Significance
The emergence of the cell-autonomous circadian oscillator is coupled with cellular differentiation. Cellular differentiation, as well as reprogramming, results in global alterations of the transcriptional program via epigenetic modification such as DNA methylation. We here demonstrate that c-Myc constitutive expression and Dnmt1 ablation disrupt the differentiation-coupled emergence of the clock from mouse ES cells (ESCs). Using these model ESCs, 484 genes were identified by global gene expression analysis as factors correlated with circadian clock development. Among them, we find that misregulation of Kpna2 (Importin-α2) during the differentiation culture of ESCs significantly impairs clock development, and KPNA2 facilitates cytoplasmic localization of PER1/2. These results suggest that the programmed gene expression network regulates the differentiation-coupled circadian clock development in mammalian cells.
Keywords: circadian clock, cellular differentiation, c-Myc, Dnmt1, Kpna2 (Importin-α2)
Abstract
The circadian clock in mammalian cells is cell-autonomously generated during the cellular differentiation process, but the underlying mechanisms are not understood. Here we show that perturbation of the transcriptional program by constitutive expression of transcription factor c-Myc and DNA methyltransferase 1 (Dnmt1) ablation disrupts the differentiation-coupled emergence of the clock from mouse ESCs. Using these model ESCs, 484 genes are identified by global gene expression analysis as factors correlated with differentiation-coupled circadian clock development. Among them, we find the misregulation of Kpna2 (Importin-α2) during the differentiation of the c-Myc-overexpressed and Dnmt1−/− ESCs, in which sustained cytoplasmic accumulation of PER proteins is observed. Moreover, constitutive expression of Kpna2 during the differentiation culture of ESCs significantly impairs clock development, and KPNA2 facilitates cytoplasmic localization of PER1/2. These results suggest that the programmed gene expression network regulates the differentiation-coupled circadian clock development in mammalian cells, at least in part via posttranscriptional regulation of clock proteins.
The circadian clock is an intrinsic time-keeping system that regulates essential physiological functions such as sleep/wake cycles, body temperature, and metabolism (1–3). In the mammalian clock system the central pacemaker resides in the suprachiasmatic nucleus of the hypothalamus, coordinating cell-autonomous molecular oscillators throughout the body to perform tissue-specific functions. The molecular oscillator consists of transcriptional/translational feedback loops of clock genes. Two transcription factors, CLOCK and BMAL1, heterodimerize and transactivate core clock genes such as the Period (Per1, 2, 3), Cryptochrome (Cry1, 2), and Rev-Erbα genes via E-box regulatory elements. PER and CRY proteins, in turn, translocate into the nucleus to suppress CLOCK/BMAL1 activity, leading to cyclic expression of these clock genes (4–9). Furthermore, REV-ERBα negatively regulates Bmal1 transcription via the RORE promoter element, thus driving antiphasic expression patterns of Bmal1 (10, 11).
Despite uncovering the emergence of the circadian rhythms that occur during development (12–14), the precise mechanism of circadian clock development in mammalian cells remains unclear. Recently it has been found that pluripotent ES cells (ESCs) do not display discernible circadian molecular oscillations, whereas in vitro-differentiated ESCs displayed robust circadian oscillations of reporter expression (15–17). Moreover, we also have shown that circadian oscillations were abolished when differentiated cells were reprogrammed to regain pluripotency by expression of reprogramming factors (Oct3/4, Sox2, Klf4, and c-Myc) (15). These results suggested that the emergence of the cell-autonomous circadian oscillator is coupled with cellular differentiation. Cellular differentiation, as well as reprogramming, results in global alterations of the transcriptional program and epigenetic modifications such as DNA methylation (18, 19). On the other hand, misregulated differentiation can lead to aberrant transdifferentiation and abnormal cellular states, such as cancer (20–22).
In this study our established in vitro circadian clock formation assay clearly shows that the constitutive expression of c-Myc and Dnmt1−/− disrupted the development of the circadian clock. Global gene expression analysis reveals that 484 genes are identified as candidate factors correlating with emergence of circadian clock oscillation. In failure of clock development, a significant increase of Kpna2, one of the candidate factors, encoding Importin-α2 and aberrant subcellular localization of PER proteins are identified as shared events. Moreover, the doxycycline (Dox)-dependent overexpression of Kpna2 during ESC differentiation results in the significant impairment of clock development. In addition, Kpna2 expression facilitates cytoplasmic accumulation of PER proteins. These observations suggest that the differentiation-coupled transcriptional program of certain genes, including Kpna2, may critically regulate circadian clock development in mammalian cells.
Results
In Vitro Assay System Investigating Differentiation-Coupled Circadian Clock Development Using Mouse ESCs.
We recently described an in vitro differentiation method to observe the development of the circadian clock from ESCs and its utility for evaluating the effect of genetic mutations on circadian clock development (15, 17). Here we use ESCs expressing mBmal1:luc (15) and PER2::Luc knock-in (PER2Luc) (23, 24) circadian reporters to investigate the developmental mechanisms of the cellular circadian clock. Although no circadian oscillation of reporter bioluminescence was detected in these ESC lines as we previously reported in other cell lines, weak circadian bioluminescence rhythms first appeared in day-14 cultures (Fig. S1). Oscillation of bioluminescence became robust on day 21 and attained maximum amplitude on day 28 (Fig. S1). Bmal1:luc and PER2Luc ESC-derived differentiated cells showed the emergence of circadian bioluminescence at almost the same time, with nearly antiphasic rhythms, concordant with their endogenous phase relationship in the mammalian clock system.
To elucidate the molecular mechanisms underlying the observed differentiation-coupled clock development, we tried to analyze two model systems in which we perturbed mouse ESC differentiation and tested their effects on the development of circadian rhythmicity using the in vitro circadian clock formation assay: (i) ESCs with Dox-conditional expression of c-Myc, and (ii) ESCs deficient in DNA metyltransferases, such as Dnmt1−/−, Dnmt3a−/−, Dnmt3b−/−, Dnmt3a−/−3b−/−, and Dnmt1−/−3a−/−3b−/− (triple knockout, TKO).
Dox-Inducible c-Myc Overexpression ESCs as a Perturbation Model for Differentiation-Coupled Transcriptional Program.
