Abstract
Purpose
The tumor suppressor p53 is known to be inactivated frequently in various cancers. In addition, germline polymorphisms in TP53 are known to affect protein function and influence risk of developing different types of cancers. In this study, we analyzed the association of TP53 Pro72Arg polymorphism with squamous cell carcinoma of oral tongue (SCCOT) and esophagus (ESCC) in India.
Methods
We assessed the distribution of TP53 Pro72Arg polymorphism in one hundred and fifteen and eighty two SCCOT and ESCC patients, respectively, with respect to one hundred and ten healthy controls from the same population. In addition, we analyzed association of the polymorphism with several clinico-pathological and molecular parameters.
Results
Pro72 allele was significantly enriched in SCCOT patients compared to the healthy control group but neither allele was enriched in ESCC. Interestingly, Pro72 allele was preferentially mutated in ESCC which was confirmed by analysis of samples heterozygous for Pro72Arg.
Conclusions
Our study revealed the association of Pro72 allele with SCCOT suggesting the effect of this polymorphism on SCCOT risk. Preferential mutation of Pro72 allele exclusively in ESCC indicates the need for further studies to understand the tissue specific effect of p53 polymorphism.
Introduction
Squamous Cell Carcinoma of the Oral Tongue (SCCOT) is a common form of head and neck squamous cell carcinoma (HNSCC) and its incidence is consistently increasing worldwide [1]. The increase in incidence has been noted predominantly in younger patients [2], [3] in whom it appears to exhibit lower association with common risk factors such as alcohol and tobacco suggesting possible genetic susceptibility [3]. SCCOT is known to be aggressive and is associated with higher rates of occult and nodal metastasis when compared to other HNSCC subtypes [4]. It is often associated with poor survival which has not improved significantly over the past four decades [5], [6]. Esophageal cancer (EC) is the sixth and eighth most common cancer in males and females respectively in India (http://www.icmr.nic.in/ncrp/report_pop_2001-04/cancer_p_based.htm). Squamous (ESCC; occurs usually in proximal and middle esophagus) and adeno (EAC; occurs mainly in distal esophagus) carcinoma are the two major EC subtypes [7]. Though ESCC was more common few decades ago, a rapid increase in EAC has been noted since the 1980s in the West [8], in parallel with an increased incidence of gastrointestinal reflex disease (GERD). GERD causes an inflammation induced pathological condition called ‘Barrett's esophagus’, which predisposes to EAC [9]. Though, a similar trend in increase of GERD has been noted in Asian countries in the last few decades, ESCC remains the predominant EC subtype [10], suggesting possibility of role of genetic factors. Due to its closer location to neck and similarities in tumorigenesis pathways, ESCC is sometimes classified with HNSCC.
Somatic inactivation of TP53 is a frequent event in most cancers whereas germ line aberrations are associated with Li-Fraumeni [11], a hereditary cancer predisposition syndrome. Common modes of p53 inactivation are point mutations, allelic loss [12] and inactivation mediated by oncoviral proteins [13]. In addition, numerous polymorphisms occur in TP53 of which a few are suggested to perturb protein function and may influence cancer susceptibility [14]. Among these, the codon 72 Pro/Arg polymorphism (rs1042522) is the most common and well-studied. The two p53 codon72 alleles encode proline (Pro72) or arginine (Arg72), located in a polyproline region present between the transactivation and the DNA binding domains and may affect the structure of the putative SH3-binding domain [15]. The Pro72 allele is known to be associated with coronary artery disease [16], higher susceptibility to endometriosis [17], primary open angle glaucoma [18], systemic lupus erythematosus (especially in Asians) [19] and ulcerative colitis [20] whereas the Arg72 allele is associated with progression of Diabetic Nephropathy [21]. More importantly, the polymorphism exhibits varying association with risk [22], [23], survival [24], [25], and treatment response [25] in several cancers in different populations.
In the current study, we assessed the frequency of Pro72Arg polymorphism in SCCOT and ESCC. Pro72 allele appeared to be significantly associated with SCCOT whereas no association with either allele was detected in ESCC. However, in ESCC, TP53 DNA binding domain mutations occurred at a significantly higher frequency in the Pro72 allele.
Materials and Methods
Patient and control samples
One hundred and fifteen and eighty two previously untreated and surgically resected SCCOT and ESCC samples respectively were collected during the period 2007 to 2013 from three hospitals in Hyderabad, India, following informed consent. Seventy two and seventy five SCCOT and ESCC patients respectively were from our earlier studies [26], [27]. Clinico-pathological details of the patients are given in Table S1. Median age and male to female ratio were 49 and 50; and 1.94 and 1 for SCCOT and ESCC respectively. Peripheral blood from one hundred and ten age and gender matched cancer free healthy individuals belonging to the same geographical region were collected.
