Significance
Pseudomonas aeruginosa is a pathogen that kills a remarkably wide range of hosts. The environmental cues that regulate P. aeruginosa virulence have remained unclear. Here, we develop a rapid imaging-based virulence assay to quantify virulence. We find that association with rigid surfaces induces virulence toward multiple hosts. Virulence induction depends on the mechanical, but not chemical, properties of the surfaces and requires the surface-exposed protein PilY1, which has homology to the mechanosensitive von Willebrand factor A domain. Specific mutation of this mechanosensitive domain is sufficient to constitutively activate virulence independent of surface attachment. Mechanosensitive virulence induction can explain how P. aeruginosa infects a broad range of hosts while tightly regulating virulence. Consistently, association with one host induces virulence toward other hosts.
Keywords: bacterial mechanosensation, PilY1, von Willebrand factor, host detection, contact regulation
Abstract
Pseudomonas aeruginosa infects every type of host that has been examined by deploying multiple virulence factors. Previous studies of virulence regulation have largely focused on chemical cues, but P. aeruginosa may also respond to mechanical cues. Using a rapid imaging-based virulence assay, we demonstrate that P. aeruginosa activates virulence in response to attachment to a range of chemically distinct surfaces, suggesting that this bacterial species responds to mechanical properties of its substrates. Surface-activated virulence requires quorum sensing, but activating quorum sensing does not induce virulence without surface attachment. The activation of virulence by surfaces also requires the surface-exposed protein PilY1, which has a domain homologous to a eukaryotic mechanosensor. Specific mutation of the putative PilY1 mechanosensory domain is sufficient to induce virulence in non–surface-attached cells, suggesting that PilY1 mediates surface mechanotransduction. Triggering virulence only when cells are both at high density and attached to a surface—two host-nonspecific cues—explains how P. aeruginosa precisely regulates virulence while maintaining broad host specificity.
The bacterium Pseudomonas aeruginosa is a metabolically versatile pathogen that inhabits diverse environments and infects a remarkable range of hosts, including mammals, insects, worms, amoeba, fungi, and other bacteria. P. aeruginosa produces a large number of secreted and cell-associated virulence factors that are redundant and multifactorial (1, 2). Many of P. aeruginosa’s virulence factors—including pyocyanin, elastase, and hydrogen cyanide—are host-nonspecific (3–5), bolstering the ability of P. aeruginosa to attack a large range of hosts. Although many of the virulence factors in P. aeruginosa have been identified, the cues that regulate their activity are less understood. Because many of the virulence factors are host-nonspecific, we explored whether virulence in P. aeruginosa is regulated by host-nonspecific cues.
Host cell membranes and cell surfaces are the first line of defense against bacterial toxins and invasion. P. aeruginosa attaches to host cell surfaces early during the infection process. The presence of a surface could thus act as a cue for P. aeruginosa, signaling the presence of a host. Surface attachment is also a critical initial step that enables the establishment of biofilms (6–8). Although biofilms are clearly important for pathogenesis, it remains unclear whether they directly promote host cell killing or mediate other important processes such as long-term colonization.
One host-nonspecific cue that could regulate virulence is the mechanical force that bacteria experience upon surface attachment. P. aeruginosa performs surface-associated behaviors (7, 8) such as swarming and twitching (9, 10), but it remains unclear whether P. aeruginosa senses the chemical or mechanical properties of surfaces. There is precedence for mechanotransduction in eukaryotes, in which surface substrate recognition is an important regulator of development and behavior (11). In prokaryotes, surface mechanical forces affect the binding affinity of cells to substrates (12, 13) and alter the rotation of flagella (14, 15). However, the effects of mechanical forces on cell behaviors other than motility are not understood, and the regulation of virulence by mechanical cues has not been explored.
Here, we show that attachment to surfaces induces P. aeruginosa to become virulent. Virulence is activated on a variety of chemically distinct abiotic and host surfaces, suggesting that mechanical cues associated with surface attachment activate virulence. We identify PilY1 as a key mediator of surface-activated virulence. PilY1 is a cell-surface–exposed protein that regulates a number of surface-associated behaviors and contains a mechanically sensitive von Willebrand Factor A (VWFa) domain. Although P. aeruginosa lacking PilY1 cannot activate virulence upon surface contact, bacteria with a specific deletion of the VWFa domain hyperactivate virulence, even in the absence of surface contact. Together, our results suggest that cells detect mechanical cues associated with surface attachment through a mechanosensitive pathway that requires the PilY1 protein. We suggest that detecting mechanical cues associated with surface attachment enables P. aeruginosa to induce virulence toward a broad range of hosts without relying upon chemical recognition of any specific host factor.
Results
Surface Attachment Rapidly Induces P. aeruginosa Virulence.
