Significance
Complex cellular phenotypes are dependent upon a large underlying network of genes. Determining the number of interacting genes, the nature of their interactions, and the robustness of their encompassing network to cellular and genomic perturbations is important for understanding how genotype determines phenotype. Here, we approached these questions in the cyanobacterial circadian gene network by knocking out circadian input kinase A (cikA), a gene involved in synchronizing and relaying temporal signals within the cell, and screening for second-site suppressor mutations that rescue the severely altered cikA-null circadian phenotype. We identified two independent mutations in sasA (Synechococcus adaptive sensor A), a known clock-associated gene, that restore normal rhythms. These results change our conception of the circadian gene network architecture and show how easily the network can adapt to severe perturbations.
Keywords: gene network, cyanobacteria, evolution
Abstract
The circadian input kinase of the cyanobacterium Synechococcus elongatus PCC 7942 (CikA) is important both for synchronizing circadian rhythms with external environmental cycles and for transferring temporal information between the oscillator and the global transcriptional regulator RpaA (regulator of phycobilisome-associated A). KOs of cikA result in one of the most severely altered but still rhythmic circadian phenotypes observed. We chemically mutagenized a cikA-null S. elongatus strain and screened for second-site suppressor mutations that could restore normal circadian rhythms. We identified two independent mutations in the Synechococcus adaptive sensor A (sasA) gene that produce nearly WT rhythms of gene expression, likely because they compensate for the loss of CikA on the temporal phosphorylation of RpaA. Additionally, these mutations restore the ability to reset the clock after a short dark pulse through an output-independent pathway, suggesting that SasA can influence entrainment through direct interactions with KaiC, a property previously unattributed to it. These experiments question the evolutionary advantage of integrating CikA into the cyanobacterial clock, challenge the conventional construct of separable input and output pathways, and show how easily the cell can adapt to restore phenotype in a severely compromised genetic network.
Second-site suppressor mutagenesis screens have been useful in untangling the genetic components and pathways that underlie complex cellular phenotypes (1). By understanding how mutations in one gene can compensate for changes in another, we can infer how those genes interact with each other and with their encompassing genetic network. We used this approach to better understand the genetic determination of circadian rhythms in the cyanobacterium Synechococcus elongatus PCC 7942. Because the core clock genes in this species exist as single copies, suppressing mutations are not buffered by paralogs, making S. elongatus ideal for this type of mutagenic assay. Approximately 15 genes have been identified that influence the circadian phenotype to various degrees, the most important of which are the kai genes: kaiA, kaiB, and kaiC. In cyanobacteria, time is kept through the central oscillator protein KaiC, whose phosphorylation state, conformation, and ATPase activity, which are mediated by interactions with KaiA and KaiB, cycle within a 24-h period (2–4). The temporal phosphorylation of KaiC can be reconstituted in vitro by simply combining the three Kai proteins and ATP, suggesting that time is kept primarily through a posttranslational mechanism (5). Although the Kai oscillator functions relatively robustly in isolation, additional proteins are necessary to synchronize the oscillations with the environment (entrainment) through the detection of cellular and environmental cues (input pathway) and to transfer temporal information from the oscillator to circadian-dependent transcriptional regulators (output pathway). One of these proteins, circadian input kinase A (CikA), has been implicated in both clock input and output.
KOs of cikA are characterized by a significantly altered circadian rhythm: an ∼2.5-h shorter period and a decrease in amplitude (6). Additionally, cikA-null cells lose their ability to reset phase after a 5-h dark pulse, strongly suggesting that CikA is involved in oscillator entrainment (6). Interactions with the oscillator are further supported by the coprecipitation of CikA with KaiC in pull-down experiments and by the modulation of the rhythmic KaiC phosphorylation pattern in a cikA-null background (7). CikA’s involvement in the output pathway is supported by its direct interaction with the global transcriptional regulator RpaA, whose temporal phosphorylation is essential for the rhythmic regulation of gene expression (8). CikA has been shown to dephosphorylate and subsequently deactivate RpaA, a function antagonistic to the kinase SasA that temporally phosphorylates RpaA (8). Although CikA belongs to the same His protein kinase family as SasA, it is not known if its kinase activity has an important role in the output pathway.
We were interested in identifying suppressor mutations that can compensate for the phenotypic consequences of a cikA deletion. Because cikA is involved in both circadian input and output, we could screen for mutations that rescue phenotypic changes in either pathway. Here, we describe two independent mutations in the sasA gene that rescue both the circadian input and output phenotypes in a cikA-null background. These results address how SasA and CikA interact, the potential role of SasA in clock entrainment, the resiliency of the circadian gene network, and the evolutionary advantage of integrating CikA into the clock.
Results
Chemical Mutagenesis and Screening.
We disrupted the cikA gene in S. elongatus PCC 7942 reporter strain AMC462 [PkaiBC::luxAB, PpsbAI::luxCDE (9)] by replacing it with a kanamycin resistance gene by homologous recombination. As previously reported, the cikA-null strain displays a number of altered circadian gene expression phenotypes (6): a 2.72-h shorter period, a dramatic decrease in amplitude, and loss of the ability to reset the phase of the rhythms after a 5-h dark pulse (Fig. 1 and Table 1). We chemically mutagenized this strain with ethyl methanesulfonate (EMS) and screened plated colonies for altered circadian rhythms (Materials and Methods). To perform this screen, we designed and built a computer-controlled turntable that enabled the determination of circadian rhythms from thousands of primary colonies at once by iteratively rotating Petri dishes under a cooled CCD camera [complete design plans and image analysis software are reported elsewhere (10)]. This device mimics the general design used that led to the identification of the kai genes by Kondo and Ishiura (11). Using this apparatus, we screened 2,052 colonies and identified eight that showed a reproducibly rhythmic circadian phenotype that differed from the cikA-null strain.
Fig. 1.
