Summary
Stromal cells within the tumor microenvironment are essential for tumor progression and metastasis. Surprisingly little is known about the factors that drive the transcriptional reprogramming of stromal cells within tumors. We report that the transcriptional regulator Heat-Shock Factor 1 (HSF1) is frequently activated in cancer-associated fibroblasts (CAFs), where it is a potent enabler of malignancy. HSF1 drives a transcriptional program in CAFs that complements, yet is completely different from, the program it drives in adjacent cancer cells. This CAF program is uniquely structured to support the malignant potential of cancer cells in a non-cell-autonomous way. Two central stromal signaling molecules—TGFβ and stromal-derived factor 1 (SDF1) – play a critical role. In early stage breast and lung cancer, high stromal HSF1 activation is strongly associated with poor patient outcome. Thus, tumors co-opt the ancient survival functions of HSF1 to orchestrate malignancy in both cell-autonomous and non-cell-autonomous ways, with far-reaching therapeutic implications.
Introduction
Cancer cells in a tumor mass are surrounded by a variety of other cell types, including immune cells, fibroblasts and endothelial cells as well as extracellular matrix (ECM) components. Taken together, these comprise the tumor microenvironment. Cells of the tumor microenvironment contribute to the hallmarks of cancer and their co-evolution with cancer cells is essential for tumor formation and progression (Bissell and Hines, 2011; Hanahan and Weinberg, 2011).
In the majority of carcinomas, the most abundant cells in the tumor microenvironment are CAFs, cancer-associated fibroblasts (Hanahan and Coussens, 2012; Hanahan and Weinberg, 2011). CAFs include myofibroblasts and reprogrammed variants of normal tissue-derived fibroblasts that are recruited by the tumor to support cancer cell proliferation, angiogenesis, invasion, metastasis and drug-resistance (Erez et al., 2010; Kalluri and Zeisberg, 2006; Olumi et al., 1999; Straussman et al., 2012; Wilson et al., 2012). CAFs support cancer cells in a non-cell-autonomous manner through secretion of ECM, chemokines, cytokines and growth factors (Lu et al., 2012; Moskovits et al., 2006; Orimo et al., 2005; Pickup et al., 2013; Siegel and Massague, 2003). The secretion of cytokines also feeds back to promote the fibroblast-to-CAF transition, through autocrine TGFβ and SDF1 signaling (Kojima et al., 2010).
Despite accumulating evidence for the non-cell-autonomous effects of CAFs on cancer cells, little is known about the transcriptional regulators that are responsible for stromal reprogramming to support tumorigenesis. That such reprogramming must occur is clear from evidence that normal fibroblasts usually constitute a tumor-restrictive environment (Bissell and Hines, 2011). In mouse models, tumor suppressors such as p53 and PTEN can act in the stroma to limit tumor growth (Lujambio et al., 2013; Moskovits et al., 2006; Trimboli et al., 2009). If tumor suppressors act in both the cancer cells and the stroma to inhibit malignancy, might there also be factors that actively support or enable malignancy in both cancer cells and in the stroma? Presumably these would not be classical oncogenes, as non-malignant stromal cells are relatively stable genetically (Qiu et al., 2008). Instead, we wondered if tumors might hijack normal physiological pathways and programs in the stroma, subverting them to enable neoplastic growth and metastatic dissemination. Here, we provide evidence for such a mechanism by investigating the stromal function(s) of Heat Shock Factor 1 (HSF1) in tumor biology.
HSF1 is a ubiquitously expressed transcription factor best known for its activation by heat (Sakurai and Enoki, 2010; Shamovsky and Nudler, 2008). Recently it has been shown to play a fundamental role in tumor biology (Dai et al., 2007; Jin et al., 2011). In a wide variety of human cancer cell lines, the depletion of HSF1 markedly reduces growth, survival and metastatic potential (Mendillo et al., 2012; Meng et al., 2010; Santagata et al., 2012; Scott et al., 2011). Hsf1 null mice develop normally, but are profoundly resistant to tumorigenesis.
The transcriptional program that is activated by HSF1 in cancer cells is surprisingly different from the program activated by classical heat-shock (Mendillo et al., 2012). In particular, it acts to support the malignant state by blunting apoptotic responses and promoting pathways that facilitate anabolic metabolism, protein folding, proliferation, invasion, and metastasis (Dai et al., 2012; Fang et al., 2012; Jin et al., 2011; Mendillo et al., 2012; Meng et al., 2010; Santagata et al., 2013; Scott et al., 2011). In humans, activation of this program by HSF1 in cancer cells is strongly associated with disease progression in patients with breast, colon, lung, and hepatocellular carcinomas (Fang et al., 2012; Mendillo et al., 2012; Santagata et al., 2011).
Clearly, HSF1 plays a central role in supporting the malignant transformation and progression of diverse cancer types. Here, we ask if it plays a complementary, and perhaps equally important, role in subverting the normally repressive activity of the stroma by converting it to a pro-tumorigenic state. We also discuss the possible evolutionary origins of HSF1-mediated cross talk between cancer and stromal cells in tumors, as well as its potential therapeutic implications.
Results
HSF1 is activated in cancer-associated fibroblasts within human tumors
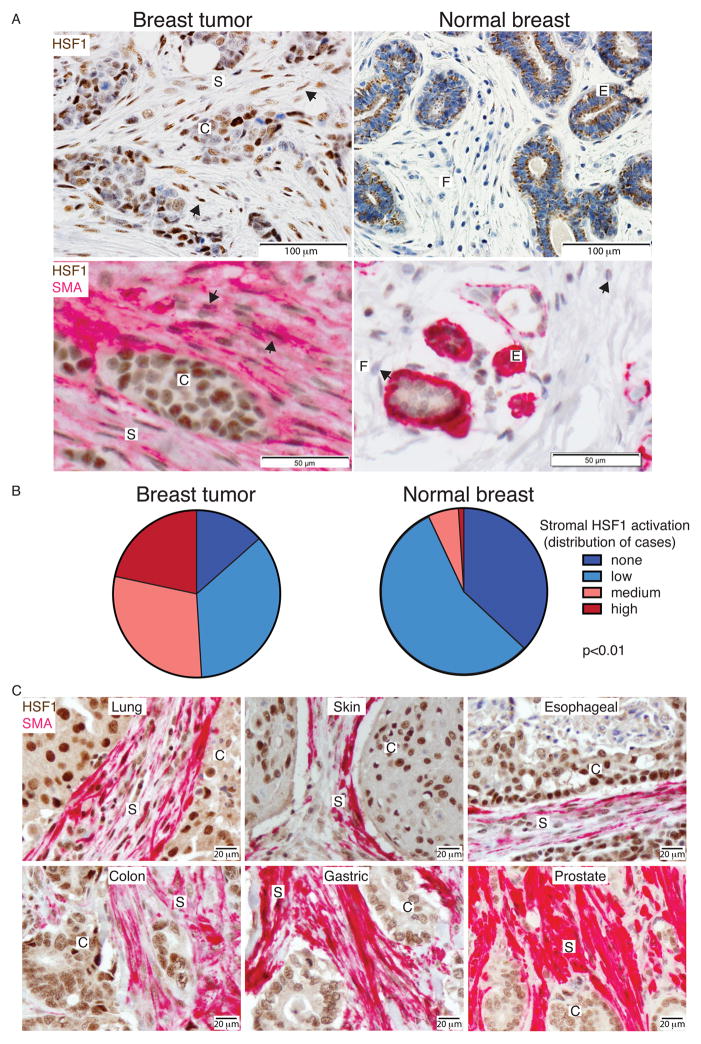
Under basal conditions in normal cells, HSF1 resides primarily in the cytoplasm. Upon activation, it accumulates in the nucleus (Morimoto, 2008; Santagata et al., 2011). To determine whether HSF1 is activated in cells of the tumor microenvironment we scored the staining intensity of this transcription factor in the nuclei of tumor-associated stroma within patient-derived breast cancer samples. Stromal cells residing in the lobules of neighboring, normal breast tissue were used for comparison. These normal cells were almost invariably low or negative for nuclear HSF1. However, strong nuclear HSF1 staining was frequently present in stromal cells situated in close proximity to malignant cells (Figure 1A upper panel and 1B).