Ethics statement
The study was approved by ethics committees of Apollo Hospitals (14/05/2005), MNJ Institute of Oncology and Regional Cancer Centre (23/09/2006 and 20/10/2008) and Indo-American Cancer Institute and Research Centre (08/08/2007) as well as Institutional bioethics committee of Centre for DNA Fingerprinting and Diagnostics (CDFD) (20/12/2009) as per modified Helsinki declaration 2005. All samples were collected following written informed consent.
Genotyping
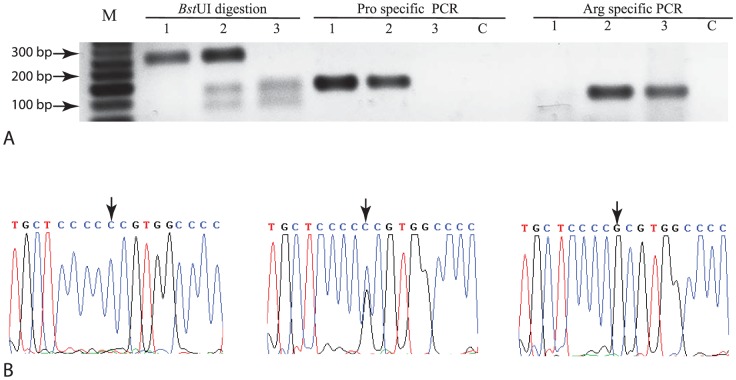
DNA was isolated from histologically confirmed normal tissue adjacent to tumor for each sample as detailed in Document S1. Genotyping of codon 72 was performed using a two-pronged approach including PCR-RFLP (Figure 1A) [28] and allele specific PCR (Figure 1A) [29] as previously described. The results were independently confirmed in twenty and fifteen SCCOT and ESCC samples respectively using bidirectional Sanger sequencing on a 3100 Genetic analyzer (ABI Inc., Foster city, CA, USA) (Figure 1B). Primer sequences are listed in Table S2.
Figure 1. Identification of the three different p53 codon 72 genotypes.
Panel A, PCR-RFLP analysis confirmed by allele specific PCR performed as described in the materials and methods section. M, 50 bp DNA ladder; 1, Pro/Pro; 2, Pro/Arg; 3, Arg/Arg; and C, No template control. Panel B, Sanger sequencing. Pro/Pro (left), Pro/Arg (middle) and Arg/Arg (right).
TP53 mutation screening
TP53 mutation screening was performed as described earlier [26]. To identify whether the Pro72 or the Arg72 allele was harboring p53 mutation in samples heterozygous for the Pro72Arg polymorphism, a long amplicon (∼2700 bp) spanning exons 4–8 of TP53 (which includes the codon 72 as well as the region encoding the DNA binding domain) was amplified (Primer pairs Arg+ and Exon 8R, Table S2) from genomic DNA and cloned into TA vector (Invitrogen, Carlsbad, CA, USA). The recombinant plasmids were screened for mutation as well as for the codon 72 polymorphism using bidirectional Sanger sequencing.
Molecular analysis
Status of p53 nuclear stabilization, TP53 mutation (only for SCCOT), EGFR over expression, microsatellite instability, β-catenin nuclear localization status (only for ESCC), HPV infection and LOH and clinico-pathological variables including age, gender, smoking, alcohol consumption, grade and pathological stage were reported in our previous studies [26], [27].
Statistical analysis
Deviation from Hardy-Weinberg equilibrium for genotype frequency for cases and controls, was analyzed using χ2 test. Odds ratio and corresponding 95% confidence intervals for disease risk were calculated. Dominant, codominant, recessive, over dominant and log additive inheritance models were tested to determine whether the SNP was associated with disease. Akaike (AIC) and Bayesian (BIC) information criterion in addition to χ2 p values were used to select the best model of inheritance. Association between different genotypes and clinico-pathological variables was assessed using χ2 or Fisher exact test as appropriate.
Results
TP53 Pro72 allele is enriched in SCCOT but not in ESCC patients
We analyzed the distribution of p53 codon 72 genotypes in SCCOT and ESCC with respect to healthy controls (Figure 1) as described in materials and methods section. 26 (23.6%), 53 (48.2%) and 31 (28.2%) control samples harbored Pro/Pro, Pro/Arg and Arg/Arg genotypes, respectively, exhibiting thereby no significant enrichment of one allele over the other (Table 1). However, Pro72 allele appeared to be significantly enriched in SCCOT compared to controls (Table 1) whereas no significant enrichment was observed in ESCC (Table 1). The genotype distributions of SCCOT and ESCC samples as well as controls were not deviating from Hardy-Weinberg equilibrium (Table 1). Co-dominant, recessive and log-additive genetic models were found to be appropriate for the inheritance of SCCOT for this SNP (Table S3A). However, lowest value of AIC (310.7) and BIC (317) for recessive model indicates it to be the best model (Table S3A). In contrast, ESCC did not exhibit association with any genetic model (Table S3B) as expected. None of several molecular and clinico-pathological variables exhibited significant association with codon 72 allele frequency in SCCOT and ESCC (Table S1).