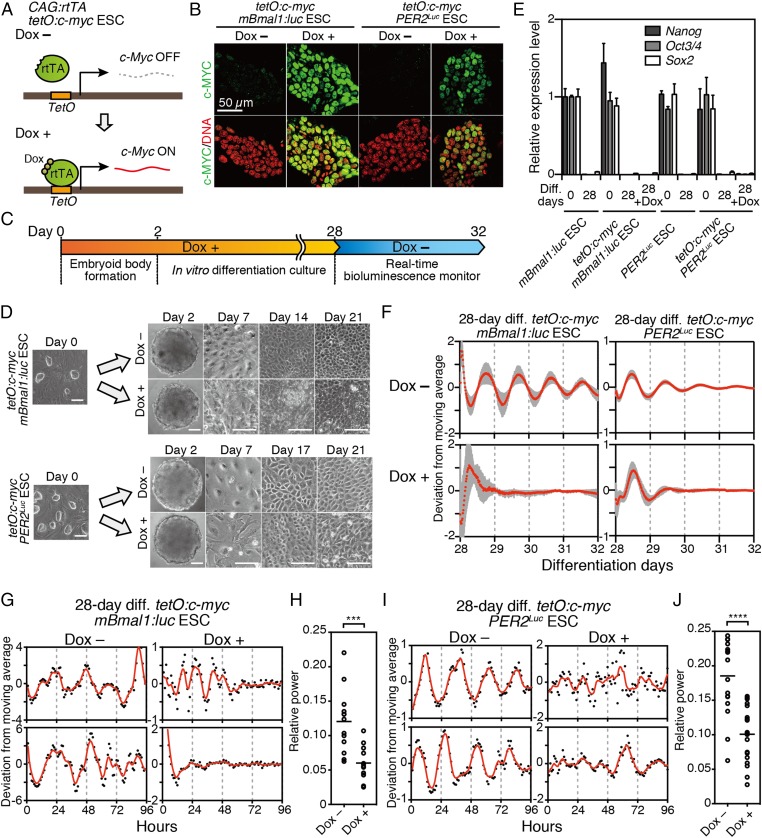
Previous studies have shown that the MYC affects global gene expression and can induce misregulation of the transcriptional program in various cell types (21, 25, 26). Therefore, overexpression of c-Myc during ESC differentiation could perturb the regulatory network of the cellular differentiation process. To test the role of c-Myc in differentiation-coupled circadian clock development, we used Dox-inducible c-Myc expression in ESCs with either mBmal1:luc or PER2Luc reporters (tetO:c-myc mBmal1:luc ESC and tetO:c-myc PER2Luc ESC) (Fig. 1A). Dox treatment of these ESCs showed constitutive expression of c-Myc mRNA and nuclear accumulation of c-MYC protein in both ESCs (Fig. 1B and Fig. S2).
Fig. 1.
Constitutive c-Myc expression across cellular differentiation prevents the development of circadian clock oscillation. (A) mBmal1:luc ESCs or PER2Luc ESCs carrying Dox-inducible c-Myc (tetO:c-myc mBmal1:luc ESCs or tetO:c-myc PER2Luc ESCs) were established using PiggyBac transposon vectors with a Dox-inducible c-Myc. (B) Immunofluorescent staining of tetO:c-myc ESCs using anti-c-MYC antibody. c-Myc was induced by addition of 40 ng/mL Dox (Left, tetO:c-myc mBmal1:luc ESCs) or 500 ng/mL Dox (Right, tetO:c-myc PER2Luc ESCs) for 1 d. (C) Schematic of method to express c-Myc constitutively. During in vitro differentiation, culture medium was changed with EFM containing 0, 40 (for tetO:c-myc mBmal1:luc ESCs), or 500 ng/mL Dox (for tetO:c-myc PER2Luc ESCs). (D) Morphological observation of in vitro 2-, 7-, 14-, and 21-d differentiated tetO:c-myc mBmal1:luc ESCs (Upper) or tetO:c-myc PER2Luc ESCs (Lower) in the absence or presence of Dox. (Scale bars, 100 µm.) (E) Quantitative RT-PCR analysis of differentiation markers, Nanog, Oct3/4 (Pou5f1), and Sox2, in ESCs or in vitro-differentiated ESCs. Data are mean ± SD (n = 3). (F) Bioluminescence rhythms of tetO:c-myc mBmal1:luc ESCs (Left) and tetO:c-myc PER2Luc ESCs (Right) after 28-d differentiation culture with or without Dox. (mean with SD, n = 12). (G) Representative single-cell bioluminescence traces of in vitro 28-d-differentiated tetO:c-myc mBmal1:luc ESCs in the absence or presence of Dox. The red line is the LOWESS curve of the fitted values. (H) FFT (Fast Fourier Transform) spectral power analysis of bioluminescences of in vitro 28-d-differentiated tetO:c-myc mBmal1:luc ESCs in the absence or presence of Dox. Bars are mean (n = 12). Student t test, ***P < 0.001. (I) Representative single-cell bioluminescence traces of in vitro 28-d-differentiated tetO:c-myc PER2Luc ESCs in the absence or presence of Dox. (J) FFT spectral power analysis of bioluminescence of in vitro 28-d-differentiated tetO:c-myc PER2Luc ESCs in the absence or presence of Dox. Bars are mean (n = 15–19). Student t test, ****P < 0.0001.
To investigate the development of circadian clock oscillation, the in vitro differentiation culture was performed for 28 d using these c-Myc inducible ESCs with (Dox+) or without Dox (Dox−) (Fig. 1C). In this condition, differentiated-cell-like morphology (Fig. 1D) and loss of Nanog, Oct3/4, and Sox2 gene expression examined by quantitative PCR using the primers indicated in Table S1 (Fig. 1E) were observed after the differentiation under both Dox+ and Dox− conditions. In the in vitro circadian clock formation assay, real-time bioluminescence analysis revealed that the mBmal1:luc- and PER2::Luc-driven circadian oscillations were abolished even after 28-d differentiation culture in Dox+ condition, whereas robust circadian oscillations were present in the Dox− condition (Fig. 1F). Likewise, at the single-cell level, constitutive expression of c-Myc also led to significant impairment of cellular clock formation both in mBmal1:luc and PER2Luc ESCs (Fig. 1 G–J). These results revealed that the sustained expression of c-Myc during ESC differentiation resulted in the disruption of circadian clock formation.
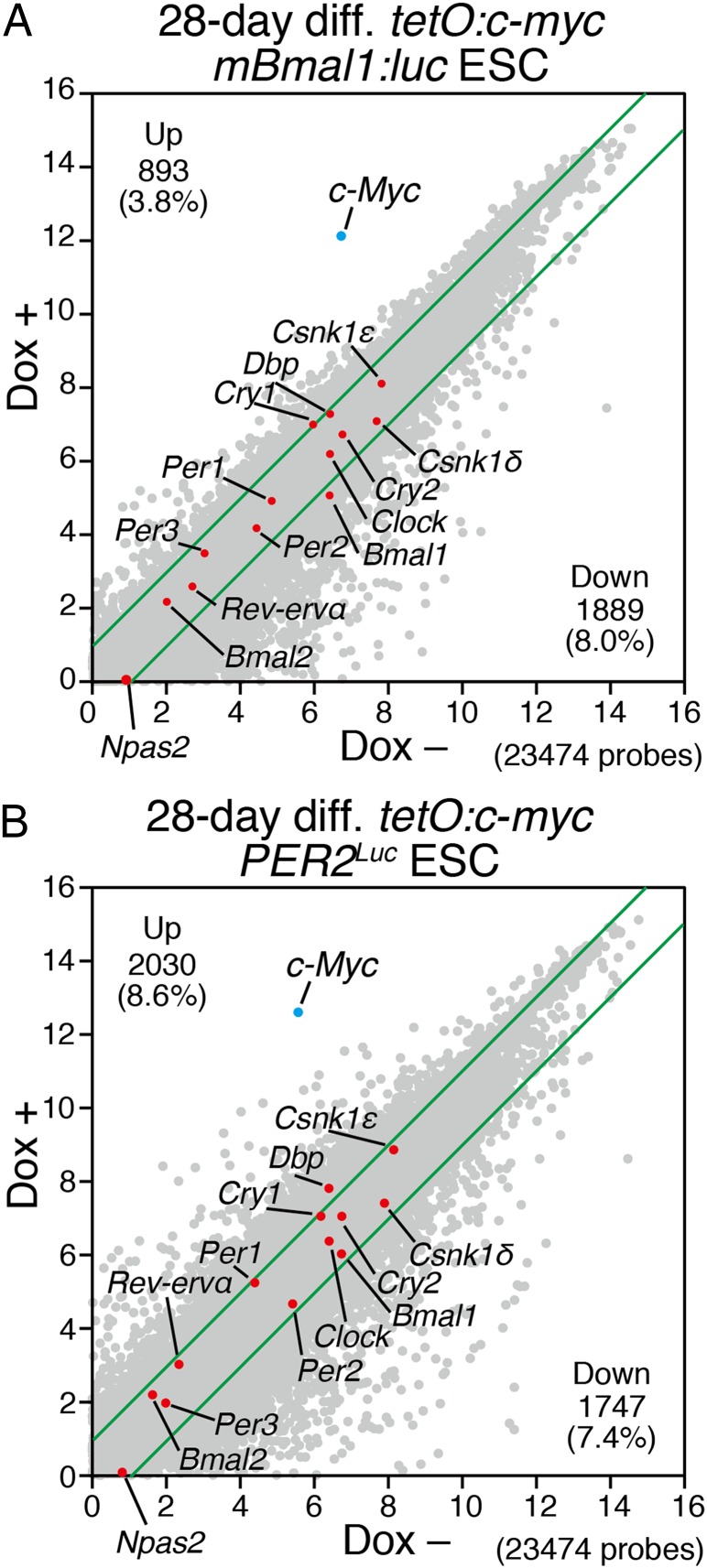
