Abstract
Nudix hydrolases comprise a large gene family of twenty nine members in Arabidopsis, each containing a conserved motif capable of hydrolyzing specific substrates like ADP-glucose and NADH. Until now only two members of this family, AtNUDX6 and AtNUDX7, have been shown to be involved in plant immunity. RPP4 is a resistance gene from a multigene family that confers resistance to downy mildew. A time course expression profiling after Hyaloperonospora arabidopsidis inoculation in both wild-type (WT) and the rpp4 mutant was carried out to identify differentially expressed genes in RPP4-mediated resistance. AtNUDX8 was one of several differentially expressed, downregulated genes identified. A T-DNA knockout mutant (KO-nudx8) was obtained from a Salk T-DNA insertion collection, which exhibited abolished AtNUDX8 expression. The KO-nudx8 mutant was infected separately from the oomycete pathogen Hpa and the bacterial pathogen Pseudomonas syringae pv. maculicola ES4326. The mutant displayed a significantly enhanced disease susceptibility to both pathogens when compared with the WT control. We observed a small, stunted phenotype for KO-nudx8 mutant plants when grown over a 12/12 hour photoperiod but not over a 16/8 hour photoperiod. AtNUDX8 expression peaked at 8 hours after the lights were turned on and this expression was significantly repressed four-fold by salicylic acid (SA). The expression of three pathogen-responsive thioredoxins (TRX-h2, TRX-h3 and TRX-h5) were downregulated at specific time points in the KO-nudx8 mutant when compared with the WT. Furthermore, KO-nudx8 plants like the npr1 mutant, displayed SA hypersensitivity. Expression of a key SA biosynthetic gene ICS1 was repressed at specific time points in the KO-nudx8 mutant suggesting that AtNUDX8 is involved in SA signaling in plants. Similarly, NPR1 and PR1 transcript levels were also downregulated at specific time points in the KO-nudx8 mutant. This study shows that AtNUDX8 is involved in plant immunity as a positive regulator of defense in Arabidopsis.
Introduction
Plants are sessile organisms that evolved remarkable signaling pathways in order to cope with several abiotic and biotic stresses such as pathogen attack. In one branch of the plant immune system there are nucleotide binding leucine-rich repeat (NB-LRR) proteins in the cell that recognize a plethora of pathogen effectors from several kingdoms and activate a cascade of signaling pathways ultimately leading to effector triggered immunity (ETI) [1], [2]. In Arabidopsis Columbia ecotype (Col-0), the resistance (R) gene RPP4 confers resistance to the oomycete pathogen Hyaloperonospora arabidopsidis (Hpa) isolates Emwa1 and Emoy2 (downy mildew), which involves multiple defense signaling components, including the NPR1 protein among others [3].
Some plant immune responses are associated with NPR1 protein conformational changes induced by redox levels [4]. NPR1 is a well-known master regulator of pathogenesis related (PR) gene expression and salicylic acid (SA) signaling [5], [6]. NPR1 protein resides in the cytoplasm as an oligomer maintained by disulphide bonds that are sensitive to redox changes [4]. Reduction of disulphide bonds cause NPR1 monomer migration to the nucleus and activation of PR gene expression. NPR1 also works upstream of SA suppressing expression of ICS1 and inhibiting SA biosynthesis in a negative feedback loop [7].
Thioredoxins (TRXs) are small cytosolic proteins that act as disulphide reductase proteins [8], [9]. In Arabidopsis, there are eight cytosolic types of TRXs, three of which have been related to pathogen attack: TRX-h2, TRX-h3 and TRX-h5 [4]. Of these, TRX-h3 is the only one constitutively expressed. Reduction of the NPR1 oligomer-to-monomer reaction is catalyzed by cytosolic TRXs by the reduction of their intermolecular disulphide bonds. Incubation of NPR1-GFP protein extracts with recombinant TRX-h5 protein can increase the amount of NPR1-GFP monomer [4].
The Nudix (nucleoside diphosphates linked to some moiety X) gene family comprises 29 homologs in Arabidopsis and is well conserved across several species and all domains of life (Eukaryotes, Prokaryotes and Archaea) [10]. Its members contain a conserved Nudix box motif “GX5EX7REVXEEXGU” that catalyzes the hydrolytic breakdown of nucleoside diphosphates linked to other moieties by cleavage of chemical bonds. Nudix hydrolases have been shown to catalyze the hydrolysis of nucleoside diphosphates such as nucleotide sugars (ADP-glucose) [11], [12], [13] and pyridine nucleotides such as NADH, NADPH and 8-oxo-GTP [13], [14], [15]. Nucleoside diphosphates are key metabolic intermediates and signaling molecules that are often toxic to the cell. It has been proposed that Nudix hydrolases may have a role as house cleaning enzymes by getting rid of toxic, excessive nucleoside diphosphate and hence maintaining normal cellular homeostasis [12], [16], [17].
Previous phylogenetic analysis of the Nudix gene family in Arabidopsis has further divided these into four subfamilies. AtNUDX8 appears in the fibroblast growth factor type Nudix enzyme (FGFTNE) subfamily in a monophyletic clade. All members in this clade have hydrolase activity towards ADP-ribose and NADH, which is important for defense responses in plants [18]. Most Nudix family members are mainly present in the cytosol (AtNUDX1 to 11 and 25) but there are also organelle-type Nudix hydrolases which localize to the chloroplast and mitochondrion (AtNUDX14, 15, 19, 23, 26 and 27) in Arabidopsis [12]. Enzyme activity has been tested in all Nudix hydrolases (AtNUDX1-AtNUDX27) in Arabidopsis. Only four genes (AtNUDX2, AtNUDX6, AtNUDX7 and AtNUDX10) showed pyrophosphohydrolase activity towards both ADP-rib and NADH in vitro [11]. The involvement of reactive oxygen species (ROS) such as H2O2 in plant defense against bacterial and fungal pathogens was previously demonstrated [19]. Intensive ROS production from NADH pools during oxidative burst might be required by several defense responses such as lignin formation, antimicrobial action and hypersensitive response during systemic acquired resistance [19]. Nudix enzymes may also play a significant regulatory role in photosynthesis in the reduction of O2 to O2 +, which is accompanied by H2O2 and OH production [17]. Indeed, mutations in house cleaning enzymes and oxidation protective enzymes could generate an imbalance in cellular homeostasis affecting several outputs in plant signaling such as pathogen defense and hormone signaling [20].
