Significance
In animals, gut microbes are essential for digestion. Here, we show that bacteria outside the gut can also play a critical role in digestion. In shipworms, wood-eating marine bivalves, endosymbiotic bacteria are found within specialized cells in the gills. We show that these endosymbionts produce wood-degrading enzymes that are selectively transported to the shipworm’s bacteria-free gut, where wood digestion occurs. Because only selected wood-degrading enzymes are transported, the shipworm system naturally identifies those endosymbiont enzymes most relevant to lignocellulose deconstruction without interference from other microbial proteins. Thus, this work expands the known biological repertoire of bacterial endosymbionts to include digestion of food and identifies previously undescribed enzymes and enzyme combinations of potential value to biomass-based industries, such as cellulosic biofuel production.
Keywords: Teredinidae, endosymbionts, symbiosis, xylotrophy, carbohydrate-active enzymes
Abstract
Bacteria play many important roles in animal digestive systems, including the provision of enzymes critical to digestion. Typically, complex communities of bacteria reside in the gut lumen in direct contact with the ingested materials they help to digest. Here, we demonstrate a previously undescribed digestive strategy in the wood-eating marine bivalve Bankia setacea, wherein digestive bacteria are housed in a location remote from the gut. These bivalves, commonly known as shipworms, lack a resident microbiota in the gut compartment where wood is digested but harbor endosymbiotic bacteria within specialized cells in their gills. We show that this comparatively simple bacterial community produces wood-degrading enzymes that are selectively translocated from gill to gut. These enzymes, which include just a small subset of the predicted wood-degrading enzymes encoded in the endosymbiont genomes, accumulate in the gut to the near exclusion of other endosymbiont-made proteins. This strategy of remote enzyme production provides the shipworm with a mechanism to capture liberated sugars from wood without competition from an endogenous gut microbiota. Because only those proteins required for wood digestion are translocated to the gut, this newly described system reveals which of many possible enzymes and enzyme combinations are minimally required for wood degradation. Thus, although it has historically had negative impacts on human welfare, the shipworm digestive process now has the potential to have a positive impact on industries that convert wood and other plant biomass to renewable fuels, fine chemicals, food, feeds, textiles, and paper products.
Shipworms are important pest organisms that burrow in wood (Fig. 1), causing extensive damage to wooden structures in marine and brackish waters, including ships, boats, piers, and fishing equipment. However, these worm-like marine mollusks also provide benefits by clearing wood debris from navigable waters and transforming this recalcitrant material into their own more readily digestible biomass (1, 2). At least one shipworm species (Lyrodus pedicellatus) has been shown to grow and reproduce normally using wood as its sole particulate food source (3). However, unlike their terrestrial herbivorous and xylophagous counterparts, whose digestive systems contain complex communities of microbes (4–9), Bankia setacea and several other shipworm species accumulate and digest wood in the cecum (Fig. 1 A and B), a region of the foregut that is devoid of any conspicuous microbial community (10).
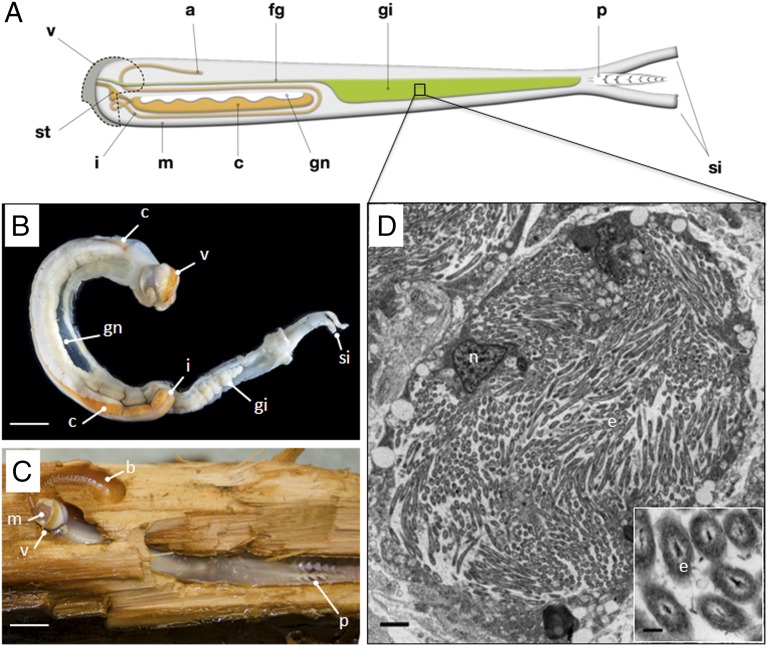
Fig. 1.
Anatomy of B. setacea. (A) Schematic diagram of B. setacea. (B) B. setacea removed from its burrow. (Scale bar: 1 cm.) (C) B. setacea exposed in wood burrow. The abrasive surfaces of the valves (shells) excavate wood particles that are ingested and transported to the cecum for digestion. a, anus; b, burrow; c, cecum; fg, food groove; gi, gill; gn, gonad; I, intestine; m, mouth; p, pallets; si, siphons; st, stomach; v, valve. (Scale bar: 1 cm.) B and C courtesy of Jeffrey Schilling, Oregon Health & Science University, Portland, OR. In B, the cecum (orange) is partially obscured by the gonad (white). (D) Transmission electron micrograph showing endosymbionts within a single bacteriocyte in the B. setacea gill. e, endosymbonts; n, nucleus. (Scale bar: 2 μm.) (Inset) Endosymbiont cells. (Scale bar: 250 nm.)
Although the cecum of B. setacea is depauperate of microorganisms, dense communities of endosymbiotic bacteria (Fig. 1D) are found in an internal region of the gill referred to as the gland of Deshayes (11–15). Culture-independent 16S rRNA gene analyses have shown that the gill endosymbiont community of L. pedicellatus is composed of several endosymbiont types that are closely related to the polysaccharide-degrading gammaproteobacterium Saccharophagus degradans (11, 12, 16, 17) (Fig. 2A and Fig. S1A). These endosymbiont types include Teredinibacter turnerae, a cellulolytic and nitrogen-fixing gammaproteobacterium that has been isolated in pure culture from the gills of many shipworm species from around the world (16, 18, 19). The metabolic capabilities displayed by T. turnerae when grown in vitro suggest two potential functions for the shipworm gill endosymbionts: (i) fixing nitrogen to supplement the host’s nitrogen-deficient diet of wood and (ii) producing hydrolytic enzymes that contribute to wood digestion (1, 18). Although the former function has been demonstrated experimentally (20), the latter has not until now.