Next we compared global transcriptional profiles by microarray analysis in cells after in vitro differentiation culture from the tetO:c-myc ESCs with (Dox+) or without Dox (Dox−). In the cells differentiated in Dox+ condition, the expression profiles of 2,782 genes [893 up-regulated (3.8%), 1,889 down-regulated (8.0%)] in tetO:c-myc mBmal1:luc ESC-derived cells and 3,777 genes [2,030 up-regulated (8.6%), 1,747 down-regulated (7.4%)] in tetO:c-myc PER2Luc ESC-derived cells were changed more than twofold (Fig. 2 A and B and Dataset S1). Interestingly, core clock gene expression remained largely unchanged, whereas the expression levels of thousands of other genes were more strongly (>twofold) affected by c-Myc during differentiation (Fig. 2 A and B). Because these assays were performed in the absence of synchronizing agents, the levels are indicative of the mean expression levels of oscillating clock genes in these cells. Therefore, the lack of dramatic changes in the expression level of core clock genes suggests that they were not responsible for the disruption of circadian clock development during ESC differentiation.
Fig. 2.
Global gene expression analysis of in vitro 28-d-differentiated ESCs with c-Myc expression. (A and B) Microarray analysis of in vitro 28-d-differentiated tetO:c-myc mBmal1:luc ESCs (A) or tetO:c-myc PER2Luc ESCs with/without Dox (B). Scatter plots of all examined gene expressions (gray), a set of core clock gene expressions (red), and c-Myc (blue) of in vitro 28-d-differentiated tetO:c-myc mBmal1:luc ESCs (A) or tetO:c-myc PER2Luc ESCs (B) compared with with/without Dox. Green lines indicate twofold up- or down-changes. All data were transformed to the log 2 base scale.
Next we examined the effect of acute overexpression of c-Myc in differentiated and clock-oscillating cells. In contrast to the developing condition, acute induction of the c-Myc gene did not abolish circadian oscillations in the differentiated condition (Fig. S3 A–C). Moreover, even after an additional 28 d with Dox (a total of 56 d), the circadian rhythm in c-Myc-expressing cells remained robust, suggesting that after the completion of circadian clock differentiation, c-Myc expression does not affect the rhythm (Fig. S3 D and E). In addition, PER2Luc mouse embryonic fibroblasts (MEFs) expressing tetO:c-Myc and CAG:rtTA showed that acute c-Myc expression did not disrupt circadian oscillations but altered the phase of PER2Luc bioluminescence in MEFs (Fig. S3 F and G). Because induction of c-Myc resulted in a 1.3- to twofold increase of expression levels in Per1, Per2, Clock, Rev-erbα, and Cry1 genes but a slight down-regulation in Bmal1 and Dbp genes (Fig. S3H), acute over-expression of c-Myc affects clock gene expression patterns but does not abolish the circadian oscillation of a preexisting clock system.
Dnmt-Deficient ESCs as Perturbation Models for Differentiation-Coupled Epigenetic Regulation.
Next, because it is known that cellular differentiation-coupled transition of the transcriptional network is regulated by programmed DNA methylation (18, 19), mouse ESCs lacking DNA methyltransferase(s) [Dnmt1−/−, Dnmt3a−/−, Dnmt3b−/−, Dnmt3a−/−3b−/−, and Dnmt1−/−3a−/−3b−/− (TKO)] (27, 28) were used as models for perturbing epigenetic regulation during differentiation.
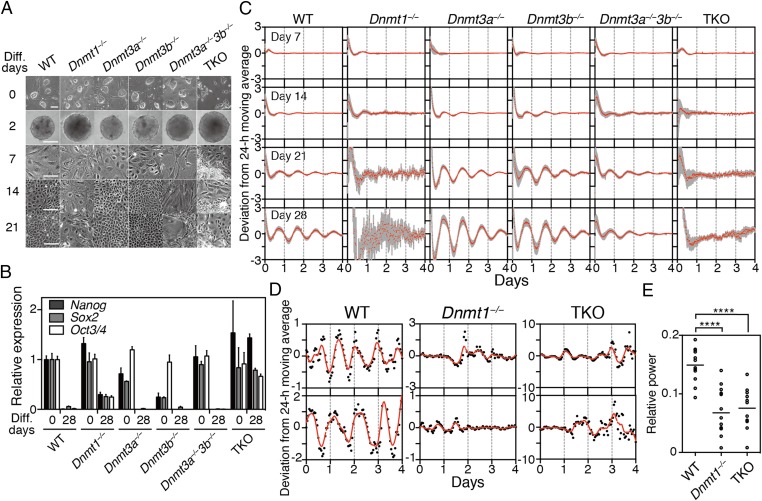
We initially observed morphological changes during the differentiation culture of WT, Dnmt1−/−, Dnmt3a−/−, Dnmt3b−/−, Dnmt3a−/−3b−/−, and TKO ESCs (Fig. 3A). To evaluate the differentiation, ESC markers such as Nanog, Sox2, and Oct3/4 were examined by quantitative PCR. Other than TKO cells, the expression of these ESC markers was dramatically reduced or lost after the 28-d differentiation culture (Fig. 3B). Weak expression of ESC markers was still detected in differentiated Dnmt1−/− cells (Fig. 3B), suggesting that Dnmt1 deficiency may result in abnormal or partial differentiation.
Fig. 3.
Disruption of differentiation-coupled circadian clock development in Dnmt1−/− ESCs. (A) Morphological changes after differentiation culture of WT, Dnmt1−/−, Dnmt3a−/−, Dnmt3b−/−, Dnmt3a−/−3b−/−, and Dnmt1−/−3a−/−3b−/− (TKO) ESCs. (B) Quantitative RT-PCR analysis of pluripotent markers, Nanog, Oct3/4 (Pou5f1), and Sox2, in ESCs or in vitro-differentiated ESCs. Data are mean ± SD (n = 3). (C) Averaged bioluminescence traces after in vitro 7-, 14-, 21-, or 28-d differentiation of indicated ESCs carrying mBmal1:luc reporters. Data, detrended by subtracting a 24-h moving average, are means with SD (n = 24). (D and E) Single-cell bioluminescence observations and FFT spectral power analysis of in vitro 28-d-differentiated indicated ESCs. The red line is the LOWESS curve of the fitted values. Each circle represents a single cell from in vitro-differentiated ESCs. Bars are mean (n = 11 or 12, one-way ANOVA followed by Bonferroni’s post hoc comparisons tests, ****P < 0.0001).