Other studies have shown that both AtNUDX6 and AtNUDX7 genes are involved in plant defense. AtNUDX6 is directly involved in the plant immune response as a positive regulator of NPR1-mediated defense [21]. KO-nudx6 plants showed suppressed levels of several SA-induced, NPR1-dependent genes and also TRX-h5 involved in SA-induced NPR1 activation. KO-nudx6 mutant also displays increased NADH levels [21]. Additionally AtNUDX6 was shown to be involved in SA signaling and its transcript levels induced by SA [21]. AtNUDX7 on the other hand is involved in plant immunity as a negative regulator of defense response. A previously studied T-DNA knockout line Atnudt7-1 exhibited an array of pleiotropic effects such as increased resistance to bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000 (avrRpt2) [22] and oomycete pathogen Hyaloperonospora arabidopsidis [23], microscopic cell death and constitutive expression of PR genes [14]. Atnudt7-1 and Atnudt7-2 knockout lines displayed a small stunted phenotype influenced by edaphic factors and high NADH and ADP-ribose levels [14]. AtNUDX7 transcript levels are SA insensitive unlike its SA-induced homolog AtNUDX6 but its transcript was shown to be induced upon Pst infection [22].
In this study, we investigated the functional role of the AtNUDX8 gene (AT5G47240), a previously uncharacterized member of the Nudix hydrolase family, in defense response upon pathogen attack in Arabidopsis. In order to perform this study, we first identified an Arabidopsis T-DNA insertion knockout mutant for AtNUDX8 (KO-nudx8) and studied disease phenotypes in response to pathogen infection and transcriptional changes observed in pathogen responsive TRXs and NPR1-dependent genes by quantitative real-time PCR. The results obtained in this study indicate that the AtNUDX8 gene is a novel element in plant immune responses against pathogen attack.
Materials and Methods
Plant Material
Seeds of WT (Col-0) and KO-nudx8 mutant from Salk T-DNA insertion collection (SALK_092325.55.50.x) were sown in pots containing Metro-Mix 360 soil for 3–4 weeks in a growth chamber. Plants were maintained at 22°C constant temperature and 50% relative humidity. Plants were grown in long day light conditions (16/8 hours of light/dark) for most experiments and a 12/12 hour photoperiod for the small stunted phenotype. For time course, quantitative real-time PCR (qRT-PCR) analysis, three to four leaves from at least three individual plants were collected from the time the lights were turned on in the growth chambers (Zeitgeber time, 0 hour) and five time points were collected (0, 4, 8, 16 and 24 hours). The samples were immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction.
Molecular analysis of insertion lines
We used a PCR approach to confirm the T-DNA insertion in the AtNUDX8 (AT5G47240) gene using T-DNA left border LBb1 primer (5′-GCGTGGACCGCTTGCTGCAACT-3′) and gene specific primers LP (5′-GCAATATCTTCGAGCAGCAAC-3′) and RP (5′-TGTTACATTTACCTTTGCGGC-3′). Positive, homozygous T-DNA insertion lines for AtNUDX8 were allowed to grow for seed collection.
Pseudomonas infection assay
Whole leaves of three-week-old plants were infiltrated with a bacterial suspension (OD600 = 0.0002) of Pseudomonas syringae pv. maculicola (Psm) ES4326 in 10 mM of MgSO4. We collected infected tissues three days after the infection. Leaf-disks of 28 mm diameter were collected per plant from 8–12 plants. Then, leaf-discs were placed in tubes containing metal beads and 500 µL of MgSO4 and ground using Geno-Grinder (SPEX). Suspensions of 20 µL were transferred to a plate containing 180 µL of MgSO4 and 10× serial dilutions were made. Aliquots of 10 µL for each serial dilution were then plated on KB medium with antibiotics and incubated at 30°C for 2–3 days after which colony forming units were counted. Statistical analysis were performed using the Student's t test as described previously [24].
Hpa Emwa1 infection assay
Arabidopsis seedlings were grown for 10 days at 18°C and 80–100% relative humidity before Hpa infection at dawn (0 hour) of growth chamber used. Ten-day-old Arabidopsis plants were spray-inoculated (5×105 spores per mL) with an asexual spore suspension of Hpa. Disease phenotypes were scored seven days post-infiltration (dpi) using trypan blue staining. A total of 60 infected cotyledons per genotype were used for sporangiophore (SPP) counting and statistical analysis were performed as described previously [25].
Time course SA treatment
Three-week-old plants were sprayed with 1 mM of salicylic acid and their leaves were collected at five different time points: when lights were turned on (0 hour), 4, 8, 16 and 24 hours. The leaves were immediately frozen in liquid nitrogen and stored at −80°C for subsequent RNA extraction by Trizol method and cDNA synthesis followed by qRT-PCR time course analysis. Mock-treated control plants were sprayed with water and collected at the same time points as SA-treated ones.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using Trizol reagent (Ambion), and at least 2 µg of RNA were used for cDNA synthesis using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Amplicons were amplified from cDNA using gene specific primers (see Table 1), and their relative gene expression values were normalized to the ACTIN2 gene (invariant control) using the ΔΔCT method [26]. Quantitative real-time PCR (qRT-PCR) was performed in a Mastercycler ep realplex S instrument (Eppendorf) using SYBR Green (Applied Biosystems) to monitor dsDNA synthesis.
Table 1. Primers used for qRT-PCR.
| Primer nomenclature* | Primer sequence (5′-3′) |
| nudx8f | TGTGGAGCAACCGATGATAA |
| nudx8r | CGCAGTACCGATGGCTTAAT |
| trxh3f | CCGTCTTTGCTGACTTAGCC |
| trxh3r | GTTGGCATTGCCTGAACTTT |
| trxh5f | GAATTGCAAGCTGTTGCTCA |
| trxh5r | CACCGACAACACGATCAATG |
| trxh2f | TAATGTGACGGCAATGCCTA |
| trxh2r | TGGCACCAATGATTCTTTCA |
| npr1f | TGCAATTGCTCTCCAACAGCTTCG |
| npr1r | GCGGCTAAAGCGCTCTTGAAGAAA |
| PR1f | CTCATACACTCTGGTGGG |
| PR1r | TTGGCACATCCGAGTC |
| ics1f | AGGTACGAGCTTTTGTCCAGA |
| ics1r | TTGACTTGGTGAACTGCAAA |
| actin2f | GGCAAGTCATCACGATTGG |
| actin2r | CAGCTTCCATTCCCACAAAC |
*f- forward primer; r- reverse primer.
NPR1 protein analysis
Total protein extracts were obtained from four-week-old Arabidopsis plant leaves either treated with SA (24 hours) or non-treated. Plant leaves were flash-frozen in liquid nitrogen and grinded using Geno-Grinder (SPEX). Cell debris was pelleted by centrifugation at 12,000 rpm for 10 min at 4°C to obtain clear protein extracts. Total protein was extracted using plant extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 5 mM EDTA, 0.1% Triton X-100, 0.2% Nonidet P-40, 6 mM beta-mercaptoethanol and 1 protease inhibitor cocktail (Roche) tablet per 100 mL of solution). Protein concentration for the various extracts were measured according to Bradford's method [27]. Aliquots of protein extract were separated on a 12% SDS-PAGE gel [28] and transferred to a nitrocellulose membrane. Western analysis was performed using an antibody against NPR1 protein as described previously [29].
