This study reports an advanced method for the generation of induced pluripotent stem cells (iPSCs) from frozen peripheral blood mononuclear cells (PBMCs) by the tetracistronic Sendai virus (SeVdp) in feeder-free conditions. Generation of iPSC does not require cytokine induction of PBMC before reprogramming, and the produced iPSCs are transgene-free and do not contain T-cell-specific genomic rearrangements. Together, these properties make this novel method a valuable tool for large-scale reprogramming of PBMCs for biobanking purposes.
Keywords: Induced pluripotent stem cells, Sendai virus, Nuclear reprogramming, Cell banking
Abstract
Generation of validated human induced pluripotent stem cells (iPSCs) for biobanking is essential for exploring the full potential of iPSCs in disease modeling and drug discovery. Peripheral blood mononuclear cells (PBMCs) are attractive targets for reprogramming, because blood is collected by a routine clinical procedure and is a commonly stored material in biobanks. Generation of iPSCs from blood cells has previously been reported using integrative retroviruses, episomal Sendai viruses, and DNA plasmids. However, most of the published protocols require expansion and/or activation of a specific cell population from PBMCs. We have recently collected a PBMC cohort from the Finnish population containing more than 2,000 subjects. Here we report efficient generation of iPSCs directly from PBMCs in feeder-free conditions in approximately 2 weeks. The produced iPSC clones are pluripotent and transgene-free. Together, these properties make this novel method a powerful tool for large-scale reprogramming of PBMCs and for iPSC biobanking.
Introduction
Reprogramming technology enables generation of induced pluripotent stem cells (iPSCs) that recapitulate human genetic diversity and pathology [1, 2]. Somatic cells can be reprogrammed into iPSCs using both integrative and nonintegrative methods [2–7]. iPSCs have been generated from a vast spectrum of somatic cells including fibroblasts [1], blood cells [8–11], and myoblasts [12].
One important foreseeable application of iPSC technology is the generation of validated iPSC biobanks, which will be instrumental for the study of pathogenic disease processes, drug discovery, and prediction of drug safety. Biobanking requires readily available somatic cells and reliable and affordable methods for the production of iPSC lines. Several research institutes have started efforts in establishing specific iPSC biobanks that can produce, store, and distribute high quality iPSCs for research purposes. It is currently unclear which method would be the most suitable for generation of iPSC biobanks. Although the majority of iPSC lines have been generated from skin fibroblasts, taking of skin biopsies and growing of fibroblasts is not compatible with a large-scale cellular biobanking program. In contrast, collection of blood from patients is a fast, routine practice and also clearly less costly than derivation of fibroblasts from skin biopsies. In addition, many research sample collections already include frozen peripheral blood mononuclear cells (PBMCs). All current protocols for generation of iPSCs from PBMCs require activation and/or expansion of cell subpopulations, which increases the time and cost of the process [8, 10, 11, 13–16].
We have recently collected PBMC samples from a cohort of more than 2,000 Finnish subjects. In this study, we used frozen PBMCs from this cohort and examined time- and cost-effective protocols suitable for large-scale PBMC reprogramming for iPSC biobanking purposes. By using the tetracistronic Sendai virus (SeVdp) containing reprogramming factors [7, 17, 18], we generated iPSC lines in feeder-free conditions in 16 days. The resulting iPSC lines were transgene-free and did not contain T-cell-specific genomic rearrangements. To our knowledge, this is the first system describing direct reprograming of PBMCs without the need for activation and/or expansion of cell subpopulations within PBMCs. This novel strategy is a valuable tool for a large-scale reprogramming of PBMCs for iPSC biobanking.
Materials and Methods
Ethical Approval
Blood samples were collected from voluntary donors of FINRISK 2012 population cohort (http://www.nationalbiobanks.fi/index.php/studies2/7-finrisk) [19], with written consent permitted by the ethical committee of the hospital district of Helsinki and Uusimaa (permit 17/13/03/00/2011).
Cell Culture
Human PBMCs were isolated by Ficoll-Paque extraction method from 8 ml of whole blood and cryopreserved in two ampoules in the gas phase of liquid nitrogen. Before starting the reprogramming experiments, frozen PBMCs were quickly thawed and allowed to recover overnight in RPMI 1640 medium (Gibco, Grand Island, NY, http://www.invitrogen.com) supplemented with 10% fetal bovine serum (FBS) (Gibco) and antibiotics. iPSC lines were cultured on mitomycin C-treated mouse feeder cells in human embryonic stem cell (hES) medium: Dulbecco’s modified Eagle’s medium/F12 with GlutaMAX, supplemented with 20% KO-serum replacement, 0.1 mM β-mercaptoethanol, 1% nonessential amino acids (all from Life Technologies, Rockville, MD, http://www.lifetech.com), and 6 ng/ml basic fibroblast growth factor (bFGF; Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com). iPSCs cultured on feeders were passaged using collagenase IV (Gibco). For feeder-free conditions, the cells were cultured on Matrigel (growth factor reduced; BD Biosciences, San Diego, CA, http://www.bdbiosciences.com) in Essential 8 medium (Life Technologies) and passaged using 0.5 mM EDTA.
For embryoid body (EB) formation, iPSCs were grown to confluence on a six-well plate and detached by collagen IV. EBs were cultured in suspension for 14 days in a low-attachment six-well plate (Corning Enterprises, Corning, NY, http://www.corning.com) in hES medium without bFGF and then plated on gelatin-coated 24-well plates (Corning) for an additional 7 days. Differentiated cells were fixed with 4% paraformaldehyde (PFA) for immunocytochemical analysis.