Figure 1. HSF1 activation in cancer-associated fibroblasts within human tumors.
(A) Tissue sections of breast resection specimens from 12 patients encompassing both invasive ductal carcinoma and neighboring normal breast lobules (in the same section) were immunostained with anti-HSF1 antibodies (brown, upper panels) or co-stained with anti-HSF1 and anti-SMA (pink) antibodies (lower panels). Representative images are shown. Arrows indicate HSF1-positive CAFs in the left panels, and HSF1-negative normal fibroblasts in the lower right panel. (B) Pie charts depict the distribution of relative nuclear HSF1 staining intensity in the stroma amongst 12 breast resection specimens with matching controls. For each specimen, 4 regions of tumor or normal tissue were evaluated. Statistical significance of the differences between normal and tumor was assessed using repeated-measures ANOVA (p=4e-13), as well as paired t-tests, followed by Bonferroni correction (p<0.01). (C) Representative images of tumor sections from patients with the indicated types of cancer co-stained for HSF1 (brown) and SMA (pink). C and S indicate cancer- or stroma-rich regions, respectively. For normal tissue, E and F indicate regions rich with epithelial cells or fibroblasts, respectively. See also Figure S1.
The morphology of the HSF1-positive stromal cells suggested that they were cancer-associated fibroblasts (CAFs). To confirm, we co-stained tumor sections for HSF1 and smooth muscle actin (SMA). SMA stains normal myoepithelial cells (Figure 1A, lower right panel). It is not present in normal fibroblasts however, and is often used as a marker for stromal CAFs (Kalluri and Zeisberg, 2006; Quante et al., 2011). We also investigated markers of two other stromal components, leukocytes (LCA) and endothelial cells (CD31). Most of the HSF1-positive stromal cells in the tumors co-stained with SMA, suggesting that they are indeed CAFs (Figure 1A, lower left panel, and Figure S1).
To test the generality of HSF1 activation in CAFs across different tumor types, we co-stained tumor sections from lung, skin, esophageal, colon, gastric and prostate carcinomas with antibodies for HSF1 and SMA (Figure 1C). In all of these, most SMA-positive CAFs also had strong nuclear HSF1 staining.
Loss of Hsf1 in fibroblasts reduces xenograft tumor growth
To explore the function of stromal HSF1 activation in a tractable model system, we analyzed xenografts of human MCF7 breast cancer cells injected subcutaneously into immunocompromised (NOD-scid) mice. As expected, xenografts recruited endogenous stromal cells from their mouse hosts to support tumor formation. These SMA-positive CAFs exhibited strong nuclear HSF1 staining (Figure S2A).
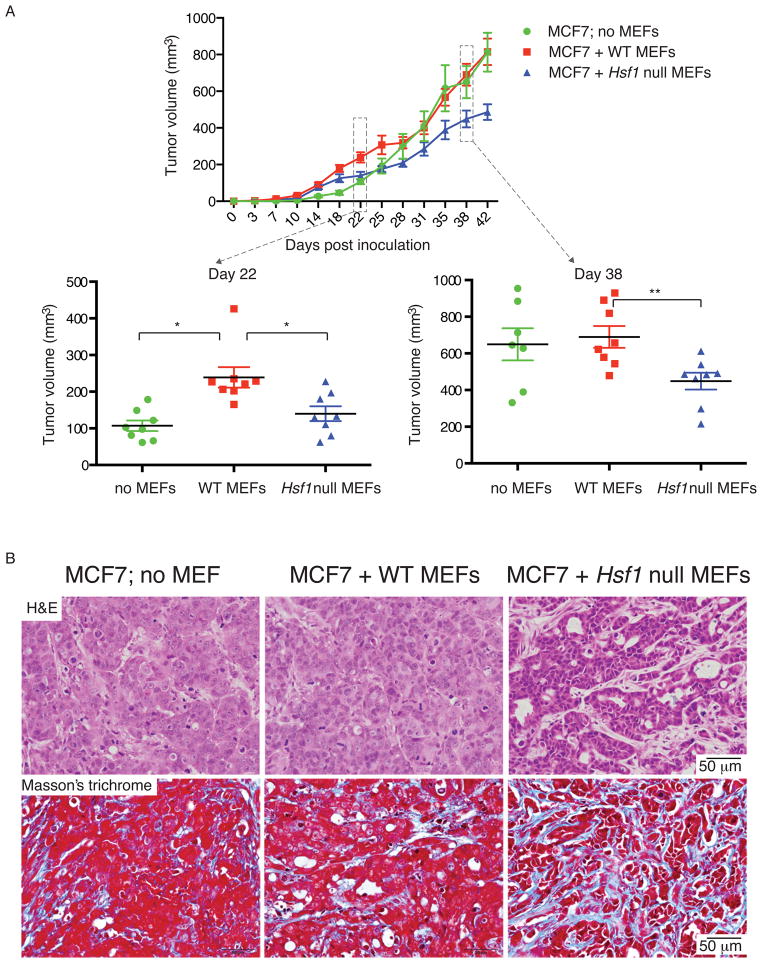
To test whether HSF1 activation in stromal cells plays a role in supporting malignant cells, we mixed primary mouse embryonic fibroblasts (MEFs) – either wild-type (WT) or Hsf1 null – with the MCF7 cancer cells and co-injected them subcutaneously into NOD-scid mice (Figure 2A). Tumors arising from MCF7 cells co-injected with Hsf1 null MEFs (blue) grew significantly more slowly than those of mice co-injected with WTMEFs (red).
Figure 2. Stromal Hsf1 status alters tumor progression and histology in human breast xenografts.
MCF7 breast cancer cells alone or mixed with WT or Hsf1 null primary MEFs were injected subcutaneously into NOD-scid mice. The experiment was repeated twice, with 4 mice per group in each experiment. (A) The mean tumor volume (total 8 per treatment group) is shown. The distribution of individual measurements is shown in the lower panels, in scatter plots for days 22 and 38 post injection. Error bars, SEM. *p<0.05, **p<0.01. (B) Mice were sacrificed when tumor burden reached size limit and the tumors were excised, fixed and stained with hematoxylin & eosin (H&E, upper panels) or Masson’s trichrome stain (lower panels). All images collected at the same magnification. Scale bar = 50μm. See also Figure S2.
MCF7 cells injected without MEFs formed tumors more slowly than MCF7 cells co-injected with WT MEFs (Figure 2A). With time, cells injected without MEFs recruited WT host stroma and the tumors grew to the same size as those formed by co-injection with WT MEFs. However, throughout the experiment, tumors formed by MCF7 cells co-injected with Hsf1 null MEFs remained significantly smaller.
To better understand this result, we excised tumors at the end of the experiment and examined their histology (Figure 2B, upper panels). Tumors from mice injected with MCF7 cells only, and tumors from mice co-injected with WT MEFs shared a poorly differentiated and sheet-like morphology typical of high-grade tumors. In contrast, co-injection of MCF7 cells and Hsf1 null MEFs produced tumors with a more differentiated, stromal-rich architecture, indicative of a less malignant phenotype. Notably, some of the stromal fibroblasts were HSF1-positive, indicating that in additon to the injected MEFs, the tumors had recruited host fibroblasts (Figure S2B). (This might explain why, even though slower, these tumors still grew.) Masson’s trichrome staining indicated that stroma-rich regions are mostly comprised of fibrous tissue and deposits of collagen (Figure 2B, lower panels). These results suggest that, in response to cancer cells, HSF1 is activated in stromal CAFs to support tumor growth. Moreover, in the absence of this HSF1-driven response, fibroblasts actually exert an inhibitory effect on tumor expansion.