Data analysis
All the measurements were performed with three biological replicates grown independently. Significant differences between data sets were evaluated using the Student's t test with GraphPad Prism software.
Results and Discussion
KO-nudx8 mutant displays a small, stunted photoperiod-dependent phenotype
In order to identify genes involved in RPP4-mediated resistance against Hpa Emwa1, a time course gene expression profiling was performed after Hpa infiltration (0 hour, 2 days post-infiltration, 4 dpi and 6 dpi) for the rpp4 mutant and WT. This microarray data was submitted in NCBI's Gene Expression Omnibus database (GEO, accession number GSE22274) [30].
We found 106 differentially expressed genes between WT and rpp4. The Athena database (http://www.bioinformatics2.wsu.edu/Athena) was used to analyse the promoter regions to identify regulatory cis-acting elements for all the differentially expressed genes. Remarkably, it was identified that the most prominent cis-element present in the promoters of 30 genes analysed was the evening element (EE). The evening element is known to be regulated by the CIRCADIAN CLOCK-ASSOCIATED 1 gene (CCA1), and the fact the EE can be found in the promoter region of some defense genes suggests a link between the circadian clock and plant defense. It was previously reported that CCA1 transcription factor can control expression of several R genes [25], establishing a link between the circadian clock and plant immunity. We obtained T-DNA insertion mutants of these genes and profiled their disease susceptibility phenotype to Hpa infection and Pseudomonas syringae pv. maculicola (Psm) ES4326 followed by functional characterizations of susceptible mutants.
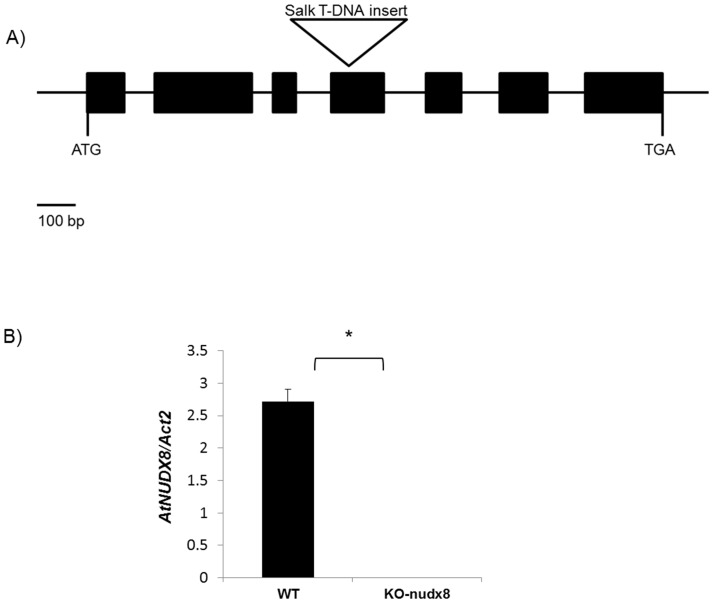
Using PCR we genotyped a Salk T-DNA insertion line (SALK_092325.55.50.x) for the AtNUDX8 gene (AT5G47240) to confirm its homozygosity. The T-DNA insertion was located in exon 4 and position +963 relative to the start codon (Fig. 1A) based on the sequence obtained from the left-border T-DNA primer of SALK_092325.55.50.x (http://signal.salk.edu/). We performed quantitative real-time PCR to measure the expression of AtNUDX8 gene in WT and homozygous KO-nudx8 mutant plants containing the T-DNA insertion, and we demonstrated that the expression of AtNUDX8 was abolished in the Salk mutant line (Fig. 1B).
Figure 1. AtNUDX8 expression is completely abolished in the KO-nudx8 mutant.
A) Diagram showing Salk T-DNA insertion (SALK_092325.55.50.x) position based on the flanking sequence. AtNUDX8 gene has seven exons (black boxes) and six introns. The T-DNA insertion is located in exon 4 and position +963 relative to the start codon. B) qRT-PCR showing relative expression of AtNUDX8 in WT and KO-nudx8 mutant plants where bars represent change in expression of AtNUDX8 transcript in the WT and knockout mutant plants relative to the internal control ACTIN2. The control was selected as a reference gene since it does not vary in the different conditions tested. Data are reported as means ±SD of three independent biological replicates. Asterisk indicates significant difference according to Student's t test (P<0.05).
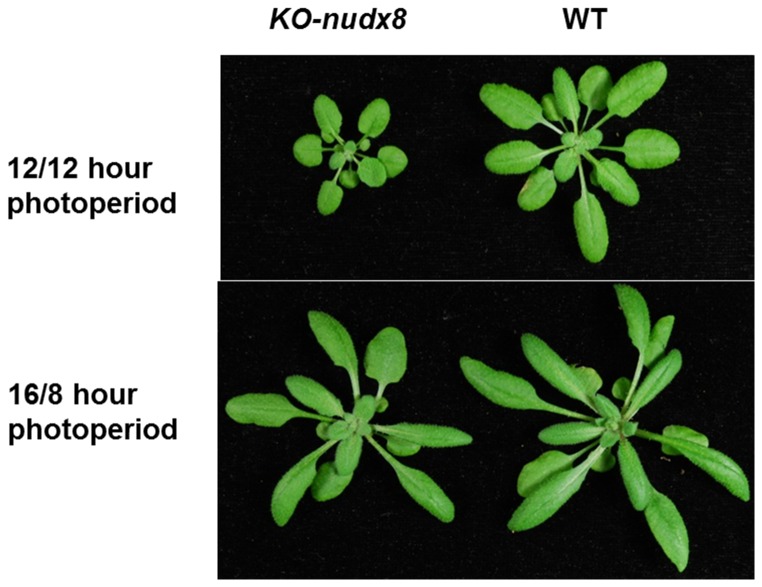
Furthermore, we observed a small stunted phenotype for KO-nudx8 plants when grown on 12/12 hour photoperiod light (Fig. 2) similar to the AtNUDX7 gene. The same phenotype was not observed for short or long day light exposures (8/16 hours or 16/8 hours of light/dark).
Figure 2. AtNUDX8 displays a small stunted phenotype.
Three-week-old WT and KO-nudx8 plants grown over a 12/12 hour (light/dark) and 16/8 hour photoperiod.