Table 1. Comparative analysis of p53 Pro72Arg polymorphism status in SCCOT, ESCC and healthy control samples.
| Genotype distributiona | Allele distribution | ||||||
| Pro/Pro | Pro/Arg | Arg/Arg | Pc | Pro72 | Arg72 | Pc | |
| Controls (110) b | 26 (23.6%) | 53 (48.2%) | 31 (28.2%) | - | 105 (47.7%) | 115 (52.3%) | - |
| SCCOT (115) b | 44 (38.3%) | 48 (41.7%) | 23 (20%) | 0.051 | 136 (59.1%) | 94 (40.9%) | 0.019 |
| ESCC (82) b | 20 (24.4%) | 46 (56.1%) | 16 (19.5%) | 0.363 | 86 (52.4%) | 78 (47.6%) | 0.361 |
χ2 test p values for deviation from Hardy-Weinberg equilibrium are 0.71, 0.18 and 0.37 for controls, SCCOT and ESCC, respectively.
Number in parenthesis indicates total number of samples.
p is from χ2 test performed for comparison of SCCOT or ESCC with controls.
TP53 mutations are exclusively observed in ESCC tumors with p53 nuclear stabilization
TP53 mutation status was reported earlier for all SCCOT samples analyzed in the previous study [26]; mutations were associated with poor disease specific survival [26] but not with several clinico-pathological parameters enumerated in materials and methods section. We screened ESCC samples for somatic mutations in exons 5–8 of TP53 that encode the DNA binding domain and are known to harbor majority of cancer associated mutations [30]. Mutations were detected in twenty nine of forty five samples exhibiting nuclear stabilization and none of thirty seven samples that did not, suggesting that absence of nuclear stabilization may be a reliable indicator of absence of TP53 DNA binding domain mutation in ESCC. A total of twenty seven (nineteen missense, three nonsense and five indels) mutations (all heterozygous) were identified. Two mutations (c.610G>T (p.E204X) and c.566delC (p. P190Lfs*57)) were detected in two samples each (Table S4). We identified three novel somatic mutations viz., c.621_639del19, c.454_466dupCCGCCCGGCACCC and c.428_432delTGCAG+ 440delG (Table S4). Detailed description of the novel mutations is given in Document S2. Mutations in ESCC were not associated with any of the clinico-pathological variables analyzed (Table S5). Proportion of transitions and transversions (Figure S1A) as well as frameshift, missense and nonsense mutations (Figure S1B) were similar to previous reports for ESCC as per the International Agency for Research on Cancer (IARC) TP53 database [15]. Interestingly, proportion of deletions was higher than reported in the TP53 database (Figure S1B). G:C>A:T transitions constituted the major TP53 mutation type (9/29; 34.48%) in ESCC samples in this study similar to the database [15]. However, frequency of C>T transitions at CpG dinucleotides was lower (10.34%) while frequency of deletions (5/29; 17.24%) was higher than reported in the database [15].
Pro72 allele harbored inactivating mutations frequently in ESCC
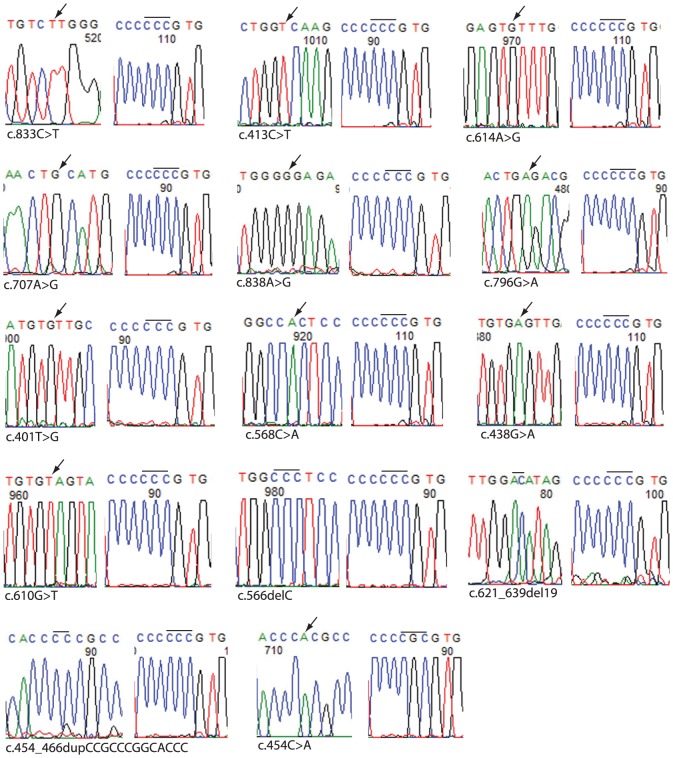
P53 DNA binding mutations were not associated with codon 72 genotype in SCCOT (p = 0.917) (Table 2). In contrast, ESCC samples with Pro/Pro (10/20; 50%) genotype were more likely to harbor mutation than samples with Arg/Arg genotype (5/16; 31.3%) (p = 0.289) (Table 2). Therefore we proceeded to determine whether Pro72 allele was more frequently mutated by analyzing samples heterozygous for Pro72Arg polymorphism. A single PCR product that included the mutation as well as the polymorphism was generated and cloned into a suitable plasmid vector and sequenced to determine whether the mutation was present in the Pro72 or the Arg72 allele. As shown in Figure 2, the mutation was preferentially located in the Pro72 allele (p = 0.0001) (Table 2), thus supporting the result obtained for samples homozygous for codon 72 and enabling us to compare Pro72Arg polymorphism and p53 mutation status for all 75 ESCC samples which revealed that mutation was significantly associated with Pro72 allele (p = 0.0018) (Table 2).