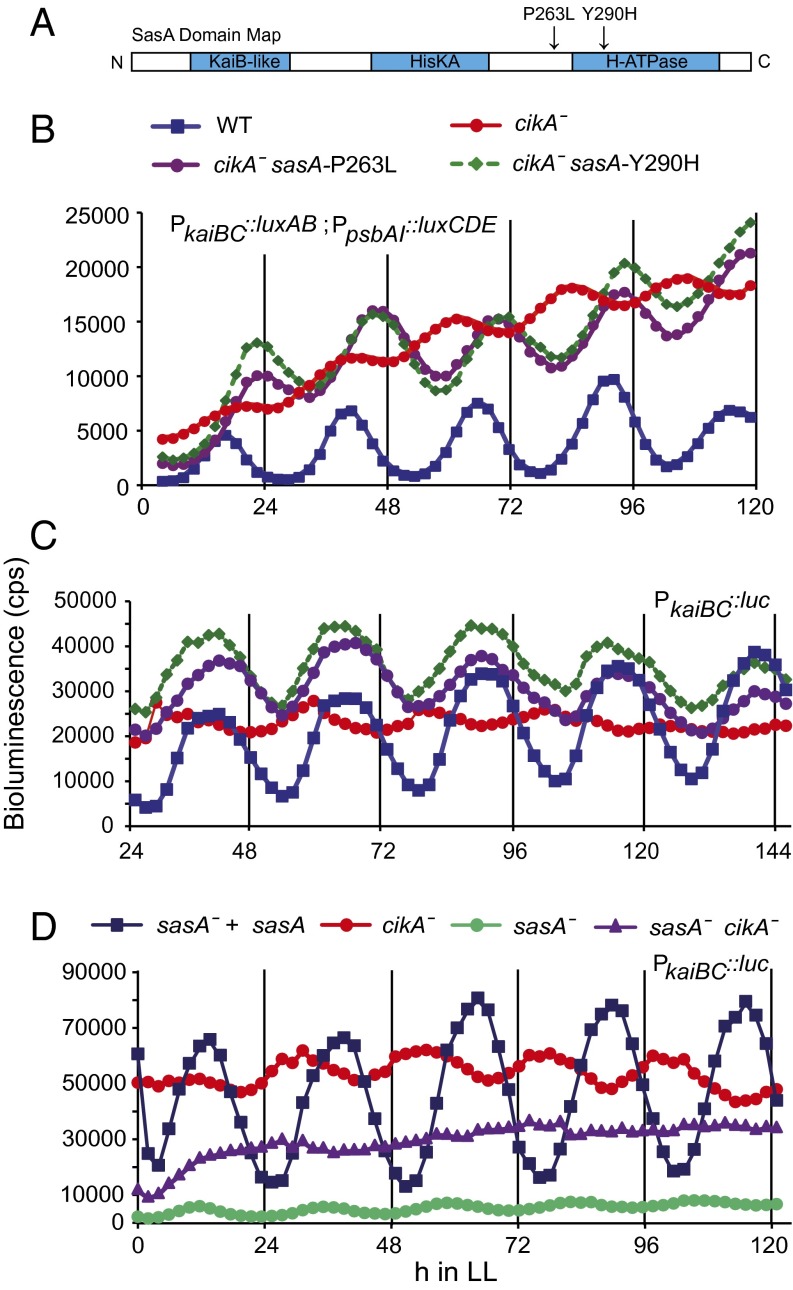
SasA P263L and Y290H mutants independently restore circadian phenotype in a cikA-null background. (A) SasA protein has three domains: a KaiB-like domain, a His kinase A dimerization/phosphoacceptor domain (HisKA), and a His kinase-like ATPase domain (H-ATPase). The arrows above the domain map denote the locations of the cikA-null suppressing sasA mutations. (B) Bioluminescence of S. elongatus PCC 7942 strains was measured every 2 h in constant light on a TopCount plate reader. Representative traces are shown for the sasA mutant cikA-null strains originally identified in the chemical mutagenesis screen. The bioluminescent reporter used for these strains was PkaiBC::luxAB, PpsbAI::luxCDE. (C) Representative traces of cleanly reconstructed sasA/cikA double mutants with a PkaiBC::luc reporter. (D) Representative traces of SasA/CikA single and double KOs with a PkaiBC::luc reporter. LL, constant light.
Table 1.
Circadian period data for CikA and SasA mutant strains
| Strain | Period, h ± SD | n |
| Circadian periods of SasA single alleles | ||
| WT(AMC541) | 25.12 ± 0.05 | 9 |
| sasA | 23.36 ± 0.34 | 9 |
| sasA + WTSasA | 25.50 ± 0.34 | 7 |
| sasA + SasA P263L | 27.87 ± 0.34 | 9 |
| sasA + SasA Y290H | 27.44 ± 0.34 | 7 |
| Circadian periods of original cikA sasA suppressor strains | ||
| WT(AMC541) | 24.76 ± 0.26 | 10 |
| cikAKmR | 22.04 ± 0.27 | 9 |
| cikA + SasA P263L | 24.08 ± 0.48 | 10 |
| cikA + SasA Y290H | 24.17 ± 0.54 | 10 |
| Circadian periods of reconstructed cikA sasA suppressor strains | ||
| WT(AMC541) | 25.12 ± 0.05 | 9 |
| cikAGmR | 24.70 ± 0.89 | 10 |
| cikA + WTSasA | 22.80 ± 0.40 | 8 |
| cikA + SasA P263L | 24.18 ± 0.76 | 10 |
| cikA + SasA Y290H | 24.44 ± 0.22 | 10 |
To identify the genetic changes that account for these phenotypic differences, we sequenced the genomes of the six mutants that showed the most dramatic changes in circadian rhythms relative to the cikA-null parent at ≥50-fold coverage. We observed as few as one and as many as seven EMS-induced mutations in these sequenced strains (Tables S1 and S2). Surprisingly, five of these genomes contained a mutation in the sasA gene. In these five genomes, there were three unique mutations: a deletion of the first base of the stop codon that resulted in a 15-residue extension of the C terminus of sasA (SasA C-ext), a single-nucleotide mutation in the putative ATPase domain that changed the Tyr at residue 290 to a His (Y290H), and a single base change upstream of the ATPase domain that altered the Pro at residue 263 to a Leu (P263L) (Fig. 1A). Two sequenced genomes had the same Y290H mutation but did not share other mutations. The P263L mutation was also observed in two sequenced genomes that did not share all polymorphisms. These colonies may either be unique mutants or sister colonies in which additional mutations were picked up by differential coverage during sequencing, because the protocol allowed cellular proliferation during a recovery period. Suspected sister colonies also exhibited similar circadian phenotypes. The sixth genome did not contain a mutation in sasA, but rather had a point mutation in the intergenic region upstream of the circadian-controlled transcriptional regulator rpaA and downstream of the crm gene (12). This mutant displayed a further disruption in bioluminescence amplitude relative to the cikA-null strain, approaching arrhythmia.
All sequenced strains had five common polymorphisms relative to the published S. elongatus PCC 7942 sequenced genome (GenBank accession no. NC_007604). Four of the five differences are also present in a resequencing of our laboratory strain, provided courtesy of Life Technologies, suggesting possible errors in GenBank accession no. NC_007604 or accumulated polymorphisms in our lab strain since it was originally sequenced. Two of these polymorphisms are also present in the S. elongatus PCC 6301 genome (GenBank accession no. NC_006576), further suggesting that they represent the WT sequence. The fifth polymorphism was not observed in any previously sequenced genome but is present in our starting strain as determined by Sanger sequencing. This C-to-A mutation in a noncoding region is unlikely to have any functional significance. A summary of all identified mutations is reported in Tables S1 and S2.
Molecular and Cellular Phenotypes of the SasA Mutants.