KO-nudx8 plants exhibit a significant increase in susceptibility to pathogen infection
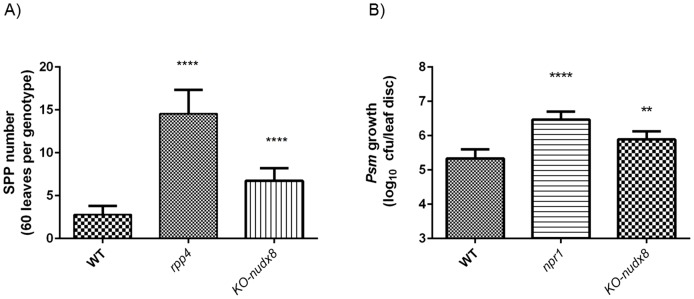
KO-nudx8 plants were significantly more susceptible to infection with oomycete pathogen Hpa Emwa1 in comparison with WT (Fig. 3A). Similarly, KO-nudx8 also exhibited a significant increase in bacterial growth of Psm ES4326 when compared to WT plants (Fig. 3B). Together, these results indicate that AtNUDX8 is involved in plant defense responses upon pathogen attack whereas plants without a functional copy of AtNUDX8 exhibit a disease susceptible phenotype. Moreover, AtNUDX8 transcript levels were found to be downregulated in the rpp4 mutant compared to WT plants from microarray data upon Hpa infection (GEO accession number GSE22274). This result indicates that the AtNUDX8 gene is involved in RPP4-mediated defense against Hpa. In addition to being susceptible to Hpa, we also demonstrated increased disease susceptibility in KO-nudx8 mutant for Psm ES4326 infection, suggesting a common defense mechanism for both types of pathogens analyzed in this study. All these factors indicate that AtNUDX8 acts as a positive regulator of defense in Arabidopsis. Members of the NB-LRR protein class can confer resistance to both Hpa and Psm. The idea that one gene in plant immunity may confer resistance against more than one pathogen type is not new [31] and it has recently been shown that pathogen effectors interact within a limited set of highly connected cellular hubs in plant immunity [32]. There is growing evidence, including for the gene presented in this study, that some defense genes are involved in plant resistance to more than one pathogen such as RPP4 and PgPR10-1 genes amongst others [31], [32].
Figure 3. KO-nudx8 mutant is more susceptible to infection against bacterial and oomycete pathogen than WT plants.
A) KO-nudx8 mutant is more susceptible to Hyaloperonospora arabidopsidis Emwa1 infection than the WT (three-fold change). Sporangiophore (SPP) count after Hpa Emwa1 infection at dawn. Ten-day-old Arabidopsis plants were inoculated (5×105 spores per mL) with asexual spore suspension of Hpa. Disease phenotypes were scored seven days post-infiltration (dpi) after trypan blue staining. B) KO-nudx8 plants are more susceptible to the bacterial pathogen Psm ES4326 than WT (0.6 fold change Log10). Infected tissue was collected three days after infection. All experiments were carried out with at least three biological replicates. Error bars represent 95% confidence intervals of log-transformed data (n = 7). Asterisks indicates significant difference according to Student's t test (P<0.05).
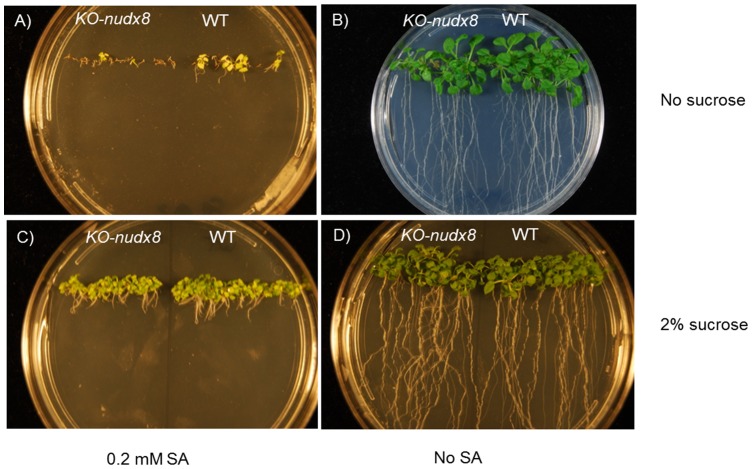
Additionally, in a similar way to the npr1 mutant, KO-nudx8 exhibited hypersensitivity to SA compared with WT control plants. Two-week-old KO-nudx8 Arabidopsis seedlings displayed impaired cotyledon germination in media with 0.2 mM SA compared to the WT control with the same conditions (Fig. 4). Control plants in Murashige and Skoog (MS) medium without SA developed normally. When sucrose was subtracted from MS, the control plants grew normally in MS media without SA, but in 0.2 mM SA lacking sucrose plates there was a marked difference between WT and KO-nudx8 plants (Fig. 4A and 4B) whereby WT plants germinated cotyledons but KO-nudx8 barely germinated or developed cotyledons. These results suggest that the AtNUDX8 gene is involved in SA signaling and also, that in nutrient poor, energy deprived conditions AtNUDX8 is required for growth and detoxification.
Figure 4. KO-nudx8 mutant hypersensitivity to SA compared to WT.
Seeds were plated on MS medium with 0.2 mM SA and without SA. KO-nudx8 and WT seedlings were grown for two weeks on A) MS, 0.2 mM SA, no sucrose, B) MS, no sucrose, C) MS, 0.2 mM SA, 2% sucrose and D) MS, 2% sucrose.
Effect of disruption of the AtNUDX8 gene in pathogen induced thioredoxins TRX-h1, TRX-h2 and TRX-h3
NPR1 conformational changes in the cytosol can influence plant immunity. These conformational changes were shown to be modulated by TRXs that promote NPR1 monomerization [4]. In order to verify the role of the AtNUDX8 gene in thioredoxin-dependent NPR1 oligomer-monomer reduction we analyzed the gene expression of three thioredoxins (TRX-h2, TRX-h3 and TRX-h5) which are known to be upregulated upon pathogen response. The transcript levels of all three pathogen-regulated thioredoxins were significantly reduced in KO-nudx8 plants at specific time points in SA-treated and untreated plants (Fig. 5A, 5B and 5C). This result indicates that AtNUDX8 can modulate expression of the three pathogen induced thioredoxins at specific time points. Further investigation is required to understand how the AtNUDX8 coding enzyme interacts with thioredoxins to modulate NPR1 conformational changes. AtNUDX8 interaction with thioredoxins could be by direct protein-protein interaction or by changing the levels of a metabolic intermediate by substrate hydrolysis that consequently will affect the redox pools in the cell. AtNUDX8 could act as a potential house-cleaning enzyme in the process of removing excessive reactive oxygen species (ROS) towards the end of a pathogen attack event. Mutations in the AtNUDX8 gene is likely to cause changes in redox homeostasis in the cell by affecting levels of a key metabolic intermediate impacting several plant physiological processes such as pathogen defense and SA biosynthesis.
