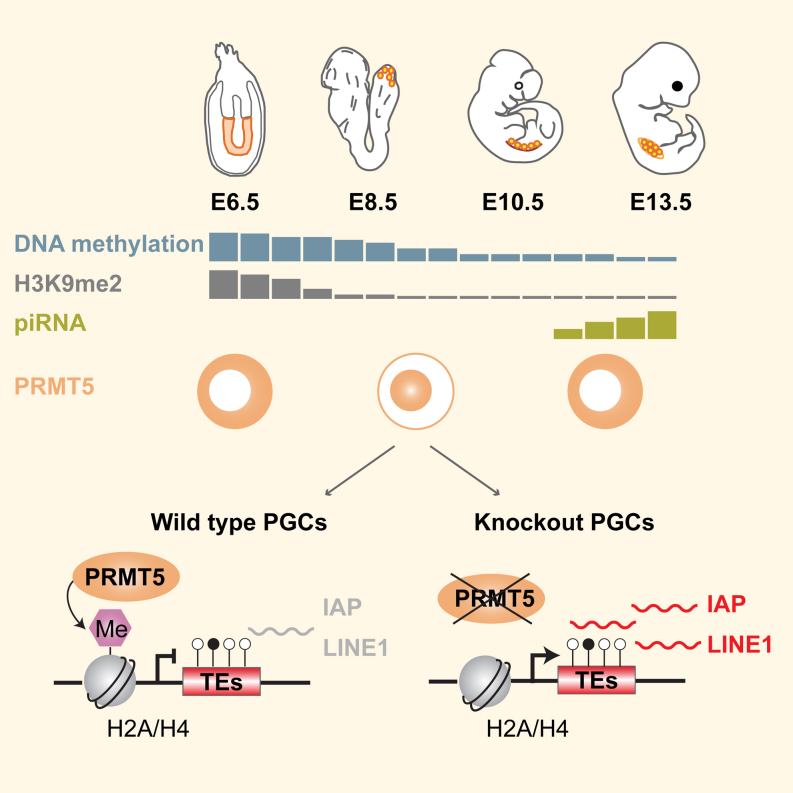
Summary
Primordial germ cells (PGCs) and preimplantation embryos undergo epigenetic reprogramming, which includes comprehensive DNA demethylation. We found that PRMT5, an arginine methyltransferase, translocates from the cytoplasm to the nucleus during this process. Here we show that conditional loss of PRMT5 in early PGCs causes complete male and female sterility, preceded by the upregulation of LINE1 and IAP transposons as well as activation of a DNA damage response. Similarly, loss of maternal-zygotic PRMT5 also leads to IAP upregulation. PRMT5 is necessary for the repressive H2A/H4R3me2s chromatin modification on LINE1 and IAP transposons in PGCs, directly implicating this modification in transposon silencing during DNA hypomethylation. PRMT5 translocates back to the cytoplasm subsequently, to participate in the previously described PIWI-interacting RNA (piRNA) pathway that promotes transposon silencing via de novo DNA remethylation. Thus, PRMT5 is directly involved in genome defense during preimplantation development and in PGCs at the time of global DNA demethylation.
Graphical Abstract
Highlights
-
•
PRMT5 is essential for primordial germ cell (PGC) development
-
•
PRMT5 translocates to the nucleus of PGCs during epigenetic reprogramming
-
•
PRMT5-dependent H2A/H4R3me2s is enriched on transposable elements in PGCs
-
•
PRMT5 loss in PGCs or preimplantation embryos activates transposable elements
Primordial germ cells undergo epigenetic reprogramming that involves DNA demethylation, yet repressed elements such as transposons are not activated. Kim et al. show that Prmt5 is essential in the suppression of transposable elements during epigenetic reprogramming in primordial germ cells and is crucial for their development.
Introduction
During the mammalian life cycle, two major epigenetic reprogramming events restore the developmental potential toward the totipotent and pluripotent states: in PGCs following their specification at embryonic days (E) 7.25–E12.5 and during preimplantation development at E0.5–E3.5, respectively (Surani et al., 2007). One key component of epigenome resetting is global DNA demethylation, which renders PGCs and early embryos vulnerable to the activation of transposable elements (TEs) that are normally repressed by DNA methylation (Walsh et al., 1998).
Specification of PGCs occurs at E7.25 in response to BLIMP1, PRDM14, and AP2γ, which also initiates epigenetic reprogramming (Magnúsdóttir et al., 2013; Nakaki et al., 2013). Notably, there is comprehensive erasure of histone H3 lysine 9 dimethyl mark (H3K9me2), followed by genome-wide DNA demethylation and erasure of genomic imprints between E8.5–E11.5 (Hajkova et al., 2008, 2002; Seisenberger et al., 2012). Genomic imprints are reestablished during gametogenesis and subsequently play an essential role during development (McGrath and Solter, 1984; Surani et al., 1984). Epigenetic reprogramming and global DNA demethylation without the erasure of imprints also occurs at ∼E0.5–E3.5 during development of blastocysts (Borgel et al., 2010).
At the onset of global DNA demethylation in PGCs, PRMT5, a highly conserved arginine methyltransferase, translocates from the cytoplasm to the nucleus at ∼E8.5, and during preimplantation development at the ∼4-cell stage (Ancelin et al., 2006; Tee et al., 2010). PRMT5 catalyzes the symmetric dimethylation of arginine residues including arginine 3 of the histones H2A and H4 (H2A/H4R3me2s), a repressive histone modification (Branscombe et al., 2001; Pal et al., 2004), and of other diverse nuclear and cytoplasmic substrates. This includes Sm proteins in neural progenitors, which are required for RNA splicing and p53 (Bezzi et al., 2013; Jansson et al., 2008; Zhao et al., 2009). Loss of PRMT5 is early embryonic lethal at ∼E6.5, and is essential for the derivation and maintenance of pluripotent ESCs (Tee et al., 2010).
In the germline, PRMT5 interacts with BLIMP1, a key regulator of PGC specification, which may facilitate its nuclear import at ∼E8.0, resulting in high levels of H2A/H4R3 methylation in PGCs (Durcova-Hills et al., 2008). At ∼E11.5, PRMT5-BLIMP1 translocate back to the cytoplasm with a consequent decrease of H2A/H4R3me2s modification, as DNA methylation reaches basal levels in PGCs (Ancelin et al., 2006). In the zygote, PRMT5 is maternally inherited, followed by activation of embryonic Prmt5 at the two- to four-cell stage. PRMT5 relocates predominantly to the nucleus in four- to eight-cell-stage embryos (Tee et al., 2010). Thus, PRMT5 resides in the nucleus in early blastomeres at the onset of global DNA demethylation that reaches basal levels in blastocysts at ∼E3.5–E4.5 (Smith et al., 2012). PRMT5 relocates back to the cytoplasm when de novo DNA methylation and maintenance resume in postimplantation epiblast cells.
DNA methylation is important for the repression of TEs, which comprise 40% of the mammalian genome; their overexpression can induce apoptosis and senescence due to their endonuclease activity and random transpositions (Belgnaoui et al., 2006; Wallace et al., 2008). Global erasure of DNA methylation in PGCs and embryos could cause activation of TEs and affect genome integrity (Burns and Boeke, 2012; Walsh et al., 1998). Of note, there is a transient upregulation of TEs at the two-cell stage during the transition from “zygote to embryo” developmental program (Fadloun et al., 2013; Peaston et al., 2004).
In the germline, a key mechanism for the repression of TEs is through Piwi-interacting small RNAs (piRNAs) acting primarily through de novo DNA methylation (Aravin et al., 2008), which is initiated at ∼E12.5. Thus, additional mechanisms for the repression of TEs are probably required in early PGCs, and during preimplantation development, to coincide with the comprehensive erasure of DNA methylation.
Here we specifically investigated the role of PRMT5 in PGCs and preimplantation embryos at the onset of DNA demethylation. We found that the H2A/H4R3me2s modification catalyzed by PRMT5 was enriched on the LINE1 and IAP TEs of early PGCs. Consequently, conditional loss of PRMT5 resulted in loss of H2A/H4R3me2s and upregulation of TEs, apoptosis of PGCs, and complete male and female sterility in otherwise viable adults. Similarly, depletion of maternally inherited and zygotic PRMT5 in preimplantation embryos caused an upregulation of IAP. In PGCs, PRMT5 relocates back to the cytoplasm at ∼E11.5, where it has a different role in piRNA-mediated silencing of TEs through methylation of PIWI proteins (Vagin et al., 2009). This study demonstrates that nuclear PRMT5 is crucial for suppressing TEs in PGCs and preimplantation embryos at the time of global DNA demethylation.
Results
Loss of PRMT5 in PGCs Results in Male and Female Sterility
We previously showed that PRMT5, which is localized in the cytoplasm of all postimplantation cells, translocates to the nucleus following PGC specification at ∼E8.0–8.5 onward (Ancelin et al., 2006), which prompted us to examine the role of nuclear PRMT5 during PGC development.
To delete PRMT5 in PGCs, we generated a Prmt5 conditional allele (Prmt5flox) and crossed Prmt5flox/flox mice with Blimp1-Cre-BAC transgenic mice (Ohinata et al., 2005) (see Figures S1A–S1D available online), and followed development of Prmt5 mutant germ cells beyond E8.5 (Figure 1A). We initially found alkaline phosphatase (AP)-positive mutant PGCs in numbers similar to those in control embryos at E8.5 (41 versus 47 at 0–4 somite stage; 81 versus 88 at 5–10 somite stage; Figure S1E). While PRMT5 was still detectable in the majority of mutant PGCs at E8.5 (89% versus 99%; Figure 1B), their levels declined progressively thereafter as they migrated to the gonads. Indeed, PRMT5 was depleted in the majority of mutant PGCs by E10.5 but, as expected, not in the surrounding somatic cells (13% versus 99%; Figure 1B). While the mutant embryos developed to apparently normal adulthood (Figures S1F and S1G and see below), both males and females were sterile, with significantly smaller testes and ovaries that lacked germ cells (Figure 1C). These observations establish unequivocally that Prmt5 is essential for the development of PGCs. We set out to investigate why PRMT5 is essential in PGCs after their specification.
