Significance
The organization of long genomes in the confined spaces of a cell requires special facilitating mechanisms. A variety of architectural proteins play key roles in these processes. The bacterial heat-unstable (HU) protein helps to condense DNA by introducing sharp turns along its pathway. The protein binds in a sequence-neutral fashion, randomly distorting linear DNA when introduced in computer-simulated structures at levels comparable to those found in the cell. The natural resistance of DNA to severe deformation, however, restricts the nonspecific protein to specific loci when the molecule is covalently closed or looped by a protein. The interplay of DNA topology and protein-induced bending provides insights into ways in which gene fragments may be organized and linked to biological function.
Keywords: DNA cyclization, lac operon, Monte Carlo simulation, nonspecific binding, protein-mediated looping
Abstract
Topological constraints placed on short fragments of DNA change the disorder found in chain molecules randomly decorated by nonspecific, architectural proteins into tightly organized 3D structures. The bacterial heat-unstable (HU) protein builds up, counter to expectations, in greater quantities and at particular sites along simulated DNA minicircles and loops. Moreover, the placement of HU along loops with the “wild-type” spacing found in the Escherichia coli lactose (lac) and galactose (gal) operons precludes access to key recognition elements on DNA. The HU protein introduces a unique spatial pathway in the DNA upon closure. The many ways in which the protein induces nearly the same closed circular configuration point to the statistical advantage of its nonspecificity. The rotational settings imposed on DNA by the repressor proteins, by contrast, introduce sequential specificity in HU placement, with the nonspecific protein accumulating at particular loci on the constrained duplex. Thus, an architectural protein with no discernible DNA sequence-recognizing features becomes site-specific and potentially assumes a functional role upon loop formation. The locations of HU on the closed DNA reflect long-range mechanical correlations. The protein responds to DNA shape and deformability—the stiff, naturally straight double-helical structure—rather than to the unique features of the constituent base pairs. The structures of the simulated loops suggest that HU architecture, like nucleosomal architecture, which modulates the ability of regulatory proteins to recognize their binding sites in the context of chromatin, may influence repressor–operator interactions in the context of the bacterial nucleoid.
DNA behaves differently in a cellular milieu than in aqueous salt solution. Proteins attach to widely spaced sites along genomic sequences in vivo and force the intervening DNA into loops much shorter in length than those expected from the natural deformational properties of the double helix. For example, the control of gene expression in Escherichia coli entails the formation of short DNA loops, some ∼100 bp steps in length, that preclude access of RNA polymerase to the signals needed to transcribe the enzymes encoded within various operons (1, 2). DNA of the same length tends to be highly extended in vitro, with very low chances of closing spontaneously into a loop (3, 4).
Aside from the proteins that hold specific sequences at the ends of short DNA loops in place, the cellular environment includes a number of other components that influence the properties of DNA loops. For example, the naturally abundant, nonspecific E. coli heat-unstable (HU) protein plays important roles both in cellular processing and in shaping the architecture of the bacterial nucleoid. The dimeric protein introduces large deformations in DNA—sharp bends, helical untwisting, and helical axis dislocation (5)—that are central to its activity. The binding of HU to an AT-rich sequence element stabilizes loops important for repression of the galactose (gal) operon (6), and its absence weakens repression of the lactose (lac) operon (7). The loss of protein also leads to nucleoid instability and growth defects (8, 9).
Our recent studies of the structures of DNA loops mediated by the Lac repressor, the tetrameric protein assembly that controls the expression of lac genes in E. coli, show how the deformations of the double helix found in known high-resolution complexes of HU with DNA compensate for the intrinsic stiffness of short DNA (10, 11). The random binding of the protein, at levels approximating those found in vivo, brings the predicted looping propensities in line with values deduced from gene-expression studies. The introduction of low levels of HU similarly enhances the formation of DNA minicircles (12), accounting for the very small rings detected in DNA ligation studies (13, 14) and the ease of cyclization determined for longer chains at different concentrations of HU (15).
Closer investigation of the structures captured in the course of these simulations and in new studies of Gal repressor-mediated looping reveals how the random binding of HU organizes the folding of DNA loops and minicircles. Here we show how the nonspecific-binding protein builds up, counter to expectations, in greater quantities and localizes at particular sites along the DNA. Moreover, the placement of HU along loops with the spacing found in the lac and gal operons precludes access to key recognition elements on DNA. The architectural protein reconfigures the pathways of the repressor-mediated DNA loops and introduces a structural barrier to operon accessibility. The DNA appears to guide the placement of proteins and to take advantage of the nonspecificity of HU binding. The predicted placement of HU accounts for the known specificity of the protein on Gal repressor-mediated loops (6) and hints of a similar role for the architectural protein in wild-type loops attached to the Lac repressor. The computations show how a protein with no discernible DNA sequence-recognizing features becomes site-specific upon loop formation and point to the statistical advantage of nonspecific protein binding in effecting DNA ring closure.
Results
Nonspecific Proteins Accumulate at Unexpected Levels and Locations on Circular DNA.
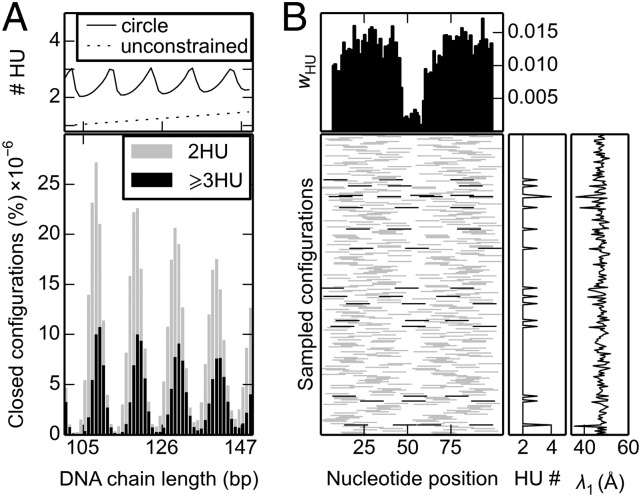
The random binding of HU has a number of surprising effects on the simulated structures of short DNA minicircles. First of all, minicircles collected from large ensembles of linear, unconstrained double-helical molecules bind more nonspecific proteins than anticipated from the imposed binding probabilities. The average number of HU dimers found on 100–150-bp cyclic DNA species exceeds the number accumulated on ensembles of spatially unconstrained chains of the same length (Fig. 1A). The enhanced levels of HU on circular DNA are especially pronounced at short DNA chain lengths and at low HU-binding levels, confined in this example to values of one randomly attached protein dimer every 100 bp of DNA.