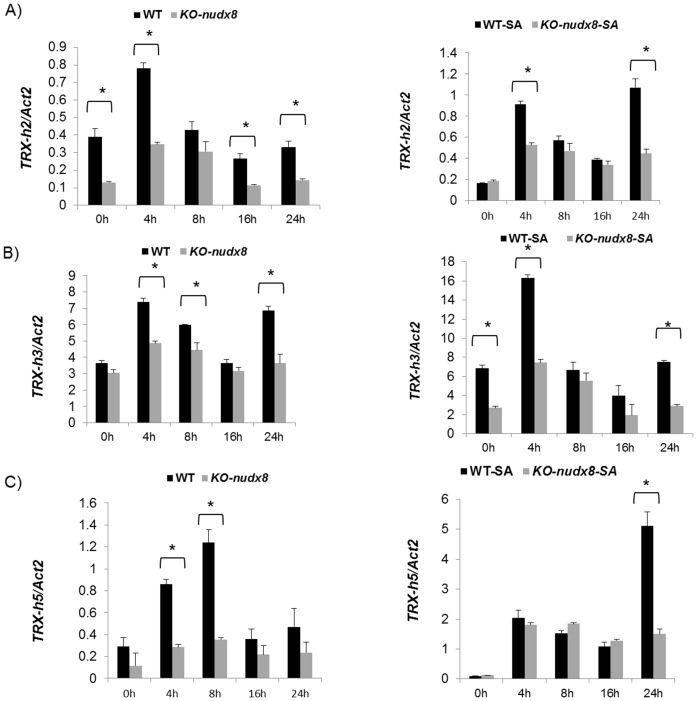
Figure 5. Pathogen responsive thioredoxins TRX-h2, TRX-h3 and TRX-h5 are downregulated in the KO-nudx8 mutant.
Time course expression analysis by qRT-PCR of pathogen responsive thioredoxins in A) TRX-h2, B) TRX-h3 and C) TRX-h5. Relative expression levels were measured from WT control and KO-nudx8 mutant with and without 1 mM SA treatment for each gene tested. Three-week-old plants were sprayed with 1 mM of SA and samples were collected at different time points. Bars represent change in relative expression for each thioredoxin in WT and KO-nudx8 mutant plants (with and without SA) relative to the internal control ACTIN2 used as a reference gene (since it does not vary under the different conditions and treatments tested). Relative expression values were normalized to ACTIN2 mRNA using the ΔΔCT method. Data are reported as means ±SD of three independent biological replicates. Asterisks indicate significant differences according to Student's t test (P<0.05). Zeitgeber time is indicated from the time the lights were turned on at 0 hour for a period of 24 hours.
AtNUDX8 expression peaks at 8 hours and is significantly repressed four-fold by Salicylic Acid
We examined AtNUDX8 transcript levels in a time course expression analysis, and we observed that this gene, unlike other Nudix homologs previously described involved in defense [21], [22], displayed a typical circadian-like pulse gene expression with expression peaking at 8 hours after the lights were turned on (Fig. 6). A time course expression profiling for AtNUDX8 from Diurnal database (http://diurnal.mocklerlab.org/) profiled similar results with AtNUDX8 expression also peaking at 8 hours (Zeitgeber time). Although AtNUDX8 transcript exhibited a rhythmic circadian-like expression, further experiments will be required to validate AtNUDX8 as a bona fide binding site for CCA1 or other circadian clock transcription factors by ChIP qRT-PCR or Yeast-One-hybrid experiments. Additionally, AtNUDX8 transcription was repressed four-fold at 8 hours in response to 1 mM SA treatment compared with mock-treated plants. Regulation of defense gene expression must be controlled between positive and negative regulators in order to avoid an overactivation of defense in plants, which would lead to impaired normal cell activity and loss of redox homeostasis in the cell.
Figure 6. AtNUDX8 expression peaks at eight hours and is repressed by SA.
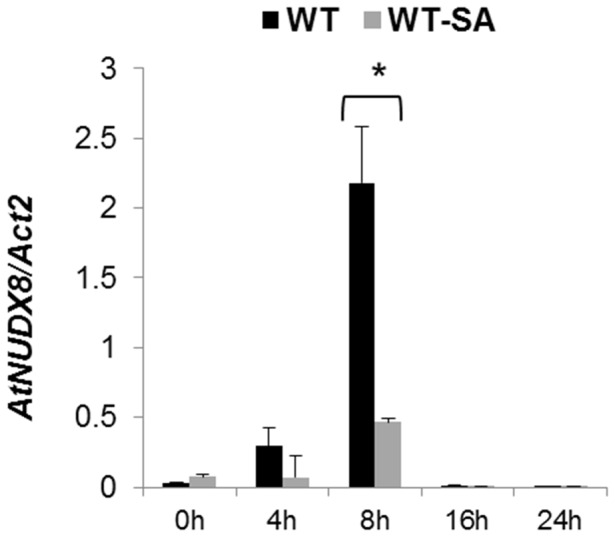
qRT-PCR analysis of AtNUDX8 relative expression levels in the leaves of WT plants without SA treatment and 1 mM SA in a 24 hour time course expression analysis. Three-week-old plants were sprayed with 1 mM of SA and water (mock control), and samples were collected at different time points. Bars represent change in expression of AtNUDX8 transcript in WT plants without SA and SA-treated relative to the internal control ACTIN2 used as reference gene (since it does not vary under the different conditions and treatments tested). Relative expression values were normalized to ACTIN2 mRNA using the ΔΔCT method. Data are reported as means ±SD of three independent biological replicates. Asterisk indicates significant difference according to Student's t test (P<0.05). Zeitgeber time is indicated from the time the lights were turned on at 0 hour for a period of 24 hours.
Effect of disruption of AtNUDX8 in NPR1-mediated defense
To study the role of AtNUDX8 in NPR1-mediated defense and the SA signaling pathway, we performed a time course expression analysis to measure the transcript levels of NPR1, PR1 and the SA biosynthetic gene ICS1 in KO-nudx8 and WT plants. NPR1 transcript levels were significantly downregulated in KO-nudx8 plants at specific time points in SA-induced plants and untreated (no SA) (Fig. 7). PR1 gene expression was also reduced significantly at 8 hours in the KO-nudx8 mutant compared to WT in SA-induced plants (Fig. 8). Similarly, expression of the ICS1 gene was significantly repressed at 4 hours in untreated plants and at 16 and 24 hours in SA-treated plants in the KO-nudx8 mutant plants compared with the WT (Fig. 9). This downregulation of ICS1 suggests that AtNUDX8 is involved in the SA signaling pathway by indirectly acting on ICS1 transcript levels. SA is synthesized from chorismate through isochorismate by the action of ICS1. The fact that AtNUDX8 acts to modulate, not only thioredoxin-dependent NPR1 oligomer-monomer formation, but also ICS1 transcript levels, indicates that the AtNUDX8 gene can impact SA levels in the plant indirectly since NPR1 has been shown to maintain SA levels. NPR1 acts not only downstream to SA but also upstream as a negative regulator of ICS1 gene expression, inhibiting SA [7], [33].