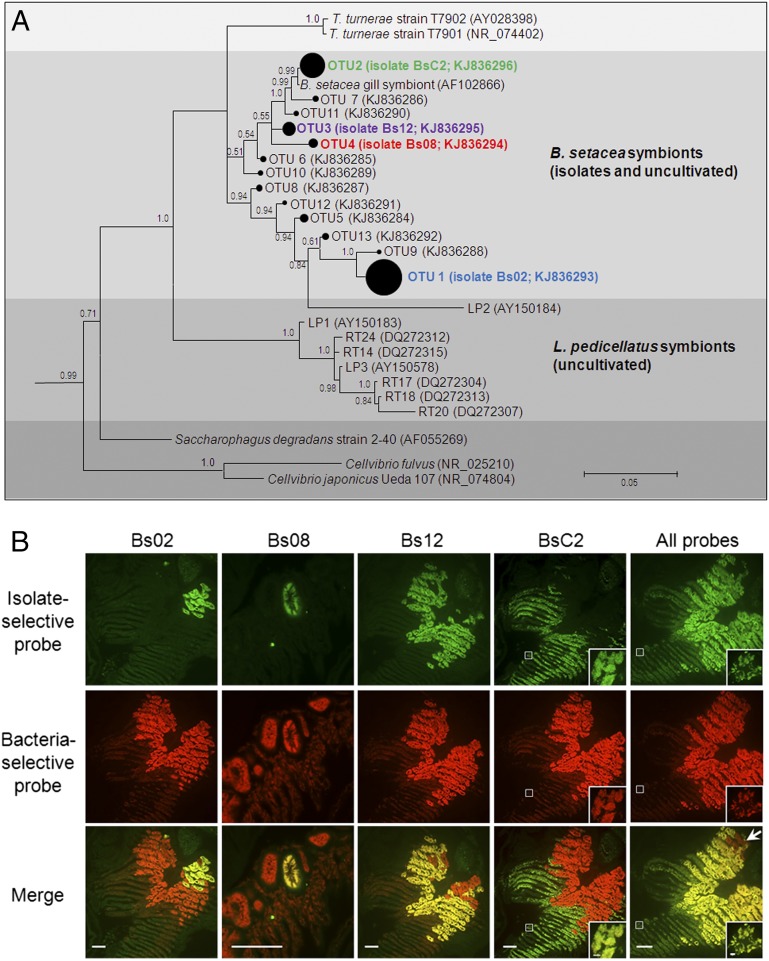
Fig. 2.
Gill endosymbiont community of B. setacea. (A) Bayesian phylogeny (16S rRNA) of cultivated and uncultivated gill endosymbionts of B. setacea (subtree from Fig. S1A). The area of closed circles is proportional to the fraction of the clone library represented by each OTU (Table S2). Posterior probabilities >0.5 are shown. (Scale bar: 0.05 substitutions per nucleotide position.) (B) FISH showing five gill tissue sections from B. setacea (columns), each dual-labeled with a bacteria-selective probe (EUB338; red) plus the indicated isolate-selective probe (green). (Scale bars: 100 μm.) Colocalization (yellow) of bacteria- and isolate-selective probes demonstrates that the four isolates account for most but not all (arrow) detectable gill endosymbionts. (Insets) Details of the boxed areas at a magnification and exposure appropriate to demonstrate the fainter but clearly detectable fluorescence in BsC2 containing bacteriocytes. (Scale bar: 5 μm.) Negative controls are shown in Fig. S1B.
Results
Phylogenetic Composition of the Gill Endosymbiont Community of B. setacea.
To begin our investigation of the role of gill endosymbionts in wood digestion, we cultivated four phylogenetically distinct bacterial strains (designated as Bs02, Bs08, Bs12, and BsC2) from the gill endosymbiont community of B. setacea, using methods similar to those methods used previously to cultivate T. turnerae (16, 18). Phylogenetic analysis of 16S rRNA gene sequences from these cellulolytic aerobes indicated that they are members of a well-supported clade that also includes T. turnerae and other, as yet uncultivated, shipworm endosymbionts (Fig. 2A and Fig. S1A). To estimate the frequency of these four isolates within the total gill endosymbiont community of B. setacea and to summarize broadly the phylogenetic composition of this community, we sequenced ∼26,000 short bacterial 16S rRNA gene fragments (∼200 bp) amplified from the gills of three specimens. Gammaproteobacteria accounted for >90% of these amplicons (Table S1). To provide greater phylogenetic resolution, we sequenced 445 near full-length amplicons (∼1,300 bp) from seven additional specimens. After removing likely chimeras, 80% of the remaining sequences fall within five operational taxonomic units (OTUs) when clustered at 99% sequence identity. The 16S rRNA gene sequences of isolates Bs02, Bs08, Bs12, and BsC2 fall within four of these five OTUs that encompass 46%, 6.8%, 3.1%, and 23.6%, respectively, of the examined clones (Table S2). All of these 16S rRNA gene sequences coalesce into a single OTU at 93% identity.
In Situ Localization of B. setacea Gill Isolates.
To detect these isolates in gill tissue, we performed FISH using four 16S rRNA-directed oligonucleotide probes, each designed to target one of the four isolates selectively. Each of the four probes hybridized with an apparently distinct subset of bacteriocytes within the gills (Fig. 2B and Fig. S1B). By comparing these hybridization patterns with those hybridization patterns observed in a probe that broadly targets the domain bacteria (21) [EUB338; Fig. 2B], we observed that, in combination, the four isolates colocalize with and account for most but not all (e.g., Fig. 2B, arrow) of the bacteria detected.
Sequence Comparisons Among Isolate Genomes and the Gill Endosymbiont Metagenome.
We next asked whether the genomes of the four isolates constitute a significant proportion of the total gill endosymbiont community metagenome. To answer this question, we sequenced (i) the metagenome of a sample of endosymbiont cells enriched by differential centrifugation from the gills of a single specimen of B. setacea and (ii) the genomes of each of the four isolates. The assembled gill endosymbiont metagenome included ∼26.5 Mbp in 38,060 scaffolds that ranged from 100 to 172,446 bp in length (Table S3). The four isolate genomes were similar in estimated size (3.8–5.4 Mbp) and coding sequence content (∼87%) (Table S4) to the estimated size and coding sequence content of T. turnerae (22) and related free-living cellulolytic gammaproteobacteria, such as S. degradans (17) and Cellvibrio japonicus (23). The nucleotide composition of the four isolate genomes, expressed as percent guanine plus cytosine content (%GC), ranged from 45.9–48 (Table S4), which is lower than that of T. turnerae (50.9%) (22) but consistent with the average %GC of the B. setacea gill endosymbiont metagenome (∼47.1%; Tables S3 and S4).
To assess the contribution of the four isolate genomes to the total gill endosymbiont community metagenome, we mapped individual reads from the metagenome sample to the four isolate genome assemblies. The results showed that a large majority (82.4%) of gill endosymbiont metagenome reads mapped to the isolate genomes (Bs02, 27.6%; Bs08; 0.29%; Bs12, 8.39%; and BsC2, 46.1%) (Table S5). By performing the inverse comparisons, we showed that 88.4%, 8%, 78.9%, and 63.2% of the sequence reads from isolates Bs02, Bs08, Bs12, and BsC2, respectively, mapped to the assembled gill endosymbiont metagenome (Table S6). In contrast, few reads from any B. setacea isolate mapped to any other B. setacea isolate genome assembly (1.16–0.4%; Table S6). Moreover, little matching sequence was observed (0.04–1.31%, at stringencies ranging from 95 to 75% identity) when metagenome sequences not assigned to the four isolates were queried against a phylogenetically broad sequence database. This database was constructed from 5,100 complete bacterial genome and plasmid sequences (ftp.ncbi.nlm.nih.gov/genomes/Bacteria/) and two eukaryotic genomes. The two eukaryotic genomes, Aplysia californica (Mollusca) and Populus trichocarpa (Viridiplantae), were chosen to aid in detection of sequences contributed by host tissue and woody food materials. At this same range of cutoff values (95–75%), the eukaryotic sequences account for a maximum of 0.007–0.02% of the total assembled metagenome. Combined with the results of the 16S rRNA sequence and hybridization analyses described above, these data demonstrate that the isolates are bona fide gill endosymbionts and that both the isolate genome sequences and the metagenome sequence are representative of the gill endosymbiont community of B. setacea.
Carbohydrate-Active Enzymes in the Isolate Genomes and Gill Endosymbiont Metagenome.