Activation of T Cells
For T-cell activation, the 24-well plates (Nunc, Rochester, NY, http://www.nuncbrand.com) were precoated with 200 μl of anti-CD3 antibody (BD Pharmingen, San Diego, CA, http://www.bdbiosciences.com; 10 μg/ml, diluted in phosphate-buffered saline [PBS]) at 37°C/5% CO2 for 4 hours. The wells were then washed three times with PBS before plating the cells. A total of 0.5 × 106 PBMCs were then plated to each well in 400 μl of RPMI 1640 medium supplemented with 10% FBS, antibiotics, 30 pg/ml human interleukin 2 (hIL-2), and anti-CD28 antibody (5 μg/ml; BD Pharmingen) to enhance T-cell activation. The cells were then incubated for 5 days in the cell incubator; during that time cell aggregates were formed, indicating successful activation of T cells.
Generation of PBMC-Derived iPSCs Using the 4V Method
Activated cells (1–3 × 105 cells) were transferred on new 24-well plates (Corning) coated with anti-CD3 antibody (5 μg/ml) in 400 μl of RPMI 1640 medium supplemented with 10% FCS, antibiotics, and 30 pg/ml hIL-2. Nonactivated cells from the same donor were plated on low-attachment tissue culture plates (Corning) without coating, and both were infected in parallel with a Cytotune iPS reprogramming kit (Life Technologies) (indicated as the 4V method elsewhere) with a multiplicity of infection (MOI) of 3, 5, or 6 for 24 hours. The cells were then plated on mitomycin C-treated mouse embryonic fibroblasts (MEFs; 3.75 × 105 cells per well of a six-well plate) in hES medium. The medium was changed every second day, and the floating cells were recovered by centrifugation.
Reprogramming of PBMCs by SeVdp
SeVdp vector was produced in the National Institute of Advanced Industrial Science and Technology (AIST) of Japan as previously described [7]. Reprogramming using SeVdp was performed by infecting 0.1–1 × 106 activated or nonactivated cells with SeVdp(KOSM)302L vector (106–3 × 107 cell infectious units/ml) at MOI 2 for 2 hours at room temperature. The cells were then plated on mitomycin C-treated MEFs (3.75 × 105 cells per well of a 6-well plate) and grown in hES medium. For feeder-free reprogramming, infected PBMCs were directly seeded on 6- or 12-well Matrigel-coated plates at a density of 0.1–1 × 106 cells per well in Essential 6 medium (Life Technologies). From day 10, the cells were cultured in Essential 8 medium. The medium was changed every second day, and the floating cells were recovered by centrifugation. For inductions using small molecule inhibitor, the reprogramming medium was supplemented with 0.25 mM sodium butyrate (NaB; Sigma-Aldrich) at day 3 after transduction.
Characterization of iPSC Clones
All iPSCs were fixed using 4% PFA and permeabilized in 0.5% Triton X-100/PBS (Sigma-Aldrich). Ultra V bloc (Thermo Fisher Scientific, Waltham, MA, http://www.thermofisher.com) was used to block unspecific antigens. Primary antibodies against TRA-1-60 (1:500; Millipore, Billerica, MA, http://www.millipore.com), NANOG (1:500; Cell Signaling Technology, Beverly, MA, http://www.cellsignal.com), SSEA4 (1:1,000; Millipore), and OCT4 (1:1,000; Santa Cruz Biotechnology Inc., Santa Cruz, CA, http://www.scbt.com) were used to detect stem cell-specific markers. Alkaline-phosphatase was detected using nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate solution (Roche Applied Science, Indianapolis, IN, https://www.roche-applied-science.com). The nuclei were stained with 4′,6-diamidino-2-phenylindole (Vectashield; Vector Laboratories, Burlingame, CA, http://www.vectorlabs.com). Live immunostaining was performed using antibody against TRA-1-60 (1:40; Millipore) diluted in E8 medium for 60 minutes in a cell culture incubator at 37°C, followed by fluorescence-conjugated secondary antibody incubation for 60 minutes. Differentiation of iPSC clones was detected by staining the EB-derived cells with markers of the three germ layers: mouse anti-β-III-tubulin (1:2,000; Covance, Princeton, NJ, http://www.covance.com), rabbit anti-α-fetoprotein (AFP) (1:500; Dako, Glostrup, Denmark, http://www.dako.com), and mouse anti-vimentin (1:2,000; Dako). Alexa Fluor 488-conjugated secondary antibodies A21203, A21206, and A21203 were from Invitrogen (Carlsbad, CA, http://www.invitrogen.com).
For whole-genome microarray and PluriTest analysis, RNA was extracted from PBMCs, iPSCs, and hES cells using an AllPrep DNA/RNA/protein kit (Qiagen, Hilden, Germany, http://www.qiagen.com) according to the manufacturer’s instructions. Illumina HT12v4 microarrays were hybridized in the Institute for Molecular Medicine Finland (FIMM) Technology Center, University of Helsinki. The raw expression data were processed with PluriTest for testing pluripotent features in iPSC lines (http://www.pluritest.org). Genetic integrity was evaluated by G-banded karyotype analysis.