Stromal HSF1 regulates cancer cell growth in vitro
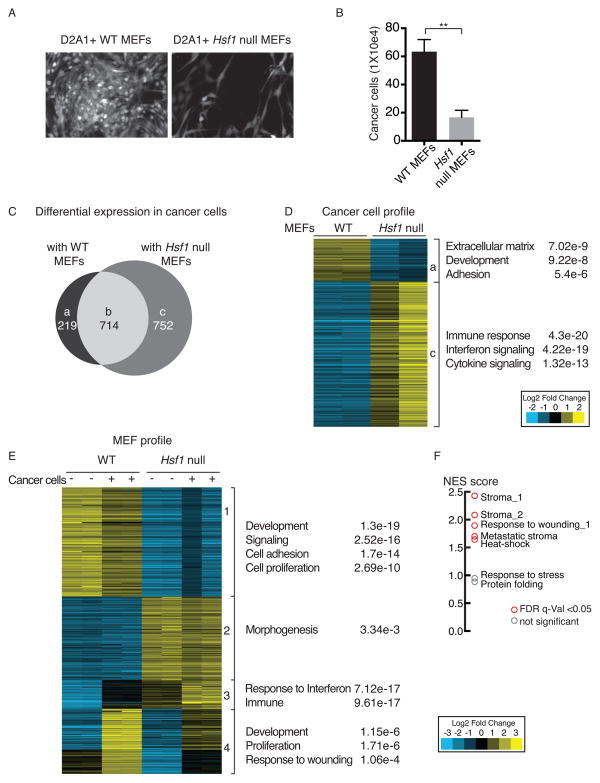
To learn how activation of HSF1 in stromal fibroblasts supports cancer cells, we plated fluorescently-labeled breast cancer cells onto feeder layers of either WT or Hsf1 null MEFs (Figure 3A & S3A). We found a higher number of cancer cells in co-cultures with WT MEFs than with Hsf1 null MEFs. This held true for several different mammary cancer cell lines (mouse D2A1, Figure 3A & B; human MCF7, human HCC38 and mouse 4T7; Figure S3B & C).
Figure 3. HSF1 in fibroblasts supports cancer cell growth by activating gene expression programs both in cancer cells and in fibroblasts.
(A–B) WT or Hsf1 null immortalized MEFs were treated with 10 μg/ml mitomycin C. D2A1 mouse mammary tumor cells stably expressing dsRed (D2A1-dsRed) were seeded on top of the MEFs and allowed to grow for 72h–96h, after which cancer cells were either visualized by fluorescent microscopy (A) or trypsinized and quantitated by flow cytometry (B). The mean of 3 independent experiments is shown. Error bars, SEM ** p<0.005. (C–D) Total RNA was purified from duplicate cultures of D2A1 cancer cells grown with or without WT or Hsf1 null MEFs and sorted as described above. RNA was hybridized to Agilent microarrays, and relative gene expression levels were analyzed using cluster 3.0. For each gene, expression in D2A1 cells grown alone was set to 1, and the relative change in expression upon co-culture with WT or Hsf1 null MEFs was calculated. (C) Overlap of genes differentially expressed in D2A1 cancer cells in the presence of WT or Hsf1 null MEFs. (D) Heat-map depicting fold change in mRNA levels of genes differentially expressed in D2A1 cells grown in co-culture with WT versus Hsf1 null MEFs (in duplicate). Gene ontology (GO) enrichment is shown to the right of the panel. Groups a & c correspond to groups a & c in panel (C). (E) WT or Hsf1 null MEFs were co-cultured with D2A1-dsRed cells as described in (A), but not treated with mitomycin C. After 72–96h, cultures were sorted and mRNA was extracted and hybridized to Agilent microarrays. MEFs cultured without D2A1 cells and processed in the same manner served as controls. Gene expression was analyzed using cluster 3.0 and the differentially expressed genes were clustered into 4 groups. Gene ontology (GO) enrichment is shown to the right of the panel. (F) Gene set enrichment analysis of genes upregulated in WT vs Hsf1 null MEFs co-cultured with cancer cells (groups 1 & 4 in panel E). Enrichment was calculated for the indicated gene sets, and is presented as normalized enrichment score (NES). Statistically significant enrichment (false discovery rate (FDR) q-value<0.05) is shown in red, non-significant enrichment is shown in gray. See also Figure S3 and Tables S1–S5.
To confirm true HSF1-dependence we used MEFs that were deleted for WT Hsf1 but expressed a tetracycline-repressible Hsf1 transgene (Bi-TetO-Hsf1). We repressed Hsf1 expression for 5 days and then co-cultured the MEFs with D2A1 cancer cells. Repression of Hsf1 resulted in decreased accumulation of cancer cells (Figure S3D). Thus, even short-term loss of Hsf1 impairs the ability of fibroblasts to support the growth of co-cultured cancer cells.
Stromal HSF1 drives a transcriptional program in cancer cells that promotes malignant phenotypes
To test the effects of co-culture on gene expression, we separated cancer cells from fibroblasts by FACS sorting, extracted RNA and hybridized it to gene expression arrays. As a point of comparison, each cell type was grown alone (without co-culture), treated and analyzed in a similar manner.
In D2A1 cancer cells, regardless of the Hsf1 status of the co-cultured MEFs, the expression of ~700 genes was altered by ≥ 2-fold following co-culture (Figure 3C, group b; Table S1). Of these, ~400 genes were upregulated and ~300 genes were downregulated. The upregulated set was enriched for genes involved in cellular differentiation, migration and extracellular matrix organization. No significant functional enrichment was found in the downregulated set.
With specific regard to the Hsf1 status of the MEFs, approximately 200 genes were upregulated in cancer cells co-cultured with WT, but not with Hsf1 null MEFs (Figure 3C, group a). This set was enriched for genes involved in extracellular matrix organization, development and adhesion (e.g. Dmp1, Dkk3, Thy1, Grem1, Sparc, Mmp2, and Mmp3, Figure 3D; Table S1). In cancer cells co-cultured with Hsf1 null MEFs, ~750 genes were uniquely upregulated (Figure 3C, group c). Pro-inflammatory cytokines (e.g. Ccl5 and Ccl8) and immune responses (e.g. the response to type 1 interferon) were most significantly enriched in this group (Figure 3D; Table S1). Thus, activation of HSF1 in the stroma helps to reprogram cancer cells in at least two important ways. In a non-cell-autonomous manner it upregulates genes in cancer cells that enhance their malignant potential and downregulates genes that would trigger host immune defense responses.
Stromal HSF1 drives a transcriptional program in fibroblasts that supports malignant cells
Next, we examined a complementary question: how does co-culture with cancer cells affect HSF1-dependent gene expression in stromal fibroblasts? Profiling of FACS-sorted MEFs showed that even in the absence of cancer cells, HSF1 regulated many genes involved in development, cell adhesion and proliferation (e.g. Fgf, Igf, Col, Lama, Snail, and Sdf1; Figure 3E, group 1; Tables S2 & S3). This suggests that HSF1 alters the basal phenotype of MEFs in culture and these alterations enhance the growth of cancer cells. In an HSF1-dependent manner, co-culture with cancer cells induced an additional cluster of genes involved in development, proliferation and response to wounding (e.g. Tgfβ1, Cxcl1, Cxcl3, and Vcam1; Figure 3E, group 4; Tables S2 & S3). Also in an HSF1-dependent manner, cancer cells induced in MEFs a striking downregulation of genes involved in cellular immune responses (e.g. Cxcl10, Bst2, and C3; Figure 3E, group 3; Tables S2 & S3). Thus, WT MEFs respond to cancer cells in a manner that supports tumor growth, whereas Hsf1 null MEFs respond in a manner likely to impede the process.
To further characterize the HSF1 stromal signature, we performed additional analyses of the genes that are differentially upregulated in WT vs Hsf1 null MEFs co-cultured with cancer cells (groups 1 & 4). We compared this list to publicly available gene-sets of stroma from human cancer patients, fibroblast wound healing responses, and the heat-shock response. Although some heat-shock-related genes were enriched, this was not the most prominent response. Rather, the HSF1 stromal signature was most highly enriched for genes previously characterized by their up-regulation in fibroblasts in response to wounding and in stromal cells isolated from human tumors (Beck et al., 2008; Chang et al., 2004; Dvorak, 1986; Karnoub et al., 2007) (Figure 3F; Table S4). We also compared this list to the HSF1-dependent gene expression signature in cancer cells (Mendillo et al., 2012) and found that these signatures were, if anything, anti-correlated (Table S4). Thus, in fibroblasts HSF1 activates a transcriptional program likely to support tumor progression, which is profoundly different from the response activated by HSF1 in the cancer cells themselves, or in cells exposed to heat.