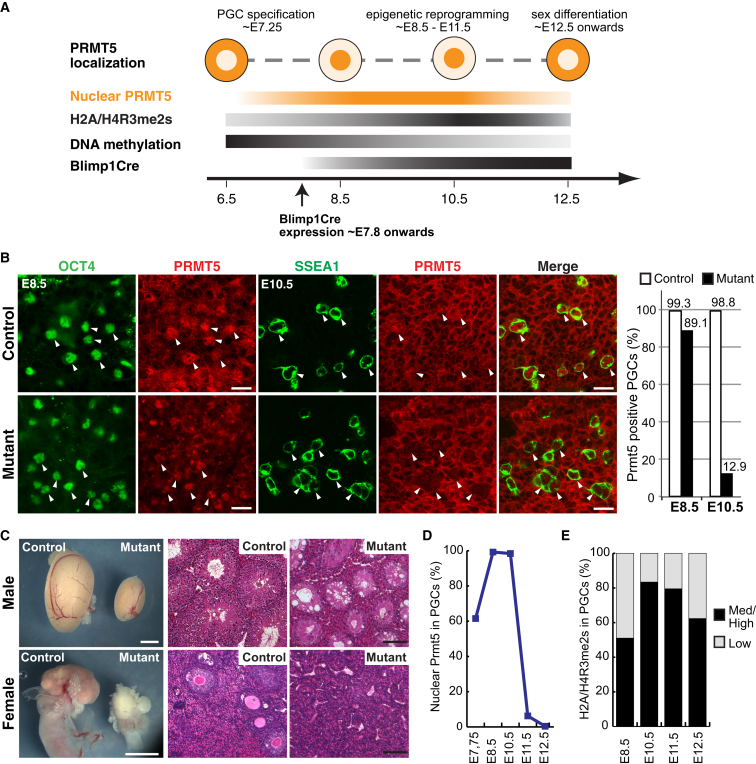
Figure 1.
Deletion of Prmt5 in the Germline using Blimp1-Cre Results in Male and Female Sterility
(A) A schematic of PGCs development (E6.5–E12.5) represents the following: nuclear-cytoplasmic translocation of PRMT5, increase of H2A/H4R3me2s modification, progressive erasure of DNA methylation, and the initiation of Blimp1-Cre expression to induce deletion of nuclear Prmt5.
(B) Detection of PRMT5 (red) by immunofluorescence (IF), and of the PGC markers OCT4 (E8.5; green) or SSEA1 (E10.5; green) in genital ridges. Right graph shows the number of PRMT5-positive PGCs (% PRMT5-positive/PGC marker-positive cells). At E8.5, 89% of mutant PGCs were PRMT5 positive (57/64), and at E10.5 13% of mutant PGCs were PRMT5 positive (9/70). Scale bar, 20 μm. The arrowheads mark PGCs.
(C) (Left panel) Testis and ovary from adult mutants (right) are considerably smaller than from control littermate (left). Scale bar, 2 mm. (Right panel) Hematoxylin and eosin staining of sections from adult testis and ovary shows a lack of sperm and oocytes in mutants. Scale bar, 100 μm. The genotype of the control is Blimp1Cre;Prmt5flox/+ and the mutant is Blimp1Cre;Prmt5flox/− in (B) and (C).
(D) The number of PGCs (in %) with nuclear PRMT5 detected by IF at E7.5–E12.5 in wild-type embryos.
(E) The number of PGCs with similar or higher level of H2A/H4R3me2s detected by IF (Med/High, black, in %) in PGCs compared to surrounding somatic cells at E8.5–E12.5.
See also Figures S1 and S2.
The Histone Modification H2A/H4R3me2s Is Lost in Prmt5 Mutant PGCs
Next, we established that the enrichment of PRMT5 in the nucleus occurred in most PGCs by ∼E8.5 (E7.75, 62%; E8.5, 99%; Figures 1D and S2A), where it persisted as the PGCs migrated and colonized the genital ridge at ∼E10.5 (98%; Figures 1D and S2A). PRMT5 relocates back to the cytoplasm after E11.5, and was rarely detected in the nucleus after E11.5 (E11.5, 6%; E12.5, 0.4%; Figures 1D and S2A). Notably, this transient presence of PRMT5 in the PGC nucleus at ∼E8.0–E11.0 occurs following erasure of H3K9me2 and coincides with comprehensive epigenetic reprogramming in PGCs, including global DNA demethylation (Hackett et al., 2013).
The translocation of PRMT5 to the nucleus is accompanied by an enrichment of the H2A/H4R3me2s modification in PGCs, which occurs progressively from E8.5 to E10.5, and persists until ∼E12.5 in PGCs (E8.5, 51%; E10.5, 83%; Figures 1E and S2B). By contrast, this chromatin modification diminished significantly and rapidly in Prmt5 mutant PGCs, and was virtually undetectable by E12.5 (E10.75; 49% versus 99%, E12.5; 4% versus 93%, Figure 2A).
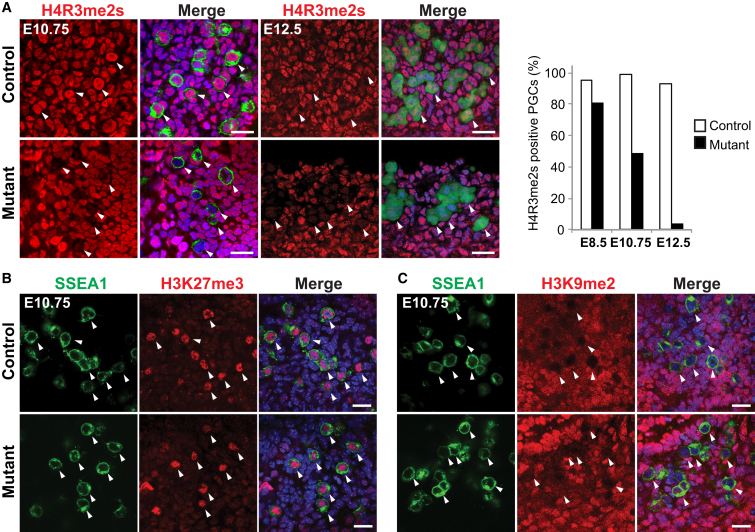
Figure 2.
Loss of Prmt5 in the Germline Results in Loss of H2A/H4R3me2s
(A) IF staining of genital ridges for H2A/H4R3me2s (red). PGCs were detected with antibodies against SSEA1 (green, E10.75) or GFP for GOF (green, E12.5). Merged images are shown with DNA stained with DAPI (blue). (Right graph) Number of PGCs labeled with H2A/H4R3me2s (in %). At E8.5, 81% (21/26) of mutant PGCs were H2A/H4R3me2s positive, which were reduced to 49% (50/102) at E10.75, and at E12.5 there were 4% (3/74) mutant PGCs that were positive for H2A/H4R3me2s.
(B) IF staining of H3K27me3 (red) in genital ridges at E10.75. PGCs were detected with SSEA1 antibody (green).
(C) IF staining of H3K9me2 (red) in genital ridges at E10.75 shows a lack of this modification in PGCs. PGCs were detected with SSEA1 antibody (green). Scale bar, 20 μm in (A)–(C). The arrowheads in (A)–(C) indicate examples of PGCs. The genotype of the control is Blimp1Cre;Prmt5flox/+, and the mutant is Blimp1Cre;Prmt5flox/− in (A)–(C).
See also Figure S2.
Next we examined if the loss of H2A/H4R3me2s in PGCs had an effect on the dynamic changes of other histone modifications during epigenetic reprogramming of PGCs; this includes the loss of H3K9me2 and an increase in the intensity of H3K27me3 (Hajkova et al., 2008). There were no detectable effects however on these modifications on a global level in mutant PGCs at E10.75–E12.5 (Figures 2B, 2C, and S2C). Other histone modifications, including H3K9ac and H3K9me3, were also unchanged in mutant PGCs at E11.5 (Figure S2C). Thus, the loss of PRMT5 in PGCs specifically affects the H2A/H4R3me2s modification, at least on a global level.
Prmt5 Mutant PGCs Undergo Apoptosis
As noted above, loss of PRMT5 in PGCs results in complete sterility in both males and females. To determine when precisely mutant PGCs are affected, we first examined the total number of PGCs by AP staining at E11.5, and found that the Prmt5 mutant PGCs were reduced in both male and female embryos (Figures 3A and S3A). Further analysis using Blimp1Cre;Prmt5flox/− mice harboring an Oct4ΔPE-GFP reporter gene (GOF) (Yeom et al., 1996) showed that 87% of the mutant PGCs had lost PRMT5 by E10.5 (Figure 1B). Moreover, flow cytometry analysis for GOF-positive cells revealed a reduction in the number of Prmt5 mutant PGCs to 59% compared to those in control littermates at E11.5 (399 versus 682; n = 10, n = 25 embryos, p < 0.05) (Figure 3B). By E15.5–E16.5, none of the Prmt5 mutant PGCs survived in either male or female embryos (Figures 3C and 3D), or in the postnatal testis (9 dpp and 6 weeks; Figures S3B–S3D). However, the somatic cells formed apparently normal seminiferous tubules in males (Figures 1C and S3B). Thus, the loss of mutant PGCs coincides with the rapid loss of H2A/H4R3me2s modification from the nucleus of Prmt5 null PGCs from ∼E10.75 onward.