Fig. 1.
Enhanced build-up and global organization of HU dimers randomly bound on DNA minicircles. (A) Average number and relative distribution of HU dimers, as a function of chain length, in closed chains generated under binding conditions of one HU per 100 bp. (B) Mosaic of the positions of bound HU on representative 105-bp rings collected under the same conditions. The average number of dimers (#HU) on chains of a given length, in A, and the likelihood wHU of finding a protein centered at a given nucleotide position, in B, are depicted, respectively, by the solid lines and histograms in the Upper plots. The greater binding occupancy on the closed chains is clear from comparison with the degree of binding in unconstrained ensembles of all (linear and cyclic) HU-bound chains of the same length (dotted line in A). The distribution of closed chains, expressed as a percentage of the simulated sample size in A, illustrates the dependence of ring closure on chain length. The number of dimers (HU#) of bound proteins and the length λ1 of the longest principal axis, plotted to the right of each entry in B, reveal the tight spatial organization of the closed duplexes, primarily elongated states with two antipodal HU dimers and occasional less extended configurations with three or more bound proteins (denoted, respectively, by the light gray and black bars in the Lower images).
Unlike the uniform placement of HU dimers on open DNA molecules, the spacing of the protein on cyclized duplexes is not uniform. That is, DNA ring closure induces a preference for HU to bind at particular sites on the closed duplex or, more precisely, at specific separation distances along the chain. For example, most of the successfully closed 105-mers adopt elongated, nearly planar configurations with two copies of HU located at the apices of the structures (Fig. 2A). Significantly, none of the 105-mers captured in the simulations binds less than two HU dimers, and a small fraction takes up three or more dimers (Fig. 1B). Elongated configurations also dominate the closed states of longer HU-bound DNA rings although structures of this type constitute a smaller proportion of the modeled configurations (SI Appendix, Fig. S3).
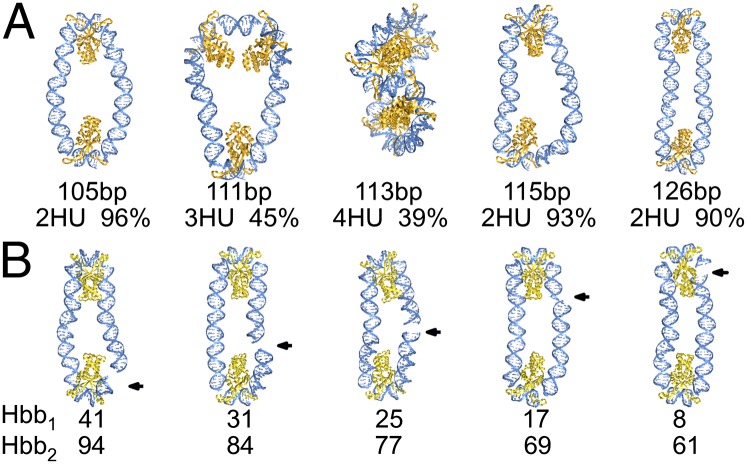
Fig. 2.
Representative molecular images, rendered in PyMOL (www.pymol.org), illustrating (A) the dominant spacings of HU on DNA minicircles of different lengths and (B) the variable sites of covalent chain closure, noted by arrows, relative to the positions of protein on 105-bp Hbb-decorated rings. Structures are aligned such that the longest principal axes are vertical, and, except for the 113-bp minicircle bearing four HU dimers, the shortest axes are perpendicular to the plane of the page. Chain lengths and populations with the depicted number of HU molecules and sequential positions of Hbb centers are denoted below the respective images. DNA is shown as a blue ribbon with attached bases, HU as a dark-gold ribbon, and Hbb as a light-gold ribbon.
In general, for minicircles with two bound HU dimers, the proteins lie at antipodal positions separated by roughly integral helical repeats. If the chain length is an even multiple of the 10.5-bp DNA repeat, the binding sites tend to be spaced midway along the chain contour. For example, the vast majority of HU dimers found on 105-bp minicircles lie five helical turns apart, i.e., distances corresponding to half the respective B-DNA contour lengths (SI Appendix, Fig. S4). The protein positioning on minicircles with an odd number of helical repeats is slightly asymmetric, with spacings equal to the two integral helical repeats that bracket half the contour length. For example, the two HU dimers on rings of 115 or 116 bp, ∼11 helical repeats of B-DNA, are separated by five or six helical turns (SI Appendix, Fig. S4).
The number and placement of HU dimers is quite different on minicircles comprising half-integral B-DNA repeats. Chains of these lengths tend to cyclize somewhat less easily (11, 12) and to take up additional proteins upon ring closure (Fig. 1A). The added proteins help to relieve the bending and torsional stress needed to bring the ends of the DNA in alignment for chain ligation. The placement of proteins on the closed rings takes advantage of both the symmetry of the system and the DNA double-helical repeat. For example, the three HU molecules associated with nearly half of the 111-bp minicircles are fairly evenly spaced on the DNA, with the DNA folded into tight triangular configurations with “sides” one and three B-DNA repeats in length (Fig. 2A). Similarly, the separation distances between the four HU molecules found in many 113-bp minicircles are roughly constant, with the DNA wrapped in a tight “figure 8,” and each lobe of the figure 8 binding two proteins separated by two or three helical turns (Fig. 2A). The ∼11° untwisting associated with each 14-bp HU-bound DNA fragment leads to a 2–3 bp shift in the lengths of protein-decorated chains most readily closed into duplexes compared with HU-free DNA, e.g., 108- or 118-bp circles with two or three HU dimers versus 105 or 115–116 bp of bare B-DNA.
