The authors demonstrated that retinal pigmented epithelial (RPE) cells can be directly reprogrammed to a neuronal fate by introduction of SOX2 protein. The RPARPAR-mediated delivery of SOX2 alone was sufficient to allow cell lineage reprogramming of both fetal and stem cell-derived RPE cells to become functional neurons.
Keywords: Lineage reprogramming, Protein transduction, Retinal pigmented epithelium, Neuron, Cell penetrating peptide
Abstract
Somatic cells can be reprogrammed to an altered lineage by overexpressing specific transcription factors. To avoid introducing exogenous genetic material into the genome of host cells, cell-penetrating peptides can be used to deliver transcription factors into cells for reprogramming. Position-dependent C-end rule (CendR) cell- and tissue-penetrating peptides provide an alternative to the conventional cell-penetrating peptides, such as polyarginine. In this study, we used a prototypic, already active CendR peptide, RPARPAR, to deliver the transcription factor SOX2 to retinal pigmented epithelial (RPE) cells. We demonstrated that RPE cells can be directly reprogrammed to a neuronal fate by introduction of SOX2. Resulting neuronal cells expressed neuronal marker mRNAs and proteins and downregulated expression of RPE markers. Cells produced extensive neurites and developed synaptic machinery capable of dye uptake after depolarization with potassium. The RPARPAR-mediated delivery of SOX2 alone was sufficient to allow cell lineage reprogramming of both fetal and stem cell-derived RPE cells to become functional neurons.
Introduction
Somatic cell lines can be reprogrammed to various cell lineages by overexpressing one or a set of defined reprogramming factors, usually transcription factors specific to target cell lineages [1–6]. For example, human fibroblasts have been reprogrammed directly to neurons via expression of BRN2, ASCL1, MYT1L, and NEUROD1 [5]. To obtain constitutive expression of reprogramming factors, retroviral or lentiviral vectors have been used that integrate into the genome of host cells. However, this can lead to insertional mutagenesis, resulting in tumorigenesis or genomic instability [7, 8]. The use of viral vectors in cell lineage reprogramming would be unsuitable for clinical applications [9]. Although nonintegrating adenoviral and episomal vectors have been used in reprogramming [10–12], there is still a small chance of transgene integration [13]. To avoid introducing exogenous genetic material into the genome of host cells, cell-penetrating peptides such as polyarginine, have been used to deliver transcription factors into cells for the purpose of reprogramming [14, 15], although the frequency of conversion is very low. There is a need for improved methods of protein-mediated reprogramming. Newly discovered C-end rule (CendR) cell- and tissue-penetrating peptides exhibit unique properties suitable for lineage reprogramming [16]. The CendR motif must be exposed at the C-terminus to activate cell internalization and tissue penetration, and the activation of a cryptic CendR motif by proteolysis can be engineered to take place in specific tissues. Several tumor-specific CendR peptides, including iRGD, LyP-1, and iNGR, have been identified and used in tumor-specific drug delivery [17–20]. The cell internalization of CendR peptides requires cell surface receptors neuropilin-1 (NRP1) and neuropilin-2 (NRP2) [16, 20]. In this study, we used RPARPAR, a CendR peptide that binds to the NRPs without activation and internalizes into several cell types [16, 20].
Retinal pigmented epithelial (RPE) cells are adjacent to the neural retina and have the potential to serve as a source of neurons for the treatment of neurodegenerative ocular diseases such as age-related macular degeneration, retinitis pigmentosa, or glaucoma. RPE cells are derived from the anterior neural plate and have been shown to retain some plasticity because they are capable of transdifferentiation to alternative fates [21, 22]. Previous studies have shown that chicken RPE cells could be reprogrammed to a neuronal state via expression of SOX2 [23], although human RPE cells have not been investigated.
SOX2 is a transcription factor that plays important roles in the determination of multiple cell lineages, including the presumptive neuroectoderm, sensory placodes, brachial arches, gut endoderm, and primordial germ cells [24–26]. SOX2 is considered a key factor of neural commitment, and this is supported by high-level SOX2 expression suppressing other lineage-determination factors, such as brachyury, during the earliest stage of embryonic differentiation toward the neural lineage in vivo [27, 28]. During development of the central nervous system and peripheral nervous system, SOX2 controls the proliferation and differentiation of fetal neural progenitor cells [29–31]. Expression of SOX2 is essential for neural progenitor cell proliferation and differentiation in the retina [32]. Overexpression of SOX2 promotes central nervous system progenitor cells, whereas deficiency of SOX2 results in cell-cycle exit followed by neuronal determination [33]. Studies of SOX2 hypomorphic or knockout mice suggested that SOX2 is required for differentiation of distinct subsets of neuronal cells, such as GABAergic neurons [33, 34]. SOX2 is also one of the Yamanaka factors required for reprogramming of induced pluripotent stem cells [4]. Moreover, a recent study showed that SOX2 can reprogram mouse and human fibroblasts to neural stem cells [35]. SOX2 has been proposed as a master regulator for reprogramming somatic cells to a neural state [36]. Together, these studies strongly suggest the importance of SOX2 in early neural differentiation and later neuronal determination.
We used a prototypic active CendR peptide, RPARPAR, to deliver the transcription factor SOX2 to RPE cells. We showed that RPE cells can be directly reprogrammed to a neuronal fate by introduction of SOX2. The RPARPAR-mediated delivery of SOX2 was sufficient to allow high levels of cell lineage reprogramming of RPE cells, both fetal and stem cell derived, to functional neurons. RPARPAR was more efficient than polyarginine, and the relatively high protein transduction rate suggests that RPARPAR may be potentially suitable for safely delivering exogenous proteins to reprogram cells from one lineage to another.
