IH, Vierkant RA, Pankratz VS, Ryan JE, Jacobson RM, Ovsyannikova IG, Poland GA. Predominant inflammatory cytokine secretion pattern in response to two doses of live rubella vaccine in healthy vaccinees. Cytokine. 2010b;50:24–9. doi: 10.1016/j.cyto.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman N, Ovsyannikova IG, Cunningham JM, Vierkant RA, Kennedy RB, Pankratz VS, Poland GA, Jacobson RM. Associations between measles vaccine immunity and single-nucleotide polymorphisms in cytokine and cytokine receptor genes. The Journal of infectious diseases. 2007;195:21–9. doi: 10.1086/510596. [DOI] [PubMed] [Google Scholar]
- Dhiman N, Ovsyannikova IG, Vierkant RA, Pankratz VS, Jacobson RM, Poland GA. Associations between cytokine/cytokine receptor single nucleotide polymorphisms and humoral immunity to measles, mumps and rubella in a Somali population. Tissue antigens. 2008;72:211–20. doi: 10.1111/j.1399-0039.2008.01097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs A, Cella M, Giurisato E, Shaw AS, Colonna M. Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155) Journal of immunology. 2004;172:3994–8. doi: 10.4049/jimmunol.172.7.3994. [DOI] [PubMed] [Google Scholar]
- Haralambieva IH, Dhiman N, Ovsyannikova IG, Vierkant RA, Pankratz VS, Jacobson RM, Poland GA. 2′-5′-Oligoadenylate synthetase single-nucleotide polymorphisms and haplotypes are associated with variations in immune responses to rubella vaccine. Human immunology. 2010;71:383–91. doi: 10.1016/j.humimm.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralambieva IH, Ovsyannikova IG, Umlauf BJ, Vierkant RA, Shane Pankratz V, Jacobson RM, Poland GA. Genetic polymorphisms in host antiviral genes: associations with humoral and cellular immunity to measles vaccine. Vaccine. 2011;29:8988–97. doi: 10.1016/j.vaccine.2011.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeyman MC, Forrest JM, Dorman DC. Cell-mediated immune response following natural rubella and rubella vaccination. Clinical and experimental immunology. 1974;17:665–71. [PMC free article] [PubMed] [Google Scholar]
- Hwang Y, Chen EY, Gu ZJ, Chuang WL, Yu ML, Lai MY, Chao YC, Lee CM, Wang JH, Dai CY, Shian-Jy Bey M, Liao YT, Chen PJ, Chen DS. Genetic predisposition of responsiveness to therapy for chronic hepatitis C. Pharmacogenomics. 2006;7:697–709. doi: 10.2217/14622416.7.5.697. [DOI] [PubMed] [Google Scholar]
- Jacobsen SJ, Ackerson BK, Sy LS, Tran TN, Jones TL, Yao JF, Xie F, Cheetham TC, Saddier P. Observational safety study of febrile convulsion following first dose MMRV vaccination in a managed care setting. Vaccine. 2009;27:4656–61. doi: 10.1016/j.vaccine.2009.05.056. [DOI] [PubMed] [Google Scholar]
- Jacobson RM, Ovsyannikova IG, Poland GA. Genetic basis for variation of vaccine response: our studies with rubella vaccine. Paediatrics & child health. 2009;19:S156–S159. doi: 10.1016/j.paed.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy RB, Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Ryan MA, Poland GA. Gender effects on humoral immune responses to smallpox vaccine. Vaccine. 2009;27:3319–23. doi: 10.1016/j.vaccine.2009.01.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy RB, Ovsyannikova IG, Vierkant RA, Jacobson RM, Poland GA. Effect of human leukocyte antigen homozygosity on rubella vaccine-induced humoral and cell-mediated immune responses. Human immunology. 2010;71:128–35. doi: 10.1016/j.humimm.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor CC, Vannberg FO, Chapman SJ, Walley A, Aucan C, Loke H, White NJ, Peto T, Khor LK, Kwiatkowski D, Day N, Scott A, Berkley JA, Marsh K, Peshu N, Maitland K, Williams TN, Hill AV. Positive replication and linkage disequilibrium mapping of the chromosome 21q22.1 malaria susceptibility locus. Genes and immunity. 2007;8:570–6. doi: 10.1038/sj.gene.6364417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein NP, Fireman B, Yih WK, Lewis E, Kulldorff M, Ray P, Baxter R, Hambidge S, Nordin J, Naleway A, Belongia EA, Lieu T, Baggs J, Weintraub E. Measles-mumps-rubella-varicella combination vaccine and the risk of febrile seizures. Pediatrics. 2010;126:e1–8. doi: 10.1542/peds.2010-0665. [DOI] [PubMed] [Google Scholar]
- Marin M, Broder KR, Temte JL, Snider DE, Seward JF. Use of combination measles, mumps, rubella, and varicella vaccine: recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR. Recommendations and reports : Morbidity and mortality weekly report. Recommendations and reports / Centers for Disease Control. 2010;59:1–12. [PubMed] [Google Scholar]