The relative frequency of configurations with three or more HU dimers increases in larger minicircles as the average number of bound HU molecules builds up with chain length (Fig. 1A and SI Appendix, Figs. S5 and S6). The different proportions of HU give rise to a slight phase shift in the computed chain-length dependence of ring closure compared with that of bare DNA (11, 12).
DNA Guides the Positioning of HU on Short Minicircles.
The nonspecificity of HU binding allows the architectural protein to accumulate at any site along DNA. The random placement and enhanced numbers of dimers on successfully closed minicircles stand out in mosaics of HU-occupied positions (Fig. 1B). Here we depict the sequential locations of bound proteins on 105-bp minicircles relative to the site of chain ligation, i.e., the terminal base pairs of the modeled chains. The random build-up of protein leads, as expected, to uniform binding-site occupancies, as measured by the average number of HU dimers centered on individual base pairs in the set of closed chains. The dip in HU occupancy at the ends and middle of the DNA arises from the implicit presence of DNA ligase on a naturally straight chain segment at the sites of ring closure and the consequent drop in bound HU at the antipodal site (see Supporting Information for details). The uniformity in global chain configuration, as measured by the lengths of the principal axes of the DNA rings (Fig. 1B and SI Appendix, Fig. S7), stems from the regular spacing between successively bound proteins in the computed structures, each introducing a significant (120° ± 10°) bend in the DNA (5).
The spacing of proteins at integral helical repeats allows the DNA to remain in the vicinity of its minimum-energy rest state. In this sense, the mechanics of DNA, i.e., the tendency for the double helix to remain naturally straight with 10.5 residues per turn, dictates the positions of proteins along the closed molecules. The proteins, in turn, determine the degree of DNA deformation, e.g., the magnitude of local bending, and thereby control the overall shape of the molecular complex. For example, substitution of the two HU dimers found in most 105-bp minicircles by the structurally related, albeit more strongly DNA-distorting HU-like protein from Borrelia burgdorferi (Hbb) (16), changes the smooth, oval pathways induced by HU to more tightly closed configurations with the protein-free DNA segments brought into nearly perfect antiparallel alignment by the ∼160° bending of DNA by Hbb (Fig. 2B and SI Appendix, Fig. S8).
Significantly, the configurations of closed circular DNA brought about by the nonspecific binding of HU remain the same regardless of simulation conditions. That is, the uptake of protein on DNA yields the same few overall folds regardless of the assumed binding probability. Changes in the likelihood of HU binding affect the fraction of minicircles with a given number of bound dimers but not the shapes of the HU-bound DNA. For example, although there is a twofold drop in the fraction of 103-bp rings bearing two HU dimers with an increase in the binding probability from one HU per 100 bp to one per 50 bp, the writhing numbers of the minicircles with specific numbers of HU dimers remain the same—e.g., average values of 0.09 ± 0.11 for the duplexes bearing two proteins and –0.12 ± 0.31 and –0.14 ± 0.31 for those bearing three proteins under the respective binding conditions. The different proportion of minicircles with two, three, and four HU dimers, however, results in a difference in the average writhing numbers for the ensembles of closed HU-bearing chains, e.g., values of 0.03 ± 0.20 and –0.12 ± 0.27 for 103-bp rings generated at the lower and higher binding probabilities. The principal axes of the short minicircles are similarly “quantized” (SI Appendix, Fig. S7). The DNA chain length and level of protein binding consequently influence the distribution of topological states, or topoisomers, of the closed molecules. The difference, ΔLk, in the linking number of the HU-decorated minicircles compared with that of a relaxed protein-free molecule of the same length, is greater in the rings that bind more HU on average. These chains are negatively supercoiled (ΔLk < 0) and less easily cyclized. The molecules that bind only two HU dimers are nearly fully relaxed with approximately zero excess link (ΔLk = 0) and are more easily closed into minicircles.
The many ways in which a nonspecific protein can attach to DNA enhance the likelihood of ring closure. The simulated formation of 100–150 bp, HU-decorated minicircles is orders of magnitude greater than the probability of covalently closing a bare DNA molecule of the same chain length (11, 12). The protein not only binds at two or more sites on the closed rings but also occupies all possible combinations of binding sites with a given spacing, thereby providing many different ways to achieve ring closure (Fig. 2B). Proteins with specific DNA recognition sites do not have this capability. Thus, under comparable conditions—e.g., similar on/off rates of DNA association and protein concentrations—a nonspecific binding protein has a statistical advantage in effecting ring closure over a protein that introduces comparable deformations of DNA at a specific binding site.
HU Reconfigures the Pathways of Wild-Type Repressor-Mediated DNA Loops.
Although the spatial constraints placed on short DNA fragments looped between the binding sites of the Lac and Gal repressors differ from those associated with ring closure (10), the propensity for DNA to adopt a naturally straight double-helical form continues to determine the sites of preferential HU build-up on the spatially constrained duplex. The positioning of the architectural protein, in turn, influences the way in which DNA attaches to the loop-mediating protein (10, 11). For example, in the absence of HU, DNA loops with the 92-bp wild-type spacing found in the lac operon tend to bind in antiparallel orientations to the binding headpieces of a V-shaped model of the Lac repressor (10, 17, 18). The looped segments make gradual U-turns with build-up of curvature on the 5′- or 3′-half of the DNA, so-called A1- or A2-type looping in which the bound DNA operators point in roughly opposing spatial directions (19) (Fig. 3 and SI Appendix, Fig. S9). The random binding of HU shifts the orientational preference such that nearly 90% of the HU-bound loops attach to the repressor in a parallel orientation (with operators running in nearly the same direction) (10). Moreover, the random binding of HU perturbs the fold of the DNA. Nearly all of the loops that closed in a parallel orientation on the Lac repressor bind two HU dimers, most of which are separated by two helical turns of straight DNA (Fig. 3 and SI Appendix, Fig. S10). These “flattened” and globally reoriented loops dominate the configurational landscape and contribute to the large increase in the J-factor of looping found in the presence vs. absence of HU (10).