Materials and Methods
Cell Culture
Human fetal RPE (hfRPE) cells were obtained from the Center of Study of Macular Degeneration, University of California Santa Barbara (UCSB). The hfRPE cells were isolated from fetal eyes of a random donor at 12 weeks of gestation, independently procured by Advanced Bioscience Resources (Alameda, CA). The RPE cells derived from human embryonic stem cell (hESC) line H14 (H14-RPE; WiCell Research Institute, Madison, WI, http://www.wicell.org) were obtained from the Center for Stem Cell Biology and Engineering at UCSB. All RPE cells were cultured on gelatin-coated six-well plates in RPE medium [37]: MEM-α modification (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com) supplemented with fetal bovine serum (5%, 15% for the first 3 days after seeding; HyClone, Logan, UT, http://www.hyclone.com), N1 (1×; Sigma-Aldrich), nonessential amino acids (1×), GlutaMAX-I (2 mM; Invitrogen, Carlsbad, CA, http://www.invitrogen.com), taurine (250 μg/ml; Sigma-Aldrich), triiodothyronine (0.013 μg/l; Sigma-Aldrich), and hydrocortisone (20 ng/ml; Sigma-Aldrich). For each passage, approximately 1 × 106 RPE cells were seeded on one six-well plate. RPE cells were cultured until pigmentation developed (usually 30–45 days after seeding), at which point they were passaged using TrypLE (Invitrogen). For each RPE cell line, a total of 2.0 × 107 passage 1 RPE cells were frozen and stored as stocks. Reprogrammed neurons were cultured on six-well plates or chamber slides coated with mouse laminin (Sigma-Aldrich) in ReNcell NSC maintenance medium (Millipore, Billerica, MA, http://www.millipore.com) without growth factor. Neuronal cells were dissociated using Accutase (Millipore) and replated to allow observation of single cells.
Preparation of Lentivirus Expressing SOX2
Using standard cloning techniques, human SOX2 cDNA was inserted into the pSIN-EF2-puro lentiviral vector. Lentivirus constructs were cotransfected into 293T cells with the packaging plasmids. Supernatants containing lentiviral particles were harvested at 2 days after transfection. The lentivirus was precipitated by centrifugation after adding PEG-IT virus precipitation solution (System Biosciences, Mountain View, CA, http://www.systembio.com). Viral titration was determined by infecting 293T cells with serial-diluted lentivirus, followed with puromycin selection and crystal violet staining.
Flow Cytometry
Human fetal RPE cells were incubated with antihuman-NRP1 mouse monoclonal antibody (Miltenyi Biotec, Bergisch Gladbach, Germany, http://www.miltenyibiotec.com) or anti-NRP2 goat polyclonal antibody (R&D Systems Inc., Minneapolis, MN, http://www.rndsystems.com), followed by incubation with fluorescein-conjugated secondary antibody. Fluorescence intensity was measured using a Guava flow cytometer (Millipore).
Phage-Binding Assay
Human fetal RPE 1914 (fRPE1914), H14-RPE, and melanoma M21 cells were cultured to 60%–90% confluence in 12-well plates and incubated with T7 coliphage expressing the RPARPAR peptide (T7-RPARPAR; 5 × 107 plaque-forming units per milliliter) or polyglycine (7G) for 2 hours at 4°C. Unbound phage was removed by washing with phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA). Cells were lysed with 1% NP-40 in Luria Broth medium. Resultant lysates were titered to determine the number of cell-bound phages.
Reverse Transcription-Polymerase Chain Reaction
Total RNA was prepared using the RNeasy Mini Plus Kit (Qiagen, Hilden, Germany, http://www.qiagen.com). The cDNA was synthesized using the Superscript First-Strand Synthesis System (Invitrogen). Polymerase chain reaction (PCR) was performed using the GoTaq Flexi DNA Polymerase System (Promega, Madison, WI, http://www.promega.com).
Synaptic Activity Assay
Reprogrammed neuronal cells were seeded on laminin-coated chamber slides in neural stem cell medium. The synaptic activities of neuronal cells were stopped by starving them in fresh-made HEPES buffered saline (HBS; 290 mOsm, 110 mM NaCl, 5.4 mM KCl) at 37°C for 60 minutes. The cultured cells were then stimulated for 5 minutes with high KCl-HBS (HBS with 55 mM NaCl and 60 mM KCl) at 37°C. Lipophilic styryl dye FM1-43 (Invitrogen) was added to cells at a final concentration of 1 μm for 1 minute. Cells were washed with PBS five times and visualized with fluorescent microscopy. Phase contrast images were superimposed with corresponding fluorescence images using Photoshop (Adobe Systems Inc., San Jose, CA, http://www.adobe.com).
Immunocytochemistry
Cultured cells were washed with PBS, fixed in 4% paraformaldehyde in PBS at 4°C for 20 minutes, washed in PBS, and incubated in PBS with 0.1% NP-40 and 3% BSA for 1 hour. Samples were then incubated with primary antibodies diluted in 3% BSA, 0.1% NP-40, and PBS at 25°C for 1 hour. After three PBS washes, the samples were incubated with appropriate secondary antibodies diluted in 3% BSA and PBS at 25°C for 1 hour. Nuclei were stained using Hoechst 33342 or DAPI (4′,6-diamidino-2-phenylindole; Sigma-Aldrich). Samples were visualized using a fluorescent microscope (Olympus, Tokyo, Japan, http://www.olympus-global.com). For anti-VAMP2 and anti-CtBP2 antibodies, the incubation was performed overnight at 4°C. For anti-NPY antibody, the incubation of primary antibody was performed in 3%BSA and PBS without NP-40. Primary antibodies are listed in supplemental online Table 1.
Preparation of Recombinant SOX2 Protein
Human SOX2 cDNA was cloned to the pET-15b vector. To generate cell-internalizing SOX2 proteins, the oligonucleotide encoding the CendR peptides RPARPAR or polyarginine with a glycine-serine linker was synthesized (Integrated DNA Technologies) and inserted at the 5′ end of the SOX2 stop codon using site-directed mutagenesis (Stratagene, La Jolla, CA, http://www.stratagene.com). All constructs were confirmed by DNA sequencing. Proteins were expressed in Escherichia coli BL21 (DE3) (Novagen, San Diego, CA, http://www.emdbiosciences.com) after induction at 30°C for 16 hours using MagicMedia E. coli Expression Medium (Invitrogen) according to the manufacturer’s instructions. Protein was purified using Ni-NTA affinity chromatography under denaturing condition and refolded overnight at 4°C in a buffer containing 0.88 M of L-arginine, 55 mM Tris-Cl, 21 mM NaCl, 0.88 mM KCl, 5 mM dithiothreitol, 1 mM reduced glutathione, 1 mM oxidized glutathione, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride. The proteins were dialyzed against PBS pH 7.4 with the addition of 360 mM NaCl and 20% glycerol (weight/volume). The molecular weight of the fusion proteins was analyzed on 4%–20% gradient sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions.
