Significance
This paper presents a system-level analysis of Wnt-signaling functions in the gene regulatory networks (GRNs) controlling pregastrular development of the sea urchin embryo. We include the spatial expression of signaling ligands, receptors, and antagonists and determine regulatory roles of Wnt signaling in local GRNs operating throughout the embryo and the control of wnt gene expression by these GRNs. Wnt signaling affects only certain cell-fate specification processes operating in particular domains without affecting others. We show that Wnt ligands function by short-range signaling between and within regulatory-state domains. We enumerate the specific functions of each expressed Wnt ligand and find that these differ, despite the expression of only one Frizzled receptor and despite partially overlapping expression patterns.
Keywords: embryonic interdomain signaling, developmental GRNs, Wnt system regulatory functions, Porcupine
Abstract
Wnt signaling affects cell-fate specification processes throughout embryonic development. Here we take advantage of the well-studied gene regulatory networks (GRNs) that control pregastrular sea urchin embryogenesis to reveal the gene regulatory functions of the entire Wnt-signaling system. Five wnt genes, three frizzled genes, two secreted frizzled-related protein 1 genes, and two Dickkopf genes are expressed in dynamic spatial patterns in the pregastrular embryo of Strongylocentrotus purpuratus. We present a comprehensive analysis of these genes in each embryonic domain. Total functions of the Wnt-signaling system in regulatory gene expression throughout the embryo were studied by use of the Porcupine inhibitor C59, which interferes with zygotic Wnt ligand secretion. Morpholino-mediated knockdown of each expressed Wnt ligand demonstrated that individual Wnt ligands are functionally distinct, despite their partially overlapping spatial expression. They target specific embryonic domains and affect particular regulatory genes. The sum of the effects of blocking expression of individual wnt genes is shown to equal C59 effects. Remarkably, zygotic Wnt-signaling inputs are required for only three general aspects of embryonic specification: the broad activation of endodermal GRNs, the regional specification of the immediately adjacent stripe of ectoderm, and the restriction of the apical neurogenic domain. All Wnt signaling in this pregastrular embryo is short range (and/or autocrine). Furthermore, we show that the transcriptional drivers of wnt genes execute important specification functions in the embryonic domains targeted by the ligands, thus connecting the expression and function of wnt genes by encoded cross-regulatory interactions within the specific regional GRNs.
The formation of spatial patterns of gene expression and the development of the body plan are controlled by gene regulatory networks (GRNs). Signaling interactions have a particular role in these networks, in that they provide the means of communication between cell-fate specification processes operating in separate cellular domains. The timing, location, and function of each signaling interaction is determined by GRN linkages that control the expression of signaling ligands and receptors as well as the expression of regulatory genes in response to a combination of signaling inputs and cell fate-specific transcription factors. Cell-fate specification GRNs active during pregastrular development of the sea urchin Strongylocentrotus purpuratus are particularly well understood. During the first 30 h of sea urchin embryogenesis, more than 15 gene-expression domains are formed, and specifically expressed regulatory genes have been identified for each domain. In most cases, the regulatory mechanisms determining their spatial expression patterns have been resolved. Thus, fairly complete GRN models have been constructed for the majority of cell-fate domains in the pregastrular stage embryo (1–6). The sea urchin GRN models at this point incorporate more than 60 regulatory genes and their interactions and cover almost the entire embryo.
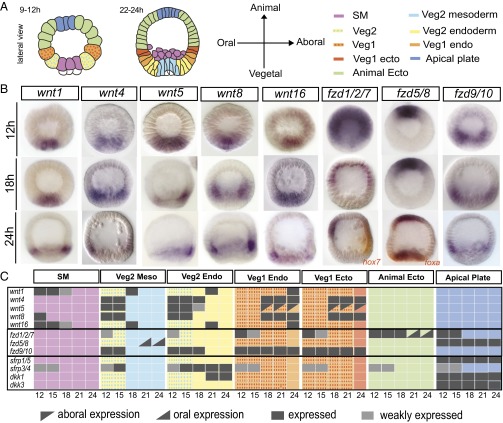
The principle organization of mesodermal, endodermal, and ectodermal cell-fate specification domains in sea urchin embryos along the animal–vegetal axis is summarized in the diagram in Fig. 1A. Cells located at the vegetal pole will become skeletogenic mesodermal cells. These cells are surrounded by the veg2 cell lineage. This lineage consists of veg2 mesodermal cells, located adjacent to skeletogenic cells and giving rise to all other mesodermal cell fates such as esophageal muscle cells, blastocoelar cells, and pigment cells, and of veg2 endoderm cells, which will form the foregut and parts of the midgut. At a further distance from the vegetal pole, but still within the vegetal half of the embryo, is the veg1 lineage, consisting of veg1 endoderm, located adjacent to veg2 endoderm and giving rise to the other parts of the midgut and the hindgut, and of veg1 ectoderm, the future perianal ectoderm. Finally, the animal half of the embryo forms exclusively ectodermal cell fates, with apical neurogenic cell fates being specified in cells at the animal pole.
Fig. 1.
Spatial expression of Wnt-signaling genes. (A) Schematic representation of early developmental stages of S. purpuratus embryos showing the spatial arrangement of regulatory-state domains. SM, skeletogenic mesoderm; “veg1” and “veg2” denote cell lineages descended from the sixth cleavage ring of eight sister cells, each giving rise to the parts of the embryo indicated in the diagrams; “veg2 mesoderm” is also known as “nonskeletogenic mesoderm”; “animal ectoderm” and “veg1” ectoderm denote both oral and aboral ectodermal domains. (B) WMISH of significantly expressed wnt and frizzled genes at selected time points (12 h, 18 h, and 24 h); additional time points for these genes and expression patterns of dkk and sfrp genes are shown in SI Appendix, Figs. S2 and S3. (C) Expression matrix for each regulatory-state domain of the examined Wnt-signaling genes, indicating whether the gen is expressed (black/gray) or not expressed (colored background) every 3 h from 12–24 h. Regulatory domains are marked by the color code used in A. Developmental stages include 12 h (early blastula), 15 h (midblastula), 18 h (hatching blastula), and 24 h (mesenchyme blastula).
The response to Wnt signaling is mediated by several alternative intracellular signaling pathways. In the canonical Wnt-signaling pathway, signaling-dependent gene expression is controlled by the transcription factor Tcf/Lef, which forms a complex with the coactivator β-catenin in cells that receive Wnt signaling but forms a complex with the corepressor Groucho in the absence of Wnt signaling (7). Transcriptional control by Tcf/Lef thus effects a Boolean readout of gene expression, mediating activation or repression of the same target genes in cells with or without Wnt signaling (8). Cis-regulatory analyses including mutation of Tcf binding sites have demonstrated direct control by Tcf in the skeletogenic GRN of S. purpuratus embryos, in which the regulatory gene directly responsive to Tcf, pmar1, operates at the top of the GRN hierarchy (1, 9, 10). Furthermore, most, if not all, regulatory genes expressed in early veg2 endoderm and/or veg1 endoderm cells of this embryo are direct targets of Tcf (2, 11, 12). Transcriptional control by Tcf not only is responsible for activation of endodermal genes in future gut cells but also is used to exclude endodermal regulatory genes from the mesoderm (3, 11, 13, 14). Furthermore, perturbation of particular Wnt ligand gene expression has shown that two additional cell-fate specification GRNs are sensitive to Wnt signaling: one operating in ectodermal cells located closest to the endoderm, which responds to Wnt5 signaling (15), and one operating neuronal specification, which is restricted to the apical domain by a mechanism dependent on Wnt signaling (16).
However, understanding the particular functions of signaling interactions requires not only knowledge of affected target genes and cellular domains but also identification of the cells producing the responsible signaling ligand. As in many other invertebrate embryos, the analysis of zygotic Wnt-signaling functions in sea urchin embryos has been complicated by the presence of maternally localized β-catenin at the vegetal pole (17). Here we determined these zygotic Wnt-signaling functions on a global scale by assessing the temporal and spatial expression of all genomically encoded genes producing Wnt ligands, Frizzled (Fzd) receptors, or potential Wnt-signaling antagonists during the pregastrular development of S. purpuratus. We have summarized these expression patterns abstractly to highlight the signal-sending and signal-receiving capacity for each cell-fate domain. We analyzed effects of interference with Wnt signaling on 172 specifically expressed regulatory genes, irrespective of the intracellular signaling pathways that might mediate this function. For a system-wide perturbation of Wnt signaling, we made use of the C59 inhibitor of Porcupine, which interferes with the secretion of Wnt ligands and thus with all Wnt-dependent processes. We show that the only GRNs affected by C59 perturbation are the two endodermal GRNs, the veg1 ectoderm GRN, and the apical neurogenic GRN. For each GRN affected by C59-mediated inhibition of Wnt signaling, we identified the responsible Wnt ligands using morpholino perturbations. Furthermore, by identifying the upstream transcription factors activating wnt gene transcription, we established functional linkages between the GRNs regulated by Wnt signaling and the GRNs controlling Wnt ligand expression. Our intent was to achieve a system-wide understanding of the roles of Wnt signaling in this phase of development and for this embryo and to generate a causal spatial regulatory analysis of Wnt-signaling inputs into the regional embryonic GRNs.