Given these conclusions, we asked whether the gene content of the endosymbiont community of B. setacea is consistent with its proposed role in degrading the plant cell wall polysaccharides (PCWPs) that are the dominant components of wood. Using the sequence-based Carbohydrate-Active Enzyme (CAZy) Database (24) (www.cazy.org) as a reference, we searched the isolate genomes and gill endosymbiont metagenome for carbohydrate-active catalytic modules and carbohydrate-binding modules (CBMs). Modules are a class of protein domains defined by their ability to function independent of the remaining protein structure. Because carbohydrate-active enzymes (CAZymes) are characteristically modular in structure, and each CAZyme can contain several modules with distinct functions, we refer to modules, rather than the proteins in which they are contained, in the following discussion. CAZy modules typically retain their functions when expressed independent of the remaining protein regions.
In each of the four isolate genomes, we identified between 83 and 128 predicted glycoside hydrolase (GH), 7–21 predicted carbohydrate esterase (CE), and 1–40 predicted polysaccharide lyase (PL) modules, as well as 89–137 predicted CBMs. The B. setacea gill endosymbiont metagenome contains 401 CBMs and 734 GH, 116 CE, and 104 PL modules (Tables S7 and S8). We note that the composition of these isolate genomes is comparable to that of other known PCWP-degradation specialists, such as S. degradans (17) and C. japonicus (23), with respect to number and diversity of catalytic modules and CBMs, predicted to bind or modify PCWP (22).
Composition of the Gill and Cecum Proteomes.
Although the gill endosymbiont community of B. setacea is rich in genes encoding PCWP-active CAZy modules, a key question is whether these genes are expressed and functional in the intact symbiosis. To answer this question, we used genome-enabled proteomic methods to detect and identify endosymbiont-encoded proteins in extracts of gill tissue and cecum contents (Dataset S1). As expected for a tissue containing bacterial cells, we identified many (102) functionally diverse endosymbiont-encoded proteins in the gill. These endosymbiont-encoded proteins include representatives of 12 of the 19 functional categories defined by the Clusters of Orthologous Groups (COG) database (25) (www.ncbi.nlm.nih.gov/COG/) (Fig. 3 and Table S9). Among these proteins, ∼11% (11 of 102) are predicted to have catalytic and/or binding activities toward cellulose or hemicellulose.
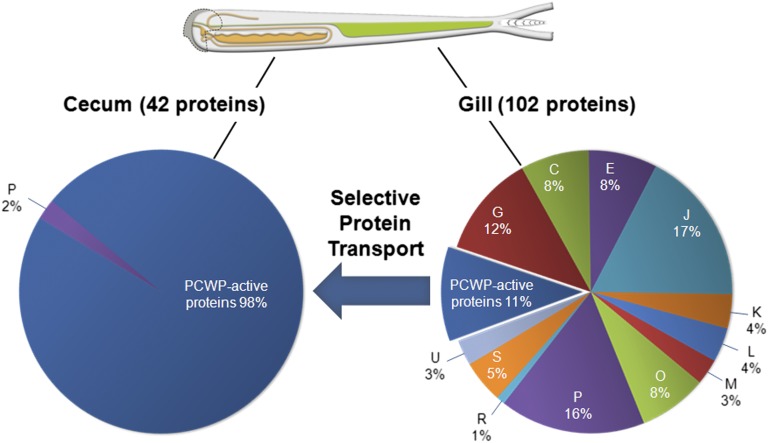
Fig. 3.
Endosymbiont-encoded proteins in gill and cecum content of B. setacea. PCWP-active proteins comprise 98% of the cecum proteome, whereas the gill proteome is functionally diverse, suggesting selective transport of proteins from gill to gut (arrow). All endosymbiont-encoded PCWP-active proteins detected in the gill proteome were also detected in the cecum proteome. COG categories are as follows: C, energy production and conversion; E, amino acid metabolism and transport; G, carbohydrate metabolism and transport; I, lipid metabolism; J, translation, ribosome structure, and biogenesis; K, transcription; L, replication, recombination, and repair; M, cell wall structure, biogenesis, and outer membrane; O, molecular chaperones and related functions; P, inorganic ion transport and metabolism; Q, secondary structure; R, general functional prediction only; U, intracellular trafficking, secretion, and vesicular transport; S, no functional prediction.
Remarkably, although the endosymbionts of B. setacea are located in its gills, numerous endosymbiont-encoded proteins were also detected in cecum, which lacks endosymbionts. Moreover, in contrast to the functionally diverse endosymbiont proteome found in the gills, nearly all [41 of 42 (∼98%)] of the endosymbiont-encoded proteins detected in the cecum are predicted to have catalytic and/or binding activity against PCWP components of wood (Fig. 3 and Table S10). These data indicate that PCWP-active proteins are selectively transported from their site of synthesis in the gills to their site of action in the cecum.
The proteins detected in the cecum contain a broad array of PCWP-active catalytic modules representing GH families 5, 6, 9, 10, 11, 45 and 53 and CE families 1, 3, 4, 6 and 15, as well as the newly described lytic oxidative cellulase (polysaccharide monooxygenase) auxiliary activity family AA10 (26). All of the PCWP-active proteins that were detected in the gills were also detected in the cecum. We note that a small fraction [six of 41 (∼15%)] of the endosymbiont-encoded proteins that were detected in the cecum contain putative catalytic modules of unknown function that are associated with CBMs predicted to bind to cellulose or xylan, suggesting novel activities against wood components.
Abundance and Catalytic Activities of Endosymbiont-Encoded Proteins in the Cecum Contents.
To evaluate the abundance of endosymbiont-encoded proteins in the cecum, we partially purified a protein fraction (fraction 11) that contained >30% of the total protein recovered from cecum contents (Fig. 4A and Fig. S2 A and B). In this fraction, we identified two dominant proteins by N-terminal amino acid sequencing and showed that these sequences matched previously identified endosymbiont-encoded proteins. These two proteins contain a GH family 5, subfamily 53 (GH5_53), and a GH family 6 module (Fig. S2C), respectively. The former is predicted to be an endo-1,4-beta-glucanase [Enzyme Commission (EC) 3.2.1.4] or cellodextrinase (EC 3.2.1.74) and is encoded in the genome of Bs02. The latter, a predicted endo-1,4-beta-glucanase (EC 3.2.1.4) or cellobiohydrolase (EC 3.2.1.91), is encoded in the Bs12 genome. The abundance of these proteins in the cecum suggests that they play important roles in wood digestion.
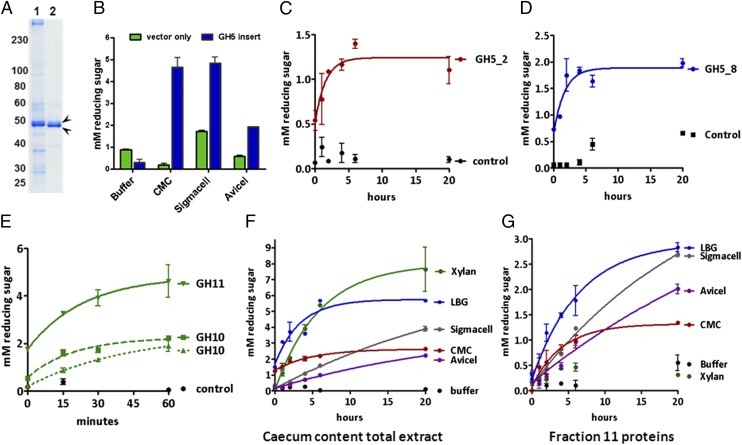
Fig. 4.