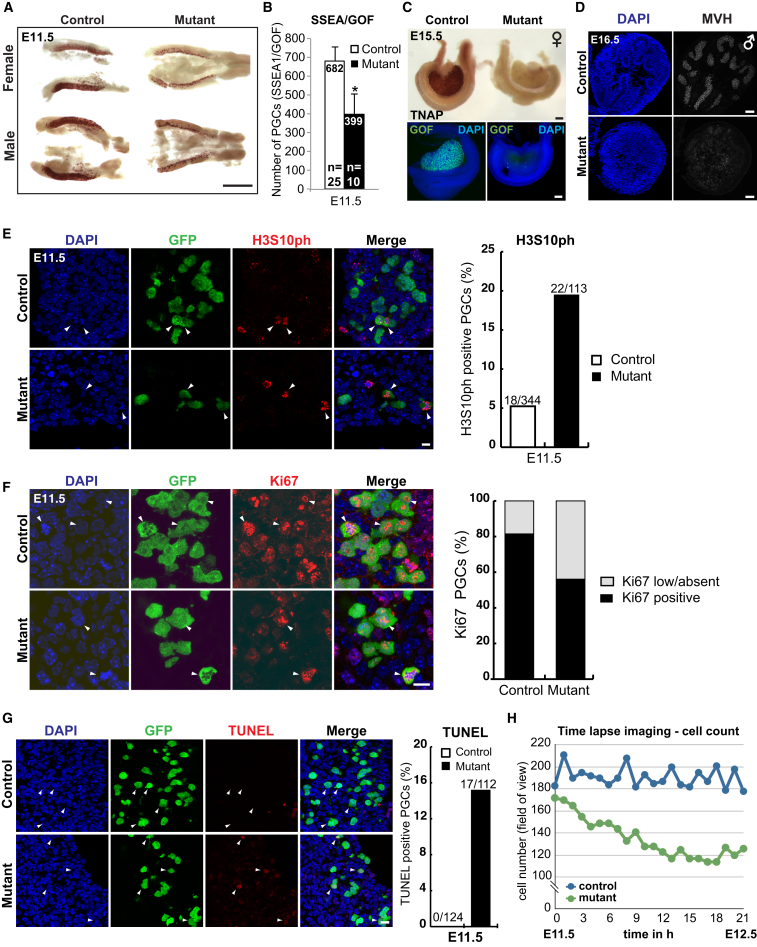
Figure 3.
Prmt5 Mutant PGCs Undergo Apoptosis
(A) Staining of E11.5 genital ridges for alkaline phosphatase (AP, seen as brown), a marker of PGCs. Scale bar, 0.5 mm.
(B) The number of PGCs at E11.5 determined by flow cytometry for SSEA1/GOF-positive cells. Shown are the mean values ± SE (Student’s t test, ∗p < 0.05).
(C) AP staining (top) and IF staining of GFP (bottom) in E15.5 female gonads. Merged images are shown with DNA stained with DAPI (blue) and PGCs stained with GFP (for GOF transgene, green). Note a lack of mutant germ cells in the gonads. Scale bar, 100 μm.
(D) IF staining of the late germ cell marker MVH (mouse vasa homolog, white) shows a lack of staining in E16.5 male mutant PGCs in gonads (bottom right hand pane). Scale bar, 50 μm.
(E) IF staining for the mitosis marker H3S10ph (red) in genital ridges at E11.5. PGCs are costained with antibodies against GFP that recognizes GOF (green). Scale bar, 10 μm. The number of PGCs that were labeled for H3S10ph is shown on the right (in %).
(F) IF staining for the mitosis marker Ki67 (red) in genital ridges at E11.5. PGCs are costained with GFP antibody that recognizes GOF (green). Scale bar, 10 μm. The number of PGCs in control and Prmt5 mutant embryos (in %) that were positive for Ki67 are shown in black; cells with low or no staining are shown in gray.
(G) TUNEL assay (red) to visualize apoptosis in genital ridges at E11.5. Scale bar, 10 μm. PGCs were costained with antibodies against GFP for GOF transgene (green). The number of PGCs that were labeled in the TUNEL assay are shown on the right (in %). The arrowheads in (E) and (F) indicate examples of PGCs.
(H) GFP-positive PGCs in the field of view during ∼24 hr time-lapse imaging of E11.5 genital ridges. Note a steady decline in the number of mutant PGCs (green) compared to controls (blue), in which the numbers fluctuated but did not change significantly. The genotype of the control is Blimp1Cre;Prmt5flox/+ and the mutant is Blimp1Cre;Prmt5flox/− in (A)–(H).
See also Figure S3, Movie S1 (Mutant), and Movie S2 (Control).
To determine the cause of the loss of PGCs in PRMT5 mutants, we examined the rate of cell proliferation and cell death at E11.5 by immunostaining of phosphorylated histone H3 Ser10 (H3S10ph), a marker of mitosis. This showed 5% of the PGCs were positive in control embryos (18/344) compared to 19% (22/113) in Prmt5 mutants (Figure 3E). Further, we stained for the proliferation marker Ki67 nuclear antigen, which is enriched in S, G2, and M phases but low in G0 and G1 (Gerdes et al., 1984). This showed that 44% (29/66) of mutant PGCs have low Ki67 staining compared to 19% (29/157) of control germ cells (Figure 3F). These results suggest that relatively more Prmt5 mutant germ cells are in G0/G1 phase or arrested in cell cycle. The small number of surviving mutant PGCs progress through the cell cycle asynchronously, which prevents detailed analysis. Nevertheless, these combined data suggest that there is a heterogeneous population of Prmt5 mutant PGCs with defects in cell-cycle regulation.
Next, we analyzed the rate of cell death by TUNEL labeling, which showed that a greater number of mutant PGCs were undergoing apoptosis from E11.5 onward compared to controls (15% versus 0%; Figure 3G). Consistently, live imaging for 24 hr of genital ridges ex vivo from E11.5 control and Prmt5 mutant embryos with the GOF reporter transgene revealed that the number of mutant PGCs declined progressively as they fragmented and disappeared (Figures 3H and S3E; Movie S1 and Movie S2). Taken together, these results suggest that the loss of PGCs in Prmt5 mutants is caused by apoptosis and cell-cycle deregulation.
DNA Damage Response Genes Are Upregulated in Prmt5 Mutant PGCs
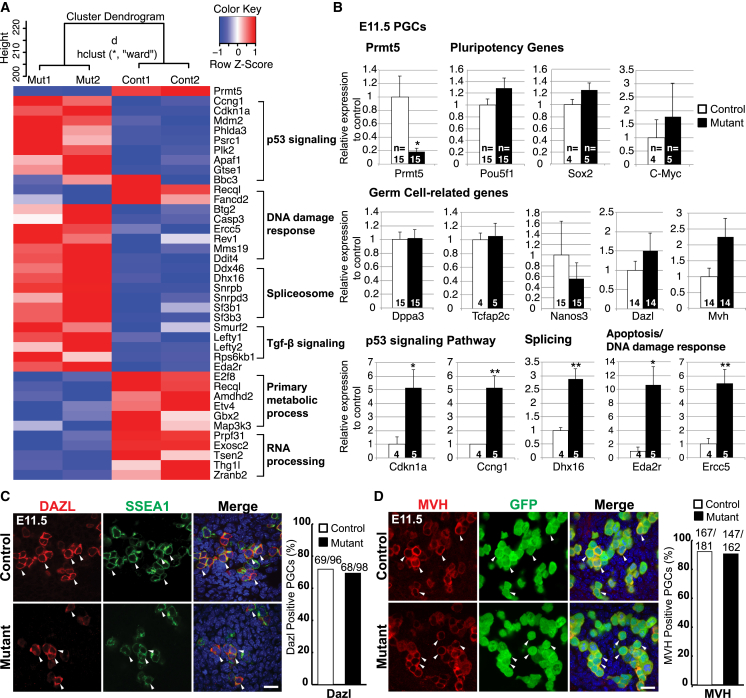
To determine the consequences of the loss of H2A/H4R3me2s modification in Prmt5 mutant PGCs, we analyzed their transcriptome by microarrays at E11.5. Surprisingly, only 32 out of 22,166 probes showed significant differential gene expression changes (adj.p < 0.05) (Table S1). We sought more comprehensive information by performing RNA-Seq analysis on control and mutant PGCs at E11.5 (Tang et al., 2010). Again, we found that only 422 genes were differentially expressed (p < 0.05), among which 201 genes were upregulated (>1.24-fold) and 221 were downregulated (<0.82-fold) (Table S2). Notably, we detected upregulation of the p53 signaling genes (Cdkn1a, Mdm2, Ccng1) and DNA damage response genes (Ercc5, Btg2, Rev1) in the Prmt5 mutant PGCs (Figure 4A; Table S1 and Table S2), while the downregulated genes were categorized to RNA processing (Prpf31, Exosc2, Zranb2) and DNA metabolic process (Fbxo18, Recql, Lig3). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed upregulation of peptidase regulator activity, DNA damage response, and p53 class mediators, as well as of the spliceosome complex in Prmt5 mutant PGCs (Figures S4A and S4B). The splicing defects and the activation of the p53-signaling pathway upon loss of PRMT5 are consistent with the observations in other cell types such as neural progenitor cells (Bezzi et al., 2013; Figure S4E). We validated the microarray and the RNA-Seq results by quantitative RT-PCR (qPCR) for selected genes in E11.5 PGCs (Figure 4B). Importantly, some key germline genes, including POU5F1 (Oct4) and Dppa3 (stella) were not affected in Prmt5 mutant PGCs (Figures 4B and S4C; Table S1 and Table S2), which we confirmed by immunostaining for OCT4, VASA, and DAZL (Figures 4C and 4D). Collectively, these results indicate that among the key effects of loss of PRMT5 in PGCs is the induction of DNA damage response and p53 signaling genes, which is consistent with their apoptotic response. Of note, the loss of the repressive H2A/H4R3me2s modification did not result in a global deregulation of transcription in PGCs.
Figure 4.
The Loss of PRMT5 in PGCs Induces p53 Signaling and DNA Damage Response Genes
(A) A heatmap of selected up- and downregulated genes in E11.5 PGCs from RNA-Seq analysis. Cluster dendogram for two biological replicates of each genotype is shown on top. Note that p53 signaling genes and DNA damage response genes are upregulated (red) in the mutant, while primary metabolic process and RNA processing genes are downregulated (blue).
(B) qRT-PCR analysis of selected genes in E11.5 PGCs (control, white; mutant, black). Shown are the mean values ± SE. Significance is shown by Student’s t test: ∗∗p < 0.01, ∗p < 0.05.