Next, in vitro circadian clock formation was assessed in Dnmt1−/−, Dnmt3a−/−, Dnmt3b−/−, Dnmt3a−/−3b−/−, and TKO mutant ESCs stably transfected with Bmal1:luc bioluminescence reporter. Dnmt1−/− ESCs failed to show normal circadian clock development, whereas robust circadian oscillations were observed in Dnmt3a−/− and Dnmt3b−/− cells after the differentiation (Fig. 3C). Double mutant Dnmt3a−/−3b−/− ESCs could also develop circadian oscillations, indicating that Dnmt3a−/− and Dnmt3b−/− are not essential for circadian clock development. Single-cell analysis of 28-d differentiated Dnmt1−/− cells confirmed that the circadian clock was abolished at the single-cell level (Fig. 3 D and E).
To investigate the mechanism underlying clock development, we determined global gene expression profiles using next-generation sequencing comparing rhythmic cells (28-d differentiation of WT, PER2Luc, Dnmt3a−/−, Dnmt3b−/−, and Dnmt3a−/−3b−/−) and nonrhythmic cells (all ESCs; 7-d differentiation of WT, PER2Luc, Dnmt1−/−, and TKO; 28-d differentiation of Dnmt1−/− and TKO). First we examined the expression profiles of core clock genes in these samples (Fig. S4). Because the gene expression analysis was performed in the absence of synchronizing agents, the obtained results represented the mean expression levels of oscillating clock genes in these cells. Interestingly, critical impairment of core clock gene expression explaining the loss of circadian clock oscillation was not observed in the nonrhythmic cells.
Identification of Candidate Genes Correlating with Differentiation-Coupled Circadian Clock Development.
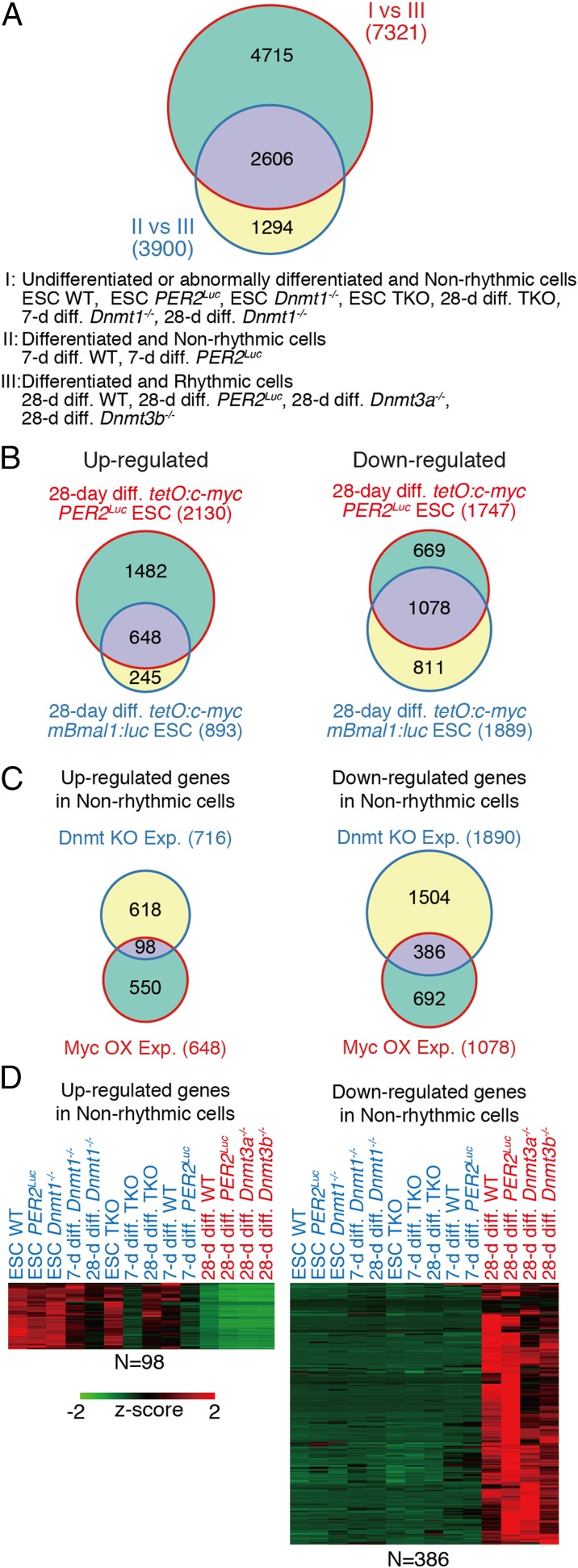
To identify potential factors commonly altered in the clock disrupted cells and ESCs, RNA-sequencing (RNA-seq) data obtained from Dnmt-deficient cells and microarray data obtained from tetO:c-myc cells were compared. First, using the RNA-seq data of Dnmt-deficient cells, 2,606 overlapping genes altered under all clock-disrupted conditions (all ESCs, differentiated Dnmt1−/− cells, differentiated TKO cells, and 7-d differentiated WT and PER2Luc ESCs) were identified from the comparison with the clock-oscillating cells (Fig. 4A and Dataset S2). Next, genes showing similar expression changes in the differentiated cells in the Dox+ of tetO:c-myc Bmal1:luc ESCs and tetO:c-myc PER2Luc ESCs were extracted. Compared with the Dox− condition, expression of 648 genes was increased in the Dox+ (c-Myc over-expressed) condition for both cell lines, and 1,078 genes were decreased in the Dox+ condition (Fig. 4B and Dataset S3). Then, these extracted gene sets were compared with the up- or down-regulated gene sets from the RNA-seq of the clock-disrupted cells. This analysis revealed 98 up-regulated and 386 down-regulated genes shared between both clock-disrupted Dnmt-deficient cells and c-Myc-overexpressed cells (Fig. 4C and Dataset S4). Heat map representation shows the different patterns of expression levels between clock-oscillating (rhythmic) cells and clock-disrupted (nonrhythmic) cells (Fig. 4D). Because the identified 484 genes (98 genes up-regulated and 386 genes down-regulated in “Non-rhythmic cells”) are candidate genes for regulating circadian clock development in mammalian cells, we next tried to validate the biological significance of this gene set as clock-development regulators. As described above, expression profiles of essential core clock genes were not predictive indicators of clock-oscillating cells. Moreover, the identified gene set did not include core clock genes (Dataset S4). Therefore, the mechanism to generate cell-autonomous circadian cycling includes additional gene networks other than the clock genes.
Fig. 4.
Identification of correlated genes with differentiation-coupled circadian clock development. (A) Venn diagrams extracting common factors correlating with disruption of circadian clock development using RNA-seq data obtained from the Dnmt-deficient ESC model systems. Groups I, II, and III indicate condition of cells as described. (B) Venn diagrams extracting up- or down-regulated genes of in vitro 28-d-differentiated tetO:c-myc mBmal1:luc ESCs with Dox against Dox-untreated conditions. (C) Venn diagrams extracting the up- or down-regulated genes in clock-disrupted conditions of Dnmt-deficient ESCs derived cells and c-Myc overexpression experiments. (D) Heat maps of the extracted “clock development correlating-module” gene set correlating with differentiation-coupled circadian clock development.