Activities of endosymbiont-encoded proteins. (A) Polyacrylamide gel showing lane 1 (extract of total cecum content) and lane 2 (fraction 11) (Fig. S2 A and B). (B) Recombinant GH5_53 module from fraction 11 hydrolyzes CMC, Sigmacell, and Avicel. Synthetic GH5_2 (KJ861970), GH5_8 (KJ861968), GH10 (KJ861985 and KJ861963), and GH11 (KJ861993) modules from cecum contents hydrolyze their predicted substrates [C, CMC (red); D, LBG (blue); and E, xylan (green)]. (F) Cecum content extract hydrolyzes xylan, CMC, Avicel (purple), Sigmacell (gray), and LBG. (G) Fraction 11 hydrolyzes Sigmacell, CMC, and LBG, but not xylan. (Error bars represent the SEM.)
We confirmed these and other predicted catalytic activities by expressing the identified catalytic modules exogenously and then testing the resulting proteins for hydrolytic activity against appropriate substrates. In these experiments, only the catalytic modules were cloned and expressed. CBMs and linker regions were omitted.
The abundant GH5_53 catalytic module identified in fraction 11, when expressed in Escherichia coli, exhibited hydrolytic activity against carboxymethylcellulose (CMC) and microcrystalline cellulose [Sigmacell (Sigma-Aldrich) and, to a lesser extent, Avicel (FMC Biopolymer)] (Fig. 4B), consistent with its predicted function as a β-1,4-endoglucanase. Catalytic modules of five additional endosymbiont-encoded proteins, when expressed by in vitro transcription/translation, also exhibited activities predicted by sequence homology. These endosymbiont-encoded proteins included one GH family 5, subfamily 2 module with activity against CMC (Fig. 4C); a GH family 5, subfamily 8 module with activity against galactomannan [locust bean gum (LBG)] (Fig. 4D); and a GH family 11 and two GH family 10 modules with activity against xylan (Fig. 4E).
Finally, we confirmed the presence of these specific catalytic activities in the cecum contents of B. setacea by measuring the hydrolytic activity of the crude cecum extract and protein fraction 11 against a variety of PCWP substrates. Crude cecum extract displayed the predicted hydrolytic activities against Sigmacell, Avicel, CMC, xylan, and LBG (Fig. 4F). Protein fraction 11 exhibited hydrolytic activity against cellulose and LBG but not xylan (Fig. 4G).
Discussion
Here, we detect endosymbiont-encoded proteins in the endosymbiont-free cecum of B. setacea and show that nearly all of these proteins have demonstrated and/or predicted activity against PCWP components of wood. Moreover, we show that these proteins are abundant in the cecum, suggesting that they play an important role in wood digestion. In contrast, we detect a functionally diverse endosymbiont proteome in the gills of B. setacea, where the endosymbionts are found, strongly suggesting that PCWP-active proteins are selectively transported from their point of synthesis by endosymbionts in the gill to their site of action in the cecum.
Although the mechanism of this selective transport is beyond the scope of this investigation, we note that the genes corresponding to all endosymbiont-encoded proteins detected in the cecum contain putative secretion signal sequences that direct their secretion into the periplasmic space. The isolate genomes include genes of the general secretion (Sec) and twin-arginine translocation (Tat) pathways, as well as type II and type VI secretion systems that can explain the transport of these proteins across the bacterial plasma membrane and outer membrane (27).
The system reported here constitutes a mode of symbiont-mediated digestion not previously described in animals. Although we are aware of no precedent in other symbioses for selective export of endosymbiont proteins to the external environment of the host, we note that endosymbiont-derived proteins have been found in the external secretions [saliva (28) and honeydew (29)] of the pea aphid Acyrthosiphum pisum. One of these proteins, GroEL, has been shown to influence the aphid’s interaction with its plant host (28). These recent observations, in combination with our own, suggest that the repertoire of endosymbiont-derived proteins may be broader than previously anticipated and could include such functions as chemical and antimicrobial defense, biological signaling, and other modifications of the host environment.
This newly described digestive strategy in B. setacea raises compelling questions. For example, how do endosymbiont-made proteins escape from the bacteriocytes that contain them and complete their journey from gill to gut? Enzyme transport between these two tissues might be explained by the presence of a conduit, such as the duct of Deshayes, a vessel with no known function that is purported to extend from the gill to the esophagus of shipworms along the inner surface of the afferent branchial vein (30, 31). However, the mechanisms that might explain the export of endosymbiont-made proteins from their intracellular location within bacteriocytes to the external environment of the host remain an open question. Such mechanisms might involve as yet undetected pores that connect the symbiont-containing vesicles within bacteriocytes to the bacteriocyte plasma membrane. Alternatively, endosymbiont proteins might be selectively transported across host cell membranes using host- or symbiont-derived secretion systems. These alternatives are the subject of ongoing investigations.
It also remains to be determined whether the remote placement of digestive bacteria in shipworms confers advantages over maintenance of an endogenous gut microbiota. We suggest that this placement eliminates competition between the host and symbionts for soluble products of digestion in the gut. Due to their intracellular location, the endosymbionts must still obtain nutrients from the host, and so compete with the host for resources. However, by virtue of their aerobic metabolism, the shipworm gill bacteria may consume less carbohydrate per unit of digestive enzymes produced than would typical gut anaerobes (32). Furthermore, the placement of these bacteria away from the gut content may allow the host greater control over the products of wood digestion, as well as their downstream transformations and fluxes.
This unique digestive strategy in shipworms may also have advantages with respect to discovery of industrial enzymes. For example, the identification of robust and efficient new enzymes for deconstruction of plant cell wall biopolymers is now considered to be among the most promising areas for innovation in the production of cellulosic biofuel (33). Because it requires few enzymes, and these enzymes are neatly translocated to the gut, the shipworm provides a simple model system in which to explore minimum enzymatic requirements and efficient enzyme mixtures for lignocellulose decomposition. This stands in contrast to cellulolytic systems in other xylotrophs and herbivores, where vast numbers of microbes and enzymes coexist in the gut (4–9), confounding the discovery of synergistic enzyme combinations that may be most useful to industry.
Methods
Specimens.
B. setacea were obtained from Puget Sound (latitude: 47.85072°, longitude: −122.33843°). Animals were maintained at 12–14 °C in seawater aquaria.
Isolation and Propagation of B. setacea Gill Endosymbionts.
Shipworms were extracted from wood and dissected to isolate gill and cecum tissue. Tissues were homogenized in ice-cold sterile seawater buffered with 50 mM Hepes (pH 8.0). Homogenates were streaked onto culture plates containing 1.0% agar in shipworm bacteria medium (SBM) (18) at pH 7 or pH 8.0 with 0.2% Sigmacell cellulose (wt/vol) added as a carbon source. Plates were incubated at 18 °C or 30 °C until colonies were observed. Selected colonies were subjected to two rounds of isolation by streaking for single colonies on plates. Isolates were propagated in liquid SBM with either 0.2% Sigmacell or 0.5% sucrose (wt/vol) as a carbon source.
16S rRNA Gene Sequencing and Analyses of B. setacea Gill Endosymbionts.
DNA was isolated from bacterial cultures using the DNeasy Blood and Tissue Kit (Qiagen) following the manufacturer’s directions. The 16S rRNA gene was amplified using 27 forward and 1,492 reverse primers, and sequenced with these primers plus four internal primers (Table S11) as described by Altamia et al. (19).
Pyrosequence Analysis of Short 16S rRNA Gene Tags (Pyrotag Analysis).