The effects of stromal HSF1 activation on cancer cells are mediated by TGFβ and SDF1 signaling
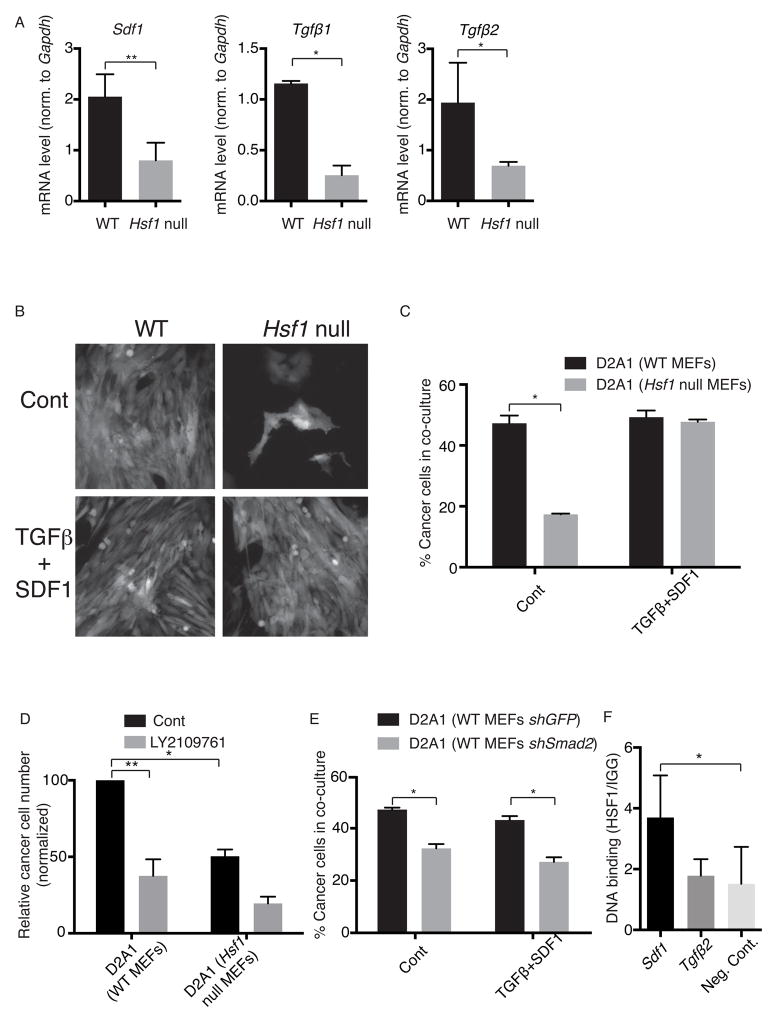
Unbiased analysis of gene-set enrichment established that TGFβ signaling was one of the top categories regulated by HSF1 in MEFs co-cultured with cancer cells (group 4 in Figure 3E; Table S5). Because TGFβ, along with SDF1, was previously found to promote CAF phenotypes (Kojima et al., 2010), we further interrogated both signaling pathways. We extracted RNA from both immortalized and 3 separate sets of primary WT or Hsf1 null MEFs and performed qPCR with primers targeting Tgfβ1, Tgfβ2, Tgfβ3 and Sdf1. This confirmed that expression levels of Sdf1, Tgfβ1 and Tgfβ2 were significantly lower in Hsf1 null MEFs than in WT MEFs, even without co-culture with cancer cells (Figure 4A & S4A).
Figure 4. TGFβ and SDF1 mediate the support of cancer cell growth by stromal HSF1.
(A) The relative expression of Sdf1, Tgfβ1 and Tgfβ2 in WT or Hsf1 null immortalized MEFs was measured by qPCR, and normalized to Gapdh. The mean of 3 experiments is shown. Error bars, SEM. (B–C) WT or Hsf1 null immortalized MEFs were co-cultured with D2A1-dsRed cells as explained in Figure 3A, in the presence or absence of 10 ng/ml TGFβ1 and 100 ng/ml SDF1. After 96h, cells were either visualized by fluorescent microscopy (B) or quantitated by flow cytometry (C). The percentage of cancer cells in co-culture is presented. The experiment was repeated 3 times, in triplicate. Representative results of one experiment are shown as mean +/− SEM. (D) Immortalized WT or Hsf1 null mitomycin-treated MEFs were pretreated, or not, with LY2109761 for 30 minutes before co-culture with D2A1-dsRed cells. Cultures were continued for 72h, with daily supplementation of LY2109761 (or not, as control), and then analyzed as in (C). The experiment was repeated 3 times, in triplicate. Results are expressed as the mean relative number of cancer cells, normalized to non-drug-treated co-cultures with WT MEFs. Error bars, SEM. (E) Immortalized WT MEFs stably expressing shRNA hairpins targeting Smad2 (shSmad2) or GFP (shGFP) were co-cultured with D2A1 cells, treated and analyzed as in (C). The percentage of cancer cells in the co-culture is presented. (F) Chromatin immunoprecipitation (IP) was performed with anti-HSF1 antibodies using material prepared from MCF7 tumor xenografts. Normal rat-IgG served as a negative IP control. IPs were analyzed by qPCR with primers targeting potential heat shock elements in mouse Sdf1 and Tgfβ2. Primers targeting an intergenic region in the mouse DNA, not expected to be amplified, were used as a negative control. The experiment was repeated twice, tumors from 3 mice were used for each experiment. Representative results from one experiment are shown as mean +/− SEM, *p<0.05, **p<0.01. See also Figure S4.
Next, we asked if TGFβ and SDF1 mediate HSF1’s stromal support of cancer cells. We added these factors as purified recombinant proteins to co-cultures of D2A1 cancer cells with Hsf1 null MEFs. Combined addition of TGFβ1 and SDF1 restored cancer cell growth to levels achieved by co-culture with WT MEFs (Figure 4B & C). Partial effects, that did not reach statistical significance, were achieved by addition of either factor alone (Figure S4B).
As a further functional test, we repressed TGFβ signaling in co-cultures by adding a TGFβ receptor type I/II (TβRI/II) dual inhibitor, LY2109761 to the media (Dituri et al., 2013). To control for direct effects on cancer cells themselves, we treated cancer cells with the inhibitor in the absence of MEFs. Treatment with LY2109761 did not affect cancer cells grown alone (Figure S4C). It did, however, significantly reduce their growth in co-culture with WT MEFs (Figure 4D; p=0.008). A smaller effect, that did not reach statistical significance, was seen in co-culture with Hsf1 null MEFs (Figure 4D; p=0.1). Taken together with the increased expression of Tgfβ and Sdf1 in WT MEFs compared to Hsf1 null MEFs (Figure 4A), these results suggest that TGFβ and SDF1 are produced and secreted by fibroblasts in an HSF1-dependent manner.
Once secreted, TGFβ and SDF1 could activate either the fibroblasts themselves, the cancer cells, or both. To investigate, we knocked down the expression of several signaling molecules downstream of TGFβ and SDF1 in either cancer cells or MEFs (see Experimental Procedures). Knock-down of Smad2, a key downstream mediator of TGFβ signaling, in WT MEFs, impaired the growth of co-cultured D2A1 cancer cells (Figure 4E & S4E). This growth defect could not be rescued by addition of recombinant TGFβ1 and SDF1 (Figure 4E). Notably, Smad2 knockdown was only effective in the MEFs. Knockdown of the same gene in the D2A1 cells themselves had no effect on cell number (Figure S4D & S4E). We conclude that HSF1 supports an autocrine TGFβ signaling loop in MEFs. As for SDF1, although we cannot discriminate whether it signals to the cancer cells or to the stroma, SDF1 expression is clearly upregulated by HSF1 in fibroblasts. Taken together, our data indicate that TGFβ and SDF1 are key mediators of the tumor-promoting activity of stromal HSF1.