Results
Spatial and Temporal Expression of wnt, fzd, Secreted Frizzled-Related Protein 1, and Dickkopf Genes.
The Wnt-signaling system encoded in the genome of S. purpuratus includes 11 wnt ligand genes and four fzd receptor genes (18). To identify wnt and fzd genes expressed during pregastrular development (12–24 h), the time courses of their expression levels were analyzed by quantitative PCR (qPCR) (SI Appendix, Fig. S1). Five wnt genes (wnt1, wnt4, wnt5, wnt8, and wnt16) are expressed in this embryo before gastrulation; only one of them, wnt16, is transcribed maternally (SI Appendix, Fig. S1 A and B). All other wnt genes are not expressed at all until gastrulation, and even then their transcript levels are very low (<100 transcripts per embryo). We cannot confirm the observation of maternal wnt6 transcripts reported earlier (19), and this conclusion is substantiated in our recent transcriptome study (20). Of the four fzd genes, fzd1/2/7, fzd5/8, and fzd9/10 were expressed at high levels (>1,000 transcripts) before 30 h; fzd4 begins to be transcribed only just before gastrulation and was not considered further in this study (SI Appendix, Fig. S1C). Four genes encoding potential inhibitors of Wnt signaling also were found to be expressed during early sea urchin embryogenesis. These are genes dkk1 and dkk3 encoding Dickkopf proteins and two genes encoding secreted Frizzled-related protein (SFRP), sfrp1/5 and sfrp3/4 (SI Appendix, Fig. S1D).
The spatial expression patterns of all expressed wnt, fzd, dkk, and sfrp genes during embryogenesis were analyzed at 3-h intervals between 12 and 24 h. Results for wnt and fzd genes at 12, 18, and 24 h postfertilization are shown in Fig. 1B, and the complete dataset is presented in SI Appendix, Figs. S2 and S3. Similar results have been established in the embryos of a related species of sea urchin, Paracentrotus lividus (21). Because this study focuses on identifying the function of Wnt signaling in the interaction between different cell-fate specification processes, we have represented individual gene-expression patterns abstractly according to their embryonic expression domain (Fig. 1C). In the following we summarize the expression patterns of genes encoding ligands, receptors, and potential antagonists of the Wnt-signaling system by embryonic regulatory-state domain (compare with Fig. 1A).
Skeletogenic mesodermal cells are the precursors of cells producing the larval skeleton and are located at the vegetal pole before they start to ingress into the blastocoel at 21 h. These cells inherit high levels of maternal nuclear β-catenin and initially express wnt1, wnt8, and wnt16 but do so only up to 15 h (Fig. 1 B and C). No wnt gene is expressed in these cells thereafter. Thus, after the degradation of maternal Fzd1/2/7 proteins, skeletogenic mesoderm cells most likely are not responsive to Wnt signaling, because no fzd transcripts are detectable in these cells.
Veg2 mesodermal cells give rise to all other (i.e., nonskeletogenic) mesodermal cell types, including esophageal muscle cells, blastocoelar immune cells, coelomic pouch cells, and pigment cells. Veg2 mesodermal cells express wnt4, wnt5, and wnt8 early in development at 12 and 15 h and transiently also express wnt1 and wnt16 at 18 h (Fig. 1 B and C). However, after 18 h and up to the onset of gastrulation, mesodermal precursor cells do not express wnt genes. Similarly, early expression of a fzd gene, fzd9/10, terminates after 15 h, and only one fzd gene, fzd5/8, is transcribed after 21 h in a subset of mesodermal cells located in the oral portion of the veg2 mesoderm. This finding is consistent with earlier results (22). No sfrp or dkk genes are expressed in Veg2 mesodermal cells after 15 h. Thus, with the exception of oral mesodermal cells, most mesodermal cells express neither wnt nor fzd genes from 18 h to gastrulation and are not likely to send or receive Wnt signaling.
Veg2 endodermal cells are the precursors of anterior endoderm, forming the foregut and the aboral midgut (3, 23). These cells derive from the veg2 cell lineage, as do the mesodermal cells discussed above. The common ancestor cells of the veg2 mesoderm and veg2 endoderm cells express wnt4, wnt5, and wnt8 genes as well as fzd9/10 at 12 and 15 h. Endodermal and mesodermal cell fates become distinct in the veg2 lineage by 18 h, and after that veg2 endodermal cells transiently express wnt1 and wnt16 at 21 h, but no wnt gene is expressed in this domain by 24 h. Expression of fzd9/10 continues until 18 h, but no receptor gene is expressed in the anterior endoderm domain after that time point. Furthermore, after 18 h, genes encoding potential Wnt-signaling inhibitors, sfrp3/4 and dkk1, start to be expressed in veg2 endodermal cells, suggesting that these cells do not depend on Wnt-signaling inputs after this time.
Veg1 endoderm cells are the precursors of posterior endoderm, eventually giving rise to the hindgut and the oral parts of the midgut. In a pattern that almost reverses the pattern in anterior endoderm precursors, the posterior endoderm domain expresses no wnt genes before 18 h, but by 24 h all five wnt genes are expressed in these cells. These cells also transcribe fzd9/10 at all time points considered, but neither sfrp nor dkk genes are expressed after the early ubiquitous expression of sfrp3/4. These results indicate that the veg1 endoderm domain is capable of responding to Wnt signaling through Fzd9/10 throughout pregastrular stages, but this domain may also contribute to Wnt signaling after 18 h.
Like the precursors of the posterior gut, Veg1 ectodermal cells derive from the veg1 lineage and ultimately give rise to ectodermal cells surrounding the anus. From 18 h until the separation of endodermal and ectodermal cell fates at 24 h, veg1 cells express wnt4, wnt5, and wnt8, and these genes, as well as fzd9/10, continue to be expressed in veg1 ectodermal cells at 24 h (Fig. 1 B and C). However, unlike the veg1 endodermal cells, veg1 ectoderm cells do not turn on expression of wnt1 and wnt16. Sfrp and dkk genes are not expressed in the veg1 lineage at pregastrular stages after 15 h. Thus, veg1 ectodermal and veg1 endodermal cells have a similar potential both to send and receive Wnt signaling.
Animal ectodermal cells include cells of various regulatory-state domains of the animal half, all of which give rise to ectodermal cell types, the stomodeal structures, and neurons of the ciliated band (24, 25). These cells express no wnt signaling gene and no sfrp or dkk genes after 15 h. However, these cells express one fzd gene, fzd1/2/7, first in all animal ectodermal cells and by 21 h exclusively in the oral ectoderm. Thus, animal ectoderm cells are capable of responding to Wnt signaling but do not themselves emit Wnt signals.
Apical plate cells are the precursors of neurogenic cells at the animal pole. These cells transcribe no wnt genes but do specifically express fzd5/8 at all pregastrular stages. Furthermore, these cells express sfrp1/5, dkk1, and dkk3 genes at all stages considered and also transiently express sfrp3/4. Based on these expression patterns, cells of the apical plate likely neither respond to nor present Wnt signals.
System-Wide Perturbation of Wnt Signaling by a Porcupine Inhibitor.
To achieve a system-wide perturbation of Wnt signaling, we made use of a recently reported small chemical inhibitor of Porcupine, a membrane-bound O-acyltransferase required for acylation of Wnt proteins. Porcupine-mediated acylation of Wnts occurs at a conserved serine residue and is necessary for the secretion of the Wnt proteins that include this serine residue. Experimental perturbation of Porcupine interferes with secretion of all Drosophila Wnts (except for WntD, which lacks the target serine residue) and all mouse and human Wnts (26–29). Small chemical inhibitors of Porcupine recently have been proposed to be efficient agents that interfere systemically with Wnt signaling in clinical applications; here we used the Porcupine inhibitor C59 (30–32). To test the efficacy of C59 in sea urchin embryos, we first assessed the phenotypes that develop in the presence of various concentrations of C59. Embryos treated with C59 showed no apparent defects in early development at pregastrular stages, and the ingression of skeletogenic cells occurs similarly as in control embryos. At the late gastrula stage, defects in gastrulation, in the development of the tripartite gut, and in the formation of skeletogenic spicules were detected in a dose-dependent manner, but the specification of mesodermal pigment cells was not affected (SI Appendix, Fig. S4A). Embryos were exposed to C59 at different concentrations; expression levels of the Tcf target genes foxa, hox11/13b, and eve were strongly reduced at 0.5 μM, but the expression of the mesodermal regulatory gene gcm remained unchanged (SI Appendix, Fig. S4B). Similarly, when expression of Porcupine was blocked by morpholino injection, embryos showed defects in gut development as well as reduced expression of the endodermal regulatory genes blimp1b, hox11/13b, and eve, but not gcm. All these results confirm both the specificity and the efficacy of C59-mediated inhibition of Wnt signaling (SI Appendix, Fig. S5 A and B). Furthermore, all five expressed Wnt ligands contain the conserved serine residue required for Porcupine-mediated acylation, and the surrounding amino acid sequences conform to a consensus sequence recently identified in Porcupine targets (33), indicating that indeed all five Wnt ligands should require Porcupine for their secretion (SI Appendix, Fig. S5C).