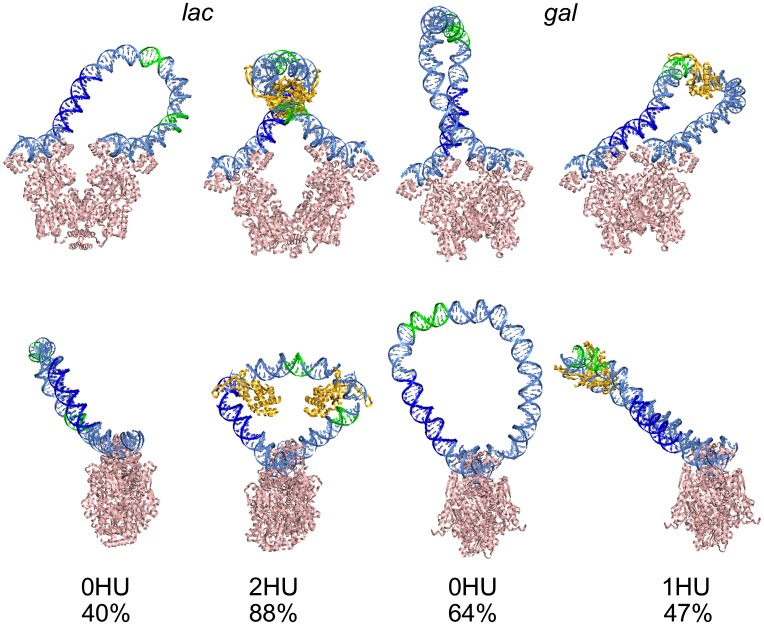
Fig. 3.
Molecular snapshots of wild-type Lac and Gal repressor-mediated DNA loops captured in the absence and presence of HU. Views perpendicular to and down the vector connecting the centers of bound operators (Upper and Lower images, respectively) on structures representative of the states that dominate the respective configurational landscapes. Color-coding of regulatory elements (CAP in deep blue, –35 and –10 sites on lac and P1/P2 on gal in green) reveals the potential structural barrier to transcription brought about by specifically placed HU dimers. Images show the changes in loop orientation and shape that accompany the uptake of HU. The numerical values denote the proportion of simulated loops in the illustrated forms.
By contrast, the 113-bp wild-type loops of bare DNA anchored to the Gal repressor preferentially adopt an ℓ-shaped configuration with a tight U-turn centered near the midpoint of the loop, so-called P1 looping (19) (Fig. 3). As with the Lac repressor, the random uptake of HU on the DNA changes the orientational preference of Gal repressor–operator association. Nearly three-fourths of the simulated HU-decorated loops attach to the protein in an antiparallel orientation and two-thirds of these loops adopt the A1 form previously found to be of lowest energy when HU is restricted to the known highest-affinity binding site (19). There is no such constraint on the site of HU occupancy in the present work. The localized build-up of HU reflects the physical properties of the DNA model and the structural deformations imposed by the associated proteins. The opposing changes in loop type with the uptake of HU on the Gal vs. Lac systems—P1 to A1 vs. A1/A2 to P1, respectively—stem from the different lengths of the DNA loops and the different locations of the anchoring points on the two proteins (SI Appendix, Fig. S1 and Table S1).
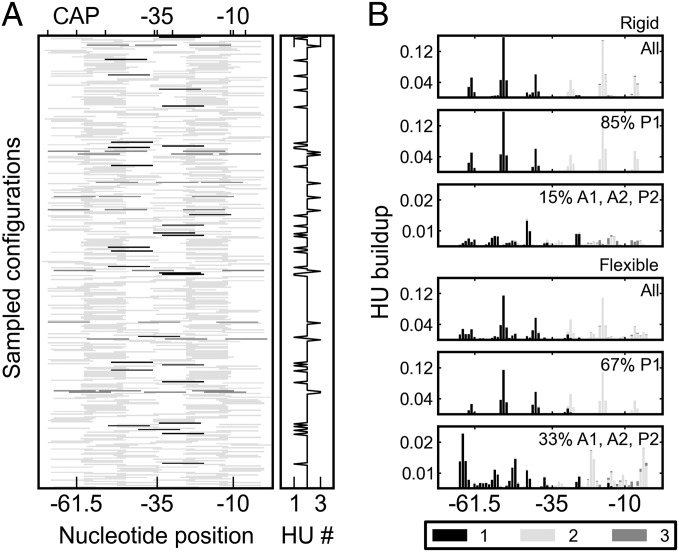
In contrast to the uniform build-up of HU on minicircles, the architectural protein accumulates at specific sites and with nearly fixed spacing along both kinds of wild-type repressor-mediated DNA loops. For example, most of the HU pairs in the 92-bp loops successfully anchored to the Lac repressor (>60%) are symmetrically positioned, with the centers of the bound HU proteins located ∼30 bp from the ends of the loop, at the operator centers (Figs. 3 and 4). Some dimer pairs occupy sites roughly a helical turn away from the dominant location; i.e., HU centers ∼20 or 40 bp from the 5′-end of the 92-bp loop, while preserving the ∼35-bp center-to-center spacing of HU (two turns of protein-free DNA flanked by the half-sites of each HU). The pathway of DNA on the Lac repressor introduces a rotational constraint on the loop not present in the minicircles. In contrast to the “free” rotation of DNA base pairs around the helical axis of a minicircle, the binding of the repressor determines which segments of DNA lie on the inner and outer surfaces of the loop. Thus, the positions of HU are restricted to sites where the bending of DNA is in phase with the repressor-induced rotational setting.