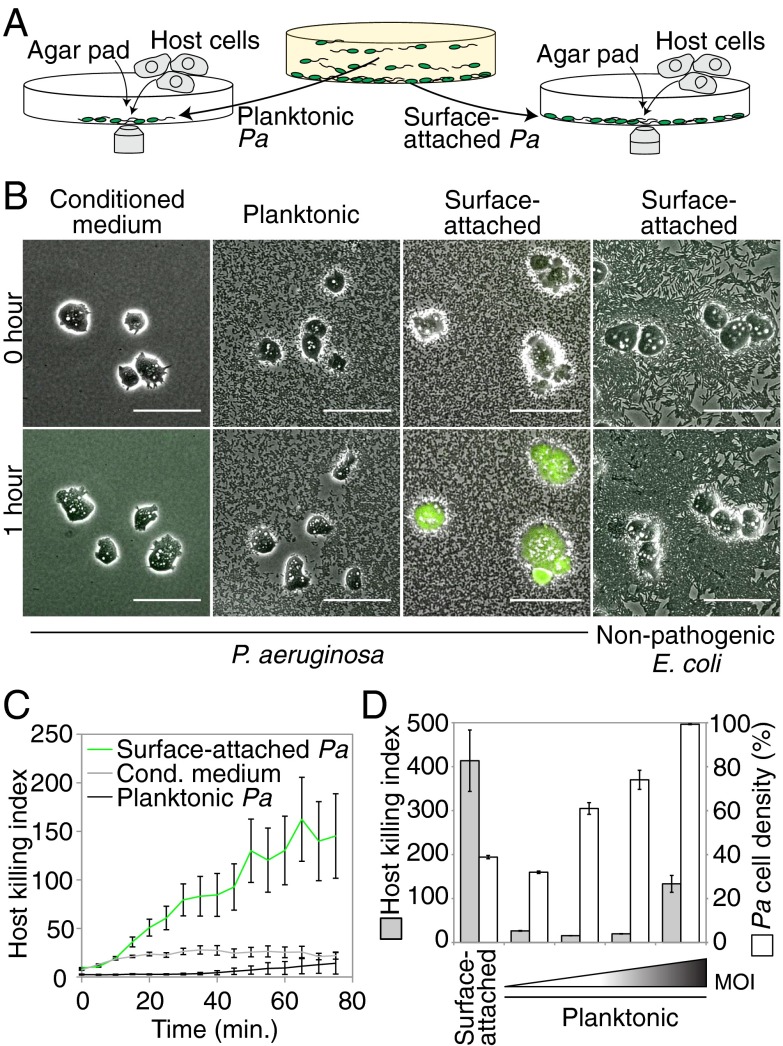
Traditional bacterial virulence assays involve prolonged exposure of bacteria to rigid surfaces such as culture plates or agar dishes and thus cannot establish the specific contribution of surface contact to virulence. We developed a virulence assay that uses single-cell fluorescence imaging to directly monitor virulence at short timescales, enabling us to separately assay the virulence of planktonic (liquid-grown) and surface-attached bacterial subpopulations in shaking cultures. We initially focused on the amoeba Dictyostelium discoideum as a model host. D. discoideum is a natural host for P. aeruginosa and is functionally similar to mammalian macrophages, exhibiting chemotaxis toward and phagocytosis of bacteria. Genetic studies have validated that the virulence factors that act on mammalian and amoeba hosts are largely identical (16, 17). We monitored virulence by mixing amoebae with planktonic or surface-attached bacteria, confining all cells to a single plane using an agar pad, and measuring cell viability using single-cell time-lapse imaging in the presence of calcein acetoxymethyl ester (calcein-AM) (Fig. 1A and SI Appendix, Fig. S1). Calcein-AM does not permeate the D. discoideum cell membrane and fluoresces when it is cleaved by intracellular esterases. Healthy D. discoideum cells are flat, motile, and produce little or no calcein-AM fluorescence, whereas unhealthy D. discoideum cells are rounded, immotile, and produce strong calcein-AM fluorescence before lysing (Fig. 1B and SI Appendix, Fig. S2A). We compute a host killing index, a measure of host cell viability, by integrating the calcein-AM fluorescence of individual amoebae, dividing by the cell area, and averaging this value over many cells (SI Appendix, Fig. S1).
Fig. 1.
Surface attachment stimulates P. aeruginosa virulence toward D. discoideum. (A) Schematic depicting a rapid imaging-based virulence assay. Planktonic or surface-attached subpopulations of P. aeruginosa (Pa) were isolated from a Petri dish, mixed with host cells, confined to a single plane using an agar pad, and imaged. (B) Composite phase contrast (grayscale) and calcein-AM fluorescence (green) images of D. discoideum (amoebae) that were mixed with conditioned medium, planktonic or glass surface-attached P. aeruginosa, or with nonpathogenic E. coli. (Scale bars: 50 µm.) (C) Host killing indexes of amoebae that were mixed with conditioned medium or with planktonic or plastic surface-attached P. aeruginosa. (D) Host killing indexes (gray bars) and corresponding cell densities (white bars) of surface-attached or planktonic P. aeruginosa at different multiplicities of infection (MOI). Bars represent the average of three independent experiments, and error bars indicate SE.
Our imaging-based virulence assay revealed a striking difference between planktonic and surface-attached P. aeruginosa. Amoebae that were mixed with planktonic P. aeruginosa flattened and exhibited robust motility and phagocytosis, but little or no calcein-AM fluorescence (Fig. 1 B and C and Movie S1). These behaviors were, in all respects, similar to those observed in the presence of nonpathogenic Escherichia coli B/r (Fig. 1B). In contrast, amoebae that were exposed to surface-attached P. aeruginosa became rounded, did not exhibit motility or phagocytosis, and typically produced calcein-AM fluorescence within 15–20 min (Fig. 1 B and C and Movie S1). We note that amoebae that are exposed to conditioned medium only (Fig. 1C) enter a starvation phase, which appears to make them more susceptible to secreted P. aeruginosa host killing factors (2–5). We confirmed the increased virulence of surface-attached P. aeruginosa using three other host viability reporters (SI Appendix, Fig. S2B). To establish the multiplicity of infection (MOI), we showed that P. aeruginosa cell density correlates with the number of P. aeruginosa cells in our imaging assays and then used P. aeruginosa density as a proxy for MOI in subsequent experiments (SI Appendix, Fig. S3). The significantly increased virulence of surface-attached P. aeruginosa cannot be attributed to differences in MOI, because surface-attached P. aeruginosa kill amoebae at significantly lower cell numbers than planktonic P. aeruginosa (Fig. 1D).
Virulence Activation Is Surface- and Host-Independent.