Real-Time Polymerase Chain Reaction
Total RNA was purified from iPSCs using NucleoSpin RNA II kit (Macherey-Nagel, Bethlehem, PA, http://www.mn-net.com). DNA was digested in a separate reaction using DNase I (Promega, Madison, WI, http://www.promega.com). Two micrograms of RNA were used for reverse transcription (RT) reaction that was performed using Moloney murine leukemia virus reverse transcriptase, random hexamers, and oligoT (all from Promega) according to the manufacturer’s instructions. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a housekeeping gene. Primer sequences (5′ → 3′) were as follows: NANOG (AAACCATGGATTTATTCCTAA and AGGAAGGATTCAGCCAGT), REX1 (TTCGTGTGTCCCTTTCAA and TTGTTCATTCTTGTTCGT), TDGF1 (GAGATGACAGCATTTGGC and GGCAGCAGGTTCTGTTTA), SeV (AGACCCTAAGAGGACGAAGACAGA and ACTCCCATGGCGTAACTCCATAG), and GAPDH (CATCCATGACAACTTTGG and GCTTCCCGTTCAGCTC).
Analysis of T-Cell Receptor β-Chain Rearrangements in iPSC Clones
Genomic DNA of iPSC clones was isolated using a Puregene kit (Qiagen) according to the manufacturer’s instructions. Before the experiments, the quality of all DNA samples was tested in the polymerase chain reaction (PCR) with a control size ladder mix provided by the TCRB Gene Clonality assay kit (Invivoscribe, San Diego, CA, http://www.invivoscribe.com). T-cell clonal DNA (IVS0004) was used as a positive control for all specific multiplex PCRs, performed according to the kit instructions (Invivoscribe). Human ES cell line H9 was used as a negative control. T-cell receptor (TCR)-β PCR products were detected in 2% agarose gel, and fragment analysis was performed in the sequencing unit of the FIMM Technology Centre by capillary electrophoresis using ABI3730 DNA analyzer. Peak Scanner software (Applied Biosystems, Foster City, CA, http://www.appliedbiosystems.com) was used for fragment size analysis.
Results
Production of PBMC-Derived iPSCs Using the 4V Method
Generation of iPSC from activated T-cells using Sendai virus (SeV) each carrying individually one of the four Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC) has previously been reported [9, 10]. We therefore reprogrammed both activated and nonactivated PBMCs (n = 3 donors) using commercially available Cytotune-iPS reprogramming kit, referred to here as the 4V method. The first iPSC-like colonies appeared at day 15. By day 21, iPSC colonies with morphology similar to embryonic stem cells (ESCs) were picked and expanded (Fig. 1). The mean reprogramming efficiency was determined to be 0.005% (n = 3, SD = 0.001) measured as the ratio of produced iPSC colonies to the starting PBMC cell number (supplemental online Fig. 1). The optimal conditions for PBMC reprogramming were determined to be 3 × 105 cells infected with MOI 3.
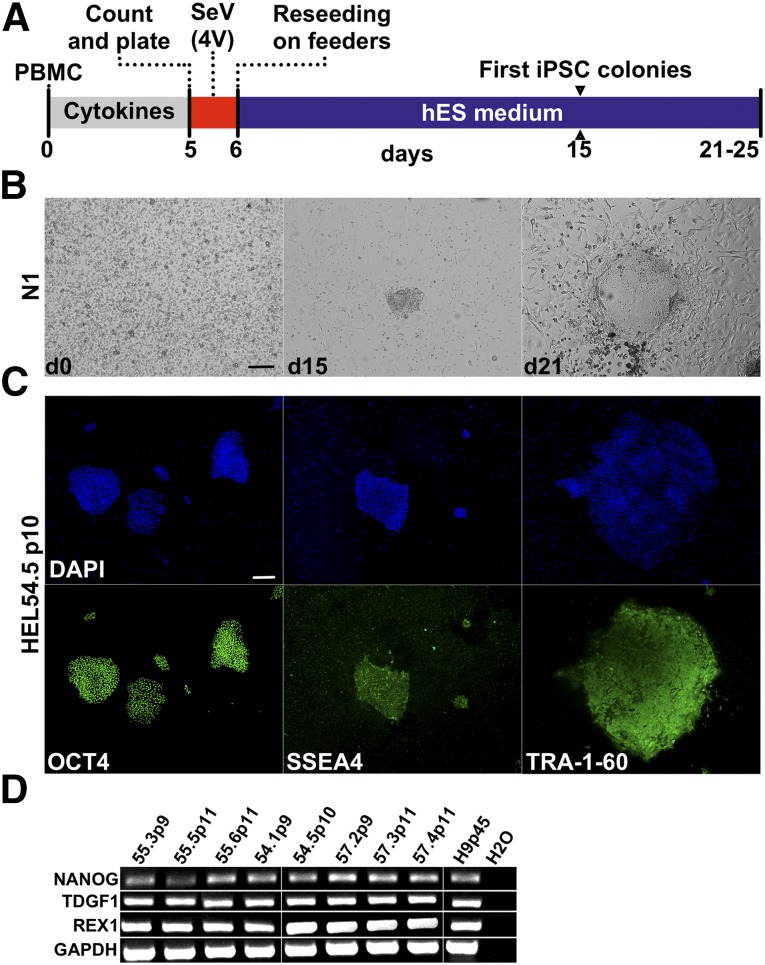
Figure 1.
Reprogramming using the 4V method. (A): Schematic presentation of reprogramming using Cytotune iPS reprogramming kit (referred to as the 4V method in the text) producing iPSCs only from the activated PBMCs. (B): Morphology of reprogrammed PBMCs at days 0, 15, and 21 from one representative donor (N1). (C): Expression of stem cell markers from one representative iPSC line (HEL54.5): OCT4 (green), SSE4 (green), and TRA-1-60 (green) and nuclear staining with DAPI (blue). (D): Reverse transcription-polymerase chain reaction from generated iPSC lines, stem cell markers: NANOG, TDGF1, REX1, and housekeeping gene GAPDH. Scale bars = 200 μm. Abbreviations: d, day; DAPI, 4′,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; hES, human embryonic stem cell; iPSC, induced pluripotent stem cell; PBMC, peripheral blood mononuclear cell; SeV (4V), Sendai virus (Cytotune).