We measured the circadian rhythms of the three cikA-null sasA double mutants identified above by detecting the temporal expression of a luciferase reporter under the control of a circadian-regulated promoter (13). Two of these mutants (Y290H and P263L) reproducibly displayed phenotypes more similar to WT than the cikA-null strain: an ∼2-h increase in period and an increase in amplitude (Fig. 1B and Table 1). In subsequent tests, SasA C-ext showed a phenotype similar to the cikA-null parent strain, with some variability in phase between measurements, and was not analyzed extensively. To determine if the rescuing effects observed for Y290H and P263L were due to the mutations in the sasA gene rather than others accumulated during mutagenesis, we reconstructed these mutants in a clean genetic background (Materials and Methods). Both reconstructed mutants displayed the same circadian phenotypes as each other and the original mutants, suggesting that they compensate for the loss of CikA (Fig. 1C). When sasA was completely deleted in the cikA-null background, we observed an arrhythmic phenotype, indicating that SasA inactivation alone is not sufficient to restore normal rhythms (Fig. 1C).
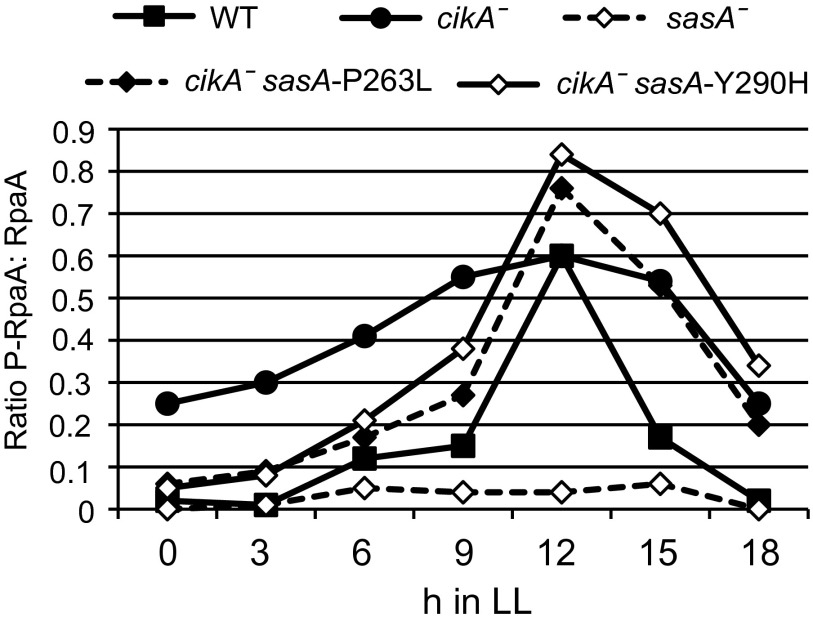
SasA and CikA have been shown to produce antagonistic effects on the phosphorylation state of the transcriptional regulator RpaA (8). We measured the ratio of phosphorylated RpaA in the cell by immunoblotting in the presence of Phos-tag reagent (Wako Chemicals USA) for each of the double mutants over 18 h (Fig. 2). As previously reported, the cikA-null strain showed a broad peak in RpaA phosphorylation with a high baseline (8). This pattern is in contrast to WT, which has a low baseline and a sharp peak at 12 h. The SasA Y290H and P263L mutations in a cikA-null background both resulted in a more normal RpaA phosphorylation cycle but had higher and broader peaks than WT, which may account for the wider peaks of luciferase expression in Fig. 1.
Fig. 2.
sasA alleles differentially affect the RpaA phosphorylation cycle. The level of phosphorylated and unphosphorylated RpaA in the original suppressor strains was measured by immunoblotting samples harvested every 3 h in constant light. Data from representative immunoblots are plotted as the ratio of phosphorylated RpaA (P-RpaA) to unphosphorylated RpaA.
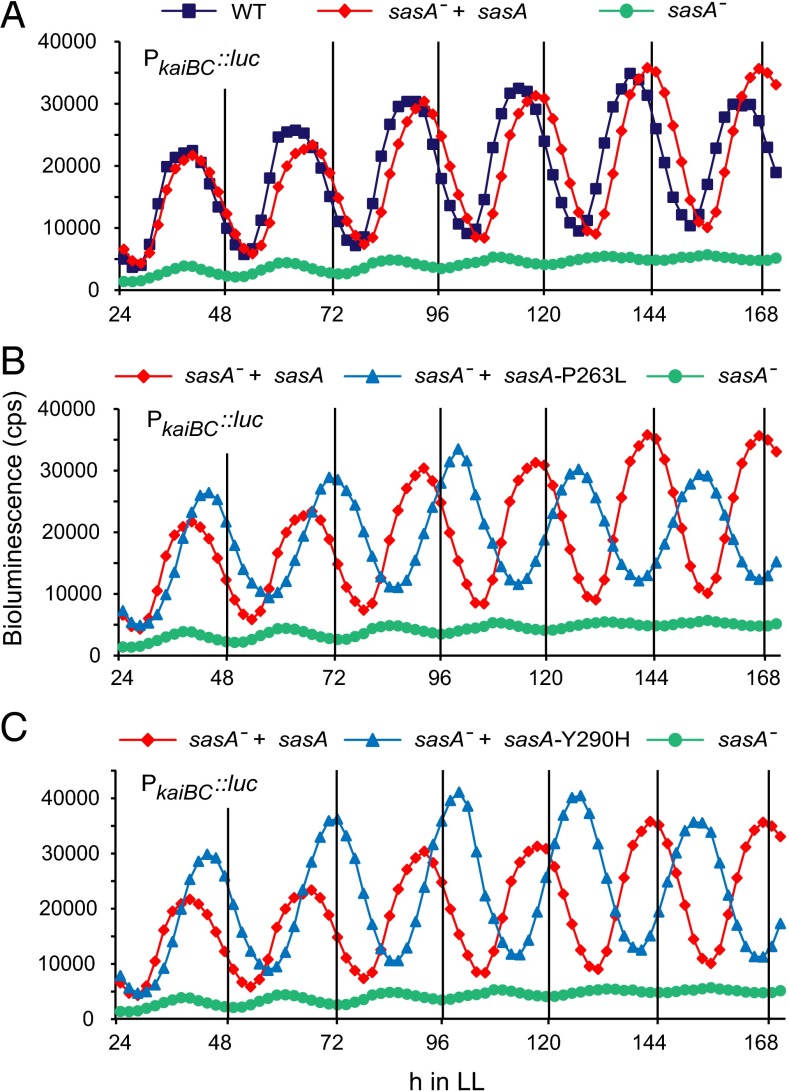
To better understand the nature of the genetic interaction between cikA and sasA, we measured the circadian rhythms of each of the single sasA mutants in an otherwise WT background (Fig. 3). When the WT sasA allele was inserted into neutral site 1 (NS1) in a sasA-null background, the strain showed a slight increase in period (20 min; Fig. 3A and Table 1). We used this reconstructed WT strain as the comparison for all other sasA alleles. The SasA Y290H and P263L mutants both had circadian periods that were 2 h longer than the reconstructed WT strain. This period increase may simply offset the 2-h decrease seen in the cikA-null mutant, yielding the WT phenotype observed in Fig. 1 and suggesting an additive genetic interaction between sasA and cikA.