Table 2. Comparative analysis of p53 mutation and Pro72Arg polymorphism status in SCCOT and ESCC samples.
| Codon 72 genotype | P53 mutation status | Significance | |
| Present | Absent | ||
| SCCOT all samples (Stratified by genotype) | |||
| Pro/Pro (22) | 07 | 15 | 0.917a |
| Pro/Arg (27) | 08 | 19 | |
| Arg/Arg (12) | 03 | 09 | |
| ESCC all samples (Stratified by genotype) | |||
| Pro/Pro (20) | 10 | 10 | 0.289a |
| Pro/Arg (46) | 14 | 32 | |
| Arg/Arg (16) | 05 | 11 | |
| ESCC samples heterozygous for codon 72 polymorphism | |||
| Pro72 | 13 | 01 | 0.000b |
| Arg72 | 01 | 13 | |
| ESCC all samples (Stratified by allele) | |||
| Pro72 | 23 | 63 | 0.0018b |
| Arg72 | 6 | 72 | |
from χ2 test.
from Fisher's exact test.
Figure 2. TP53 mutation identified in Pro72 or Arg72 allele in fourteen ESCC samples heterozygous for p53 codon 72 polymorphism.
Electrophoretograms showing mutation (left) and codon 72 polymorphism (right) for each of the fourteen samples are shown. Location of missense and deletion mutations are indicated by arrows and bars, respectively. Proline (CCC) or Arginine (CGC) codons are indicated by a bar.
Interestingly, mutations expected to cause p53 truncation (nonsense and frameshift) were exclusively identified in the Pro72 allele (8/8, 100%) in ESCC samples. Though, one deletion and one complex mutation (of total six mutations) were identified in Arg72 allele, both were not expected to alter reading frame and thus may not cause protein truncation. Only 16.67% of TP53 mutations in SCCOT [26] (as compared to 27.59% in ESCC) were expected to result in p53 truncation, further suggesting differences in biology of SCCOT and ESCC with respect to p53 function.
Discussion
Several studies conducted on mixture of Head and Neck squamous cell carcinoma (HNSCC) samples did not detect any significant association with TP53 codon 72 polymorphism [31]–[33] perhaps due to heterogeneity of tumor subtypes. Studies on tumors of specific HNSCC site and/or molecular subtype did however reveal association with polymorphism in different ethnicities [34]–[37]. To the best of our knowledge, this is the first specific case control study undertaken to find the association between TP53 Pro72Arg polymorphism and SCCOT. There are conflicting reports on the association of Pro72Arg polymorphism with ESCC risk with studies reporting significant association with Pro72 [22], [23], [38], [39], Arg72 [40] or neither [41], [42]. Interestingly, the effect may vary within the same population [22], [40]. Our analysis revealed that Pro72 allele was significantly associated with SCCOT but not with ESCC, perhaps reflecting differences in biology of SCCOT and ESCC among Indians. Of note, a recent study suggested significant difference in p53 function between Arg72 and Pro72 allele. However this difference was tissue specific [43]. It was also suggested that Pro72 allele could be more efficient in causing cell cycle arrest [44] whereas Arg72 allele was shown to be more efficient in inducing apoptosis and localization to mitochondria [45]. Though it has been shown that Arg72 allele could be more efficiently targeted by HPV protein E6 [29], we did not detect any significant association in this study.
Similar to our results, another study from India also reported absence of TP53 mutations in ESCC tumors without nuclear stabilization [46], unlike reports from other countries (54–55) perhaps indicating a feature exclusively associated with Indian population. In addition, the ESCC TP53 somatic mutation spectrum detected in this study is similar to the observations made earlier from India [47]. Lower proportion of G:C>A:T transitions at CpG dinucleotides (attributed to spontaneous deamination of cytosine) identified in ESCC samples in this study is in line with previous studies from India [47]. Higher frequency of G:C>A:T transitions at non-CpG sites can be attributed to alkylating agents in food and environment [48]. Of note, nitrosamines in alcoholic beverages [49] and processed meat [50] are known to increase the risk of esophageal and stomach cancer, respectively. ESCC studies from Iran [51] as well as southern Brazil [52] reported an association of habit of drinking hot tea with G:C>A:T transitions.