Fig. 4.
Localized HU build-up on simulated Lac repressor-mediated DNA loops. (A) Mosaic of HU-binding sites on representative wild-type fragments anchored to the headpieces of a rigid, V-shaped Lac repressor assembly. (B) Distributions of binding-site occupancy on loops attached to the rigid repressor and to a flexible model that allows for opening between the two dimeric halves of the repressor. The entries in B are shaded to denote the positions occupied by the first (black), second (light gray), and third (dark gray) proteins on the simulated structures. The same color-coding is used to distinguish the number of HU dimers on the sampled chains in A. The protein build-up in B is depicted separately for the dominant (P1) loops, the secondary (A1, A2, P2) loops, and the composite (All) set of structures. The centers of the regulatory elements on the lac operon—the CAP and (–35, –10) promoter sites—are labeled.
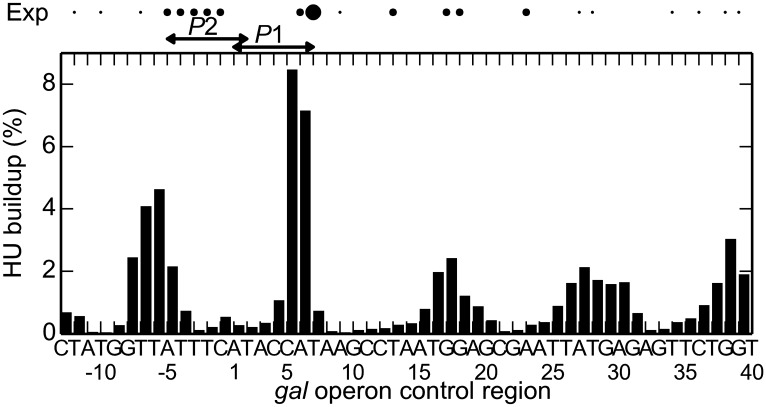
The positioning of HU is similarly localized on wild-type Gal repressor-mediated loops. Moreover, the dominant site of protein uptake coincides with the site of highest HU occupancy detected upon conversion of the protein to a chemical “nuclease”—position +6.5 nearer the 3′- than the 5′-end of the loop (6)—and the predicted sites of secondary protein uptake occur in the vicinity of the sites of less frequently observed binding affinity (Fig. 5). The dominant mode of binding occurs almost exclusively in the context of an A1 loop with a single HU dimer (SI Appendix, Fig. S12) and, as noted previously (6), impedes access to the overlapping gal promoters. The predicted secondary binding sites near the ends of the loop occur in the context of a P2 loop with two symmetrically positioned HU dimers. Simulated loops of 113 bp attach to the Lac repressor in a P1 orientation with two symmetrically bound dimers (SI Appendix, Fig. S13).
Fig. 5.
Comparison of the calculated build-up of HU on wild-type Gal repressor-mediated DNA loops with the binding sites detected by Aki and Adhya through biochemical footprinting (6). The observed data represent the sites of DNA cutting found upon conversion of HU to a chemical nuclease. The predicted values represent the frequency of HU uptake centered about a given location along the loop; i.e., the modeled protein–DNA complex is symmetrically placed about the site. Thus, the presence of HU on the dominant site impedes access to the P1 promoter. The dots of increasing size at the top depict the HU-binding centers of increasing affinity extracted through the cleavage of DNA. The depicted sequence—53 of the 113 bp between operator centers—spans the sites reported in ref. 6.
Allowance in the simulations for large-scale opening of the Lac repressor, along the lines suggested by various experiments (e.g., refs. 20, 21), changes the above picture somewhat. “Flattened” HU-decorated loops anchored in parallel orientations on the repressor continue to dominate the configurational landscape if the repressor is allowed to open, although with lesser probability than if attached to the rigid, V-shaped structure (Fig. 4B). The symmetrical placement of HU, however, limits the opening of the repressor (10). By contrast, all of the 92-bp loops captured in the absence of HU on a deformable repressor bind in a parallel orientation to a highly opened protein structure, and, of these, most adopt an extended, smoothly curved pathway free of DNA self contacts (10, 17, 18), i.e., without the crossing of the loop found in the ℓ-shaped HU-free configurations formed on the rigid, V-shaped model (SI Appendix, Figs. S9 and S10). In this sense, the addition of HU has the same dramatic effect on the configuration of the DNA loop as that found when the repressor is kept rigid, although the types of overall folding differ.
HU Introduces a Structural Barrier to lac Operon Accessibility.
The simulated structures of DNA loops anchored to the Lac repressor and decorated by randomly bound HU dimers (Fig. 3) differ substantially from all models of the protein–DNA assembly offered to date (17, 18, 22–27). The loops adopt a broader and different range of configurational states than heretofore anticipated and hint of an indirect functional role of HU in lac-gene repression. The presence of HU allows the DNA to adopt spatial pathways normally inaccessible to the stiff, naturally straight duplex. As noted above, the tendency of DNA to remain straight directs the placement of the HU dimers and thus the sites of sharp, localized, protein-induced deformation. The consequent changes in overall DNA folding, in turn, reposition key regulatory elements on the lac operon relative to the ends of the repressor-mediated loop. These elements include the 22-bp site of catabolic activator protein (CAP) binding, located adjacent to the operator-bound DNA at the 5′-end of the loop, and the hexameric –35 and –10 promoter elements, located, respectively, in the middle and near the 3′-end of the loop. The known propensity of HU to remain in the vicinity of its binding sites (28) is expected to preclude access of other molecules to any regulatory elements decorated by HU upon loop closure.