Results
Reprogramming Human RPE Cells to Neurons Using Lentivirus
Because earlier studies showed that chicken RPE cells could be reprogrammed to a neuronal state via expression of SOX2 [23, 38], we sought to determine whether human RPE cells could be reprogrammed in a similar fashion. A lentiviral construct containing a puromycin resistance gene and human SOX2 cDNA was generated. To ensure that any neurons we detected in our reprogramming efforts were derived from RPE cells that had been successfully reprogrammed and not from preexisting, contaminating neurons, we generated homogeneous populations of hfRPE cells or RPE cells derived from the hESC line H14. We expanded these cells using media that favor RPE growth [39] and assayed passage 2 RPE cells for the presence of neural markers. Little or no neuronal or neural stem cell marker expression was detected by immunocytochemical analysis. A small percentage of the RPE cells were positive for nestin or PAX6, consistent with an immature RPE phenotype (supplemental online Fig. 1).
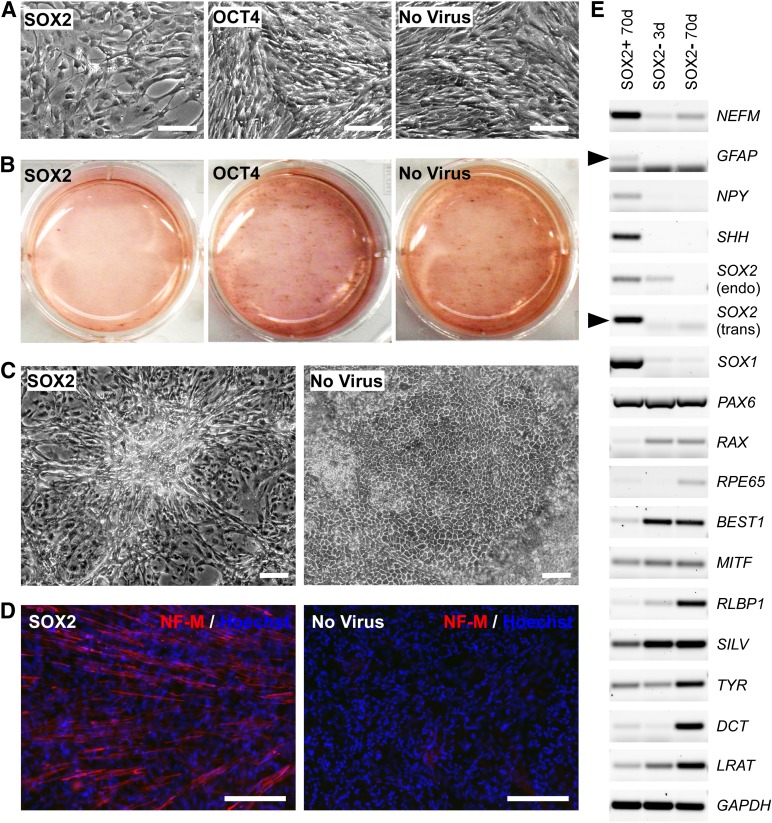
Passage 2 hfRPE cells (1 × 106 cells on a 6-well plate) were transduced with the SOX2 vector at a multiplicity of infection of 2.5–7.5. As controls, RPE cells were either transduced with a vector expressing human OCT4 cDNA, another gene associated with pluripotentcy, or cells were not transduced with virus. RPE cells transduced with SOX2 proliferated more slowly and contained significantly less pigmentation compared with control cells (Fig. 1A, 1B). After 50 days, the SOX2 infected cells exhibited neuron-like morphologies, forming asterisk-shaped cell clusters, whereas control RPE cells (either transduced with OCT4 or not transduced), displayed normal RPE cobblestone-like morphology (Fig. 1C). RPE cells transduced with SOX2 showed robust staining for neurofilament-M relative to control cells (Fig. 1D). Reverse transcription PCR (RT-PCR) analysis revealed that at 70 days after infection, RPE cells transduced with SOX2 showed increased expression of neuronal genes, such as NEFM, NPY, SHH, and SOX1, whereas expression of RPE-specific genes, such as RAX, RPE65, BEST1, MITF, RLBP1, SILV, TYR, DCT, and LRAT, was decreased (Fig. 1E). Expression of the astrocyte/radial glial cell marker GFAP was also increased in the SOX2-transduced cells, indicative of some glial differentiation. Interestingly, the exogenous SOX2 induced expression of the endogenous SOX2 gene (Fig. 1E), which may reflect the presence of some neural progenitor cells.
Figure 1.
Human fetal retinal pigmented epithelial (hfRPE) cells are reprogrammed to neurons using lentiviral constructs. (A): Representative images of hfRPE cells transduced with SOX2 lentivirus and control cell cultures at 5 days after infection, showing proliferation differences. (B): Representative whole-well images showing pigmentation of cell cultures at 50 days after infection. (C): Representative images at 50 days after infection showing hfRPE cells transduced with SOX2 lentivirus exhibiting neuron-like morphology, forming asterisk-shaped cell clusters, whereas control cells maintained RPE cobble-stone morphology. (D): Immunofluorescent staining using anti-NF-M antibody showed a high level of NF-M expression (red) in SOX2-transduced RPE cells at 70 days after infection. (E): Reverse transcription polymerase chain reaction analysis of SOX2-transduced RPE cells at 70 days after infection using neuronal markers and RPE markers (black arrowheads indicate correct size bands, whereas the smaller bands are free primers). Scale bars = 200 μm. Abbreviations: d, days; NF-M, neurofilament-M.