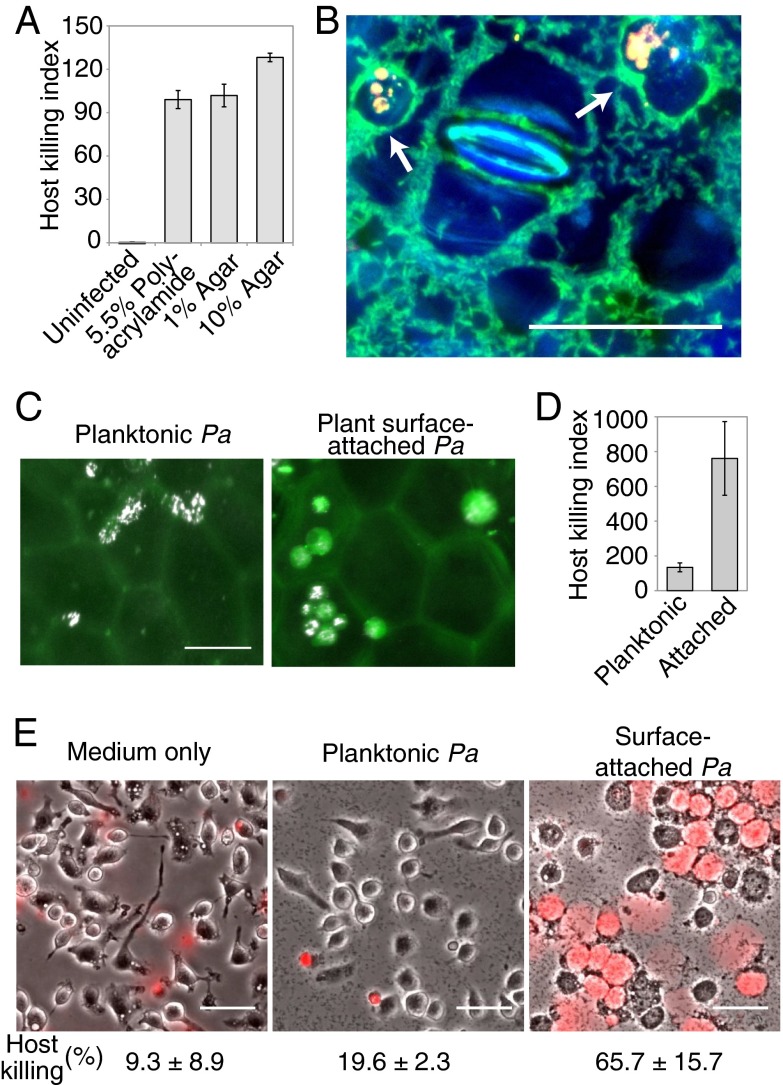
Bacteria could sense surfaces by detecting chemical or mechanical features of the surface. To differentiate these possibilities, we assayed a range of chemically distinct rigid surfaces. P. aeruginosa virulence was similarly activated by glass (Fig. 1B), plastic (Fig. 1C), polyacrylamide (Fig. 2A), and agar (Fig. 2A) surfaces. Thus, virulence activation requires a mechanically rigid surface, but does not depend on the surface’s specific chemical composition.
Fig. 2.
Virulence activation is surface- and host-independent. (A) Host killing indexes of P. aeruginosa cells that were attached to polyacrylamide or agar surfaces. Error bars indicate the range of two independent experiments. (B) Assay to test virulence of P. aeruginosa (green) that attach to the surface of a plant leaf (surface and stoma in blue) using amoebae as hosts (arrows point to cell compartments labeled yellow by uptake of fluorescent beads). (C and D) Amoebae (white) were mixed with planktonic (C, Left) or plant surface-attached (C, Right) P. aeruginosa cells (not visible in image), imaged on the plant leaf surface (faint green from leaf autofluorescence), and assessed for viability by using calcein-AM (green), which was used to compute host killing indexes (D). Error bars indicate the SD of three independent experiments. (E) Composite images of phase contrast (grayscale) and propidium iodide fluorescence (red) of mouse macrophages that were exposed to medium only (Left) or to planktonic (Center) or surface-attached (Right) P. aeruginosa for 3 h. The fraction of macrophages with propidium iodide fluorescence after 4.5 h of exposure is given. (Scale bars: 50 µm.)
Because P. aeruginosa infects a broad range of host types (18), surface contact could serve as a nonspecific cue for host infection. In such a model, P. aeruginosa mechanically detects the presence of a host and broadly activates virulence factors, predicting that surface contact with one type of host induces virulence factors that kill other hosts. We tested this prediction by culturing P. aeruginosa cells on the surface of one host, a pothos plant (Epipremnum aureum), and monitoring the virulence of P. aeruginosa toward a second host, D. discoideum (Fig. 2 B–D). Attachment to the surface of a pothos leaf stimulated virulence toward D. discoideum (Fig. 2 B–D), demonstrating that surface contact is a host-nonspecific cue that induces P. aeruginosa virulence.
The mechanical cues associated with surface contact are not host-specific, suggesting that surfaces could also stimulate P. aeruginosa virulence toward hosts other than amoebae. We therefore compared the viability of mouse macrophages in the presence of planktonic and surface-attached P. aeruginosa. To quantify macrophage health, we used propidium iodide, a nucleic acid dye that cannot permeate healthy cells. Uninfected macrophages and macrophages that were mixed with planktonic P. aeruginosa were largely motile and displayed no propidium iodide fluorescence (Fig. 2E). In contrast, surface-attached P. aeruginosa were more virulent and caused the majority of macrophages to lyse (Fig. 2E).
Quorum Sensing Is Necessary but Not Sufficient for Surface-Activated Virulence.