To the best of our knowledge, this is the only study to evaluate status of TP53 mutation in Pro/Arg heterozygotes, which is expected to provide more accurate information on extent of association since both alleles are present in the same genetic background. We observed that p53 DNA binding domain mutations were predominantly detected in Pro72 allele (compared to Arg72 allele) in ESCC whereas neither allele was preferentially mutated in SCCOT. Several studies on various cancers reported possible association of p53 DNA binding domain mutation with Arg72 allele [53], [54] while a few reported otherwise [24], [55]. Perhaps, the association may vary according to ethnicity [24]. Mutated p53 Arg72 is suggested to be more effective compared to p53 Pro72 in binding and inactivating p73, a p53 homologue that can transactivate p53 targets [56]. In addition, previous studies also showed that mutated Pro72 allele may have higher potential to inactivate p53 itself [57]. Of note, ESCC exhibits frequent inactivation of p73 through loss of heterozygosity [58], thereby perhaps explaining higher frequency of mutation in Pro72 allele causing inactivation of p53.
Interestingly, we observed protein truncating mutations associated exclusively with the Pro72 allele in ESCC. Of note, truncation mutations are expected to completely abrogate p53 function while missense mutations can be expected to retain partial/altered activity. In addition, unlike truncation mutation, missense mutations in p53 DNA binding domain may alter the ability of p53 to bind to DNA and transactivate target genes but may not affect p53 functions exclusive to other domains of protein [59]. Thus complete inactivation of Pro72 might be more tumorigenic than Arg72 in ESCC.
Conclusion
This study has revealed effect of TP53 Pro72 allele in increasing the risk of SCCOT and suggested that SCCOT may have biological difference with other forms of HNSCC. A unique feature of this study was determination of mutation status in samples heterozygous for p53 Pro72Arg polymorphism, which enabled us to conclude that Pro72 allele was indeed preferentially mutated in ESCC. Our results support previous observations suggesting different behavior of the p53 Pro72Arg alleles in different cancer types and ethnicities and suggest distinct molecular function of p53 Arg72 and p53 Pro72 with respect to associated mutation, in ESCC. Further studies on other cancers should be conducted to analyze the association of polymorphism and mutation. Our results can be extended to analyze the effect of polymorphism and mutation on patient survival and response to chemo/radiotherapies. Molecular and functional studies on mutant p53 in Pro72 and Arg72 background could possibly elucidate differential activity of p53. Finally, to elucidate the effect of polymorphism on mutation, studies using animal models may help in understanding the role of this polymorphism in tumorigenesis.
Supporting Information
Comparative analysis of distribution of ESCC TP53 mutations in India with the IARC TP53 database at nucleotide level (Panel A), DNA level (Panel B) and protein level (Panel C). Unclassified and silent mutations reported in the database were omitted from this analysis. The p.V143_W146delinAV mutation was not included in the analysis (in Panel C) due to complexity of its effect at protein level.
(EPS)
(DOCX)
(DOC)
(DOCX)
(DOC)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Dr. Mukta Srinivasulu, of the MNJ Institute of Oncology & Regional Cancer Centre, Hyderabad, Dr. MohanaVamsy Chigurupati, Drs. Sujith Patnaik, Subramanyeshwar Rao and NarasimhaRaju Kalidindi of Indo-American Cancer Hospital & Research Institute, Hyderabad, Drs. Swarnalata Gowrishankar and Umanath K Nayak of the Apollo Hospitals, Hyderabad, for their help in sample collection. We thank Dr. Kumarasamy Thangaraj, Centre for Cellular & Molecular Biology, Hyderabad, for collection of DNA samples of healthy individuals. RSRA and RP are registered PhD students of Manipal University, Karnataka, India and University of Hyderabad, respectively. RSRA and RP are thankful to University Grants Commission (UGC), Govt. of India and Indian Council for Medical Research (ICMR), Govt. of India, respectively for junior and senior research fellowships.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