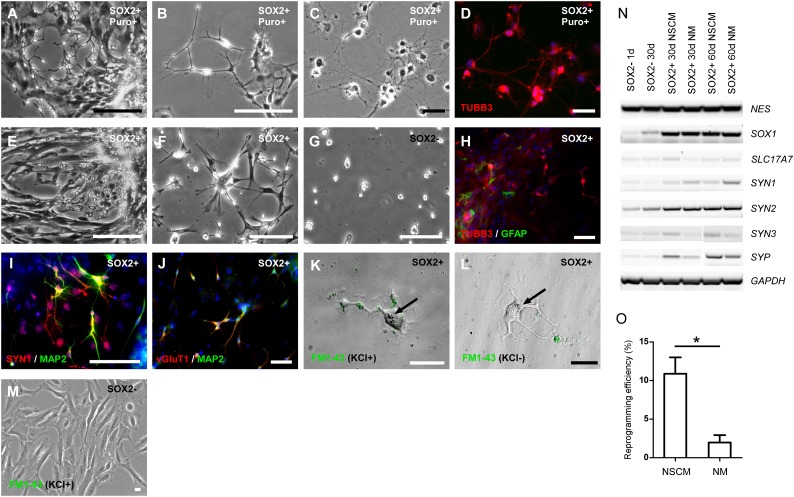
To isolate transduced cells, RPE cells were infected with the virus and subjected to selection for puromycin resistance (Fig. 2; supplemental online Fig. 2). These cells displayed small cell bodies with multiple β3-tubulin-positive neurites radiating outward from the somata, especially if grown on laminin-1 in neural stem cell media (Fig. 2F). This neuronal morphology was observed in multiple SOX2-transduced RPE cultures derived from three hfRPE cell lines (fRPE1914, fRPE1916, and fRPE3909) and one hESC-derived RPE cell line (H14P89-RPE) (supplemental online Fig. 3). In addition, some GFAP-positive cells were observed (Fig. 2H).
Figure 2.
Characterization of induced neurons selected using puromycin resistance or neural stem cell medium after SOX2 lentiviral transduction. (A–D): Human fetal retinal pigmented epithelial (hfRPE) cells (fRPE1914) transduced with SOX2 and selected by puromycin. (A): Representative image of RPE cells transduced with SOX2 that were cultured in RPE medium with puromycin for 15 days. Scale bar = 400 μm. (B): Representative image of RPE cells transduced with SOX2 selected by puromycin for 30 days. Scale bar = 100 μm. (C, D): Representative images of hfRPE cells at 90 days after transduction with SOX2, showing typical neuronal morphology (C) and immunofluorescent staining showing expression of a neuronal marker, TUBB3 (red) (D). Scale bars = 50 μm. (E–J): RPE cells transduced with SOX2 and cultured in neural stem cell (NSC) medium. (E–G): Human fetal RPE cells (fRPE1914). (E): Representative image of RPE cells transduced with SOX2 and cultured in NSC medium for 30 days. Scale bars = 400 μm. (F): RPE cells from Figure 2E attached and proliferating on laminin-coated plates at 24 hours after plating. (G): Fewer normal RPE cells attached on laminin-coated plates at 48 hours after plating. Scale bar = 200 μm. (H–J): Human embryonic stem cell-derived (hESC-derived) RPE cells transduced with SOX2 and cultured in NSC medium. (H): Immunofluorescent staining of hESC-derived RPE cells transduced with SOX2, using antibodies against a neuronal marker (TUBB3, red), and glial marker (GFAP, green) at 40 days after infection. (I): Immunofluorescent staining using antibodies against SYN1 (red) and MAP2 (green) at 60 days after infection. (J): Immunofluorescent staining using antibodies against vGluT1 (red) and MAP2 (green) at 60 days after infection. Scale bars = 50 μm. (K–M): Potassium-stimulated uptake of FM1-43 at 90 days after infection. (K): After addition of potassium, neuron-like cells were labeled with FM1-43 in a punctate pattern. (L): Without potassium stimulation, no FM1-43 uptake was detected. (M): Potassium-stimulated FM1-43 uptake was not detected in parental RPE cells. Black arrows: pigmented particles inside induced neuronal cells. Scale bars = 20 μm. (N): Reverse transcription polymerase chain reaction analysis of SOX2-transduced RPE cells at different time points and in different cell culture media. (O): Comparison of reprogramming efficiency in different cell culture media. Student’s t test, n = 3, p = .01956. ∗, p < .05. Abbreviations: d, days; NSCM, neural stem cell medium; NM, neuronal medium (Neurobasal with N2); Puro, puromycin.
Selection Conditions for Protein-Mediated Reprogramming
Because puromycin selection cannot be used following protein-mediated reprogramming, we tested several cell culture media and substrates for their ability to favor the growth of neuronal cell populations in RPE cells transduced with SOX2. We found that NSC media and mouse laminin-1 substrates selectively favored the growth of neuronal cells (Fig. 2E, 2F, 2H; supplemental online Fig. 2F–2H). These conditions did not fully support the propagation of RPE cells (Fig. 2G). We also compared reprogramming efficiency in two different media, NSC medium and neuronal medium (Neurobasal with N2), and found that the reprogramming efficiency in NSC medium is higher than in neuronal medium (10.89 ± 3.67% vs. 1.97% ± 1.67%, n = 3, p = .01956) (Fig. 2O). RT-PCR analysis indicated that reprogramming NSC medium induced higher expression levels of many neuronal markers, such as SLC17A7 (vGluT1), SYN3 and SYP (Fig. 2N).
To determine whether the reprogrammed cells had functional synaptic machinery, cultures were stimulated with high potassium and stained using the lipophilic styryl dye FM1-43, which is taken up by neurons during the endocytotic recycling of synaptic vesicles (Fig. 2I). Without potassium stimulation, reprogrammed neurons failed to internalize FM1-43 (Fig. 2J). Because both fetal and adult RPE cells can phagocytose exogenous particles, we also tested parental RPE cells using FM1-43. With or without potassium stimulation, RPE cells were unable to internalize FM1-43, indicating an absence of synaptic machinery (Fig. 2K) [40, 41]. Furthermore, pigment particles were observed in some reprogrammed neuronal cells, providing additional supporting evidence that these neuronal cells were reprogrammed from pigmented RPE cells (Fig. 2I, 2J; black arrows).