Close examination of the simulated loops reveals HU-induced changes in both the global configuration and the local DNA microenvironment. As noted above, the two symmetrically placed dimers on the dominant HU-bound loops introduce a large-scale change of shape compared with the pathways taken by DNA in the HU-free loops (Fig. 3). The chain comes in closer contact with the Lac repressor than in the absence of HU, and the loop reorients with respect to the operators. The intervening base pairs also twist such that the major-groove edges of two of the six base pairs in the –35 recognition element tend to face inward. In the absence of HU, these edges, which are recognized by the σ-factor of the RNA polymerase holoenzyme, have a greater propensity to face toward the exterior of the ℓ-shaped loops attached in the same orientation to a rigid Lac repressor structure. The architectural proteins thus change the rotational setting of the intervening DNA.
Notably, the fixed HU dimer sites overlap the CAP, –35, and –10 elements (Figs. 3 and 4). One dimer preferentially builds up on the 3′-half of the CAP site, and the second occupies one or the other of the promoter elements. For example, almost all of the loops attached to the Lac repressor in the dominant parallel orientation have one HU molecule that covers part of the CAP site, and nearly 70% of these loops have an HU that covers all or part of the –35 or –10 hexamer. The wrapping of DNA on the surface of the HU dimer precludes access of other proteins to any base pairs in contact with the architectural protein. The presence of HU may thus add a structural barrier to the initiation of gene transcription. There is no such barrier in the Lac repressor-mediated DNA loop structures previously predicted to occur in vivo (24).
Discussion
The decoration of linear DNA by nonspecific architectural proteins produces a long, randomly crumpled double-helical molecule upon arbitrary uptake of low levels of the deformation-inducing ligands (29). Here we show, using the HU protein, how ordinary topological constraints placed on short fragments of DNA change the molecular disorder expected of DNA bound by a nonspecific protein into tightly organized 3D structures. Even though the simulated association of HU with DNA is random, the ligands build up at regularly spaced intervals along closed duplexes. Because of the rotational symmetry of DNA, ring-closure constraints have almost no effect on the specificity of the protein for any particular binding site. The computed distribution of HU on the minicircles reflects long-range mechanical correlations between bound species rather than the recognition of a given sequence. In other words, the ligands appear to recognize molecular shape rather than unique chemical features of the DNA base pairs. Here that shape is the stiff, naturally straight pathway of short, double-helical DNA, which also seems to direct the elongated, elliptical shapes of small, torsionally stressed DNA minicircles captured in cryo electron microscopic images (30). The many ways in which a protein like HU can effect cyclization point to the advantage of its nonspecificity on the enhancement of DNA ring closure compared with sequence-specific ligands that bind with similar affinity to a single site.
The rotational setting imposed on DNA by proteins such as the Lac and Gal repressors, which mediate DNA loop formation, enhances the specificity of HU binding. That is, the nonspecific ligands build up at particular sites on the constrained duplex. The preferential build-up shows, in turn, how a nonspecific ligand might assume a functional role in a particular spatial setting. As noted above, the placement of HU on both Gal and Lac repressor-mediated loops with the wild-type operator spacing precludes access to key recognition elements on DNA. The localized build-up of architectural proteins on DNA loops helps to understand why the stable, site-specific binding of HU to DNA in the gal repressosome requires binding of the Gal repressor to both operators (6). The HU binds to any site on a simulated chain if the ends are free but to specific sites if the ends are held in place.
A variety of experimental data support the structures of HU-bound DNA reported here. The predicted structures are ones that dominate the configurational landscapes (10–12) found to account successfully for the looping propensities of DNA deduced from gene expression (7, 31, 32) and single-molecule (33, 34) studies and the probabilities of DNA ring closure measured at different chain lengths under various HU concentrations (3, 15) (SI Appendix, Fig. S2). The models also support the build-up of HU on Lac repressor-mediated loops found in recent tethered-particle motion studies (35). Significantly, the simulated build-up of HU on wild-type Gal repressor-mediated DNA loops matches the observed localization of the protein in biochemical footprinting studies (6).
Earlier treatments of HU-bound DNA loops (24, 36) were phenomenological in the sense that the effects of HU on chain configuration were subsumed in the parameters of simplified models fitted to the data. By contrast, the present work takes direct account of the structures and fluctuations of successive DNA base pairs in the free and protein-bound state, the random binding of HU at various concentration levels, and the structures of the Lac and Gal repressors. The current model, however, omits direct treatment of electrostatic interactions that might bias the conformation and spacing of proteins on DNA. The current treatment, nevertheless, could prove useful in the design of new, more direct experiments to test the effects of HU on spatially constrained DNA, e.g., single-molecule studies of the energy transfer between judiciously labeled fluorophores on short, HU-decorated DNA loops. Atomic coordinates of modeled structures are available upon request.
Finally, structures like those reported here suggest that HU architecture, like nucleosomal architecture, which modulates the ability of regulatory proteins to recognize their binding sites in the context of chromatin (37), may influence repressor–operator interactions in the context of the bacterial nucleoid. In principle, the operators can bind both repressor and HU proteins if appropriately oriented, e.g., with the major-groove edge of the CG base-pair step at the center of the operator facing the HU surface and the minor-groove edge in contact with the repressor. The shared-binding motif hints, in turn, of a potential mechanism of repressor opening. The simulated structures show, for example, that the accommodation of HU near one or both ends of the DNA loop requires Lac repressor opening. Subtle changes in operator structure, which contribute to the simulated looping propensities of DNA in the absence of HU (38), may also affect the uptake and placement of the architectural protein.
Methods
Spatially Constrained DNA Structures.