A previous study reported that the gene expression signature of ESCs was dissected into three functional modules: core-pluripotency factors (ESC-Core), polycomb repressive complex factors (ESC-PRC), and Myc-related factors (ESC-Myc) (25). Because circadian clock development is also closely correlated with cellular differentiation from ESCs, we compared our identified gene set with these ESC-related modules. Interestingly, the identified gene set did not overlap greatly with the factors of the ESC-Core module, ESC-PRC module, and even ESC-Myc module (Fig. S5). These results suggest that circadian clock development-related factors may form an independent functional module from other ESC-related modules.
Abnormal Cytoplasmic Accumulation of PER Proteins in Clock-Disrupted Cells and Undifferentiated ESCs.
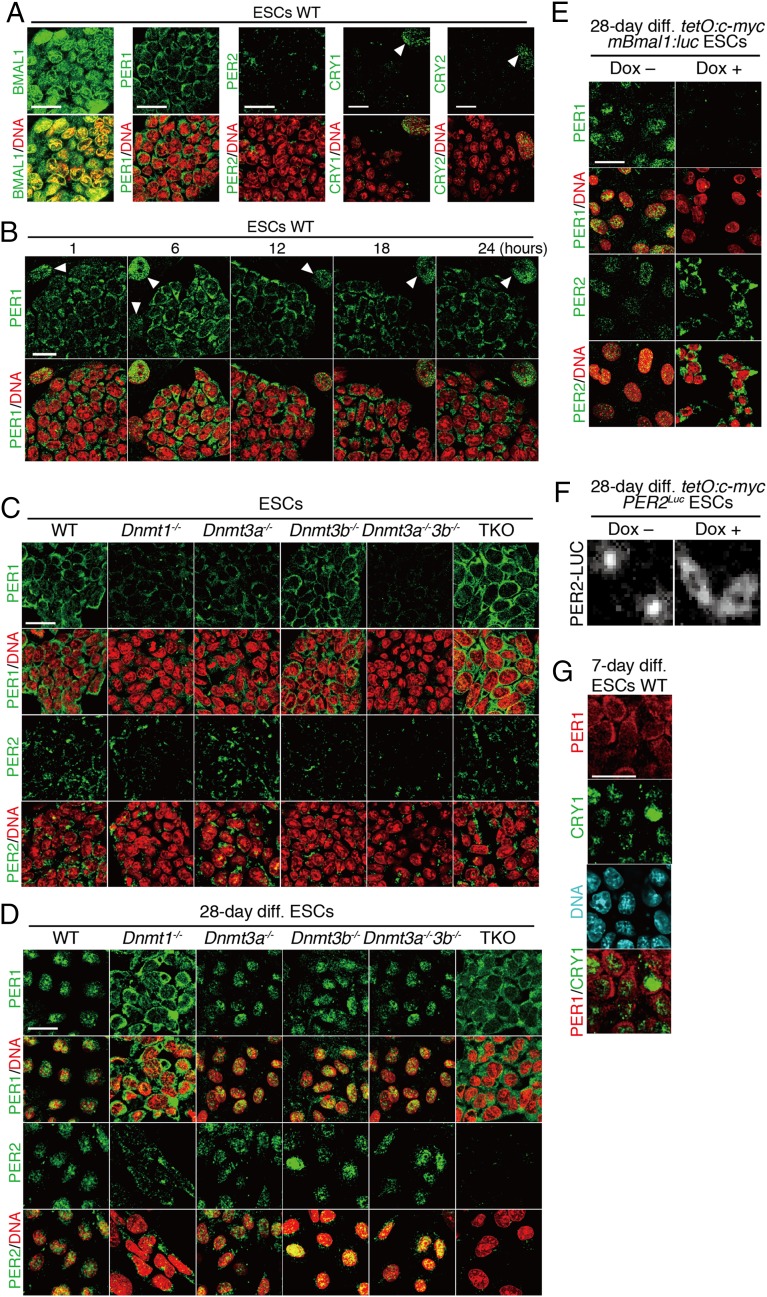
To survey the underlying mechanism for the loss of oscillation in ESCs as well as Dnmt1−/− and c-Myc-overexpressed ESC-derived cells, immunofluorescence staining of endogenous circadian clock proteins was performed. Interestingly, in ESCs, PER1 protein was located exclusively in the cytoplasm, although a nuclear dominant expression pattern was observed in MEFs (Fig. 5A and Fig. S6). PER2 signals were weak, consistent with low-level expression of Per2 mRNA, as described above. To rule out the circadian variation of PER1 subcellular localization, we investigated the temporal profile of the PER1 expression pattern. Immunofluorescence analysis revealed that the PER1 protein in ESCs was expressed exclusively in the cytoplasm throughout the day (Fig. 5B). Because temporal accumulation of PER proteins in the nucleus is believed to be essential for circadian clock oscillation, the cytoplasmic expression of PER1 is a possible reason why circadian clock oscillation is not exhibited in ESCs.
Fig. 5.
Cytoplasmic PERs in clock-disrupted conditions. (A) Immunofluorescence study of endogenous BMAL1, PER1, PER2, CRY1, and CRY2 proteins in WT ESCs. (B) Temporal expression pattern of PER1 protein in WT ESCs after medium change. Arrowheads indicate nucleus of feeder cells. (C) Immunofluorescence of PER1 and PER2 proteins in Dnmt-deficient ESCs (WT, Dnmt1−/−, Dnmt3a−/−, Dnmt3b−/−, Dnmt3a−/−3b−/−, and TKO). (D) Immunofluorescence of PER1 and PER2 proteins in in vitro 28-d-differentiated Dnmt-deficient ESCs. (E) Immunofluorescence of PER1 and PER2 proteins in in vitro 28-d-differentiated tetO:c-myc mBmal1:luc ESCs with/without Dox. (F) Bioluminescence of PER2-Luciferase in in vitro 28-d-differentiated tetO:c-myc PER2Luc ESCs with/without Dox. (G) Immunofluorescence double staining of PER1 and CRY1 in vitro 7-d-differentiated WT ESCs. (Scale bars, 25 µm.)
Next, we also examined the expression patterns of PER proteins in Dnmt-deficient ESCs and 28-d differentiated cells. All Dnmt-deficient ESCs also showed cytoplasmic accumulation of PER proteins, which was essentially identical to the expression pattern of WT ESCs (Fig. 5C). Clock-oscillating differentiated cells exhibited a predominant nuclear localization of PER proteins (Fig. 5D). However, clock-disrupted cells such as Dnmt1−/− and TKO cells showed cytoplasmic localization of PERs even after the 28-d differentiation culture, similar to ESCs (Fig. 5D).
Intriguingly, immunofluorescence staining of PER2 in tetO:c-myc mBmal1:luc ESCs after the differentiation culture also exhibited the cytoplasmic accumulation of PER2 in the Dox+ condition, whereas nuclear localization occurred in the Dox− condition (Fig. 5E) In tetO:c-myc PER2Luc ESCs, we found that PER2::Luc fusion proteins localized exclusively in the cytoplasm when the cells were differentiated in the Dox+ condition, whereas PER2::Luc was predominantly nuclear in localization in the Dox− condition (Fig. 5F). Moreover, we investigated the expression of PER1 and CRY1 in 7-d differentiated WT ESCs. Notably, CRY1 protein already expressed in the nuclei of 7-d differentiated ESCs, but PER1 was still predominantly accumulated in the cytoplasm (Fig. 5G). These findings strongly indicate that the regulatory mechanisms of cytoplasmic accumulation of PERs occur in the clock-less conditions.