Reprogramming Human RPE Cells to Neurons Using Recombinant Proteins
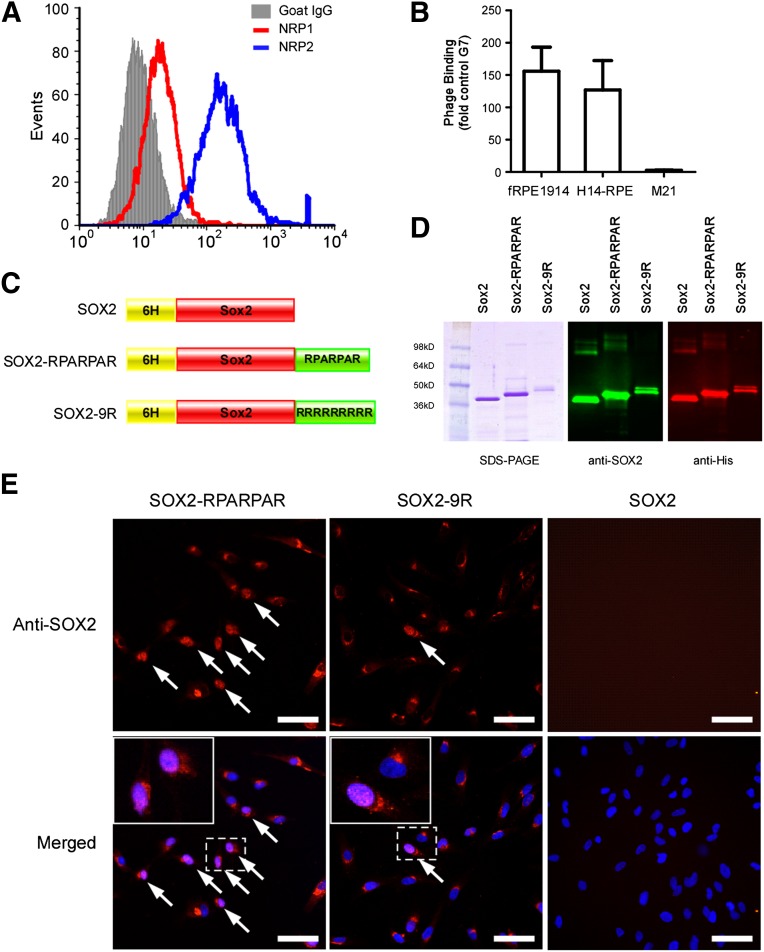
The cellular internalization of the RPARPAR peptide depends on two cell surface receptors, NRP1 and NRP2. To confirm that these receptors are expressed in RPE cells, flow cytometric analysis was performed. Approximately 98% of hfRPE cells expressed high levels of NRP2, and less than 50% of hfRPE cells expressed moderate levels of NRP1 (Fig. 3A). The binding of the RPARPAR peptide to RPE cells was also analyzed using a phage-binding assay. The T7 phage displaying the RPARPAR peptide bound to both hfRPE cells and hESC-derived RPE cells approximately 150-fold higher than to control phage expressing heptaglycine peptide (Fig. 3B). Melanoma M21 cells, which do not express NRP1 and NRP2, did not bind T7-RPARPAR (Fig. 3B).
Figure 3.
Preparation and characterization of SOX2 recombinant proteins. (A): Flow cytometry analysis of the expression of NRP1 and NRP2 in human fRPE1914 cells. (B): Binding of RPARPAR phage to RPE cells. The M21 cell line, which does not express NRP1 or NRP2, was used as a negative control. The results are normalized by the phage expression levels of seven glycines. (C): Schematic of three SOX2 recombinant proteins. (D): Purified recombinant proteins were characterized using SDS-PAGE, Coomassie Blue staining, and immunoblotting. (E): Representative images of human fRPE1914 cells internalizing recombinant proteins fused with different peptides after 16 hours of incubation. Arrows: nucleolus-localized SOX2 protein. Scale bars = 50 μm. Abbreviations: fRPE, fetal retinal pigmented epithelial cells; G7, seven glycines; NRP1, neuropilin-1; NRP2, neuropilin-2; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis.
To generate SOX2 protein fused with the cell-penetrating peptides, human SOX2 cDNA was cloned into a bacterial expression vector, pET-15b, and fused with either the RPARPAR peptide or nona-arginine (9R) at the C-terminus (Fig. 3C). The recombinant proteins were purified under denaturing conditions using 8M urea, refolded, and then subjected to SDS-PAGE analysis. Coomassie Blue staining and immunoblotting showed the expected bands for SOX2 at 36.6 kDa, for SOX2-RPARPAR at 38.9 kDa, and for SOX2-9R at 40.7 kDa (Fig. 3D). To test for internalization, hfRPE cells were incubated with recombinant proteins and immunofluorescent stained for SOX2. Both SOX2-RPARPAR and SOX2-9R proteins were detected in the intracellular space, including cytosol, perinuclear, and nuclear regions (Fig. 3E). This distribution is not consistent with phagocytosis. Interestingly, SOX2-RPARPAR showed higher nuclear localization (20%–40%) than SOX2-9R (about 10%), suggesting that the RPARPAR-fused protein may have an internalization and nuclear localization advantage (Fig. 3E; supplemental online Fig. 5).