Fig. 3.
SasA P263L and Y290H substitutions result in a 2-h period increase. Data were collected and plotted as in Fig. 1C. (A) Bioluminescence traces for the sasA− + sasAWT strain, where the sasA gene was disrupted at its endogenous locus and a WT allele was inserted into NS1. This strain is used as the WT reference in B and C. (B) Traces for the sasA-P263L mutant inserted in NS1 in a sasA-null background. (C) Same as in B but with sasA-Y290H. All strains contain the PkaiBC::luc reporter.
SasA Point Mutants Restore Phase-Resetting Phenotype in CikA KOs.
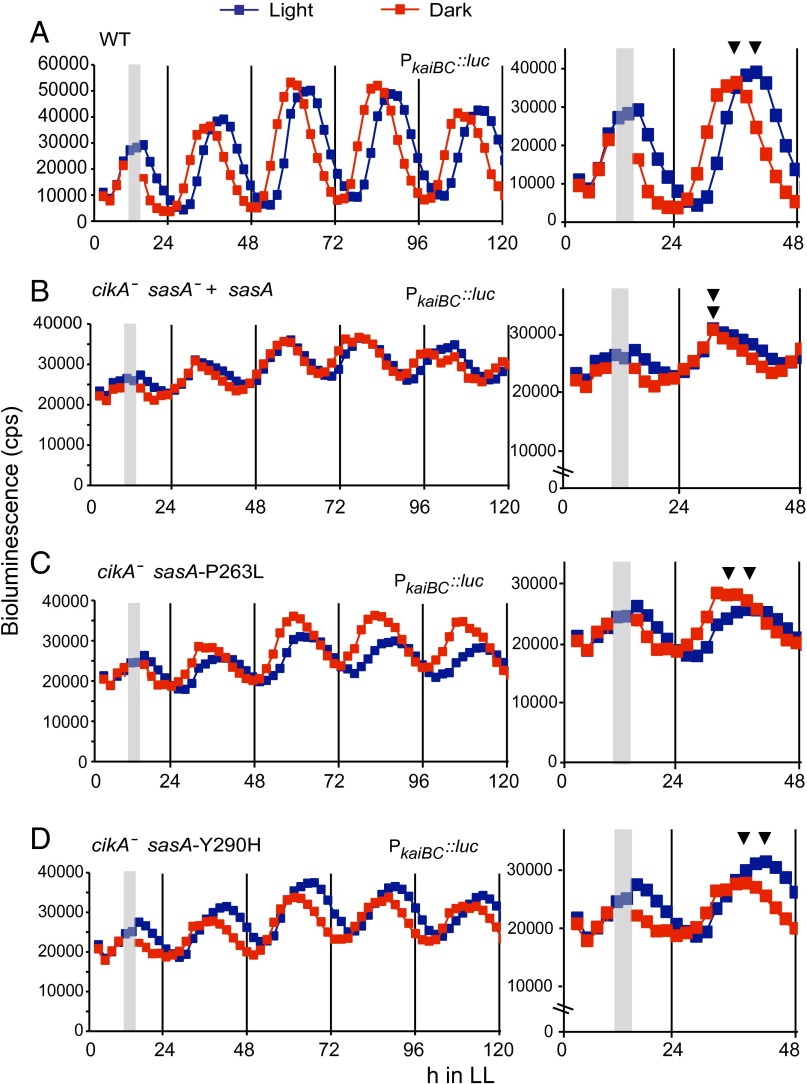
CikA was initially identified as part of the circadian input pathway because when knocked out, the cell does not reset the phase of rhythm after a period in the dark (6). We tested the ability of both of the reconstructed double mutants to reset circadian phase after a 5-h dark pulse that was delivered 8 h after entry into the light (Fig. 4). The reconstructed WT strain that carried a WT sasA allele in a cikA+ background showed an ∼4-h phase shift and exhibited no shift in a cikA-null background, as expected (Fig. 4 A and B). Both SasA Y290H and SasA P263L restored the ability to reset the phase of the rhythm in the absence of CikA (Fig. 4 C and D). WT phase resetting was also observed for the single sasA mutants in a cikA+ background as expected.
Fig. 4.
SasA P263L and Y290H substitutions restore phase resetting in a cikA-null background. Temporal recordings of bioluminescence were taken on cultures kept in constant light (blue squares) and given a 5-h dark pulse 8 h after first exposure to light (red squares). A gray bar signifies the duration of the dark pulse. The peak of the bioluminescence measurement immediately after the dark pulse is marked (▼) to show differences in phase. (A–D) Genotype of each strain is given above the respective trace. All strains contain the PkaiBC::luc reporter. Representative traces are shown.
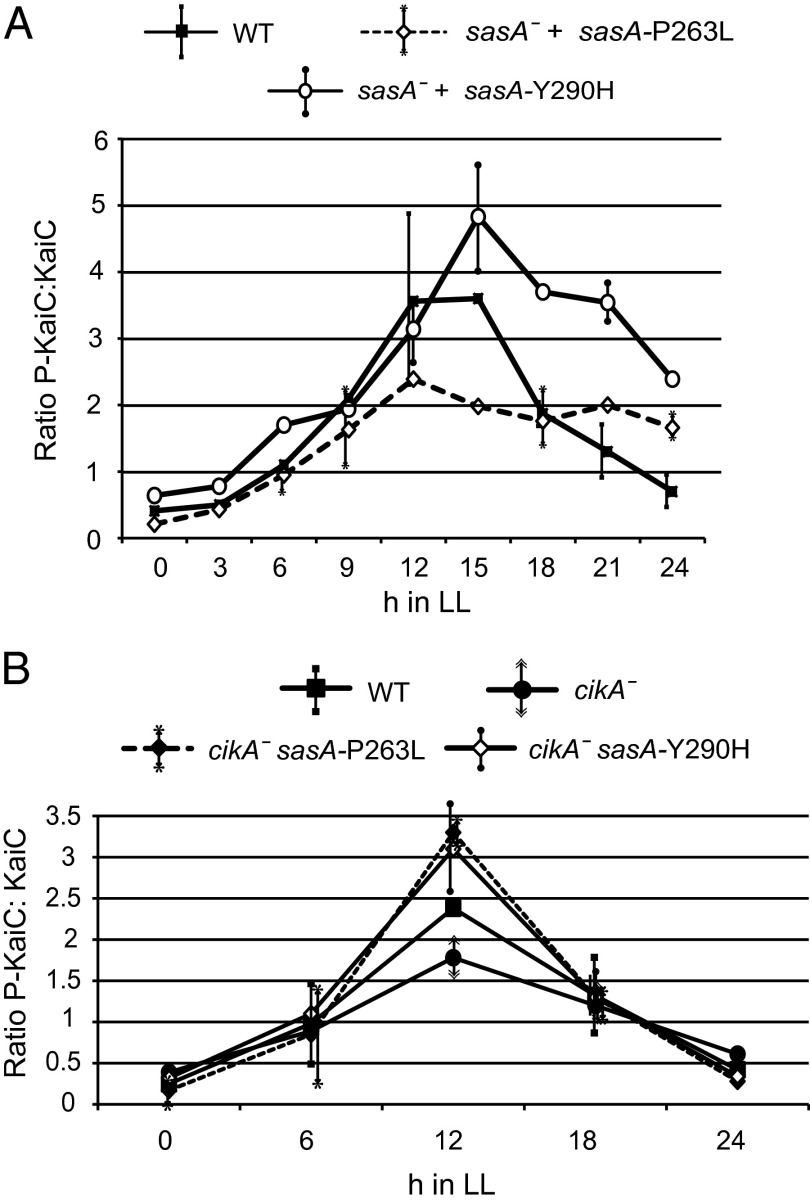
KaiC phosphorylation has been implicated in phase resetting (14), and SasA has been shown to interact directly with KaiC through a KaiB-like domain at the N terminus (15), but it has not previously been associated with clock entrainment. We examined the phosphorylation rhythms of KaiC in strains that carry each of the sasA alleles (Fig. 5A and Tables 2 and 3). Interestingly, the mutants showed different effects on KaiC phosphorylation even though they had the same phenotypes: The KaiC phosphorylation peak was higher with SasA Y290H and lower with SasA P263L than when SasA was WT. These differences in KaiC phosphorylation were not observed between SasA Y290H and SasA P263L in a cikA-null background, where both mutants had similar KaiC phosphorylation profiles that peaked higher than in cikA-null and WT (Fig. 5B). It is unclear why KaiC is hypophosphorylated in the SasA P263L mutant in a cikA+ background but hyperphosphorylated in a cikA-null background or why the P263L mutant retests in both situations. These data suggest that the phosphorylation state of KaiC is not solely responsible for restoration of resetting.