HSF1 directly binds HSEs of the Sdf1 gene in stromal cells
Next we asked whether TGFβ and SDF1 are direct transcriptional targets of HSF1. A bioinformatic search for HSF1-binding elements (HSEs) in genes of the TGFβ and SDF1 signaling pathways confirmed that the Tgfβ2 and Sdf1 genes themselves contain canonical HSEs. No HSEs were found in Tgfβ1 or any of the downstream signaling molecules mentioned above. To determine whether HSF1 directly regulates Tgfβ2 and Sdf1 expression in CAFs, we performed chromatin immunoprecipitation (ChIP) using anti-HSF1 antibodies and extracts prepared from MCF7 tumor xenografts. To focus specifically on the supporting mouse stromal cells, and not the human cancer cells, we performed quantitative PCR (qPCR) using primers flanking potential HSF1 binding sites that were specific to the mouse genes (Figure S4F). Primers for an intergenic region served as a negative control. Sdf1 was significantly amplified from stromal (mouse) DNA bound by HSF1 (Figure 4F). No significant amplification was detected for Tgfβ2. Together with the effects of HSF1 seen on expression of these genes, these data suggest that regulation of Tgfβ by HSF1 may be indirect. However, HSF1 directly binds and activates Sdf1.
HSF1 activation in breast cancer stroma is associated with poor patient outcome
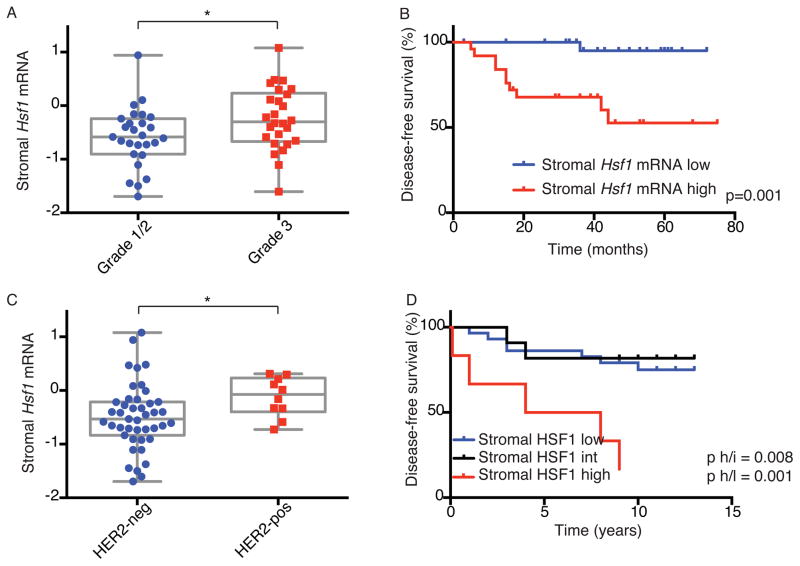
Our findings in mouse xenografts and in vitro co-culture models indicate that stromal HSF1 contributes to tumor progression. To evaluate whether stromal HSF1 contributes to disease progression in human cancers, we first asked whether Hsf1 mRNA levels in the stroma correlate with disease outcome. We looked for this association in a publicly available mRNA dataset from 53 pure tumor stroma samples obtained from patients with primary breast tumors (stromal cells were separated from cancer cells by laser microdissection (Finak et al., 2008)).
In this dataset, high Hsf1 levels significantly correlated with increased tumor grade (Figure 5A) and poorer patient outcome (Figure 5B). We further asked whether high stromal Hsf1 expression is associated with a specific breast cancer subtype. No significant association was found with estrogen receptor (ER, Figure S5A) or progesterone receptor (PR, Figure S5B) expression. (The number of triple negative tumors in this cohort was too small to determine a possible association with HSF1 expression). Hsf1 expression was, however, significantly higher in HER2-positive tumors as compared to HER2-negative tumors (Figure 5C).
Figure 5. Increased HSF1 activation in the stroma is associated with decreased survival in breast cancer patients.
(A–C) Analysis of Hsf1 mRNA expression levels in the stroma of 53 breast cancer patients from (Finak et al., 2008). (A) The association between Hsf1 expression and tumor grade is presented in a box & whiskers plot. (B) Kaplan-Meier (KM) analysis of patients stratified by Hsf1 expression. (C) The correlation between Hsf1 expression and HER2 status is presented in a box & whiskers plot. (D) Breast cancer resections from 46 early-stage patients were stained with anti-HSF1 antibodies and scored for HSF1 protein activation (relative nuclear staining intensity) in the stroma by immunohistochemistry. Association of stromal HSF1 activation with disease-free survival was assessed by KM analysis. *p<0.05. See also Figure S5 and Table S6.
HSF1 is often activated post-transcriptionally without a change in its mRNA level. To provide an indepenent assessment of the importance of its activation in breast cancer stroma, we assembled a new breast cancer cohort to evaluate HSF1 activation at the protein level by immunohistochemistry (IHC). We examined a total of 46 samples from patients with early stage breast cancer (Table S6), for whom we had both appropriate tissue sections as well as a minimum of 8 years of continuous clinical followup. Tumor sections were scored in a blinded manner for nuclear HSF1 staining intensity in the cancer cells and in the stromal cells.
We found markedly reduced disease-free survival, as well as overall survival, in patients whose tumors had high stromal HSF1 activation (Figure 5D & S5C). In this cohort, HSF1 activation in stromal cells was correlated with HSF1 activation in the cancer cells (p=0.01, Chi-square test). Indeed, high HSF1 activation in the cancer cells also corellated with lower overall survival, consistent with our previous findings (Mendillo et al., 2012; Santagata et al., 2011). However, the association with patient outcome was weaker in cancer cells than in the stroma (Figure S5D & E). Moreover, in a multivariate model considering the independent contributions of HSF1 activation in the cancer cells and in the stroma to overall survival, only stromal HSF1 (and not cancer-cell HSF1), was a significant predictor of survival (Table S6). Stromal HSF1 was also an independent, significant predictive factor in a multivariate model considering various clinicopathologic factors (Table S6). The significant association of stromal HSF1 activation with poor patient outcome seen in two independent cohorts using very different methodologies suggests that stromal HSF1 could be a useful, independent prognostic indicator in breast cancer.
HSF1 activation in early-stage lung cancer stroma is associated with poor outcome
Might stromal HSF1 serve as a potential prognostic marker in other tumor types? Our initial survey of human cancers showed that HSF1 is activated in the CAFs of many tumor types, including lung, colon, skin, esophageal, gastric and prostate (Figure 1C & 6A). Of these tumor types, we had access to a cohort of lung cancer patients with appropriate tissue samples for stromal assessment of HSF1, together with clinical follow-up data. Encouraging the analysis of this data set, pilot testing of human non-small cell lung cancer (NSCLC) cell lines (A549 & H1703), showed that these lines grew more robustly when co-cultured with WT MEFs than when co-cultured with Hsf1 null MEFs (Figure S6A).
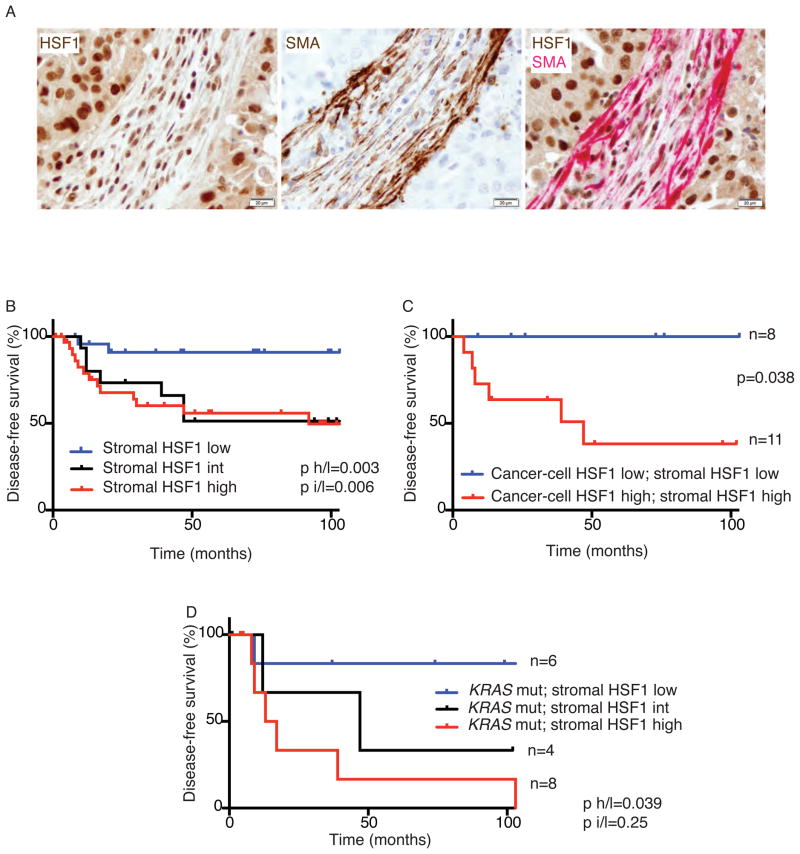
Figure 6. Increased HSF1 activation in the stroma is associated with decreased survival in lung cancer patients.