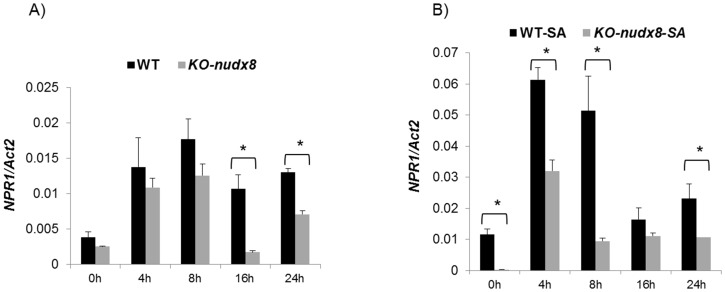
Figure 7. Time course analysis showing NPR1 gene downregulation in KO-nudx8 mutant in relation to WT plants.
qRT-PCR analysis of NPR1 relative expression levels in a 24 hour time course in the leaves of WT plants and KO-nudx8 mutant. A) No SA treatment, B) 1 mM SA treatment. Expression of NPR1 is downregulated in several time points in the KO-nudx8 mutant compared to WT. Three-week-old plants were sprayed with 1 mM of SA and samples were collected at different time points. Bars represent change in expression of NPR1 transcript in the WT and KO-nudx8 plants relative to the internal control ACTIN2 used as reference gene (since it does not vary under the different conditions and treatments tested). Relative expression values were normalized to ACTIN2 mRNA using the ΔΔCT method. Data are reported as means ±SD of three independent biological replicates. Asterisks indicate significant differences according to Student's t test (P<0.05). Zeitgeber time is indicated from the time the lights were turned on at 0 hour for a period of 24 hours.
Figure 8. Time course analysis showing PR1 downregulation at 8 hours in the KO-nudx8 mutant.
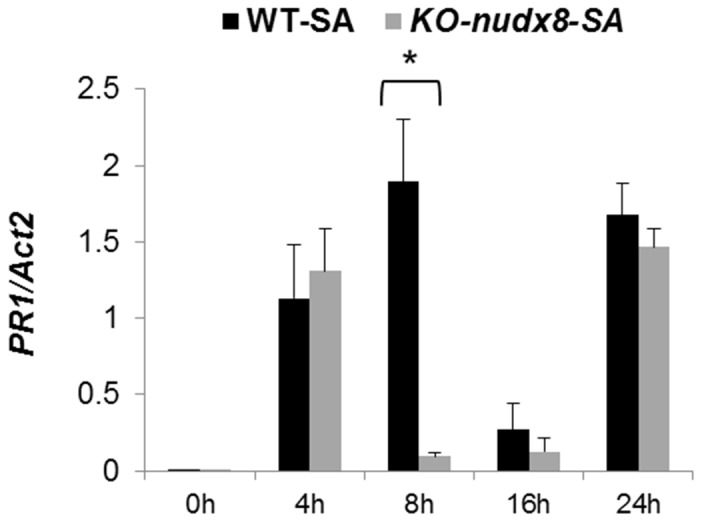
qRT-PCR analysis of PR1 relative expression levels in a 24 hour time course in the leaves of WT plants and KO-nudx8 mutant treated with 1 mM SA. Expression of PR1 was downregulated at 8 hours in the KO-nudx8 mutant compared to WT. Three-week-old plants were sprayed with 1 mM of SA and samples were collected at different time points. Bars represent change in expression of PR1 transcript in the WT and KO-nudx8 plants relative to the internal control ACTIN2 used as reference gene (since it does not vary under the different conditions and treatments tested). Relative expression values were normalized to ACTIN2 mRNA using the ΔΔCT method. Data are reported as means ±SD of three independent biological replicates. Asterisk indicates significant difference according to Student's t test (P<0.05). Zeitgeber time is indicated from the time the lights were turned on at 0 hour for a period of 24 hours.
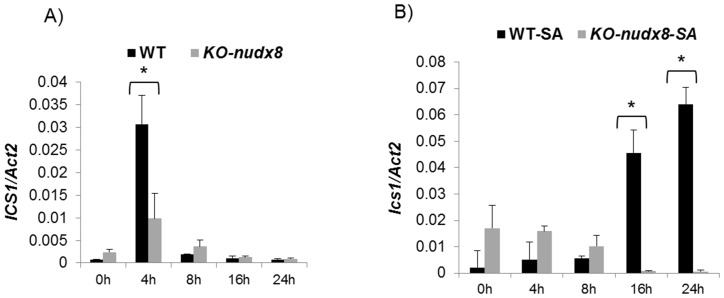
Figure 9. Time course analysis showing ICS1 gene downregulation in the KO-nudx8 mutant in relation to WT plants.
A) No SA treatment, B) 1 mM SA treatment. ICS1 expression peaks at 4 hours (no SA) and is downregulated in the KO-nudx8 mutant in specific time points for both SA-untreated and SA-treated plants. Three-week-old plants were sprayed with 1 mM of SA and leaf samples were collected at different time points. Bars represent change in expression of ICS1 transcript in the WT and KO-nudx8 plants relative to the internal control ACTIN2 used as reference gene (since it does not vary under the different conditions and treatments tested). Relative expression values were normalized to ACTIN2 mRNA using the ΔΔCT method. Data are reported as means ±SD of three independent biological replicates. Asterisk indicates significant difference according to Student's t test (P<0.05). Zeitgeber time is indicated from the time the lights were turned on at 0 hour for a period of 24 hours.
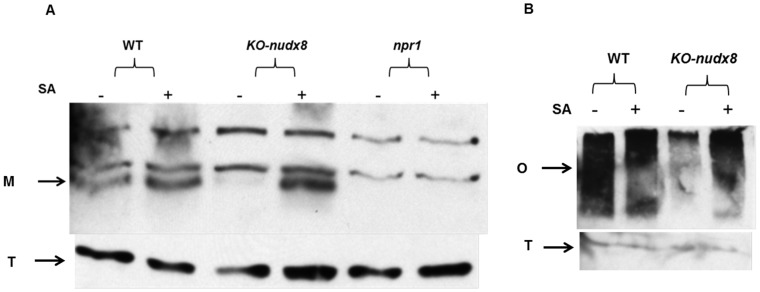
Furthermore, we did not detect NPR1 protein in the KO-nudx8 mutant without SA treatment (Fig. 10A), but we were able to detect with SA treatment. The fact that NPR1 protein was detected in the SA-treated KO-nudx8 mutant indicates that other pathways and genes can compensate for AtNUDX8 loss in SA-dependent NPR1 signaling. A similar result was observed for NPR1 in non-reducing conditions with a marked decrease in NPR1 oligomer for the KO-nudx8 mutant compared to the WT plants without SA treatment (Fig. 10B). No difference in oligomer level was observed upon SA treatment.