All iPSC clones (n = 8) stained positive for stem cell markers OCT4, SSEA4, and TRA-1-60 as detected by immunofluorescence and expressed NANOG, TDGF1, and REX1 as detected by RT-PCR (Fig. 1). Notably, iPSC colonies were only detected from the preactivated PBMCs, and no colonies appeared from the nonactivated PBMCs directly infected with the 4V method, indicating that activated T cells are the most probable target of SeV infection.
Reprogramming of PBMCs by Tetracistronic SeVdp
The use of polycistronic SeV enables balanced expression of all transgenes, which increases reprogramming efficiency [7, 20]. Therefore, we next reprogrammed PBMCs using replication-defective and persistent SeV (SeVdp), which accommodates all four Yamanaka factors in a single vector [7]. PBMCs from three donors were infected with or without preceding cell activation and seeded on feeder cells. The first iPSC-like colonies appeared on plates originating from nonactivated PBMCs already at day 6, and by day 16, the colonies were large enough for passaging (supplemental online Fig. 1). Mean reprogramming efficiency was found to be 0.014% ± 0.006% (mean ± SD of two separate experiments with three different donors), which is higher compared with the 4V method (supplemental online Fig. 1D). A total of nine iPSC clones were further characterized. All iPSC lines expressed stem cell markers as determined by immunofluorescence and RT-PCR and were able to differentiate into cells derivative of all three germ layers by spontaneous differentiation (data not shown).
Generation of iPSCs Directly From PBMCs in Feeder-Free Conditions
Encouraged by the efficient reprogramming of PBMCs without cell activation on feeders, we performed SeVdp-mediated inductions from three different donors in feeder-free conditions (Fig. 2A). The first iPSC-like colonies were observed firmly attached to the culture plates already at day 6 after induction (Fig. 2B). At this time point, cells were rounded with clear cell-to-cell boundaries. By day 16, cells adhered to each other and formed large ESC-like colonies with distinct borders containing tightly packed cells with high nucleus/cytoplasm ratios and prominent nucleoli. The colonies stained positively for alkaline phosphatase and were visualized by live immunostaining using an antibody against TRA-1-60 (Fig. 2B). Mean reprogramming efficiency was determined to be 0.011% ± 0.006% (n = 6), being similar to SeVdp-mediated induction on feeder cells.
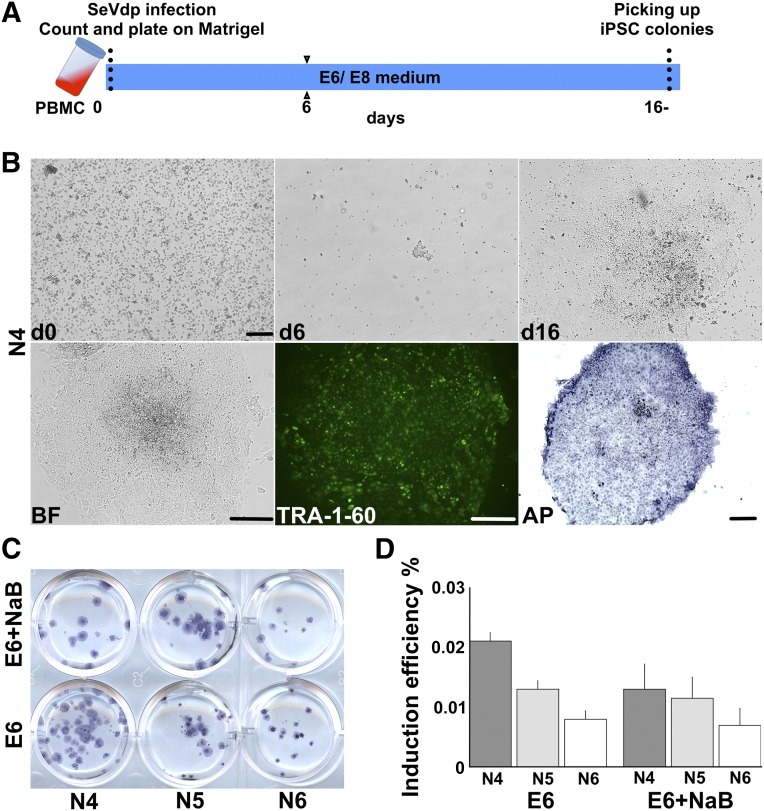
Figure 2.
Reprogramming of PBMCs in a feeder-free conditions by SeVdp. (A): Schematic representation of PBMC reprogramming using SeVdp, which produces iPSCs from the intact PBMCs. (B): Morphology of reprogrammed PBMCs at days 0, 6, and 16. BF, TRA-1-60 (green), and AP staining (blue) of bona fide iPSCs at day 16, shown from one representative induction (N4). (C, D): Reprogramming efficiency of PBMCs of three donors (N4, N5, and N6) with and without NaB was determined using AP staining. Induction efficiency is shown as percentages (y-axis). Scale bars = 200 μm. Abbreviations: AP, alkaline phosphatase; BF, bright field; d, day; iPSC, induced pluripotent stem cell; NaB, sodium butyrate; PBMC, peripheral blood mononuclear cell; SeVdp, tetracistronic Sendai virus.
It has been previously reported that addition of NaB to the reprogramming cocktail improves the retrovirus-mediated reprogramming efficiency of fibroblasts and myoblasts [12, 21]. Therefore, we tested whether the addition of NaB could further enhance the formation of iPSCs in feeder-free conditions. However, the addition of NaB to the reprogramming cocktail did not increase the mean reprogramming efficiency, which was found to be 0.01% ± 0.004% (n = 6) (Fig. 2C, 2D).