- Metcalf CJ, Lessler J, Klepac P, Cutts F, Grenfell BT. Impact of birth rate, seasonality and transmission rate on minimum levels of coverage needed for rubella vaccination. Epidemiology and infection. 2012;140:2290–301. doi: 10.1017/S0950268812000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell LA, Tingle AJ, Decarie D, Shukin R. Identification of rubella virus T-cell epitopes recognized in anamnestic response to RA27/3 vaccine: associations with boost in neutralizing antibody titer. Vaccine. 1999;17:2356–65. doi: 10.1016/s0264-410x(99)00040-7. [DOI] [PubMed] [Google Scholar]
- Ovsyannikova IG, Dhiman N, Haralambieva IH, Vierkant RA, O’Byrne MM, Jacobson RM, Poland GA. Rubella vaccine-induced cellular immunity: evidence of associations with polymorphisms in the Toll-like, vitamin A and D receptors, and innate immune response genes. Human genetics. 2010a;127:207–21. doi: 10.1007/s00439-009-0763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Haralambieva IH, Dhiman N, O’Byrne MM, Pankratz VS, Jacobson RM, Poland GA. Polymorphisms in the vitamin A receptor and innate immunity genes influence the antibody response to rubella vaccination. The Journal of infectious diseases. 2010b;201:207–13. doi: 10.1086/649588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Haralambieva IH, Kennedy RB, Pankratz VS, Vierkant RA, Jacobson RM, Poland GA. Impact of cytokine and cytokine receptor gene polymorphisms on cellular immunity after smallpox vaccination. Gene. 2012a;510:59–65. doi: 10.1016/j.gene.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Haralambieva IH, Vierkant RA, O’Byrne MM, Jacobson RM, Poland GA. The association of CD46, SLAM and CD209 cellular receptor gene SNPs with variations in measles vaccine-induced immune responses: a replication study and examination of novel polymorphisms. Human heredity. 2011a;72:206–23. doi: 10.1159/000331585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Jacobson RM, Vierkant RA, Jacobsen SJ, Pankratz VS, Poland GA. The contribution of HLA class I antigens in immune status following two doses of rubella vaccination. Human immunology. 2004;65:1506–15. doi: 10.1016/j.humimm.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Ovsyannikova IG, Jacobson RM, Vierkant RA, Jacobsen SJ, Pankratz VS, Poland GA. Human leukocyte antigen class II alleles and rubella-specific humoral and cell-mediated immunity following measles-mumps-rubella-II vaccination. The Journal of infectious diseases. 2005;191:515–9. doi: 10.1086/427558. [DOI] [PubMed] [Google Scholar]
- Ovsyannikova IG, Jacobson RM, Vierkant RA, O’Byrne MM, Poland GA. Replication of rubella vaccine population genetic studies: validation of HLA genotype and humoral response associations. Vaccine. 2009a;27:6926–31. doi: 10.1016/j.vaccine.2009.08.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Poland GA. Human leukocyte antigen haplotypes in the genetic control of immune response to measles-mumps-rubella vaccine. The Journal of infectious diseases. 2006;193:655–63. doi: 10.1086/500144. [DOI] [PubMed] [Google Scholar]
- Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Poland GA. Consistency of HLA associations between two independent measles vaccine cohorts: a replication study. Vaccine. 2012b;30:2146–52. doi: 10.1016/j.vaccine.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Ryan JE, Vierkant RA, O’Byrne MM, Pankratz VS, Jacobson RM, Poland GA. Influence of host genetic variation on rubella-specific T cell cytokine responses following rubella vaccination. Vaccine. 2009b;27:3359–66. doi: 10.1016/j.vaccine.2009.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Vierkant RA, Pankratz VS, Jacobson RM, Poland GA. Extended LTA, TNF, LST1 and HLA gene haplotypes and their association with rubella vaccine-induced immunity. PloS one. 2010c;5:e11806. doi: 10.1371/journal.pone.0011806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Vierkant RA, Pankratz VS, Jacobson RM, Poland GA. Human leukocyte antigen genotypes in the genetic control of adaptive immune responses to smallpox vaccine. The Journal of infectious diseases. 2011b;203:1546–55. doi: 10.1093/infdis/jir167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz VS, Vierkant RA, O’Byrne MM, Ovsyannikova IG, Poland GA. Associations between SNPs in candidate immune-relevant genes and rubella antibody levels: a multigenic assessment. BMC immunology. 2010;11:48. doi: 10.1186/1471-2172-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reef SE, Plotkin SA. Rubella vaccine. In: Plotkin SA, Orenstein WA, Offit PA, editors. Vaccines. 6. Elsevier/Saunders; Edinburgh: 2013. p. xix.p. 1550. [Google Scholar]