Figure 10. NPR1 protein detection in protein extracts from WT and KO-nudx8 mutant plants.
Total protein was extracted from WT, KO-nudx8 and npr1 mutant plants with protein extraction buffer. Leaf samples were collected for each genotype from 1 mM SA treated (+) and water treated (−) samples. Proteins were analyzed by reducing and non-reducing SDS-PAGE and western blotting using anti-NPR1 (A and B respectively). Western blot shows NPR1 monomer protein detection (M) and NPR1 oligomer (O). NPR1 is absent in the KO-nudx8 mutant without SA treatment grown in long day conditions. Total protein loading control (T) for each sample is shown at the bottom. Zeitgeber time is indicated from the time the lights were turned on at 0 hour for a period of 24 hours.
A previous study [21] demonstrated a role for an AtNUDX8 paralog, AtNUDX6, in NPR1-mediated defense and SA biosynthesis. Interestingly AtNUDX6 belongs to the same Nudix subfamily as AtNUDX8, grouping together in a monophyletic clade [18]. It has been shown that both AtNUDX6 and AtNUDX7 genes are positive and negative regulators of defense response respectively [21], [22]. Our findings indicate a positive role for the AtNUDX8 gene in NPR1-mediated defense in Arabidopsis in a similar way to AtNUDX6, both antagonizing AtNUDX7's negative role in defense. Indeed, both AtNUDX8 and AtNUDX6 genes may act synergistically to promote plant immunity through the regulation of NPR1 conformation in the cytosol albeit by slightly different mechanisms. Contrarily to AtNUDX6, which is induced upon SA treatment, AtNUDX8 was repressed up to four-fold by SA when its expression was at its peak at 8 hours (see Fig. 5), indicating slightly different regulatory functions for these family member homologs upon SA signaling. A fine-tuning of both positive and negative regulation of defense related genes is required in order to avoid excessive activation of defense reaction upon pathogen infection.
Furthermore, we demonstrated in this study that AtNUDX8 transcript was significantly induced by ABA treatment (Fig. 11) indicating a possible functional role in abiotic stress responses such as osmotic stress, drought and salinity. Further investigations will be required to assess the involvement of the AtNUDX8 gene in such events. Microarray data from the eFP Arabidopsis browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) corroborates this hypothesis as it shows that AtNUDX8 transcription is upregulated under several abiotic stress parameters such as osmotic stress, salt, drought, genotoxic and wounding.
Figure 11. AtNUDX8 transcript levels are induced by ABA.
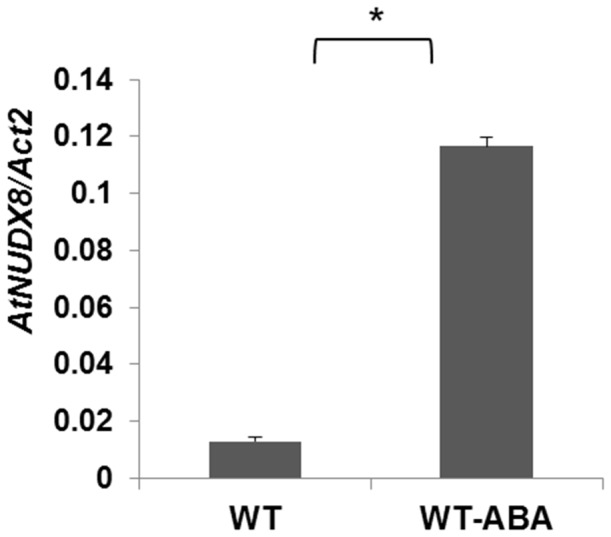
qRT-PCR analysis of AtNUDX8 relative expression levels in the leaves of WT plants treated with 50 µM of ABA (WT-ABA) and untreated (WT). Three-week-old plants were sprayed with 50 µM ABA and samples were collected at different time points. Bars represent change in expression of AtNUDX8 transcript in the untreated (WT) and ABA-treated plants (24 hours) relative to the internal control ACTIN2 used as reference gene (since it does not vary under the different conditions and treatments tested). Relative expression values were normalized to ACTIN2 mRNA using the ΔΔCT method. Data are reported as means ±SD of three independent biological replicates. Asterisk indicates significant difference according to Student's t test (P<0.05).
Conclusions
This work lays the foundation for the study of a novel, previously uncharacterized member of the Nudix hydrolases, AtNUDX8 gene, in plant immunity and SA signaling in Arabidopsis. Plants without a functional copy of the AtNUDX8 gene were significantly more susceptible to infection than WT plants against Hpa and Psm ES4326 pathogen infection, indicating a positive role in plant defense for the AtNUDX8 gene. Additionally, KO-nudx8 plants displayed a small, stunted phenotype when grown over a 12/12 hour photoperiod (light/dark) but not over a 16/8 hour photoperiod. KO-nudx8 plants also displayed a phenotype of SA hypersensitivity compared to the WT control when grown in SA-containing media. All the three pathogen-responsive TRXs (TRX-h2, TRX-h3 and TRX-h5) were downregulated at specific time points in the KO-nudx8 mutant indicating that AtNUDX8 may have an important function in the oligermerization state of NPR1 indirectly by the modulation of pathogen responsive TRXs. We also demonstrated that KO-nudx8 mutant displayed significantly lower NPR1, PR1 and ICS1 transcription levels at specific time points compared with the WT indicating that AtNUDX8 plays a role in NPR1-dependent defense pathway and SA signaling. Collectively, these results demonstrate that the AtNUDX8 gene is involved in RPP4-mediated resistance by the modulation of pathogen responsive TRXs that, in turn, could cause changes in NPR1 reduction state. The precise mechanism of how AtNUDX8 modulates TRXs remains unknown. We speculate that AtNUDX8 is an important element linking redox or energy metabolism and biotic stress signaling by affecting levels of a yet unknown metabolic intermediate by substrate hydrolysis.
AtNUDX8 exhibited a typical circadian-like rhythmic expression peaking at 8 hours after the lights were turned on indicating a possible involvement of AtNUDX8 in light regulation or the circadian clock but further experiments will be necessary to determine AtNUDX8 as a bona fide target for core clock transcription factors such as CCA1 transcription factor. Indeed, out of the three Nudix members shown to be involved in plant immune responses thus far (AtNUDX8, AtNUDX6 and AtNUDX7), only AtNUDX8 has EE binding motifs on its promoter region.