Six iPSC clones generated from three different donors were picked at day 16 and further propagated in feeder-free conditions for at least 10 passages. All iPSC lines expressed stem cell markers TRA-1-60, SSEA4, and OCT4 as determined by immunocytochemistry (Fig. 3A). Stem cell markers NANOG, TDGF1, and REX1 were also expressed in all iPSC lines as determined by RT-PCR (Fig. 3B). In addition, all iPSC lines were able to form EBs and spontaneously differentiate into derivatives of all three germline lineages analyzed by neuroectodermal (TUJ1), endodermal (AFP), and mesodermal (VIMENTIN) marker expression (Fig. 3C), indicating their pluripotent nature. The pluripotent nature of the produced iPSC lines (n = 5) was further confirmed using PluriTest [22] (Fig. 3D).
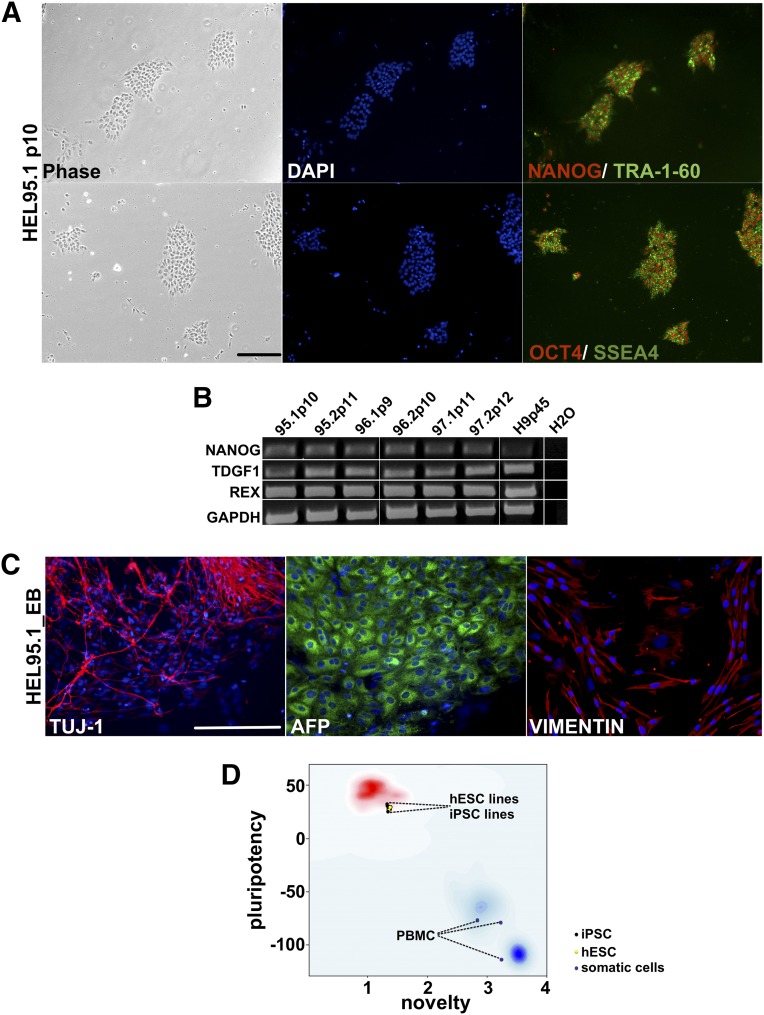
Figure 3.
Characterization of iPSC lines generated from PBMCs in feeder-free conditions. (A): Morphology of generated iPSC lines (phase), nuclear staining DAPI (blue), stem cell markers: NANOG (red), OCT4 (red), TRA-1-60 (green), and SSEA4 (green). (B): Reverse transcription-polymerase chain reaction of PBMC-derived iPSC lines. The stem cell markers were NANOG, TDGF1, and REX1, and housekeeping gene GAPDH. (C): Spontaneous differentiation of PBMC-derived iPSCs. Shown are immunostainings of neuronal TUJ1 (red), endodermal AFP (green), and mesodermal VIMENTIN (red) lineage markers, and nuclear staining DAPI (blue); data shown are from one representative iPSC line (HEL95.1). (D): Pluripotent transcriptional profile measured in PluriTest. iPSC lines (n = 5) and hES cell lines (n = 2) qualified as a pluripotent as determined by the pluripotency and novelty scores. Donor PBMCs (n = 3) are plotted in the previously referenced somatic cells (blue cloud). Scale bar = 200 μm. Abbreviations: AFP, α-fetoprotein; DAPI, 4′,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; hES, human embryonic stem cell; iPSC, induced pluripotent stem cell; PBMC, peripheral blood mononuclear cell.
Clearance of SeV Vectors and Origin of iPSC Lines
Ideally, the produced iPSC lines are transgene-free. Two of eight iPSC clones (25%) generated using 4V were still found to be positive for SeV episome. On the contrary, all iPSC clones generated by SeVdp (n = 15) were completely clear of viral sequences by passage 10 (supplemental online Fig. 1E, 1F). In comparison, we also analyzed the clearance of viral sequences from fibroblast-derived iPSC clones (n = 59 iPSC lines generated from 23 donors) reprogrammed using the 4V method. Similarly to the PBMC-derived iPSCs, SeV episomes were still found to be present in 16 of 59 (27%) of the fibroblast-derived iPSC lines by passage 14 (supplemental online Fig. 1F). Further passaging of fibroblast-derived iPSCs did not completely clear the SeV episome, because 7 of 16 of iPSC lines (44%) remained positive for SeV sequences also at later passages (p14–21).