(C) IF staining of DAZL (late germ cell marker, red) in genital ridges at E11.5. PGCs were detected with antibodies against SSEA1 (green). Merged images are shown with DNA stained with DAPI (blue). The ratio of DAZL-positive PGCs in SSEA1-positive PGCs (in %) was determined (controls, white; mutants, black).
(D) IF staining of MVH (late germ cell marker, red) in genital ridges at E11.5. The ratio of MVH-positive PGCs in GFP positive (green) PGCs (in %) was determined. Scale bar, 20 μm in (C) and (D). The arrowheads in (C) and (D) indicate examples of PGCs. The genotype of the control is Blimp1Cre;Prmt5flox/+, and the mutant is Blimp1Cre;Prmt5flox/− in (A)–(D).
Transposable Elements Are Upregulated in Prmt5 Mutant PGCs
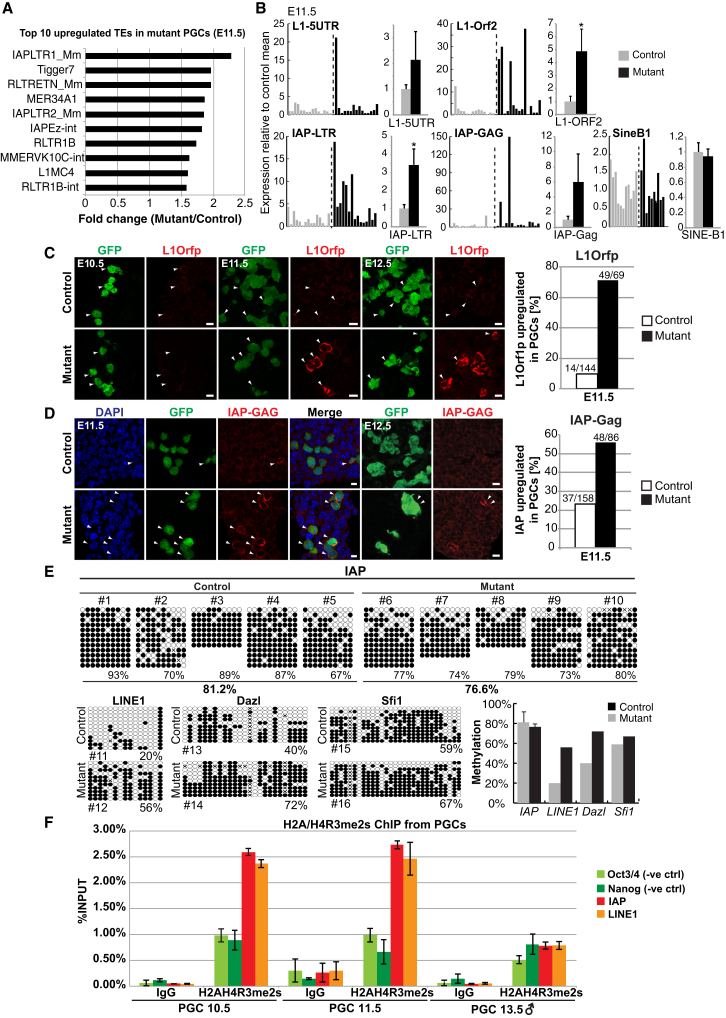
PGCs are likely to be vulnerable to the expression of TEs, especially during major epigenetic changes, including global DNA demethylation and the loss of H3K9me2, both of which are involved in the repression of TEs (Di Giacomo et al., 2013; Hajkova et al., 2002). Overexpression of TEs can have major genome-destabilizing effects in association with abnormal cell-cycle progression (Belgnaoui et al., 2006). TEs-induced mutations could also be detrimental for subsequent development, especially if it occurs repeatedly during the subsequent germline cycles. As Prmt5 mutant PGCs showed upregulation of DNA damage-response genes, we examined the RNA-Seq data of E11.5 PGCs and found that IAP-LTR1 is one of the most highly upregulated TEs in Prmt5 mutant PGCs (Figure 5A; Table S3). IAP-LTR1 is among the most active and abundant class of IAP elements whose expression can compromise genome integrity through random mutations following transpositions (Qin et al., 2010).
Figure 5.
Transposable Elements Are Upregulated in the Absence of PRMT5 in PGCs
(A) Top ten upregulated TEs in E11.5 mutant PGCs identified in the RNA-seq analysis. Mutant PGC values were compared to control PGC values and are shown as fold change (Mutant/Control).
(B) Single-cell qPCR analysis of TEs as indicated from E11.5 control (gray) and mutant (black) FACS-sorted PGCs (GOF/SSEA1). Each bar represents a single cell. The data are combined as mean values ± SE of all single cells as shown in the chart on the right. Significance is shown by Student’s t test, ∗p < 0.05.
(C) IF staining of L1Orf1p (red) in E10.5–E12.5 genital ridges and quantification of PGCs that shows an increase of L1Orf1p fluorescence intensity. PGCs were detected with GFP antibody for GOF (green). Scale bar, 10 μm.
(D) IF staining of IAP-GAG (red) in E11.5 and E12.5 genital ridges and quantification of PGCs that show an increase in IAP-GAG fluorescence intensity. Scale bar, 10 μm. The arrowheads in (C) and (D) indicate examples of PGCs.
(E) DNA methylation changes in IAPs determined by bisulfite sequencing with FACS-sorted PGCs (GOF/SSEA1) from E11.5 genital ridges from five control (#1–#5) and mutant embryos (#6–#10). DNA methylation of E11.5 PGCs from one littermate control and mutant embryo were tested for the LINE1, Dazl and Sfi1 locus (#11–#16). The right graph shows the average level of methylation ± SD at the IAP, LINE1, Dazl, and Sfi1 loci.
(F) ChIP with wild-type PGCs from E10.5, E11.5, and E13.5 male using a H2A/H4R3me2s antibody and IgG, respectively. ChIP-qPCR results ± SE are shown. The Oct4 and Nanog locus served as negative controls. Data are from biological duplicates. The genotype of the control is Blimp1Cre;Prmt5flox/+, and the mutant is Blimp1Cre;Prmt5flox/− in (A)–(E).
Since the PGC population is developmentally heterogeneous, and the H2A/H4R3me2s modification is lost in ∼49% of mutant PGCs by E10.75, we examined the expression of TEs by qRT-PCR analysis on single-cell cDNAs from PGCs. Remarkably, despite the heterogeneity of expression levels of TEs in single PGCs, the bulk of Prmt5 mutant PGCs at E11.5 showed an ∼3-fold upregulation of IAP-LTR (p < 0.05). This suggests that among the mutant PGCs, some of them have very high levels of IAP-LTR expression. In addition, the open reading frame (ORF)2 of LINE-1 which encodes an endonuclease and a reverse transcriptase showed ∼5-fold higher expression in mutant PGCs (p < 0.05) compared to controls (Figure 5B). The LINE-1 5′ UTR and IAP-GAG were apparently not significantly affected, which may be due to technical reasons following the generation of 3′ biased cDNAs during single-cell reverse transcription amplification (Tang et al., 2010). The expression of short interspersed nonrepetitive elements B1 (SINE-B1) was also seemingly not affected (p = 0.85; Figure 5B). Notably, in E12.5 PGCs, we detected a 6-fold increase of LINE-1 ORF2 in E12.5 mutant PGCs (Figure S5A). However, the levels of IAP-GAG seemed to be less affected, probably owing to the heterogeneity among mutant cells such that the surviving E12.5 mutant PGCs might represent a minor population that have yet to exhibit the effects of the Prmt5 mutation (Figure S5A). Indeed, the development of individual PGCs does not occur synchronously, within or between embryos.
Next, we looked for the presence of L1ORF1p and IAP-GAG proteins by immunostaining of E11.5 PGCs with specific antibodies. This showed a significant increase in the levels of L1ORF1p in a substantial number of Prmt5 mutant PGCs compared to the control (71% versus 10%; Figure 5C). The levels of L1ORF1p expression in Prmt5 mutant PGCs increased progressively from E10.5 to E12.5 (Figure 5C). In addition, IAP-GAG was more abundant in mutant PGCs at E11.5 (56% versus 23%; Figure 5D) and in surviving mutant germ cells at E12.5. Wild-type PGCs rarely expressed detectable levels of IAP-GAG (Figure 5D). This shows that both the IAP and LINE-1 elements are derepressed in mutant PGCs, which coincides with the progressive loss of the H2A/H4R3me2s modification.
To test whether this upregulation of TEs expression and proteins has any consequence, we stained E12.5 genital ridges for γH2AX, which is a marker for DNA double-strand breaks. We found that the mutant PGCs that contained bright and greater number of γH2AX nuclear foci compared to control (Figure S5B). Accordingly, the mean intensity of γH2AX in mutant GFP-positive PGCs was 1.3-fold higher than in control. Combined with the upregulation of DNA damage response genes in mutant PGCs, this result suggests that the upregulation of TEs may cause DNA lesions in Prmt5 mutant germ cells (Figures S4A and S5B).
We asked if the loss of PRMT5 has an effect on DNA methylation of repetitive elements in PGCs. While the global erasure of DNA methylation in PGCs reaches basal levels by E13.5, some repetitive elements such as IAP are only partially demethylated (Hackett et al., 2013). There was little effect on the status of DNA methylation of IAP and LINE1 elements in Prmt5 mutant PGCs by bisulphite genomic sequencing of E11.5 PGCs, except for a slightly higher extent of demethylation of IAP in mutant compared to wild-type PGCs (average level 76% versus 81%; n = 5, Figure 5E). There was no reduction in DNA methylation of LINE1 in Prmt5 mutant PGCs, suggesting that the loss of DNA methylation was not a major cause for the elevation of IAP and LINE-1 expression (Figure 5E). Indeed, the expression of LINE1 was not upregulated despite high levels of demethylation in wild-type PGCs. There was also no detectable effect on the expression of DNA methyltransferase 1 (DNMT1) in Prmt5 mutant PGCs at E11.5 (Figure S5C). This indicates a lack of significant correlation between overall changes in DNA methylation levels and the expression of LINE1 and IAP, which is also not due to any additional loss of DNA methylation from these elements in Prmt5 mutant PGCs. This supports the possibility that the translocation of PRMT5 from the cytoplasm to the nucleus at ∼E8.5, and the associated H2A/H4R3me2s modification detected in the PGC nuclei, might have a role in the repression of LINE1 and IAP.