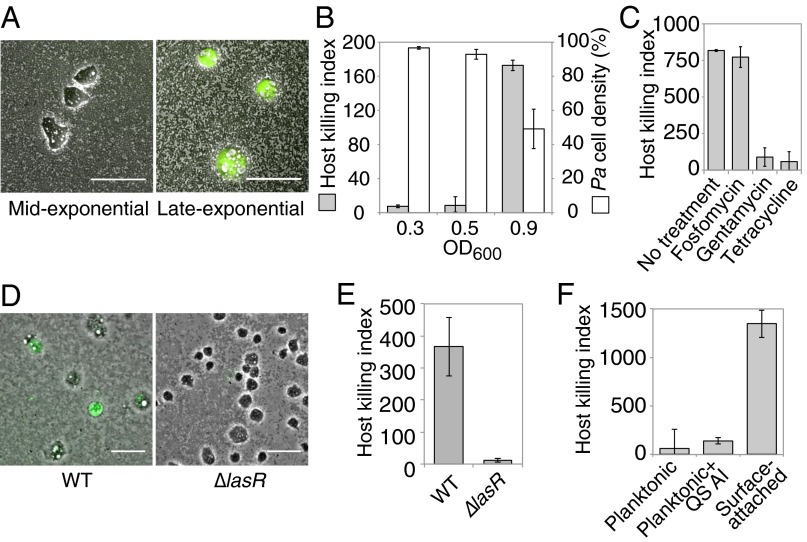
There are at least two explanations for the increased virulence of surface-attached bacteria relative to their planktonic counterparts. Bacteria could develop virulence once they encounter a surface, or surface adhesion could enrich for a subpopulation of cells that are already highly virulent. By isolating P. aeruginosa from different growth phases, allowing them to surface-attach for 1 h, and then exposing them to amoebae to assay their virulence, we found that virulence dramatically increases between midexponential (OD600 = 0.5) and late-exponential (OD600 = 0.9) growth phases (Fig. 3 A and B). The number of P. aeruginosa cells attached to the surface was lower for late-exponential cultures (Fig. 3B), indicating that virulence is not due to an increase in MOI. Although some surface-activated virulence was also observed after <1 h of attachment, the host killing was heterogeneous (SI Appendix, Fig. S4A). Because virulence was most activated at the late-exponential growth phase and host killing was homogeneous after 1 h of attachment, we performed the rest of our virulence assays using cells from this growth phase and after at least 1 h of surface attachment. We found that P. aeruginosa treated with fosfomycin, an antibiotic that inhibits growth and division but does not block protein synthesis, became virulent after a similar duration (Fig. 3C), confirming that virulence development is not a result of increased bacterial growth or division. In contrast, inhibitors of protein synthesis such as gentamycin and tetracycline blocked induction of virulence by surface attachment (Fig. 3C). Once the bacteria had become virulent, treatment with gentamycin no longer inhibited virulence (SI Appendix, Fig. S4B), indicating that new protein synthesis is required for the development of virulence, but not for the subsequent process of killing host cells.
Fig. 3.
Contact-mediated virulence is rapidly induced and requires quorum sensing but not growth. (A and B) Composite phase contrast and calcein-AM fluorescence images (A) and host killing indexes (gray bars) and corresponding P. aeruginosa cell densities (white bars; B; see SI Appendix, SI Materials and Methods for details) for P. aeruginosa that were surface-attached for 1 h at midexponential (OD600 = 0.3 or 0.5) or late-exponential (OD600 = 0.9) growth phases and mixed with amoebae. (C) Host killing indexes for P. aeruginosa that were treated with antibiotics during surface attachment at late-exponential phase. (D and E) Composite phase contrast and calcein-AM fluorescence images (D) and host killing indexes (E) for WT and ΔlasR (quorum-sensing defective) cells. (F) Host killing indexes of planktonic P. aeruginosa cells that were supplemented with DMSO or quorum-sensing autoinducers (QS AI). Bars represent the average of at least two independent experiments, and error bars indicate SD. (Scale bars: 50 µm.)
The development of virulence in late-exponential phase, when cell density is high, suggested that quorum sensing could influence surface-activated virulence. To examine the role of quorum sensing, we assayed a ΔlasR mutant. This mutation significantly disrupted surface-activated virulence (Fig. 3 D and E), indicating that quorum sensing is necessary for surface-activated virulence. However, two lines of evidence suggest that quorum sensing is not sufficient to activate virulence in the absence of a surface. First, high-density (quorum-sensing–activated) planktonic P. aeruginosa cells did not lyse amoebae (Fig. 1 B–D). Second, hyperactivating quorum sensing by administering the LasR and RhlR autoinducers N-(3-oxododecanoyl)-l-homoserine lactone (3OC12-HSL) and N-butyryl-homoserine lactone (C4-HSL), respectively (19, 20) (SI Appendix, Fig. S5), did not activate virulence in planktonic bacteria (Fig. 3F). The finding that quorum sensing is necessary but not sufficient for surface-activated virulence is consistent with the hypothesis that surface activation of virulence also requires detection of a mechanical stimulus. Furthermore, these results explain why planktonic cells were not virulent when they were placed on surfaces with agar pads for 1 h (Fig. 1C), because the agar pads were made with fresh medium and thus diluted the quorum-sensing activator.
Surface Detection Is Mediated by PilY1, a Putative Mechanosensor.