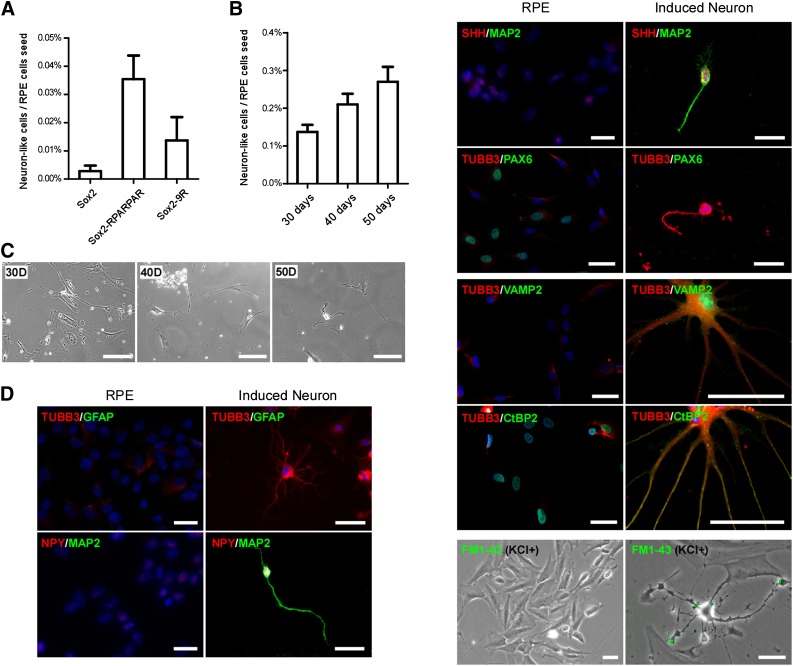
The half-life of recombinant proteins relevant to reprogramming in mammalian cells is about 12–24 hours [14, 15]. In our study, recombinant SOX2 proteins internalized by hfRPE cells were still detectable after 48 hours (supplemental online Fig. 6). In our initial protein reprogramming experiments, recombinant SOX2 proteins were added to RPE cells daily for 30 days, and neuron-like cells were quantified (Fig. 4A). Using the NSC media on laminin-1-coated wells, cells with RPE morphology were lost, but cells with neuron-like morphology persisted. Cultures treated with SOX2-RPARPAR generated approximately 0.04% neuron-like cells, whereas SOX2-9R treatment generated 0.015%. Although some variation was observed between different hfRPE cell lines, SOX2-RPARPAR consistently demonstrated a higher percentage of neuron-like cells. The addition of recombinant protein every other day increased the percentage of neuron-like cells generated by the SOX2-RPARPAR protein to 0.3%, after 50 days of treatment (Fig. 4B). The resulting neuronal phenotype was stable for at least 30 days after cessation of SOX2 protein application.
Figure 4.
Reprogramming human fetal RPE (hfRPE) cells to neurons using recombinant SOX2 proteins. (A): Efficiency of hfRPE cells to be reprogrammed to neuron-like cells after recombinant proteins was added to the media every 24 hours for 30 days. (B): Efficiency of hfRPE cells to be reprogrammed by adding SOX2-RPARPAR recombinant protein every 48 hours for different time courses. (C): Representative images of hfRPE (fRPE1914) cells during reprogramming to neuron-like cells after 30, 40, and 50 days in culture with SOX2-RPARPAR protein. Scale bars = 100 μm. (D): Representative images of hfRPE (fRPE1914) cells reprogrammed to neuron-like cells expressing neuronal markers, but not an RPE marker (PAX6), using SOX2-RPARPAR protein. Scale bars = 50 μm. Abbreviations: D, days; RPE, retinal pigmented epithelial cells.
The resultant cells expressed multiple neuronal markers, including TUBB3, MAP2, NPY, SHH, NES, VAMP2 (synaptobrevin 2), and CtBP2, and had various neuronal morphologies. Conversely, these cells did not express an early RPE marker, PAX6 (Fig. 4D). In addition, the axon terminals of these neuron-like cells stained positive for FM1-43 after potassium stimulation, suggesting that they developed synaptic machinery within 2 months.
Discussion
We demonstrated that the introduction of a single transcription factor, SOX2, is sufficient to convert human RPE cells to functional neurons. The resulting neuronal phenotype was stable for at least 30 days after cessation of SOX2 protein addition. We note that expression of SOX2 via lentivirus led to induction of endogenous SOX2 (Fig. 1E), so, as in induced pluripotent stem cell reprogramming, it may be possible to achieve a stable reprogrammed state by the transient addition of SOX2 protein. The phenotype of the neuronal cells generated is unclear. Although Ma et al. [23] saw retinal ganglion cell gene expression in SOX2 expressing chicken RPE cells, we were unable to detect retinal neuron markers Islet 1/2 or protein kinase C (data not shown). Interestingly, we did not detect vGAT in reprogrammed cells using RT-PCR or immunocytochemistry. This suggests that there are no GABAergic neurons in the reprogrammed population. It may be necessary to introduce additional neural transcription factors to obtain more defined populations of neurons in human cell reprogramming. Future studies will be required to define neuronal phenotypes via electrophysiological characterization and analysis of global transcription patterns.
We demonstrated that the RPARPAR peptide facilitated SOX2-mediated cell lineage reprogramming with higher efficiency than a traditional cell-penetrating peptide, polyarginine. The higher efficiency may result because the internalization pathways used by the two peptides are different. RPARPAR-mediated cell internalization uses the CendR endocytosis pathway activated by specific cell-surface receptors, whereas the mechanisms of polyarginine-mediated internalization include direct translocation through plasma membrane and endocytosis followed by endosomal escape into cytoplasm. The contribution of the former process increases when the payload is of low molecular weight (typically 5–10 kDa), and the internalization of large payload exhibits uptake by the latter process [42]. For general experimental conditions and common chemical properties of the peptides and payloads involved, the direct translocation should be much less efficient than endocytosis [43]. However, the exact endocytic pathways exploited by polyarginine peptide are not well identified and are likely to be dependent on the type and density of the peptide as well as the size and composition of the associated payload [42]. Moreover, it also has been reported that cell transduction of correctly folded green fluorescent protein (GFP) fused with the TAT peptide (a cell-internalization peptide from the HIV-1 TAT protein that is similar to the polyarginine peptide in how it gains entry) results in a high intracellular level of GFP-TAT protein but with a significant loss of GFP emission [44]. This suggests that transduction across the cellular membrane facilitated by the polyarginine or arginine-rich peptides, such as the TAT peptide, may partially or completely unfold proteins. Taken together, our observation that the CendR delivery pathway is compatible with the delivery of reprogramming factors suggests the possibility of in vivo reprogramming using systemic administration of target tissue-specific CendR fusion proteins.