The configurations of HU-decorated DNA chains capable of closing into minicircles or looping between the headpieces of the Lac repressor assembly were obtained by direct Monte Carlo simulation of double-helical structures and subsequent identification of chains with terminal base pairs in the desired spatial disposition. The DNA is treated at the level of base-pair steps, using six rigid-body parameters to specify the arrangements of successive base pairs (39, 40) and a potential that allows for elastic deformations of the long, thin molecule from the canonical B-DNA structure (4). The model, however, ignores sequence-dependent variations in double-helical structure and/or deformability that might modulate the rotational setting of spatially constrained structures (i.e., direct the edges of the inward- and outward-facing base pairs) and omits direct treatment of electrostatics. The polyelectrolyte properties of DNA are treated indirectly through the choice of elastic parameters, and the proteins are neutral. The pathway of protein-free DNA is constructed, one base-pair step at a time, from randomly sampled sets of rigid-body parameters subject to the elastic potential. The protein-bound DNA is treated in terms of the rigid-body parameters of DNA found in available protein-DNA structures (5, 16). The proteins are included as “side groups” of the DNA; i.e., the atoms of protein are expressed in the reference frame of one of the bases in the bound molecular complexes. See Supporting Information for full details.
Nonspecific Protein Binding.
Each site of possible HU binding on a simulated protein-free duplex is visited, and the protein is placed on the basis of the assumed binding probability (12). One of the HU-DNA step motifs is selected at random and introduced at the accepted binding sites, thereby altering the configuration of DNA. The same procedure is used to introduce Hbb. The latter protein, although known to bind sequence-specifically to DNA (41), is used here to illustrate the effects of sharp protein-induced bending on ring closure. Successfully closed chains are discarded if the distances between pairs of Cα atoms on protein and base-pair centers on DNA fall below standard steric limits (12). The ease of DNA loop formation is estimated from the number of simulated configurations of a linear molecule with terminal base pairs positioned so as to overlap the base pairs at the ends of the operators bound to the selected repressor (10). The arrangement of operators is based on the relative disposition of the two dimeric halves of the tetrameric assembly.
Repressor Models.
The simulations include the looping of DNA between the headpieces of V-shaped models of the Lac and Gal repressors (SI Appendix, Fig. S1) obtained by superposition of the structures of well-resolved protein fragments (17, 19) as well as the ease of placing the double helix between the ends of a Lac tetramer subject to large-scale opening. The looping computations also take account of the four distinct orientations of DNA operator sequences on the repressor assemblies (19). Although the computations do not specify the level of DNA supercoiling, the topological properties of the successfully closed pathways—i.e., the writhing, twisting, and linking numbers of the protein-DNA assemblies—are determined from the positions of the constituent base pairs (42). The principal axes of the closed DNA structures are obtained from a covariance analysis of the coordinates of the base-pair centers (see Supporting Information for further details).
Supplementary Material
Acknowledgments
We thank Dr. Victor Zhurkin for sharing his model of the Gal repressor–operator assembly. W.K.O. also thanks the Aspen Center for Physics for providing a stimulating environment to carry out parts of this work. This work has been generously supported by the US Public Health Service under research Grant GM34809 and instrumentation Grant RR022375 and by the Human Frontiers in Science Program under Grant RGP0051/2009. M.A.G. acknowledges support from a US Department of Education Graduate Assistance in Areas of National Need Fellowship, and L.C. acknowledges computational resources from XSEDE startup TG-140034.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405016111/-/DCSupplemental.
References
- 1.Adhya S. Multipartite genetic control elements: Communication by DNA loop. Annu Rev Genet. 1989;23:227–250. doi: 10.1146/annurev.ge.23.120189.001303. [DOI] [PubMed] [Google Scholar]
- 2.Müller-Hill B. The lac Operon. Walter de Gruyter; Berlin: 1996. pp. 66–67. [Google Scholar]
- 3.Du Q, Smith C, Shiffeldrim N, Vologodskaia M, Vologodskii A. Cyclization of short DNA fragments and bending fluctuations of the double helix. Proc Natl Acad Sci USA. 2005;102(15):5397–5402. doi: 10.1073/pnas.0500983102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Czapla L, Swigon D, Olson WK. Sequence-dependent effects in the cyclization of short DNA. J Chem Theory Comput. 2006;2(3):685–695. doi: 10.1021/ct060025+. [DOI] [PubMed] [Google Scholar]
- 5.Swinger KK, Lemberg KM, Zhang Y, Rice PA. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003;22(14):3749–3760. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aki T, Adhya S. Repressor induced site-specific binding of HU for transcriptional regulation. EMBO J. 1997;16(12):3666–3674. doi: 10.1093/emboj/16.12.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker NA, Kahn JD, Maher LJ., III Bacterial repression loops require enhanced DNA flexibility. J Mol Biol. 2005;349(4):716–730. doi: 10.1016/j.jmb.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 8.Rouvière-Yaniv J, Yaniv M, Germond JE. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell. 1979;17(2):265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- 9.Malik M, Bensaid A, Rouviere-Yaniv J, Drlica K. Histone-like protein HU and bacterial DNA topology: Suppression of an HU deficiency by gyrase mutations. J Mol Biol. 1996;256(1):66–76. doi: 10.1006/jmbi.1996.0068. [DOI] [PubMed] [Google Scholar]
- 10.Czapla L, Grosner MA, Swigon D, Olson WK. Interplay of protein and DNA structure revealed in simulations of the lac operon. PLoS ONE. 2013;8(2):e56548. doi: 10.1371/journal.pone.0056548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olson WK, Grosner MA, Czapla L, Swigon D. Structural insights into the role of architectural proteins in DNA looping deduced from computer simulations. Biochem Soc Trans. 2013;41(2):559–564. doi: 10.1042/BST20120341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czapla L, Swigon D, Olson WK. Effects of the nucleoid protein HU on the structure, flexibility, and ring-closure properties of DNA deduced from Monte Carlo simulations. J Mol Biol. 2008;382(2):353–370. doi: 10.1016/j.jmb.2008.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodges-Garcia Y, Hagerman PJ, Pettijohn DE. DNA ring closure mediated by protein HU. J Biol Chem. 1989;264(25):14621–14623. [PubMed] [Google Scholar]