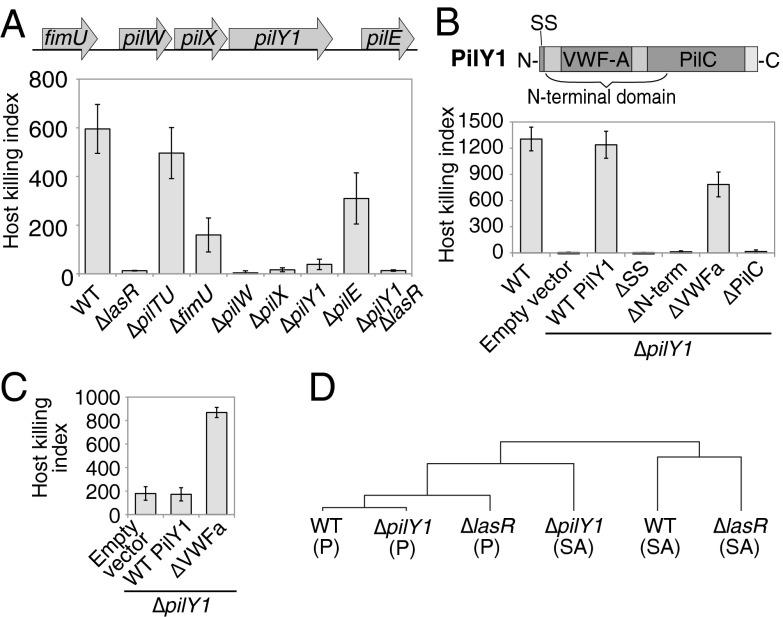
Which genes mediate the ability to sense surfaces and activate virulence? Although flagella are regulated by mechanical forces (14, 15), surface-activated virulence does not require flagella (SI Appendix, Fig. S6), suggesting that P. aeruginosa uses a previously uncharacterized mechanosensory system. Because many signaling pathways autoregulate gene expression upon activation, we performed gene-expression profiling and operon analysis of planktonic and surface-attached P. aeruginosa to identify candidate surface signaling pathways. Consistent with previous studies suggesting that P. aeruginosa virulence is multifactorial (1) and our finding that virulence induction requires new protein synthesis (Fig. 3C), attachment to a surface for 1 h led to the induction of multiple genes previously associated with virulence (SI Appendix, Table S1). Because candidate surface sensors should be present on the bacterial cell surface, our attention was drawn to one of the most highly activated operons in surface-attached P. aeruginosa (SI Appendix, Fig. S7 A and B), which encoded the cell-surface–associated protein PilY1 (21, 22) and the minor pilin proteins FimU, PilW, PilX, and PilE. The minor pilins and PilY1 have been primarily characterized as pilus biogenesis factors (22–24), and mutations in these genes cause defects in type IV pilus production (23, 25, 26). We found that ΔpilY1, ΔpilW, and ΔpilX mutants were defective for surface-activated virulence toward amoebae, whereas ΔfimU and ΔpilE partially retained virulence (Fig. 4A). Importantly, the lack of pili cannot explain the reduced virulence of ΔpilY1, ΔpilW, and ΔpilX in our assay because other mutants that lack type IV pili, such as ΔpilB and ΔpilC mutants, remain virulent (SI Appendix, Fig. S6). Minor pilins are incorporated into type IV pili by a process that requires PilD (27), but ΔpilD mutants also retained virulence (SI Appendix, Fig. S6). Furthermore, although pilus mutants are partially defective in surface attachment (6), these mutants retained virulence in our assays, indicating that virulence is independent of pilus-mediated surface attachment. Together, our results suggest that PilY1 and the minor pilins regulate surface-activated virulence independently of pilus assembly and function.
Fig. 4.
PilY1 regulates surface-activated virulence and gene expression through a mechanosensory VWFa domain. (A) Amoebae host killing indexes of surface-attached pilY1 operon P. aeruginosa mutants. (B) Host killing indexes of surface-attached PilY1 mutants containing a deletion of the N-terminal signal sequence (SS) or deletion of the N-terminal, VWFa, or PilC domains (schematic of PilY1 domains at top). (C) Host killing indexes of planktonic (non–surface-attached) P. aeruginosa cells containing empty vector or expressing WT or VWFa-domain–deleted PilY1. All bars are the average of three independent experiments, and error bars represent SD. (D) Hierarchical clustering tree (correlation values are given in SI Appendix, Fig. S7D) for transcriptional profiles from microarrays for WT, ΔlasR, and ΔpilY1 cells for either surface-attached (SA) or planktonic (P) P. aeruginosa cells for genes that were activated by at least fourfold by surface attachment in overnight cultures.
PilY1 has a domain that shares homology with the mechanosensitive VWFa domain (28) and could consequently serve as a sensor of the mechanical cues associated with surface contact. To characterize the mechanism of PilY1 activity, we performed a structure–function analysis of the domains required for PilY1 surface sensing. The N-terminal region of PilY1 encodes a signal sequence and a VWFa domain, whereas the C-terminal region encodes a PilC domain (28). Deletion of the signal sequence, the whole N-terminal region, or the PilC domain abrogated surface-induced virulence, mimicking the loss of the full-length PilY1 protein, whereas VWFa deletion mutants retained virulence on surfaces (Fig. 4B). We hypothesized that the deletion of the putatively mechanosensitive VWFa domain places PilY1 in a constitutively active state. As predicted, the VWFa domain deletion induced virulence even in planktonic P. aeruginosa cells (Fig. 4C). Given that PilY1 is present on the bacterial cell surface, up-regulated upon surface contact, necessary for surface-activated virulence, and contains a domain homologous to a known mechanosensor whose mutation hyperactivates virulence in the absence of surface contact, we propose that the VWFa domain of PilY1 is responsible for surface detection and that the specific loss of this domain constitutively activates the surface-detection response.