(A) Lung cancer resections from 5 patients were stained with anti-HSF1 (brown), anti-SMA (brown) or a combination of both antibodies (HSF1 in brown; SMA in red). Representative images are shown. Scale bar = 20μm (B–C) Lung cancer resections from 72 patients with Stage I disease were stained with anti-HSF1 antibodies and scored for HSF1 activation in the stromal cells and in the cancer cells. (B) HSF1 stromal scores are correlated with disease-free survival by KM analysis. (C) KM analysis of disease-free survival for patients with concordant high or low HSF1 scores in both stromal cells and cancer cells. (D) Stromal HSF1 levels in KRAS mutant tumors (n=18) from the lung cancer cohort correlate with disease-free survival by KM analysis. See also Figure S6 and Table S7.
A total of 72 samples from patients with stage I non-small cell lung adenocarcinoma (Table S7) (Sholl et al., 2010) were scored in a blinded manner for HSF1 activation (nuclear staining intensity) in cancer cells and stromal cells. Patients with stage I NSCLC have a 5-year survival of 60–70% (Goldstraw et al., 2007). Stromal HSF1 activation did not correlate with demographic factors such as age, sex or smoking status (Table S7). It did, however, show a significant correlation with patient outcome.
Disease-free survival was significantly shorter in lung cancer patients whose tumors had either high or intermediate HSF1 activity in the stroma (Figure 6B). A similar trend was found for survival of patients with high HSF1 in the cancer cells (Figure S6B). In this cohort, HSF1 activation in the cancer cells did not correlate with HSF1 activation in the stroma (p=0.28, Chi-square test). We therefore asked if evaluation of HSF1 activation in both cell types could improve our ability to predict patient outcome. Although the number of patients is small, it is striking that there was not a single recurrence in any of the patients that had low HSF1 activity in both the cancer cells and in the stroma over the course of followup (Figure 6C).
To assess the independent contributions to outcome of increased HSF1 activation in cancer cells versus stromal cells, we fitted a multivariate Cox proportional hazards regression model to recurrence-free survival, considering stromal HSF1 activation separately from cancer-cell activation. Cancer-cell HSF1 was not independent from stroma in its association with disease progression. However, as in our breast cancer cohort, stromal HSF1 activation was significantly and independently associated with disease-free survival (Table S7).
To further refine our analysis, we genotyped the collection of tumor samples for the most commonly mutated oncogenes in lung adenocarcinoma, KRAS and EGFR (Pillai and Ramalingam, 2014), and tested the association of HSF1 activation and disease outcome with different mutations. In the 52 samples from our cohort that were successfully genotyped, KRAS mutations, but not EGFR mutations, correlated with poor disease-free survival (Figure S6C & S6D). We found no correlation between HSF1 activation (in either cancer cells or stroma) and KRAS or EGFR mutations per se. We did, however, find a significant association between high activity of HSF1 in the stroma and poor outcome in patients with KRAS mutant tumors (Figure 6D). Moreover, stromal HSF1 (but not cancer-cell HSF1) was an independent predictor of progression-free survival in several multivariate models considering KRAS and EGFR mutational status as well as clinicopathologic factors (Table S7). These clinical association data suggest that HSF1 status could serve as a promising independent prognostic marker in lung cancer as well as breast cancer.
Discussion
For cancer cells to proliferate, invade and metastasize, they must recruit and reprogram non-malignant stromal cells. We find that HSF1 activation is a key factor in the transcriptional reprogramming of the stroma from a tumor-repressive environment to a supportive one. At least two central signaling pathways in the tumor microenvironment are empowered by HSF1 — pathways mediated by TGFβ and by SDF1. Establishing the relevance of our experimental findings to human disease, HSF1 was activated in the stroma of a wide variety of human cancers and this activation correlated strongly with poor outcome in both lung and breast cancer.
Our work establishes a role for stromal HSF1 in tumor biology that is distinct, yet highly complementary to, its recently established role in malignant cells. HSF1 has historically been viewed as a stress-activated transcription factor. In tumors, stromal and cancer cells alike must cope with a variety of potentially lethal challenges, including oxidative stress, nutrient-deprivation and protein misfolding. Yet neither the cancer-HSF1 program we previously reported in malignant cells (Mendillo et al., 2012), nor the stromal-HSF1 program we report here, is a simple reflection of these inevitable stresses.
The cancer-HSF1 program supports the malignant life style of cancer cells in a multitude of ways, including direct effects on cell cycle, DNA repair, anabolic metabolism and proliferation (Jin et al., 2011; Mendillo et al., 2012; Meng et al., 2010; Santagata et al., 2013). The stromal-HSF1 program, drives pathways that are of specific benefit to the malignant elements within the tumor. These pathways facilitate angiogenesis, ECM organization, adhesion and migration.
Clearly, HSF1 is capable of driving highly divergent transcriptional programs depending on the cellular context. One feature of these programs, which we have begun to unravel, is the way HSF1 responses are coordinated between cancer cells and stroma. We have found that TGFβ and SDF1 are two extracellular mediators of the HSF1 program in CAFs. While it was previously recognized that these proteins, when secreted by CAFs, enhance the pro-tumorigenic phenotype (Kojima et al., 2010), the factors responsible for their upregulation were unknown. HSF1 has been shown to directly bind to HSEs in the genes of several chemokines (Henderson and Kaiser, 2013; Maity et al., 2011) during heat-shock. Conversely, HSF1 can be activated by exposure to cytokines such as TGFβ and IL-1β in vitro (Sasaki et al., 2002). Taking these observations together, we suggest that reciprocal interactions between secreted cytokines and intracellular HSF1 programs that are normal responses to fever and infection have been co-opted by diverse cell types in tumors to fuel the malignant state.
But how did such non-cell-autonomous HSF1 programs evolve? The HSF1-dependent heat-shock response has traditionally been conceived as an internally-driven cellular response to proteotoxic stress. However, recent work in C. elegans has established that HSF1 can be activated in a non-cell-autonomous manner. Acute stresses detected by thermosensory neurons can orchestrate HSF1-dependent heat-shock responses throughout the animal. This coordinated response benefits the organism as a whole (Prahlad et al., 2008). Similarly, in tumors, cancer cells induce the activation of HSF1 in the stroma, and this activation benefits the tumor as a whole (albeit to the detriment of the patient). But in addition to this, in stroma the HSF1-regulated program itself is non-cell-autonomous. It results in secretion of factors that act to enhance the survival and proliferation of neighboring cancer cells. We suggest that the interplay between HSF1 responses in cancer cells and stroma have their origins in ancient biological mechanisms that act to promote the survival of multicellular organisms in a non-cell-autonomous way.