H2A/H4R3me2s Is Enriched at IAP and LINE-1 in PGCs
The enrichment of the H2A/H4R3me2s repressive modification in PGCs might have a specific role in the repression of TEs, as this mark is generally implicated in gene repression (Xu et al., 2010). This modification was detected by immunofluorescence, concomitantly with the translocation of PRMT5 to the nucleus at ∼E8.5 in PGCs (Figures 1D, 1E, S2A, and S2B).
To check for the presence of the H2A/H4R3me2s modification on TEs, we carried out chromatin immunoprecipitation (ChIP) analysis for this mark on LINE1 and IAP in PGCs. Indeed, we found that both LINE1 and IAP are enriched for the H2A/H4R3me2s modification at E10.5–E11.5 in PGCs (Figure 5F). Subsequently, it is lost in male PGCs at E13.5 following translocation of PRMT5 to the cytoplasm at ∼E11.5. The specificity of this modification was evident by comparison with the negative control regions at the Oct4 and Nanog loci with a highly diminished signal. Further, LINE1 and IAP are not enriched for the modification in male PGCs at E13.5. It is technically not possible to similarly analyze Prmt5 mutant PGCs due to their extremely low numbers (∼400 PGCs/embryo at E11.5, Figure 3B). We did not, however, detect the H2A/H4R3me2s modification in mutant PGCs by immunofluorescence at E12.5 (Figure 2A), which indirectly implicates the loss of this modification on TEs. The combined data indicate that the enrichment of H2A/H4R3me2s modification in PGCs coincides with the entry of PRMT5 into the nucleus during global DNA demethylation in ∼E8.5–E11.5 PGCs. Notably, the H2A/H4R3me2s enrichment on LINE1 and IAP precedes expression of piRNAs and PIWI proteins in male germ cells at E12.5–E15.5 (Aravin et al., 2008; Kuramochi-Miyagawa et al., 2004). As discussed later, loss of cytoplasmic PRMT5 in early postimplantation epiblast has a negligible effect on TE expression owing to the resumption of DNA methylation in these cells (see later).
In summary, the loss of H2A/H4R3me2s in Prmt5 mutant PGCs occurs at E10.5–E11.5 followed by upregulation of LINE1 and IAP elements at E11.5–E12.5 (Figures 2A and 5B–5D). Our results suggest that PRMT5 and the histone modification H2A/H4R3me2s are required for the suppression of TEs during extensive reprogramming, including loss of H3K9me2 and global DNA demethylation in PGCs.
PRMT5 Represses Transposable Elements during Preimplantation Development
In principle, it is possible that PRMT5 might have a similar role elsewhere during global DNA demethylation of the genome. We took the opportunity to test this idea during another major epigenetic reprogramming event through development of the mouse zygote to the blastocyst stage, which is accompanied by global DNA demethylation (Smith et al., 2012). Our observation on PGCs prompted us to examine the role of maternally inherited and zygotic PRMT5 during preimplantation development (Tee et al., 2010).
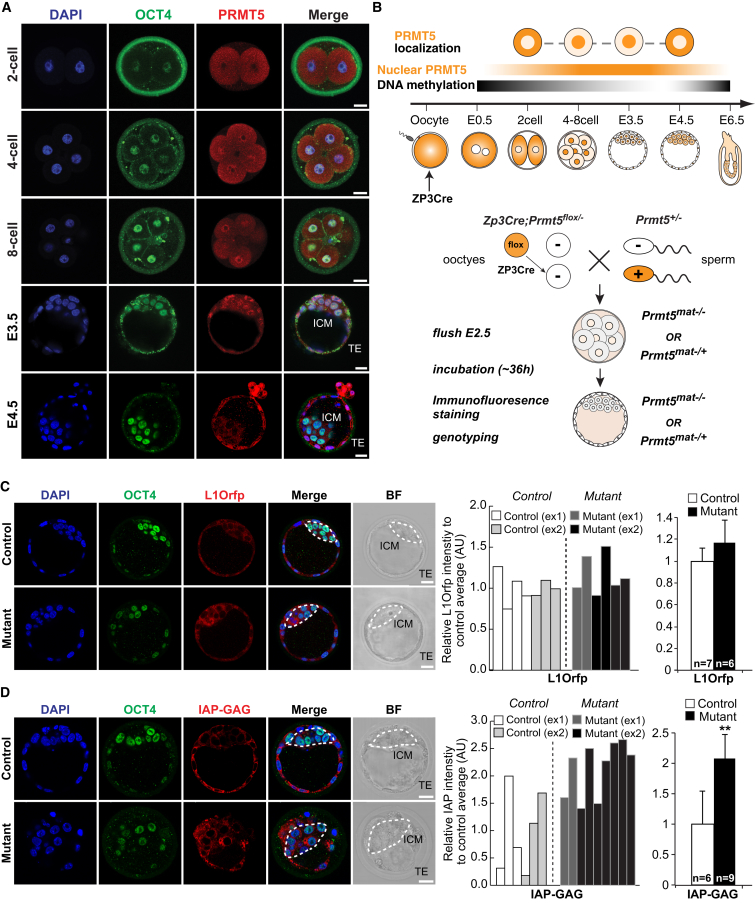
In preimplantation embryos, PRMT5 is present in the cytoplasm of the zygote and two-cell-stage embryos, but it localizes to the nucleus at the four- to eight-cell stage and persists there until the early E3.5 blastocyst stage (Figure 6A). Concomitantly with the nuclear localization of PRMT5, we could detect enrichment of H2A/H4R3me2s in the nucleus, particularly in eight- to sixteen-cell-stage embryos; prior to this, there are relatively low levels of this modification detectable at the two-cell stage (Figure S6A). Thereafter, PRMT5 is mostly cytoplasmic in the ∼E4.5 epiblast cells of developing embryos, concurrently with the initiation of de novo DNA methylation in epiblast cells, although it remains in the nucleus of trophectoderm (TE) cells that are relatively hypomethylated (Chapman et al., 1984).
Figure 6.
Prmt5 Is Required for the Suppression of TEs in Preimplantation Embryos
(A) IF staining of OCT4 (green) and PRMT5 (red) in wild-type two-cell to blastocyst-stage preimplantation embryos (stages are indicated). Note that PRMT5 is detected in the nucleus from four-cell-stage embryos. TE, trophectoderm; ICM, inner cell mass; scale bars, 20 μm.
(B) Schematic diagram of the subcellular localization of PRMT5 (shown in orange) and the level of nuclear PRMT5 during preimplantation development (top). Zp3Cre is expressed during oocyte maturation. The mating scheme shows how the maternal-zygotic Prmt5 knockout embryos were generated followed by the experimental outline (bottom).
(C) IF staining of OCT4 (green) and L1Orf1p (red) in control (Prmt5mat−/+) and maternal-zygotic Prmt5 mutant (Prmt5mat−/−) preimplantation embryos. The dashed line indicates the ICM region. Scale bars, 20 μm. The fluorescence intensity of L1Orf1p in preimplantation embryos was determined using seven control and six mutant embryos. The data are from two independent experiments, and the relative L1Orf1p intensity of each embryo compared to the mean intensity of control embryos is shown. The right graph shows the mean intensity ± SD of all embryos. n, number of embryos.
(D) IF staining of OCT4 (green) and IAP-GAG (red) in control (Prmt5mat−/+) and maternal-zygotic Prmt5 mutant (Prmt5mat−/−) preimplantation embryos. The ICM region is indicated by the dashed line. Scale bars, 20 μm. The fluorescence intensity of IAP-GAG in preimplantation embryos was determined in six control embryos and nine mutant embryos. The data are from two independent experiments, and the relative IAP-GAG intensity of each embryo compared to the mean intensity of control embryos is shown. The right graph shows the mean intensity ± SD of all embryos. n, number of embryos; significance was tested with the Student’s t test, ∗∗p < 0.01.
See also Figure S6.
We had previously shown that embryonic development following the loss of zygotic PRMT5 after an intercross between Prmt5+/− mice, was morphologically indistinguishable from wild-type E3.5 blastocysts (Tee et al., 2010). Consistently, zygotic Prmt5 mutant blastocysts have OCT4-positive cells in the inner cell mass (ICM) and CDX2-positive TE cells (Figure S6B). Notably, there were no significant changes in the expression of IAP and LINE1 elements in zygotic Prmt5 mutant embryos at the eight-cell stage and blastocysts (Figures S6C–S6E).
We considered that the maternally inherited PRMT5 could to some extent compensate for the zygotic loss of PRMT5 during preimplantation development (Figure S6F). We therefore depleted the oocytes of the maternally inherited PRMT5, by generating an oocyte-specific Prmt5 mutant female of Prmt5flox/− with Zp3-Cre (Zp3, zona pellucida glycoprotein 3) that is active in maturing oocytes (Figure 6B) (de Vries et al., 2000). The loss of maternally inherited PRMT5 in oocytes did not have an effect on their properties or development, since we found these females to be fertile when mated with the wild-type males (Figures S1G and S6G).
Next we examined the development of preimplantation embryos that are depleted of both maternally inherited as well as zygotic PRMT5 by mating Zp3Cre;Prmt5flox/- mice with Prmt5+/− males (Figure 6B). These embryos were collected at E2.5 and cultured for 2 days in vitro. Mutant embryos (Prmt5mat−/−) showed slightly higher L1Orf1p expression, which was not significant compared to controls (Prmt5mat−/+) (Figure 6C, 1:1.2, p = 0.19). However, IAP-GAG expression in the ICM of blastocysts was significantly elevated in Prmt5 mutant embryos (Prmt5mat−/−) (1:2.1, p < 0.01) (Figure 6D). At E4.5 in vivo, mutant embryos (Prmt5mat−/−) have a reduced number of cells compared to control (Prmt5mat−/+), and they do not form of a blastocoel and appear to collapse, which likely causes early embryonic lethality (Figure S6H). This suggests that PRMT5 also plays an important role in the suppression of specific TEs in preimplantation embryos during DNA demethylation.