DNA was isolated from gill tissue using the Wizard gDNA Purification Kit (Promega) following the manufacturer’s protocol for Gram-negative bacteria, with Proteinase K (New England Biolabs) added to the extraction buffer (final concentration of 2 mg/mL). To generate microbial community profiles, the V6–V8 region of the 16S rRNA locus was PCR-amplified using the 454 adaptor-added 16S primer set, 926-F and 1392-R (Table S11). This pyrotag primer set amplifies 16S rRNA sequences from bacteria and Archaea, as well as 18S rRNA sequences from Eucarya. PCR amplicons were sequenced by the Department of Energy Joint Genome Institute (DOE-JGI), using the Roche Diagnostics 454 GS Titanium technology as previously described (34). Sequences were analyzed through the Pyrotagger computational pipeline (pyrotagger.jgi-psf.org) for quality trimming, clustering into OTUs based on 97% sequence identity, and taxonomically assigned by BLASTn against the Greengenes database (35, 36). Eukaryote OTUs were excluded from subsequent analysis.
16S rRNA Clone Library Sequencing and Analysis.
DNA was extracted from the gills of each of seven specimens of B. setacea, and PCR amplification of near full-length 16S rRNA genes was performed using Easy-A High-Fidelity PCR Master Mix (Agilent Technologies). For bacteria, 16S rRNA amplification primers 27-F′ and 1391-R were used, whereas 4a-F and 1391-R were used for Archaea (37) (Table S11). Three replicate PCR reactions were performed, PCR products were pooled and ligated into the pCR4-TOPO vector (TOPO TA Cloning Kit; Life Technologies), and the plasmids were transformed into One Shot TOP10 Electrocomp (Life Technologies) E. coli cells. Approximately 384 clones per library were picked and grown in selective media for sequencing using a BigDye Sequencing Kit (Applied Biosystems) according to the manufacturer’s instructions. The reactions were purified using Solid Phase Reversible Immobilization magnetic beads (Agencourt Biosciences Corp.) and analyzed on an ABI PRISM 3730 (Applied Biosystems) capillary DNA sequencer. The bidirectional 16S rRNA gene sequence reads were end-paired and trimmed for PCR primer sequence and quality. Chimeric sequences were removed using usearch61 (38, 39) for de novo filtering and ChimeraSlayer for reference sequence-guided filtering (40). The remaining nonchimeric sequences and reference 16S rRNA sequences (Bs02, Bs08, Bs12, and BsC2) were clustered de novo into OTUs at 90–99% similarity (in 1% increments) using UCLUST (38) as part of the QIIME 1.8.0 pipeline (41). All OTUs were annotated against Greengenes taxonomy (35, 36) using the the Ribosomal Database Project (RDP) Classifier 2.2 (42). Sequences were aligned using ClustalW2 (43) and phylogenetic trees were constructed using RAxML (44).
Phylogenetic Analysis of B. setacea Isolates and Uncultivated Endosymbionts.
The 16S rDNA sequences of B. setacea isolates, and cloned 16S sequences as described above, were aligned with sequences from selected reference Gammaproteobacteria using the Geneious Aligner implemented in Geneious (v.6.1.6; Biomatters; www.geneious.com). The alignment was masked to omit regions where sequence length variation prevented unambiguous alignment, and all sequences were trimmed to equal lengths. The Akaike information criterion in jModelTest (45) was used to identify the best-fit model of evolution for the final 1,233-bp alignment (E. coli positions 111–1,353). The phylogenetic positions of the B. setacea isolates were determined by Bayesian inference analysis using MrBayes (46) implemented in Geneious. Five million Markov chain Monte Carlo iterations were used, utilizing the best-fit nucleotide substitution model GTR + I + Γ with subsampling every 2,000 generations, discarding the first 20% of the samples as burn-in. Thiomicrospira crunogena was specified as the outgroup.
FISH.
Oligodeoxynucleotide probes were designed to target the 16S rRNA of each of the four symbiont isolates (Table S11). The Probe Match search tool (rdp.cme.msu.edu/probematch/search.jsp) in the RDP was used to evaluate the selectivity of the symbiont-targeted probes. The probe target sequences in Bs08, Bs12, and Bs02 genomes were unique. The probe target for BsC2 matched several unidentified environmental 16S rRNA sequence records. Negative control probes were designed to contain two mismatches to the target (Table S11, bold). The symbiont-specific positive and negative control probes were labeled at the 5′ end with Alexa Fluor 488 (Integrated DNA Technologies, Inc.). A probe broadly targeting the domain bacteria (21) (EUB338; Table S11) was labeled at the 5′ end with Alexa Fluor 594. Tissues were fixed in 4% (wt/vol) paraformaldehyde in seawater, embedded in paraffin, and sectioned as previously described (12). Optimal hybridization conditions were predicted using the mathFISH program (mathfish.cee.wisc.edu/) (47). Probes were diluted in hybridization buffer [30% formamide (vol/vol), 1 M NaCl, 20 mM Tris (pH 7.4), 0.01% SDS] to a final concentration of 500 nM each and incubated with tissue sections on slides for 2 h at 35 °C. Hybridization was followed by a single wash in 20 mM Tris (pH 7.4), 112 mM NaCl, 10 mM EDTA, and 0.01% SDS for 30 min at 35 °C. Slides were rinsed in water, mounted in Vectashield (Vector Laboratories), and examined by fluorescence microscopy. Digital images were collected, and deconvolution and colocalization of the fluorescent labels were performed using Volocity software (PerkinElmer).
Preparation of the Enriched Gill Endosymbiont Community Metagenomic DNA Sample.
Gill tissue from a single specimen of B. setacea was rinsed and homogenized in sterile SBM. Host cells and tissue were sedimented by two low-speed spins at 284 × g and 640 × g, each for 5 min at 4 °C. Bacteria were then sedimented at 4,790 × g for 5 min at 4 °C. The degree of bacterial enrichment was evaluated by epifluorescence microscopy using a DNA-specific stain (DAPI). DNA was isolated from the final pellet using the Wizard gDNA Purification Kit.
Sequencing, Assembly, and Aannotation.
Metagenomic DNA extracted from the gill endosymbiont community sample was sequenced at the DOE-JGI using a combination of Illumina GAIIx (Illumina, Inc.) and 454 GS Titanium technology technologies. Two Illumina libraries and three 454 libraries (∼72 million reads combined) were generated and used to assemble the metagenome. The %GC was determined for each set of metagenome reads using custom scripts (jgi_seq_stats.pl) and averaged to calculate the overall GC content. Illumina reads were assembled using Velvet (48), with a range of substring lengths (Kmers) from 21 to 31 by steps of 2. The best assembly of these six reads was selected by manual inspection. Contigs larger than 2 Kbp in length were decomposed into 1.8-Kbp shreds with 500-bp overlap with leading and following shreds. Shredded Velvet contigs were combined with 454 and Sanger reads and assembled using Newbler (Roche Diagnostics), utilizing a minimum overlap length of 60 and a minimum identity of 98%. Standard Illumina shotgun libraries were constructed for Bs02, Bs08, and Bs12 genomes (Illumina TruSeq) and sequenced to ∼250-, 300-, and 900-fold genome coverage, respectively, using the Illumina GAIIx platform (49). Illumina mate-paired reads from an ∼300-bp insert library were generated and sequenced at 2 × 76 bp for Bs02 and Bs08, whereas 2 × 100-bp reads were produced for Bs12. Illumina sequencing data were assembled with Velvet version 1.0.19 (48) to obtain draft quality genomes and were annotated by the DOE-JGI (50).
The genome of BsC2 was sequenced at New England Biolabs on a PacBio RSII instrument (Pacific Biosciences) using single-molecule real-time sequencing (SMRT) sequencing methodology (51, 52). SMRTbell template libraries were prepared as previously described (53, 54). SMRT sequencing was carried out on the PacBio RSII instrument using standard protocols for large insert SMRTbell libraries. Sequencing reads were processed, mapped, and assembled via the Pacific Biosciences SMRT Analysis pipeline (www.pacbiodevnet.com/SMRT-Analysis/Software/SMRT-Pipe) using the Hierarchical Genome Assembly Process protocol (55). All genome sequences were submitted to the DOE-JGI for annotation (50).