MDB received Indian Council for Medical Research (ICMR), Govt. of India (Grant No. 5/13/129/2009-NCD-III), Council of Scientific and Industrial Research (CSIR), Govt. of India (Grant No. 27(265)/12/EMR-II), Department of Biotechnology, Govt. of India (Grant No. BT/PR11873/MED/30/173/2009) and a core grant from the Department of Biotechnology, Govt. of India to Centre for DNA Fingerprinting and Diagnostics (CDFD). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rethman MP, Carpenter W, Cohen EE, Epstein J, Evans CA, et al. (2010) Evidence-based clinical recommendations regarding screening for oral squamous cell carcinomas. J Am Dent Assoc 141:509–520. [DOI] [PubMed] [Google Scholar]
- 2. Moore SR, Johnson NW, Pierce AM, Wilson DF (2000) The epidemiology of tongue cancer: a review of global incidence. Oral Dis 6:75–84. [DOI] [PubMed] [Google Scholar]
- 3. Myers JN, Elkins T, Roberts D, Byers RM (2000) Squamous cell carcinoma of the tongue in young adults: increasing incidence and factors that predict treatment outcomes. Otolaryngol Head Neck Surg 122:44–51. [DOI] [PubMed] [Google Scholar]
- 4. Bello IO, Soini Y, Salo T (2010) Prognostic evaluation of oral tongue cancer: means, markers and perspectives (I). Oral Oncol 46:630–635. [DOI] [PubMed] [Google Scholar]
- 5. Zini A, Czerninski R, Sgan-Cohen HD (2010) Oral cancer over four decades: epidemiology, trends, histology, and survival by anatomical sites. J Oral Pathol Med 39:299–305. [DOI] [PubMed] [Google Scholar]
- 6. Hammarstedt L LY, Marklund L, Dalianis T, Munck-Wikland E, Ye W. (2011) Differential survival trends for patients with tonsillar, base of tongue and tongue cancer in Sweden. Oral Oncol 47:636–641. [DOI] [PubMed] [Google Scholar]
- 7. Siewert JR, Stein HJ, Feith M, Bruecher BL, Bartels H, et al. (2001) Histologic tumor type is an independent prognostic parameter in esophageal cancer: lessons from more than 1,000 consecutive resections at a single center in the Western world. Ann Surg 234:360–369 discussion – [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Devesa SS, Blot WJ, Fraumeni JF Jr. (1998) Changing patterns in the incidence of esophageal and gastric carcinoma in the United States. Cancer 83:2049–2053. [PubMed] [Google Scholar]
- 9. Haggitt RC, Tryzelaar J, Ellis FH, Colcher H (1978) Adenocarcinoma complicating columnar epithelium-lined (Barrett's) esophagus. Am J Clin Pathol 70:1–5. [DOI] [PubMed] [Google Scholar]
- 10. Ho KY (2011) From GERD to Barrett's esophagus: is the pattern in Asia mirroring that in the West? J Gastroenterol Hepatol 26:816–824. [DOI] [PubMed] [Google Scholar]
- 11. Hainaut P, Hollstein M (2000) p53 and human cancer: the first ten thousand mutations. Adv Cancer Res 77:81–137. [DOI] [PubMed] [Google Scholar]
- 12. Baker SJ, Preisinger AC, Jessup JM, Paraskeva C, Markowitz S, et al. (1990) p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res 50:7717–7722. [PubMed] [Google Scholar]
- 13. Tommasino M, Accardi R, Caldeira S, Dong W, Malanchi I, et al. (2003) The role of TP53 in Cervical carcinogenesis. Hum Mutat 21:307–312. [DOI] [PubMed] [Google Scholar]
- 14. Whibley C, Pharoah PD, Hollstein M (2009) p53 polymorphisms: cancer implications. Nat Rev Cancer 9:95–107. [DOI] [PubMed] [Google Scholar]
- 15. Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, et al. (2007) Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat 28:622–629. [DOI] [PubMed] [Google Scholar]
- 16. Gloria-Bottini F, Banci M, Saccucci P, Nardi P, Scognamiglio M, et al. (2012) p53 codon 72 polymorphism and coronary artery disease: evidence of association with left ventricular ejection fraction. Am J Med Sci 343:127–130. [DOI] [PubMed] [Google Scholar]
- 17. Jia S, Xu L, Chan Y, Wu X, Yang S, et al. (2012) p53 codon 72 polymorphism and endometriosis: a meta-analysis. Arch Gynecol Obstet 285:1657–1661. [DOI] [PubMed] [Google Scholar]
- 18. Lin HJ, Chen WC, Tsai FJ, Tsai SW (2002) Distributions of p53 codon 72 polymorphism in primary open angle glaucoma. Br J Ophthalmol 86:767–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee YH, Bae SC, Choi SJ, Ji JD, Song GG (2012) Associations between the p53 codon 72 polymorphisms and susceptibility to systemic lupus erythematosus and rheumatoid arthritis: a meta-analysis. Lupus 21:430–437. [DOI] [PubMed] [Google Scholar]
- 20. Vaji S, Salehi Z, Aminian K (2011) Association of p53 codon 72 genetic polymorphism with the risk of ulcerative colitis in northern Iran. Int J Colorectal Dis 26:235–238. [DOI] [PubMed] [Google Scholar]
- 21.Kankova K (2014) Association of the Arg72Pro Polymorphism in p53 with Progression of Diabetic Nephropathy in T2DM Subjects. Journal of Nephrology & Therapeutics 04.