Origin of iPSC Lines
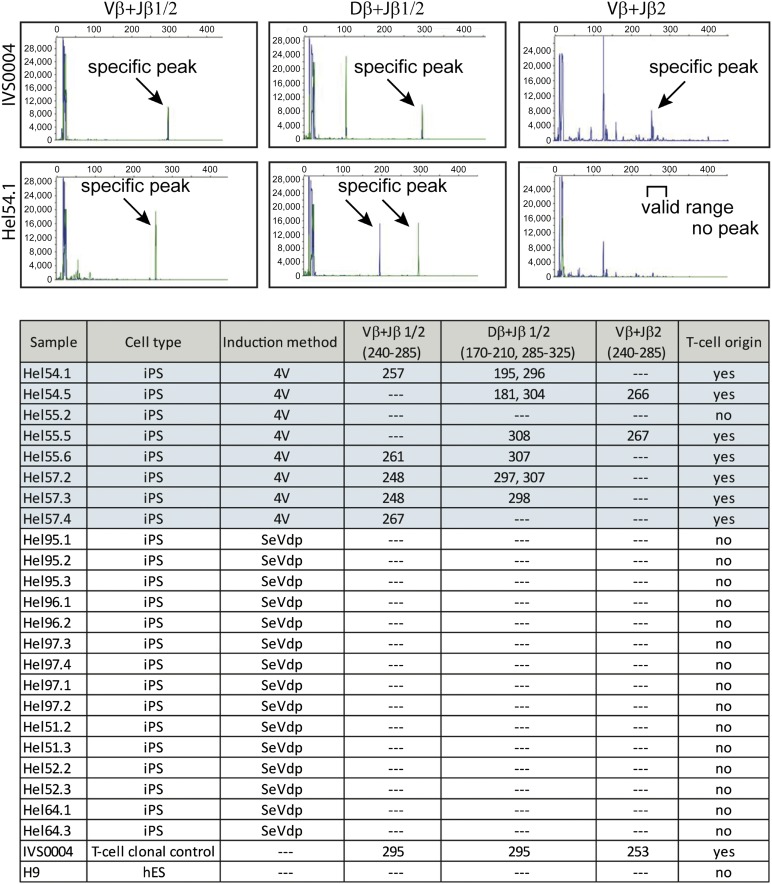
The fact that the two SeV systems infect activated and nonactivated PBMCs with different efficiency suggests that the iPSCs originate from different cell types present in PBMCs. Reprogramming with the 4V method was completely dependent on preceding T-cell activation, suggesting that the produced iPSC clones originate from T lymphocytes. T-cell origin can be analyzed by detecting the specific TCR rearrangements naturally occurring during T-cell development. We performed a multiplex PCR of the genomic TCR-β region of the iPSC lines produced with 4V (n = 8 clones) and with SeVdp (n = 15 clones) and analyzed the results by the gel electrophoresis (supplemental online Fig. 2) and by capillary electrophoresis using ABI3730 DNA analyzer (Fig. 4). The majority of the iPSC clones (7 of 8) produced by the 4V method were positive for TCR-β rearrangements indicating their T-cell origin, whereas none of the SeVdp-induced clones (n = 15) showed TCR-β rearrangements (Fig. 4).
Figure 4.
Origin of iPSCs. Multiplex polymerase chain reaction fragment size analysis of T-cell receptor β region was performed by capillary electrophoresis using ABI3730 DNA analyzer. Specific peak profiles are shown for positive clonal T-cell control (IVS0004) and for one representative iPSC line (HEL54.1) produced by the 4V method. Fragment sizes for all iPSC clones are indicated in the table. DNA from hES cells (H9) was used as a negative control for the experiments. Abbreviations: 4V, Sendai virus (Cytotune); hES, human embryonic stem cell; iPSC, induced pluripotent stem cell; SeVdp, tetracistronic Sendai virus.
Discussion
Retroviral vectors have mostly been used in the delivery of inducing transgenes into somatic cells during reprogramming. However, retro- and lentivirus vectors are integrated into the parental cell genome. Integration may result in undesirable changes such as malignancy in the produced iPSC lines [23] or reactivation of transgenes during iPSC differentiation [24]. In addition, there is a minimal risk of production of replication- competent HIV by homologous recombination events if lentivirus vectors are used for the induction of HIV-positive donor cells [25] that may be present in the large sample cohorts. HIV testing prior to induction is ethically questionable and increases the costs of reprogramming. For these reasons, nonintegrative methods are more suitable for large-scale reprogramming.
Reprogramming of blood cells in a large scale has been problematic because of the inefficient delivery of transgenes into terminally differentiated blood cells. This problem has been bypassed by inducing more immature blood cell precursors or by activating proliferation of different subpopulations present in PBMCs [8, 10, 11, 13, 14]. Besides raising the costs and time required for iPSC production, stimulation of cell proliferation may result in generation of several iPSC lines from the same parental cell population that may lead to distortion of data obtained from parallel iPSC clones.