Role of PRMT5 in Embryonic Stem Cells in “2i” Culture Conditions
We among others showed that ESCs when cultured in a chemically defined condition using inhibitors of GSK3 and MEK (2i) (Ying et al., 2008) have a hypomethylated status compared to ESCs in serum (Leitch et al., 2013). We tested whether PRMT5 has a similar role in suppressing TEs in ESCs cultured in 2i medium. PRMT5 in ESCs cultured in standard medium with serum is almost exclusively in the cytoplasm (Tee et al., 2010), but we found that the transfer of ES to 2i medium resulted in the detection of PRMT5 in the nucleus, consistent with DNA hypomethylation (Leitch et al., 2013), albeit only in ∼20%–30% of the ESCs. It is possible that nuclear-cytoplasmic translocation occurs dynamically in all individual cells (Figure S7A).
To examine the consequences of loss of PRMT5 in ESCs in “2i,” we crossed the Prmt5flox/flox mice to R26Cre-ERT (R26Cre) mice and derived ESCs in 2i condition (Figure S7B; Vooijs et al., 2001). Following treatment of cells with 4-hydroxytamoxifen (OHT), the floxed Prmt5 allele was efficiently excised by R26Cre after 24 hr, followed by a decline in the Prmt5 mRNA levels (∼0.18-fold) (Figures S7B and S7E). These R26Cre;Prmt5flox/flox cells died 5 days after OHT treatment, while control EtOH treatment of R26Cre;Prmt5flox/flox or OHT-treated heterozygous line (R26Cre;Prmt5flox/+) did not show any detectable defects (Figure S7C). While we detected upregulation of IAP-Gag (∼1.8-fold) and not of L1Orfp (∼1.3-fold) in the mutant cells (Figure S7E), there was also no detectable increase in IAP-GAG protein (mean intensity, 1:1.02, Figure S7F). Indeed, we also neither detected any significant decrease in the global H2A/H4R3me2s modification, nor specifically on the TEs (Figures S7G and S7H). It is possible that Prmt7, a related family member, might compensate for the loss of Prmt5. Since we detected splicing defects in mutant ESCs, this might account for the death of these ESCs (Figure S7D; Bezzi et al., 2013).
Loss of Cytoplasmic PRMT5 in Early Postimplantation Epiblast Cells
Development of the postimplantation epiblast is accompanied by DNA methylation, when PRMT5 translocates to the cytoplasm. To check whether the predominantly cytoplasmic PRMT5 in epiblast has any role in the repression of TEs, we examined these cells that were depleted of PRMT5 specifically at E5.5–E6.5 by mating Prmt5flox/flox mice with Sox2-Cre transgenic mice (Hayashi et al., 2002) (Figure S7I). Examination of these cells that were depleted of PRMT5 at E6.5 showed no significant expression of IAP-GAG by immunostaining (Figure S7J). Furthermore, qRT-PCR analysis of dissected epiblasts at E7.5 confirmed that both IAP and LINE1 showed no significant change in their expression in the Prmt5 mutant epiblast (Figure S7K). However, as with the mutant ESCs, we detected splicing defects in the mutant epiblast cells (Figure S4E), which is consistent with a previous report (Bezzi et al., 2013).
Loss of PRMT5 did not, however, affect specification of PGCs, since we detected PGCs in E7.75 mutant embryos as judged by the detection of PGC markers, AP2γ and TNAP (Figures S7L and S7M). These results suggest that DNA methylation rather than PRMT5 in the postimplantation epiblast cells is likely to repress TEs. Thus, loss of cytoplasmic PRMT5 has no marked effect on the expression of TEs.
Discussion
This study demonstrates that PRMT5 is indispensable during extensive resetting of the epigenome in early PGCs, and in preimplantation development. In both instances, PRMT5 relocates from the cytoplasm to the nucleus concomitantly with the onset of comprehensive DNA demethylation. Notably, there is also comprehensive erasure of H3K9me2 following specification of PGCs (Hajkova et al., 2008), which has been shown to suppress LINE1 during spermatogenesis (Di Giacomo et al., 2013). These epigenetic changes in the DNA and chromatin in early PGCs are conducive for the expression of TEs prior to the piRNA biosynthesis; nuclear PRMT5 seems to have a role in the suppression of TEs at this time.
PRMT5 catalyzes the repressive chromatin modification H2A/H4R3me2s, which is detected on the LINE1 and IAP in PGCs. Loss of PRMT5 results in the loss of H2A/H4R3me2s and upregulation of IAP and LINE1, suggesting that nuclear PRMT5 has a role in the repression of TEs. Similarly, loss of maternally inherited and zygotic PRMT5 results in upregulation of TEs. In contrast, loss of cytoplasmic PRMT5 in the postimplantation epiblast did not cause a significant upregulation of TEs, where DNA methylation is likely involved in their repression (Walsh et al., 1998). The nuclear PRMT5-dependent mechanism of TEs repression in the germline differs from the previously described repression of TEs by small RNA-induced silencing, by DNA methylation, or constitutive repressive histone modifications (Kuramochi-Miyagawa et al., 2008; Rowe et al., 2010). Notwithstanding, germline-specific loss of nuclear PRMT5 results in complete sterility of adult males and females. In mammals, TEs are more active in the germline compared to somatic lineages, and the loss of their suppression could affect fertility, as shown in mutants of piRNA components and Tex19.1 (Carmell et al., 2007; Öllinger et al., 2008). This is especially likely at the time of global DNA demethylation and loss of H3K9me2 that could make PGCs more susceptible to misregulation of TEs. Their deregulation could result in deleterious effects, including high rate of illegitimate pairing between nonhomologous chromosomes during meiosis, which triggers an apoptotic checkpoint (Zamudio and Bourc’his, 2010), especially in female PGCs that enter meiosis at E13.5.
The onset of global DNA demethylation is a significant feature of PGCs and preimplantation embryos, but what precisely triggers the relocation of PRMT5 from the cytoplasm to the nucleus on both these occasions is unknown. PRMT5 has three nuclear export signals but no nuclear localization signal (Gu et al., 2012), suggesting that specific mediators might be involved in the nuclear import, acting together with a block on the export signal. These mediators could include BLIMP1 (a key regulator of PGC specification) since PRMT5-BLIMP1 interaction was reported previously (Ancelin et al., 2006), as are AJUBA and SNAIL in the case of U2OS cells (Hou et al., 2008). The targeting of PRMT5 to specific loci such as TEs remains to be elucidated.
Global DNA demethylation in actively dividing PGCs is potentially conducive for TE expression and transpositions, which would increase the vulnerability of PGCs to transpositions (Burns and Boeke, 2012). However, there is no evidence to suggest that DNA demethylation and H2A/H4R3me2s modification are functionally interdependent in PGCs. For example, H2A/H4R3me2s modification was still detectable on “imprinted” regions after the erasure of DNA methylation in PGCs (Henckel et al., 2012). Furthermore, while loss of PRMT5 and the H2A/H4R3me2s modification did not affect DNA methylation of LINE1, expression of LINE1 was elevated in Prmt5 mutant PGCs. We propose that the H2A/H4R3me2s mark accompanies and compensates for the reduction in DNA methylation and the loss of H3K9me2 to repress TEs in PGCs. Moreover, the H2A/H4R3me2s modification can function regardless of DNA demethylation to repress gene expression (Henckel et al., 2012), potentially by inhibiting RNA polymerase II binding as they show inverse relationship on silenced loci in primary erythroid progenitors (Zhao et al., 2009).
The repressive H3K9me2 histone modification has been reported to suppress the LINE1 expression with piRNA pathway during spermatogenesis until late zygotene stages, which can compensate for defective DNA methylation and piRNA biogenesis in Mili mutant mice (Di Giacomo et al., 2013). Notably, the H3K9me2 modification is globally erased shortly after the specification of PGCs at E7.5 and is absent thereafter until E12.5 (Hajkova et al., 2008). Thus the nuclear PRMT5-mediated repressive H2A/H4R3me2s modification might be even more critical for the repression of TEs in early PGCs. The H3K27me3 histone modification was also reported to be enriched on retrotransposons at E13.5 PGCs (Ng et al., 2013). We did not detect any obvious difference in the H3K27me3 modification in Prmt5 mutant PGCs at E11.5 compared to control. This suggests that this modification might function independently for the suppression of TEs, but it evidently does not compensate for the loss of H2A/H4R3me2s at E11.5 in mutant PGCs.
There is a transient (but not sustained) developmentally programmed expression of TEs in the ∼2- to 4-cell-stage embryos, which is important for the “oocyte to embryo” transition. Some TEs act as alternative promoters and exons for the expression of key host genes with significant roles in development (Peaston et al., 2004; Fadloun et al., 2013). The transient expression of LINE-1 and IAP at this time is unlike the aberrant expression of LINE1 and IAP in Prmt5 mutant PGCs with potentially detrimental consequences for the germline, and thereafter for embryonic development. While there is relatively low H2A/H4R3me2s modification at the two-cell stage, following relocation of PRMT5 from the cytoplasm at ∼4-cell stage, this modification becomes detectable from the 8- to 16-cell stage, which coincides with the ongoing DNA demethylation (Smith et al., 2012). The maternal/zygotic PRMT5 plays an important role when the DNA methylation reaches its lowest levels at the blastocyst stage, at which point there is an upregulation of IAPs in the mutant. However, there is no significant effect on LINE-1 expression, which might be regulated by an alternative mechanism (Fadloun et al., 2013).
It is known that KAP1 and ESET can suppress TEs through the H3K9me3 and H4K20me3 modifications (Matsui et al., 2010; Rowe et al., 2010). While detailed analysis of these modifications at TEs of mutant PGCs is not possible, there were no detectable changes on the global level in H3K9me3 or H4K20me3, suggesting that the loss of H2A/H4R3me2s at TEs is not compensated by other histone modifications. On the other hand, loss of the H2A/H4R3me2s repressive modification in Prmt5 mutant PGCs resulted in only a few significant overall transcriptional changes. This could be because the other repressive histone modifications might compensate for the loss of H2A/H4R3me2s (Matsui et al., 2010; Rowe et al., 2010). Therefore, the key role of the PRMT5 catalyzed nuclear H2A/H4R3me2s modification in PGCs is evidently in the repression of TEs in PGCs.