- 22. Hong Y, Miao X, Zhang X, Ding F, Luo A, et al. (2005) The role of P53 and MDM2 polymorphisms in the risk of esophageal squamous cell carcinoma. Cancer Res 65:9582–9587. [DOI] [PubMed] [Google Scholar]
- 23. Cai L, Mu LN, Lu H, Lu QY, You NC, et al. (2006) Dietary selenium intake and genetic polymorphisms of the GSTP1 and p53 genes on the risk of esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 15:294–300. [DOI] [PubMed] [Google Scholar]
- 24. Katkoori VR, Jia X, Shanmugam C, Wan W, Meleth S, et al. (2009) Prognostic significance of p53 codon 72 polymorphism differs with race in colorectal adenocarcinoma. Clin Cancer Res 15:2406–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dong HJ, Fang C, Wang L, Fan L, Xu J, et al. (2014) TP53 Pro72 allele potentially increases the poor prognostic significance of TP53 mutation in chronic lymphocytic leukemia. Med Oncol 31:908. [DOI] [PubMed] [Google Scholar]
- 26. Adduri RS, Kotapalli V, Gupta NA, Gowrishankar S, Srinivasulu M, et al. (2014) P53 nuclear stabilization is associated with FHIT loss and younger age of onset in squamous cell carcinoma of oral tongue. BMC Clin Pathol 14:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pandilla R, Kotapalli V, Gowrishankar S, Chigurupati M, Patnaik S, et al. (2013) Distinct genetic aberrations in oesophageal adeno and squamous carcinoma. Eur J Clin Invest 43:1233–1239. [DOI] [PubMed] [Google Scholar]
- 28. Xu Y, Yao L, Ouyang T, Li J, Wang T, et al. (2005) p53 Codon 72 polymorphism predicts the pathologic response to neoadjuvant chemotherapy in patients with breast cancer. Clin Cancer Res 11:7328–7333. [DOI] [PubMed] [Google Scholar]
- 29. Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, et al. (1998) Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature 393:229–234. [DOI] [PubMed] [Google Scholar]
- 30. Joerger AC, Fersht AR (2008) Structural biology of the tumor suppressor p53. Annu Rev Biochem 77:557–582. [DOI] [PubMed] [Google Scholar]
- 31. Hamel N, Black MJ, Ghadirian P, Foulkes WD (2000) No association between P53 codon 72 polymorphism and risk of squamous cell carcinoma of the head and neck. Br J Cancer 82:757–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McWilliams JE, Evans AJ, Beer TM, Andersen PE, Cohen JI, et al. (2000) Genetic polymorphisms in head and neck cancer risk. Head Neck 22:609–617. [DOI] [PubMed] [Google Scholar]
- 33. Summersgill KF, Smith EM, Kirchner HL, Haugen TH, Turek LP (2000) p53 polymorphism, human papillomavirus infection in the oral cavity, and oral cancer. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 90:334–339. [DOI] [PubMed] [Google Scholar]
- 34. Ji X, Neumann AS, Sturgis EM, Adler-Storthz K, Dahlstrom KR, et al. (2008) p53 codon 72 polymorphism associated with risk of human papillomavirus-associated squamous cell carcinoma of the oropharynx in never-smokers. Carcinogenesis 29:875–879. [DOI] [PubMed] [Google Scholar]
- 35. Sousa H, Santos AM, Catarino R, Pinto D, Vasconcelos A, et al. (2006) Linkage of TP53 codon 72 pro/pro genotype as predictive factor for nasopharyngeal carcinoma development. Eur J Cancer Prev 15:362–366. [DOI] [PubMed] [Google Scholar]
- 36. Tiwawech D, Srivatanakul P, Karaluk A, Ishida T (2003) The p53 codon 72 polymorphism in Thai nasopharyngeal carcinoma. Cancer Lett 198:69–75. [DOI] [PubMed] [Google Scholar]
- 37. Tsai MH, Lin CD, Hsieh YY, Chang FC, Tsai FJ, et al. (2002) Prognostic significance of the proline form of p53 codon 72 polymorphism in nasopharyngeal carcinoma. Laryngoscope 112:116–119. [DOI] [PubMed] [Google Scholar]
- 38. Lee JM, Lee YC, Yang SY, Shi WL, Lee CJ, et al. (2000) Genetic polymorphisms of p53 and GSTP1, but not NAT2, are associated with susceptibility to squamous-cell carcinoma of the esophagus. Int J Cancer 89:458–464. [DOI] [PubMed] [Google Scholar]
- 39. Shao Y, Tan W, Zhang S (2008) P53 gene codon 72 polymorphism and risk of esophageal squamous cell carcinoma: a case/control study in a Chinese population. Dis Esophagus 21:139–143. [DOI] [PubMed] [Google Scholar]
- 40. Yang W, Zhang Y, Tian X, Ning T, Ke Y (2008) p53 Codon 72 polymorphism and the risk of esophageal squamous cell carcinoma. Mol Carcinog 47:100–104. [DOI] [PubMed] [Google Scholar]
- 41. Pantelis A, Pantelis D, Ruemmele P, Hartmann A, Hofstaedter F, et al. (2007) p53 Codon 72 polymorphism, loss of heterozygosity and high-risk human papillomavirus infection in a low-incidence German esophageal squamous cell carcinoma patient cohort. Oncol Rep 17:1243–1248. [PubMed] [Google Scholar]