Comparing the Gill Endosymbiont Community Metagenome and the Genomes of the Endosymbiont Isolates.
To evaluate the level of agreement between the metagenome and individual isolate genomes, we mapped shotgun metagenome reads to each isolate genome. We used a reference index constructed from the combined set of all four isolate genome scaffolds using bowtie2-build (default parameters). Metagenome reads from the 454 platform in standard flowgram format (sff) file format were converted to FASTQ format (Sanger-format quality scores) using the utility sff2fastq (github.com/indraniel/sff2fastq). Metagenome reads from each library were mapped separately to the combined isolate reference index with Bowtie 2 (56) using the “very-sensitive” preset with other parameters set to default (using unpaired and paired for 454 and Illumina input FASTQ files, respectively). The parameters specified by the very-sensitive preset are: −D (consecutive seed extension attempts) 20, −R (maximum number of times Bowtie reseeds reads with repetitive seeds) 3, −N (mismatches in the seed alignment) 0, −L (length of seed alignment) 20, −i (function defining the interval between seeds) S, 1.0, 0.50 [f(x) = 1 + 0.5 * √(x), where x is the read length]. The percentage of reads mapping to each isolate genome is the number of reads from all five libraries mapping to the scaffolds belonging to each isolate genome divided by the total number of reads for the five libraries.
To calculate the percentage of each isolate genome to which shotgun metagenome reads mapped, we concatenated and then converted the Bowtie2 alignments for each library from Sequence Alignment/Map (SAM) to the binary version of SAM (BAM) format, and we used the SAMtools depth utility (57) to determine per-base coverage levels. The percentage of each isolate genome mapped by metagenome reads is the number of base pairs with at least one read mapping divided by the total number of base pairs in the reference index for that isolate.
To calculate the percentage of reads from each isolate that map to each of the remaining three isolate genomes and the metagenome assembly, we followed two procedures. For Bs02, Bs08, and Bs12, we mapped each set of isolate reads to each isolate genome and the metagenome assemblies as described above. For the BsC2 genome, the aligner bbmap (sourceforge.net/projects/bbmap/) was used. Reference databases were built for each of the isolate genomes and the metagenome scaffolds. The BsC2 PacBio reads were converted to FASTA format using a simple grep command, and alignments were performed with the command mapPacBio8k. Parameters were set to default, with the fastareadlen parameter set to 500.
Potential sources of sequences in the endosymbiont metagenome not accounted for by the isolate genomes were explored by searching sequences against a database built from the bacterial genomes collection of the National Center for Biotechnology Information (NCBI; ftp.ncbi.nlm.nih.gov/genomes/Bacteria/). The four B. setacea isolate genomes were added to this database to ensure that any hits to the NCBI collection had no better match in one of the isolate genomes. One mollusk, A. californica (GenBank accession no. 683478), and one woody plant genome, P. trichocarpa (GenBank accession no. 314288), were also added to this database to help identify sequences potentially derived from the genome of B. setacea or from ingested wood. To facilitate the identification of distant homologs, longer metagenome contigs were broken into 5,000-bp query sequences and identity thresholds were relaxed to 95 to 75%.
Liquid Chromatography/Tandem MS.
Proteins were extracted from cecum contents and gill tissues by boiling in 1% SDS. Gill tissues were homogenized or sonicated before extraction. Detergents were removed either by acetone precipitation or by passage over a Detergent Removal Spin column (Thermo Fisher Scientific). The samples were subject to reverse-phase liquid chromatography (LC) separation using a 100 × 1-mm, 1,000-Å pore PLRP-S column (Higgins Analytical). Fractions containing protein were dried, pooled, resuspended in digestion buffer, and digested with trypsin.
Tryptic peptides were analyzed by online nanoelectrospray ionization (ESI) tandem MS (MS/MS) using an Agilent 6330 Ion Trap mass spectrometer with an integrated C18 Chip/nano-ESI interface as described previously (58). The MS/MS data were analyzed using Spectrum Mill 3.03 (Agilent Technologies). Peptides generated by tryptic digest were searched against a peptide database consisting of all peptides predicted by in silico digestion of predicted proteins encoded in the gill endosymbiont metagenome of B. setacea and in the genomes of Bs02, Bs08, Bs12, and BsC2 (58). All peptide identifications were validated using a reverse database search. The stringency of protein identifications was adjusted to a threshold of less than 1% false-positive results using a reverse database target decoy search. Multiple spectra were required for all positive protein identifications. Proteins scoring greater than 20 were considered valid identifications.
Annotation of Cecum Proteins.
Among the 42 endosymbiont-encoded proteins detected in cecum samples, 38 could be traced to complete ORFs and four to truncated reading frames in one of the four isolate genomes and/or in the gill endosymbiont community metagenome. The protein sequences were subjected to a BLAST analysis against a library of sequences built with the individual modules (GH, PL, CE, glycosyl transferase, and CBM) classified in the CAZy database (24) (www.cazy.org) The GH5 sequences were assigned to subfamilies defined as in the study by Aspeborg et al. (59). After mapping the CAZy modules onto the shipworm cecum sequences, the S-rich intermodule linker sequences were mapped to identify the remaining modules. When these remaining modules showed homology to Pfam domains, they were assigned the Pfam accession number. Otherwise they were assigned to an X-module family (modules of unknown function).
N-Terminal Sequencing of Abundant Cecum Proteins.
Cecum contents were collected from 12 shipworms and pooled, and the proteins were extracted overnight at 4 °C into 50 mM sodium acetate (pH 5.5), containing 0.01% Tween 80. Extracted proteins were exchanged into 10 mM Tris (pH 8.2) and 10 mM NaCl by ultrafiltration in a 10,000-molecular weight cutoff (MWCO) Vivaspin concentrator (Viva Products). The cecum protein extract was fractionated over an anion exchange column (Q column; Qiagen) using a 0–1 M NaCl gradient in 10 mM Tris⋅HCl (pH 8.2). Fractions were collected at a rate of 2 mL per minute, and eluted proteins were detected by A at 280 nm. The protein concentrations of the cecum content extract and the protein-containing Q column fractions were determined against a standard curve (75–12.5 μg/mL BSA) using a Bio-Rad Protein Assay Kit 1 (Bio-Rad Laboratories, Inc.). Eluted proteins were analyzed by SDS/PAGE and Coomassie staining (Simply Blue SafeStain; Life Technologies). Proteins in fraction 11, which contained the largest detected absorbance peak, were resolved by SDS/PAGE, transferred to a PVDF membrane (MiniProBlott Membranes; Applied Biosystems), and Coomassie-stained. The two most prominent protein bands were excised from the membrane, and their N-terminal sequences (11–12 aa) were determined by Edman degradation on a Procise 494 sequencer (Applied Biosystems). Endosymbiont-encoded proteins in the total cecum extract and in fraction 11 were also identified by LC/MS/MS as described above.
Cloning and Expression of the Dominant GH5_53 Catalytic Module from Fraction 11.
To facilitate expression, catalytic modules of interest were cloned and expressed independently, with CBMs and linker regions omitted. Because it is highly unlikely that the elimination of these protein regions would result in the gain of a function not present in the intact native protein, the demonstration of activity in the exogenously expressed catalytic module alone is strong evidence that the observed activity is also characteristic of the full-length protein.