The misregulation of p53 signaling genes and the apoptosis in Prmt5 mutant PGCs is potentially a combinatorial consequence of splicing defects and TEs-induced DNA damage response (Belgnaoui et al., 2006; Gasior et al., 2006). The splicing defects have been observed in other instances, such as neural progenitor cells (Bezzi et al., 2013), as we also did in the Prmt5 mutant ESCs and in postimplantation epiblast cells, which are ascribed to the cytoplasmic PRMT5. In contrast, upregulation of TEs is a PGC- and preimplantation embryo-specific defect in Prmt5 mutant. The expression of TEs following the loss of nuclear PRMT5, however, is in addition to the effects of the loss of cytoplasmic PRMT5 in ESCs, postimplantation epiblast cells, and neuronal progenitors (Bezzi et al., 2013). Notably, the p53 signaling-related genes including Cdkn1a (also known as p21) are among the top differentially expressed genes in Prmt5 mutant PGCs; consistently, in Dnmt1 null fibroblasts, IAP upregulation following global DNA demethylation also results in the activation of Cdkn1a and causes p53-dependent apoptosis (Jackson-Grusby et al., 2001). Thus, activation of TEs could contribute to the activation of the p53-signaling pathway. Due to subfertility of p53 mutant female mice and the small litter size of Blimp1Cre;Prmt5flox mice (∼4 embryos/litter, Figure S1G), it is not possible to investigate the mutant Prmt5 PGCs in the p53 null background (Hu et al., 2007).
It is known that there is a dramatic increase in DNA methylation after implantation of the blastocysts, as observed in the postimplantation epiblast cells (Borgel et al., 2010). Thus, it is possible that DNA methylation suppresses TEs in epiblast cells, and subsequently in most somatic tissues. Consistent with this hypothesis, PRMT5 is predominantly in the cytoplasm of the postimplantation epiblast cells, where it is likely to have other roles such as in the regulation of alternative splicing. Indeed, deletion of cytoplasmic PRMT5 in postimplantation epiblast cells does not significantly affect the expression of TEs, suggesting that DNA methylation is sufficient to suppress TEs in these cells. On the other hand, the lack of upregulation of IAP-GAG in Prmt5 mutant hypomethylated ESCs in 2i is consistent with the presence of the H2A/H4R3me2s modification, which could be attributed to Prmt7. It is likely that, as in the postimplantation epiblast, cytoplasmic PRMT5 in ESCs may have a role in alternative splicing.
In PGCs, PRMT5 relocates to the cytoplasm at ∼E11.5, resulting in a loss of enrichment of H2A/H4R3me2s at TEs in male PGCs at ∼E13.5, which indicates a link between the enzyme and the repressive mark (Figures 5F and S2A). There is initiation of piRNAs biosynthesis after global DNA demethylation from E8.5–E11.5 PGCs, and the expression of Mili, which is relatively low in E10.5 PGCs, increases from E12.5 onward (Figure S7N). PIWI proteins, MILI and MIWI2, become detectable at E12.5 and E15.5, respectively (Aravin et al., 2008; Kuramochi-Miyagawa et al., 2004). Interestingly, expression of Mili and Miwi2 themselves may be directly linked to DNA demethylation, which thus couples both nuclear PRMT5 and piRNA biogenesis to epigenetic reprogramming in PGCs (Hackett et al., 2012). Importantly, while a loss of MILI and MIWI2 results in postnatal germ cell defects (Carmell et al., 2007), there is comprehensive and dramatic loss of early PGCs following loss of PRMT5.
The translocation of PRMT5 to the cytoplasm after ∼E11.5 plays yet another distinct role as it methylates murine PIWI family proteins, which is essential for piRNA biogenesis and the silencing of TEs (Vagin et al., 2009). Notably, the suppression of TEs by PRMT5 through PIWI protein has also been shown in planarians and Drosophila and is therefore evolutionarily conserved (Kirino et al., 2010; Rouhana et al., 2012). In mice, the mechanism of piRNA-based repression of TEs involves DNA methylation (Aravin et al., 2008). By contrast, nuclear PRMT5 is required in E8.5–E11.5 for the repression of TEs through the H2A/H4R3me2s repressive chromatin modification; a mutation in Prmt5 results in loss of early PGCs in both male and female embryos before the onset of meiosis (Figure 7). The role of DNA methylation in the suppression of TEs in early PGCs is also unlikely, since both the de novo and maintenance DNA methylation enzymes are repressed in E8.5–E11.5 PGCs (Hackett et al., 2013; Figure S5D). Furthermore, the repression of TEs by PRMT5-mediated H2A/H4R3me2s was seen without a significant effect on DNA methylation. In conclusion, our study reveals a role for nuclear PRMT5 in protecting the genome by silencing TEs at a critical time during comprehensive epigenetic reprogramming and global DNA demethylation in early PGCs and preimplantation embryos in mice.
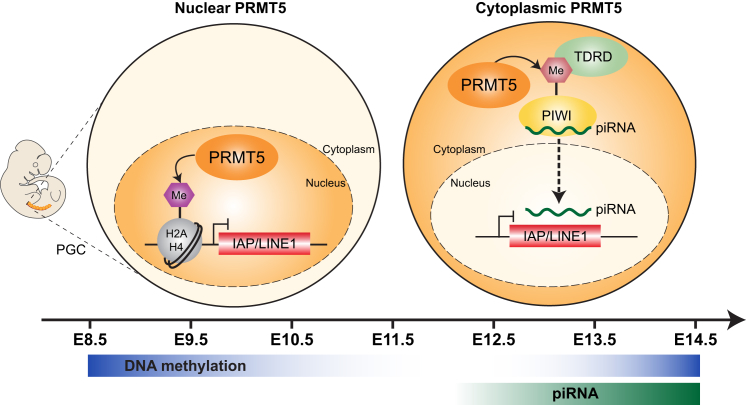
Figure 7.
Model of the Suppression of IAP and LINE1 by PRMT5 in PGCs
PRMT5 has a dual function during epigenetic reprogramming of PGCs: when global DNA demethylation starts (E8.5–E10.5), PRMT5 translocates to the nucleus, and the nuclear PRMT5 catalyzes the H2A/H4R3me2s modification to suppress IAP and LINE1 (left). After global DNA demethylation at E12.5, expression of Mili starts to become detectable. PRMT5 translocates to the cytoplasm at ∼E11.5 to coincide with the onset of the expression of PIWI proteins. Cytoplasmic PRMT5 is required to methylate PIWI proteins. This methylated arginine residue recruits Tudor domain proteins to facilitate piRNA biogenesis, which in turn are required for the silencing of IAP and LINE1 (right).
See also Figure S7.
Experimental Procedures
Detailed experimental procedures are provided in the Supplemental Information.
Targeted Disruption of Prmt5 Locus and Generation of Cell-Type-Specific Prmt5 Knockout Mice
The Prmt5 targeting vector was constructed by inserting Pgk-neo flanked by frt site and the 5′ loxP site the sixth intron of Prmt5. 3′ loxP site to PGK-neo cassette was inserted into the seventh intron of Prmt5 (Figure S1). The targeting vector was linearized, electroporated into E14Tg2a ESCs (129/Ola). Detailed procedure of screening and generation of mice is provided in the Supplemental Information. All husbandry and experiments involving mice were carried out according to the local ethics committee and were performed in a facility designated by the Home Office.
Whole-Mount Immunofluorescence Staining
Embryos from timed mating were dissected and processed for immunostaining as described previously (Ohinata et al., 2005). Images were acquired using confocal microscope (Olympus, Leica) and analyzed with ImageJ software.
Author Contributions
S.K. designed the project, performed experiments, analyzed the data, and wrote the manuscript. U.G. performed experiments, analyzed data, and contributed to the manuscript. J.J.Z., J.A.H., D.C., S.B., C.L., and R.S. performed experiments and analyzed data. S.D. and G.E.A. analyzed RNA-seq data. M.A.S. designed the project and wrote the manuscript.
Acknowledgments
We thank Wee-Wei Tee for help on initial project design, Nigel Miller for flow cytometry, Julien Bauer and Charles Bradshow for bioinformatics analysis, Dang Vinh Do and Anne Turberfield for sharing preliminary data, Alex Bortvin and Bryan R. Cullen for generously sharing antibodies, Kaiqin Lao and Applied Biosystems for RNA-Seq run, and members of the Surani lab for helpful discussions. U.G. was supported by a Marie Skłodowska Curie Intra-European Fellowship. J.J.Z. was a recipient of a Wellcome Trust PhD Studentship (RG44593). This research was supported by grants from the Wellcome Trust to M.A.S. (WT096738).
Accession Numbers
All RNA-seq data files are available for download from NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE60875.
Supplemental Information
List of up- and downregulated genes in female mutant PGCs (genotype, Blimp1Cre;Prmt5flox/−;GOF) compared to control (Blimp1Cre;Prmt5flox/+;GOF ) are shown. Related to Figure 4.
Green fluorescent cells are GOF-positive PGCs. Related to Figure 3.
Green fluorescent cells are GOF-positive PGCs. Related to Figure 3.