- 42. Peixoto Guimaraes D, Hsin Lu S, Snijders P, Wilmotte R, Herrero R, et al. (2001) Absence of association between HPV DNA, TP53 codon 72 polymorphism, and risk of oesophageal cancer in a high-risk area of China. Cancer Lett 162:231–235. [DOI] [PubMed] [Google Scholar]
- 43. Azzam GA, Frank AK, Hollstein M, Murphy ME (2011) Tissue-specific apoptotic effects of the p53 codon 72 polymorphism in a mouse model. Cell Cycle 10:1352–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Siddique MM, Balram C, Fiszer-Maliszewska L, Aggarwal A, Tan A, et al. (2005) Evidence for selective expression of the p53 codon 72 polymorphs: implications in cancer development. Cancer Epidemiol Biomarkers Prev 14:2245–2252. [DOI] [PubMed] [Google Scholar]
- 45. Dumont P, Leu JI, Della Pietra AC 3rd, George DL, Murphy M (2003) The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 33:357–365. [DOI] [PubMed] [Google Scholar]
- 46. Gaur D, Arora S, Mathur M, Nath N, Chattopadhaya TK, et al. (1997) High prevalence of p53 gene alterations and protein overexpression in human esophageal cancer: correlation with dietary risk factors in India. Clin Cancer Res 3:2129–2136. [PubMed] [Google Scholar]
- 47. Mir MM, Dar NA, Gochhait S, Zargar SA, Ahangar AG, et al. (2005) p53 mutation profile of squamous cell carcinomas of the esophagus in Kashmir (India): a high-incidence area. Int J Cancer 116:62–68. [DOI] [PubMed] [Google Scholar]
- 48. Groenen PJ, Busink E (1988) Alkylating activity in food products–especially sauerkraut and sour fermented dairy products–after incubation with nitrite under quasi-gastric conditions. Food Chem Toxicol 26:215–225. [DOI] [PubMed] [Google Scholar]
- 49. Walker EA, Castegnaro M, Garren L, Toussaint G, Kowalski B (1979) Intake of volatile nitrosamines from consumption of alcohols. J Natl Cancer Inst 63:947–951. [PubMed] [Google Scholar]
- 50. Larsson SC, Bergkvist L, Wolk A (2006) Processed meat consumption, dietary nitrosamines and stomach cancer risk in a cohort of Swedish women. Int J Cancer 119:915–919. [DOI] [PubMed] [Google Scholar]
- 51. Abedi-Ardekani B, Kamangar F, Sotoudeh M, Villar S, Islami F, et al. (2011) Extremely high Tp53 mutation load in esophageal squamous cell carcinoma in Golestan Province, Iran. PLoS One 6:e29488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Putz A, Hartmann AA, Fontes PR, Alexandre CO, Silveira DA, et al. (2002) TP53 mutation pattern of esophageal squamous cell carcinomas in a high risk area (Southern Brazil): role of life style factors. Int J Cancer 98:99–105. [DOI] [PubMed] [Google Scholar]
- 53. Bergamaschi D, Gasco M, Hiller L, Sullivan A, Syed N, et al. (2003) p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell 3:387–402. [DOI] [PubMed] [Google Scholar]
- 54. Schneider-Stock R, Mawrin C, Motsch C, Boltze C, Peters B, et al. (2004) Retention of the arginine allele in codon 72 of the p53 gene correlates with poor apoptosis in head and neck cancer. Am J Pathol 164:1233–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hu Y, McDermott MP, Ahrendt SA (2005) The p53 codon 72 proline allele is associated with p53 gene mutations in non-small cell lung cancer. Clin Cancer Res 11:2502–2509. [DOI] [PubMed] [Google Scholar]
- 56. Marin MC, Jost CA, Brooks LA, Irwin MS, O'Nions J, et al. (2000) A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet 25:47–54. [DOI] [PubMed] [Google Scholar]
- 57. Tada M, Furuuchi K, Kaneda M, Matsumoto J, Takahashi M, et al. (2001) Inactivate the remaining p53 allele or the alternate p73? Preferential selection of the Arg72 polymorphism in cancers with recessive p53 mutants but not transdominant mutants. Carcinogenesis 22:515–517. [DOI] [PubMed] [Google Scholar]
- 58. Cai YC, Yang GY, Nie Y, Wang LD, Zhao X, et al. (2000) Molecular alterations of p73 in human esophageal squamous cell carcinomas: loss of heterozygosity occurs frequently; loss of imprinting and elevation of p73 expression may be related to defective p53. Carcinogenesis 21:683–689. [DOI] [PubMed] [Google Scholar]
- 59. Miyaki M, Iijima T, Yasuno M, Kita Y, Hishima T, et al. (2002) High incidence of protein-truncating mutations of the p53 gene in liver metastases of colorectal carcinomas. Oncogene 21:6689–6693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparative analysis of distribution of ESCC TP53 mutations in India with the IARC TP53 database at nucleotide level (Panel A), DNA level (Panel B) and protein level (Panel C). Unclassified and silent mutations reported in the database were omitted from this analysis. The p.V143_W146delinAV mutation was not included in the analysis (in Panel C) due to complexity of its effect at protein level.
(EPS)
(DOCX)
(DOC)
(DOCX)
(DOC)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.