One of the clear advantages of using a polycistronic viral vector for transgene delivery is the synchronized expression of all induction factors in the target cells. Reprogramming efficiency of PBMCs using SeVdp was detected to be on average 0.01%, which was considerably higher compared with the efficiency obtained here by the 4V method (0.005%). The power of providing all transgenes in a single virus (SeVdp) compared with the 4V system (Cyto-Tune) has previously been shown in fibroblast reprogramming [7]. The efficiency of SeVdp-mediated reprogramming was lower than what has been reported for the DNA plasmid-based reprogramming of PBMCs (0.06%) [13]. However, the high efficiency of DNA plasmid-mediated induction was achieved by adding the EBNA1 and inhibition of TP53 by small hairpin RNA to the reprogramming cocktail, in addition to stimulating cell growth by cytokine activation [13]. Although downregulation of TP53 is temporal during reprogramming [26, 27], it is possible that transient downregulation of TP53 is favorable for cells with genomic aberrations, which would otherwise be eliminated during induction, increasing the probability of acquiring iPSC clones with poor genomic quality [28]. High reprogramming efficiency of PBMCs has recently also been obtained with a modified set of DNA plasmids containing a strong spleen focus-forming virus promoter and antiapoptotic factor BCL-XL, making this method also noteworthy for induction of blood cells [15]. However, episomal DNA plasmids have been shown to undergo occasional genomic integration, resulting in laborious screening of the generated iPSC lines [29].
Importantly, iPSC clones generated by the SeVdp method were all clear of exogenic sequences by passage 10, even without active elimination of replicons. Clearance of SeVdp genome is semiautomatic in response to induction of mir302 in generated nascent iPSC colonies. If required, the removal of transgenes can be enhanced by small interfering RNA treatment [7, 30]. Using this method, we were able to produce iPSC clones from low amounts of PBMCs with minimal amount of virus. We were also able to shorten the generation time of iPSC clones to 16 days, which is approximately 1 week less than with the 4V method. The SeVdp induction did not require activation steps of PBMC subpopulations with cytokines, thus reducing the time and the cost of reprogramming. Importantly, PBMCs were also reprogrammed and maintained in feeder-free conditions, which is a prerequisite for any iPSC biobanking strategy.
Mononuclear cells extracted from peripheral blood contain cells from both lymphoid and nonlymphoid origin. Cytokine stimulation can activate the cell fate or growth and direct induction to the specific cell type. As expected, and also previously reported, activation of T cells was a prerequisite for reprogramming with 4V and resulted in iPSC clones carrying specific TCR rearrangements [10]. However, the iPSC lines should represent the genomic DNA of the donor, lacking T- and B-cell-related DNA rearrangements. The SeVdp-mediated method used in this study did not require cytokine treatment, and the iPSC clones were shown to originate from cells other than T-lymphocytes. Target specificity of SeV vectors is restricted primarily by the initial phase of infection, e.g., fusion between virus envelope and cell membrane [31]. As a result, SeV can infect monocytes and CD4(−)/CD8(−) T cells, but not B cells and CD4(+) T cells [31, 32]. Therefore, it is likely that SeVdp-mediated iPSC clones analyzed in this study originate from cells other than T or B cells and therefore do not carry major genetic rearrangements found in TCR and immunoglobulin regions. The fact that SeVdp virus preferred nonlymphoid cells as its target in the intact PBMC pool to the increased number of T cells in the activated PBMC sample suggests that in addition to virus efficiency, different cell sensitivity also contributes to the reprogramming power.
Nonlymphoid cells of PBMCs have previously been targeted by cytokine induction both by DNA plasmids and viral vectors [8, 15]. It was recently shown that iPSCs generated from human finger prick using the 4V system do not contain T-cell-specific genomic rearrangements [33]. However, extensive expansion of PBMCs with cytokines is required prior to infection with SeV carrying the Yamanaka reprogramming factors.
Conclusion
Our study demonstrates that PBMCs can be efficiently reprogrammed without cytokine activation directly from PBMCs in feeder-free conditions in just 16 days. The generated iPSC lines are transgene-free and do not contain genomic rearrangement. Thus, our reprogramming system has potential to advance large-scale reprogramming of PBMCs into iPSCs for biobanking purposes.
Supplementary Material
Acknowledgments
The SeVdp vector was produced in the National Institute of Advanced Industrial Science and Technology (AIST), and is available upon request (mahito-nakanishi@aist.go.jp). This work was supported by the Academy of Finland (Grant 271884), the Sigrid Jusélius Foundation, and the Research Funds of the Helsinki University Central Hospital. We thank Anne Nyberg, Seija Puomilahti, Jarkko Ustinov, and Eila Korhonen for excellent technical assistance. We also thank Jarno Honkanen (Department of Vaccination and Immune Protection, National Institute for Health and Welfare, Helsinki, Finland) for the protocol for T-cell activation. We thank the participants of the FINRISK 2012 study. K.N. is currently affiliated with the Laboratory of Gene Regulation, Faculty of Medicine, University of Tsukuba, Tsukuba, Ibaraki, Japan.