The emerging picture indicates an active role for Nudix hydrolases in plant immunity and future work will be necessary to clarify our understanding of, not only AtNUDX8 gene, but also other Nudix family members in biotic and also abiotic stress such as drought, osmotic stress and wounding amongst others. These results argue for potential biotechnological applications of the AtNUDX8 gene in commercial plant systems for pathogen resistance and possibly abiotic stress tolerance, although further work will be required to assess the functional role of AtNUDX8 in abiotic stress responses. Future experiments such as enzyme activity assays will be important to know which substrate the AtNUDX8 enzyme affects. Global expression profile studies between KO-nudx8 mutant and WT will help to identify differentially expressed genes and pathways regulated by AtNUDX8. Additionally, differences in metabolite signatures detected by mass spectrometry between KO-nudx8 and WT plants will be useful in helping to identify which cell metabolites are affected by the AtNUDX8 enzyme.
Acknowledgments
We would like to thank all members of the Dong lab for the assistance with this work, especially Dr. Wei Wang and Dr. Shui Wang for helpful discussions. Also, Dr. Clarissa Boschiero, Mun Keat Looi and Chistopher O'Mara for critical review of the manuscript.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
This work was supported by the Hargitt Fellowship. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jones JDG, Dangl JL (2006) The plant immune system. Nature 444:323–329. [DOI] [PubMed] [Google Scholar]
- 2. Spoel SH, Dong X (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12:89–100. [DOI] [PubMed] [Google Scholar]
- 3. Van der Biezen EA, Freddie CT, Kahn K, Parker JE, Jones JD (2002) Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signaling components. Plant J 29:439–451. [DOI] [PubMed] [Google Scholar]
- 4. Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, et al. (2008) Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321:952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, et al. (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486:228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mou Z, Fan W, Dong X (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113:935–944. [DOI] [PubMed] [Google Scholar]
- 7. Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 417:562–565. [DOI] [PubMed] [Google Scholar]
- 8. Gelhaye E, Rouhier N, Navrot N, Jacquot JP (2005) The plant thioredoxin system. Cell Mol Life Sci 62:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sweat TA, Wolpert TJ (2007) Thioredoxin h5 is required for victorin sensitivity mediated by a CC-NBS-LRR gene in Arabidopsis . Plant Cell 19:673–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kraszewska E (2008) The plant Nudix hydrolase family. Acta Biochim Pol 55:663–771. [PubMed] [Google Scholar]
- 11. Muñoz FJ, Baroja-Fernández E, Morán-Zorzano MT, Alonso-Casajús N, Pozueta-Romero J (2006) Cloning, expression and characterization of a Nudix hydrolase that catalyzes the hydrolytic breakdown of ADP-glucose linked to starch biosynthesis in Arabidopsis thaliana. Plant Cell Physiol 47:926–934. [DOI] [PubMed] [Google Scholar]
- 12. Bessman MJ, Frick DN, O'Handley SF (1996) The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem 271:25059–25062. [DOI] [PubMed] [Google Scholar]
- 13. Ogawa T, Yoshimura K, Miyake H, Ishikawa K, Ito D, et al. (2008) Molecular characterization of organelle-type Nudix hydrolases in Arabidopsis . Plant Physiol 148:1412–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jambunathan N, Penaganti A, Tang Y, Mahalingam R (2010) Modulation of redox homeostasis under suboptimal conditions by Arabidopsis nudix hydrolase 7. BMC Plant Biol 10:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoshimura K, Ogawa T, Ueda Y, Shigeoka S (2007) AtNUDX1, an 8-oxo-7,8-dihydro-2′-deoxyguanosine 5′-triphosphate pyrophosphohydrolase, is responsible for eliminating oxidized nucleotides in Arabidopsis . Plant Cell Physiol 48:1438–1449. [DOI] [PubMed] [Google Scholar]
- 16. Xu WL, Dunn CA, Jones CR, D'Souza G, Bessman MJ (2004) The 26 nudix hydrolases of Bacillus cereus, a close relative of Bacillus anthracis. J Biol Chem 279:24861–24865. [DOI] [PubMed] [Google Scholar]
- 17. Ogawa T, Ueda Y, Yoshimura K, Shigeoka S (2005) Comprehensive analysis of cytosolic Nudix hydrolases in Arabidopsis thaliana . J Biol Chem 280:25277–25283. [DOI] [PubMed] [Google Scholar]
- 18. Gunawardana D, Likic V, Gayler KR (2009) A comprehensive bioinformatics analysis of the Nudix superfamily in Arabidopsis thaliana. Comp Funct Genomics 2009:820381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patykowski J, Urbanek H (2003) Activity of Enzymes Related to H2O2 Generation and metabolism in leaf apoplastic fraction of tomato leaves infected with Botrytis cinerea . J Phytopathol 151:153–161. [Google Scholar]
- 20. Volkert MR, Elliott NA, Housman DE (2000) Functional genomics reveals a family of eukaryotic oxidation protection genes. Proc Natl Acad Sci USA 97:14530–14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ishikawa K, Yoshimura K, Harada K, Fukusaki E, Ogawa T, et al. (2010) AtNUDX6, an ADP-ribose/NADH pyrophosphohydrolase in Arabidopsis, positively regulates NPR1-dependent salicylic acid signaling. Plant Physiol 152:2000–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jambunathan N, Mahalingam R (2006) Analysis of Arabidopsis growth factor gene 1 (GFG1) encoding a nudix hydrolase during oxidative signaling. Planta 224:1–11. [DOI] [PubMed] [Google Scholar]
- 23. Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, et al. (2006) Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18:1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Durrant WE, Wang S, Dong X (2007) Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc Natl Acad Sci USA 104:4223–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang W, Barnaby JY, Tada Y, Li H, Tör M, et al. (2011) Timing of plant immune responses by a central circadian regulator. Nature 470:110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. [DOI] [PubMed] [Google Scholar]
- 27. Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. [DOI] [PubMed] [Google Scholar]
- 28. Laemmli UK (1970) Cleavage of structural proteins during the assembly of the Head of Bacteriophage T4. Nature 227:680–685. [DOI] [PubMed] [Google Scholar]
- 29. Fan W, Dong X (2002) In vivo interaction between NPR1 and Transcription Factor TGA2 leads to Salicylic Acid-Mediated gene activation in Arabidopsis . Plant Cell 14:1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Edgar R, Domrachev M, Lash A (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee OR, Kim Y-J, Balusamy SRD, Khorolragchaa A, Sathiyaraj G, et al. (2012) Expression of the ginseng PgPR10-1 in Arabidopsis confers resistance against fungal and bacterial infection. Gene 506:85–92. [DOI] [PubMed] [Google Scholar]
- 32. Mukhtar MS, Carvunis A-R, Dreze M, Epple P, Steinbrenner J, et al. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333:596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang X, Chen S, Mou Z (2010) Nuclear localization of NPR1 is required for regulation of salicylate tolerance, isochorismate synthase 1 expression and salicylate accumulation in Arabidopsis . J Plant Physiol 167:144–148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.