References
- Ancelin K., Lange U.C., Hajkova P., Schneider R., Bannister A.J., Kouzarides T., Surani M.A. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 2006;8:623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Sachidanandam R., Bourc’his D., Schaefer C., Pezic D., Toth K.F., Bestor T., Hannon G.J. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgnaoui S.M., Gosden R.G., Semmes O.J., Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 2006;6:13. doi: 10.1186/1475-2867-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezzi M., Teo S.X., Muller J., Mok W.C., Sahu S.K., Vardy L.A., Bonday Z.Q., Guccione E. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev. 2013;27:1903–1916. doi: 10.1101/gad.219899.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgel J., Guibert S., Li Y., Chiba H., Schübeler D., Sasaki H., Forné T., Weber M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 2010;42:1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- Branscombe T.L., Frankel A., Lee J.H., Cook J.R., Yang Z., Pestka S., Clarke S. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem. 2001;276:32971–32976. doi: 10.1074/jbc.M105412200. [DOI] [PubMed] [Google Scholar]
- Burns K.H., Boeke J.D. Human transposon tectonics. Cell. 2012;149:740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell M.A., Girard A., van de Kant H.J.G., Bourc’his D., Bestor T.H., de Rooij D.G., Hannon G.J. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Chapman V., Forrester L., Sanford J., Hastie N., Rossant J. Cell lineage-specific undermethylation of mouse repetitive DNA. Nature. 1984;307:284–286. doi: 10.1038/307284a0. [DOI] [PubMed] [Google Scholar]
- de Vries W.N., Binns L.T., Fancher K.S., Dean J., Moore R., Kemler R., Knowles B.B. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis. 2000;26:110–112. [PubMed] [Google Scholar]
- Di Giacomo M., Comazzetto S., Saini H., De Fazio S., Carrieri C., Morgan M., Vasiliauskaite L., Benes V., Enright A.J., O’Carroll D. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol. Cell. 2013;50:601–608. doi: 10.1016/j.molcel.2013.04.026. [DOI] [PubMed] [Google Scholar]
- Durcova-Hills G., Tang F., Doody G., Tooze R., Surani M.A. Reprogramming primordial germ cells into pluripotent stem cells. PLoS ONE. 2008;3:e3531. doi: 10.1371/journal.pone.0003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadloun A., Le Gras S., Jost B., Ziegler-Birling C., Takahashi H., Gorab E., Carninci P., Torres-Padilla M.E. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 2013;20:332–338. doi: 10.1038/nsmb.2495. [DOI] [PubMed] [Google Scholar]
- Gasior S.L., Wakeman T.P., Xu B., Deininger P.L. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J., Lemke H., Baisch H., Wacker H.H., Schwab U., Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J. Immunol. 1984;133:1710–1715. [PubMed] [Google Scholar]
- Gu Z., Li Y., Lee P., Liu T., Wan C., Wang Z. Protein arginine methyltransferase 5 functions in opposite ways in the cytoplasm and nucleus of prostate cancer cells. PLoS ONE. 2012;7:e44033. doi: 10.1371/journal.pone.0044033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett J.A., Reddington J.P., Nestor C.E., Dunican D.S., Branco M.R., Reichmann J., Reik W., Surani M.A., Adams I.R., Meehan R.R. Promoter DNA methylation couples genome-defence mechanisms to epigenetic reprogramming in the mouse germline. Development. 2012;139:3623–3632. doi: 10.1242/dev.081661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett J.A., Sengupta R., Zylicz J.J., Murakami K., Lee C., Down T.A., Surani M.A. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P., Erhardt S., Lane N., Haaf T., El-Maarri O., Reik W., Walter J., Surani M.A. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- Hajkova P., Ancelin K., Waldmann T., Lacoste N., Lange U.C., Cesari F., Lee C., Almouzni G., Schneider R., Surani M.A. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Lewis P., Pevny L., McMahon A.P. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 2002;119(Suppl 1):S97–S101. doi: 10.1016/s0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Henckel A., Chebli K., Kota S.K., Arnaud P., Feil R. Transcription and histone methylation changes correlate with imprint acquisition in male germ cells. EMBO J. 2012;31:606–615. doi: 10.1038/emboj.2011.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z., Peng H., Ayyanathan K., Yan K.P., Langer E.M., Longmore G.D., Rauscher F.J., 3rd The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol. Cell. Biol. 2008;28:3198–3207. doi: 10.1128/MCB.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Feng Z., Teresky A.K., Levine A.J. p53 regulates maternal reproduction through LIF. Nature. 2007;450:721–724. doi: 10.1038/nature05993. [DOI] [PubMed] [Google Scholar]
- Jackson-Grusby L., Beard C., Possemato R., Tudor M., Fambrough D., Csankovszki G., Dausman J., Lee P., Wilson C., Lander E., Jaenisch R. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Jansson M., Durant S.T., Cho E.C., Sheahan S., Edelmann M., Kessler B., La Thangue N.B. Arginine methylation regulates the p53 response. Nat. Cell Biol. 2008;10:1431–1439. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- Kirino Y., Vourekas A., Sayed N., de Lima Alves F., Thomson T., Lasko P., Rappsilber J., Jongens T.A., Mourelatos Z. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA. 2010;16:70–78. doi: 10.1261/rna.1869710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S., Kimura T., Ijiri T.W., Isobe T., Asada N., Fujita Y., Ikawa M., Iwai N., Okabe M., Deng W. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S., Watanabe T., Gotoh K., Totoki Y., Toyoda A., Ikawa M., Asada N., Kojima K., Yamaguchi Y., Ijiri T.W. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch H.G., McEwen K.R., Turp A., Encheva V., Carroll T., Grabole N., Mansfield W., Nashun B., Knezovich J.G., Smith A. Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 2013;20:311–316. doi: 10.1038/nsmb.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnúsdóttir E., Dietmann S., Murakami K., Günesdogan U., Tang F., Bao S., Diamanti E., Lao K., Gottgens B., Azim Surani M. A tripartite transcription factor network regulates primordial germ cell specification in mice. Nat. Cell Biol. 2013;15:905–915. doi: 10.1038/ncb2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Leung D., Miyashita H., Maksakova I.A., Miyachi H., Kimura H., Tachibana M., Lorincz M.C., Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- McGrath J., Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- Nakaki F., Hayashi K., Ohta H., Kurimoto K., Yabuta Y., Saitou M. Induction of mouse germ-cell fate by transcription factors in vitro. Nature. 2013;501:222–226. doi: 10.1038/nature12417. [DOI] [PubMed] [Google Scholar]
- Ng J.H., Kumar V., Muratani M., Kraus P., Yeo J.C., Yaw L.P., Xue K., Lufkin T., Prabhakar S., Ng H.H. In vivo epigenomic profiling of germ cells reveals germ cell molecular signatures. Dev. Cell. 2013;24:324–333. doi: 10.1016/j.devcel.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Ohinata Y., Payer B., O’Carroll D., Ancelin K., Ono Y., Sano M., Barton S.C., Obukhanych T., Nussenzweig M., Tarakhovsky A. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- Öllinger R., Childs A.J., Burgess H.M., Speed R.M., Lundegaard P.R., Reynolds N., Gray N.K., Cooke H.J., Adams I.R. Deletion of the pluripotency-associated Tex19.1 gene causes activation of endogenous retroviruses and defective spermatogenesis in mice. PLoS Genet. 2008;4:e1000199. doi: 10.1371/journal.pgen.1000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S., Vishwanath S.N., Erdjument-Bromage H., Tempst P., Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaston A.E., Evsikov A.V., Graber J.H., de Vries W.N., Holbrook A.E., Solter D., Knowles B.B. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Qin C., Wang Z., Shang J., Bekkari K., Liu R., Pacchione S., McNulty K.A., Ng A., Barnum J.E., Storer R.D. Intracisternal A particle genes: Distribution in the mouse genome, active subtypes, and potential roles as species-specific mediators of susceptibility to cancer. Mol. Carcinog. 2010;49:54–67. doi: 10.1002/mc.20576. [DOI] [PubMed] [Google Scholar]
- Rouhana L., Vieira A.P., Roberts-Galbraith R.H., Newmark P.A. PRMT5 and the role of symmetrical dimethylarginine in chromatoid bodies of planarian stem cells. Development. 2012;139:1083–1094. doi: 10.1242/dev.076182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe H.M., Jakobsson J., Mesnard D., Rougemont J., Reynard S., Aktas T., Maillard P.V., Layard-Liesching H., Verp S., Marquis J. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Seisenberger S., Andrews S., Krueger F., Arand J., Walter J., Santos F., Popp C., Thienpont B., Dean W., Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Z.D., Chan M.M., Mikkelsen T.S., Gu H., Gnirke A., Regev A., Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani M.A., Barton S.C., Norris M.L. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308:548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- Surani M.A., Hayashi K., Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Nordman E., Li B., Xu N., Bashkirov V.I., Lao K., Surani M.A. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat. Protoc. 2010;5:516–535. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee W.W., Pardo M., Theunissen T.W., Yu L., Choudhary J.S., Hajkova P., Surani M.A. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2010;24:2772–2777. doi: 10.1101/gad.606110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin V.V., Wohlschlegel J., Qu J., Jonsson Z., Huang X., Chuma S., Girard A., Sachidanandam R., Hannon G.J., Aravin A.A. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vooijs M., Jonkers J., Berns A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2001;2:292–297. doi: 10.1093/embo-reports/kve064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace N.A., Belancio V.P., Deininger P.L. L1 mobile element expression causes multiple types of toxicity. Gene. 2008;419:75–81. doi: 10.1016/j.gene.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C.P., Chaillet J.R., Bestor T.H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Xu X., Hoang S., Mayo M.W., Bekiranov S. Application of machine learning methods to histone methylation ChIP-Seq data reveals H4R3me2 globally represses gene expression. BMC Bioinformatics. 2010;11:396. doi: 10.1186/1471-2105-11-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom Y.I., Fuhrmann G., Ovitt C.E., Brehm A., Ohbo K., Gross M., Hübner K., Schöler H.R. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamudio N., Bourc’his D. Transposable elements in the mammalian germline: a comfortable niche or a deadly trap? Heredity (Edinb) 2010;105:92–104. doi: 10.1038/hdy.2010.53. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Rank G., Tan Y.T., Li H., Moritz R.L., Simpson R.J., Cerruti L., Curtis D.J., Patel D.J., Allis C.D. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat. Struct. Mol. Biol. 2009;16:304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of up- and downregulated genes in female mutant PGCs (genotype, Blimp1Cre;Prmt5flox/−;GOF) compared to control (Blimp1Cre;Prmt5flox/+;GOF ) are shown. Related to Figure 4.
Green fluorescent cells are GOF-positive PGCs. Related to Figure 3.
Green fluorescent cells are GOF-positive PGCs. Related to Figure 3.