Fig. 5.
KaiC phosphorylation is elevated in sasA-Y290H. The ratio of phosphorylated to unphosphorylated KaiC was measured by immunoblotting every 3 h in constant light for each of the sasA mutants in a cikA+ background (A) and every 6 h in a cikA− background (B). Averages for each time point ± SD are shown.
Table 2.
KaiC phosphorylation cycle in WT background
| Average ratio P-KaiC/KaiC ± SD | |||
| Light, h | WTSasA | SasA P263L | SasA Y290H |
| 0 | 0.41 ± 0.08 | 0.21 ± 0.03 | 0.64 ± 0.17 |
| 3 | 0.50 ± 0.04 | 0.43 ± 0.14 | 0.78 ± 0.07 |
| 6 | 1.10 ± 0.54 | 0.95 ± 0.20 | 1.70 ± 0.01 |
| 9 | 2.10 ± 0.10 | 1.63 ± 0.60 | 1.94 ± 0.03 |
| 12 | 3.56 ± 1.33 | 2.39 ± 0.16 | 3.14 ± 0.48 |
| 18 | 1.87 ± 0.18 | 1.76 ± 0.43 | 3.70 ± 0.07 |
| 21 | 1.30 ± 0.42 | 2.00 ± 0.07 | 3.54 ± 0.30 |
| 24 | 0.70 ± 0.25 | 1.66 ± 0.19 | 2.39 ± 0.04 |
Table 3.
KaiC phosphorylation cycle in cikA-null background
| Average ratio P-KaiC/KaiC ± SD | ||||
| Light, h | WTSasA | cikA | cikA SasA P263L | cikA SasA Y290H |
| 0 | 0.25 ± 0.13 | 0.39 ± 0.05 | 0.17 ± 0.16 | 0.32 ± 0.04 |
| 6 | 0.98 ± 0.08 | 0.89 ± 0.07 | 0.85 ± 0.59 | 1.06 ± 0.08 |
| 12 | 2.40 ± 0.11 | 1.78 ± 0.23 | 3.30 ± 0.16 | 3.13 ± 0.54 |
| 18 | 1.30 ± 0.47 | 1.20 ± 0.21 | 1.30 ± 0.18 | 1.33 ± 0.23 |
| 24 | 0.42 ± 0.00 | 0.61 ± 0.04 | 0.28 ± 0.08 | 0.34 ± 0.03 |
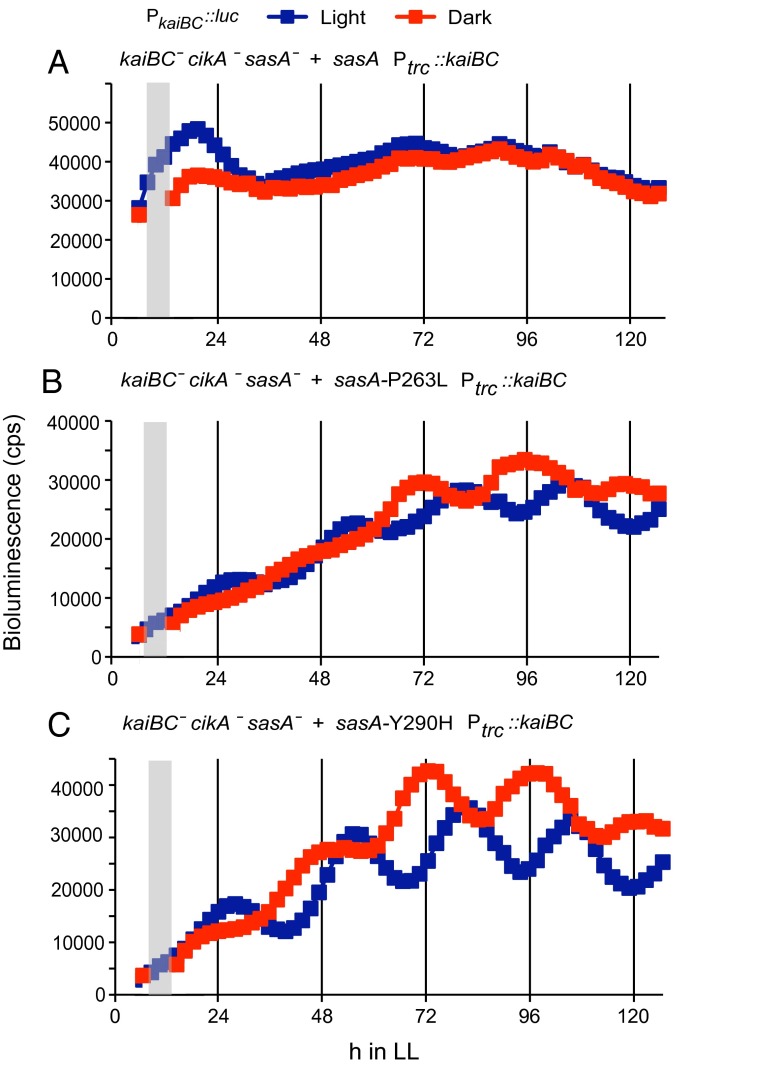
Because SasA has previously been attributed only to the output pathway, another possibility is that the restoration of phase resetting by SasA P263L and SasA Y290H could be caused by alteration of RpaA transcriptional feedback on the kaiBC operon (16). To test this hypothesis, we measured phase resetting of sasA alleles in a cikA-null background, where the kaiBC locus is driven exclusively by an inducible trc promoter that lacks an RpaA binding site and eliminates direct transcriptional feedback. These strains were able to reset phase after administration of a 5-h dark pulse (Fig. 6), suggesting that the point mutations in SasA are not affecting entrainment directly through RpaA-dependent regulation of kaiBC. It is possible that this phenotype can be explained by the altered regulation of some other target of RpaA. Because the RpaA phosphorylation cycle in the original double mutants is similar to that of WT (Fig. 2), we do not expect significant differences in the regulation of RpaA genomic targets.