The complementary but distinct roles of HSF1 in cancer and stromal cells of tumors have both diagnostic and therapeutic implications. From a diagnostic perspective, assessing HSF1 in both stromal and cancer cells might help to guide treatment choices in early stage cancers, especially lung cancer, where currently there are no reliable markers for gauging malignant potential other than tumor size. The increased surveillance of patients at high risk to develop lung cancer is creating an acute need for markers that can predict which early-stage tumors are most likely to progress, in order to avoid over-treatment and its associated morbidities. The wide spread activation of stromal HSF1 in diverse cancers suggests it might be a useful biomarker in other tumor types as well, as we have shown for breast cancer. From a therapeutic perspective, the dependence of even the most robust cancers on supporting stromal cells, and the relative genetic stability of the stroma, make HSF1 an attractive target for intervention in both cancer cells and stroma (Bissell and Hines, 2011; Luo et al., 2009; Place et al., 2011; Saturno et al., 2013; Whitesell and Lindquist, 2009). As we and others have suggested, the nearly unthwartable ability of advanced cancers to evolve resistance to virtually every available therapy makes it attractive to target normal biological networks that have been co-opted to support malignancy, rather than relying solely on the targeting of mutated malignant drivers.
Experimental procedures
Cell Culture
D2A1, 4T7, MCF7 and MEFs were cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (FBS). HCC38, A549 and H1703 cells were cultured in RPMI with 10% FBS. For co-culture, immortalized MEFs were plated at near confluency, and 24h later treated, where indicated, with 10 μg/ml mitomycin C (Sigma) for 2h and washed with PBS. Cancer cells were seeded on top of the MEFs (1:5 ratio of cancer cells:MEFs), and allowed to grow for 72h–96h. Where indicated, MEFs were incubated with LY2109761 (1 μM, Selleck chemicals) for 30 minutes before seeding of cancer cells. The same concentration of inhibitor was then added daily. Recombinant TGFβ1 (R&D systems, 240-B-002) and SDF1 (R&D systems, 460-SD-010) were added to co-cultures at the indicated concentrations once, when co-culture was started.
Bi-Tet-Hsf1 MEFs
Bi-Tet-Hsf1 MEFs were constructed as explained in Extended Experimental Procedures. Where indicated, cells were treated with 2 μg/ml doxycycline to inhibit Hsf1 expression.
Flowcytometry
For expression profiling, co-cultures were sorted using a FACS-Aria (BD-biosciences) instrument, as explained in Extended Experimental Procedures. For all other experiments, a Guava EasyCyte (Millipore) cytometer was used, 10000 cells/sample were analyzed and the fraction of cancer cells was calculated using FlowJo 8.8.7 software.
Gene Expression Analysis
Duplicate RNA samples were reverse transcribed and hybridized to duplicate SurePrint Agilent microarrays (Agilent, G4852A). Data were analyzed using Cluster, GOrilla, and MSigDB, and visualized using Java TreeView (Details in Extended Experimental Procedures). Microarray raw data was deposited in a public database (Gene Expression Omnibus (GEO) accession GSE56252).
shRNA knockdown of genes in the TGFβ and SDF1 signaling networks
The following genes were stably knocked down, in D2A1 cells and in MEFs: Smad2, Smad3, Smad4, TgfβR2 (Details in Extended Experimental Procedures).
Xenografts
MCF7 cells (1×106) were inoculated subcutaneously in the right inguinal region of each mouse. Where indicated, 1×106 MCF7 cells were mixed with 3×106 wt or Hsf1 null primary MEFs prior to injection. Tumor growth was monitored by caliper measurements twice weekly. Mice were sacrificed and tumors were excised when volume reached 1.5 cm3 or overlying skin became ulcerated. Half the resected tissue was flash frozen for ChIP and half fixed in 10% formalin, processed using standard methods, cut into 5mm sections and immunostained as described below.
ChIP-qPCR
Flash-frozen tumor xenografts (0.5 cm3 each) were pulverized, fixed in formalin and processed as described previously (Lee et al., 2006; Mendillo et al., 2012). anti-HSF1 (Thermoscientific, RT-629-PABX) was used to IP HSF1, and normal rat IgG (Jackson ImmunoResearch Laboratories, 012-000-003) was used as control. qPCR was performed using the primers listed in Extended Experimental Procedures.
Immunohistochemistry of tissues, scoring and patient outcome analysis
Paraffin blocks and tissue microarrays were retrieved, processed, stained and scored as described in Extended Experimental Procedures. Outcome analysis was performed on 46 breast cancer patients, and 72 lung cancer patients. Time to progression of disease and overall survival were estimated by the Kaplan-Meier (KM) method using GraphPad Prism 6 software. Unless indicated otherwise the log-rank test was used to assess statistical significance. All statistical tests were two-sided, p<0.05 was considered statistically significant. Multivariate Cox proportional hazards regression analysis was performed using the coxph function in the survival package in R (http://www.r-project.org/).
EGFR and KRAS Genotyping
Total nucleic acid was extracted from formalin-fixed, paraffin-embedded surgical specimens of the lung cohort described above using a modified FormaPure System (Agencourt Bioscience Corporation, Beverly, MA). SNaPshot mutational analysis of a panel of cancer genes that included EGFR and KRAS was performed using primers listed in the Extended Experimental Procedures as previously described (Dias-Santagata et al., 2010).
Stromal Hsf1 mRNA profiling and patient outcome analysis
Stromal gene expression profiling data and clinical outcome for 53 breast cancers were obtained from GEO (GSE9014) and the Finak et al. study (Finak et al., 2008). Analysis was performed as explained in Extended Experimental Procedures. Survival distributions in the stromal Hsf1-low and stromal Hsf1-high patients were compared by KM analysis. The associations between Hsf1 expression, tumor grade and molecular subtype are presented by box & whisker plots. Statistical significance was assessed with the log-rank test using GraphPad Prism 6. All statistical tests were two-sided, p<0.05 was considered significant.
Supplementary Material
Highlights.
Reprogramming of cancer-associated fibroblasts by HSF1 enables malignant progression
Distinct transcriptional programs are driven by HSF1 in stromal vs. malignant cells
HSF1 activation in tumor stroma is strongly associated with poor patient outcome
Dual roles in both tumor cells and stroma make HSF1 an attractive anticancer target
Acknowledgments
We thank I. Barrasa, G. Bell, S. Gupta, and T. DiCesare for bioinformatic analysis and graphical assistance. We thank M. Duquette and A. Topolszky (WIBR), K. Lynch (MGH) and A. Tuvar (Rabin Medical Center) for technical assistance. We thank C.K. Dai for cloning Bi-Tet-Hsf1, and R. Jaenisch and Y. Freyzon for construction of Bi-Tet-Hsf1 transgenic mice. We thank K. Polyak, P. Gupta, D. Pincus, L. Clayton and members of the Lindquist lab for helpful discussions and comments. We thank R. Weinberg for the gift of cell-lines and for helpful discussions. RSS was supported by the Human Frontiers Science Program, the Fulbright Program and the Israel National Postdoctoral Award Program for Women in Science. SS is supported by the Jared Branfman Sunflowers for Life Fund, the V Foundation and by NIH grant K08 NS064168. MLM was supported by the National Cancer Institute of the National Institutes of Health under Award Number K99CA175293. LW was supported by the Komen Foundation, Grant KG110450 and by the J&J COSAT focused funding program. SL is an investigator of the Howard Hughes Medical Institute. Support for this study was also provided by the Alexander and Margaret Stewart Trust and the Koch Center at MIT. DDS is a consultant for Bioreference Labs, submitted a patent application (pending) for the SNaPshot methods described here, has licensed SNapshot technology and receives royalties from Bioreference Labs.