The effect of inhibiting Wnt signaling by C59 treatment on regulatory gene expression in all embryonic domains was detected by Nanostring nCounter analysis using a probe set targeting 208 genes, including 172 genes encoding transcription factors. Sea urchin embryos were treated with C59 starting at 1 h postfertilization, and gene-expression levels were determined at 12, 15, 18, 21, and 24 h. Results for a few selected genes expressed in each of the seven embryonic domains are shown in Fig. 2; the complete data are listed in SI Appendix, Table S1. A tabular summary of experimental evidence for Wnt-signaling effects on all specific target genes addressed in this study can be found in SI Appendix, Table S2. Of the 172 regulatory genes monitored in this experiment, 147 were expressed in at least at one stage during the developmental time interval considered (SI Appendix, Table S1). Treatment with C59 resulted in the down-regulation of 16 regulatory genes; 10 of these genes are components of endoderm GRNs, and five are components of veg1 ectoderm GRNs; the expression domains of gbx at 24 h have not yet been resolved. In addition, expression of nine regulatory genes was up-regulated in embryos upon exposure to C59; eight of these genes are known to be expressed in the apical neurogenic domain, and the expression pattern of acsc is not known. The effects of C59 treatment on the activity of GRNs specific to the individual embryonic regulatory-state domains are summarized in the following sections.
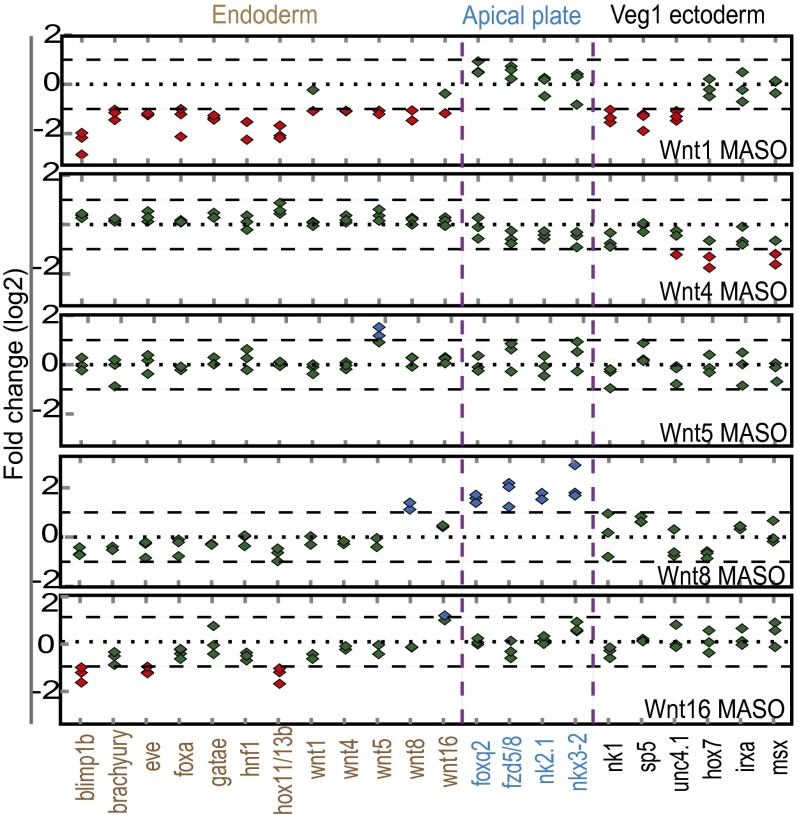
Fig. 2.
Effects of inhibiting Porcupine-dependent Wnt ligand secretion by C59 treatment. Embryos were treated with C59 or DMSO (control) at 1 h postfertilization. Transcripts of each gene were measured by Nanostring nCounter analysis using a probe set detecting expression of 172 regulatory genes at five successive times (abscissa). Shown are fold differences of transcript abundance in embryos treated with C59 compared with control embryos. Each diamond represents one of three experimental repeats, shown in red if down-regulated more than twofold upon treatment with C59, in blue if up-regulated more than twofold, or in green if unchanged. The dotted line indicates a ratio of 1 (experimental/control), and the dashed lines indicate a twofold envelope of significance. Note that the ordinate scale is log2. Genes expressed at low levels (<100 transcripts per embryo) in treated and/or control embryos are not shown. A representative set of genes is shown for each regulatory-state domain. The colored bar on the right of each column indicates the spatial-expression domain(s) at 24 h. The color code is as in Fig. 1. Complete results are shown in SI Appendix, Table S1.
Skeletogenic and veg2 mesodermal GRNs.
Regulatory genes expressed in skeletogenic cells include alx1, pmar1, dri, erg, ets1/2, tbr, tel, and tgif (1). Expression of these genes is not changed in embryos exposed to C59, except for tgif, which shows reduced expression levels at 24 h, when it is expressed in veg2 endoderm as well (SI Appendix, Table S1). In veg2 mesodermal cells, the specifically expressed regulatory genes include gcm, gatae, gatac, e2f3, erg, ese, ets1/2, hex, prox1, scl, shr2, six1/2, and z166 (6). Of these, only gatae shows reduced expression levels upon C59 treatment, again at a time when it also is expressed in veg2 endodermal cells. Expression of no other mesodermal regulatory genes is affected by C59 (SI Appendix, Table S1), indicating that mesodermal cell fates do not require Wnt-signaling inputs during pregastrular development. This finding is in agreement with the absence of wnt and fzd gene expression in most mesodermal cells after 18 h. [Note that although oral veg2 mesoderm cells later express frz5/8 (Fig. 1 B and C), expression of the canonical oral mesoderm regulatory genes prox1, gatac, erg, ese, and scl is impervious to C59].
Veg2 endodermal GRN.
By 18 h, the expression of all regulatory genes of the anterior endoderm GRN that are transcribed exclusively in veg2 endoderm cells at this time, namely, blimp1b, foxa, hox11/13b, brachyury, and krl/z13 (2, 3), is down-regulated in embryos treated with C59 (Fig. 2 and SI Appendix, Table S1). Two additional regulatory genes, myc and soxc, are expressed in veg2 endoderm cells at 18 h, but their whole-embryo expression levels are not affected by C59 treatment, possibly because these genes also are transcribed in cells of nonendodermal fates. Thus, most, if not all, regulatory genes specifically expressed in veg2 endodermal cells are down-regulated in C59-treated embryos. This result is consistent with the observed expression of wnt and fzd genes in these cells up to 18 h. Furthermore, previous cis-regulatory evidence demonstrated that transcription of most genes of the early endoderm GRN is controlled by Tcf/β-catenin (reviewed in ref. 2). However, the initial expression of regulatory genes in endodermal cells before 15 h is not affected by C59, and indeed, treating embryos with C59 only after 15 h has an effect on regulatory gene expression at 24 h similar to the effect seen with the addition of C59 at 1 h after fertilization (SI Appendix, Fig. S6A). For comparison, in embryos in which maternal as well as zygotic accumulation of β-catenin is inhibited by the injection of mRNA encoding dominant negative cadherin, the expression of endodermal genes is affected at all time points considered, starting at 9 h (SI Appendix, Fig. S7). These results indicate that, because of the presence of maternal β-catenin, secreted Wnt ligands are not required for regulatory gene expression in endodermal precursor cells before 15 h. A similar observation was made in mouse embryos, in which the earliest developmental processes do not require Porcupine-dependent Wnt secretion (28).
Veg1 endodermal GRN.
By 15 h, the regulatory gene eve is expressed throughout the veg1 lineage, and by 21–24 h its product is responsible for activating the expression of regulatory genes specific to the future posterior endoderm, including hox11/13b, brachyury, and hnf1 (3). The expression of all four genes is down-regulated in embryos treated with C59. Thus, the veg1 endoderm GRN is activated right at the time (24 h) when all wnt genes and fzd9/10, but no dkk or sfrp genes, are expressed in the same cells, and its operation depends on Wnt signaling.