Kpna2 Functions as a Critical Factor Regulating Differentiation-Coupled Circadian Clock Development.
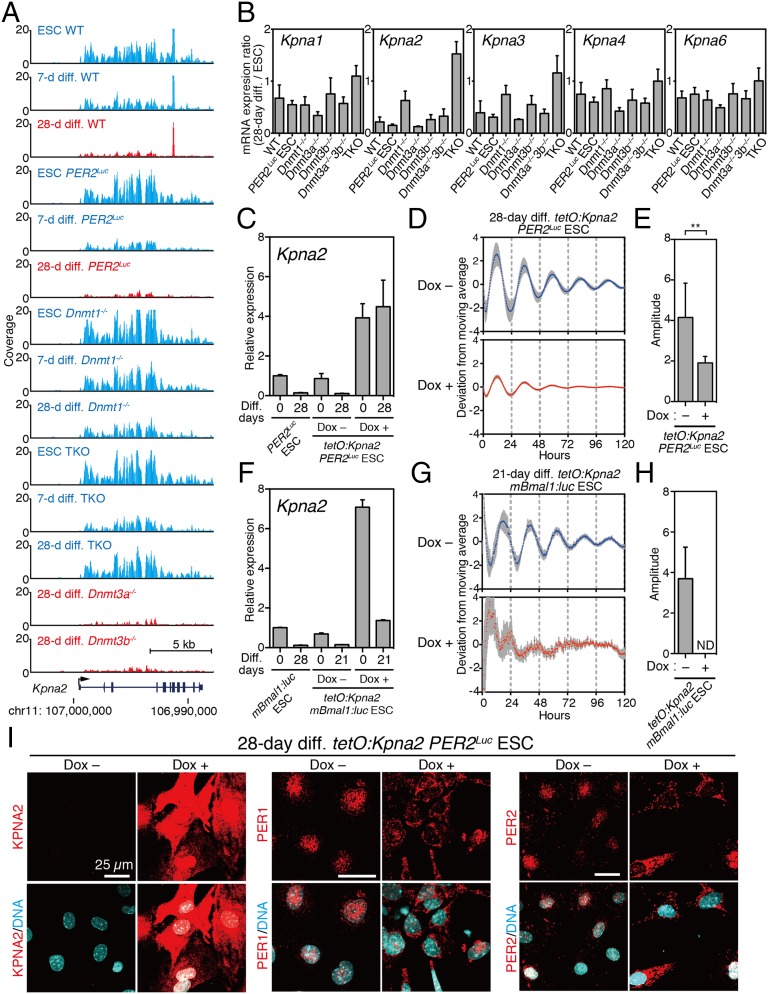
Within the gene set correlating with circadian clock development, the Kpna2 gene was identified as an up-regulated gene in all of the clock-disrupted cells, including ESCs (Fig. 6A). The Kpna2 gene encodes the Importin-α2 protein known to regulate the nuclear translocation of transcription factors, and the switching of the expression of the Importin-α subtype from α2 to α1 is known to play an important role in ESC differentiation (29, 30). Therefore, we first analyzed the ratio of Kpna expression in differentiated cells against ESCs. In WT ESCs, the ratio of Kpna2 expression in differentiated cells was dramatically reduced, whereas the expression level of Kpna2 remained higher in Dnmt1−/− and TKO cells even after 28-d differentiation culture conditions (Fig. 6B). Immunofluorescence of endogenous KPNA2 protein confirmed that the strong expression of KPNA2 was observed in WT and all Dnmt-deficient ESCs. Strikingly, strong expression of KPNA2 protein was also seen in 28-d differentiated Dnmt1−/− and Dnmt TKO cells (Fig. S7 A and B). These results suggest that sustained high expression of Kpna2 may be a causal factor blocking circadian clock development.
Fig. 6.
Constitutive Kpna2 expression during ESC differentiation inhibits the development of circadian rhythm. (A) University of California, Santa Cruz (UCSC) genome browser view of Kpna2 locus detected as a candidate factor is shown in nonrhythmic cells (blue) and rhythmic cells (red). (B) Quantitative RT-PCR analysis of Kpna1, Kpna2, Kpna3, Kpna4, and Kpna6 in in vitro 28-d-differentiated ESCs relative to undifferentiated ESCs. Data are mean ± SD (n = 3). (C) Relative gene expression of Kpna2 in tetO:Kpna2 ESCs or in vitro 28-d differentiation with/without Dox (500 ng/mL). PER2Luc ESC represents a control ESC. Data are mean ± SD (n = 3). (D) Averaged bioluminescence traces of in vitro 28-d-differentiated tetO:Kpna2 ESCs with (red) or without (blue) Dox treatment. Data detrended by subtracting a 24-h moving average are mean with SD (n = 6). (E) PER2Luc-driven bioluminescence of in vitro 28-d-differentiated tetO:Kpna2 ESCs with or without Dox (Student t test, **P < 0.01). (F) Relative gene expression of Kpna2 in tetO:Kpna2 Bmal1:luc ESCs or in vitro 28-d differentiation with/without Dox (500 ng/mL). Data are mean ± SD (n = 3). (G) Averaged bioluminescence traces of in vitro 28-d-differentiated tetO:Kpna2 Bmal1:luc ESCs with (red) or without (blue) Dox treatment. Data detrended by subtracting a 24-h moving average are mean with SD (n = 6). (H) Bmal1:luc-driven bioluminescence of in vitro 28-d-differentiated tetO:Kpna2 ESCs with or without Dox (Student t test, **P < 0.01). (I) Immunofluorescence study against KPNA2, endogenous PER1, and endogenous PER2 in 28-d-differentiated tetO:Kpna2 PER2Luc ESCs. DNA was stained by Hoechst 33342.
Next we examined subcellular localization of transiently expressed PER2 with or without EGFP-KPNA2 coexpression in MEFs. Coexpression of EGFP-KPNA2 significantly accelerated cytoplasmic accumulation of PER2 protein (Fig. S8 A and B), whereas CRY2 coexpressed with EGFP-KPNA2 was exclusively localized in the nucleus (Fig. S8 C and D). Thus, the observed immunofluorescence indicated an increase of cytoplasmic localization of PER2 by coexpression of EGFP-KPNA2 and supported the involvement of Kpna2 in the mechanism of circadian clock development.
Next, to test the effect of Kpna2 expression on the development of the circadian clock during ESC differentiation, we created Dox-inducible Kpna2 overexpression PER2Luc (tetO:Kpna2 PER2Luc) ESCs. The expression of Kpna2 after Dox treatment was observed in a dose-dependent fashion in these ESCs (Fig. 6C). We subsequently performed in vitro circadian clock formation assays using differentiation cultures with (Dox+) or without Dox (Dox−). Bioluminescence analysis revealed significant reduction of the PER2Luc oscillatory amplitude in the 28-d differentiated cells with Kpna2 overexpression (Dox+) (Fig. 6 D and E). A similar tendency was also observed in a mBmal1:luc reporter read-out system. TetO:Kpna2 mBmal1:luc ESCs differentiated with or without Dox revealed that the constitutive overexpression of Kpna2 severely suppressed the mBmal1:luc-driven circadian clock oscillation (Fig. 6 F–H), suggesting that high-level expression of Kpna2 during ESC differentiation actually obstructed the circadian clock development. Importantly, cytoplasmic accumulation of endogenous PER1 and PER2 proteins was dramatically increased in the Dox+ condition compared with the Dox− condition (Fig. 6I).