Fig. 6.
Restoration of phase resetting by SasA alleles does not require RpaA-dependent transcriptional feedback. SasA variants expressed in a cikA-null background, in which the kaiBC operon was deleted from its native locus and expressed ectopically from the trc promoter, exhibit phase shifts in response to a dark pulse. (A) cikA-null; representative traces are shown. (B and C) SasA P263L and SasA Y290H; averages of three traces each are shown. Temporal recordings of bioluminescence were taken on cultures kept in constant light (blue squares) and given a 5-h dark pulse 6 h after first exposure to light (red squares). A gray bar signifies the duration of the dark pulse. All strains contain the PkaiBC::luc reporter.
Discussion
The impact of a cikA KO on circadian rhythms is one of the most extreme yet still rhythmic phenotypes observed, suggesting that CikA is an integral component of the circadian gene network in S. elongatus. It is surprising that the cell can compensate for its loss to both the input and output pathways with a single-nucleotide mutation in the sasA gene. Complex phenotypes have been shown to be fairly insensitive to random genomic manipulations in many systems (17, 18). This mutational robustness is often the result of congruently evolved pathways within the genetic network that buffer the effects of individual mutations (18). One possibility is that the sasA mutations are uncovering a cryptic mechanism with functional redundancy by relaxing its suppression. This scenario may be true for the input pathway through an increased contribution of KaiA to clock entrainment.
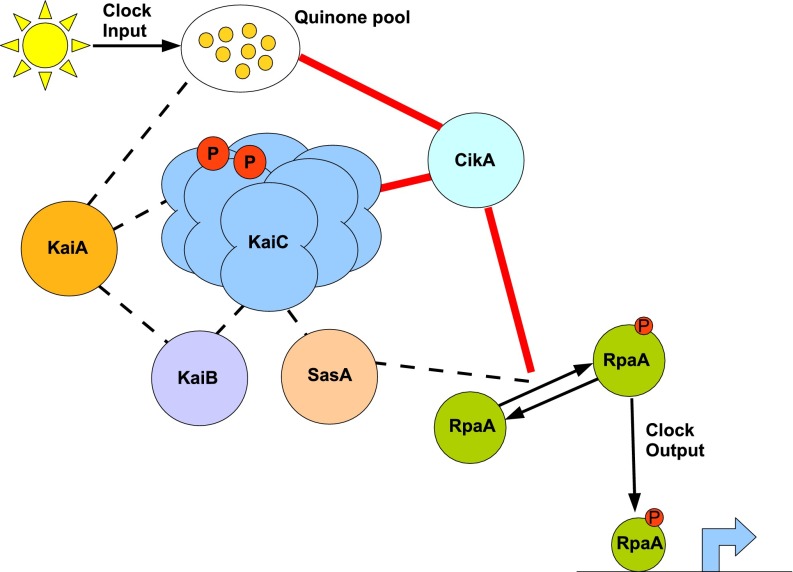
Like CikA, KaiA has been shown to bind oxidized quinone, a major redox-responsive cofactor of the photosynthetic apparatus, which can directly reset the phase of the KaiABC oscillator in vitro (19). Because SasA and KaiB compete for the same binding site on KaiC (20), these mutations in SasA may affect KaiB-KaiC binding kinetics, and subsequently alter the temporal association of KaiA with the oscillator. An increased interaction between KaiA and KaiC could account for the hyperphosphorylation of KaiC in the cikA-null SasA mutant strains (Fig. 5B). Because SasA modulates the phosphorylation state of the transcription factor RpaA, restoration of the phase-resetting phenotype could be explained by changes in the transcriptional feedback of RpaA on the kaiBC operon. The persistence of phase resetting when this feedback loop is broken (Fig. 6) suggests that the impact of the SasA mutants on clock input is not mediated directly by clock output through RpaA. That is not to say that these mutants do not affect clock output. We expect that the SasA mutations are restoring WT output properties (period and amplitude) by altering the autophosphorylation efficiency of SasA, which subsequently modulates the temporal phosphorylation of RpaA (Fig. 2) and the circadian regulation of gene expression. Regardless of the mechanism, these mutations clearly enable a simpler network architecture that preserves normal circadian properties in the absence of cikA (Fig. 7).
Fig. 7.
SasA substitutions may restore phenotype by altering the temporal association of multiple clock components in a highly connected network. A model of core components of the cyanobacterial circadian clock network is shown. Lines indicate known interactions between S. elongatus clock proteins. Red lines represent connections that are lost in a cikA KO, and dashed lines represent interactions that may be directly or indirectly altered by sasA mutants. P, phosphate group.
These results question the evolutionary advantage of adding cikA to the cyanobacterial clock. Relative to the other clock proteins, CikA is poorly conserved across, and absent from, many cyanobacteria (21). Admittedly, circadian rhythms have not been characterized in most of these species, although diel behaviors are widespread (22–25). A reduced network that lacks kaiA and cikA has been shown to produce strong diel rhythms in the cyanobacteria Prochlorococcus marinus MED4, suggesting that CikA is indeed not necessary for daily timing (21, 26). However, these rhythms are less robust than those rhythms driven by the S. elongatus clock, because they dampen quickly in constant light (27). Interestingly, the highly conserved Tyr at residue 290 in SasA is a Phe in Prochlorococcus (28), which may result in SasA functionally similar to the Y290H mutant identified in this study.
Phylogenetic data suggest that SasA evolved before CikA (21, 28). The conservation of the KaiB-like domain in SasA homologs suggests a conserved interaction with KaiC and the circadian gene network. The S. elongatus CikA is one of the most unusual cyanobacterial orthologs. It lacks a bilin-binding cysteine in its GAF domain and a phosphorylatable aspartate in its receiver domain, both of which are highly conserved among these respective domains (6), suggesting that the network can tolerate drastic functional mutations to this protein. The current state of the S. elongatus clock system may be an interesting evolutionary intermediate in the integration of a new gene into an existing genetic network. The ability of the cell to restore phenotype easily in a cikA KO implies that there is functional redundancy between genes. CikA may have been initially coopted by the network through a redundancy in function, and subsequently evolved to fill a specialized role. Over time, functional redundancies in other genes may decay because of a decrease in selective pressure, increasing the advantage of maintaining cikA. We expect that if CikA is the newest addition to the circadian gene network, it may be that its loss is the easiest for the network to adjust to, because evolved specialization is incomplete. If correct, this assumption would predict that a sasA deletion would be much more difficult to suppress by mutations in another gene.