Veg1 ectoderm GRNs.
Regulatory gene expression in these veg1 ectodermal cells shows varied effects upon treatment with C59 (Fig. 2 and SI Appendix, Table S1). Thus, expression of several regulatory genes is affected, as indicated by lower levels of nk1 and unc4.1 transcripts (34). Other transcripts, such as msx, are present at decreased levels only at a stage when their expression is restricted to veg1 ectodermal cells; later, when these transcripts also are expressed widely in animal aboral ectoderm cells, transcript levels are comparable to those in control embryos (SI Appendix, Fig. S8) (5). These results suggest that veg1 ectoderm GRNs are at least partially affected by Wnt signaling. However, the majority of regulatory genes are not exclusive to veg1 ectoderm cells, and it is not possible to assess the extent of this regulatory input by quantitative measurements of gene-expression levels. The observed effect of C59 on gene expression in veg1 ectodermal cells is consistent with the presence of Wnt ligands and Fzd receptors in these cells and with previous reports of Wnt signaling affecting gene expression in veg1 ectoderm (15).
Animal ectoderm GRNs.
Regulatory genes expressed in animal ectoderm cells mostly are not affected by C59 perturbation, e.g., foxg, not, and gsc [oral animal ectoderm (4)], emx [lateral animal ectoderm (5)], and hmx, hox7, dlx, and ets4 [aboral animal ectoderm (35)]. However, the transcription of sp5, which is expressed in veg1 ectoderm and in the oral animal ectoderm domain (SI Appendix, Fig. S8), is strongly down-regulated at 18 h and up to the onset of gastrulation by C59 treatment (Fig. 2). Thus, specification of most animal ectoderm cell fates does not depend on Wnt-signaling inputs, but the expression of one particular transcription factor appears to be regulated by Wnt signaling in these cells.
Apical ectoderm GRNs.
Expression levels of regulatory genes transcribed in cells of the neurogenic apical ectoderm, such as foxq2 (36), foxj1 (37), fez (38), zic (39), hbn, and nk2.1 (40), are up-regulated in embryos treated with C59 (Fig. 2 and SI Appendix, Table S1). Earlier observations showed that absence of Wnt signaling leads to an increase in the expression levels of regulatory genes associated with neurogenic fate in adjacent animal ectoderm cells (16, 36). In embryos exposed to C59 at 1 h postfertilization, the earliest effect on regulatory gene expression, on foxq2 and nk2.1, is observed at 15 h. The effects of adding C59 at 12 h showed effects on apical gene expression similar to adding this drug at 1 h, whereas C59 treatment after 15 h on showed much weaker effects, and no effects were observed when C59 was added at 18 h (SI Appendix, Fig. S6B). These results indicate that the Wnt signal critical for suppression of neurogenic fate in animal ectoderm cells is secreted after 12 h and before 18 h. As described above, the neurogenic apical domain expresses no Wnt-signaling ligand but expresses sfrp1/5, sfrp3/4, dkk1, and dkk3, which encode potential Wnt-signaling inhibitors. These observations thus indicate that the neurogenic fate is suppressed by Wnt signaling and that, in turn, the apical neurogenic GRN ensures the expression of Wnt-signaling antagonists.
Taken together, these results indicate that the anterior (veg2) and posterior (veg1) endodermal GRNs are broadly activated by Wnt signaling in pregastrular embryos and that some regulatory genes of the veg1 ectoderm GRNs also receive positive Wnt-signaling inputs. All three domains express only one Fzd receptor gene, fzd9/10, but express several genes encoding Wnt ligands, which individually or together could be responsible for the observed effects on regulatory gene expression. In addition, Wnt signaling appears to antagonize apical neurogenic fate, and the wnt genes expressed closest to the animal ectoderm are wnt4, wnt5, and wnt8, expressed in veg1 ectodermal cells. Determining if these different Wnt ligands execute similar or overlapping functions and whether they can substitute for one another or operate in entirely distinct ways requires the perturbation of the gene expression of individual Wnt ligands.
Wnt Ligands Execute Distinct Functions.
To distinguish the functional contribution of each Wnt ligand, we individually perturbed the expression of the five Wnts that could be responsible for the effects observed in embryos treated with C59. Embryos were injected with morpholino antisense oligonucleotides (MASOs) blocking the translation of Wnt1, Wnt4, Wnt5, Wnt8, or Wnt16, and gene-expression levels in these embryos compared with control embryos were analyzed at 24 h for a set of regulatory genes that represent each of the four embryonic expression domains affected by C59 treatment.
The results summarized in Fig. 3 demonstrate that each Wnt ligand affects the expression of a specific set of regulatory genes. Thus, injection of Wnt1MASO broadly affected the expression of all endodermal regulatory genes tested and also decreased the expression levels of some regulatory genes expressed in the veg1 ectoderm (nk1, sp5, and unc4.1), but other examined veg1 ectodermal genes and neurogenic apical genes were not affected. Wnt1 MASO also weakly affected the expression of all other wnt genes (Fig. 3). Embryos treated with Wnt4 MASO exhibited marginally decreased expression levels of unc4.1, hox7, and msx in veg1 ectodermal cells, but Wnt4 MASO had no effect on genes expressed in either the endoderm or the apical neurogenic plate. Injection of Wnt5 MASO did not result in any changes in regulatory gene expression except for an up-regulation of wnt5 transcripts. In embryos injected with Wnt8 MASO, transcript levels of all neurogenic apical genes were increased, but genes expressed in endoderm and veg1 ectoderm were not affected. Wnt16 MASO selectively decreased the transcript levels of blimp1b, eve, and hox11/13b in the veg2 and veg1 endoderm domains but did not affect genes specific to the neurogenic apical plate and veg1 ectodermal cells.
Fig. 3.
Morpholino perturbation of individual Wnt ligands and effects on regulatory gene expression. Embryos were injected with morpholinos targeting the expression of Wnt1, Wnt4, Wnt5, Wnt8, or Wnt16 or with randomized control morpholinos. Expression levels of regulatory genes in morpholino-injected embryos were analyzed by qPCR at 24 h, except for three genes that were analyzed at earlier developmental stages: hox7 (21 h) and irxa and msx (18 h). The genes selected for this analysis are those demonstrated to respond to C59 treatment (see text). Shown are ratios (log2) of expression levels in Wnt-specific morpholino-injected embryos to expression levels in control morpholino-injected embryos. Symbols and color coding are as in Fig. 2.
Taken together, these results indicate that four of the five Wnt ligands indeed execute specific functions in the regulation of transcription factor gene expression that cannot be performed by other Wnt ligands, despite the largely overlapping expression of all five wnt genes. Taken together, the effects of perturbing the expression of individual Wnts largely correspond to the effects observed in C59-treated embryos. Thus, the expression of every gene tested here that was affected by C59 treatment also was affected by the perturbation of at least one Wnt ligand. Similarly, each embryonic regulatory-state domain in which C59 treatment was observed to have altered gene expression also was affected by at least one of the Wnt ligands expressed at these developmental stages, as summarized in the following sections.
Veg2 endoderm.
By 18 h, the expression of blimp1b, foxa, hox11/13b, and brachyury in the veg2 endoderm is affected by C59 treatment. The responsible Wnts could be Wnt4 and Wnt5, which are expressed in these cells up to 18 h; Wnt8, which is produced in these cells from 12–15 h; or Wnt1 and Wnt16, which are expressed in the adjacent veg2 mesodermal cells at 18 h and in veg2 endodermal cells at 21 h. Wnt4, Wnt5, and Wnt8 morpholinos do affect gene expression in veg2 endoderm cells. However, Wnt1 and Wnt16 signaling clearly is required to activate gene expression in these cells by short-range signaling. Injection of Wnt1 MASO results in lower expression of all tested regulatory genes (blimp1b, foxa, and gatae) expressed in veg2 endoderm cells at 24 h. The expression of blimp1b also is affected by Wnt16 MASO, indicating that this gene requires both Wnt-signaling inputs for normal expression.
Veg1 endoderm.