Our data implicate PilY1 and LasR as two master regulators of P. aeruginosa virulence. To determine the effectors that are regulated by each pathway and whether the pathways have overlapping targets, we examined the transcriptional profiles of WT, ΔpilY1, and ΔlasR. The profiles revealed that PilY1 and LasR regulate distinct targets (SI Appendix, Fig. S7C and Tables S2 and S3). For example, expression of the pilY1 operon was not significantly changed in the ΔlasR mutant, and expression of the major quorum-sensing controlled genes was not significantly altered in the ΔpilY1 mutant (SI Appendix, Tables S2 and S3). Furthermore, comparing the transcriptional profiles revealed that surface-attached ΔpilY1 cells were more similar to planktonic WT cells (R = 0.77) than to surface-attached WT cells (R = 0.69) (Fig. 4D and SI Appendix, Fig. S7D). Genes up-regulated by surface attachment in WT are also significantly less induced in ΔpilY1 (SI Appendix, Table S1). Unlike ΔpilY1, surface-attached ΔlasR more closely resembled attached WT (R = 0.76) than planktonic WT (R = 0.39) cells (Fig. 4D and SI Appendix, Fig. S7D). Together, transcriptional profiling suggests that PilY1 is required for the bulk of the surface-induced virulence response and that P. aeruginosa still senses surface contact in the absence of LasR.
If PilY1 is a general sensor for surface contact, its loss should disrupt a wide range of surface-regulated behaviors. Indeed, PilY1 regulates many surface-associated behaviors such as swarming, twitching, cyclic diguanosine monophosphate (c-di-GMP) signaling, and biofilm formation (28–30). These surface-associated behaviors could be mediated by the PilY1-dependent transcriptional response to surface attachment identified in this work (SI Appendix, Table S1).
Multiple Redundant Virulence Factors Likely Function Downstream of Surface Detection.
To understand which genes are activated downstream of surface detection, we investigated the role of previously implicated virulence factors and regulators in surface-activated virulence toward D. discoideum using P. aeruginosa mutants defective in type III secretion, type VI secretion, flagella, type IV pili, fimbriae, exopolysaccharide, quorum sensing, c-di-GMP signaling, two-component signaling, chemotaxis, sigma factor control, or secreted effectors (SI Appendix, Fig. S8). With the exception of mutants of the Las quorum-sensing system characterized above (Fig. 3 D and E), none of the 42 mutants assayed significantly disrupted surface-activated virulence. The lack of a pronounced virulence defect in any one virulence factor mutant supports the hypothesis that P. aeruginosa induces multiple virulence factors that function in a redundant manner (1, 2).
Although type III secretion was not required for virulence in our assay, which was performed over a short timescale (hours) at moderate MOI, type III secretion was previously shown to be important for P. aeruginosa virulence toward D. discoideum when assayed over the course of days at high MOI (16). We suggest that these results indicate that type III secretion contributes to later stages of pathogenesis but is not necessary for the early stages of host cell killing that follow host cell contact.
C-di-GMP is an established regulator of surface-associated behaviors and biofilm formation (31). We were thus surprised that roeA and sadC, which encode two of the diguanylate cyclases that synthesize c-di-GMP and are responsible for biofilm formation (32), were dispensable for surface-activated virulence (SI Appendix, Fig. S9). Surface-activated virulence also remained intact in WspA and WspR mutants (SI Appendix, Fig. S8), which have also been implicated in surface-activated c-di-GMP signaling (7, 8). The independence of surface-activated virulence from biofilm formation was further supported by the lack of a virulence defect in the algR and pelA exopolysaccharide mutants (SI Appendix, Fig. S8). Our results thus suggest that c-di-GMP signaling and biofilm formation are not necessary for surface-activated P. aeruginosa to kill host cells. Biofilm formation may contribute to host killing in a manner that is redundant with other virulence factors and facilitate pathogenesis by promoting other pathogenesis-associated behaviors such as persistence within a host.
Discussion
Bacteria Regulate Their Behaviors in Response to Their Mechanical Environment.
What types of environmental signals do cells detect? Historically, studies of cell behavior have focused on chemical signals, such as nutrients and signaling molecules. Our study suggests that bacteria also detect mechanical signals associated with growth in different environments and use mechanical sensing of surface contact to induce virulence. In mammalian cells, the mechanical detection of substrates is an established driver of development and behavior (11). Because regulatory systems in bacteria generally have reduced complexity compared to their eukaryotic counterparts, the identification and characterization of a bacterial virulence “touch sensor” may represent a simplified model by which cells sense mechanical forces and transduce them into biochemical signals.
The importance of mechanosensing to the biology of P. aeruginosa is underscored by the fact that it regulates virulence, one of the most complex and highly regulated of all bacterial behaviors. Although many of the P. aeruginosa virulence factors have been identified, the signaling cues that regulate the expression of virulence factors remain less clear, in part because virulence regulation is performed by complex networks involving hundreds of components (1, 2). Surface detection appears to be a master regulator in the virulence-regulation hierarchy for P. aeruginosa, because surface-attached cells activate virulence toward both unicellular eukaryotic and mammalian hosts.
How does P. aeruginosa detect mechanical cues associated with surface attachment? The transition from planktonic growth to surface attachment involves a significant change in the mechanical properties of the growth environment in our experiments, because surface-attached cells are subjected to large shear forces generated by the movement of fluid across the attachment surface. We propose a model in which membrane-associated PilY1 (21, 22, 28) mechanically detects surfaces by mediating contact between the outer membrane and the surface substrate. Shear forces exerted on surface-attached cells shift PilY1 into an active stretched state. Supporting its role as a surface-attachment mechanosensor, PilY1 is a surface adhesin (21) and contains a putatively mechanosensitive VWFa domain that is stretched by shear force (33, 34). Because PilY1 is found on the cell surface, a periplasmic protein and/or an inner-membrane–associated protein are likely required to transduce the surface-attachment signal into the cell. In the future, it will be important to determine whether this transduction is achieved through known mechanisms, such as two-component signaling or cAMP induction, or through a novel, as-yet-uncharacterized mechanism. In addition, the existence of other surface-responsive systems, including flagella (14, 15) and the chemosensory-type Wsp system (7, 8), which do not appear to activate virulence, suggests that multiple signaling pathways distinguish different aspects of the mechanical environment.