A DNA fragment corresponding to the predicted GH5_53 module of the most prominent protein observed in fraction 11 (KJ861991) was PCR-amplified from the genome of Bs02 using primers GH5-F and GH5-R (Table S12) and Phusion DNA polymerase (New England Biolabs). Vector pFCM21 (60) was digested with FspI and HindIII. Both the vector and PCR fragment were purified using a Qiagen PCR Purification Kit and recombined by Gibson assembly (61), using Gibson Assembly Master Mix (New England Biolabs) to generate pFCM21-GH5_53. The assembly reaction was transformed into competent NEB10β cells (New England Biolabs). Sanger sequencing was used to verify the DNA sequence of the insert. E. coli strain BL21 was transformed with either pFCM21-GH5_53 or the empty vector and grown in LB supplemented with ampicillin (100 μg/mL) at 30 °C and then induced with isopropyl β-D-1-thiogalactopyranoside (IPTG) at concentrations of 0, 10, 30, and 100 μM overnight at 30 °C. As a negative control, BL21 cells were transformed with empty pFCM21 vector and induced with 30 μM IPTG. Harvested cells were incubated in 2 mL of TES buffer [0.5 M sucrose, 0.2 M Tris⋅HCl (pH 8.0), 0.5 mM EDTA] for 30 min on ice, followed by 2 mL of ice-cold water and 30 min of gentle shaking on ice. The resulting spheroplasts were precipitated by centrifugation at 12,000 × g for 20 min at 4 °C, and the supernatants constituting the periplasmic fraction were collected. Total protein concentration in the periplasmic fraction was estimated by OD280.
Expression of Catalytic Modules of Endosymbiont-Encoded Cecum Proteins by in Vitro Transcription/Translation.
Gene sequences encoding catalytic modules from five predicted PCWP-active proteins were amplified by PCR using primers shown in Table S12 and expressed using an in vitro transcription–translation system (PURExpress; New England Biolabs) following the manufacturer’s directions. Expressed GH10 and GH11 catalytic modules were further purified by removal of the ribosomes by ultrafiltration (100,000 MWCO), and the remaining His-tagged kit components were removed by incubation with Ni-NTA resin (Qiagen). The purified proteins were quantified by comparison with BSA standards using densitometry.
Activity of Endosymbiont-Encoded Cecum Proteins Against PCWP Substrates.
Native, synthetic, and recombinant proteins from the cecum of B. setacea were tested for catalytic activity against PCWP substrates, including 1% CMC, 5% (wt/vol) Avicel, 5% (wt/vol) Sigmacell, 1% birchwood xylan, and 5% (wt/vol) LBG. Substrates were dissolved or suspended in 50 mM sodium citrate (pH 6.2). Five to 10 μL of proteins was added to 40–45 μL of substrate or buffer (as a negative control) to give a final volume of 50 μL. Digestions were conducted at 30 °C. Concentrations of purified GH10 and GH11 catalytic modules and dihydrofolate reductase (DHFR)-positive control template were between 0.1 and 0.2 μg/mL. Periplasmic fractions from E. coli containing the recombinant GH5_53 module and the control fraction were 2 and 1.2 mg/mL, respectively. For proteins expressed in the PURExpress system but not purified away from the reaction components, 5 μL of the reaction mixture was used and results were compared with a PUREexpress reaction mixture in which the DHFR template had been expressed. Samples were collected onto dry ice to stop the reaction, insoluble substrates were removed by centrifugation, and reducing sugars were detected using the Nelson–Somogyi assay (62). Glucose was used to generate a standard curve. Assays were performed in duplicate.
Statistical Analysis.
Enzymatic data were analyzed using Graphpad Prism. Optical density readings were converted to millimolar glucose concentrations by comparison with glucose standards fit to a sigmoidal dose–response curve with variable slope. Enzymatic activity measured over time was fit to a one-phase decay curve.
Supplementary Material
Acknowledgments
We thank E. Vannier and S. Vollmer for critical reading of the manuscript. This research was supported by National Science Foundation Grants IOS-0920540, IOS-1442676, IOS-1258090, and IOS-1442759 (to D.L.D.) and Grant OCE-0963010; Office of Science of the US Department of Energy Contract DE-AC02-05CH11231; National Institutes of Health Grant 1U01TW008163 (to M.G.H.); the Francis Goelet Charitable Lead Trust (D.L.D. and K.H.S); and New England Biolabs.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. KJ836284–KJ836296 (16S rRNA sequences), KJ861955–KJ861995 (cecum protein sequences), and KJ943269–KJ943359 (gill protein sequences)]. The Bankia setacea gill endosymbiont metagenome and the isolate genomes are publically available in the Joint Genome Institute’s Integrated Microbial Genomes database (img.jgi.doe.gov/cgi-bin/mer/main.cgi) under the taxon object ID numbers 2070309010 (metagenome), 2503982003 (Bs02), 2503982003 (Bs08), 2170459028 (Bs12), and 2531839719 (BsC2). B. setacea isolates Bs02, Bs08, Bs12, and BsC2 are archived in the Ocean Genome Resource biorepository (www.northeastern.edu/marinescience/ogl/) (accession nos. Sr00161, Sr00167, Sr00288, and Sr00311, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413110111/-/DCSupplemental.
References
- 1.Distel DL. 2003. The biology of marine wood boring bivalves and their bacterial endosymbionts. Wood Deterioration and Preservation, ACS Symposium Series, eds Goodell B, Nicholas DD, Schultz TP (American Chemical Society, Washington, DC), Vol 845, pp 253–271.