- 14.Paull TT, Haykinson MJ, Johnson RC. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993;7(8):1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 15.Czapla L, Peters JP, Rueter EM, Olson WK, Maher LJI., III Understanding apparent DNA flexibility enhancement by HU and HMGB proteins: Experiment and simulation. J Mol Biol. 2011;409(2):278–289. doi: 10.1016/j.jmb.2011.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mouw KW, Rice PA. Shaping the Borrelia burgdorferi genome: Crystal structure and binding properties of the DNA-bending protein Hbb. Mol Microbiol. 2007;63(5):1319–1330. doi: 10.1111/j.1365-2958.2007.05586.x. [DOI] [PubMed] [Google Scholar]
- 17.Swigon D, Coleman BD, Olson WK. Modeling the Lac repressor-operator assembly: The influence of DNA looping on Lac repressor conformation. Proc Natl Acad Sci USA. 2006;103(26):9879–9884. doi: 10.1073/pnas.0603557103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swigon D, Olson WK. Mesoscale modeling of multi-protein-DNA assemblies: The role of the catabolic activator protein in Lac-repressor-mediated looping. Int J Non-linear Mech. 2008;43(10):1082–1093. doi: 10.1016/j.ijnonlinmec.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geanacopoulos M, Vasmatzis G, Zhurkin VB, Adhya S. Gal repressosome contains an antiparallel DNA loop. Nat Struct Biol. 2001;8(5):432–436. doi: 10.1038/87595. [DOI] [PubMed] [Google Scholar]
- 20.Taraban M, et al. Ligand-induced conformational changes and conformational dynamics in the solution structure of the lactose repressor protein. J Mol Biol. 2008;376(2):466–481. doi: 10.1016/j.jmb.2007.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haeusler AR, et al. FRET studies of a landscape of Lac repressor-mediated DNA loops. Nucleic Acids Res. 2012;40(10):4432–4445. doi: 10.1093/nar/gks019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis M, et al. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science. 1996;271(5253):1247–1254. doi: 10.1126/science.271.5253.1247. [DOI] [PubMed] [Google Scholar]
- 23.Balaeff A, Mahadevan L, Schulten K. Modeling DNA loops using the theory of elasticity. Phys Rev E Stat Nonlin Soft Matter Phys. 2006;73(3 Pt 1):031919. doi: 10.1103/PhysRevE.73.031919. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, McEwen AE, Crothers DM, Levene SD. Analysis of in-vivo LacR-mediated gene repression based on the mechanics of DNA looping. PLoS ONE. 2006;1:e136. doi: 10.1371/journal.pone.0000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goyal S, et al. Intrinsic curvature of DNA influences LacR-mediated looping. Biophys J. 2007;93(12):4342–4359. doi: 10.1529/biophysj.107.112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Towles KB, Beausang JF, Garcia HG, Phillips R, Nelson PC. First-principles calculation of DNA looping in tethered particle experiments. Phys Biol. 2009;6(2):025001. doi: 10.1088/1478-3975/6/2/025001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.La Penna G, Perico A. Wrapped-around models for the lac operon complex. Biophys J. 2010;98(12):2964–2973. doi: 10.1016/j.bpj.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham JS, Johnson RC, Marko JF. Concentration-dependent exchange accelerates turnover of proteins bound to double-stranded DNA. Nucleic Acids Res. 2011;39(6):2249–2259. doi: 10.1093/nar/gkq1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: Formation of flexible hinges and rigid filaments. Proc Natl Acad Sci USA. 2004;101(18):6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lionberger TA, et al. Cooperative kinking at distant sites in mechanically stressed DNA. Nucleic Acids Res. 2011;39(22):9820–9832. doi: 10.1093/nar/gkr666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker NA, Kahn JD, Maher LJ., III Effects of nucleoid proteins on DNA repression loop formation in Escherichia coli. Nucleic Acids Res. 2007;35(12):3988–4000. doi: 10.1093/nar/gkm419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bond LM, Peters JP, Becker NA, Kahn JD, Maher LJ., III Gene repression by minimal lac loops in vivo. Nucleic Acids Res. 2010;38(22):8072–8082. doi: 10.1093/nar/gkq755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han L, et al. Concentration and length dependence of DNA looping in transcriptional regulation. PLoS ONE. 2009;4(5):e5621. doi: 10.1371/journal.pone.0005621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson S, Lindén M, Phillips R. Sequence dependence of transcription factor-mediated DNA looping. Nucleic Acids Res. 2012;40(16):7728–7738. doi: 10.1093/nar/gks473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boedicker JQ, Garcia HG, Johnson S, Phillips R. DNA sequence-dependent mechanics and protein-assisted bending in repressor-mediated loop formation. Phys Biol. 2013;10(6):066005. doi: 10.1088/1478-3975/10/6/066005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saiz L, Vilar JM. Multilevel deconstruction of the in vivo behavior of looped DNA-protein complexes. PLoS ONE. 2007;2(4):e355. doi: 10.1371/journal.pone.0000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sahu G, et al. p53 binding to nucleosomal DNA depends on the rotational positioning of DNA response element. J Biol Chem. 2010;285(2):1321–1332. doi: 10.1074/jbc.M109.081182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colasanti AV, et al. Weak operator binding enhances simulated lac repressor-mediated DNA looping. Biopolymers. 2013;99(12):1070–1081. doi: 10.1002/bip.22336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu XJ, Olson WK. 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31(17):5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu X-J, Olson WK. 3DNA: A versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc. 2008;3(7):1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobryn K, Naigamwalla DZ, Chaconas G. Site-specific DNA binding and bending by the Borrelia burgdorferi Hbb protein. Mol Microbiol. 2000;37(1):145–155. doi: 10.1046/j.1365-2958.2000.01981.x. [DOI] [PubMed] [Google Scholar]
- 42.Clauvelin N, Olson WK, Tobias I. Characterization of the geometry and topology of DNA pictured as a discrete collection of atoms. J Chem Theory Comput. 2012;8(3):1092–1107. doi: 10.1021/ct200657e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.