Cells of the future posterior endoderm express fzd9/10 between 12 and 24 h, wnt4, wnt5, and wnt8 from 18–24 h, and wnt1 and wnt16 at 24 h. In addition, Wnt signaling may occur from the adjacent veg2 endoderm cells, which transcribe wnt ligand genes between 12 and 21 h, as discussed just above. In C59-treated embryos, expression of eve in veg1 endoderm precursor cells is reduced after 15 h. As in the veg2 endoderm GRN, gene expression in veg1 endoderm cells is not affected by morpholinos targeting Wnt4, Wnt5, or Wnt8 expression but depends on Wnt1 and Wnt16 signaling. Again, injection of Wnt1MASO shows the broadest effect, reducing expression levels of brachyury, hox11/13b, hnf1, and eve, whereas injection of Wnt16 MASO decreased the expression levels of only hox11/13b and eve. Interestingly, Wnt1 and Wnt16 not only execute overlapping functions in the two endodermal GRNs but also are expressed in very similar patterns that are consistent with these functions. At 18 h, when the expression of foxa and blimp1b is restricted to veg2 endoderm, wnt1 and wnt16 expression is detected exclusively in the adjacent veg2 mesoderm. By 21 h, however, wnt1 and wnt16 transcripts are detected in anterior (veg2) endoderm cells for a brief period, and at this time point the expression of their target genes hox11/13b, brachyury, and hnf1 is induced in the adjacent veg1 posterior endoderm cells. Thus, when expressed in anterior endoderm, Wnt1 and Wnt16 again function as short-range signaling ligands, now activating expression of posterior endoderm regulatory genes in adjacent veg1 cells.
Veg1 ectoderm.
Expression of regulatory genes such as nk1 and unc4.1 in veg1 ectodermal cells is weakly affected by C59 treatment from 18 h on. Wnt4, wnt5, and wnt8 are expressed in the veg1 lineage from 18 h on, and fzd9/10 is expressed in these cells at all stages considered. In addition, by 24 h, wnt1 and wnt16 are expressed in the neighboring veg1 endoderm domain. Injection of embryos with Wnt-specific morpholinos revealed that only Wnt1 and Wnt4 affect gene expression in veg1 ectoderm cells; morpholinos against Wnt5, Wnt8, and Wnt16 showed no effect by 24 h (Fig. 3). Wnt1 and Wnt4 signaling in the veg1 ectoderm domain therefore occurs by short-range and perhaps also by intradomain signaling. Embryos injected with Wnt1 MASO showed reduced expression levels of nk1, unc4.1, and sp5, whereas Wnt4MASO affected the expression of hox7 and msx genes. Thus, as in the endodermal GRNs, two Wnt ligands regulate gene expression in the veg1 ectoderm, but in this domain they affect separate sets of regulatory genes. These results differ from those obtained in another sea urchin species, Lytechinus variegatus, in which Wnt5 was shown to activate regulatory genes, including irxa and nk1, in the veg1 ectoderm domain (15). However, in S. purpuratus, when Wnt5 expression was knocked down using two separate morpholinos, no change in ectodermal gene-expression levels was observed except for an increase in wnt5 transcripts. This result implies that Wnt5 signaling is not required in this domain, although it might function redundantly together with other Wnts. In the S. purpuratus embryo, Wnt4 regulates the expression of nk1, but irxa expression was not affected by the perturbation of any individual Wnt signal, even though its expression is moderately affected by C59 treatment at 18 h. This interspecies difference thus could reflect a relatively recent change in the identity of the Wnt ligand used for activation of regulatory genes in the veg1 ectoderm GRNs.
Apical ectoderm.
The earliest increase in expression of regulatory genes specific to the apical neurogenic fate upon C59 treatment was observed at 15 h. Because no Wnt ligand is expressed in the animal half, the Wnt signal responsible for this effect must derive from the vegetal half of the embryo. The vegetal cells closest to the animal ectoderm, the veg1-lineage cells, express wnt4, wnt5, and wnt8 from 18 h on, and before that, at 12–15 h, these same genes are expressed in the veg2 lineage. Injection of Wnt8 MASO at fertilization results in elevated expression levels of all apical regulatory genes tested. This result is consistent with a previous report on the function of Wnt8 in restricting the expression of genes associated with the neurogenic apical fate (16). Our results show that blocking the expression of Wnt1, Wnt4, Wnt5, or Wnt16 had no effect on the expression of apical-specific genes. Wnt8 is the earliest and most abundantly transcribed wnt gene, and at 12–15 h, when the embryo consists of only a little over a hundred cells, there are still only few cells separating Wnt8-expressing vegetal cells and the cells in which expression of neurogenic apical regulatory genes is to be prevented. Thus, in summary, these experiments revealed that the Wnt-signaling function responsible for limiting the apical neurogenic domain is executed solely by Wnt8 and is a very early process, probably operating in late cleavage.
Wnt-Dependent Spatial-Patterning Functions.
To assess the consequences Wnt signaling on embryonic patterning of regional regulatory states, we studied the spatial disposition of gene-expression domains in embryos in which Wnt signaling is inhibited. As an initial assessment, embryos treated with C59 from 1–24 h were fixed at 24 h and stained by whole-mount in situ hybridization (WMISH) with eight probes against regulatory genes specific for different expression domains throughout the embryo (SI Appendix, Fig. S9A). Compared with control embryos, the expression domains of alx1 in the skeletogenic mesoderm cells and of delta in the veg2 mesodermal cells are not affected by C59. However, expression of foxa, marking veg2 endoderm cells, is restricted to fewer cells in C59-treated embryos than in control embryos. The boundary usually separating veg2 endoderm (foxa) and veg1 endoderm (hox11/13b) fate appears to have shifted to the vegetal pole, and the expression of hox11/13b at least partially overlaps that of foxa in embryos treated with C59. Similarly, most gene-expression boundaries are shifted toward the vegetal pole, including the boundaries of the foxq2 expression domain at the animal pole.
To determine which Wnt signal is responsible for the vegetal shift of most expression domains (except for the mesodermal domains), we tested the spatial expression of these genes in embryos injected with Wnt morpholinos. The results, as summarized in SI Appendix, Fig. S10 (for data see SI Appendix, Fig. S9B), indicate not only that interfering with the expression of Wnt1 decreases the expression levels of foxa, hox11/13b, and eve but also that these genes are expressed in fewer cells and that these cells are located closer to the vegetal pole than in control embryos. The vegetal boundaries of gene expression of lim1 (veg1 ectoderm) and particularly that of emx (animal ectoderm) also are shifted toward the vegetal pole in Wnt1MASO-injected embryos, as is consistent with the previous observation that emx expression in veg1 cells is repressed downstream of Eve (5). However, Wnt1 MASO does not affect the boundary between emx and foxq2 expression domains. The effects seen with Wnt16MASO are similar but weaker. Embryos injected with morpholinos blocking either Wnt4 or Wnt5 expression do not appear to affect the patterning of the gene-expression domains assessed here. In embryos injected with Wnt8MASO, on the other hand, only the boundary between emx and foxq2 expression domains, which were shown to repress each other (5), is shifted vegetally, leading to an expansion of the apical neurogenic domain and confirming that the up-regulation of foxq2 expression levels in Wnt8MASO-injected embryos is caused by the de-repression of apical regulatory genes within animal ectoderm cells. This result leads to the prediction that Wn8 activates the expression of an early-acting repressor of foxq2 in the animal ectoderm. The observation that Wnt8 signaling is required for cell-fate decisions in the animal ectoderm is consistent with the expression of fzd1/2/7 in all cells of this domain between 12 and 18 h, the time when Wnt-signaling input is required to repress apical cell fates (SI Appendix, Fig. S6B), and by the absence of potential Wnt-signaling antagonists that could interfere with Wnt8 signaling. The boundaries between gene-expression domains within the vegetal half of the embryo are not affected by Wnt8 signaling. These results indicate that gene-expression patterning occurs independently in the vegetal and animal half of the sea urchin embryo.
Control of wnt Gene Expression by Cell-Fate Specification GRNs.
Even though the early expression of wnt genes occurs in dynamic spatial patterns, all five wnt genes are expressed in the veg1 endoderm by 24 h, and some also are expressed in the veg1 ectoderm (Fig. 1C). This finding raises the question of how the transcription of these genes is regulated by the cell-fate specific GRNs operating in these cells. Moreover, we would like to know if the regulatory mechanisms that control their expression are similar, given the overlapping expression patterns of wnt4, wnt5, and wnt8, as well as of wnt1 and wnt16, throughout early sea urchin development (Fig. 1C; for a direct comparison of the expression patterns of wnt genes and relevant regulatory genes, see SI Appendix, Fig. S11). The earliest specification of the veg1 cell lineage, the precursor of veg1 endoderm and veg1 ectoderm, is controlled by eve, a regulatory gene exclusively expressed in veg1 cells by 15 h (2). Eve contributes to the activation of hox11/13b in veg1 endoderm, which is the earliest regulatory gene operating in the GRN underlying the specification of posterior endoderm fate (3). Perturbation of the pan-veg1 transcription factor Eve by injection of Eve MASO resulted in decreased expression of wnt1, wnt4, wnt5, and wnt16, as is consistent with their expression in veg1 cells, although expression of wnt8 in these same cells was not affected (SI Appendix, Fig. S12). However, blocking the expression of the endoderm-specific Hox11/13b transcription factor with morpholinos affects only the transcription of wnt1 and wnt16, but not that of the other wnt genes, as might be expected from the exclusive expression of wnt1 and wnt16 in veg1 endodermal cells at 24 h (SI Appendix, Fig. S12). Blocking the expression of other endodermal regulatory factors—FoxA, Blimp1b, Brachyury, or GataE—did not affect the expression of the five wnt genes (SI Appendix, Fig. S13). Thus, the expression of wnt1 and wnt16 probably is regulated directly by both Hox11/13b and Eve, and the transcription of wnt4 and wnt5 is activated downstream of Eve but is independent of Hox11/13b. Late Wnt8 expression is controlled by a separate mechanism, which recently was shown to involve the pan-ectodermal regulator SoxB1 (5).