Author Contributions
R.T. and A.K.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; J.W.: collection and/or assembly of data, final approval of manuscript; K.N. and M.O.: conception and design, final approval of manuscript, generation of SeVdp vector; M.N.: conception and design, manuscript writing, final approval of manuscript, generation of SeVdp vector; V.S.: provision of study material or patients, final approval of manuscript; A.J. and T.O.: financial support, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Okita K, Nakagawa M, Hyenjong H, et al. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 4.Yu J, Hu K, Smuga-Otto K, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weltner J, Anisimov A, Alitalo K, et al. Induced pluripotent stem cell clones reprogrammed via recombinant adeno-associated virus-mediated transduction contain integrated vector sequences. J Virol. 2012;86:4463–4467. doi: 10.1128/JVI.06302-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fusaki N, Ban H, Nishiyama A, et al. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishimura K, Sano M, Ohtaka M, et al. Development of defective and persistent Sendai virus vector: A unique gene delivery/expression system ideal for cell reprogramming. J Biol Chem. 2011;286:4760–4771. doi: 10.1074/jbc.M110.183780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loh YH, Hartung O, Li H, et al. Reprogramming of T cells from human peripheral blood. Cell Stem Cell. 2010;7:15–19. doi: 10.1016/j.stem.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seki T, Yuasa S, Fukuda K. Generation of induced pluripotent stem cells from a small amount of human peripheral blood using a combination of activated T cells and Sendai virus. Nat Protoc. 2012;7:718–728. doi: 10.1038/nprot.2012.015. [DOI] [PubMed] [Google Scholar]
- 10.Seki T, Yuasa S, Oda M, et al. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell. 2010;7:11–14. doi: 10.1016/j.stem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Staerk J, Dawlaty MM, Gao Q, et al. Reprogramming of human peripheral blood cells to induced pluripotent stem cells. Cell Stem Cell. 2010;7:20–24. doi: 10.1016/j.stem.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trokovic R, Weltner J, Manninen T, et al. Small molecule inhibitors promote efficient generation of induced pluripotent stem cells from human skeletal myoblasts. Stem Cells Dev. 2013;22:114–123. doi: 10.1089/scd.2012.0157. [DOI] [PubMed] [Google Scholar]
- 13.Okita K, Yamakawa T, Matsumura Y, et al. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells. 2013;31:458–466. doi: 10.1002/stem.1293. [DOI] [PubMed] [Google Scholar]
- 14.Dowey SN, Huang X, Chou BK, et al. Generation of integration-free human induced pluripotent stem cells from postnatal blood mononuclear cells by plasmid vector expression. Nat Protoc. 2012;7:2013–2021. doi: 10.1038/nprot.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su RJ, Baylink DJ, Neises A, et al. Efficient generation of integration-free iPS cells from human adult peripheral blood using BCL-XL together with Yamanaka factors. PLoS One. 2013;8:e64496. doi: 10.1371/journal.pone.0064496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye L, Muench MO, Fusaki N, et al. Blood cell-derived induced pluripotent stem cells free of reprogramming factors generated by Sendai viral vectors. Stem Cells Translational Medicine. 2013;2:558–566. doi: 10.5966/sctm.2013-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wakao H, Yoshikiyo K, Koshimizu U, et al. Expansion of functional human mucosal-associated invariant T cells via reprogramming to pluripotency and redifferentiation. Cell Stem Cell. 2013;12:546–558. doi: 10.1016/j.stem.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Nishimura T, Kaneko S, Kawana-Tachikawa A, et al. Generation of rejuvenated antigen-specific T cells by reprogramming to pluripotency and redifferentiation. Cell Stem Cell. 2013;12:114–126. doi: 10.1016/j.stem.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Vartiainen E, Laatikainen T, Peltonen M, et al. Thirty-five-year trends in cardiovascular risk factors in Finland. Int J Epidemiol. 2010;39:504–518. doi: 10.1093/ije/dyp330. [DOI] [PubMed] [Google Scholar]
- 20.Nakanishi M, Otsu M. Development of Sendai virus vectors and their potential applications in gene therapy and regenerative medicine. Curr Gene Ther. 2012;12:410–416. doi: 10.2174/156652312802762518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mali P, Chou BK, Yen J, et al. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28:713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller FJ, Schuldt BM, Williams R, et al. A bioinformatic assay for pluripotency in human cells. Nat Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okita K, Yamanaka S. Induced pluripotent stem cells: Opportunities and challenges. Philos Trans R Soc Lond B Biol Sci. 2011;366:2198–2207. doi: 10.1098/rstb.2011.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toivonen S, Ojala M, Hyysalo A, et al. Comparative analysis of targeted differentiation of human induced pluripotent stem cells (hiPSCs) and human embryonic stem cells reveals variability associated with incomplete transgene silencing in retrovirally derived hiPSC lines. Stem Cells Translational Medicine. 2013;2:83–93. doi: 10.5966/sctm.2012-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connolly JB. Lentiviruses in gene therapy clinical research. Gene Ther. 2002;9:1730–1734. doi: 10.1038/sj.gt.3301893. [DOI] [PubMed] [Google Scholar]
- 26.Kawamura T, Suzuki J, Wang YV, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong H, Takahashi K, Ichisaka T, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.González F, Georgieva D, Vanoli F, et al. Homologous recombination DNA repair genes play a critical role in reprogramming to a pluripotent state. Cell Reports. 2013;3:651–660. doi: 10.1016/j.celrep.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okita K, Matsumura Y, Sato Y, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 30.Kawagoe S, Higuchi T, Otaka M, et al. Morphological features of iPS cells generated from Fabry disease skin fibroblasts using Sendai virus vector (SeVdp) Mol Genet Metab. 2013;109:386–389. doi: 10.1016/j.ymgme.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Watabe A, Yamaguchi T, Kawanishi T, et al. Target-cell specificity of fusogenic liposomes: Membrane fusion-mediated macromolecule delivery into human blood mononuclear cells. Biochim Biophys Acta. 1999;1416:339–348. doi: 10.1016/s0005-2736(98)00238-7. [DOI] [PubMed] [Google Scholar]
- 32.Eguchi A, Kondoh T, Kosaka H, et al. Identification and characterization of cell lines with a defect in a post-adsorption stage of Sendai virus-mediated membrane fusion. J Biol Chem. 2000;275:17549–17555. doi: 10.1074/jbc.M910004199. [DOI] [PubMed] [Google Scholar]
- 33.Tan HK, Toh CX, Ma D, et al. Human finger-prick induced pluripotent stem cells facilitate the development of stem cell banking. Stem Cells Translational Medicine. 2014;3:586–598. doi: 10.5966/sctm.2013-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.