The regulation of virulence by mechanical cues has not been explored in other bacteria. However, PilY1 is conserved across multiple gamma- and beta-proteobacteria, including several broad-host–spectrum pathogens such as Burkholderia and Acinetobacter species (SI Appendix, Fig. S10). We thus suggest that PilY1 could be part of a general mechanism by which pathogens use mechanosensation to regulate virulence in a host-nonspecific manner.
Requiring both Mechanosensation and Quorum Sensation Enables Host-Independent Virulence Regulation.
Because surface attachment is one of the initial steps of host interaction, activation of virulence at this early stage suggests that P. aeruginosa initially weakens the host to establish an environment that is suitable for long-term growth (Fig. 5A). In support of this model, we found that biofilm formation genes are dispensable for virulence activation, suggesting that long-term colonization acts downstream of surface-activated virulence. In a mammalian host setting, early phase virulence may be useful for killing host cells that provide the first line of defense, such as neutrophils and macrophages. Once colonization has been firmly established, virulence activation may no longer be necessary for maintaining the community. Indeed, clinical isolates from long-term infections of human lungs lose many of their virulence factors (35).
Fig. 5.
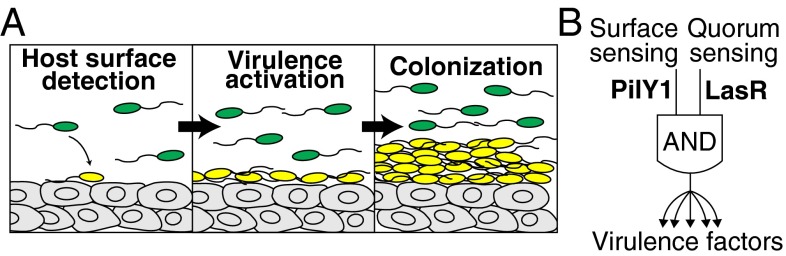
Surface detection is a host-nonspecific signal that activates virulence. (A) Schematic of proposed model in which bacterial cells detect host surfaces through mechanical cues during initial attachment. The induction of virulence through this detection mechanism establishes an environment that is suitable for bacterial colonization of the host. (B) An ‘AND’ gate model for surface-activated virulence in which both surface sensing (mediated by PilY1) and quorum sensing (mediated by LasR) are required for activating virulence.
Our findings indicate that activating P. aeruginosa virulence requires two conditions to be met: Bacterial cell density must be high, and a rigid surface must be detected. In a simplified model, the quorum- and surface-sensing pathways can be described as a coincidence detector or an “AND” logic gate whose output results in the activation of virulence genes (Fig. 5B). P. aeruginosa activates virulence in response to attachment to surfaces that have a broad range of properties, including porosity, stiffness, and chemical composition (Figs. 1 B and C and 2 A–D). Together, surface sensing and quorum sensing thus form a signaling network that is host-independent. Because surface activation of virulence is a developmental process that requires the induction of a large portion of the genome (SI Appendix, Table S1), the transition to a virulent state is energetically expensive. The requirement that two distinct conditions be met ensures that cells commit to activating virulence only when they can effectively kill host cells. Surface sensing and quorum sensing are both host-independent signaling pathways that work together to tightly regulate expression of virulence factors while maintaining the ability to target a wide diversity of host cell types. Coupled with host-specific responses, these pathways may facilitate the ubiquitous and far-reaching pathogenesis of P. aeruginosa. Given the redundancy of P. aeruginosa’s multiple virulence factors, targeting PilY1-mediated mechanosensation as a global virulence regulator represents an attractive therapeutic strategy for future exploration.
Materials and Methods
P. aeruginosa PA14 cells were grown in LB or PS:DB medium at 37 °C. D. discoideum (amoeba) AX3 cells were grown axenically in PS medium. Planktonic or surface-attached P. aeruginosa cells were mixed with amoebae, imaged using fluorescence microscopy, and analyzed using our own software to compute host killing indexes. Mouse macrophage J774A.1 cells were grown at 37 °C with 5% CO2 in DMEM. Details of media, strain construction, virulence assays, cell-density quantification, microscopy, image analysis, quorum-sensing induction, biofilm density quantification, and microarray analysis are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank T. Moser for help with the macrophage death assay, and B. Bassler and C. O’Loughlin for the gift of the quorum sensing autoinducers. This work was supported by the National Institutes of Health (NIH) Grants 1DP2OD004389 (to Z.G.) and R37-AI83256-06 (to G.A.O.), NIH Postdoctoral Fellowship F32AI095002 (to A.S.), National Science Foundation Grant 1330288 (to Z.G.), and a Human Frontiers in Science Program grant (to G.A.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The raw microarray data have been deposited in the Princeton University MicroArray (PUMA) database, puma.princeton.edu (accession no. 20141009PA14).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415712111/-/DCSupplemental.