Footnotes
Author Contributions
RSS, SL and LW conceived the project, designed experiments and wrote the paper. RSS performed all mouse and cell culture experiments, analyzed data and made the Figures. SS performed all patient-sample staining and scoring and provided clinical guidance. LMS assembled the lung cancer patient cohort, performed sample scoring and provided clinical outcome data. IBA and SMS assembled the breast cancer patient cohort and provided clinical outcome data. MLM analyzed microarray data, and MK assisted. AHB performed statistical analysis of patient data. DDS performed genotyping of lung cancer patient samples.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beck AH, Espinosa I, Gilks CB, van de Rijn M, West RB. The fibromatosis signature defines a robust stromal response in breast carcinoma. Lab Invest. 2008;88:591–601. doi: 10.1038/labinvest.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell MJ, Hines WC. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, Chi JT, van de Rijn M, Botstein D, Brown PO. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Santagata S, Tang Z, Shi J, Cao J, Kwon H, Bronson RT, Whitesell L, Lindquist S. Loss of tumor suppressor NF1 activates HSF1 to promote carcinogenesis. J Clin Invest. 2012;122:3742–3754. doi: 10.1172/JCI62727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias-Santagata D, Akhavanfard S, David SS, Vernovsky K, Kuhlmann G, Boisvert SL, Stubbs H, McDermott U, Settleman J, Kwak EL, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. Embo Mol Med. 2010;2:146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dituri F, Mazzocca A, Peidro FJ, Papappicco P, Fabregat I, De Santis F, Paradiso A, Sabba C, Giannelli G. Differential Inhibition of the TGF-beta Signaling Pathway in HCC Cells Using the Small Molecule Inhibitor LY2157299 and the D10 Monoclonal Antibody against TGF-beta Receptor Type II. Plos One. 2013;8:e67109. doi: 10.1371/journal.pone.0067109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315:1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- Erez N, Truitt M, Olson P, Arron ST, Hanahan D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer Cell. 2010;17:135–147. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- Fang F, Chang R, Yang L. Heat shock factor 1 promotes invasion and metastasis of hepatocellular carcinoma in vitro and in vivo. Cancer. 2012;118:1782–1794. doi: 10.1002/cncr.26482. [DOI] [PubMed] [Google Scholar]
- Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V, Sobin L. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2:706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Henderson B, Kaiser F. Do reciprocal interactions between cell stress proteins and cytokines create a new intra-/extra-cellular signalling nexus? Cell Stress Chaperones. 2013 doi: 10.1007/s12192-013-0444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Moskophidis D, Mivechi NF. Heat shock transcription factor 1 is a key determinant of HCC development by regulating hepatic steatosis and metabolic syndrome. Cell Metab. 2011;14:91–103. doi: 10.1016/j.cmet.2011.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- Kojima Y, Acar A, Eaton EN, Mellody KT, Scheel C, Ben-Porath I, Onder TT, Wang ZC, Richardson AL, Weinberg RA, et al. Autocrine TGF-beta and stromal cell-derived factor-1 (SDF-1) signaling drives the evolution of tumor-promoting mammary stromal myofibroblasts. Proc Natl Acad Sci U S A. 2010;107:20009–20014. doi: 10.1073/pnas.1013805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Akkari L, Simon J, Grace D, Tschaharganeh DF, Bolden JE, Zhao Z, Thapar V, Joyce JA, Krizhanovsky V, et al. Non-cell-autonomous tumor suppression by p53. Cell. 2013;153:449–460. doi: 10.1016/j.cell.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity TK, Henry MM, Tulapurkar ME, Shah NG, Hasday JD, Singh IS. Distinct, gene-specific effect of heat shock on heat shock factor-1 recruitment and gene expression of CXC chemokine genes. Cytokine. 2011;54:61–67. doi: 10.1016/j.cyto.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM, Fraenkel E, Ince TA, Whitesell L, Lindquist S. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150:549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Gabai VL, Sherman MY. Heat-shock transcription factor HSF1 has a critical role in human epidermal growth factor receptor-2-induced cellular transformation and tumorigenesis. Oncogene. 2010;29:5204–5213. doi: 10.1038/onc.2010.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovits N, Kalinkovich A, Bar J, Lapidot T, Oren M. p53 Attenuates cancer cell migration and invasion through repression of SDF-1/CXCL12 expression in stromal fibroblasts. Cancer Res. 2006;66:10671–10676. doi: 10.1158/0008-5472.CAN-06-2323. [DOI] [PubMed] [Google Scholar]
- Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59:5002–5011. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Pickup M, Novitskiy S, Moses HL. The roles of TGFbeta in the tumour microenvironment. Nat Rev Cancer. 2013;13:788–799. doi: 10.1038/nrc3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RN, Ramalingam SS. Advances in the diagnosis and treatment of non-small cell lung cancer. Mol Cancer Ther. 2014;13:557–564. doi: 10.1158/1535-7163.MCT-13-0669. [DOI] [PubMed] [Google Scholar]
- Place AE, Jin Huh S, Polyak K. The microenvironment in breast cancer progression: biology and implications for treatment. Breast Cancer Res. 2011;13:227. doi: 10.1186/bcr2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlad V, Cornelius T, Morimoto RI. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 2008;320:811–814. doi: 10.1126/science.1156093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Hu M, Sridhar A, Opeskin K, Fox S, Shipitsin M, Trivett M, Thompson ER, Ramakrishna M, Gorringe KL, et al. No evidence of clonal somatic genetic alterations in cancer-associated fibroblasts from human breast and ovarian carcinomas. Nat Genet. 2008;40:650–655. doi: 10.1038/ng.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quante M, Tu SP, Tomita H, Gonda T, Wang SS, Takashi S, Baik GH, Shibata W, Diprete B, Betz KS, et al. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell. 2011;19:257–272. doi: 10.1016/j.ccr.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H, Enoki Y. Novel aspects of heat shock factors: DNA recognition, chromatin modulation and gene expression. FEBS J. 2010;277:4140–4149. doi: 10.1111/j.1742-4658.2010.07829.x. [DOI] [PubMed] [Google Scholar]
- Santagata S, Hu R, Lin NU, Mendillo ML, Collins LC, Hankinson SE, Schnitt SJ, Whitesell L, Tamimi RM, Lindquist S, et al. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc Natl Acad Sci U S A. 2011;108:18378–18383. doi: 10.1073/pnas.1115031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S, Mendillo ML, Tang YC, Subramanian A, Perley CC, Roche SP, Wong B, Narayan R, Kwon H, Koeva M, et al. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science. 2013;341:1238303. doi: 10.1126/science.1238303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S, Xu YM, Wijeratne EM, Kontnik R, Rooney C, Perley CC, Kwon H, Clardy J, Kesari S, Whitesell L, et al. Using the heat-shock response to discover anticancer compounds that target protein homeostasis. Acs Chem Biol. 2012;7:340–349. doi: 10.1021/cb200353m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Sato T, Yamauchi N, Okamoto T, Kobayashi D, Iyama S, Kato J, Matsunaga T, Takimoto R, Takayama T, et al. Induction of heat shock protein 47 synthesis by TGF-beta and IL-1 beta via enhancement of the heat shock element binding activity of heat shock transcription factor 1. J Immunol. 2002;168:5178–5183. doi: 10.4049/jimmunol.168.10.5178. [DOI] [PubMed] [Google Scholar]
- Saturno G, Valenti M, De Haven Brandon A, Thomas GV, Eccles S, Clarke PA, Workman P. Combining Trail with PI3 Kinase or HSP90 inhibitors enhances apoptosis in colorectal cancer cells via suppression of survival signaling. Oncotarget. 2013;4:1185–1198. doi: 10.18632/oncotarget.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KL, Nogueira C, Heffernan TP, van Doorn R, Dhakal S, Hanna JA, Min C, Jaskelioff M, Xiao Y, Wu CJ, et al. Proinvasion metastasis drivers in early-stage melanoma are oncogenes. Cancer Cell. 2011;20:92–103. doi: 10.1016/j.ccr.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamovsky I, Nudler E. New insights into the mechanism of heat shock response activation. Cell Mol Life Sci. 2008;65:855–861. doi: 10.1007/s00018-008-7458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl LM, Barletta JA, Yeap BY, Chirieac LR, Hornick JL. Sox2 protein expression is an independent poor prognostic indicator in stage I lung adenocarcinoma. Am J Surg Pathol. 2010;34:1193–1198. doi: 10.1097/PAS.0b013e3181e5e024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimboli AJ, Cantemir-Stone CZ, Li F, Wallace JA, Merchant A, Creasap N, Thompson JC, Caserta E, Wang H, Chong JL, et al. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature. 2009;461:1084–1091. doi: 10.1038/nature08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L, Lindquist S. Inhibiting the transcription factor HSF1 as an anticancer strategy. Expert Opin Ther Tar. 2009;13:469–478. doi: 10.1517/14728220902832697. [DOI] [PubMed] [Google Scholar]
- Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, Peng J, Lin E, Wang Y, Sosman J, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.