Thus, in all three cases, e.g., [wnt1 + wnt 16], [wnt4 + wnt5], and [wnt8], the wnt genes are wired into the GRNs they control. The GRN circuitry, which is summarized in Fig. 4, explains the spatial and temporal expression pattern of wnt genes and indicates a remarkable relationship between the transcriptional control of wnt genes and their downstream functions. For instance, eve expression is specific to all cells of the veg1 lineage from 15 h on, and the expression of wnt4 and wnt5 in these same cells, under the control of Eve, is first observed shortly after that time point, at 18 h, and continues to 24 h. In turn, Wnt4 activates gene expression in veg1 ectodermal cells, where Eve continues to be expressed. Thus Eve plays a dual role in the specification of veg1 ectoderm: It directly represses genes of the animal ectoderm in these cells, and it activates the expression of wnt4 in veg1 endoderm and veg1 ectoderm, leading to the activation of genes of the veg1 ectodermal GRN.
Fig. 4.
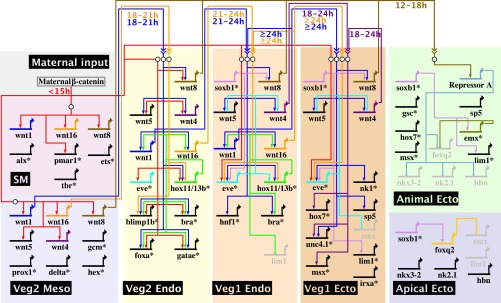
Model of regulatory interactions between wnt genes and domain-specific GRNs. This diagram summarizes the regulatory interactions between wnt genes and the regulatory genes composing the cell-fate specification GRNs operating in embryonic regulatory-state domains and the regulatory inputs from maternal β-catenin. The GRNs shown omit known input and output linkages at most regulatory genes, as indicated by asterisks at the gene name. The time windows during which Wnt-signaling inputs are active are indicated below the regulatory linkages.
In veg1 endoderm cells, expression of hox11/13b initiates at 21–24 h under the control of Eve, and transcription of wnt1 and wnt16, activated by Hox11/13b, starts at 24 h. Their expression is not observed in veg1 ectoderm, where Hox11/13b is not expressed, although eve is. Because signaling by Wnt1 and Wnt16 also affects the expression of hox11/13b and eve, these results indicate that expression of Hox11/13b, a crucial upstream transcription factor in the posterior endoderm GRN, is controlled by a positive feedback circuit between wnt1, wnt16, and hox11/13b, thus ensuring specification of the posterior endoderm cell fate (Fig. 4). Curiously, this circuit also might be responsible for the expression of wnt1, wnt16, and hox11/13b at earlier time points. At 18 h, when hox11/13b expression becomes sensitive to Wnt signaling according to the C59 experiments, hox11/13b is transcribed specifically in veg2 endoderm, and wnt1 and wnt16 are transcribed in adjacent veg2 mesoderm. By 21 h, wnt1 and wnt16 are expressed in veg2 endoderm, controlled by Hox11/13b, and the transcription of hox11/13b is activated in adjacent veg1 endoderm cells. By 24 h, the expression of all three genes has terminated in veg2 endoderm by a mechanism involving the auto-repression of Hox11/13b (3), and the entire positive feedback circuit is operative exclusively in veg1 endoderm cells. Thus, the sequence begins as an inductive relay, in which veg2 mesoderm Wnt1 and Wnt16 first activate the hox11/13b gene in veg2 endoderm, and then veg2 endoderm Hox11/13b activates the wnt1 and wnt16 genes in veg2 endoderm, whereupon Wnt1 and Wnt16 activate the hox11/13b gene in Veg1 endoderm. In veg1 endoderm their mutual positive feedback locks in the circuit. The existence of a signaling interaction between the veg2 endoderm GRN and the veg1 endoderm GRN, which is responsible for activating hox11/13b transcription in veg1 endoderm cells after 21 h, was predicted earlier (3). Here we identify this signal to be both wnt1 and wnt16, which are expressed in veg2 endoderm cells at 21 h under the control of Hox11/13b and which are required for hox11/13b transcription in the adjacent veg1 endoderm domain, thus fulfilling all criteria for the predicted signal.
Finally, the expression of wnt8 in the veg1 endoderm and veg1 ectoderm domains is regulated differently from that of wnt4 and wnt5, even though these three genes show the same expression pattern. By 24 h, wnt8 expression is activated by SoxB1, a transcription factor that is present throughout the animal half of the embryo as well as in the veg1 lineage and that functions as an activator of many ectodermal regulatory genes (5, 41). SoxB1 thus activates the specification GRNs in animal ectoderm cells and controls the expression of the signaling ligand that is required to exclude an alternative fate in these cells.
Thus, in summary, even though all five wnt genes are expressed in veg1 endoderm, only wnt1 and wnt16, which affect the activity of the endoderm GRN, are also controlled by the endoderm GRN regulator Hox11/13b (see network in Fig. 4). Conversely, the wnt genes that affect the activity of the veg1 ectoderm GRN (wnt4 and perhaps wnt5) are independent of Hox11/13b expression and instead are controlled by the veg1 regulator Eve. wnt8, which causes the expression of a repressor of apical neurogenic genes in the animal ectoderm, is activated by the animal ectoderm factor SoxB1.
Discussion
We show here the results of a system-wide analysis of the functions of Wnt signaling in the regulation of the cell-fate specification GRNs operating across the entire sea urchin embryo during pregastrular development. Our approach was to analyze in every regulatory-state domain of this embryo the spatial expression of all genes involved in intercellular Wnt signaling, encoding Wnt ligands, Frizzled receptors, and possible antagonists such as Sfrp and Dkk proteins. We addressed the overall function of Wnt signaling in developmental GRNs during the first 24 h of embryogenesis by using the C59 Porcupine inhibitor, which interferes globally with the secretion of Wnt-signaling ligands. This perturbation affects the expression of specific regulatory genes that in the GRNs lie downstream of Wnt signal inputs. The contribution of each Wnt-signaling ligand to the regulation of expression of the genes affected by C59 treatment was analyzed further by individually blocking the expression of each Wnt ligand by the injection of antisense morpholinos. Our results lead to overall conclusions regarding the functions of the Wnt-signaling system in an entire embryo and how these functions are mediated by the linkages between the Wnt-signaling system and the GRNs controlling the developmental process. This study traverses a large-scale developmental process during which diverse regulatory-state domains are established throughout the embryo.
Specific Functions of Wnt Signaling in the Embryonic GRNs.
Only 5 of the 11 Wnt ligands encoded in the S. purpuratus genome are expressed during pregastrular development. Their spatial expression, even though dynamically changing, is confined at all times to at most two adjacent gene-expression domains. The remainder of the embryo expresses no wnt genes. The spatial expression of Frizzled receptors occurs in broad contiguous domains. After 18 h, all cells express no more than one Frizzled receptor, and most cells of mesodermal fate express none. The system-level analysis of Wnt ligand and receptor expression patterns yields revealing insights into the functions of Wnt signaling in the whole process of early sea urchin development. For example, we see that the Wnt-signaling system can have nothing to do with specification of most mesodermal fates, because, except for the oral mesoderm after 21 h, no mesodermal cells express Wnt-signaling receptors. This prediction is confirmed in the C59 experiment in which no effects on mesodermal gene expression were recorded in the face of global knockdown of zygotic Wnt signaling. Although Wnt signaling plays important roles in endoderm specification, as shown by the C59 and Wnt morpholino experiments, the expression patterns indicate that after about 21 h this role must be restricted to the posterior endoderm, because transcripts of wnt ligands and receptors cease to be detectable in the anterior endoderm by this time, and the expression of the potential antagonists sfrp3/4 and dkk1 is activated. On the other hand, these expression patterns imply the importance of Wnt signaling in the posterior endoderm, where all five wnt ligands are expressed at 24 h and where fzd9/10 continues to be expressed at all times considered. The same arguments also apply to the veg1 ectoderm, where the expression of Wnt ligands and receptors but of no antagonists is consistent with the observed Wnt-signaling function. In the animal ectoderm the presence of a receptor marks the potential to receive Wnt signals, although the absence of Wnt ligand expression precludes these domains as sources of Wnt signaling. Indeed, these cells receive Wnt8 signaling inputs from more vegetally located cells, leading to the exclusion of apical neurogenic fate, but Wnt signaling is not required for activation of ectodermal GRNs. Finally, the apical neurogenic domain does not express Wnt ligands but does express a Wnt receptor and also several antagonists that protect it from Wnt8 signaling. When viewed in detail at a system level, the expression patterns thus confer a remarkably accurate set of predictions that illuminate which domains of the embryo engage in Wnt signaling.