References
- 1.Lee DG, et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006;7(10):R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feinbaum RL, et al. Genome-wide identification of Pseudomonas aeruginosa virulence-related genes using a Caenorhabditis elegans infection model. PLoS Pathog. 2012;8(7):e1002813. doi: 10.1371/journal.ppat.1002813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau GW, Hassett DJ, Ran H, Kong F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med. 2004;10(12):599–606. doi: 10.1016/j.molmed.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Morihara K, Tsuzuki H, Oka T, Inoue H, Ebata M. Pseudomonas aeruginosa elastase. Isolation, crystallization, and preliminary characterization. J Biol Chem. 1965;240:3295–3304. [PubMed] [Google Scholar]
- 5.Blumer C, Haas D. Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch Microbiol. 2000;173(3):170–177. doi: 10.1007/s002039900127. [DOI] [PubMed] [Google Scholar]
- 6.O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30(2):295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 7.Güvener ZT, Harwood CS. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol. 2007;66(6):1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Connor JR, Kuwada NJ, Huangyutitham V, Wiggins PA, Harwood CS. Surface sensing and lateral subcellular localization of WspA, the receptor in a chemosensory-like system leading to c-di-GMP production. Mol Microbiol. 2012;86(3):720–729. doi: 10.1111/mmi.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Partridge JD, Harshey RM. Swarming: Flexible roaming plans. J Bacteriol. 2013;195(5):909–918. doi: 10.1128/JB.02063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burrows LL. Pseudomonas aeruginosa twitching motility: Type IV pili in action. Annu Rev Microbiol. 2012;66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 11.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 12.Thomas WE, Trintchina E, Forero M, Vogel V, Sokurenko EV. Bacterial adhesion to target cells enhanced by shear force. Cell. 2002;109(7):913–923. doi: 10.1016/s0092-8674(02)00796-1. [DOI] [PubMed] [Google Scholar]
- 13.Lecuyer S, et al. Shear stress increases the residence time of adhesion of Pseudomonas aeruginosa. Biophys J. 2011;100(2):341–350. doi: 10.1016/j.bpj.2010.11.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lele PP, Hosu BG, Berg HC. Dynamics of mechanosensing in the bacterial flagellar motor. Proc Natl Acad Sci USA. 2013;110(29):11839–11844. doi: 10.1073/pnas.1305885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li G, et al. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol Microbiol. 2012;83(1):41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pukatzki S, Kessin RH, Mekalanos JJ. The human pathogen Pseudomonas aeruginosa utilizes conserved virulence pathways to infect the social amoeba Dictyostelium discoideum. Proc Natl Acad Sci USA. 2002;99(5):3159–3164. doi: 10.1073/pnas.052704399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alibaud L, et al. Pseudomonas aeruginosa virulence genes identified in a Dictyostelium host model. Cell Microbiol. 2008;10(3):729–740. doi: 10.1111/j.1462-5822.2007.01080.x. [DOI] [PubMed] [Google Scholar]
- 18.Rahme LG, et al. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268(5219):1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 19.Pearson JP, et al. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91(1):197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochsner UA, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92(14):6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heiniger RW, Winther-Larsen HC, Pickles RJ, Koomey M, Wolfgang MC. Infection of human mucosal tissue by Pseudomonas aeruginosa requires sequential and mutually dependent virulence factors and a novel pilus-associated adhesin. Cell Microbiol. 2010;12(8):1158–1173. doi: 10.1111/j.1462-5822.2010.01461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bohn YS, et al. Multiple roles of Pseudomonas aeruginosa TBCF10839 PilY1 in motility, transport and infection. Mol Microbiol. 2009;71(3):730–747. doi: 10.1111/j.1365-2958.2008.06559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alm RA, Hallinan JP, Watson AA, Mattick JS. Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol Microbiol. 1996;22(1):161–173. doi: 10.1111/j.1365-2958.1996.tb02665.x. [DOI] [PubMed] [Google Scholar]
- 24.Winther-Larsen HC, et al. A conserved set of pilin-like molecules controls type IV pilus dynamics and organelle-associated functions in Neisseria gonorrhoeae. Mol Microbiol. 2005;56(4):903–917. doi: 10.1111/j.1365-2958.2005.04591.x. [DOI] [PubMed] [Google Scholar]
- 25.Russell MA, Darzins A. The pilE gene product of Pseudomonas aeruginosa, required for pilus biogenesis, shares amino acid sequence identity with the N-termini of type 4 prepilin proteins. Mol Microbiol. 1994;13(6):973–985. doi: 10.1111/j.1365-2958.1994.tb00489.x. [DOI] [PubMed] [Google Scholar]
- 26.Alm RA, Mattick JS. Identification of two genes with prepilin-like leader sequences involved in type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J Bacteriol. 1996;178(13):3809–3817. doi: 10.1128/jb.178.13.3809-3817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giltner CL, Habash M, Burrows LL. Pseudomonas aeruginosa minor pilins are incorporated into type IV pili. J Mol Biol. 2010;398(3):444–461. doi: 10.1016/j.jmb.2010.03.028. [DOI] [PubMed] [Google Scholar]
- 28.Kuchma SL, et al. Cyclic-di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa: The pilY1 gene and its impact on surface-associated behaviors. J Bacteriol. 2010;192(12):2950–2964. doi: 10.1128/JB.01642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orans J, et al. Crystal structure analysis reveals Pseudomonas PilY1 as an essential calcium-dependent regulator of bacterial surface motility. Proc Natl Acad Sci USA. 2010;107(3):1065–1070. doi: 10.1073/pnas.0911616107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuchma SL, Griffin EF, O’Toole GA. Minor pilins of the type IV pilus system participate in the negative regulation of swarming motility. J Bacteriol. 2012;194(19):5388–5403. doi: 10.1128/JB.00899-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7(4):263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 32.Merritt JH, et al. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. MBio. 2010;1(4):e00183-10. doi: 10.1128/mBio.00183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moake JL, Turner NA, Stathopoulos NA, Nolasco LH, Hellums JD. Involvement of large plasma von Willebrand factor (vWF) multimers and unusually large vWF forms derived from endothelial cells in shear stress-induced platelet aggregation. J Clin Invest. 1986;78(6):1456–1461. doi: 10.1172/JCI112736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J, Zhang CZ, Zhang X, Springer TA. A mechanically stabilized receptor-ligand flex-bond important in the vasculature. Nature. 2010;466(7309):992–995. doi: 10.1038/nature09295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith EE, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA. 2006;103(22):8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.