Our observation that reprogramming efficiency using protein transduction (0.3%) is lower than when using lentiviral vectors (11%) is supported by observations in other studies [14, 15, 45]. The reason may be low levels of delivery to the nucleus or improper folding or post-translational modification in a bacterial protein expression system. Using a mammalian expression system might improve the reprogramming efficiency. We also observed that individual RPE cell lines showed variable plasticity using both virus- and protein-mediated reprogramming. Among three hfRPE cell lines, fRPE1914 showed the highest reprogramming efficiency. More interestingly, the glial cell marker GFAP was detected in the reprogrammed cell populations from one of the hfRPE cell lines and from the hESC-derived RPE cell line, sometimes appearing in spherical aggregates (supplemental online Fig. 4), suggesting that a population of the RPE cells was reprogrammed to glial cell states. This raises the question of whether RPE cells were converted to neural progenitor or stem cells and then subsequently differentiated to glial cells. Ring et al. recently showed that fibroblasts can be directly reprogrammed into neural stem cells via expression of SOX2 [35]. We detected an increase in endogenous SOX2 expression after lentiviral transduction (Fig. 1E), which is a marker of neural progenitors. We also noted a small percentage of GFAP-positive cells, which might be of radial glial/astrocytic lineage. However, we did not observe proliferation or neural stem cell morphology in the SOX2-transduced RPE cells. Reprogramming fibroblasts with SOX2 resulted in NSCs, and the different results obtained might be due to the difference in the parental cell type. It is known that RPE cells have a limited ability to expand [46], and this might hinder reprogramming to a replicative state. It has been reported that reprogramming using SOX2 and other transcription factors results in NSCs with different potential, depending on the starting cell type and the transcription factor used [36]. In normal development, expression of SOX2 is turned off during differentiation of neurons, and it may be that transient expression in RPE cells results in a transient neural stem cell state, followed by neuronal differentiation.
RPE cells are adjacent to the neural retina and might provide a convenient source of neural cells for the treatment of ocular disease. Recently, Temple et al. [47] showed that hfRPE cells can be coaxed to neuronal phenotypes using specific growth factors in vitro. It might be possible to combine selective media formulations, growth factors, small molecules, and cell-internalizing peptides to generate specific neural cell types.
Conclusion
This study demonstrates that genetic overexpression or transduction of recombinant SOX2 is sufficient to convert hfRPE and hESC-derived RPE cells to neuronal cells. Importantly, the reprogramming efficiency using RPARPAR peptide-mediated SOX2 transduction was about threefold higher than that using the conventional cell-penetrating peptide, polyarginine. As this technology improves, proteins modified with cell-internalizing peptides may provide a virus-free methodology for directly reprogramming cells for research and, possibly, for clinical use.
Supplementary Material
Acknowledgments
We thank Dr. David Buchholz and Cassidy Hinman for preparing hfRPE cells, Chelsea Presbrey for preparing H14 RPE cells, Dr. Sourav Banerjee for help with the synaptic activity assay, Gabriel Luna for help with microscopy and critical reading of the manuscript, Dr. Teisha Rowland for critical reading of the manuscript, and Dr. Sherry Hikita and all of the staff at the Center for Stem Cell Biology and Engineering at the University of California Santa Barbara. This work was supported by the Garland Initiative for Vision. Q.H. was a fellow of the California Institute for Regenerative Medicine (CIRM). This work was supported by CIRM Grants DR1-01444, CL1-00521-1, T3-00009, and FA1-00616-1. A portion of this work was supported by the Institute for Collaborative Biotechnologies through Grant W911NF-09-0001 from the U.S. Army Research Office. The content of the information does not necessarily reflect the position or the policy of the government, and no official endorsement should be inferred.
Author Contributions
Q.H.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; R.C.: conception and design, collection and assembly of data, data analysis and interpretation; T.T.: collection and assembly of data, data analysis and interpretation; E.R.: conception and design, financial support, final approval of manuscript; D.O.C.: conception and design, financial support, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
D.O.C. is a cofounder of Regenerative Patch Technologies and has uncompensated employment, intellectual property rights, and ownership interest. T.T. has uncompensated ownership interest in CendR Inc. E.R. is an uncompensated inventor, uncompensated consultant, and uncompensated shareholder for CendR Therapeutics Inc.
References
- 1.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 2.Nutt SL, Heavey B, Rolink AG, et al. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 3.Berninger B, Costa MR, Koch U, et al. Functional properties of neurons derived from in vitro reprogrammed postnatal astroglia. J Neurosci. 2007;27:8654–8664. doi: 10.1523/JNEUROSCI.1615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 5.Vierbuchen T, Ostermeier A, Pang ZP, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie H, Ye M, Feng R, et al. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 7.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 8.Ben-David U, Benvenisty N, Mayshar Y. Genetic instability in human induced pluripotent stem cells: Classification of causes and possible safeguards. Cell Cycle. 2010;9:4603–4604. doi: 10.4161/cc.9.23.14094. [DOI] [PubMed] [Google Scholar]