- 2.Turner RD. 1966. A Survey and Illustrated Catalogue of the Teredinidae (Mollusca: Bivalvia) (Museum of Comparative Zoology, Harvard University, Cambridge, MA)
- 3.Gallager SM, Turner RD, Berg CJ. Physiological aspects of wood consumption, growth, and reproduction in the shipworm Lyrodus pedicellatus quatrefages (Bivalvia, Teredinidae) J Exp Mar Biol Ecol. 1981;52(1):63–77. [Google Scholar]
- 4.Brune A. Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol. 2014;12(3):168–180. doi: 10.1038/nrmicro3182. [DOI] [PubMed] [Google Scholar]
- 5.Cardoso AM, et al. Metagenomic analysis of the microbiota from the crop of an invasive snail reveals a rich reservoir of novel genes. PLoS ONE. 2012;7(11):e48505. doi: 10.1371/journal.pone.0048505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hess M, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331(6016):463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 7.Scully ED, et al. Metagenomic profiling reveals lignocellulose degrading system in a microbial community associated with a wood-feeding beetle. PLoS ONE. 2013;8(9):e73827. doi: 10.1371/journal.pone.0073827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi W, et al. Comparative genomic analysis of the microbiome [corrected] of herbivorous insects reveals eco-environmental adaptations: Biotechnology applications. PLoS Genet. 2013;9(1):e1003131. doi: 10.1371/journal.pgen.1003131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDonald R, Schreier HJ, Watts JE. Phylogenetic analysis of microbial communities in different regions of the gastrointestinal tract in Panaque nigrolineatus, a wood-eating fish. PLoS ONE. 2012;7(10):e48018. doi: 10.1371/journal.pone.0048018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betcher MA, et al. Microbial distribution and abundance in the digestive system of five shipworm species (Bivalvia: Teredinidae) PLoS ONE. 2012;7(9):e45309. doi: 10.1371/journal.pone.0045309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luyten YA, Thompson JR, Morrill W, Polz MF, Distel DL. Extensive variation in intracellular symbiont community composition among members of a single population of the wood-boring bivalve Lyrodus pedicellatus (Bivalvia: Teredinidae) Appl Environ Microbiol. 2006;72(1):412–417. doi: 10.1128/AEM.72.1.412-417.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Distel DL, Beaudoin DJ, Morrill W. Coexistence of multiple proteobacterial endosymbionts in the gills of the wood-boring Bivalve Lyrodus pedicellatus (Bivalvia: Teredinidae) Appl Environ Microbiol. 2002;68(12):6292–6299. doi: 10.1128/AEM.68.12.6292-6299.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Distel DL, DeLong EF, Waterbury JB. Phylogenetic characterization and in situ localization of the bacterial symbiont of shipworms (Teredinidae: Bivalvia) by using 16S rRNA sequence analysis and oligodeoxynucleotide probe hybridization. Appl Environ Microbiol. 1991;57(8):2376–2382. doi: 10.1128/aem.57.8.2376-2382.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popham J. Further observations of the gland of Deshayes in the teredo Bankia australis. Veliger. 1974;18(1):55–59. [Google Scholar]
- 15.Popham J, Dickson M. Bacterial associations in the teredo Bankia australis (Lamellibranchia, Mollusca) Mar Biol. 1973;19(4):338–340. [Google Scholar]
- 16.Distel DL, Morrill W, MacLaren-Toussaint N, Franks D, Waterbury J. Teredinibacter turnerae gen. nov., sp. nov., a dinitrogen-fixing, cellulolytic, endosymbiotic gamma-proteobacterium isolated from the gills of wood-boring molluscs (Bivalvia: Teredinidae) Int J Syst Evol Microbiol. 2002;52(Pt 6):2261–2269. doi: 10.1099/00207713-52-6-2261. [DOI] [PubMed] [Google Scholar]
- 17.Weiner RM, et al. Complete genome sequence of the complex carbohydrate-degrading marine bacterium, Saccharophagus degradans strain 2-40 T. PLoS Genet. 2008;4(5):e1000087. doi: 10.1371/journal.pgen.1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waterbury JB, Calloway CB, Turner RD. A cellulolytic nitrogen-fixing bacterium cultured from the gland of deshayes in shipworms (bivalvia: teredinidae) Science. 1983;221(4618):1401–1403. doi: 10.1126/science.221.4618.1401. [DOI] [PubMed] [Google Scholar]
- 19.Altamia MA, et al. Genetic differentiation among isolates of Teredinibacter turnerae, a widely occurring intracellular endosymbiont of shipworms. Mol Ecol. 2014;23(6):1418–1432. doi: 10.1111/mec.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lechene CP, Luyten Y, McMahon G, Distel DL. Quantitative imaging of nitrogen fixation by individual bacteria within animal cells. Science. 2007;317(5844):1563–1566. doi: 10.1126/science.1145557. [DOI] [PubMed] [Google Scholar]
- 21.Amann RI, et al. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56(6):1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang JC, et al. The complete genome of Teredinibacter turnerae T7901: An intracellular endosymbiont of marine wood-boring bivalves (shipworms) PLoS ONE. 2009;4(7):e6085. doi: 10.1371/journal.pone.0006085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeBoy RT, et al. Insights into plant cell wall degradation from the genome sequence of the soil bacterium Cellvibrio japonicus. J Bacteriol. 2008;190(15):5455–5463. doi: 10.1128/JB.01701-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(Database issue):D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tatusov RL, et al. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29(1):22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink VG. Novel enzymes for the degradation of cellulose. Biotechnol Biofuels. 2012;5(1):45. doi: 10.1186/1754-6834-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Natale P, Brüser T, Driessen AJ. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane—Distinct translocases and mechanisms. Biochim Biophys Acta. 2008;1778(9):1735–1756. doi: 10.1016/j.bbamem.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 28.Chaudhary R, Atamian HS, Shen Z, Briggs SP, Kaloshian I. GroEL from the endosymbiont Buchnera aphidicola betrays the aphid by triggering plant defense. Proc Natl Acad Sci USA. 2014;111(24):8919–8924. doi: 10.1073/pnas.1407687111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabri A, et al. Proteomic investigation of aphid honeydew reveals an unexpected diversity of proteins. PLoS ONE. 2013;8(9):e74656. doi: 10.1371/journal.pone.0074656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saraswathy M. Observations on the structure of the shipworms, Nausitora hedleyi, Teredo furcifera and Teredora pricesae (Bivalvia: Teredinidae) Trans R Soc Edinb Earth Sci. 1971;68(14):508–562. [Google Scholar]
- 31.Sigerfoos CP. Natural history, organization and late development of the Teredinidae or shipworms. Bulletin of the Bureau of Fisheries. 1908;37:191–231. [Google Scholar]
- 32.Chen X, Alonso AP, Allen DK, Reed JL, Shachar-Hill Y. Synergy between (13)C-metabolic flux analysis and flux balance analysis for understanding metabolic adaptation to anaerobiosis in E. coli. Metab Eng. 2011;13(1):38–48. doi: 10.1016/j.ymben.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Harris PV, Xu F, Kreel NE, Kang C, Fukuyama S. New enzyme insights drive advances in commercial ethanol production. Curr Opin Chem Biol. 2014;19:162–170. doi: 10.1016/j.cbpa.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 34.Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: Pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12(1):118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 35.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6(3):610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Werner JJ, et al. Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J. 2012;6(1):94–103. doi: 10.1038/ismej.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lane DJ. In: 16S/23S rRNA Sequencing. Nucleic Acid Techniques in Bacterial Systematics. Stachebrandt E, Goodfellow M, editors. Wiley; Chichester, UK: 1991. [Google Scholar]
- 38.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 39.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haas BJ, et al. Human Microbiome Consortium Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21(3):494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 44.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 45.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 47.Yilmaz LS, Parnerkar S, Noguera DR. mathFISH, a web tool that uses thermodynamics-based mathematical models for in silico evaluation of oligonucleotide probes for fluorescence in situ hybridization. Appl Environ Microbiol. 2011;77(3):1118–1122. doi: 10.1128/AEM.01733-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5(4):433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- 50.Mavromatis K, et al. The DOE-JGI Standard Operating Procedure for the Annotations of Microbial Genomes. Stand Genomic Sci. 2009;1(1):63–67. doi: 10.4056/sigs.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 52.Korlach J, et al. Real-time DNA sequencing from single polymerase molecules. Methods Enzymol. 2010;472:431–455. doi: 10.1016/S0076-6879(10)72001-2. [DOI] [PubMed] [Google Scholar]
- 53.Clark TA, et al. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 2012;40(4):e29. doi: 10.1093/nar/gkr1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Travers KJ, Chin CS, Rank DR, Eid JS, Turner SW. A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic Acids Res. 2010;38(15):e159. doi: 10.1093/nar/gkq543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chin CS, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10(6):563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 56.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swaim CL, Anton BP, Sharma SS, Taron CH, Benner JS. Physical and computational analysis of the yeast Kluyveromyces lactis secreted proteome. Proteomics. 2008;8(13):2714–2723. doi: 10.1002/pmic.200700764. [DOI] [PubMed] [Google Scholar]
- 59.Aspeborg H, Coutinho PM, Wang Y, Brumer H, 3rd, Henrissat B. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5) BMC Evol Biol. 2012;12:186. doi: 10.1186/1471-2148-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Londer YY, Giuliani SE, Peppler T, Collart FR. Addressing Shewanella oneidensis “cytochromome”: The first step towards high-throughput expression of cytochromes c. Protein Expr Purif. 2008;62(1):128–137. doi: 10.1016/j.pep.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 61.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 62.Nelson N. A photometric adaptation of the Somogyi method for the determination of glucose. J Biol Chem. 1944;153:375–380. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.