In the sea urchin embryo the key important function of Wnt signaling in this developmental phase consists of their impact on the endodermal GRNs, because the expression of a majority of regulatory genes in these networks is activated by Wnt signaling (2, 3). However, contrary to previous assumption, the earliest Tcf-dependent expression of endodermal regulatory genes and also the initiation of the skeletogenic GRN do not depend on Wnt ligand signaling but are mediated by the presence of maternal β-catenin which ab initio is localized specifically in cells of the skeletogenic and veg2 lineages (17). Thus, despite the early expression of wnt genes starting at 7 h in this embryo and the presence of maternal Fzd1/2/7, Wnt signaling is not required for early gene expression, because the earliest effects upon treatment with the Porcupine inhibitor C59 are observed only at 15–18 h. This finding is a strong conclusion which depends on our and other’s evidence that C59 is highly efficient in blocking an essential step in Wnt signaling. In this work a convincing argument is that the sum total of individual Wnt morpholino effects equals the effect of C59 treatment, as we show for every domain affected by this perturbation. Outside the endodermal GRNs, only a few regulatory genes expressed in veg1 ectoderm cells require activation by Wnt signaling. In animal ectoderm cells, Wnt signaling is additionally required to repress activation of the apical neurogenic GRN, but this function probably is also confined to activation of a yet unknown early repressor of apical gene transcription. The specification of all mesodermal cell fates and of all apical cell fates occurs independently of Wnt signaling in this embryo during pregastrular development. Thus, a comprehensive view of Wnt-signaling system function reveals only a small set of target genes, and these belong to precisely confined GRNs of the embryo. Where the Wnt-signaling system does impact the embryonic GRNs, it does so by the use of characteristic circuitry affecting many nodes of the GRN, in which cross-regulatory interactions ensure continued expression of the active Wnt ligands.
Our results indicate that in this embryo, and at the stages we consider here, Wnt signaling functions by short-range intercellular interactions. Thus, each Wnt ligand affects gene expression either in cells of the same domain, where it is also expressed, or in cells of an immediately adjacent domain. Examples of the latter, which constitute short-range inductive signaling effects, are, as shown in Fig. 4, the response of veg2 endoderm to Wnt signals emitted by the veg2 mesoderm (Wnt1 and Wnt16), the response of veg1 endoderm to Wnt signals emitted by veg2 endoderm (Wnt1 and Wnt16), and the response of veg1 ectoderm to signals emitted by veg1 endoderm (Wnt1). In every case in which Wnt-signaling ligands affect an adjacent domain, this domain is located on the animal side of the domain of Wnt ligand expression. The mechanism underlying this vegetal-to-animal polarity is enigmatic. The polarity clearly is not caused by a layered or oriented expression of diverse Frizzled receptors, because their spatially simple patterns of expression excludes this possibility. A remarkable aspect of Wnt signaling revealed in this study is that it repeatedly functions in an intradomanic manner, so that cells within a given regulatory-state domain both receive and emit the Wnt signal. This feature corresponds to the output of community-effect circuitry, which in the cases examined relies on regulation of the signaling ligand gene by the signal transduction system that it activates in responding cells (42). The import of this circuitry is to ensure the homogeneity of regulatory states among cells of a given domain by establishing a positive intercellular feedback throughout the domain. In this work we find intradomain community-effect signaling specifically in the veg1 endodermal domain, where the feedback consists of the activation of wnt1 and wnt16 by Hox11/13b, and Wnt1 and Wnt16 in turn activate hox11/13b (via Tcf/β-catenin input) (Fig. 4). Similarly, wnt4 is expressed within the veg1 ectoderm and also activates gene expression within this domain.
Remarkably, the control of expression of all five relevant wnt genes is tightly correlated with their respective functions. Thus, the wnt genes that operate in the endodermal GRNs also are transcribed under the control of endodermal regulators. Similarly, Wnt8, which operates on the animal ectoderm GRN, is transcriptionally controlled by an animal ectoderm regulator. The wnt genes that operate in the veg1 GRNs are controlled by Eve, which regulates gene expression in Veg1 ectoderm. Partly, this pattern would follow de facto from the participation of these ligands in intradomain signaling, but intradomain signaling cannot be the complete explanation, because some of the relevant signaling is inductive. What emerges is that particular regulatory cassettes link transcriptional control of Wnt signaling to the cell-fate specification GRNs they regulate. The implication of this circuitry is that the temporal coexpression of the Wnt-signaling ligands and their target genes, as well as the spatial proximity of signal-sending and signal-receiving domains, is ensured by the common control of wnt genes and their target regulatory genes by the same cell fate-specific transcriptional regulators.
The Specificity Conundrum in the Wnt-Signaling System.
An important question is how specificity of Wnt signaling is mediated, and this issue is a general one, given the multiplicity of wnt and fzd genes encoded in all animal genomes. Although our study was not particularly designed to address this question, our results may hint at the basic design principle of this signaling system. Most important, in the context of GRN analysis, is the question of how Wnt signaling in different embryonic domains or different developmental phases leads to the regulation of specific, different sets of target genes. At the cis-regulatory level, the causal explanation for which genes are expressed in response to Wnt signaling must rely on combinatoriality in the regulation of gene transcription. Tcf, the transcription factor activated by Wnt signaling, always operates together with other transcription factors, some of which are expressed exclusively in cells of a given fate at a particular developmental time. We see here, as specific examples, that most regulatory genes activated in endodermal precursor cells by Wnt1 signaling also are activated by Hox11/13b, whereas Wnt1 signaling in veg1 ectoderm leads to expression of nk1, which also is a target gene of Not and Lim1 transcription factors expressed in these cells. Thus, the impact of Wnt signaling on gene expression in cells of different fates causally depends on the regulatory state in the signal-receiving cells. Note here that all gene-expression domains regulated by Wnt1 signaling express the same Frizzled receptor, Fzd9/10, and therefore, for the early sea urchin embryo, we can exclude the idea that the difference in target gene sets depends on the utilization of diverse Fzd receptors.
An additional level of specificity must be invoked to account for the selective interaction of given Wnt ligands with given domains. For example, all five Wnt ligands are expressed in veg1 endoderm cells, but only Wnt1 and Wnt16 are required for GRN function in this domain. Similarly, Wnt4 affects veg1 ectoderm cells, but Wnt16 and Wnt8 do not. In each of these domains specific Wnt ligands are required for signaling response, and that domain is blind to the presence of the other Wnt ligands. Moreover, veg1 endoderm and veg1 ectoderm respond to different Wnt ligands, despite the expression of the same receptor, Fzd9/10. This difference in response points directly at a Wnt ligand recognition function in addition to the Fzd9/10 receptor present on these cells. An explanation could be the differential expression in the diverse embryonic domains of Wnt coreceptors such as the LDL receptor-related protein family in these cells (for review see ref. 43).
Materials and Methods
The methods used in this work are provided in SI Appendix, Supplemental Materials and Methods. There we briefly detail procedures for gene cloning and WMISH; C59 treatments; mRNA and morpholino injection and RNA preparation; and quantitative measurements of transcript prevalence by qPCR and Nanostring Ncounter analysis.
Supplementary Material
Acknowledgments
We thank Prof. Mike Collins (University of California, Los Angeles) for helpful discussions and for bringing to our attention the small-molecule antagonist C59. This research was supported by National Institutes of Health Grant HD-037105 and the Lucille P. Markey Charitable Trust.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419141111/-/DCSupplemental.