- 9.Nienhuis AW, Dunbar CE, Sorrentino BP. Genotoxicity of retroviral integration in hematopoietic cells. Mol Ther. 2006;13:1031–1049. doi: 10.1016/j.ymthe.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Stadtfeld M, Nagaya M, Utikal J, et al. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu J, Chau KF, Vodyanik MA, et al. Efficient feeder-free episomal reprogramming with small molecules. PLoS One. 2011;6:e17557. doi: 10.1371/journal.pone.0017557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J, Hu K, Smuga-Otto K, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okita K, Nakagawa M, Hyenjong H, et al. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 14.Zhou H, Wu S, Joo JY, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim D, Kim CH, Moon JI, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teesalu T, Sugahara KN, Kotamraju VR, et al. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc Natl Acad Sci USA. 2009;106:16157–16162. doi: 10.1073/pnas.0908201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alberici L, Roth L, Sugahara KN, et al. De novo design of a tumor-penetrating peptide. Cancer Res. 2013;73:804–812. doi: 10.1158/0008-5472.CAN-12-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugahara KN, Teesalu T, Karmali PP, et al. Coadministration of a tumor-penetrating peptide enhances the efficacy of cancer drugs. Science. 2010;328:1031–1035. doi: 10.1126/science.1183057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugahara KN, Teesalu T, Karmali PP, et al. Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer Cell. 2009;16:510–520. doi: 10.1016/j.ccr.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth L, Agemy L, Kotamraju VR, et al. Transtumoral targeting enabled by a novel neuropilin-binding peptide. Oncogene. 2012;31:3754–3763. doi: 10.1038/onc.2011.537. [DOI] [PubMed] [Google Scholar]
- 21.Zhao S, Thornquist SC, Barnstable CJ. In vitro transdifferentiation of embryonic rat retinal pigment epithelium to neural retina. Brain Res. 1995;677:300–310. doi: 10.1016/0006-8993(95)00163-k. [DOI] [PubMed] [Google Scholar]
- 22.Yan RT, Li X, Wang SZ. Photoreceptor-like cells in transgenic mouse eye. Invest Ophthalmol Vis Sci. 2013;54:4766–4775. doi: 10.1167/iovs.13-11936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma W, Yan RT, Li X, et al. Reprogramming retinal pigment epithelium to differentiate toward retinal neurons with Sox2. Stem Cells. 2009;27:1376–1387. doi: 10.1002/stem.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avilion AA, Nicolis SK, Pevny LH, et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood HB, Episkopou V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech Dev. 1999;86:197–201. doi: 10.1016/s0925-4773(99)00116-1. [DOI] [PubMed] [Google Scholar]
- 26.Yabuta Y, Kurimoto K, Ohinata Y, et al. Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biol Reprod. 2006;75:705–716. doi: 10.1095/biolreprod.106.053686. [DOI] [PubMed] [Google Scholar]
- 27.Thomson M, Liu SJ, Zou LN, et al. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Oron E, Nelson B, et al. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 29.Pevny LH, Nicolis SK. Sox2 roles in neural stem cells. Int J Biochem Cell Biol. 2010;42:421–424. doi: 10.1016/j.biocel.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 30.Wegner M, Stolt CC. From stem cells to neurons and glia: A Soxist’s view of neural development. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 31.Graham V, Khudyakov J, Ellis P, et al. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 32.Taranova OV, Magness ST, Fagan BM, et al. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cavallaro M, Mariani J, Lancini C, et al. Impaired generation of mature neurons by neural stem cells from hypomorphic Sox2 mutants. Development. 2008;135:541–557. doi: 10.1242/dev.010801. [DOI] [PubMed] [Google Scholar]
- 34.Ferri AL, Cavallaro M, Braida D, et al. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131:3805–3819. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- 35.Ring KL, Tong LM, Balestra ME, et al. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11:100–109. doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maucksch C, Jones KS, Connor B. Concise review: The involvement of SOX2 in direct reprogramming of induced neural stem/precursor cells. Stem Cells Translational Medicine. 2013;2:579–583. doi: 10.5966/sctm.2012-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Q, Friedrich AM, Johnson LV, et al. Memory in induced pluripotent stem cells: Reprogrammed human retinal-pigmented epithelial cells show tendency for spontaneous redifferentiation. Stem Cells. 2010;28:1981–1991. doi: 10.1002/stem.531. [DOI] [PubMed] [Google Scholar]
- 38.Wang SZ, Ma W, Yan RT, et al. Generating retinal neurons by reprogramming retinal pigment epithelial cells. Expert Opin Biol Ther. 2010;10:1227–1239. doi: 10.1517/14712598.2010.495218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rowland TJ, Blaschke AJ, Buchholz DE, et al. Differentiation of human pluripotent stem cells to retinal pigmented epithelium in defined conditions using purified extracellular matrix proteins. J Tissue Eng Regen Med. 2013;7:642–653. doi: 10.1002/term.1458. [DOI] [PubMed] [Google Scholar]
- 40.Amaral E, Guatimosim S, Guatimosim C. Using the fluorescent styryl dye FM1-43 to visualize synaptic vesicles exocytosis and endocytosis in motor nerve terminals. Methods Mol Biol. 2011;689:137–148. doi: 10.1007/978-1-60761-950-5_8. [DOI] [PubMed] [Google Scholar]
- 41.Bolte S, Talbot C, Boutte Y, et al. FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J Microsc. 2004;214:159–173. doi: 10.1111/j.0022-2720.2004.01348.x. [DOI] [PubMed] [Google Scholar]
- 42.MacEwan SR, Chilkoti A. Harnessing the power of cell-penetrating peptides: Activatable carriers for targeting systemic delivery of cancer therapeutics and imaging agents. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2013;5:31–48. doi: 10.1002/wnan.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katayama S, Nakase I, Yano Y, et al. Effects of pyrenebutyrate on the translocation of arginine-rich cell-penetrating peptides through artificial membranes: Recruiting peptides to the membranes, dissipating liquid-ordered phases, and inducing curvature. Biochim Biophys Acta. 2013;1828:2134–2142. doi: 10.1016/j.bbamem.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 44.Schwarze SR, Hruska KA, Dowdy SF. Protein transduction: Unrestricted delivery into all cells? Trends Cell Biol. 2000;10:290–295. doi: 10.1016/s0962-8924(00)01771-2. [DOI] [PubMed] [Google Scholar]
- 45.Cho HJ, Lee CS, Kwon YW, et al. Induction of pluripotent stem cells from adult somatic cells by protein-based reprogramming without genetic manipulation. Blood. 2010;116:386–395. doi: 10.1182/blood-2010-02-269589. [DOI] [PubMed] [Google Scholar]
- 46.Croze RH, Buchholz DE, Radeke MJ, et al. ROCK inhibition extends passage of pluripotent stem cell-derived retinal pigmented epithelium. Stem Cells Translational Medicine. 2014;3:1066–1078. doi: 10.5966/sctm.2014-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salero E, Blenkinsop TA, Corneo B, et al. Adult human RPE can be activated into a multipotent stem cell that produces mesenchymal derivatives. Cell Stem Cell. 2012;10:88–95. doi: 10.1016/j.stem.2011.11.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.