There could be several advantages for acquiring CikA, including a stronger coupling between cellular redox state and the clock (7) or a tighter regulation of the RpaA phosphorylation state through its antagonistic relationship with SasA (8), but these explanations remain speculative without comparing circadian properties across a range of evolved network architectures. Regardless, we are intrigued by the malleability of this network and the ease with which a complex phenotype can be restored. Clock input and output have traditionally been treated as two linear and separable pathways in cyanobacteria. Given this construct, it is difficult to imagine that a single mutation in SasA could alter both of these properties simultaneously. These findings begin to dissolve the boundaries between the canonical input and output pathways, and suggest that input and output may be dependent to some degree upon the interaction kinetics of all clock proteins with KaiC and each other. The observation that SasA, a protein previously considered to be solely involved in clock output, can affect entrainment through an output-independent pathway is significant (Fig. 6) and suggests this type of highly connected, binding association-dependent network. Evolutionarily, this network architecture may be more resilient to genetic mutations, because the loss of one gene would not completely break a pathway but could be compensated for by a number of mutations that restore interaction kinetics. It is difficult to distinguish if robustness in this network is a byproduct of the absorption of CikA into the network as described above (congruent robustness) or if there has been selective pressure to maintain a highly integrated network with functional redundancy (adaptive robustness) (29). Additional second-site suppressor mutagenesis screens on the other clock genes may help us to resolve this distinction and to understand the full resiliency of the network.
Materials and Methods
Mutagenesis and Screening.
The cikA-null strain was derived from S. elongatus PCC 7942 strain AMC462 [PkaiBC::luxAB, PpsbAI::luxCDE (9)] by replacing cikA with a kanamycin resistance gene by homologous recombination with the plasmid AM2953. EMS mutagenesis of this strain was performed according to the method of Kondo et al. (30). After mutagenesis, cells were grown to stationary phase under low light, diluted 1:1,000, and plated on BG-11 agar plates + kanamycin. The luxAB and luxCDE operons in the parent strain allow the circadian output to be determined by measuring the circadian regulation of the luciferase gene (13). We used a computer-controlled turntable to rotate nine different Petri dishes iteratively into a dark chamber for 2 h, where the luminescence of individual colonies was detected by a Pixis 1024 CCD camera (Princeton Instruments) (10). This turntable is similar to the one described by Kondo and Ishiura (11). Colonies that showed a rhythmic but altered phenotype relative to the cikA-null strain were streaked in larger patches and retested to improve the quality of the circadian data by amplifying the signal. From the CCD images, we calculated the relative luciferase expression over time using image analysis tools in the R EBImage library (10, 31).
Genome Sequencing and Analysis.
Mutants selected for sequencing were grown in 10 mL of BG-11 media to stationary phase. A sample (3 mL) of culture was collected by centrifugation at 4,300 × g for 5 min, and resuspended in 0.1 mL of BG-11 in a siliconized tube. These samples were fragmented by sonication using a Bioruptor Standard (Diagenode) at high setting and for 10 cycles of 30 s on/off. RNaseA (5 μL) (Qiagen) was added and incubated at 37 °C for 1 h. To purify genomic DNA, an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1) (Invitrogen) was added to the sample. The sample was shaken three times for 15 s over 3 min and spun at 4 °C for 10 min, and the top aqueous phase was collected. To this fraction was added sodium acetate to 10% (wt/vol), 3 vol of ethanol, and 5 μL of 5 mg/mL glycogen. This mixture was frozen overnight at −80 °C to facilitate precipitation. Samples were collected at 4 °C, washed with ethanol, spun down again, allowed to air-dry, and rehydrated in 30 μL of water. The standard Illumina genomic library preparation was used to prepare the samples for sequencing (TruSeq; Illumina, Inc.). Samples were run on an Illumina HiSeq2000 DNA sequencer. Reads were mapped against the S. elongatus PCC 7942 genome (GenBank accession NC_007604), and polymorphisms were identified using the program breseq (32).
Assaying Reconstructed Mutants.
sasA alleles were amplified from EMS-mutagenized cikA-null strains by colony PCR and cloned into EcoRI-linearized plasmid AM2991 (NS1, spectinomycin/streptomycin resistance) by GeneArt seamless assembly (Life Technologies). These constructs were introduced into sasA-null S. elongatus PCC 7942 strains that contained either the PkaiBC::luc or PkaiBC::luxAB, PpsbAI::luxCDE reporter. Transformed strains were patched on solid BG-11 medium, transferred to liquid BG-11 supplemented with appropriate antibiotics, grown for 2 d in constant light at 30 °C, and transferred to 96-well plates as described previously (33). Plates were entrained with three 12-h light/12-h dark cycles at 30 °C and placed in a TopCount luminescence counter (PerkinElmer) for bioluminescence monitoring. To assay for phase-resetting ability, a duplicate plate of strains was transferred to the dark for 5 h at 8 h after entry into the light and then replaced after the dark pulse. Ptrc::kaiBC strains (RpaA feedback-independent) were grown in liquid BG-11 supplemented with appropriate antibiotics and 40 mM isopropyl β-d-1-thiogalactopyranoside. Strains were entrained and assayed as above, and phase-resetting ability was assessed by administering a 5-h dark pulse at 6 h after entry into constant light. Bioluminescence data from the TopCount were analyzed with the Biological Rhythms Analysis Software System (BRASS; millar.bio.ed.ac.uk/PEBrown/BRASS/BrassPage.htm). Strains and plasmids used in this study are listed in Tables S3 and S4, respectively. Artifactual spikes in bioluminescence were manually corrected in traces shown in Fig. 6.
For immunoblotting, SDS/PAGE gels were prepared with Phos-tag reagent (Wako Chemicals USA) and whole-cell protein extracts were prepared as described previously (12). RpaA and KaiC were detected using appropriate antisera as described by Boyd et al. (12) and Dong et al. (34). Densitometric analysis was performed using ImageJ software (National Institutes of Health).
Supplementary Material
Acknowledgments
We thank Mark Paddock and Kenyon Applebee for helpful comments, Sarah Munchel for preparing and sequencing genomic libraries, and Andrian Gutu and Erin O’Shea for providing plasmid HS 1. This work was supported by the W. M. Keck Foundation and by Army Research Office Grant W911NF-13-1-0097 (to R.J.G.), NIH Fellowship GM097977-01A1 (to R.K.S.), and NIH Grant R01GM062419 (to S.S.G.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419902111/-/DCSupplemental.