- Romporn S, Hirankarn N, Tangkijvanich P, Kimkong I. Association of IFNAR2 and IL10RB genes in chronic hepatitis B virus infection. Tissue antigens. 2013;82:21–5. doi: 10.1111/tan.12133. [DOI] [PubMed] [Google Scholar]
- S G, K OR. Meta-Analysis. In: KJ R, S G, T L, editors. Modern Epidemiology. 3. Lippincott WIlliams and Wilkins; 2008. p. 652. [Google Scholar]
- Sakisaka T, Takai Y. Biology and pathology of nectins and nectin-like molecules. Current opinion in cell biology. 2004;16:513–21. doi: 10.1016/j.ceb.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Sawcer S, Hellenthal G, Pirinen M, Spencer CC, Patsopoulos NA, Moutsianas L, Dilthey A, Su Z, Freeman C, Hunt SE, Edkins S, Gray E, Booth DR, Potter SC, Goris A, Band G, Oturai AB, Strange A, Saarela J, Bellenguez C, Fontaine B, Gillman M, Hemmer B, Gwilliam R, Zipp F, Jayakumar A, Martin R, Leslie S, Hawkins S, Giannoulatou E, D’Alfonso S, Blackburn H, Martinelli Boneschi F, Liddle J, Harbo HF, Perez ML, Spurkland A, Waller MJ, Mycko MP, Ricketts M, Comabella M, Hammond N, Kockum I, McCann OT, Ban M, Whittaker P, Kemppinen A, Weston P, Hawkins C, Widaa S, Zajicek J, Dronov S, Robertson N, Bumpstead SJ, Barcellos LF, Ravindrarajah R, Abraham R, Alfredsson L, Ardlie K, Aubin C, Baker A, Baker K, Baranzini SE, Bergamaschi L, Bergamaschi R, Bernstein A, Berthele A, Boggild M, Bradfield JP, Brassat D, Broadley SA, Buck D, Butzkueven H, Capra R, Carroll WM, Cavalla P, Celius EG, Cepok S, Chiavacci R, Clerget-Darpoux F, Clysters K, Comi G, Cossburn M, Cournu-Rebeix I, Cox MB, Cozen W, Cree BA, Cross AH, Cusi D, Daly MJ, Davis E, de Bakker PI, Debouverie M, D’Hooghe MB, Dixon K, Dobosi R, Dubois B, Ellinghaus D, Elovaara I, Esposito F, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–9. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha S, Wiener HW, Aissani B, Song W, Shendre A, Wilson CM, Kaslow RA, Tang J. Interleukin-10 (IL-10) pathway: genetic variants and outcomes of HIV-1 infection in African American adolescents. PloS one. 2010;5:e13384. doi: 10.1371/journal.pone.0013384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicotte H, Rider DN, Poland GA, Dhiman N, Kocher JP. SNPPicker: high quality tag SNP selection across multiple populations. BMC bioinformatics. 2011;12:129. doi: 10.1186/1471-2105-12-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2002;64:479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- Storey JD. The Positive False Discovery Rate: A Bayesian Interpretation and the q-Value. The Annals of Statistics. 2003;31:2013–2035. doi: 10.2307/3448445. [DOI] [Google Scholar]
- Till A, Rosenstiel P, Krippner-Heidenreich A, Mascheretti-Croucher S, Croucher PJ, Schafer H, Scheurich P, Seegert D, Schreiber S. The Met-196 -> Arg variation of human tumor necrosis factor receptor 2 (TNFR2) affects TNF-alpha-induced apoptosis by impaired NF-kappaB signaling and target gene expression. The Journal of biological chemistry. 2005;280:5994–6004. doi: 10.1074/jbc.M411541200. [DOI] [PubMed] [Google Scholar]
- Vesikari T, Becker T, Gajdos V, Fiquet A, Thomas S, Richard P, Baudin M. Immunogenicity and safety of a two-dose regimen of a combined measles, mumps, rubella and varicella live vaccine (ProQuad((R))) in infants from 9 months of age. Vaccine. 2012;30:3082–9. doi: 10.1016/j.vaccine.2012.02.062. [DOI] [PubMed] [Google Scholar]
- Vesikari T, Buimovici-Klein E. Lymphocyte responses to rubella antigen and phytohemagglutinin after administration of the RA 27/3 strain of live attenuated rubella vaccine. Infection and immunity. 1975;11:748–53. doi: 10.1128/iai.11.4.748-753.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welzel TM, Morgan TR, Bonkovsky HL, Naishadham D, Pfeiffer RM, Wright EC, Hutchinson AA, Crenshaw AT, Bashirova A, Carrington M, Dotrang M, Sterling RK, Lindsay KL, Fontana RJ, Lee WM, Di Bisceglie AM, Ghany MG, Gretch DR, Chanock SJ, Chung RT, O’Brien TR. Variants in interferon-alpha pathway genes and response to pegylated interferon-Alpha2a plus ribavirin for treatment of chronic hepatitis C virus infection in the hepatitis C antiviral long-term treatment against cirrhosis trial. Hepatology. 2009;49:1847–58. doi: 10.1002/hep.22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Jin B. A novel interface consisting of homologous immunoglobulin superfamily members with multiple functions. Cellular & molecular immunology. 2010;7:11–9. doi: 10.1038/cmi.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, Tom I, Ivelja S, Refino CJ, Clark H, Eaton D, Grogan JL. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nature immunology. 2009;10:48–57. doi: 10.1038/ni.1674. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