However, as seen by the still-remaining nuclear staining of PER2 immunofluorescence in part of the cells of Dox+ condition (Fig. 6I), the effect of KPNA2 on the subcellular localization of PER proteins may be incomplete. Because the physical interaction between KPNA2 and PER2 was very weak (Fig. S9), abnormal cytoplasmic accumulation of PER proteins in clock-less cells, including ESCs, may be an indirect effect of KPNA2. In addition, although the amplitude of the circadian clock was dramatically reduced, the differentiation-coupled PER2Luc oscillation was still detectable even in cells of the Dox+ condition (Fig. 6D). This suggests that other factors for circadian clock development in addition to Kpna2 remain to be elucidated. Altogether, our results reveal that the misregulation of Kpna2, which was identified as a candidate factor through global gene expression analysis, resulted in the abnormal cytoplasmic accumulation of PER proteins and impairment of differentiation-coupled circadian clock development.
Discussion
We recently showed that circadian clock development is closely linked with the cellular differentiation processes (15). Because cellular differentiation is regulated by global epigenetic and transcriptional programs (31), we examined c-Myc-overexpressing ESCs and Dnmt-deficient ESCs as model systems perturbing the cellular differentiation program to understand the molecular mechanisms for circadian clock development. Cellular differentiation with constitutive expression of c-Myc disrupted the circadian clock development from mouse ESCs. Moreover, Dnmt1 was found to be essential for circadian clock development during in vitro differentiation of ESCs. Through global gene expression analysis using the c-Myc-overexpressed and Dnmt-deficient ESC models, we identified a gene set correlating cellular differentiation-coupled circadian clock development in mammalian cells. Among the identified genes, we discovered that Kpna2 encoding Importin-α2 protein functioned as a critical factor regulating the differentiation-coupled switching of subcellular localization patterns of PER proteins and circadian clock development in mammalian cells.
In this study, our model system revealed that the ectopic expression of c-Myc critically affected the developmental process of the circadian clock and abolished molecular oscillations even after 28 d of culture differentiation of ES cells. One possible mechanism is the direct effect of c-MYC on E-box enhancer elements competing with BMAL1/CLOCK. Previously it has been reported that MYC can recognize the BMAL1/CLOCK E-box enhancer and up-regulate E-box-driven clock genes (32, 33). In this study we examined global gene expression profiles with or without c-Myc expression during ESC differentiation. Interestingly, clock gene expression profiles were not dramatically changed even in the c-Myc-overexpressed condition (Fig. 2). On the other hand, MYC is known to play a fundamental role in global gene expression and can induce misregulation of the transcriptional program in various cell types (21, 25, 26). Consistently, the expression of thousands of genes was dramatically changed in the differentiated cells with c-Myc overexpression. Thus, our investigation using c-Myc inducible mouse ESCs indicates an indirect role of c-Myc in preventing circadian clock development as opposed to directly abolishing clock gene expression.
In this study we show that Dnmt1 is essential for the differentiation-coupled development of circadian clock oscillations. In contrast, even Dnmt3a−/−3b−/− ESCs were able to exhibit circadian clock oscillations after differentiation culture, suggesting that Dnmt3a and Dnmt3b are not essential for circadian clock development. Dnmt3a was recently reported to affect age-related alteration of circadian behavioral rhythms (34), suggesting that Dnmt3a and/or Dnmt3b may modulate circadian rhythms under certain conditions. In comparison, DNMT1 plays an essential role in the differentiation of mammalian cells by maintaining the epigenetic landscape of DNA methylation, and disturbance of its function during the cellular differentiation critically affects global transcriptional programs and following alteration of cell states (19, 22). Thus, the findings shown in this study indicate that circadian clock development likely shares fundamental mechanisms with cellular differentiation.
Through the analysis of our mutant ESC models, we found the misregulation of Kpna2 expression, reported as one of the key factors regulating ESC differentiation (30), as a common factor affecting circadian clock development. In ESCs as well as the differentiated cells that failed to generate clock oscillation, significantly higher expression of Kpna2 gene was observed compared with cells with functional clock. Our investigation also reveals that sustained ectopic expression of Kpna2 during ESC differentiation suppressed circadian clock development. Recently it has been reported that subtype switching of Importin-α2 (KPNA2) expression to α1 (KPNA1) is observed during differentiation of ESCs, and Importin-α2 negatively regulates the nuclear import of Oct6 to inhibit ESC differentiation (29, 30). Our study revealed that KPNA2 acts as a key factor controlling circadian clock development. This mechanistic link between ESC differentiation and clock development strongly suggests that circadian clock development and cellular differentiation share a common pathway and may mutually regulate their process in mammalian cells.
In ESCs as well as differentiated c-Myc-overexpressed and Dnmt1−/− ESCs, we found that the PER proteins did not localize in the nucleus; rather they exclusively accumulated in the cytoplasm. Because PER translocation to the nucleus in critical for the generation of circadian oscillations in mammals, this defect may underlie the lack of a functional clock in these cells (6, 9, 35–37). We also demonstrate that the overexpression of Kpna2 in differentiating ESCs makes the subcellular localization pattern of PER2 shift much more cytoplasmic. These findings are compatible with the results obtained through the immunofluorescence studies showing exclusively cytoplasmic accumulation of PER proteins as a common feature in clock-disrupted conditions and ESCs. Thus, it is conceivable that the misregulation of Kpna2 expression via the disturbance of epigenetic and transcriptional programs during differentiation plays a distinct role in blocking circadian clock development. On the other hand, overexpression of Kpna2 does not completely abolish the clock development or PER2 nuclear localization. These results therefore indicate that additional mechanisms also regulate clock development.
Taken together, our studies using c-Myc-expressing ESCs as well as Dnmt1−/− ESCs show that the misregulation of the differentiation-coupled transcriptional program may critically affect not only the conversion of the cell state but also the development of the cellular circadian clock during cellular differentiation processes. Our findings suggest that Kpna2 functions as a key factor for clock development by modulating subcellular localization patterns of PER proteins. Of course, because there are hundreds of misregulated genes other than Kpna2, multiple pathways or mechanisms are likely to contribute to circadian clock development during differentiation. Our study highlights the importance of “clock development regulating factors” regulated by the differentiation-coupled transcriptional program in the emergence of the circadian clock in developing mammalian cells.
Materials and Methods
Cell Culture.
Two ESC lines (KY1.1 mentioned as WT in text and PER2Luc ESC) were used (15, 23, 24). In the PER2Luc ESCs the endogenous PER2 gene is replaced by a fusion of PER2 and firefly luciferase gene, and an additional SV40 polyadenylation site was inserted downstream of the luciferase (23). Dnmt1−/−, Dnmt3a−/−, Dnmt3b−/−, Dnmt3a−/−3b−/−, and TKO ESCs (27, 38) were kindly provided from Dr. Masaki Okano (RIKEN, Kobe, Japan). These ESC lines were cultured on a feeder layer of mitomycin C-treated primary MEFs in an ES medium containing Glasgow Minimum Essential Medium (Wako) supplemented with 15% (vol/vol) FBS (HyClone), 0.1 mM MEM nonessential amino acids (Nacalai Tesque), 0.1 mM 2-mercaptoethanol (Sigma), 1,000 U/mL of leukemia inhibitory factor (LIF), and 100 U/mL of penicillin–streptomycin (Nacalai Tesque). For preparation of PER2Luc knock-in MEFs (24) or WT MEFs, embryos were collected at embryonic day 15.5 (E15.5). After removal of the head and visceral tissues, the remaining bodies were washed in a fresh PBS and minced, and the isolated cells were maintained in embryonic fibroblast medium (EFM) as mentioned below.