References
- 1.Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci USA. 2008;105(16):5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peter IS, Davidson EH. The endoderm gene regulatory network in sea urchin embryos up to mid-blastula stage. Dev Biol. 2010;340(2):188–199. doi: 10.1016/j.ydbio.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peter IS, Davidson EH. A gene regulatory network controlling the embryonic specification of endoderm. Nature. 2011;474(7353):635–639. doi: 10.1038/nature10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li E, Materna SC, Davidson EH. New regulatory circuit controlling spatial and temporal gene expression in the sea urchin embryo oral ectoderm GRN. Dev Biol. 2013;382(1):268–279. doi: 10.1016/j.ydbio.2013.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li E, Cui M, Peter IS, Davidson EH. Encoding regulatory state boundaries in the pregastrular oral ectoderm of the sea urchin embryo. Proc Natl Acad Sci USA. 2014;111(10):E906–E913. doi: 10.1073/pnas.1323105111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Materna SC, Ransick A, Li E, Davidson EH. Diversification of oral and aboral mesodermal regulatory states in pregastrular sea urchin embryos. Dev Biol. 2013;375(1):92–104. doi: 10.1016/j.ydbio.2012.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Range RC, Venuti JM, McClay DR. LvGroucho and nuclear beta-catenin functionally compete for Tcf binding to influence activation of the endomesoderm gene regulatory network in the sea urchin embryo. Dev Biol. 2005;279(1):252–267. doi: 10.1016/j.ydbio.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 8.Peter IS, Faure E, Davidson EH. Predictive computation of genomic logic processing functions in embryonic development. Proc Natl Acad Sci USA. 2012;109(41):16434–16442. doi: 10.1073/pnas.1207852109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliveri P, Davidson EH, McClay DR. Activation of pmar1 controls specification of micromeres in the sea urchin embryo. Dev Biol. 2003;258(1):32–43. doi: 10.1016/s0012-1606(03)00108-8. [DOI] [PubMed] [Google Scholar]
- 10.Smith J, Davidson EH. Regulative recovery in the sea urchin embryo and the stabilizing role of fail-safe gene network wiring. Proc Natl Acad Sci USA. 2009;106(43):18291–18296. doi: 10.1073/pnas.0910007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de-Leon SB, Davidson EH. Information processing at the foxa node of the sea urchin endomesoderm specification network. Proc Natl Acad Sci USA. 2010;107(22):10103–10108. doi: 10.1073/pnas.1004824107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith J, Kraemer E, Liu H, Theodoris C, Davidson E. A spatially dynamic cohort of regulatory genes in the endomesodermal gene network of the sea urchin embryo. Dev Biol. 2008;313(2):863–875. doi: 10.1016/j.ydbio.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croce JC, McClay DR. Dynamics of Delta/Notch signaling on endomesoderm segregation in the sea urchin embryo. Development. 2010;137(1):83–91. doi: 10.1242/dev.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sethi AJ, Wikramanayake RM, Angerer RC, Range RC, Angerer LM. Sequential signaling crosstalk regulates endomesoderm segregation in sea urchin embryos. Science. 2012;335(6068):590–593. doi: 10.1126/science.1212867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McIntyre DC, Seay NW, Croce JC, McClay DR. Short-range Wnt5 signaling initiates specification of sea urchin posterior ectoderm. Development. 2013;140(24):4881–4889. doi: 10.1242/dev.095844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Range RC, Angerer RC, Angerer LM. Integration of canonical and noncanonical Wnt signaling pathways patterns the neuroectoderm along the anterior-posterior axis of sea urchin embryos. PLoS Biol. 2013;11(1):e1001467. doi: 10.1371/journal.pbio.1001467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development. 1999;126(2):345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- 18.Croce JC, et al. A genome-wide survey of the evolutionarily conserved Wnt pathways in the sea urchin Strongylocentrotus purpuratus. Dev Biol. 2006;300(1):121–131. doi: 10.1016/j.ydbio.2006.08.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Croce J, et al. Wnt6 activates endoderm in the sea urchin gene regulatory network. Development. 2011;138(15):3297–3306. doi: 10.1242/dev.058792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu Q, Cameron RA, Davidson EH. Quantitative developmental transcriptomes of the sea urchin Strongylocentrotus purpuratus. Dev Biol. 2014;385(2):160–167. doi: 10.1016/j.ydbio.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robert N, Lhomond G, Schubert M, Croce JC. A comprehensive survey of wnt and frizzled expression in the sea urchin Paracentrotus lividus. Genesis. 2014;52(3):235–250. doi: 10.1002/dvg.22754. [DOI] [PubMed] [Google Scholar]
- 22.Croce J, Duloquin L, Lhomond G, McClay DR, Gache C. Frizzled5/8 is required in secondary mesenchyme cells to initiate archenteron invagination during sea urchin development. Development. 2006;133(3):547–557. doi: 10.1242/dev.02218. [DOI] [PubMed] [Google Scholar]
- 23.Ransick A, Davidson EH. Late specification of Veg1 lineages to endodermal fate in the sea urchin embryo. Dev Biol. 1998;195(1):38–48. doi: 10.1006/dbio.1997.8814. [DOI] [PubMed] [Google Scholar]
- 24.Yaguchi S, Yaguchi J, Angerer RC, Angerer LM, Burke RD. TGFβ signaling positions the ciliary band and patterns neurons in the sea urchin embryo. Dev Biol. 2010;347(1):71–81. doi: 10.1016/j.ydbio.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Angerer LM, Yaguchi S, Angerer RC, Burke RD. The evolution of nervous system patterning: Insights from sea urchin development. Development. 2011;138(17):3613–3623. doi: 10.1242/dev.058172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herr P, Basler K. Porcupine-mediated lipidation is required for Wnt recognition by Wls. Dev Biol. 2012;361(2):392–402. doi: 10.1016/j.ydbio.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Biechele S, Cox BJ, Rossant J. Porcupine homolog is required for canonical Wnt signaling and gastrulation in mouse embryos. Dev Biol. 2011;355(2):275–285. doi: 10.1016/j.ydbio.2011.04.029. [DOI] [PubMed] [Google Scholar]
- 28.Biechele S, Cockburn K, Lanner F, Cox BJ, Rossant J. Porcn-dependent Wnt signaling is not required prior to mouse gastrulation. Development. 2013;140(14):2961–2971. doi: 10.1242/dev.094458. [DOI] [PubMed] [Google Scholar]
- 29.Najdi R, et al. A uniform human Wnt expression library reveals a shared secretory pathway and unique signaling activities. Differentiation. 2012;84(2):203–213. doi: 10.1016/j.diff.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen B, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5(2):100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lum L, Clevers H. Cell biology. The unusual case of Porcupine. Science. 2012;337(6097):922–923. doi: 10.1126/science.1228179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Proffitt KD, et al. Pharmacological inhibition of the Wnt acyltransferase PORCN prevents growth of WNT-driven mammary cancer. Cancer Res. 2013;73(2):502–507. doi: 10.1158/0008-5472.CAN-12-2258. [DOI] [PubMed] [Google Scholar]
- 33.Rios-Esteves J, Haugen B, Resh MD. Identification of key residues and regions important for porcupine-mediated Wnt acylation. J Biol Chem. 2014;289(24):17009–17019. doi: 10.1074/jbc.M114.561209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li E, Materna SC, Davidson EH. Direct and indirect control of oral ectoderm regulatory gene expression by Nodal signaling in the sea urchin embryo. Dev Biol. 2012;369(2):377–385. doi: 10.1016/j.ydbio.2012.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ben-Tabou de-Leon S, Su YH, Lin KT, Li E, Davidson EH. Gene regulatory control in the sea urchin aboral ectoderm: Spatial initiation, signaling inputs, and cell fate lockdown. Dev Biol. 2013;374(1):245–254. doi: 10.1016/j.ydbio.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yaguchi S, Yaguchi J, Angerer RC, Angerer LM. A Wnt-FoxQ2-nodal pathway links primary and secondary axis specification in sea urchin embryos. Dev Cell. 2008;14(1):97–107. doi: 10.1016/j.devcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 37.Tu Q, Brown CT, Davidson EH, Oliveri P. Sea urchin Forkhead gene family: Phylogeny and embryonic expression. Dev Biol. 2006;300(1):49–62. doi: 10.1016/j.ydbio.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 38.Yaguchi S, et al. Fez function is required to maintain the size of the animal plate in the sea urchin embryo. Development. 2011;138(19):4233–4243. doi: 10.1242/dev.069856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Materna SC, Howard-Ashby M, Gray RF, Davidson EH. The C2H2 zinc finger genes of Strongylocentrotus purpuratus and their expression in embryonic development. Dev Biol. 2006;300(1):108–120. doi: 10.1016/j.ydbio.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 40.Howard-Ashby M, et al. Identification and characterization of homeobox transcription factor genes in Strongylocentrotus purpuratus, and their expression in embryonic development. Dev Biol. 2006;300(1):74–89. doi: 10.1016/j.ydbio.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 41.Kenny AP, Oleksyn DW, Newman LA, Angerer RC, Angerer LM. Tight regulation of SpSoxB factors is required for patterning and morphogenesis in sea urchin embryos. Dev Biol. 2003;261(2):412–425. doi: 10.1016/s0012-1606(03)00331-2. [DOI] [PubMed] [Google Scholar]
- 42.Bolouri H, Davidson EH. The gene regulatory network basis of the “community effect,” and analysis of a sea urchin embryo example. Dev Biol. 2010;340(2):170–178. doi: 10.1016/j.ydbio.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Amerongen R, Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009;136(19):3205–3214. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.