References
- 1.Forsburg SL. The art and design of genetic screens: Yeast. Nat Rev Genet. 2001;2(9):659–668. doi: 10.1038/35088500. [DOI] [PubMed] [Google Scholar]
- 2.Ishiura M, et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281(5382):1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 3.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci USA. 2002;99(24):15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Golden SS, Canales SR. Cyanobacterial circadian clocks—Timing is everything. Nat Rev Microbiol. 2003;1(3):191–199. doi: 10.1038/nrmicro774. [DOI] [PubMed] [Google Scholar]
- 5.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308(5720):414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 6.Schmitz O, Katayama M, Williams SB, Kondo T, Golden SS. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science. 2000;289(5480):765–768. doi: 10.1126/science.289.5480.765. [DOI] [PubMed] [Google Scholar]
- 7.Ivleva NB, Gao T, LiWang AC, Golden SS. Quinone sensing by the circadian input kinase of the cyanobacterial circadian clock. Proc Natl Acad Sci USA. 2006;103(46):17468–17473. doi: 10.1073/pnas.0606639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutu A, O’Shea EK. Two antagonistic clock-regulated histidine kinases time the activation of circadian gene expression. Mol Cell. 2013;50(2):288–294. doi: 10.1016/j.molcel.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katayama M, Tsinoremas NF, Kondo T, Golden SS. cpmA, a gene involved in an output pathway of the cyanobacterial circadian system. J Bacteriol. 1999;181(11):3516–3524. doi: 10.1128/jb.181.11.3516-3524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shultzaberger RK, Paddock ML, Katsuki T, Greenspan RJ, Golden SS. High throughput and quantitative approaches for measuring circadian rhythms in cyanobacteria using bioluminescence. Methods Enzymol. 2014 doi: 10.1016/bs.mie.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondo T, Ishiura M. Circadian rhythms of cyanobacteria: Monitoring the biological clocks of individual colonies by bioluminescence. J Bacteriol. 1994;176(7):1881–1885. doi: 10.1128/jb.176.7.1881-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyd JS, Bordowitz JR, Bree AC, Golden SS. An allele of the crm gene blocks cyanobacterial circadian rhythms. Proc Natl Acad Sci USA. 2013;110(34):13950–13955. doi: 10.1073/pnas.1312793110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo T, et al. Circadian rhythms in prokaryotes: Luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 1993;90(12):5672–5676. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiyohara YB, Katayama M, Kondo T. A novel mutation in kaiC affects resetting of the cyanobacterial circadian clock. J Bacteriol. 2005;187(8):2559–2564. doi: 10.1128/JB.187.8.2559-2564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwasaki H, et al. A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell. 2000;101(2):223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- 16.Markson JS, Piechura JR, Puszynska AM, O’Shea EK. Circadian control of global gene expression by the cyanobacterial master regulator RpaA. Cell. 2013;155(6):1396–1408. doi: 10.1016/j.cell.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dixon SJ, Costanzo M, Baryshnikova A, Andrews B, Boone C. Systematic mapping of genetic interaction networks. Annu Rev Genet. 2009;43:601–625. doi: 10.1146/annurev.genet.39.073003.114751. [DOI] [PubMed] [Google Scholar]
- 18.Harrison R, Papp B, Pál C, Oliver SG, Delneri D. Plasticity of genetic interactions in metabolic networks of yeast. Proc Natl Acad Sci USA. 2007;104(7):2307–2312. doi: 10.1073/pnas.0607153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim YI, Vinyard DJ, Ananyev GM, Dismukes GC, Golden SS. Oxidized quinones signal onset of darkness directly to the cyanobacterial circadian oscillator. Proc Natl Acad Sci USA. 2012;109(44):17765–17769. doi: 10.1073/pnas.1216401109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tseng R, et al. Cooperative KaiA-KaiB-KaiC interactions affect KaiB/SasA competition in the circadian clock of cyanobacteria. J Mol Biol. 2014;426(2):389–402. doi: 10.1016/j.jmb.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baca I, Sprockett D, Dvornyk V. Circadian input kinases and their homologs in cyanobacteria: Evolutionary constraints versus architectural diversification. J Mol Evol. 2010;70(5):453–465. doi: 10.1007/s00239-010-9344-0. [DOI] [PubMed] [Google Scholar]
- 22.Pokrovsky OS, Shirokova LS. Diurnal variations of dissolved and colloidal organic carbon and trace metals in a boreal lake during summer bloom. Water Res. 2013;47(2):922–932. doi: 10.1016/j.watres.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 23.Hamisi M, Díez B, Lyimo T, Ininbergs K, Bergman B. Epiphytic cyanobacteria of the seagrass Cymodocea rotundata: Diversity, diel nifH expression and nitrogenase activity. Environ Microbiol Rep. 2013;5(3):367–376. doi: 10.1111/1758-2229.12031. [DOI] [PubMed] [Google Scholar]
- 24.Ottesen EA, et al. Pattern and synchrony of gene expression among sympatric marine microbial populations. Proc Natl Acad Sci USA. 2013;110(6):E488–E497. doi: 10.1073/pnas.1222099110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi T, Ilikchyan I, Rabouille S, Zehr JP. Genome-wide analysis of diel gene expression in the unicellular N(2)-fixing cyanobacterium Crocosphaera watsonii WH 8501. ISME J. 2010;4(5):621–632. doi: 10.1038/ismej.2009.148. [DOI] [PubMed] [Google Scholar]
- 26.Axmann IM, et al. Biochemical evidence for a timing mechanism in Prochlorococcus. J Bacteriol. 2009;191(17):5342–5347. doi: 10.1128/JB.00419-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holtzendorff J, et al. Genome streamlining results in loss of robustness of the circadian clock in the marine cyanobacterium Prochlorococcus marinus PCC 9511. J Biol Rhythms. 2008;23(3):187–199. doi: 10.1177/0748730408316040. [DOI] [PubMed] [Google Scholar]
- 28.Dvornyk V, Deng HW, Nevo E. Structure and molecular phylogeny of sasA genes in cyanobacteria: Insights into evolution of the prokaryotic circadian system. Mol Biol Evol. 2004;21(8):1468–1476. doi: 10.1093/molbev/msh106. [DOI] [PubMed] [Google Scholar]
- 29.de Visser JA, et al. Perspective: Evolution and detection of genetic robustness. Evolution. 2003;57(9):1959–1972. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 30.Kondo T, et al. Circadian clock mutants of cyanobacteria. Science. 1994;266(5188):1233–1236. doi: 10.1126/science.7973706. [DOI] [PubMed] [Google Scholar]
- 31.Pau G, Fuchs F, Sklyar O, Boutros M, Huber W. EBImage—An R package for image processing with applications to cellular phenotypes. Bioinformatics. 2010;26(7):979–981. doi: 10.1093/bioinformatics/btq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461(7268):1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 33.Mackey SR, Ditty JL, Clerico EM, Golden SS. Circadian Rhythms. Humana; Totowa, NJ: 2007. pp. 115–129. [Google Scholar]
- 34.Dong G, et al. Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell. 2010;140(4):529–539. doi: 10.1016/j.cell.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.