COS-7 cells were cultured in high-glucose DMEM supplemented with 10% (vol/vol) FBS.
In Vitro Differentiation.
In vitro differentiation of ESCs was performed as described recently (17). Briefly, after ESCs were trypsinized and feeder cells were removed, embryoid bodies (EBs) were generated by harvesting 2,000 cells and seeding them onto low-attachment 96-well plates (Lipidure Coat, NOF) in a differentiating medium without LIF supplementation (EFM), which was composed of a high-glucose DMEM (Nacalai Tesque) containing 10% FBS, 1 mM sodium pyruvate (Nacalai Tesque), 0.1 mM nonessential amino acids, GlutaMax-I (Invitrogen), 100 µM β-mercaptoethanol, and 100 U/mL penicillin–streptomycin. Two day later, EBs were plated onto gelatin-coated tissue culture 24-well plates and grown for several additional weeks. EFM was changed every 1–2 d.
For constitutive expression of c-Myc or Kpna2 during in vitro differentiation of ESCs carrying Dox-inducible c-Myc or Kpna2, EFM containing Dox (Invitrogen) was changed every 1 to 2 d. For a real-time monitoring analysis, Dox was removed by washing three times with EFM.
Detailed methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank H. Inokawa, Y. Tsuchiya, and Y. Minami (Kyoto Prefectural University of Medicine) for their valuable discussions and technical support; M. Okano (RIKEN) for providing Dnmt knockout ESCs; K. Tashiro, M. Nakano, and T. Sato (Genome Center of Kyoto Prefectural University of Medicine) for their technical support for RNA-seq; and Y. Yoneda (Osaka University) and H. Tei (Kanazawa University) for providing materials. The authors were supported in part by a Japan Science and Technology Agency Precursory Research for Embryonic Science and Technology program (K.Y.), and grants-in-aid for scientific research from the Japan Society for the Promotion of Science (to K.Y. and Y.U.), the Takeda Science Foundation (to K.Y.), the Robert A. Welch Foundation [Grant AU-1731 (to Z.C.)], and the National Institutes of Health [Grant R01AG045828 (to Z.C.)]. J.S.T. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray and RNA-sequencing data reported in this paper have been deposited in Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE61096 and GSE61184).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419272111/-/DCSupplemental.
References
- 1.Lowrey PL, Takahashi JS. Genetics of circadian rhythms in Mammalian model organisms. Adv Genet. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bass J. Circadian topology of metabolism. Nature. 2012;491(7424):348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 3.Masri S, Sassone-Corsi P. The circadian clock: A framework linking metabolism, epigenetics and neuronal function. Nat Rev Neurosci. 2013;14(1):69–75. doi: 10.1038/nrn3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King DP, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89(4):641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gekakis N, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280(5369):1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 6.Kume K, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98(2):193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 7.Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286(5440):768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 8.Yagita K, Tamanini F, van Der Horst GT, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292(5515):278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- 9.Yagita K, et al. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 2002;21(6):1301–1314. doi: 10.1093/emboj/21.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 11.Ukai-Tadenuma M, et al. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell. 2011;144(2):268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 12.Reppert SM, Schwartz WJ. Maternal suprachiasmatic nuclei are necessary for maternal coordination of the developing circadian system. J Neurosci. 1986;6(9):2724–2729. doi: 10.1523/JNEUROSCI.06-09-02724.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis FC, Gorski RA. Development of hamster circadian rhythms: Role of the maternal suprachiasmatic nucleus. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1988;162(5):601–610. doi: 10.1007/BF01342635. [DOI] [PubMed] [Google Scholar]
- 14.Amano T, et al. Expression and functional analyses of circadian genes in mouse oocytes and preimplantation embryos: Cry1 is involved in the meiotic process independently of circadian clock regulation. Biol Reprod. 2009;80(3):473–483. doi: 10.1095/biolreprod.108.069542. [DOI] [PubMed] [Google Scholar]
- 15.Yagita K, et al. Development of the circadian oscillator during differentiation of mouse embryonic stem cells in vitro. Proc Natl Acad Sci USA. 2010;107(8):3846–3851. doi: 10.1073/pnas.0913256107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kowalska E, Moriggi E, Bauer C, Dibner C, Brown SA. The circadian clock starts ticking at a developmentally early stage. J Biol Rhythms. 2010;25(6):442–449. doi: 10.1177/0748730410385281. [DOI] [PubMed] [Google Scholar]
- 17.Umemura Y, et al. An in vitro ES cell-based clock recapitulation assay model identifies CK2α as an endogenous clock regulator. PLoS ONE. 2013;8(6):e67241. doi: 10.1371/journal.pone.0067241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40(5):499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454(7200):49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hikichi T, et al. Transcription factors interfering with dedifferentiation induce cell type-specific transcriptional profiles. Proc Natl Acad Sci USA. 2013;110(16):6412–6417. doi: 10.1073/pnas.1220200110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152(6):1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohnishi K, et al. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell. 2014;156(4):663–677. doi: 10.1016/j.cell.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Chen Z, et al. Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc Natl Acad Sci USA. 2012;109(1):101–106. doi: 10.1073/pnas.1118034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo SH, et al. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101(15):5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143(2):313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141(3):432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 28.Sakaue M, et al. DNA methylation is dispensable for the growth and survival of the extraembryonic lineages. Curr Biol. 2010;20(16):1452–1457. doi: 10.1016/j.cub.2010.06.050. [DOI] [PubMed] [Google Scholar]
- 29.Yasuhara N, et al. Triggering neural differentiation of ES cells by subtype switching of importin-alpha. Nat Cell Biol. 2007;9(1):72–79. doi: 10.1038/ncb1521. [DOI] [PubMed] [Google Scholar]
- 30.Yasuhara N, et al. Importin alpha subtypes determine differential transcription factor localization in embryonic stem cells maintenance. Dev Cell. 2013;26(2):123–135. doi: 10.1016/j.devcel.2013.06.022. [DOI] [PubMed] [Google Scholar]
- 31.Holmberg J, Perlmann T. Maintaining differentiated cellular identity. Nat Rev Genet. 2012;13(6):429–439. doi: 10.1038/nrg3209. [DOI] [PubMed] [Google Scholar]
- 32.Levens DL. Reconstructing MYC. Genes Dev. 2003;17(9):1071–1077. doi: 10.1101/gad.1095203. [DOI] [PubMed] [Google Scholar]
- 33.Kondratov RV, Shamanna RK, Kondratova AA, Gorbacheva VY, Antoch MP. Dual role of the CLOCK/BMAL1 circadian complex in transcriptional regulation. FASEB J. 2006;20(3):530–532. doi: 10.1096/fj.05-5321fje. [DOI] [PubMed] [Google Scholar]
- 34.Azzi A, et al. Circadian behavior is light-reprogrammed by plastic DNA methylation. Nat Neurosci. 2014;17(3):377–382. doi: 10.1038/nn.3651. [DOI] [PubMed] [Google Scholar]
- 35.van der Horst GT, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398(6728):627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 36.Yagita K, et al. Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes Dev. 2000;14(11):1353–1363. [PMC free article] [PubMed] [Google Scholar]
- 37.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418(6901):935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 38.Lei H, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122(10):3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.